Abstract
Axonal transport involves kinesin motors trafficking cargo along microtubules that are rich in microtubule-associated proteins (MAPs). Much attention has focused on the behavior of kinesin-1 in the presence of MAPs, which has overshadowed understanding the contribution of other kinesins such as kinesin-2 in axonal transport. We have previously shown that, unlike kinesin-1, kinesin-2 in vitro motility is insensitive to the neuronal MAP Tau. However, the mechanism by which kinesin-2 efficiently navigates Tau on the microtubule surface is unknown. We hypothesized that mammalian kinesin-2 side-steps to adjacent protofilaments to maneuver around MAPs. To test this, we used single-molecule imaging to track the characteristic run length and protofilament switching behavior of kinesin-1 and kinesin-2 motors in the absence and presence of two different microtubule obstacles. Under all conditions tested, kinesin-2 switched protofilaments more frequently than kinesin-1. Using computational modeling that recapitulates run length and switching frequencies in the presence of varying roadblock densities, we conclude that kinesin-2 switches protofilaments to navigate around microtubule obstacles. Elucidating the kinesin-2 mechanism of navigation on the crowded microtubule surface provides a refined view of its contribution in facilitating axonal transport.
Keywords: Kinesin, Tau, Microtubule, Protofilament, Motility
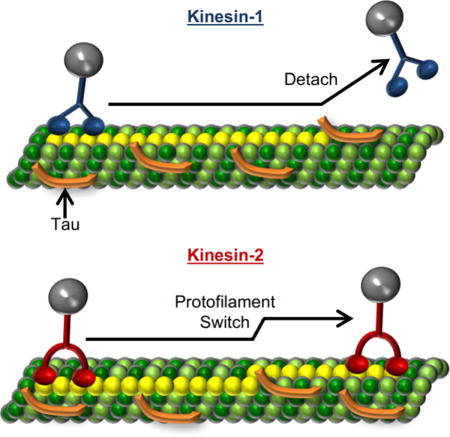
Graphical abstract
The molecular motor, kinesin-2, participates in trafficking cargo during axonal transport, but little is known about its behavior when encountering microtubule-associated proteins (MAPs), such as Tau. Using single-molecule imaging and computational modeling, we demonstrate that kinesin-2, unlike kinesin-1, is capable of switching protofilaments to navigate around Tau obstacles on the microtubule surface. The ability of kinesin-2 to navigate around Tau obstacles provides a new role for the molecular motor in trafficking cargo during axonal transport.
INTRODUCTION
Axonal transport is fundamental for the development and maintenance of neurons, where cargo is trafficked lengthy distances in the axon. Kinesin motors facilitate the transport of cargo along complex microtubule landscapes, which contain a variety of microtubule-associated proteins, such as the neuronal protein Tau, implicated in the spatio-temporal regulation of cargo delivery1–4. Much of the previous work of the regulation of cargo transport by Tau has focused on kinesin-1, where in vitro single-molecule experiments showed that Tau attenuates kinesin-1 motility in a concentration dependent manner1,4,5. However, the effects of Tau on the motility of other kinesin family members is largely unknown. Recently, we demonstrated that the run length of a related transport motor, kinesin-2, was insensitive to Tau, and determined that this property was due to the longer neck-linker of this motor compared to that of kinesin-16. The mechanism by which kinesin-2 is capable of successfully navigating Tau obstacles remains unclear, however.
There is limited knowledge regarding the structural conformation of Tau obstacles on the microtubule surface. Cyro-EM evidence is ambiguous, suggesting that Tau can lie longitudinally and/or laterally on the microtubule surface7,8. In addition, Tau is not a stationary obstacle; it transitions between static and diffusive states on the microtubule surface3,9 and will form multi-Tau complexes or patches3. Depending on the conformation and diffusional behavior of Tau, kinesin-2 encountering a Tau obstacle could: 1) side-step around it (switching protofilaments), 2) step over it, or 3) wait for Tau to transition from a static to a diffusive state and resume stepping. As previous modeling indicates that kinesin-2 has a fivefold higher probability of finding an off-protofilament axis binding site on the microtubule surface compared to kinesin-16, we hypothesize that kinesin-2 side-steps to adjacent protofilaments to navigate Tau obstacles on the microtubule surface. To test this hypothesis and distinguish between these three possible mechanisms, we tracked the motility of individual Qdot-labeled kinesin-1 and kinesin-2 motors along microtubules to observe protofilament switches on the microtubule surface in the absence and presence of two different roadblocks: Tau and monomeric rigor kinesin. As monomeric rigor kinesin is too large to step over and is a long lived static obstacle that will not diffuse away like Tau3,9, it allows a specific examination of the frequency that kinesin-1 and kinesin-2 are able to sidestep around obstacles. We find kinesin-2 switches protofilaments at a higher frequency compared to kinesin-1 and, using computational modeling, conclude kinesin-2 switches protofilaments to navigate microtubule obstacles. This behavior illuminates the different strategies that motors use to navigate the crowed microtubule lattice during axonal transport.
RESULTS
Kinesin-2 can navigate around small but not large roadblocks
Previously, we demonstrated at high Tau stoichiometries (1 Tau to 5 tubulin dimers, 2.5 fold higher than physiological levels10) the kinesin-2 characteristic run length was unimpeded, unlike kinesin-1 (Fig. 1 A)6. To test our hypothesis that kinesin-2, with its longer neck-linker, switches protofilaments to navigate Tau obstacles, we used a long lived static microtubule obstacle, rigor kinesin or RK (dwell time ≥ 1.76 ± 0.54 minutes, Fig. S1). If RK obstacles on a given protofilament block kinesin stepping11, then we presume that a motor would need to switch protofilaments to navigate around the obstacles. Using TIRF microscopy we observed the single-molecule motility of eGFP-labeled kinesin-1 and kinesin-2 motors in the presence of RK. With stoichiometries of 1 RK to 12.5 tubulin dimers (referred to as Low RK), the kinesin-1 run length decreased by 68%, matching the 65% decrease in the presence of 1 Tau to 5 tubulin dimers (referred to as Low Tau) (Fig. 1, Table 1). In contrast, at this Low RK level the kinesin-2 characteristic run length did not change (Fig. 1, Table 1), which supports our hypothesis that kinesin-2 switches protofilaments to navigate Tau.
FIGURE 1.
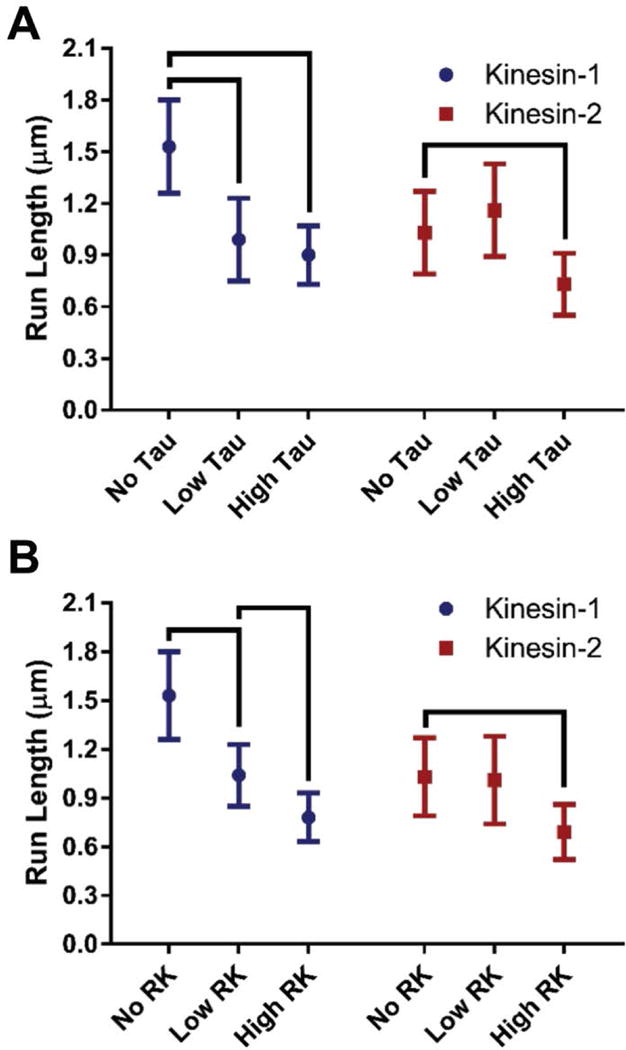
Characteristic run length comparisons between kinesin-1 and kinesin-2 in the absence and presence of microtubule obstacles. Two concentrations of (A) Tau or (B) rigor kinesin (RK) obstacles were used. Characteristic run length values were calculated from 38, the error bars represent the 99% confidence interval and the brackets represent significance between groups (p-value < 0.01) 38. (See Materials and Methods for further details.)
