Abstract
Background: Ovarian carcinoma is a highly lethal gynecological malignancy due to its frequent relapses and adoption of chemoresistance. To develop new biomarkers for disease progression in ovarian carcinoma, CSCs, which are considered to contribute to disease relapse and metastasis, were isolated from human ovarian carcinoma tissues, and differentially expressed microRNAs (miRNAs) in CSCs were identified and assessed the clinical implication of expression of these miRNAs.
Methods: Primary cancer cells derived from human ovarian carcinomas were cultured and spheroid-forming cells (SFCs) were isolated. Profiles of miRNA expression in CSC-like SFCs were identified by miRNA microarray and the results were validated by quantitative real-time RT-PCR (qRT-PCR). We also assessed the correlations between miRNA expression levels and clinicopathological parameters in ovarian carcinomas.
Results: Five miRNAs (miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b) were significantly dysregulated in CSC-like SFCs compared with primary cancer cells. The qRT-PCR showed that miR-5703 and miR-1246 expression was significantly higher in ovarian cancer cells than in normal control cells, whereas the miR-424-5p level was significantly lower. Decreased expression of miR-424-5p was significantly associated with distant metastasis in high stage (stage IIII & IV) carcinomas (35.5% vs. 72.2%, respectively, p=0.013)
Conclusion: Taken together, miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b are useful markers for enriching ovarian CSCs. Decreased expression of miR-424-5p in ovarian carcinoma might be a putative biomarker for distant metastasis in ovarian carcinoma.
Keywords: Ovarian carcinoma; Cancer stem cell; microRNA; miR-424-5p, Prognosis.
Introduction
Ovarian carcinoma is the most lethal gynecological malignancy in women 1. Its high mortality rate is due to difficulties in early detection and disease recurrence. Despite the good response to first-line chemotherapies such as taxol and platinum-based combination therapy, the majority of patients with advanced disease become refractory to conventional chemotherapeutic agents 2, resulting in disease recurrence and distant metastasis. However, the mechanisms involved in disease recurrence and distant metastasis remain unclear.
Increasing evidence indicates that cancer stem cells (CSCs) comprise a small population of cells within a heterogeneous tumor mass. They are the most tumorigenic and resistant to chemotherapy 3-7. CSCs express distinct surface markers and contribute to heterogeneous tumor growth. Thus, they resemble the functional hallmarks of genuine stem cells; namely, the ability to self-renew, modulate and balance the differentiation of various cell types 8-12. In addition, CSCs can survive from conventional treatments and contribute to disease recurrence, resulting in more chemoresistant and aggressive tumors 13. However, the molecular mechanisms that regulate CSCs are poorly understood.
Recent discoveries in the biology of microRNAs (miRNAs) have provided new insights on the genes implicated in tumorigenesis and CSC modulation 14. miRNAs are small non-coding RNAs that inhibit the expression of multiple genes at the post-transcriptional level via partial base pairing to the 3′untranslated region (UTR) of their targets. miRNAs regulate various cellular processes, including the cell cycle 15 and apoptosis 16. Furthermore, aberrant miRNA expression promotes tumorigenesis and metastasis of various human cancers, including ovarian carcinoma 17, 18. Gene expression studies have identified hundreds of dysregulated miRNAs in cancer cells, and their control of oncogenes and tumor suppressors has been demonstrated by functional studies 19. Thus, understanding the miRNA expression levels and their clinical significance may provide insight into the pathobiology of ovarian carcinoma on a molecular level and reveal new prognostic biomarkers and therapeutic targets.
Here, we isolated putative CSCs from fresh tissues of high-grade ovarian serous carcinoma (OSC)s using spheroid-forming assay. Differentially expressed miRNAs in CSCs were identified by high-throughput miRNA microarray analysis. We also studied their expression patterns in high-grade human OSC samples using qRT-PCR, and identified correlations between their expression levels and clinicopathological parameters to assess the clinical impact of differentially expressed miRNAs in OSCs, the most prevalent and lethal type of ovarian cancer 20 .
Materials and Methods
Patients and tissue collection
To quantify the miRNA expression levels by real-time PCR, fresh and formalin-fixed-paraffin-embedded (FFPE) tissues from 59 high-grade OSCs were obtained from patients who underwent an oophorectomy at the Bundang CHA Medical Center. The fallopian tubes from patients who underwent a hysterectomy with salpingectomy due to benign leiomyoma served as a control. The histological diagnosis and clinical stage were determined using the World Health Organization classification system. The samples were histopathologically graded by a pathologist at the Department of Pathology according to a two-tiered grading system and tumor staging was carried out according to a tumor-node-metastasis staging system. Each sample was assigned to either the chemosensitive or chemoresistant group according to the responsiveness of patients to first-line chemotherapy (taxol and platinum-based combination therapy) after surgery based on the NCCN guidelines. This study was approved by the Ethics Committee of the Bundang CHA Medical Center. Informed consent was obtained from each patient before surgery.
Primary ovarian carcinoma cell culture and isolation of spheroid-forming cells (SFCs)
Procedures were performed as described by Kwon et al. 21. To isolate and characterize CSCs, high-grade OSC cells isolated from the ovary of 4 patients who underwent oophorectomy were primarily cultured and a spheroid formation assay was performed using cultured primary ovarian carcinoma cells.
