Abstract
Although great strides have been made in understanding the complex bacterial community inhabiting the human oral cavity, for a variety of (mainly technical) reasons the ecological contributions of oral fungi, viruses, phages, and the candidate phyla radiation (CPR) group of ultra-small bacteria have remained understudied. Several recent reports have illustrated the diversity and importance of these organisms in the oral cavity, while TM7x and Candida albicans have served as crucial paradigms for CPR species and oral fungi, respectively. A comprehensive understanding of the oral microbiota and its influence on host health and disease will require a holistic view that emphasizes interactions among different residents within the oral community, as well as their interaction with the host.
Keywords: oral microbiome, phage, fungi, candidate phyla radiation, meta-omics
Overlooked contributors: ultra-small bacteria, fungi, and phage play a significant role in the ecology of the human oral microbiome
Since the initial discovery of bacteria from the oral cavity by Antonie van Leeuwenhoek in the 18th century [1], the human oral microbiota has become the model system for studying multispecies microbial communities [2–4]. These indigenous microbes must engage and co-evolve with their neighbors and hosts, as well as adapt to diverse and rapidly fluctuating conditions. Despite this, the microbial composition is relatively stable [5] and also displays community-level functions such as colonization resistance [6]. These characteristics require a complex level of interspecies communication, which necessitates the full arsenal of current technologies in order to wholly appreciate. As a result, until recently the study of the oral microbiota has mainly focused on bacteria due to their relative high abundance, easy detection, and cultivability. It is now apparent that the reductionist approach of studying communities via their individual components, although useful, cannot adequately describe the complex relationships that exist. Major technological and analytical advances, particularly in cultivation-independent detection methods, have made it possible to investigate communities as a whole by using and combining systems-based approaches. These studies have revealed an exceedingly more diverse oral microbial world than what was anticipated based on culture-dependent investigation. We now know that the oral cavity not only harbors enormously diverse bacterial species, but also is home to a multitude of yet-to-be cultured ultra-small bacteria belonging to the newly classified “Candidate Phyla Radiation” (CPR) group [7], as well as fungi and viruses. Compared to the traditional bacterial microbiome (bacteriome), knowledge regarding these “rare” (fungi), and “tiny” (CPR group bacteria and virus/phage) residents is lagging behind. This article will briefly review the hurdles accounting for the lag in the study of these three oral microbial sub-groups, and the techniques used to overcome these obstacles. Subsequently, discussion will cover the recently described interactions between the CPR, fungi, and viruses and their bacterial hosts and neighbors, as well as their impact on the overall oral microbiome and health of the human host—an exciting and rapidly emerging new frontier in the study of the host-associated microbiome. The interspecies interactions between established bacterial species will not be discussed in this review, as there is already an extensive amount of literature covering this topic. The authors refer the interested reader to several recent reviews for useful summaries on these relationships [2–4, 8].
From reductionism to holism: Systems-level understanding of human oral microbiome
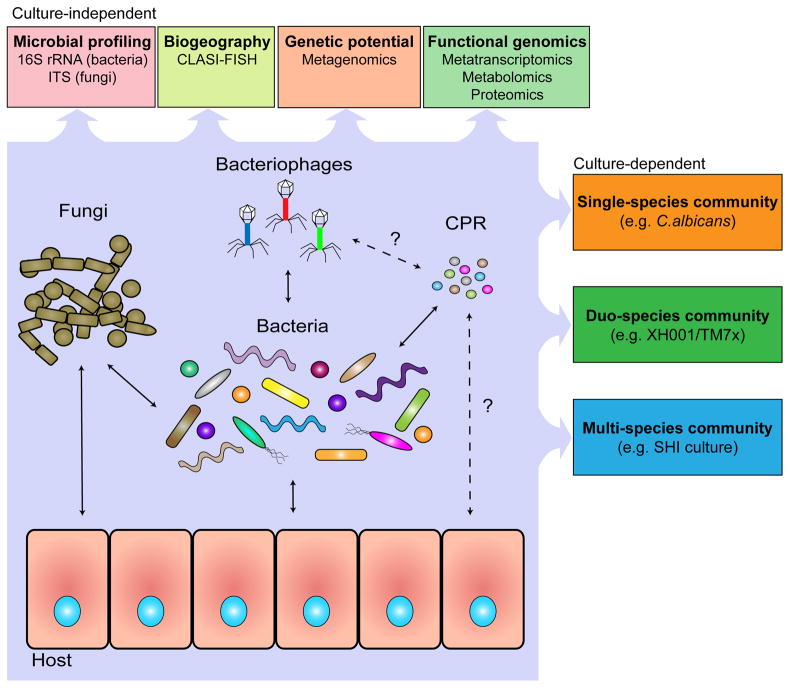
Investigations into microbial communities have traditionally relied on the study of individually isolated organisms in order to make inferences about the entire community. While this approach has greatly advanced microbiology in the past 400 years, the reductionist method cannot be used to sufficiently explain the complexities of the community structure. Recognizing that the whole is more than the simple sum of its parts, modern microbiologists are learning to apply “systems thinking” to take a holistic approach to investigations. Research has transitioned from culture-dependent studies of a single species to complex in vitro multispecies communities [9, 10] as well as the culture-independent characterization of entire in vivo microbiota, and from analyzing individual gene expression to meta-omics analysis. Microbiology is experiencing a new movement that emphasizes interactions between different elements within a community (Fig. 1).
Figure 1: Key Figure. A systemic approach to the investigation of human oral microbiome.
A holistic understanding of human microbiome requires a systemic approach to better study the interactions within and between different oral microbial groups (bacteria, CPR, fungi and virus), their impact on microbial physiology, community ecology, and host homeostasis. State-of-the-art, culture-depended methods, coupled with traditional culture-dependent approaches allow researchers to better identify the characteristics of the microbiome, such as the prevalence and biogeography of all species present, as well as the interactions and metabolic pathways critical to the community and the effects of the community on the host. The application of these approaches has revealed a highly complex oral microbial community with sophisticated and dynamic interspecies interactions, which shape and define the community structure as well as functions. Within the oral microbiome, bacterial-bacterial, bacterial-fungal and bacterial-phage interactions and their impact on host have been extensively documented (solid arrows). Meanwhile, the improved cultivation approach, together with crucial genetic information obtained using culture-independent method resulted in the discovery of distinct bacterial-CPR interactions which could potentially prove to be common in host-associated microbiota (dotted arrows). Furthermore, culture-independent metagenomic analysis also suggested putative interspecies interactions, such as the presence of phage specifically targeting CPR members and direct interaction between CPR and the host (dashed arrows).