Table 1.
Summary of experimental and simulated kinesin-1 and kinesin-2 characteristic run lengths in the absence and presence of Tau and rigor kinesin obstacles
| Microtubule Obstacle | Nobs | Nexperiment,control | Characteristic Run Length (μm) | p-value | |
|---|---|---|---|---|---|
|
|
|||||
| Kinesin-1 Experimental | – | 591 | – | 1.53 ± 0.27 | – |
| Low Tau | 243 | 129 | 0.99 ± 0.24 | < 0.01 | |
| High Tau | 498 | 95 | 0.90 ± 0.17 | < 0.01 | |
| Low RK | 624 | 156 | 1.04 ± 0.19 | < 0.01 | |
| High RK | 436 | 67 | 0.78 ± 0.15 | < 0.01 | |
|
| |||||
| Kinesin-1 Simulated | – | 1000 | – | 1.55 ± 0.13 | ≥ 0.01† |
| Low RK | 1000 | 229 | 1.14 ± 0.10 | < 0.01 | |
| High RK | 1000 | 67 | 0.79 ± 0.07 | < 0.01 | |
|
| |||||
| Kinesin-2 Experimental | – | 300 | – | 1.03 ± 0.24 | – |
| Low Tau | 325 | – | 1.16 ± 0.27 | ≥ 0.01 | |
| High Tau | 293 | 189 | 0.73 ± 0.18 | < 0.01 | |
| Low RK | 296 | – | 1.01 ± 0.27 | ≥ 0.01 | |
| High RK | 290 | 147 | 0.69 ± 0.17 | < 0.01 | |
|
| |||||
| Kinesin-2 Simulated | – | 1000 | – | 1.06 ± 0.10 | ≥ 0.01† |
| Low RK | 1000 | 408 | 0.85 ± 0.08 | < 0.01 | |
| High RK | 1000 | 85 | 0.60 ± 0.06 | < 0.01 | |
See Materials and Methods for details of simulations. Nobs is the number of observed run lengths and Nexperiment,control is the number of run lengths required to determine significance at the 99% confidence (see Materials and Methods for additional information). The error represents the 99% confidence interval and the p-values were calculated relative to the motor condition in the absence of obstacles 38. P-value < 0.01 was considered significant.
Represents the p-value between the simulated characteristic run lengths relative to the experimental characteristic run lengths
We expected a higher concentration of Tau or RK would reduce the kinesin-1 characteristic run length further while the kinesin-2 characteristic run length would remain unchanged. To test our hypothesis, we raised the microtubule obstacle concentration for both Tau (1 Tau to 3 tubulin dimers, referred as High Tau, which is over 4-fold higher than the physiological level) and RK (1 RK to 7.5 tubulin dimers, referred to as High RK). Interestingly, we found that in the presence of the High Tau concentration, the kinesin-1 characteristic run length was not statistically different than at the Low Tau concentration (0.90 ± 0.17 μm vs 0.99 ± 0.24 μm, respectively) (Fig. 1 A, Table 1). However, the kinesin-2 characteristic run length at High Tau levels was sensitive to Tau obstacles compared to Low Tau levels (0.73 ± 0.18 μm vs 1.16 ± 0.27 μm, respectively) (Fig. 1 A, Table 1). These results demonstrated that Tau does not interact with kinesin motors as 1:1 steric blockers at High Tau concentrations. It has been shown that at higher concentrations, Tau multimerizes to form islands or “patches” on the microtubule lattice3. Due to multiple Tau-labeled molecules per diffraction-limited spot (~50 molecules), we cannot measure the difference in patch size between Low Tau and High Tau by fluorescence. However, it was previously shown at the single-molecule level (1 Tau to 3000 Tubulin dimers and 1 to 300) that the amount of Tau present in patch sizes of 2-3 Tau molecules on the microtubule surface was concentration dependent3. By extension of this observation, we posit increasing the concentration of Tau on the microtubule surface increases patch size. If this were the case, increasing the Tau concentration would not increase the total number of obstacles, but instead would increase the size of the obstacle the kinesin motors encounter. The finding that High Tau levels did not further reduce the kinesin-1 run length was consistent with Tau forming patches at higher concentrations. Also, the observation that the kinesin-2 run lengths were reduced only at High Tau levels indicates that the mechanism that kinesin-2 utilizes to navigate around roadblocks at Low Tau levels breaks down when larger patches of Tau form.
As expected for a 1:1 steric blocker of kinesin-1 motility, High RK levels further reduced the kinesin-1 characteristic run length to 0.78 ± 0.15 μm (Fig. 1 B, Table 1). The kinesin-2 characteristic run length also decreased at High RK levels compared to Low RK levels (0.69 ± 0.17 μm vs 1.01 ± 0.27 μm, respectively; Fig 1 B, Table 1). Overall, these results demonstrate that the mechanism by which kinesin-2 navigates road blocks can be broken down either by increasing the number of obstacles to a very high level, or by increasing the physical size and microtubule affinity of the obstacles.
Kinesin-2 switches protofilaments four-fold more frequently than kinesin-1
Because the kinesin-2 characteristic run length decreased at High Tau concentrations, we tested how frequently kinesin-2 motors switched protofilaments under these conditions. To improve the spatial and temporal resolution, motors were functionalized by a C-terminal Qdot and kinesin stepping was observed along microtubules decorated with either Tau or RK obstacles (Fig. 2 A). Although protofilament switching causes only a small (~6 nm) lateral displacement of the kinesin motor domain, the C-terminal Qdot amplifies the later displacement (~25 nm), enabling detection of protofilament switches (Figs. 2 B & S3). A single-particle tracking algorithm12 was used to track kinesin trajectories (Fig. 2 C & E) and detect protofilament switches (Fig. 2 D & F) (detection software FIESTA and Tdetector1 are described in Materials and Methods).
FIGURE 2.
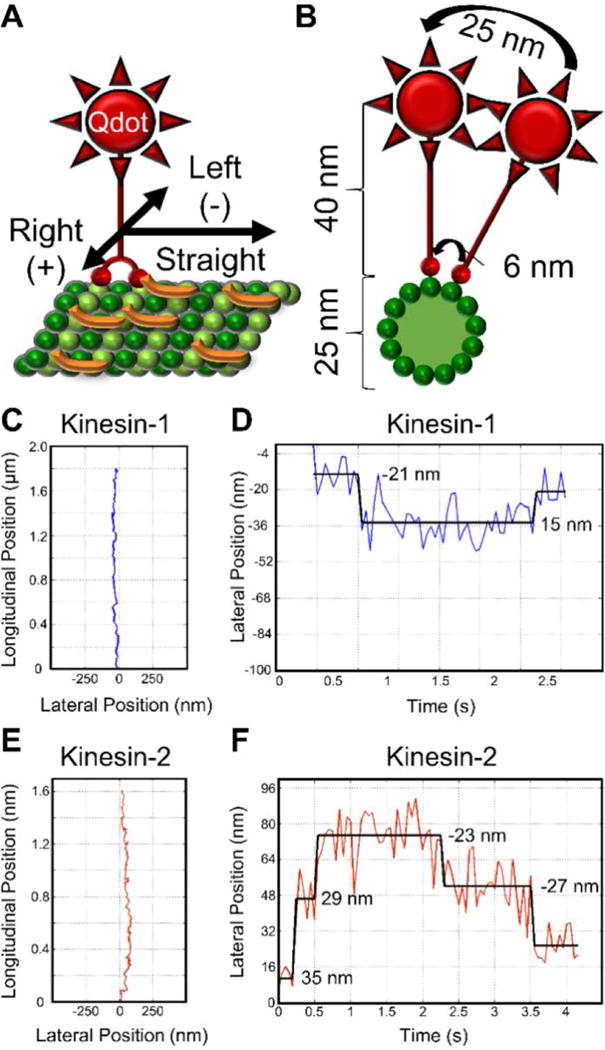
Experimental design and example kinesin protofilament switch trajectories. (A) Schematic of Qdot labeled kinesin motor (red) stepping along a Tau (orange) decorated microtubule (green). Left and right displacements are represented by negative and positive numbers, respectively. (B) Cross-section of panel A, illustrating the large lateral displacement of the C-terminally positioned Qdot relative to the motor domain displacement, which enables reliable detection of protofilament switches. (C) Example kinesin-1 trajectory longitudinal position vs lateral position and (D) kinesin-1 lateral position vs time (1.1 protofilament switches per micron). (E) Example kinesin-2 trajectory longitudinal position vs lateral position and (F) kinesin-2 lateral position vs time (2.5 protofilament switches per micron). In panels D and F, numbers represent lateral displacement and black lines represent the fit from the step finding algorithm.