Tumors were mechanically dissected into small pieces and enzymatically digested into single cell suspensions in Ca2+/Mg2+-free PBS containing 50 U/ml collagenase A (Roche, Pleasanton, CA, USA) at 37ºC for 1 h. Cells were incubated with Ber-EP4-coated magnetic Dynabeads (Life Technologies, Carlsbad, CA, USA) for 30 min to select epithelial cells, which were then cultured in RPMI medium (Gibco, Grand Island, NY, USA) containing 10% fetal bovine serum, 1% penicillin-streptomycin, and 20 ng/ml epidermal growth factor (Life Technologies). For the spheroid formation assay, single cells were plated on Corning ultra-low-binding 6-well culture plates (Corning, NY, USA) at a density of 1 × 103 cells/cm2 in serum-free DMEM/F12 medium (Life Technologies) containing 20 ng/ml epidermal growth factor (Life Technologies), 10 ng/ml basic fibroblast growth factor, 0.4% bovine serum albumin, and 5 µg/ml insulin (all from Sigma-Aldrich, St. Louis, MO, USA). The formation of spheroids, each containing approximately 50-100 cells, was assessed at 7 days after cell plating. The morphology and evidence for enriching CSCs in spheroid-forming cells were shown in our previous report 21 by demonstrating the constant spheroid-forming efficiency of 2.17 ~2.37% in the subsequent generation, and upregulation of stem cell markers, such as ALDH1, CD24, CD44, CD133, and Sox2.
RNA extraction
RNA was isolated from cultured primary ovarian carcinoma cells and SFCs using TRIzol reagent (Life Technologies), as instructed by the manufacturer. Total RNA from FFPE tissues was isolated using the miRNeasy FFPE kit (QIAGEN, Hilden, Germany), as instructed by the manufacturer. RNA purity and concentration were determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Wilmington, DE, USA).
miRNA microarray analysis
The miRNA expression profiles of four independent primary OSC cell cultures and their corresponding SFC cultures were compared using the Agilent Human miRNA 8 × 60K platform (Agilent, Inc., Santa Clara, CA, USA) based on the miRBase software package (v18.0). miRNA microarray analysis was performed as instructed by the manufacturer. In brief, 100 ng of total RNA per sample was labeled with cyanine 3-pCp (Cy3) using the miRNA Complete Labeling and Hyb kit (Agilent, Inc.). Cy3-labeled RNAs were hybridized to a miRNA microarray containing 1887 human miRNAs. The miRNA microarray was then scanned using the Agilent G2600D microarray scanner. Raw data for the same gene in primary OSCs and SFCs were summarized in the Agilent Feature Extraction software package (v11.0.1.1), which generated the gene view file and provided expression data for each gene probed on the array. Array data were filtered using gIsGeneDetected = 1 for all samples (1: detected). Logarithmically-transformed miRNA gtotalGeneSignal values were normalized using the quantile method. The comparative analysis of results from primary cancer cells and SFCs was based on -fold changes. All data analysis and visualization of differentially expressed genes were conducted using the R statistical language software package (v. 2.15.0).
Bioinformatics analysis
To identify relevant target genes, severely dysregulated miRNAs showing ≥ 2.0 fold change or < 0.5 fold with a P-value < 0.05 based on microarray data were included in the analysis. Identification of miRNA target genes was performed using Target Scan 7.1(http://www.targetscan.org) and miRDB (http://mirdb.org/miRDB/, v4.0). The list of target genes was further reduced by selecting those genes targeted by five or more miRNAs.
Bioinformatics analysis for gene set enrichment was performed using the DAVID database (DAVID Functional Annotation Bioinformatics Microarray Analysis, http://david.abcc.ncifcrf.gov/) 22 and KEGG database. The KEGG pathways were regarded to be significant when the adjusted P-value by Benjamini-Hochberg procedure was less than 0.05.
Quantitative real-time polymerase chain reaction (qRT-PCR)
The miRNA expression levels were measured by qRT-PCR using the TaqMan microRNA reverse transcription kit (Applied Biosystems, Foster City, CA, USA) and a Bio-Rad CFX96 real-time PCR detection system (Bio-Rad Laboratories, Hercules, CA, USA). The thermal cycling conditions used were recommended by the manufacturer. RNU48 served as the control for miRNA normalization. All PCR reactions were run in duplicate and relative miRNA expression levels were calculated using the 2-ΔΔCt method.
Statistical analysis
Statistical analysis was performed using the SPSS software package (IBM Analytics, Armonk, NY, USA). Correlations between miRNA expression levels and clinicopathological parameters were evaluated using the chi-squared test. Differences in miRNA expression levels between groups were expressed as the mean ± standard deviation (SD). Data were analyzed using Student's t-test and P-values < 0.05 were considered statistically significant.
Results
Clinicopathological characteristics of chemosensitive and chemoresistant patients
To validate the miRNA microarray results, 59 high-grade OSC samples were subjected to qRT-PCR. The clinicopathological characteristics of the patients are summarized in Table 1. Of 59 OSC patients, 39 cases (66.1%) were chemosensitive, and 20 cases (33.9%) were chemoresisant. The mean age of chemosensitive and chemoresistant patients was 54.7 and 57.2 years, respectively. Approximately 25.6% (10/39) of chemosensitive patients were diagnosed with stage I or II cancer, whereas 74.4% (29/39) of patients had stage III or IV cancer. All chemoresistant patients were diagnosed with stage III or IV cancer. Nodal metastasis was identified in 43.6% (17/39) of chemosensitive patients and in 70% (14/20) of chemoresistant patients. Distal metastasis was identified in 25.6% (10/39) and 40% (8/20) of patients with chemosensitive and chemoresistant carcinomas, respectively. Disease recurrence was observed in 59% (23/39) and 85% (17/20) of chemosensitive and chemoresistant patients, respectively.
Table 1.