One of the fundamental questions in the study of the microbiome is: What species are present, and in what relative abundance? To that end, sequencing of the bacterial 16S rRNA gene has yielded invaluable information on the bacterial components of the oral microbiome and has allowed for the creation of the Human Oral Microbiome Database (HOMD) [11]. With this resource, researchers have access to a curated database of oral microbial sequences, streamlining analysis of 16S sequencing data. However, extensive databases such as HOMD do not yet exist for microbes other than bacteria, which can hamper complete analysis and identification efforts. This knowledge gap is consequential; as discussed in the following sections, the fungal and viral components of the oral microbiome play significant roles in modulation of the oral community and its contribution to host disease.
Compared to the bacterial component of the microbiome, the fungal component, or mycobiome, as it has been termed, has remained relatively understudied. Several challenges have contributed to this lag: fungi are relatively rare (<0.1% of the microbiome, based on cfu), genetic material from fungi can be difficult to isolate, and many fungal species are uncultivable using current methods. Compounding these issues further is the fact that nomenclature of fungal species and annotation of fungal genomes is confusing, and the few existing databases are fraught with redundancies and errors [12–14]. Although there is now known to be a correlation between fungi and several diseases, including inflammatory bowel syndrome, Crohn’s Disease, chronic respiratory diseases, and Hepatitis B, this association was not discovered until the concepts of dysbiosis and ecological diseases had been established [12, 15, 16]. These paradigm shifts in the study of microbiology, along with shotgun metagenomic sequencing and sequencing of the internal transcribed spacer (ITS) region of rRNA genes in fungi (analogous to 16S rRNA sequencing in bacteria), have finally enabled recent research to give the human mycobiome the attention that it deserves.
Although viruses and phages are the most abdundant biological entities on earth [17], like the mycobiome, the viral component of the human microbiome (virome) has been understudied compared to the bacterial element. Viruses have been especially ignored in the context of their effects on bacteria and the human microbiome in health, as there is no overt indication of their existence in the absence of symptoms. As major technical advancements have brought the human virome to light, several critical and unanticipated observations have been described. It is now known that healthy humans carry a large diversity of both eukaryotic viruses and prokaryotic phages [17–19]. Furthermore, human blood, once thought to be sterile in healthy individuals, was found to contain both virus and phage [17, 20]. A recent review by Lecuit et al. metaphorically described the human virome as an iceberg, in which the visible tip was representative of the viruses causing symptoms (Ebola, HIV, influenza, etc.), which are relatively well-studied and small in number. Below the surface is the vast portion of the human virome that does not cause symptoms, is poorly studied, and the consequences of which are largely unknown [21]. Despite research and technological advances, there remain several challenges to studying the “bulk of the iceberg”. Although virions outnumber host cells roughly 10:1, viral nucleic acids are thought to make up less than 0.1% of the total nucleic acids and thus can easily be overlooked [22]. In addition, virions themselves, particularly those of unknown viruses, are difficult to purify [22]. Even after quality genome sequences are obtained, viral sequences may be completely novel and may encode proteins with no known homology, making analysis and annotation extremely difficult if not impossible [22]. Finally, identifying the host range and tropism of a novel virus can prove greatly challenging. Various techniques for overcoming these barriers are being developed and have been recently reviewed [23, 24]. As momentum for the study of the human virome grows, it adds an additional layer of complexity to the microbiome.
In addition to investigating presence and abundance, community sequencing data is useful in identifying potentially interacting species through co-occurrence or co-exclusion data. By analyzing the oral mycobiome of HIV patients, Mukherjee et al. [25] were able to identify an antagonistic relationship between two fungal species based on their anti-correlation. This type of data can provide hypotheses for further physiological testing of potential interactions between organisms. By simultaneously sequencing the bacterial, fungal, and viral communities, novel inter-kingdom interactions can be identified [26].
To fully understand the relationships between the inhabitants of the oral microbial community, it is not enough to identify each organism; the complex spatial and structural organization must also be taken into account. Recently, Mark Welch et al. [27] combined sequencing data with spectral fluorescence imaging that allowed for the direct visualization of bacteria within dental plaque. By specifically labeling the most prevalent bacteria, the technique of CLASI-FISH (combinatorial labeling and spectral imaging - fluorescence in situ hybridization) revealed a highly organized structure as well as novel interbacterial interactions (Fig. 1). Close proximity between organisms may suggest a metabolic or physiological relationship or dependency. This knowledge is important for understanding the microbial ecology as a whole and can aid in the discovery and cultivation of novel species.
Advanced sequencing techniques are capable of generating information on the functional activity of a microbial community. Metagenomics, which entails sequencing the entirety of the DNA from a given sample, can provide information not only on what organisms are present, but also on their functional potential through analysis of metabolic pathway genes and the use of protein-coding sequence databases [28, 29]. However, DNA sequences alone do not provide any information on actual gene expression and microbial activity. Current studies in the oral cavity, and beyond, that combine metatranscriptomics and metagenomics have shown that in many cases, the set of genes that are expressed is more important in predicting health vs. disease than the species that are present [30, 31].
Meta-metabolomics and -proteomics can complement sequencing data by providing higher-level functional information. Interacting organisms in a community commonly have intertwined metabolisms through cross-feeding or specialization of function. The most extreme examples of this include parasitic organisms such as TM7x, discussed in depth in the following section, which fully rely on the metabolic pathways of the host organism [32]. Combining systems-level analyses that generate functional information with the knowledge of abundance and spatial location can provide powerful insight into the intricate microbial network of the oral microbiome, its relationship to the human host, and its potential to influence health and disease (Fig. 1).
Interaction between oral CPR organisms and their bacterial hosts
Unlike fungi and viruses, the Candidate Phyla Radiation (CPR) group of bacterial organisms was discovered only recently. This revelation has had an enormous impact on our view of the diversity of life on earth [7]. The previously unknown CPR group of organisms contains more than 35 phyla, and may comprise greater than 15% of domain bacteria! [7, 33, 34] CPR organisms have been found in diverse ecological niches, and share characteristics such as ultra-small cell size, the presence of 16S rRNA gene self-splicing introns, archaeal-specific RuBisCO genes [35], and reduced genomes with the apparent absence of genes encoding a CRISPR/Cas bacteriophage defense-system [36]. Traits of this group also include the absence of several biosynthesis and metabolic pathways, including the electron transport chain, tricarboxylic acid cycle, amino acid and membrane biosynthesis pathways, and various ribosomal subunits [32, 33, 37–40]. Due to these shared properties, CPR organisms are predicted to be obligate-symbionts. Direct evidence of their suspected symbiotic lifestyle, as well as knowledge regarding their physiology, their interaction with hosts, and their potential role in shaping the microbial community, is lacking due to their recalcitrance to in vitro cultivation.