In the absence of roadblocks, kinesin-2 switched protofilaments four-fold more frequently than kinesin-1 (Fig. 3 A, Table 2). These results are consistent with predictions from our previous modeling work6. The kinesin-1 protofilament switching frequency was unchanged in the presence of both Low and High Tau levels (Fig. 3 A, Table 2). Unexpectedly, the kinesin-2 protofilament switch frequency, which was hypothesized to increase with higher levels of Tau, was also unchanged in the presence of both Low and High Tau levels (Fig. 3 A, Table 2).
FIGURE 3.
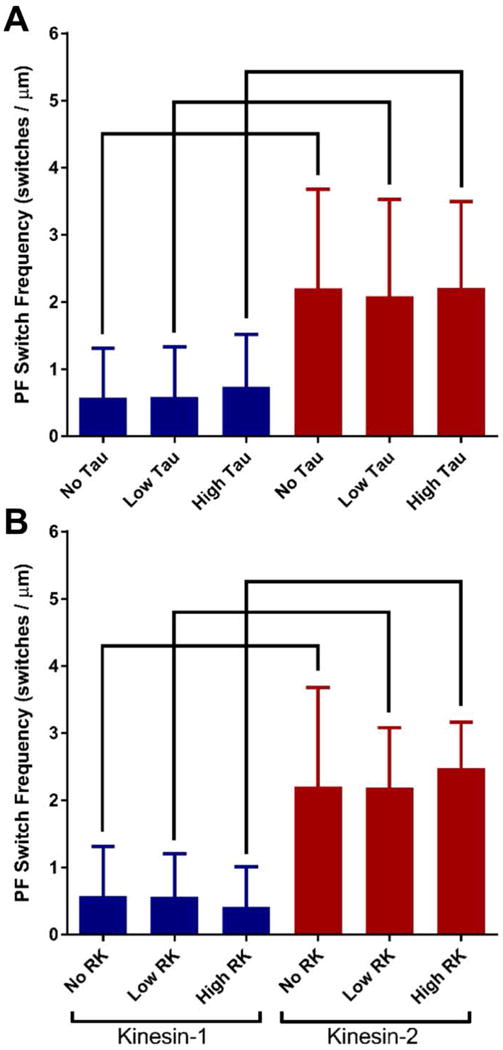
Kinesin-1 and kinesin-2 mean protofilament (PF) switching frequencies in the absence and presence of microtubule obstacles. Two concentrations of (A) Tau or (B) rigor kinesin (RK) obstacles are used. Kinesin-1 values are in blue and kinesin-2 values are in red. Error bars represent standard deviation and brackets represent significance between groups using a nonparametric ANOVA followed by a Dunn’s Multiple Comparisons Test (p-value < 0.05).
TABLE 2.
Summary of kinesin protofilament (PF) switching in the absence and presence of Tau and rigor kinesin obstacles
| Microtubule Obstacle | # Trajectories (avg length, μm) | Mean ± SD PF Switch Frequency (PF Switches / μm) | Total # Left PF Switches | Total # Right PF Switches | Median Left PF Switch Displace-ment (nm) | Median Right PF Switch Displace-ment (nm) | |
|---|---|---|---|---|---|---|---|
|
|
|||||||
| Kinesin-1 | – | 30 (1.5) | 0.57 ± 0.74 | 18 | 18 | 30 | 32 |
| Low Tau | 33 (1.4) | 0.59 ± 0.74 | 16 | 20 | 21 | 23 | |
| High Tau | 31 (1.4) | 0.74 ± 0.78 | 24 | 20 | 26 | 26 | |
| Low RK | 36 (1.8) | 0.57 ± 0.64 | 24 | 21 | 34 | 30 | |
| High RK | 44 (1.8) | 0.41 ± 0.60 | 21 | 22 | 28 | 33 | |
|
| |||||||
| Kinesin-2 | – | 31 (1.5) | 2.20* ± 1.47 | 57 | 55 | 34 | 38 |
| Low Tau | 30 (1.3) | 2.08* ± 1.44 | 46 | 44 | 35 | 31 | |
| High Tau | 30 (1.6) | 2.21* ± 1.28 | 53 | 52 | 40 | 38 | |
| Low RK | 30 (2.2) | 2.19* ± 0.89 | 74 | 72 | 38 | 39 | |
| High RK | 32 (2.0) | 2.48* ± 0.68 | 76 | 81 | 38 | 38 | |
The mean (SD, standard deviation) protofilament (PF) switch frequency for each condition was calculated from a distribution made up of the PF switch frequencies of each trajectory. The median left/right protofilament switch displacement was calculated from a distribution made up of the left/right displacements for each experimental condition. A nonparametric ANOVA was applied to all distributions and a Dunn’s Multiple Comparisons Test was used to determine significance (p-value < 0.05) among groups.
Represents statistical difference from the kinesin-1 experimental condition.
In the presence of RK obstacles, at either Low or High levels, the kinesin-1 protofilament switch frequency was unchanged just as in the presence of Tau obstacles (Fig. 3 B, Table 2). Similarly, the kinesin-2 protofilament switch frequency did not change in the presence of RK obstacles (Fig. 3 B, Table 2). Both Tau and RK obstacles did not measurably increase kinesin-1 and kinesin-2 protofilament switch frequencies. Additionally, for both motors under all conditions, there was no left/right switching bias and the left/right median lateral displacements during protofilament switches were not statistically different under any of the conditions tested (Fig. S6, Table 2).
A model in which encountering an obstacle does not change the probability of switching protofilaments is insufficient to explain the kinesin-2 measurements at Low RK levels
The protofilament switching data suggest that kinesin-2 does not switch protofilaments more frequently in the presence of obstacles. However, the kinesin-2 run length is not affected at Low RK levels, which seems paradoxical. To test these seemingly conflicting results, we simulated kinesin-1 and kinesin-2 stepping along microtubules decorated with RK/static obstacles using a simple stochastic model that allows kinesin to switch protofilaments with an intrinsic frequency that does not change as a function of obstacle concentration (see Materials and Methods). Briefly, a randomly generated run length was produced from experimentally determined characteristic run length distributions for kinesin-1 and kinesin-2. Motors walked along a seven protofilament-wide microtubule that was as long as the run length (Fig. 4 A) and each motor randomly switched protofilaments at its respective experimentally determined frequency. A run length was terminated prematurely if the motor ran into a RK/static obstacle. Microtubules were randomly decorated with obstacles at a density that produced simulated kinesin-1 characteristic run lengths that matched the experimentally derived kinesin-1 characteristic run lengths in the presence of Low/High RK levels (1.04 μm and 0.78 μm, respectively; Fig. 4 B).
FIGURE 4.