The clinicopathological characteristics of high-grade ovarian serous carcinoma (n = 59)
| Patients | Chemosensitive | Chemoresistant | ||
|---|---|---|---|---|
| Age, years | 55.5±11.1 | 54.7±11.2 | 57.2±10.9 | |
| Stage | I / II | 10 (16.9%) | 10 (25.6%) | 0 (0.0%) |
| III / IV | 49 (83.1%) | 29 (74.4%) | 20 (100%) | |
| Nodal metastasis | Absent | 28 (47.5%) | 22 (56.4%) | 6 (30.0%) |
| Present | 31 (52.5%) | 17 (43.6%) | 14 (70.0%) | |
| Distant metastasis | Absent | 41 (69.5%) | 29 (74.4%) | 12 (60.0%) |
| Present | 18 (30.5%) | 10 (25.6%) | 8 (40.0%) | |
| Recurrence | Absent | 19 (32.2%) | 16 (41.0%) | 3 (15.0%) |
| Present | 40 (67.8%) | 23 (59.0%) | 17 (85.0%) | |
| Total | 59 | 39 (66.1%) | 20 (33.9%) |
miRNA expression profiles of SFCs identified by miRNA microarray and qRT-PCR analyses
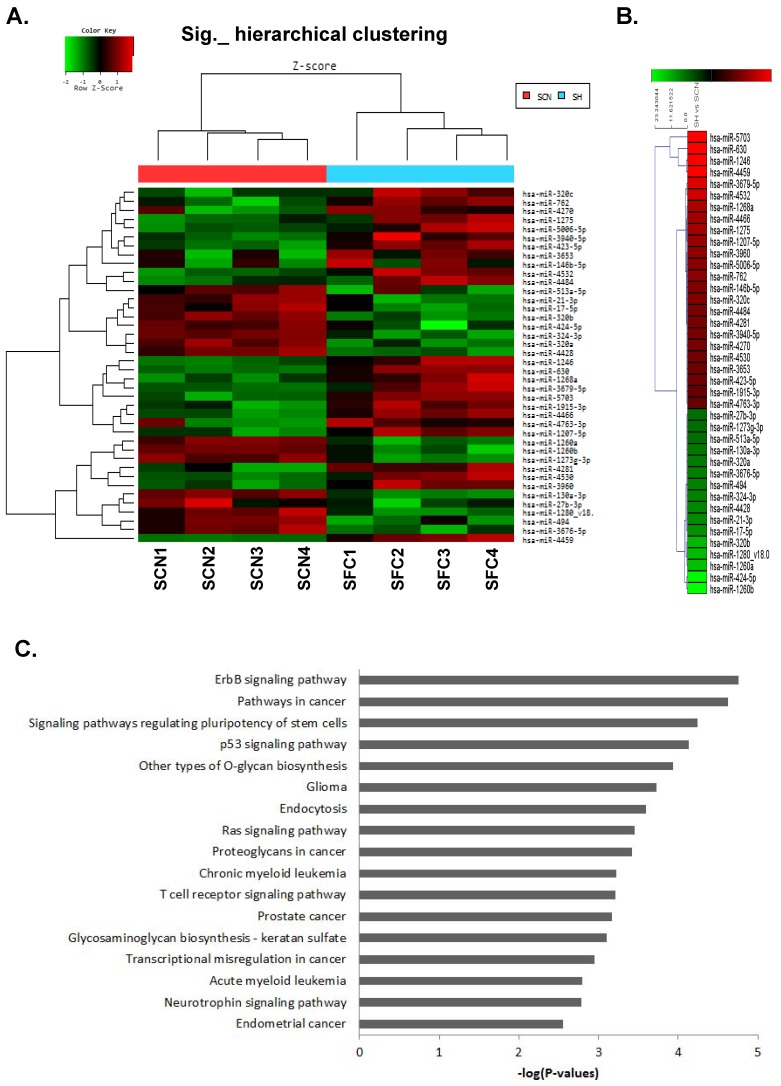
The miRNA expression profiles of CSC-like SFCs and their corresponding primary cancer cells were examined by miRNA microarray analysis (Fig. 1A).
Figure 1.
MicroRNA microarray analysis of spheroid-forming cells (SFCs) and primary cancer cells from cultured ovarian serous carcinoma (OSC) cells. (A) Hierarchical clustering of 37 significantly altered miRNAs with a ≥2.0-fold difference in expression between OSC SFCs and primary cancer cells. The clustered expression data are displayed on a heat map, with individual tumors and miRNAs listed on x- and y-axes, respectively. The x-axis indicates four SFC samples and their respective primary cancer cell samples. (B) Twenty-four miRNAs were significantly upregulated, whereas 13 miRNAs were downregulated, in the four SFC samples when compared with the control. Individual blocks are color-coded to indicate changes in miRNA expression: red indicates upregulation, green indicates downregulation, and black indicates no change. SCN, cultured primary cancer cell; SFC, spheroid-forming cells. (C) KEGG pathway analysis of genes targeted by the dysregulated miRNAs in CSC-like SFCs compared with primary OSC cells. Genes targeted by five or more miRNAs were selected for pathway enrichment analysis. The enrichment score of each pathway is expressed as -log (P-value).
The functional roles played by dysregulated miRNAs identified by miRNA microarray analysis were further analyzed. We summarized the expression levels of 37 miRNAs significantly altered with a ≥2-fold increase or decrease in CSC-like cells compared with their corresponding parental cancer cells from highest to lowest (Table 2). The hierarchical clustering of these 37 significantly altered miRNAs is shown in Figure 1B. In addition, target genes were predicted from 37 miRNAs (24 upregulated and 13 downregulated) using miRDB (http://mirdb.org/miRDB/, v4.0), PicTar-Vert (http://pictar.mdc-berlin.de/), and Target Scan (http://www.targetscan.org) databases. In total, 1762 genes were used for functional annotation analysis. After KEGG pathway enrichment analysis, the ErbB signaling pathway, various cancer-related pathways, and pathways regulating stem cell pluripotency were the highest ranked out of 17 pathways identified. The pathways for specific cancers were associated with glioma, chronic myeloid leukemia, prostate cancer, acute myeloid leukemia, and endometrial cancer. The significantly related signaling pathways were ErbB-, p53-, Ras, T-cell receptor, and neurotrophin-signaling pathways (Fig. 1C).