TM7x: the first cultivated CPR species uncovers an obligate, epiparasitic interaction with an Actinomyces host
The human microbiome project established that three CPR phyla, Gracilibacteria (GN02), Absconditabacteria (SR1) and Saccharibacteria (TM7), are frequently detected in multiple human body sites, including the oral cavity [41, 42]. Among the three human-associated CPR phyla, TM7 is the most well studied due to its association with multiple mucosal diseases such as vaginosis, inflammatory bowel disease, halitosis, and periodontitis [43–47]. TM7 is particularly prevalent in the oral cavity. Although commonly at an abundance of ~1% [48], an increase in abundance of TM7 to as high as 21% was detected in patients with various types of periodontitis [49, 50]. The positive association between oral CPR members and oral diseases [51, 52], highlights the capacity of ultra-small bacteria to contribute to human disease, as well as modulate the human immune response [32].
A species of TM7, TM7x (strain TM7x HOT_952) was recently isolated from the human oral cavity together with its host bacterium, Actinomyces odontolyticus actinosynbacter (strain XH001) [32, 53]. This is to date the only member of the entire CPR group that has been successfully cultivated and stably maintained in vitro. Initial studies indicate that TM7x is an obligate epiparasite that lives on the surface of its bacterial host, XH001. Like other CPR members, TM7x has an ultra-small cell size (200–300 nm) and a reduced genome lacking essential pathways (~700 genes) [32, 33]. Further studies also revealed that the epiparasitic relationship between the two species is dynamic, and can change depending on environmental conditions such as nutrient availability and oxygen concentration (Box 1). Interestingly, infection of macrophages with TM7x-associated XH001 led to a significant reduction in the production of cytokines, compared with cytokine production in macrophages infected with XH001 alone, suggesting that interactions between TM7x and XH001 are more complex than a simple metabolic dependency, and may modulate host response to oral microbiota [32, 54].
Box 1. The relationship between TM7x and Actinomyces odontolyticus actinosynbacter (XH001): a prototype of the parasitic lifestyle of CPR organisms.
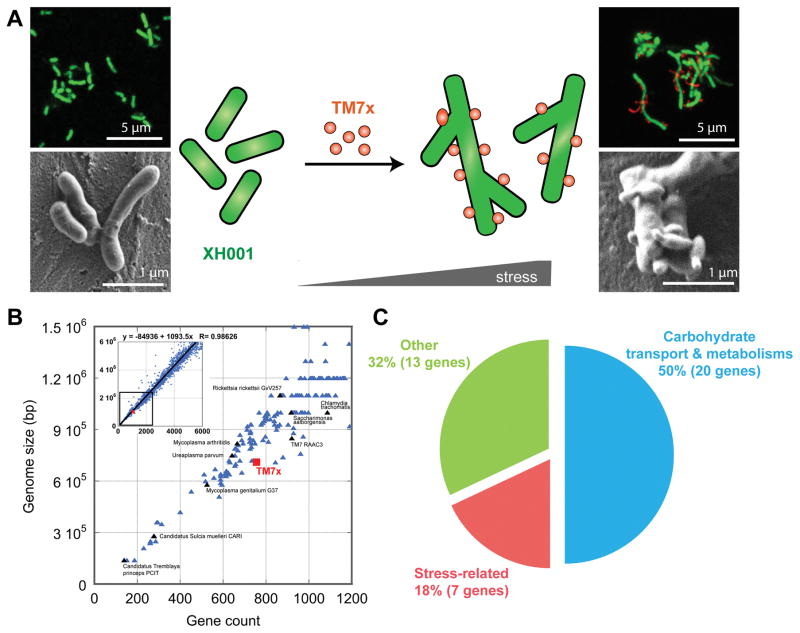
TM7x was recently isolated from the human oral cavity as an obligate epibiont that lives on the surface of its bacterial host, XH001, representing thus far the only in vitro cultivated and stably maintained member of the CPR group [32, 89]. Isolation of pure TM7x is achieved by separating TM7x cells from host XH001 via the repeated passage of the co-culture through a 28-gauge needle, followed by filtering of the mixture through a 0.45-μm filter to collect TM7x. Isolated TM7x fails to grow, but maintains its ability to re-establish parasitic association with naïve XH001 [32, 89]. During symbiotic growth under nutrient-replete conditions, TM7x mainly grows attached to the host surface as cocci with bud-like protrusions, while XH001 displays an elongated cell structure compared to the short rod-shaped naïve XH001. However, under starvation or other stress conditions, TM7x induces additional morphological changes in the host, such as clubbed ends and swollen cell body, and eventually results in host cell lysis. TM7x displays multiple cell morphotypes, ranging from cocci to short rods to filaments [89] (Fig. IA).
Whole genome sequencing revealed that the TM7x genome has a high coding density and ranks smallest among the bacteria found to date in the human body, in both genome size (705,138 bp) and gene count (699 coding sequences) (Fig. IB). TM7x lacks genes necessary for de novo biosynthesis of any essential amino acid, while its host XH001 has complete biosynthesis pathways. Transcriptomics revealed a genome-wide response of XH001 when it is associated with TM7x, with about 340 XH001 genes differentially regulated. Among the most up-regulated genes are those related to carbohydrate transport and metabolism, as well as general stress response (Fig. IC). Genomic and transcriptomic analysis in combination with the phenotypic data supports the hypothesis that TM7x employs a parasitic rather than mutualistic/commensal epibiotic relationship with XH001 [89]. This epiparasitic relationship is likely a life style adopted by many CPR members, which share the characteristics of small cell size and a reduced genome. Further genomic, transcriptomic, metabolomic, and pathogenesis investigations of TM7x/XH001 will lead to a better understanding of this intriguing relationship and shed light on the ecological and clinical impact of CPR organisms.
Figure I. Interaction between first cultivated CPR member, TM7x and its bacterial host XH001.
(A) Both fluorescence in situ hybridization and scanning electron microscopy revealed XH001 commonly has short-rod cell shape, while TM7x induces long and hyphal morphology in XH001 by inducing the stress-response genes. TM7x also displayed elongated and bud-like protrusions, in addition to cocci morphology [89]. (B) TM7x has small genome size with reduced gene count consistent with the observed lifestyle as an obligate symbiont [89]. (C) Functional classes of the forty most significantly up-regulated genes in the transcriptomic studies in XH001 upon TM7x binding [89].
Potential impact of CPR-bacterial host interaction on microbial ecology
The obligate epiparasitic relationship between TM7x and XH001 represents a novel, while likely common, interspecies interaction in oral microbiota, as suggested by the presence of a significant CPR population [55]. The distinct interspecies interactions between CPR organisms and their hosts may have a considerable impact on oral microbial ecology at various levels, ranging from direct reciprocal effects of the two species on physiology and pathogenicity, to indirect influence on the overall structure and function of the oral microbiota. It is becoming increasingly clear that the CPR group of organisms interact with the conventional microbiome, and play a critical, yet very poorly understood, role in the development of the human microbiota and disease [56, 57]. As impetus for the study of ultra-small bacteria increases, a more detailed comprehension of the relationship between TM7x and XH001 will provide a valuable prototype for understanding the unique lifestyle of the CPR group, and will shed long-awaited light on the biology of “bacterial dark matter”.