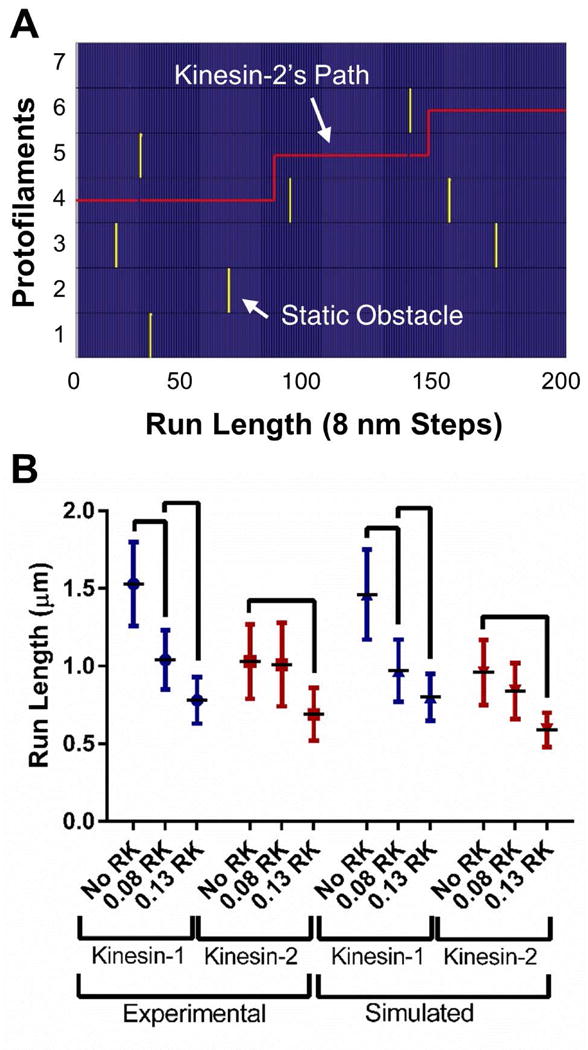
Computer simulation of kinesin-1 and kinesin-2 trajectories switching protofilaments. (A) Example kinesin-2 trajectory (red line) walking 1.6 μm (200 steps) on a seven protofilament wide microtubule (blue) navigating a field of static obstacles (yellow). (B) Experimentally observed and simulated characteristic run lengths in the presence of RK. Characteristic run length values were calculated from 38, the error bars represent the 99% confidence interval and the brackets represent significance between groups (p-value < 0.01) 38. (See Supporting Material for further details.)
The simulated obstacle density for the Low RK level was 1.75 static obstacles/μm or 0.2% (on a seven protofilament wide microtubule with 125 tubulin dimers/protofilament/μm, see Materials and Methods). The density rose to 5.25 obstacles/μm or 0.6% for the High RK level. Both of these values agree well with experimentally determined densities, 0.6% and 0.9% respectively (Fig. S2 and Table S1; see Supporting Materials for further details). We then simulated kinesin-2 trajectories with No RK, Low RK (0.2%) or High RK (0.6%) to test whether the intrinsic protofilament switching frequency (the switching frequency in the absence of obstacles) of kinesin-2 was sufficient to navigate Low and High RK/static obstacles. The simulated kinesin-2 characteristic run length at Low RK did not reproduce the experimentally observed kinesin-2 characteristic run length (Fig. 4 B, Table 1). Because the kinesin-2 intrinsic protofilament switch frequency alone could not replicate the experimental characteristic run length data at the Low RK level, we conclude that the side-stepping mechanism must work in such a way that encountering an obstacle increases the probability of switching protofilaments. Additionally, this model result shows that the lower characteristic run length of kinesin-2 alone, compared to kinesin-1, is insufficient to explain its reduced sensitivity to obstacles. It is important to note that the decrease in the simulated kinesin-2 characteristic run length at 0.2% demonstrates that the concentration of microtubule obstacles used in our experiments was sufficiently high that the kinesin-2 motors were encountering obstacles.
The lack of a measurable difference in the kinesin-2 protofilament switch frequencies on Low and High obstacle densities was unexpected. To ensure that our detection software can resolve protofilament switches if they occur more frequently than our measured values, we calculated the upper limit for switch frequency detection. If the limit is too close to our measured frequencies, the software could be artificially capping the protofilament switch frequencies. We found that for the frame rate and positional resolution in the experiments the calculated limit of detection for kinesin-1 and kinesin-2 were 2.3 and 4.7 protofilament switches/μm, respectively (Figs. S4 & S5; see Supporting Materials for further details). Hence, the average kinesin-1 and kinesin-2 protofilament switch frequencies were less than or equal to half the detection limit, which ensures our measurements were not capped.
At physiologically-relevant obstacle concentrations, a small increase in protofilament switching frequency is sufficient to explain the observed run lengths
Our first model showed that intrinsic protofilament switching alone was insufficient to explain the experimental data. Hence, encountering an obstacle must increase the probability of switching protofilaments. We therefore next consider a model that differentiates between intrinsic (not induced by an obstacle) and induced protofilament switches and test what number of induced sidesteps are necessary to explain the kinesin-2 run length data at Low and High RK levels. We posit that during each step, kinesin must select between taking a forward step, taking a sidestep to switch protofilaments or detach. This leads to the following system of equations:
Where RL0 is the characteristic run length in the absence of roadblocks, and P0 denotes probabilities in the absence of roadblocks. The value of P0(sidestep) is directly measured (Fig. 3, Table 2), and the values for P0(step) and P0(det) can be thus be calculated (Table 3). We next posit that when kinesin encounters an obstacle or roadblock, forward stepping is prohibited so the motor must then select between only sidestepping or detaching. Thus, we define the probability of taking an induced sidestep at a roadblock as:
TABLE 3.
Analytical model predicting the probability of switching protofilaments at various RK concentrations and comparison to experimental data
| P0 (step) | P0 (sidestep) | P0 (det) | PRB (sidestep) | RK Level | P (obstacle) | Ptot (sidestep) | Expected PF Switches/μm | Measured PF Switches/μm | |
|---|---|---|---|---|---|---|---|---|---|
|
|
|||||||||
| Kinesin-1 | 0.990 | 0.0047 | 0.0053 | 0.47 | No RK | 0.000 | 0.0047 | 0.57 | 0.57 |
|
| |||||||||
| Low RK | 0.002 | 0.0056 | 0.69 | 0.57 | |||||
|
| |||||||||
| High RK | 0.006 | 0.0075 | 0.92 | 0.41 | |||||
|
|
|||||||||
| Kinesin-2 | 0.947 | 0.0180 | 0.0079 | 0.69 | No RK | 0.000 | 0.0180 | 2.20 | 2.20 |
|
| |||||||||
| Low RK | 0.002 | 0.0194 | 2.36 | 2.19 | |||||
|
| |||||||||
| High RK | 0.006 | 0.0221 | 2.70 | 2.48 | |||||
Analytical model describing kinesin protofilament (PF) switch frequency by incorporating both intrinsic and induced sidesteps when encountering an obstacle or roadblock. The model replicates the kinesin-2 frequency data, but not the kinesin-1, indicating kinesin-2 switches protofilaments during an encounter. See results section above for analytical expression. P0(step) is the probability of taking a forward step in the absence of obstacles, P0(sidestep) is the experimentally observed intrinsic sidestepping frequency, P0(det) is the detachment rate per step, PRB(sidestep) is the probability of an induced sidestep during an obstacle encounter, P(obstacle) is the simulated obstacle density, and Ptot(sidestep) is the total probability of switching protofilaments. Expected PF Switches / μm is calculated by dividing the total probability of switching PFs by the tubulin dimer spacing of 0.0082 μm.
Since the probability of encountering a roadblock obstacle, P(obstacle), is known (see above and Table S1), we can therefore derive an analytical expression for the total probability of protofilament switching:
Where the first term represents the intrinsic sidesteps (those that occur when a roadblock is not present), and the second term represents induced sidesteps (those that occur due to a roadblock). We find that the expected number of protofilament switches per micron is in good agreement with the measured number of protofilament switches per micron for kinesin-2 at all RK concentrations (Table 3). This is consistent with our result that adding roadblocks does not measurably increase the total number of sidesteps for kinesin-2. Hence, we conclude that roadblocks do indeed induce sidesteps in kinesin-2, unlike kinesin-1, and that this induced sidestepping is a mechanism kinesin-2 employs to navigate crowded microtubules.