Table 2.
List of differentially expressed microRNAs in cancer stem cell-like spheroid-forming cells compared with primary cancer cells
| miRNA | Fold Change | P-value | Chromosome |
|---|---|---|---|
| Up-regulated | |||
| hsa-miR-5703 | 29.223 | 0.014 | 2 |
| hsa-miR-630 | 19.996 | 0.008 | 15q24.1 |
| hsa-miR-1246 | 13.709 | 0.024 | 2q31.1 |
| hsa-miR-4459 | 12.681 | 0.008 | 5 |
| hsa-miR-3679-5p | 4.387 | 0.035 | 2 |
| hsa-miR-4532 | 4.157 | 0.005 | 20 |
| hsa-miR-1268a | 3.366 | 0.017 | 15q11.2 |
| hsa-miR-4466 | 3.201 | 0.013 | 6 |
| hsa-miR-1275 | 3.166 | 0.02 | 6 |
| hsa-miR-1207-5p | 3.038 | 0.041 | 8 |
| hsa-miR-3960 | 2.829 | 0.026 | 9 |
| hsa-miR-5006-5p | 2.789 | 0.028 | 13 |
| hsa-miR-762 | 2.762 | 0.034 | 16 |
| hsa-miR-146b-5p | 2.602 | 0.023 | 10q24.32 |
| hsa-miR-320c | 2.585 | 0.043 | 18 |
| hsa-miR-4484 | 2.444 | 0.046 | 10 |
| hsa-miR-4281 | 2.405 | 0.002 | 5 |
| hsa-miR-3940-5p | 2.361 | 0.046 | 19 |
| hsa-miR-4270 | 2.357 | 0.023 | 3 |
| hsa-miR-4530 | 2.239 | 0.028 | 19 |
| hsa-miR-3653 | 2.2 | 0.023 | 22 |
| hsa-miR-423-5p | 2.17 | 0.023 | 17 |
| hsa-miR-1915-3p | 2.032 | 0.031 | 10 |
| hsa-miR-4763-3p | 2.003 | 0.018 | 22 |
| Down-regulated | |||
| hsa-miR-27b-3p | -2.145 | 0.036 | 9 |
| hsa-miR-1273g-3p | -2.153 | 0.005 | 1 |
| hsa-miR-513a-5p | -2.159 | 0.033 | chrX |
| hsa-miR-130a-3p | -2.357 | 0.006 | 11 |
| hsa-miR-320a | -2.454 | 0.002 | 8p21.3 |
| hsa-miR-3676-5p | -2.495 | 0.009 | 17 |
| hsa-miR-494 | -2.558 | 0.012 | 14q32.31 |
| hsa-miR-324-3p | -2.604 | 0.006 | 17 |
| hsa-miR-4428 | -2.645 | 0.003 | 1 |
| hsa-miR-21-3p | -2.757 | 0.041 | 17 |
| hsa-miR-17-5p | -2.843 | 0.05 | 13 |
| hsa-miR-320b | -3.634 | 0.001 | 1 |
| hsa-miR-424-5p | -4.905 | 0.045 | chrX |
miRNA: microRNA.
Of 37 miRNAs, we chose five miRNAs whose levels were the most increased (miR-5703, miR-630 and miR-1246) and the most decreased (miR-320b and miR-424-5p) for further validation with human OSC samples. To further explore the functional distribution of these five miRNAs, we performed genome-wide miRNA target prediction databases. The major target genes of each miRNA involved in oncogenesis are summarized in Table 3.
Table 3.
The putative target genes of the most highly dysregulated miRNAs in spheroid- forming cells compared with primary cancer cells.