Bacterial-fungal interactions in the oral cavity
Two landmark metagenomic studies using culture-independent identification methods showed that the healthy oral cavity contains a much more diverse array of fungal species than previously thought [58, 59]. Ghannoum et al. reported that the healthy oral cavity was home to more than 75 genera of fungi, with Candida, Cladosporium, Aureobasidium, and Aspergillus the most abundant, found in 25–75% of subjects [58]. These findings were confirmed by the subsequent Depuy et al. study, which used further refined protocols to illustrate the diversity of the healthy oral mycobiome and significant variation in the fungal species present between individuals with good oral health [59]. The Depuy study also concluded that in addition to the genera identified by the Ghannoum study, Malassezia spp. are a frequent member of the healthy oral mycobiome [59].
Despite being numerically underrepresented, the larger cell size of fungal species in the human microbiota indicates that they contribute a proportionally larger amount of the biomass. This greater cell size and ability to generate filamentous hyphae also places fungi in a position to create a structural “skeleton” for fungal-bacterial multispecies biofilms. In addition, as eukaryotes, fungal species stimulate the host immune system in a distinct manner with disparate immunological outcomes compared to that of their bacterial neighbors [12]. Indeed, there is a growing body of evidence supporting the importance of immunological stimulation by fungi, in addition to bacteria, for development of a robust host immune system [12, 15, 60, 61]. The fact that a small number of fungi can have a disproportionally large effect on the microbiota has led to fungal species being brought forward as potential “keystone species” [62, 63]. While this is indeed a tempting hypothesis, further ecological studies will be needed to confirm the status of fungal species as true keystones. This concept has most notably been proposed in the case of Candida albicans, which, due to its nearly ubiquitous nature and the ease of its cultivability, is an exception among the mycobiome in that it is well studied in the context of its inter-kingdom interactions and its importance at large to the microbiological community [62, 63]. The known interactome of C. albicans and its relation to disease has been reviewed in-depth [64], and several characterized relationships between C. albicans and oral bacteria are summarized in Box 2. The capability of interspecies interactions between C. albicans and Streptococcus mutans or Streptococcus oralis to exacerbate the severity of oral candidiasis or dental caries, respectively, as well as cooperation between C. albicans and Fusobacterium nucleatum to evade the host immune system, highlight the importance of inter-kingdom interactions in the pathogenesis of what are increasingly recognized as polymicrobial diseases. Going forward, the authors suggest that C. albicans and its partner bacteria will serve as a paradigm of fungal-bacterial interactions as new technologies improve the study of the role of the diverse group of fungal species that inhabit the human mouth.
Box 2. Oral Fungal-bacterial interactions and disease: C. albicans as a paradigm.
A wide array of interactions involving oral bacteria and C. albicans have been described, and range from mutualistic to competitive, with various positive and negative impacts on host immune response and health. In addition to the three examples discussed briefly below, we refer the reader to the following reviews [64, 90–93].
C. albicans and S. oralis
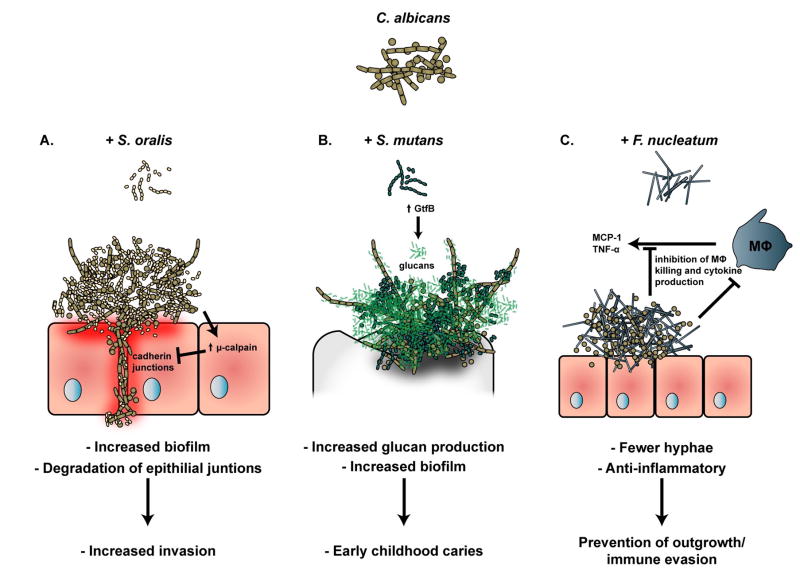
Although historically considered commensal organisms, the Mitis group of streptococci, including Streptococcus oralis, has been shown to cooperate with C. albicans to increase the severity of oral candidiasis [63, 94, 95]. S. oralis and C. albicans synergistically increase the level of μ-calpain, a proteolytic enzyme that degrades the E-cadherin from oral epithelial junctions [96]. This amplified structural breakdown enhances the ability of C. albicans to invade oral and esophageal mucosa during S. oralis co-infection [97] (Fig. IIA).
C. albicans and S. mutans
C. albicans is frequently associated with the cariogenic organism Streptococcus mutans. Although there is some conflict in the literature as to the virulence of C. albicans in the context of dental caries [98, 99], a recent report suggests that the differences observed may be due to differing growth media and conditions between the studies [100]. Regardless, several studies have confirmed that both C. albicans and S. mutans grow more robustly in a dual-species biofilm than either species alone, and that the presence of C. albicans induces changes in the expression of several S. mutans virulence factors [98, 99, 101]. C. albicans is thought to provide structural support to dental plaque by avidly binding the S. mutans exoenzyme GtfB, responsible for generation of the exopolysaccaride matrix, furnishing an agglutinative skeletal structure for biofilm growth [102]. The considerable increase in carious lesions observed in a Streptococcus mutans-C. albicans dual species rodent caries model, combined with frequent isolation of high numbers of both C. albicans and S. mutans from children with severe early childhood caries, suggests that at least under certain conditions, C. albicans contributes significantly to the capacity of the dental plaque community to cause disease [98, 103, 104] (Fig. IIB).
C. albicans and F. nucleatum
Recent work showed that a direct physical interaction between C. albicans and F. nucleatum reduced both the growth of C. albicans as well as its capacity to produce hyphae, which is more associated with invasive candidal outgrowth than the yeast form [54]. While other species have been shown to inhibit the yeast-hyphae transition in C. albicans, this report was the first describing a contact-dependent and specific mechanism [54]. The attenuation of C. albicans hyphae caused by this interaction actually increased the survival of C. albicans in the presence of macrophages, and reduced the production of proinflammatory cytokines by co-infected macrophages. Therefore, the intriguing working hypothesis is that C. albicans and F. nucleatum work together to prevent their own outgrowth from drawing excessive attention from the host immune system, promoting a commensal rather than a dysbiotic and pathogenic way of life [54] (Fig. IIC).