DISCUSSION
The goal of this study was to elucidate how kinesin-2 navigates Tau obstacles on the microtubule surface. We hypothesized kinesin-2, due to its longer neck-linker, switches protofilaments to navigate around Tau obstacles. By comparing its characteristic run length to kinesin-1, we robustly demonstrated that kinesin-2 successfully navigates rigor kinesin obstacles (Fig. 1 B, Table 1). Because of the expectation that RK obstacles are too large for the motors to step over, this data suggests that kinesin-2 switches protofilaments to navigate around them. We then directly observed protofilament switching by both kinesin-1 and kinesin-2 and found that kinesin-2 switches protofilaments more frequently than kinesin-1 both under control conditions and in the presence of either Tau or RK obstacles. Though we observed the kinesin-2 protofilament switch frequency was independent of obstacle concentration, our analytical model indicates kinesin-2 switches protofilaments to navigate obstacles while kinesin-1 fails to reliably switch protofilaments. The number of induced protofilament switches kinesin-2 makes is relatively small compared to the high intrinsic protofilament switch frequency. Thus, even though the kinesin-2 protofilament switch frequency is going up in response to obstacles, the relativity high intrinsic protofilament switch frequency is much larger than the number of induced protofilament switches. We conclude these data are consistent with kinesin-2 switching protofilaments to navigate around obstacles and that the encounter increases the probability of switching protofilaments.
The kinesin-2 protofilament switch frequency was hypothesized to be higher due to its longer and thus more flexible neck-linker, compared to kinesin-1. Elsewhere, it was demonstrated that kinesin-8, which has the same 17 amino acid neck-linker length as kinesin-2, switches protofilaments more readily13 than kinesin-1 (14 amino acid neck-linker)14. Brunnbauer et al. al demonstrated that lengthening the kinesin-1 neck-linker length induces protofilament switching15. These studies are consistent with our premise that the longer kinesin-2 neck-linker is responsible for the higher protofilament switch frequency. Brunnbauer et al. also found that the composition of the region C-terminal to the neck-liner, the neck-coil, was influential to the efficiency of wild-type kinesin-2 protofilament switching15. It should be noted that our kinesin-2 construct differs from wild-type kinesin-2. Wild-type kinesin-2 contains two heterodimeric kinesin heavy chains (kif3A/B or kif3A/C), where our construct contains a homodimeric kif3A/A motor domain and neck-linker fused to a Drosophila kinesin-1 coiled-coil (this construct has previously been demonstrated to have comparable run length, velocity and load dependent properties as wild-type16–18). Thus, our kinesin-2 construct has a more stable neck-coil, which likely does not experience the full flexibility that the wild-type kinesin-2 experiences. As such, wild-type kinesin-2 may have an even higher protofilament switch frequency and enable the motor to side-step when encountering an obstacle.
Of much interest was the observation that kinesin-1 switches protofilaments. This result was unexpected, as kinesin-1 was originally thought to only step along a single protofilament as demonstrated through microtubule gliding filament assays14,15,19. However, using single-molecule imaging Schneider et al. recently demonstrated that in the presence of obstacles kinesin-1 does occasionally switch protofilaments without left/right bias20. In that work, the dependence of run length on obstacle concentration indicated that kinesin-1 was inefficient at navigating microtubule obstacles20. Our observations that kinesin-1 occasionally switches protofilaments without left/right bias, and that the kinesin-1 run length decreases in the presence of microtubule obstacles are consistent with the findings of Schneider et al. Based on our previous model6, we expected a leftward protofilament switching bias for kinesin-2; however, our results indicate no such bias. Our previous model assumed that the motor domain searching for a binding site started with a slight leftward-bias based on previous microtubule gliding assay results21. This leftward shift observed may only exist as a load dependent property in the gliding filament assay, however. There is precedent for the behavior – dynein has an apparent leftward protofilament switching bias in microtubule gliding assays22,23, but has a rightward protofilament switching bias in single-molecule assays24,25.
Taking our observations together: 1) kinesin-2 is more efficient than kinesin-1 at navigating Tau and RK obstacles, and 2) kinesin-2 switches protofilaments more frequently than kinesin-1. These results are consistent with kinesin-2 switching protofilaments as a primary mechanism to navigate around microtubule obstacles. However, we cannot rule out other mechanisms contributing to kinesin-2 navigation. For instance, navigation may depend on a protein-protein interaction between kinesin-1 and Tau that could alter the motor’s motile behavior. We observed that the kinesin-1 velocity decreased at Low Tau concentrations, whereas the kinesin-2 velocity did not (Fig. S7). By comparison, the velocity of kinesin-1 with a different Tau isoform was independent of concentration (Fig. S7). This result indicates that kinesin-1, and possibly kinesin-2, may interact with Tau isoforms differently to affect their navigation. A second possible mechanism is the diffusion of motors on the microtubule surface. Kinesin-2 was demonstrated, in the presence of ADP, to have a mean microtubule dissociation rate of 1/0.7 s26. During this weakly-bound state, diffusion may occur if the leading head cannot bind on the same protofilament as the rear head if an obstacle is present. If we assume a diffusion coefficient of 0.065 μm2/s for kinesin27, then the motor would be expected to diffuse 300 nm in 0.7 s (x2 = (2Dt)). This distance is sufficient to briefly move around obstacles. So both motors may employ other navigational mechanisms in addition to protofilament switching to efficiently move along complex microtubule landscapes.
From the perspective of a cargo such as endosomes or other vesicles, the benefit associated with two directionally similar kinesin motors has the potential for utilizing their different functions to achieve optimal temporal and spatial cargo positioning. Hendricks et al. estimated that purified late endosomes/lysosomes have, on average, 1.7 kinesin-2 motors per vesicle, which was 3.9 fold more than kinesin-128. With so few kinesin-1 motors per vesicle, kinesin-1 would likely be interrupted in the presence of Tau. This was demonstrated by Xu et al. and Vershinin et al., who showed that the motility of synthetic beads, coupled to at least three kinesin-1 motors, was interrupted at a stoichiometry of less than 1 Tau per 10 tubulin dimers4,29. Thus, the ratio of kinesin motors on endosomes/lysosomes suggests kinesin-2 plays an important role in trafficking these cargos. For example, both motors may work together to navigate obstacles. Compared to kinesin-2, which readily dissociates under load18, kinesin-1 processivity is less affected by hindering loads(30–32). Thus, kinesin-1 may tow the vesicle until it is disrupted, at which time multiple kinesin-2 motors may aid in navigating the obstacles until kinesin-1 can reattach. A second example involves using kinesin-2 motors in regions that have a high density of obstacles. Prevo et al. demonstrated, in C. elegans cilia, the importance of heterotrimeric kinesin-2 in transporting IFT trains through the obstacle-ridden ciliary base and transition zone, which in the absence of kinesin-2 severely limited the number of intraflagellar transport (IFT) trains found in the cilia33. Once the IFT trains successfully navigated the obstacle-ridden transition zone, kinesin-2 handed off the cargo to the faster Osm3 motors for the rest of their journey. Like other cellular compartments, such as the axon, kinesin-2 could be used to navigate through the obstacle dense region of the axon hillock and initial segment, which is comparable to the dense obstacle regions of the transition zone in cilia. Afterward, kinesin-1 could take over for the duration of the journey. By understanding the underlying mechanisms of single motors, we can better understand the regulation of multi-motor complexes in intracellular cargo transport.
MATERIALS AND METHODS
Protein expression and purification
All Kinesin constructs contained Drosophila kinesin-1 neck and stalk domains (residues 346–559) fused with either a C-terminal eGFP/hexahistidine tag (expressed and purified as previously published in16 or a C-terminal Biotinylated Avi tag. Biotinylation was performed intracellularly in BL21 (DE3) bacteria (New England Biolabs, Ipswich, MA) as previously published34. Kinesin-1 constructs contained the N-terminal motor domain and neck-linker from Drosophila KIF5 (residues 1-345), while kinesin-2 constructs contained the motor domain and neck-linker from mouse KIF3A (residues 1-359). The rigor kinesin obstacle was a modified monomeric Rattus norvegicus KIF5C motor domain (residues 1-354), which was a generous gift from Dr. Kathy Trybus. A T93N point mutation, to prevent ATP hydrolysis, and a C-terminal FLAG-Tag were introduced by QuikChange II XL site-directed mutagenesis (Stratagene, La Jolla, CA). Rigor kinesin was expressed in BL21-CodonPlus(DE3)-RP E. coli cells (Stratagene, La Jolla, CA) using the isopropyl 1-thio-β-D-galactopyranoside-inducible pET vector system (Novagen, Madison, WI) and purified with the FLAG® monoclonal antibody (Sigma-Aldrich, St Louis, MO). The 3RS Tau isoform, which was a generous gift from Dr. Stephen King, was expressed and purified as previously described3. Tau and rigor kinesin concentrations were determined using the Bicinchoninic Acid Protein (BCA) Assay (Pierce, Rockford, IL) using desalted, lyophilized, 3RS-Tau or BSA, respectively, for standards. Samples were dialyzed against BRB80 (80 mM PIPES, pH 6.9 at room temperature, 1 mM EGTA, and 1 mM MgCl2). Tubulin was isolated from bovine brain (obtained from Vermont Livestock & Slaughter, Ferrisburgh, VT), using high molarity PIPES buffer (1M PIPES, pH 6.9 at room temperature, 10 mM MgCl2, and 20 mM EGTA) as previously described35. Tubulin concentration was determined using a spectrophotometer (extinction coefficient at A280 nm = 115,000 M−1 * cm−1).