| ID | Fold change | P-value | Chr | Genes | |
|---|---|---|---|---|---|
| hsa-miR-5703 | 29.223 | 0.014 | 2 | SYNPO2L | synaptopodin 2 like |
| PDE1C | phosphodiesterase 1C | ||||
| GFRA1 | GDNF family receptor alpha 1 | ||||
| ADAM19 | ADAM metallopeptidase domain 19 | ||||
| DAB2 | Dab, mitogen-responsive phosphoprotein, homolog 2 (Drosophila) | ||||
| ENTPD1 | ectonucleoside triphosphate diphosphohydrolase 1 | ||||
| hsa-miR-630 | 19.996 | 0.008 | 15q24.1 | DOCK8 | dedicator of cytokinesis 8 |
| CD200 | CD200 molecule | ||||
| F3 | coagulation factor III, tissue factor | ||||
| NEXN | nexilin (F actin binding protein) | ||||
| CARD16 | caspase recruitment domain family member 16 | ||||
| hsa-miR-1246 | 13.709 | 0.024 | 2q31.1 | VEPH1 | ventricular zone expressed PH domain containing 1 |
| TNFRSF8 | tumor necrosis factor receptor superfamily member 8 | ||||
| FGL2 | fibrinogen like 2 | ||||
| MARCH3 | membrane associated ring-CH-type finger 3 | ||||
| FRMD6 | FERM domain containing 6 | ||||
| EDA | ectodysplasin A | ||||
| PTPLAD2 | protein tyrosine phosphatase-like A domain containing 2 | ||||
| hsa-miR-424-5p | -4.905 | 0.045 | X | USP42 | ubiquitin specific peptidase 42 |
| SLC5A3 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 | ||||
| AXIN2 | axin 2 | ||||
| FECH | ferrochelatase | ||||
| PTPRD | protein tyrosine phosphatase, receptor type, D | ||||
| PHKA1 | phosphorylase kinase, alpha 1 (muscle) | ||||
| FAM81A | family with sequence similarity 81 member A | ||||
| NTRK2 | neurotrophic tyrosine kinase, receptor, type 2 | ||||
| BCAS1 | breast carcinoma amplified sequence 1 | ||||
| TMEM100 | Transmembrane Protein 100 | ||||
| ITGA10 | integrin subunit alpha 10 | ||||
| PDE3B | phosphodiesterase 3B | ||||
| UNC5B | unc-5 netrin receptor B(UNC5B) | ||||
| DLL1 | delta-like 1 (Drosophila)(DLL1) | ||||
| PTHLH | parathyroid hormone-like hormone(PTHLH) | ||||
| TMEM215 | transmembrane protein 215(TMEM215) | ||||
| hsa-miR-320b | -3.634 | 0.001 | 1 | SLC5A3 | solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 |
| PCDHA2 | protocadherin alpha 2 | ||||
| SLC2A12 | solute carrier family 2 member 12 | ||||
| RNPC3 | RNA binding region (RNP1, RRM) containing 3 | ||||
| ZNF780A | zinc finger protein 780A | ||||
| DLX1 | distal-less homeobox 1 | ||||
| RUFY2 | RUN and FYVE domain containing 2 | ||||
| TMEM100 | transmembrane protein 100 | ||||
| ST6GAL2 | ST6 beta-galactosamide alpha-2,6-sialyltranferase 2 | ||||
| BAALC | brain and acute leukemia, cytoplasmic | ||||
| ETV1 | ETS variant 1 | ||||
| GRIN2A | glutamate ionotropic receptor NMDA type subunit 2A |
chr: chromosome.
miR-5703, miR-1246, and miR-424-5p expression levels were significantly altered in OSCs
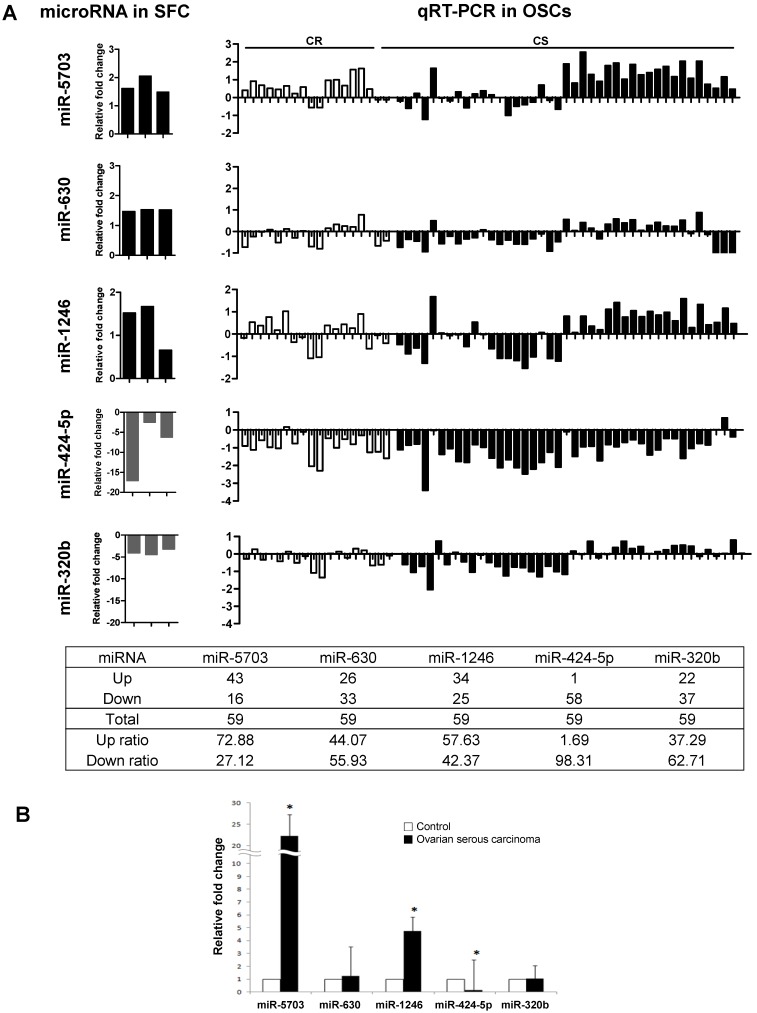
miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b expression was measured in 59 human OSC samples by qRT-PCR (Fig. 2). The control was used with normal fallopian tubes, which are known to be the origin of OSC 23-25. As shown in Figure 2A, the expression profiles of miR-5703 (upregulated), miR-424-5p, and miR320b (both downregulated) were similar to SFCs. Compared with that in the control, expression of miR-5703 was upregulated in 73% (42/59) and miR-424-5p was downregulated in 98% (58/59) of OSC samples, respectively. In addition, miR-5703 (22.23-fold), miR-1246 (4.75-fold), and miR-424-5p (0.17-fold) expression levels were significantly altered in OSC samples than in the control (Fig. 2B). However, there was no significant difference between chemosensitive and chemoresistant groups.
Figure 2.