Figure II. Three examples of inter-kingdom interactions of C. albicans and their effects on the human host.
(A.) Biofilms of C. albicans and S. oralis grow to a greater density than that of either species grown alone. In addition, the dual-species biofilm up-regulated the host protease μ-calpain, which degrades cadherin junctions leading to increased tissue invasion by C. albicans and S. oralis [96, 97]. (B.) Biofilms containing both C. albicans and S. mutans grow to a greater density than that of either species grown alone and are associated with development of severe early childhood caries, due to a C. albicans-mediated up-regulation of S. mutans virulence factors including glucan production [98, 103]. (C.) When grown in co-culture with F. nucleatum, C. albicans is greatly inhibited in its ability to form hyphae. Further, the reduction in hyphae appears to circumvent killing and cytokine production by macrophage, indicating that the co-culture may promote evasion of the host immune system [54].
The human oral virome
In recent years, several studies have pioneered the application of next-generation sequencing techniques to elucidate of the human oral virome in several contexts including health, following long-term antibiotic use, and in association with periodontal disease [19, 65–68]. The oral virome of healthy individuals contains both eukaryotic viruses and bacteriophage. The bacteriophages are vastly greater in number, and thus will be the primary focus of this section of the review. The human oral virome is highly varied between individuals and surprisingly stable over time, even compared to the oral bacteriome [18, 65, 69]. Furthermore, sharing of living environment may have an important role in determining the ecology of the human oral virome, as members of the same household had remarkably similar viromes [66]. Herpesviriade, Papillomaviridae, and Anelloviridae are among the most common eukaryotic virus families present, and are asymptomatic in healthy individuals [68]. Several studies have examined the association of Herpesviruses, such as herpes simplex virus, human cytomegalovirus, and Epstein-Barr virus, with periodontal disease, however, a clear narrative regarding the influences of eukaryotic viruses on oral microbial diseases has yet to emerge [70].
Bacteriophages modulate the oral microbiome through multiple mechanisms
The majority of the identified viruses in the oral cavity are bacteriophages, with Siphoviridae, Myoviridae, and Podoviridae as the most common phage families identified. Using homologous sequences to predict host range, Pride et al identified putative phages of numerous genera covering all of the major bacterial phyla found in the oral cavity, including CPR members such as TM7 [18]. Intriguingly, the relative abundances of salivary phages and their respective putative bacterial hosts showed both direct and inverse relationships, indicating that mutualistic and antagonistic co-evolutionary relationships between oral phage and their bacterial hosts exist [18].
As in many other microbial communities, oral phages are thought to play a significant role in shaping the oral microbiome [19]. The Siphoviridae are largely lysogenic, which from a community standpoint suggests an establishment of a dynamic equilibrium with associated host species, and also provides a huge opportunity for the transfer of genetic information. Horizontal gene transfer is thought to be particularly important in the oral cavity, as the massive diversity of organisms and large amount of extracelluar DNA in biofilm matrices gives the resident species opportunity to acquire a vast array of genes [71]. Recent work has shown that phage-mediated mobile genetic elements are critical for the spread of antibiotic resistance in the oral cavity [72, 73]. It is currently estimated that 60–70% of known bacterial genomes contain prophages, a number that is likely to increase as novel phages are sequenced. These prophages are not passive, but rather, they are active participants in shaping the bacterial community composition and biogeography, which in turn have an effect on the human host and its immune system [72, 74–76].
The Myoviridae and Podoviridae are predominantly lytic, and rapidly eliminate their susceptible hosts. Phages are now thought to account for 20–80% of bacterial death, and therefore represent a profoundly significant bacterial limiting factor [76]. In a classic Red Queen dynamic, the genetic arms race invoked by coevolution of phage and host bacteria likely serves to prevent novel bacteria and phage from establishing in the oral community [75]. Several recent studies have explored CRISPRs transcribed in the oral cavity and their protective effects against invading phage [69, 77, 78]. On the other hand, recent investigations suggest that oral phage may employ anti-CRISPR proteins to inactivate the bacterial immune system, preventing the eradication of phage by bacteria, and allowing phage to persist within the oral microbiome [79, 80]. Lytic phages have been shown to kill bacterial species that pass a particular population threshold, preventing outgrowth and dysbiosis. This “Kill the Winner” dynamic, as well as a number of other phage-bacteria dynamics, both antagonistic and mutualistic, have been described, and have been shown to result in either positive or negative outcomes for the human host [24, 75, 76, 81]. A recent study by Ly et al. showed that the human oral phage ecology in dental plaque is associated with oral health status, with significantly higher abundance of lytic myoviruses in the subgingival plaque of individuals with periodontal disease, suggesting that phage play a critical role in modulating the microbial community and contributing to disease development [19].
Phage therapy revisited
The increase in attention given to the study of phage modulation of the bacterial community, coupled with the rising tide of antibiotic resistance, has led to a resurgence of interest in phage therapy. Phage therapy does present a number of significant advantages compared to conventional antibiotic treatment: virtually limitless diversity of potential phage and the ability of phage to rapidly overcome resistance in situ [82, 83]. However, the capacity of phage therapy to effectively penetrate biofilms, where neighbors and polysaccharide barriers may block phage receptors, remains an area for further study and progress and is a particularly relevant issue in the oral cavity [84]. Additionally, while the specificity of phage allows it to minimally damage non-target species, leaving the overall community intact, it requires rapid identification of the target in question, as well as the target’s susceptibility to phage-mediated killing. Finally, phage therapy is likely to face significant regulatory headwinds, with issues concerning standardization, intellectual property, and safety [83, 85]. Several recent reports have identified phages in the oral cavity that infect F. nucleatum, S. mutans, and Neisseria meningitidis, opening the possibility of therapeutic phage development against these oral pathogens [86–88]. In the future, it is likely that both antibiotics and phage will play a role in battling pathogenic infections, as supported by a number of recent studies showing synergistic effects of combined antibiotic and phage therapies (reviewed in [83]).
Just as recognition of the influence of the bacterial microbiota on the immune system and human health recast the tenets of microbial ecology, the interactions of viruses and phage with bacteria, as well as with the human host, are likely to revolutionize thinking about the virome and microbiome at large. Phages clearly represent an additional level of balance required for eubiosis that is still poorly understood. As the complexity of the phage-bacterial interactome is unraveled, it is likely that new mechanisms of phage resistance and bacterial immunity will be discovered and will increase our understanding of the microscopic and sub-microscopic world.