Fluorescent-labeling of Tau and monomeric rigor kinesin
3RS Tau protein was incubated with a 10 fold molar excess of dithiothreitol (DTT) for two hours at room temperature and DTT was removed using a 2 mL 7K MWCO Zeba™ Spin Desalting Column (Pierce, Rockford, IL). Tau was then incubated in a 10 fold molar excess of Alexa Fluor 532-C5 maleimide (Life Technologies, Grand Island, NY) for an additional 2 hours at room temperature, and excess fluorophore was removed using a second desalting column. Labeling efficiency of Tau was determined by comparing the concentration of fluorophore to protein. Tau concentration was determined with the BCA assay and the dye concentration was determined using an extinction coefficient of 81,000 M−1*cm−1 at 531 nm (Alexa Fluor 532) in a Beckman DU® 640 spectrophotometer (Beckman Coulter, Brea, CA). The 3RS-isoform was labeled at C233 and the labeling efficiency was determined to be 85%. Rigor kinesin protein was labeled similarly except it was incubated with DTT for two hours on ice followed with Alexa Fluor 532-C5 maleimide at 4°C overnight. Rigor kinesin was labeled at any of four possible solvent-exposed cysteines (C7, C66, C169 and C296)36 and the labeling efficiency was determined to be 95%.
Microtubule preparation and labeling
Tubulin was thawed on ice and centrifuged at 350,000 × g for 20 minutes at 4°C before polymerization. Tubulin was then supplemented with 1 mM GTP (Sigma-Aldrich, St Louis, MO). For labeled microtubules, unlabeled GTP tubulin was mixed with rhodamine-labeled tubulin (Cytoskeleton, Inc., Denver, CO) at 100:1, respectively. Tubulin was polymerized for 20 minutes at 37°C and stabilized with a final concentration of 20 μM paclitaxel (Sigma-Aldrich, St Louis, MO). For experiments completed in the presence of Tau, tubulin polymerization was performed as described above, without rhodamine-labeled tubulin. Microtubules were incubated with Alexa 532 labeled 3RS-Tau at a ratio of either 1:5 Tau to tubulin or 1:3 Tau to tubulin at 37°C for an additional 20 minutes. The samples were centrifuged at room temperature for 20 minutes at 16,000 × g and the pellet was resuspended at room temperature in Motility Assay Buffer (MAB) (10 mM PIPES, pH 7.4 at room temperature, 50 mM potassium acetate, 4 mM magnesium acetate, 1 mM EGTA, supplemented with 10 mM DTT, an oxygen scavenger system (5.8 mg/mL glucose, 0.045 mg/mL catalase and 0.067 mg/mL glucose oxidase) (Sigma-Aldrich, St Louis, MO)). All microtubules were diluted in MAB. 20 μM paclitaxel was supplemented into all solutions containing paclitaxel microtubules.
In vitro single-molecule TIRF assay
Flow chambers were prepared by adhering ARTUS shims (Englewood, NJ) with Norland Optical Adhesive (Cranbury, NJ) to siliconized glass cover slips. Samples were prepared by incubating the flow chamber with monoclonal anti-β III (neuronal) antibodies (Sigma-Aldrich, St Louis, MO) at 33 μg/mL in MAB for 5 minutes or NEM myosin (used during protofilament switching experiments), which was a generous gift from Dr. Kathy Trybus. The chambers were washed and blocked with 0.5 mg/mL of bovine serum albumin (BSA) in MAB for an additional 2 minutes before the addition of 1 μM of the desired microtubule preparation. After an 8 minute incubation, the chambers were washed with MAB followed by 50 pM of the desired kinesin construct (with 1 mM or 0.1 mM ATP), which was added just prior to image acquisition for all experimental conditions examined. It should be noted that due to differences in buffer conditions our observed kinesin run lengths in MAB were different than previously measured values in BRB80 with the same constructs16.
For experiments completed in the presence of rigor kinesin, tubulin polymerization was performed as described above, without rhodamine-labeled tubulin. After the microtubule incubation and wash in the flow cell (as described above), a 5 minute incubation with Alexa 532 labeled monomeric rigor kinesin-1 (at either 1:12.5 rigor kinesin to tubulin or 1:7.5 rigor kinesin to tubulin dimers) was then added followed by another MAB wash. Finally, 50 pM of the desired kinesin construct in MAB, supplemented with desired concentration of ATP, was added just prior to image acquisition.
For Qdot labeled kinesin motors, C-terminal biotinylated kinesin constructs were incubated with a fivefold higher concentration of streptavidin coated 655 Qdots for 30 minutes on ice in MAB and supplemented with desired concentration of ATP.
Total internal reflection fluorescence microscopy (TIRFM) was performed at room temperature using a Nikon Eclipse Ti-U inverted microscope equipped with a 100X plan apochromatic objective lens (1.49 NA), an auxiliary 1.5X magnification and a custom “Micro Optic Fiber Launch TIRFM” (patent pending) as described in37. Images were obtained using an XR/Turbo-Z intensified 10-bit camera running Piper Control software v2.3.39 (Stanford Photonics, Palo Alto, CA). Kinesin-eGFP and Qdot 655 labeled kinesin constructs were excited with a 473 nm argon laser and imaged through emission 525/50 and 655/70 band-pass filters, respectively. Alexa 532-labeled 3RS-Tau, Alexa 532 labeled rigor kinesin and rhodamine-labeled tubulin were excited with a 532 nm argon laser and imaged through an emission 605/70 band-pass filter. The pixel resolution was 93.0 nm, kinesin-eGFP movies were acquired at 5 Hz and Qdot 655 labeled kinesin movies were acquired at 20 Hz. Representative movies of the single-molecule TIRF assay are found in the Supporting Material (Movies S1–S4). All experimental conditions tested were performed at least three times.
Data analysis
Run length motility data was collected with eGFP kinesin constructs and measured using the MTrackJ plug-in for ImageJ software, version 1.46r (National Institutes of Health, Bethesda, MD) and track lengths were measured using the segmented line tool in ImageJ. Average velocity values reported are the mean and standard deviation. Characteristic run length measurements, significance testing and power calculations were calculated as previously described38. Briefly, power calculations determined the minimum number of measurements, Nexperiment,control, required to ensure statistical significance at the 99% confidence level using the following expression: Nexperiment,control ≥ , where is the larger of the two compared characteristic run lengths and is the observed characteristic run length difference between two conditions38. For detecting the frequency and lateral displacement of protofilament switches, movies of Qdot-labeled kinesin constructs were analyzed in MATLAB (MATLAB 2012b, MathWorks, Natick, MA) using FIESTA39 to track particle positions and Tdetector112 to identify statistically significant lateral displacements. The X-Y resolution for protofilament switch data was calculated by tracking 20 stationary 655 Qdots over 1,000 frames acquired at 20 Hz. The standard deviations from the X-Y position tracking for the 20 Qdots were averaged to calculate an X-Y resolution of 14.7 nm. Movie simulations with point spread functions were used to test the minimum distance required to detect a single protofilament switch with experimentally derived Qdot intensity and background noise (see Supporting Material).