Comparison of miRNA expression levels (miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b) in OSCs using two approaches; namely, miRNA microarray analysis and qRT-PCR. (A) The left panel shows the relative miRNA expression levels determined by miRNA microarray analysis. Five miRNAs were dysregulated in CSC-like SFCs when compared with primary cancer cells. The right panel shows relative miRNA expression levels determined by qRT-PCR. The expression profiles of miR-5703 (upregulated), miR-424-5p, and miR320b (both downregulated) were similar between OSC and SFC samples. (B) The expression levels of candidate miRNAs in OSCs relative to fallopian tubes (control). miR-5703 and miR-1246 expression levels were significantly higher in OSCs than in fallopian tubes, whereas the miR-424-5p expression level was lower in OSCs than in the control. Data are presented as the mean ± SD. (*P < 0.05).
Decreased expression of miR-424-5p is associated with distant metastasis in high stage OSCs
Correlations between miRNA expression levels and clinicopathological parameters, including clinical stage, nodal and distant metastasis, chemoresistance, and disease recurrence were identified only for the high stage OSCs (n=49), because other clinicopathological parameters are related with clinical stage. Decreased expression of miR-424-5p (<0.1-fold) was significantly associated with distant metastasis (35.5% vs. 72.2%, respectively, p=0.013) (Table 4). However, there was no correlation between the aberrant expression of other miRNAs and clinical stage, nodal metastasis, or disease recurrence.
Table 4.
Relationship between microRNA expression and clinicopathologic parameters in ovarian serous carcinomas with stage III & IV
| hsa-miR-5703† | hsa-miR-630† | hsa-miR-1246† | hsa-miR-424-5p§ | hsa-miR-320b§ | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| up-regulated | P-value | up-regulated | P-value | up-regulated | P-value | down-regulated | P-value | down-regulated | P-value | |||||
| Age | ||||||||||||||
| <55 | 17/25 | 0.686 | 6/25 | 0.138 | 14/25 | 0.058 | 12/25 | 0.889 | 3/25 | 0.957 | ||||
| ≥55 | 15/24 | 2/24 | 7/24 | 12/24 | 3/24 | |||||||||
| Nodal metastasis | ||||||||||||||
| Absent | 15/20 | 0.236 | 4/20 | 0.563 | 9/20 | 0.801 | 10/20 | 0.906 | 3/20 | 0.625 | ||||
| Present | 17/29 | 4/29 | 12/29 | 14/29 | 3/29 | |||||||||
| Distant metastasis | ||||||||||||||
| Absent | 22/31 | 0.275 | 5/31 | 0.961 | 15/31 | 0.305 | 11/31 | 0.013* | 4/31 | 0.854 | ||||
| Present | 10/18 | 3/18 | 6/18 | 13/18 | 2/18 | |||||||||
| Recurrence | ||||||||||||||
| Absent | 6/12 | 0.200 | 3/12 | 0.350 | 5/12 | 0.924 | 6/12 | 0.935 | 2/12 | 0.591 | ||||
| Present | 26/37 | 5/37 | 16/37 | 18/37 | 4/37 | |||||||||
hsa-miR-5703, hsa-miR-630 and hsa-miR-1246 upregulation of microRNAs was determined when the level of expression is 2.5-fold or more†; and hsa-miR-424-5p, hsa-miR-320b, 0.1 or less§. Pearson's chi-squared test (*, P < 0.05)
Discussion
In spite of relatively small population (0.01-1.0%) of CSCs, CSCs contribute disease recurrence because these cells are resistant to conventional treatments that target rapidly dividing cells, such as chemotherapy and radiotherapy 13, 26. Therefore, identifying CSCs and characterizing their molecular phenotype may provide insight into new therapeutic strategies for lethal human cancers such as ovarian carcinoma. From this point of view, ovarian CSCs are considered to be involved in disease relapse 5 as well as cancer development and chemoresistance; thus, CSC-specific molecular changes may be disease biomarkers or novel therapeutic targets 12.
A growing body of evidence indicates that miRNAs regulate most human genes involved in critical biological processes, including tumorigenesis, progression, and therapy resistance. miRNAs also control CSC self-renewal and differentiation, according to several studies that reported differentially expressed miRNAs in CSCs of various human cancers 22, 27, 28. Recently, the dysregulation of miR-200a, miR-199a and miR-214 was reported in CD133 (+) and CD44 (+) ovarian CSCs 29. We previously demonstrated that miR-23b, miR-27b, miR-424, and miR-503 expression is dysregulated in ovarian cancer cells positive for ALDH1 30, a biomarker of CSCs. However, there are few data regarding miRNA expression profiles in ovarian CSCs.
To identify predictive biomarkers for disease recurrence and poor prognosis, and to provide new treatment strategies for life-threatening ovarian carcinoma, we identified ovarian CSC-specific miRNAs using high-throughput miRNA microarray and qRT-PCR. We also assessed the clinical implications of CSC-specific miRNAs by evaluating correlations between differentially expressed miRNAs and clinicopathological parameters. We found that miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b were aberrantly expressed in CSC-like SFCs. We also demonstrated that miR-5703 and miR-1246 expression increased, and that miR-424-5p expression decreased, significantly in OSCs compared with the control, indicating that these miRNAs are involved in the development of OSCs. We then analyzed correlations between miRNA expression levels and various clinicopathologic parameters to identify putative prognostic biomarkers as for the high stage OSCs (Table 4). Regarding that resistance to chemotherapy is due mainly to CSCs, and that miRNAs are responsible for the maintenance and regulation of CSCs properties and drug resistance by several recent studies 31, we postulated that chemoresistance is associated with changes in miR-5703, miR-1246 and miR-424-5p expression. However, we could not find evidence to support this postulation in our analysis. Thus, further studies are required to understand the roles of these miRNAs in human ovarian carcinoma.