Concluding remarks
Our ability to identify and isolate oral microbial residents and decipher their extensive interactions has rapidly increased within the past decade. However, our understanding of the oral microbiota as an ecological system with diversified, interactive entities is still in its infancy (see Outstanding Questions). A comprehensive understanding of the oral microbiota and its influence on host health and disease requires a holistic view that emphasizes interactions among different residents within the community as well as their interaction with the host. Such a deep systems-level understanding at the molecular level demands expansion of our knowledge from bacterial interspecies interactions to inter-kingdom interactions which include CPR, fungi, and viruses—a challenging task that necessitates a systems-level approach combining conventional culture-dependent methods with state-of-the-art culture-independent molecular techniques. Continued expansion of such information in the near future will greatly improve our understanding of oral microbial physiology, pathogenesis and ecology, as well as our ability to diagnose and treat microbial infections.
Acknowledgments
This work was supported in part by grants from the National Institute of Health (NIH-1-R01-DE020102, NIH-1-R01-DE023810, and NIH-1-R01-DE026186-01), National Institute of Health National Research Service Award Postdoctoral Fellowship F32DE025548-01 (B.B.) and NIDCR T90 (DE022734) training award to (M.A.). The authors declare that WS is an employee of C3 Jian, Inc., which has licensed technologies from UC Regents that could be indirectly related to this research project.
Glossary
- Bacteriome
the bacterial component of the microbiome.
- Candidate Phyla Radiation (CPR)
a recently identified, but highly abundant (>15% of bacteria) group of ultra-small (extremely small bacteria with sizes in the nanometer range, compared to classic bacterial size on the micron scale) bacteria with reduced genomes and unusual ribosomes.
- Obligate-symbiont
organism that absolutely requires a host species to survive and replicate.
- Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system
a prokaryotic adaptive immune system that provides resistance to foreign genetic elements such as those from plasmids and phage.
- Culture-independent detection methods
methods of detection that do not require laboratory growth or isolation of the organism(s).
- Dysbiosis
compositional shift in the population of a particular microbial community that promotes development of an inflammatory or disease state.
- Keystone species
a species with an elevated capacity to influence a microbial community relative to its abundance.
- Lysogenic
viral life cycle in which the viral nucleic acid is integrated into the host genome, and dissemination of viral DNA occurs through usual host reproduction.
- Lytic
viral life cycle in which progeny virions are assembled inside of the infected host and eventually cause lysis of the host and subsequent spread of virus.
- Mycobiome
the fungal component of the microbiome.
- Phage therapy
the use of phage to treat pathogenic bacterial infections.
- Prophage
a bacteriophage viral genome that has integrated into the host genome.
- Red Queen Hypothesis
the concept that organisms must constantly adapt and evolve to survive competition with other organisms that are also constantly adapting and evolving
- Tropism
the range of host cells and/or growth conditions for an organism
- Virion
a single viral particle
- Virome
the viral component of the microbiome.
References
- 1.Gest H. The discovery of microorganisms by Robert Hooke and Antoni Van Leeuwenhoek, fellows of the Royal Society. Notes Rec R Soc Lond. 2004;58(2):187–201. doi: 10.1098/rsnr.2004.0055. [DOI] [PubMed] [Google Scholar]
- 2.Kuramitsu HK, et al. Interspecies interactions within oral microbial communities. Microbiol Mol Biol Rev. 2007;71(4):653–70. doi: 10.1128/MMBR.00024-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–37. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 4.Kolenbrander PE, et al. Oral multispecies biofilm development and the key role of cell-cell distance. Nat Rev Microbiol. 2010;8(7):471–80. doi: 10.1038/nrmicro2381. [DOI] [PubMed] [Google Scholar]
- 5.Belstrom D, et al. Temporal Stability of the Salivary Microbiota in Oral Health. PLoS One. 2016;11(1):e0147472. doi: 10.1371/journal.pone.0147472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X, et al. The social structure of microbial community involved in colonization resistance. ISME J. 2014;8(3):564–74. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hug LA, et al. A new view of the tree of life. Nat Microbiol. 2016;1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 8.Guo L, et al. Intercellular communications in multispecies oral microbial communities. Front Microbiol. 2014;5:328. doi: 10.3389/fmicb.2014.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edlund A, et al. An in vitro biofilm model system maintaining a highly reproducible species and metabolic diversity approaching that of the human oral microbiome. Microbiome. 2013;1(1):25. doi: 10.1186/2049-2618-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edlund A, et al. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 2015;9(12):2605–19. doi: 10.1038/ismej.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dewhirst FE, et al. The human oral microbiome. J Bacteriol. 2010;192(19):5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cui L, et al. The human mycobiome in health and disease. Genome Med. 2013;5(7):63. doi: 10.1186/gm467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delhaes L, et al. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community--implications for therapeutic management. PLoS One. 2012;7(4):e36313. doi: 10.1371/journal.pone.0036313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seed PC. The human mycobiome. Cold Spring Harb Perspect Med. 2015;5(5):a019810. doi: 10.1101/cshperspect.a019810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizzetto L, et al. Richness and diversity of mammalian fungal communities shape innate and adaptive immunity in health and disease. Eur J Immunol. 2014;44(11):3166–81. doi: 10.1002/eji.201344403. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen LD, et al. The lung mycobiome: an emerging field of the human respiratory microbiome. Front Microbiol. 2015;6:89. doi: 10.3389/fmicb.2015.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abeles SR, Pride DT. Molecular bases and role of viruses in the human microbiome. J Mol Biol. 2014;426(23):3892–906. doi: 10.1016/j.jmb.2014.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pride DT, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. 2012;6(5):915–26. doi: 10.1038/ismej.2011.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ly M, et al. Altered oral viral ecology in association with periodontal disease. MBio. 2014;5(3):e01133–14. doi: 10.1128/mBio.01133-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Breitbart M, Rohwer F. Method for discovering novel DNA viruses in blood using viral particle selection and shotgun sequencing. Biotechniques. 2005;39(5):729–36. doi: 10.2144/000112019. [DOI] [PubMed] [Google Scholar]