Stochastic modeling of protofilament switching
Kinesin-1 and kinesin-2 stepping along microtubules decorated with static obstacles was simulated in MATLAB (MATLAB 2015b, MathWorks, Natick, MA). Microtubules were generated by a large, two-dimensional sparse array (i.e. array containing mostly 0-valued cells, where 1-valued cells represented permanent/static obstacles and generated stochastically) with an x-axis representing 8-nm steps along a microtubule protofilament and a y-axis corresponding to a seven protofilament wide microtubule, which effectively represents the available kinesin binding sites in a flow cell chamber. Kinesin run lengths were stochastically generated based off the cumulative frequency distribution C(x) = 1 − e(−x/xo), where x is the calculated run length, xo is the characteristic run length (1.53 μm for kinesin-1 and 1.03 μm for kinesin-2). Microtubule lengths were the same length as the simulated run length trajectory to eliminate premature termination. The density of static obstacles used was calibrated by finding the obstacle density at which the simulated kinesin-1 characteristic run length matched the experimental kinesin-1 characteristic run length in the presence of rigor kinesin obstacles. The kinesin motor trajectories were terminated after their calculated trajectory or at the intersection of their trajectory and a static obstacle. The average experimentally determined kinesin-1 or kinesin-2 protofilament switching frequency, in the absence of obstacles, was applied to the respective simulated motor trajectory. For each simulated condition, sample size of n = 1,000, the characteristic run length and significance testing were calculated as previously described 38.
Supplementary Material
FIGURE S1 Duration of rigor kinesin binding events on microtubules. (A) Example kymograph of Alexa 532 rigor kinesin obstacles on a microtubule. Scale bars are 3 μm. (B) Cumulative frequency plot (black curve) of rigor kinesin binding durations observed on the microtubule during movie acquisitions. Red dashed line is the cumulative Gaussian fit where the average time from the fit was 1.76 ± 0.54 min (n = 124).
FIGURE S2 Example kymograph of two labeled rigor kinesins (green) on a single microtubule. Experimental conditions were 200 nM rigor kinesin and 1.5 uM microtubules (a rigor kinesin to tubulin ratio of 1 to 7.5, High). Scale bar is 4 μm.
FIGURE S3 Experimental design and predictions for protofilament switching. (A) Schematic of microtubule cross-section (green) illustrating the larger lateral displacement of the C-terminally positioned Qdot (red) over the smaller lateral displacement of the kinesin motor domain. (B) Illustrating x versus y position (nm) of the cross-section of a microtubule (green) and the trajectory (red) of a kinesin stepping around the microtubule in the x versus y position (nm). From this perspective, the z-position is stepping along a microtubule, which is into the page. The forbidden region (grey) would not be accessible to a motor as the distance between the microtubule and the slide is too small for a motor to successfully walk. (C) Illustrating radians versus x position (nm), which is the camera’s perspective of the motor switching protofilaments as it records a projection of the x-y position. (D) Illustrating the radians versus x displacement (nm), which represents the lateral distance of the motor traveling around the microtubule. The lateral displacement that cannot be detected using the single particle tracking program is shaded in yellow.
FIGURE S4 Determining the detection limit of the single particle tracking program for kinesin-1 and kinesin-2 protofilament switch frequencies. (A) Kinesin-1 simulated values (blue curve) analyzed by the tracking program compared to extrapolated theoretical curve (black). (B) Kinesin-2 simulated values (red curve) analyzed by the tracking program compared to extrapolated theoretical curve (black). Error bars represent 95% confidence intervals. Dashed lines are the fits to the data and solid lines represent the 95% confidence intervals to the fit. The arrows and values represent the detection limit or the point at which the tracked simulated data set (red or blue) deviated from the 95% confidence band of the extrapolated data set (black).
FIGURE S5 Kinesin-1 (blue) and kinesin-2 (red) average protofilament (PF) switching frequencies at either 1 mM or 0.1 mM ATP in the absence of microtubule obstacles. N=30 to 33 for all conditions. A nonparametric ANOVA followed by a Dunn’s Multiple Comparisons Test was used to determine significance between groups (p-value < 0.05).
FIGURE S6 Kinesin-1 and kinesin-2 lateral (protofilament) displacements in the absence and presence of microtubule obstacles. Two concentrations of (A) Tau or (B) rigor kinesin (RK) obstacles are used. Tau and rigor kinesin obstacle ratios are relative to tubulin dimer concentration. L = left protofilament switches and R = right protofilament switches. Median lateral displacements with the interquartile range displayed. Kinesin-1 values are in blue and kinesin-2 values are in red. A nonparametric ANOVA followed by a Dunn’s Multiple Comparisons Test was used to determine significance (p-value < 0.05). No statistical differences between lateral displacements of different experimental conditions were found. (Note: lateral distances were slightly larger than the predicted (≤ 2 fold change). This is likely due to additional distance not accounted for between the neck and coil-1 of the rod domain. See section above.)
TABLE S1 Summary of rigor kinesin (RK) density along microtubules (MT)
Acknowledgments
The authors thank Dr. David Warshaw and Guy Kennedy for equipment, operational support and helpful discussions. Also, Dr. Jason Stumpff and Dr. M. Yusuf Ali for helpful discussions during preparation of the manuscript. No conflicts of interest.
GRANTS
This work was supported by the National Institutes of Health (R01-GM-101066 to C.L.B. and R01-GM-076476 to W.O.H.).
Footnotes
SUPPORTING CITATIONS
References 40–44 appear in the Supporting Material.
References
- 1.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319(5866):1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kanaan NM, Morfini G, Pigino G, et al. Phosphorylation in the amino terminus of tau prevents inhibition of anterograde axonal transport. Neurobiology of aging. 2012;33(4):826, e815–830. doi: 10.1016/j.neurobiolaging.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McVicker DP, Hoeprich GJ, Thompson AR, Berger CL. Tau interconverts between diffusive and stable populations on the microtubule surface in an isoform and lattice specific manner. Cytoskeleton (Hoboken) 2014 doi: 10.1002/cm.21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vershinin M, Carter BC, Razafsky DS, King SJ, Gross SP. Multiple-motor based transport and its regulation by Tau. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(1):87–92. doi: 10.1073/pnas.0607919104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McVicker DP, Chrin LR, Berger CL. The Nucleotide-binding State of Microtubules Modulates Kinesin Processivity and the Ability of Tau to Inhibit Kinesin-mediated Transport. J Biol Chem. 2011;286(50):42873–42880. doi: 10.1074/jbc.M111.292987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoeprich GJ, Thompson AR, McVicker DP, Hancock WO, Berger CL. Kinesin’s Neck-Linker Determines its Ability to Navigate Obstacles on the Microtubule Surface. Biophysical journal. 2014;106(8):1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA. MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. The Journal of cell biology. 2002;157(7):1187–1196. doi: 10.1083/jcb.200201048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Santarella RA, Skiniotis G, Goldie KN, et al. Surface-decoration of microtubules by human tau. Journal of molecular biology. 2004;339(3):539–553. doi: 10.1016/j.jmb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 9.Hinrichs MH, Jalal A, Brenner B, Mandelkow E, Kumar S, Scholz T. Tau protein diffuses along the microtubule lattice. J Biol Chem. 2012;287(46):38559–38568. doi: 10.1074/jbc.M112.369785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morfini G, Pigino G, Mizuno N, Kikkawa M, Brady ST. Tau binding to microtubules does not directly affect microtubule-based vesicle motility. J Neurosci Res. 2007;85(12):2620–2630. doi: 10.1002/jnr.21154. [DOI] [PubMed] [Google Scholar]
- 11.Telley IA, Bieling P, Surrey T. Obstacles on the microtubule reduce the processivity of Kinesin-1 in a minimal in vitro system and in cell extract. Biophysical journal. 2009;96(8):3341–3353. doi: 10.1016/j.bpj.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Deffenbaugh NC, Anderson CT, Hancock WO. Molecular counting by photobleaching in protein complexes with many subunits: best practices and application to the cellulose synthesis complex. Molecular biology of the cell. 2014;25(22):3630–3642. doi: 10.1091/mbc.E14-06-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bormuth V, Nitzsche B, Ruhnow F, et al. The highly processive kinesin-8, Kip3, switches microtubule protofilaments with a bias toward the left. Biophysical journal. 2012;103(1):L4–6. doi: 10.1016/j.bpj.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitzsche B, Ruhnow F, Diez S. Quantum-dot-assisted characterization of microtubule rotations during cargo transport. Nature nanotechnology. 2008;3(9):552–556. doi: 10.1038/nnano.2008.216. [DOI] [PubMed] [Google Scholar]