However, we revealed for the first time that miR-424-5p was associated with distant metastasis of OSC. Tumor metastasis is a multi-step process that involves many critical factors because tumor heterogeneity and the mechanisms underlying distant metastasis may be completely different, even where primary tumors have similar clinical manifestations and histological types. CSCs are considered to be one of the causative factors of distant metastasis 32. As for miR-424-5p, several studies reported the role of miR-424 in different physiological and pathological conditions. For example, expression of miR-424 is higher in distant metastasis than in primary mouse and human pancreatic neuroendocrine tumors 33. Another study for non-small cell lung cancers shows that upregulation of miR-424 is related to a poor prognosis 34. On the other hand, lower expression of miR-424 is positively correlated with advanced clinical stage, lymph node metastasis, and other poor prognostic parameters in cervical cancers 35, indicating that it functions as a tumor suppressor. The present study also showed that expression of miR-424-5p was significantly lower in OSCs than in the control, and decreased expression of miR-424-5p significantly correlated with distant metastasis. Taken together, the function of miR-424-5p is cell type- or context-specific, and it may function as a tumor suppressor and could be a potential predictive biomarker for distant metastasis in OSCs.
As for miR-5703, which was upregulated in OSCs, its roles in tumor development and progression remain unclear. To predict the roles of miR-5703 in tumor development and progression, we searched for predicted target genes associated with apoptosis and tumor suppression using the web-based computational programs PicTar and Target Scan. ADAM19, a putative miR-5703 target gene, is a member of the Notch pathway. Its dysregulation results in neoplastic proliferation and stem cell regulation in many human cancers 36, 37. The cleavage of the Notch transmembrane domain by ADAM proteases, which are multi-functional, multi-domain proteins involved in cell adhesion, cell signaling, and the proteolytic processing of other transmembrane proteins results in activation of the Notch pathway 38, 39. Recent studies show that several ADAMs are highly expressed in cancer cells and tissues 40. Of these, ADAM19 is highly expressed in human primary brain tumors and its upregulation is associated with increased invasiveness 41. However, the function of ADAM19 in other cancers remain unclear. Therefore, further studies are necessary to determine whether miR-5703 induces ovarian cancer cell growth or migration/invasion, and whether ADAM19 is a direct target gene of miR-5703.
MiR-1246 was reported to promote migration and invasion of hepatocellular carcinoma 42. Another study demonstrated that miR-1246, a putative target of the p53 transcription factor, inhibits Down syndrome-associated DYRK1A, thereby activating NFAT and leading to tumorigenesis 43. Serum miR-1246 was reportedly a novel diagnostic and prognostic biomarker of esophageal squamous cell carcinoma 44. The further study is needed to investigate the role of miR-1246 in this subset of ovarian cancer.
In summary, we found that miR-5703, miR-630, miR-1246, miR-424-5p, and miR-320b were dysregulated in CSC-like cells of OSC. Of these, decreased expression of miR-424-5p was associated with distant metastasis of OSCs. Our results indicate that these microRNAs are putative CSC markers, and that miR-424-5p is a potential predictive biomarker for poor prognosis and a possible therapeutic target in OSC.
Acknowledgments
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health &Welfare, Republic of Korea (grant number: HI16C1559).
References
- 1.Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin. 2002;52(1):23–47. doi: 10.3322/canjclin.52.1.23. [DOI] [PubMed] [Google Scholar]
- 2.McGuire WP, Hoskins WJ, Brady MF, Kucera PR, Partridge EE, Look KY, Clarke-Pearson DL, Davidson M. Cyclophosphamide and cisplatin versus paclitaxel and cisplatin: a phase III randomized trial in patients with suboptimal stage III/IV ovarian cancer (from the Gynecologic Oncology Group) Semin Oncol. 1996;23(5 Suppl 12):40–47. [PubMed] [Google Scholar]
- 3.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 4.Kim CF, Dirks PB. Cancer and stem cell biology: how tightly intertwined? Cell Stem Cell. 2008;3(2):147–150. doi: 10.1016/j.stem.2008.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N, Abubaker K, Findlay J, Quinn M. Cancerous ovarian stem cells: obscure targets for therapy but relevant to chemoresistance. J Cell Biochem. 2013;114(1):21–34. doi: 10.1002/jcb.24317. [DOI] [PubMed] [Google Scholar]
- 6.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nature reviews Cancer. 2005;5(4):275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 7.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324(5935):1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conic I, Dimov I, Tasic-Dimov D, Djordjevic B, Stefanovic V. Ovarian epithelial cancer stem cells. ScientificWorldJournal. 2011;11:1243–1269. doi: 10.1100/tsw.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalerba P, Clarke MF. Cancer stem cells and tumor metastasis: first steps into uncharted territory. Cell Stem Cell. 2007;1(3):241–242. doi: 10.1016/j.stem.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 10.Mor G, Yin G, Chefetz I, Yang Y, Alvero A. Ovarian cancer stem cells and inflammation. Cancer Biol Ther. 2011;11(8):708–713. doi: 10.4161/cbt.11.8.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, La Noce M, Laino L, De Francesco F, Papaccio G. Cancer stem cells in solid tumors: an overview and new approaches for their isolation and characterization. FASEB J. 2013;27(1):13–24. doi: 10.1096/fj.12-218222. [DOI] [PubMed] [Google Scholar]