- 21.Lecuit M, Eloit M. The human virome: new tools and concepts. Trends Microbiol. 2013;21(10):510–5. doi: 10.1016/j.tim.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bikel S, et al. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput Struct Biotechnol J. 2015;13:390–401. doi: 10.1016/j.csbj.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards RA, et al. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40(2):258–72. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogilvie LA, Jones BV. The human gut virome: a multifaceted majority. Front Microbiol. 2015;6:918. doi: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee PK, et al. Oral mycobiome analysis of HIV-infected patients: identification of Pichia as an antagonist of opportunistic fungi. PLoS Pathog. 2014;10(3):e1003996. doi: 10.1371/journal.ppat.1003996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Norman JM, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160(3):447–60. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mark Welch JL, et al. Biogeography of a human oral microbiome at the micron scale. Proc Natl Acad Sci U S A. 2016;113(6):E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oh J, et al. Temporal Stability of the Human Skin Microbiome. Cell. 2016;165(4):854–66. doi: 10.1016/j.cell.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 30.Yost S, et al. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 2015;7(1):27. doi: 10.1186/s13073-015-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jorth P, et al. Metatranscriptomics of the human oral microbiome during health and disease. MBio. 2014;5(2):e01012–14. doi: 10.1128/mBio.01012-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He X, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci U S A. 2015;112(1):244–9. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain Bacteria. Nature. 2015;523(7559):208–11. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 34.Attar N. Bacterial evolution: CPR breathes new air into the tree of life. Nat Rev Microbiol. 2016;14(6):332–3. doi: 10.1038/nrmicro.2016.63. [DOI] [PubMed] [Google Scholar]
- 35.Wrighton KC, et al. RubisCO of a nucleoside pathway known from Archaea is found in diverse uncultivated phyla in bacteria. ISME J. 2016;10(11):2702–2714. doi: 10.1038/ismej.2016.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burstein D, et al. Major bacterial lineages are essentially devoid of CRISPR-Cas viral defence systems. Nat Commun. 2016;7:10613. doi: 10.1038/ncomms10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell JH, et al. UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. Proc Natl Acad Sci U S A. 2013;110(14):5540–5. doi: 10.1073/pnas.1303090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rinke C, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499(7459):431–7. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 39.Luef B, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 40.Solden L, et al. The bright side of microbial dark matter: lessons learned from the uncultivated majority. Curr Opin Microbiol. 2016;31:217–26. doi: 10.1016/j.mib.2016.04.020. [DOI] [PubMed] [Google Scholar]
- 41.Segata N, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13(6):R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou Y, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14(1):R1. doi: 10.1186/gb-2013-14-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fredricks DN, et al. Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med. 2005;353(18):1899–911. doi: 10.1056/NEJMoa043802. [DOI] [PubMed] [Google Scholar]
- 44.Soro V, et al. Axenic culture of a candidate division TM7 bacterium from the human oral cavity and biofilm interactions with other oral bacteria. Appl Environ Microbiol. 2014;80(20):6480–9. doi: 10.1128/AEM.01827-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kianoush N, et al. Bacterial profile of dentine caries and the impact of pH on bacterial population diversity. PLoS One. 2014;9(3):e92940. doi: 10.1371/journal.pone.0092940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuehbacher T, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57(Pt 12):1569–76. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 47.Paster BJ, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183(12):3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinig MM, et al. Prevalence of bacteria of division TM7 in human subgingival plaque and their association with disease. Appl Environ Microbiol. 2003;69(3):1687–94. doi: 10.1128/AEM.69.3.1687-1694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rylev M, et al. Microbiological and immunological characteristics of young Moroccan patients with aggressive periodontitis with and without detectable Aggregatibacter actinomycetemcomitans JP2 infection. Mol Oral Microbiol. 2011;26(1):35–51. doi: 10.1111/j.2041-1014.2010.00593.x. [DOI] [PubMed] [Google Scholar]
- 50.Liu B, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7(6):e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Griffen AL, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6(6):1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeshita T, et al. Discrimination of the oral microbiota associated with high hydrogen sulfide and methyl mercaptan production. Sci Rep. 2012;2:215. doi: 10.1038/srep00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLean JS, et al. Draft Genome Sequence of Actinomyces odontolyticus subsp. actinosynbacter Strain XH001, the Basibiont of an Oral TM7 Epibiont. Genome Announc. 2016;4(1) doi: 10.1128/genomeA.01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bor B, et al. Morphological and physiological changes induced by contact-dependent interaction between Candida albicans and Fusobacterium nucleatum. Sci Rep. 2016;6:27956. doi: 10.1038/srep27956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Camanocha A, Dewhirst FE. Host-associated bacterial taxa from Chlorobi, Chloroflexi, GN02, Synergistetes, SR1, TM7, and WPS-2 Phyla/candidate divisions. J Oral Microbiol. 2014:6. doi: 10.3402/jom.v6.25468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7(5):1016–25. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162(2 Pt A):22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghannoum MA, et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 2010;6(1):e1000713. doi: 10.1371/journal.ppat.1000713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupuy AK, et al. Redefining the human oral mycobiome with improved practices in amplicon-based taxonomy: discovery of Malassezia as a prominent commensal. PLoS One. 2014;9(3):e90899. doi: 10.1371/journal.pone.0090899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Underhill DM, Iliev ID. The mycobiota: interactions between commensal fungi and the host immune system. Nat Rev Immunol. 2014;14(6):405–16. doi: 10.1038/nri3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oever JT, Netea MG. The bacteriome-mycobiome interaction and antifungal host defense. Eur J Immunol. 2014;44(11):3182–91. doi: 10.1002/eji.201344405. [DOI] [PubMed] [Google Scholar]
- 62.Janus MM, et al. Candida albicans in Multispecies Oral Communities; A Keystone Commensal? Adv Exp Med Biol. 2016;931:13–20. doi: 10.1007/5584_2016_5. [DOI] [PubMed] [Google Scholar]
- 63.Diaz PI, et al. Fungal-bacterial interactions and their relevance to oral health: linking the clinic and the bench. Front Cell Infect Microbiol. 2014;4:101. doi: 10.3389/fcimb.2014.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Allison DL, et al. Candida-Bacteria Interactions: Their Impact on Human Disease. Microbiol Spectr. 2016;4(3) doi: 10.1128/microbiolspec.VMBF-0030-2016. [DOI] [PubMed] [Google Scholar]