- 12.Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006;124(6):1111–1115. doi: 10.1016/j.cell.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 13.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 14.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nature reviews Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435(7043):839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 16.Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15(6):341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- 17.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nature reviews Cancer. 2006;6(4):259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 18.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67(18):8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 19.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 20.Vang R, Shih Ie M, Kurman RJ. Ovarian low-grade and high-grade serous carcinoma: pathogenesis, clinicopathologic and molecular biologic features, and diagnostic problems. Adv Anat Pathol. 2009;16(5):267–282. doi: 10.1097/PAP.0b013e3181b4fffa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwon AY, Kim GI, Jeong JY, Song JY, Kwack KB, Lee C, Kang HY, Kim TH, Heo JH, An HJ. VAV3 Overexpressed in Cancer Stem Cells Is a Poor Prognostic Indicator in Ovarian Cancer Patients. Stem Cells Dev. 2015;24(13):1521–1535. doi: 10.1089/scd.2014.0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, Dirbas FM, Somlo G, Pera RA, Lao K, Clarke MF. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karst AM, Drapkin R. Ovarian cancer pathogenesis: a model in evolution. J Oncol. 2010;2010:932371. doi: 10.1155/2010/932371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leeper K, Garcia R, Swisher E, Goff B, Greer B, Paley P. Pathologic findings in prophylactic oophorectomy specimens in high-risk women. Gynecol Oncol. 2002;87(1):52–56. doi: 10.1006/gyno.2002.6779. [DOI] [PubMed] [Google Scholar]
- 25.Medeiros F, Muto MG, Lee Y, Elvin JA, Callahan MJ, Feltmate C, Garber JE, Cramer DW, Crum CP. The tubal fimbria is a preferred site for early adenocarcinoma in women with familial ovarian cancer syndrome. Am J Surg Pathol. 2006;30(2):230–236. doi: 10.1097/01.pas.0000180854.28831.77. [DOI] [PubMed] [Google Scholar]
- 26.Hambardzumyan D, Becher OJ, Holland EC. Cancer stem cells and survival pathways. Cell Cycle. 2008;7(10):1371–1378. doi: 10.4161/cc.7.10.5954. [DOI] [PubMed] [Google Scholar]
- 27.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, Qin LX, Yang W, Wang HY, Tang ZY, Croce CM, Wang XW. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50(2):472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, Huang Y, Hu X, Su F, Lieberman J, Song E. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131(6):1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 29.Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim SW, Kim YT. MicroRNA profiling of a CD133(+) spheroid-forming subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med Genomics. 2012;5:18. doi: 10.1186/1755-8794-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park YT, Jeong JY, Lee MJ, Kim KI, Kim TH, Kwon YD, Lee C, Kim OJ, An HJ. MicroRNAs overexpressed in ovarian ALDH1-positive cells are associated with chemoresistance. J Ovarian Res. 2013;6(1):18. doi: 10.1186/1757-2215-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garg M. Emerging role of microRNAs in cancer stem cells: Implications in cancer therapy. World J Stem Cells. 2015;7(8):1078–1089. doi: 10.4252/wjsc.v7.i8.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang JC. Cancer stem cells: Role in tumor growth, recurrence, metastasis, and treatment resistance. Medicine (Baltimore) 2016;95(1 Suppl 1):S20–25. doi: 10.1097/MD.0000000000004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olson P, Lu J, Zhang H, Shai A, Chun MG, Wang Y, Libutti SK, Nakakura EK, Golub TR, Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23(18):2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donnem T, Fenton CG, Lonvik K, Berg T, Eklo K, Andersen S, Stenvold H, Al-Shibli K, Al-Saad S, Bremnes RM, Busund LT. MicroRNA signatures in tumor tissue related to angiogenesis in non-small cell lung cancer. PLoS One. 2012;7(1):e29671. doi: 10.1371/journal.pone.0029671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32(8):976–987. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]
- 36.Bolos V, Grego-Bessa J, de la Pompa JL. Notch signaling in development and cancer. Endocrine reviews. 2007;28(3):339–363. doi: 10.1210/er.2006-0046. [DOI] [PubMed] [Google Scholar]
- 37.Ranganathan P, Weaver KL, Capobianco AJ. Notch signalling in solid tumours: a little bit of everything but not all the time. Nature reviews Cancer. 2011;11(5):338–351. doi: 10.1038/nrc3035. [DOI] [PubMed] [Google Scholar]
- 38.Fortini ME. Notch and presenilin: a proteolytic mechanism emerges. Current opinion in cell biology. 2001;13(5):627–634. doi: 10.1016/s0955-0674(00)00261-1. [DOI] [PubMed] [Google Scholar]
- 39.Zolkiewska A. ADAM proteases: ligand processing and modulation of the Notch pathway. Cellular and molecular life sciences: CMLS. 2008;65(13):2056–2068. doi: 10.1007/s00018-008-7586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mochizuki S, Okada Y. ADAMs in cancer cell proliferation and progression. Cancer science. 2007;98(5):621–628. doi: 10.1111/j.1349-7006.2007.00434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wildeboer D, Naus S, Amy Sang QX, Bartsch JW, Pagenstecher A. Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. Journal of neuropathology and experimental neurology. 2006;65(5):516–527. doi: 10.1097/01.jnen.0000229240.51490.d3. [DOI] [PubMed] [Google Scholar]
- 42.Sun Z, Meng C, Wang S, Zhou N, Guan M, Bai C, Lu S, Han Q, Zhao RC. MicroRNA-1246 enhances migration and invasion through CADM1 in hepatocellular carcinoma. BMC cancer. 2014;14:616. doi: 10.1186/1471-2407-14-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liao JM, Zhou X, Zhang Y, Lu H. MiR-1246: a new link of the p53 family with cancer and Down syndrome. Cell Cycle. 2012;11(14):2624–2630. doi: 10.4161/cc.20809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, Ikeda N, Isozaki Y, Maruyama T, Akanuma N, Komatsu A, Jitsukawa M, Matsubara H. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108(3):644–652. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]