- 65.Abeles SR, et al. Human oral viruses are personal, persistent and gender-consistent. ISME J. 2014;8(9):1753–67. doi: 10.1038/ismej.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robles-Sikisaka R, et al. Association between living environment and human oral viral ecology. ISME J. 2013;7(9):1710–24. doi: 10.1038/ismej.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abeles SR, et al. Effects of Long Term Antibiotic Therapy on Human Oral and Fecal Viromes. PLoS One. 2015;10(8):e0134941. doi: 10.1371/journal.pone.0134941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wylie KM, et al. Metagenomic analysis of double-stranded DNA viruses in healthy adults. BMC Biol. 2014;12:71. doi: 10.1186/s12915-014-0071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Naidu M, et al. Characterization of bacteriophage communities and CRISPR profiles from dental plaque. BMC Microbiol. 2014;14:175. doi: 10.1186/1471-2180-14-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stein JM, et al. Failure to detect an association between aggressive periodontitis and the prevalence of herpesviruses. J Clin Periodontol. 2013;40(1):1–7. doi: 10.1111/jcpe.12021. [DOI] [PubMed] [Google Scholar]
- 71.Roberts AP, Kreth J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:124. doi: 10.3389/fcimb.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lugli GA, et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. 2016;18(7):2196–213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 73.Manrique P, et al. Healthy human gut phageome. Proc Natl Acad Sci U S A. 2016;113(37):10400–5. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Debarbieux L. Bacterial sensing of bacteriophages in communities: the search for the Rosetta stone. Curr Opin Microbiol. 2014;20:125–30. doi: 10.1016/j.mib.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 75.De Paepe M, et al. Bacteriophages: an underestimated role in human and animal health? Front Cell Infect Microbiol. 2014;4:39. doi: 10.3389/fcimb.2014.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mills S, et al. Movers and shakers: influence of bacteriophages in shaping the mammalian gut microbiota. Gut Microbes. 2013;4(1):4–16. doi: 10.4161/gmic.22371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhou H, et al. CRISPRs provide broad and robust protection to oral microbial flora of gingival health against bacteriophage challenge. Protein Cell. 2015;6(7):541–5. doi: 10.1007/s13238-015-0182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lum AG, et al. Global transcription of CRISPR loci in the human oral cavity. BMC Genomics. 2015;16:401. doi: 10.1186/s12864-015-1615-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pawluk A, et al. Inactivation of CRISPR-Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat Microbiol. 2016;1(8):16085. doi: 10.1038/nmicrobiol.2016.85. [DOI] [PubMed] [Google Scholar]
- 80.Maxwell KL. Phages Fight Back: Inactivation of the CRISPR-Cas Bacterial Immune System by Anti-CRISPR Proteins. PLoS Pathog. 2016;12(1):e1005282. doi: 10.1371/journal.ppat.1005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wahida A, et al. The Janus-Face of Bacteriophages across Human Body Habitats. PLoS Pathog. 2016;12(6):e1005634. doi: 10.1371/journal.ppat.1005634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Betts A, et al. Back to the future: evolving bacteriophages to increase their effectiveness against the pathogen Pseudomonas aeruginosa PAO1. Evol Appl. 2013;6(7):1054–63. doi: 10.1111/eva.12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Torres-Barcelo C, Hochberg ME. Evolutionary Rationale for Phages as Complements of Antibiotics. Trends Microbiol. 2016;24(4):249–56. doi: 10.1016/j.tim.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 84.Abedon ST. Ecology of Anti-Biofilm Agents II: Bacteriophage Exploitation and Biocontrol of Biofilm Bacteria. Pharmaceuticals (Basel) 2015;8(3):559–89. doi: 10.3390/ph8030559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Young R, Gill JJ. MICROBIOLOGY. Phage therapy redux--What is to be done? Science. 2015;350(6265):1163–4. doi: 10.1126/science.aad6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Machuca P, et al. Isolation of a novel bacteriophage specific for the periodontal pathogen Fusobacterium nucleatum. Appl Environ Microbiol. 2010;76(21):7243–50. doi: 10.1128/AEM.01135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Aljarbou AN, Aljofan M. Genotyping, Morphology and Molecular Characteristics of a Lytic Phage of Neisseria Strain Obtained from Infected Human Dental Plaque. Journal of Microbiology. 2014;52(7):609–618. doi: 10.1007/s12275-014-3380-1. [DOI] [PubMed] [Google Scholar]
- 88.Dalmasso M, et al. Isolation of a Novel Phage with Activity against Streptococcus mutans Biofilms. PLoS One. 2015;10(9):e0138651. doi: 10.1371/journal.pone.0138651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bor B, et al. Phenotypic and Physiological Characterization of the Epibiotic Interaction Between TM7x and Its Basibiont Actinomyces. Microb Ecol. 2016;71(1):243–55. doi: 10.1007/s00248-015-0711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shirtliff ME, et al. Cross-kingdom interactions: Candida albicans and bacteria. FEMS Microbiol Lett. 2009;299(1):1–8. doi: 10.1111/j.1574-6968.2009.01668.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Krom BP, et al. Candida and other fungal species: forgotten players of healthy oral microbiota. J Dent Res. 2014;93(5):445–51. doi: 10.1177/0022034514521814. [DOI] [PubMed] [Google Scholar]
- 92.Xu H, et al. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol Oral Microbiol. 2014;29(3):99–116. doi: 10.1111/omi.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wright CJ, et al. Microbial interactions in building of communities. Mol Oral Microbiol. 2013;28(2):83–101. doi: 10.1111/omi.12012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cavalcanti IM, et al. Interactions between Streptococcus oralis, Actinomyces oris, and Candida albicans in the development of multispecies oral microbial biofilms on salivary pellicle. Mol Oral Microbiol. 2016 doi: 10.1111/omi.12154. [DOI] [PubMed] [Google Scholar]
- 95.Cavalcanti IM, et al. Interkingdom cooperation between Candida albicans, Streptococcus oralis and Actinomyces oris modulates early biofilm development on denture material. Pathog Dis. 2016;74(3) doi: 10.1093/femspd/ftw002. [DOI] [PubMed] [Google Scholar]
- 96.Xu H, et al. Streptococcus oralis and Candida albicans Synergistically Activate mu-Calpain to Degrade E-cadherin From Oral Epithelial Junctions. J Infect Dis. 2016;214(6):925–34. doi: 10.1093/infdis/jiw201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Diaz PI, et al. Synergistic interaction between Candida albicans and commensal oral streptococci in a novel in vitro mucosal model. Infect Immun. 2012;80(2):620–32. doi: 10.1128/IAI.05896-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Falsetta ML, et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect Immun. 2014;82(5):1968–81. doi: 10.1128/IAI.00087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Willems HM, et al. Candida albicans in oral biofilms could prevent caries. Pathog Dis. 2016;74(5) doi: 10.1093/femspd/ftw039. [DOI] [PubMed] [Google Scholar]
- 100.Arzmi MH, et al. Polymicrobial biofilm formation by Candida albicans, Actinomyces naeslundii, and Streptococcus mutans is Candida albicans strain and medium dependent. Med Mycol. 2016;54(8):856–64. doi: 10.1093/mmy/myw042. [DOI] [PubMed] [Google Scholar]
- 101.Sztajer H, et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 2014;8(11):2256–71. doi: 10.1038/ismej.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hwang G, et al. Binding Force Dynamics of Streptococcus mutans-glucosyltransferase B to Candida albicans. J Dent Res. 2015;94(9):1310–7. doi: 10.1177/0022034515592859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koo H, Bowen WH. Candida albicans and Streptococcus mutans: a potential synergistic alliance to cause virulent tooth decay in children. Future Microbiol. 2014;9(12):1295–7. doi: 10.2217/fmb.14.92. [DOI] [PubMed] [Google Scholar]
- 104.Thomas A, et al. Association of Oral Candida albicans with Severe Early Childhood Caries - A Pilot Study. J Clin Diagn Res. 2016;10(8):ZC109–12. doi: 10.7860/JCDR/2016/19387.8357. [DOI] [PMC free article] [PubMed] [Google Scholar]