- 15.Brunnbauer M, Dombi R, Ho TH, Schliwa M, Rief M, Okten Z. Torque generation of kinesin motors is governed by the stability of the neck domain. Molecular cell. 2012;46(2):147–158. doi: 10.1016/j.molcel.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Shastry S, Hancock WO. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Current biology : CB. 2010;20(10):939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muthukrishnan G, Zhang Y, Shastry S, Hancock WO. The processivity of kinesin-2 motors suggests diminished front-head gating. Current biology : CB. 2009;19(5):442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreasson JO, Shastry S, Hancock WO, Block SM. The Mechanochemical Cycle of Mammalian Kinesin-2 KIF3A/B under Load. Current biology : CB. 2015 doi: 10.1016/j.cub.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ray S, Meyhofer E, Milligan RA, Howard J. Kinesin follows the microtubule’s protofilament axis. The Journal of cell biology. 1993;121(5):1083–1093. doi: 10.1083/jcb.121.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider R, Korten T, Walter WJ, Diez S. Kinesin-1 Motors Can Circumvent Permanent Roadblocks by Side-Shifting to Neighboring Protofilaments. Biophysical journal. 2015;108(9):2249–2257. doi: 10.1016/j.bpj.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yajima J, Cross RA. A torque component in the kinesin-1 power stroke. Nature chemical biology. 2005;1(6):338–341. doi: 10.1038/nchembio740. [DOI] [PubMed] [Google Scholar]
- 22.Can S, Dewitt MA, Yildiz A. Bidirectional helical motility of cytoplasmic dynein around microtubules. eLife. 2014;3:e03205. doi: 10.7554/eLife.03205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitra A, Ruhnow F, Nitzsche B, Diez S. Impact-Free Measurement of Microtubule Rotations on Kinesin and Cytoplasmic-Dynein Coated Surfaces. PloS one. 2015;10(9):e0136920. doi: 10.1371/journal.pone.0136920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nature cell biology. 2010;12(12):1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126(2):335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen GY, Arginteanu DF, Hancock WO. Processivity of the Kinesin-2 KIF3A Results From Rear-Head Gating and Not Front-Head Gating. J Biol Chem. 2015 doi: 10.1074/jbc.M114.628032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu H, Ali MY, Bookwalter CS, Warshaw DM, Trybus KM. Diffusive movement of processive kinesin-1 on microtubules. Traffic. 2009;10(10):1429–1438. doi: 10.1111/j.1600-0854.2009.00964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hendricks AG, Perlson E, Ross JL, Schroeder HW, 3rd, Tokito M, Holzbaur EL. Motor Coordination via a Tug-of-War Mechanism Drives Bidirectional Vesicle Transport. Current biology : CB. 2010 doi: 10.1016/j.cub.2010.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, King SJ, Lapierre-Landry M, Nemec B. Interplay between Velocity and Travel Distance of Kinesin-based Transport in the Presence of Tau. Biophysical journal. 2013;105(10):L23–25. doi: 10.1016/j.bpj.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svoboda K, Schmidt CF, Schnapp BJ, Block SM. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365(6448):721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 31.Yildiz A, Tomishige M, Vale RD, Selvin PR. Kinesin walks hand-over-hand. Science. 2004;303(5658):676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 32.Schnitzer MJ, Visscher K, Block SM. Force production by single kinesin motors. Nature cell biology. 2000;2(10):718–723. doi: 10.1038/35036345. [DOI] [PubMed] [Google Scholar]
- 33.Prevo B, Mangeol P, Oswald F, Scholey JM, Peterman EJ. Functional differentiation of cooperating kinesin-2 motors orchestrates cargo import and transport in C. elegans cilia. Nature cell biology. 2015 doi: 10.1038/ncb3263. [DOI] [PubMed] [Google Scholar]
- 34.Mickolajczyk KJ, Deffenbaugh NC, Ortega Arroyo J, Andrecka J, Kukura P, Hancock WO. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castoldi M, Popov AV. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif. 2003;32(1):83–88. doi: 10.1016/S1046-5928(03)00218-3. [DOI] [PubMed] [Google Scholar]
- 36.Rice S, Lin AW, Safer D, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402(6763):778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 37.Previs MJ, Beck Previs S, Gulick J, Robbins J, Warshaw DM. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 2012;337(6099):1215–1218. doi: 10.1126/science.1223602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson AR, Hoeprich GJ, Berger CL. Single-molecule motility: statistical analysis and the effects of track length on quantification of processive motion. Biophysical journal. 2013;104(12):2651–2661. doi: 10.1016/j.bpj.2013.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruhnow F, Zwicker D, Diez S. Tracking single particles and elongated filaments with nanometer precision. Biophysical journal. 2011;100(11):2820–2828. doi: 10.1016/j.bpj.2011.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deshpande A, Win KM, Busciglio J. Tau isoform expression and regulation in human cortical neurons. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2008;22(7):2357–2367. doi: 10.1096/fj.07-096909. [DOI] [PubMed] [Google Scholar]
- 41.RCoreTeam. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2016. [Google Scholar]
- 42.Kerssemakers J, Howard J, Hess H, Diez S. The distance that kinesin-1 holds its cargo from the microtubule surface measured by fluorescence interference contrast microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(43):15812–15817. doi: 10.1073/pnas.0510400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crevel IM, Nyitrai M, Alonso MC, Weiss S, Geeves MA, Cross RA. What kinesin does at roadblocks: the coordination mechanism for molecular walking. The EMBO journal. 2004;23(1):23–32. doi: 10.1038/sj.emboj.7600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Howard J, Spudich JA. Is the lever arm of myosin a molecular elastic element? Proceedings of the National Academy of Sciences of the United States of America. 1996;93(9):4462–4464. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Duration of rigor kinesin binding events on microtubules. (A) Example kymograph of Alexa 532 rigor kinesin obstacles on a microtubule. Scale bars are 3 μm. (B) Cumulative frequency plot (black curve) of rigor kinesin binding durations observed on the microtubule during movie acquisitions. Red dashed line is the cumulative Gaussian fit where the average time from the fit was 1.76 ± 0.54 min (n = 124).
FIGURE S2 Example kymograph of two labeled rigor kinesins (green) on a single microtubule. Experimental conditions were 200 nM rigor kinesin and 1.5 uM microtubules (a rigor kinesin to tubulin ratio of 1 to 7.5, High). Scale bar is 4 μm.
FIGURE S3 Experimental design and predictions for protofilament switching. (A) Schematic of microtubule cross-section (green) illustrating the larger lateral displacement of the C-terminally positioned Qdot (red) over the smaller lateral displacement of the kinesin motor domain. (B) Illustrating x versus y position (nm) of the cross-section of a microtubule (green) and the trajectory (red) of a kinesin stepping around the microtubule in the x versus y position (nm). From this perspective, the z-position is stepping along a microtubule, which is into the page. The forbidden region (grey) would not be accessible to a motor as the distance between the microtubule and the slide is too small for a motor to successfully walk. (C) Illustrating radians versus x position (nm), which is the camera’s perspective of the motor switching protofilaments as it records a projection of the x-y position. (D) Illustrating the radians versus x displacement (nm), which represents the lateral distance of the motor traveling around the microtubule. The lateral displacement that cannot be detected using the single particle tracking program is shaded in yellow.
FIGURE S4 Determining the detection limit of the single particle tracking program for kinesin-1 and kinesin-2 protofilament switch frequencies. (A) Kinesin-1 simulated values (blue curve) analyzed by the tracking program compared to extrapolated theoretical curve (black). (B) Kinesin-2 simulated values (red curve) analyzed by the tracking program compared to extrapolated theoretical curve (black). Error bars represent 95% confidence intervals. Dashed lines are the fits to the data and solid lines represent the 95% confidence intervals to the fit. The arrows and values represent the detection limit or the point at which the tracked simulated data set (red or blue) deviated from the 95% confidence band of the extrapolated data set (black).
FIGURE S5 Kinesin-1 (blue) and kinesin-2 (red) average protofilament (PF) switching frequencies at either 1 mM or 0.1 mM ATP in the absence of microtubule obstacles. N=30 to 33 for all conditions. A nonparametric ANOVA followed by a Dunn’s Multiple Comparisons Test was used to determine significance between groups (p-value < 0.05).
FIGURE S6 Kinesin-1 and kinesin-2 lateral (protofilament) displacements in the absence and presence of microtubule obstacles. Two concentrations of (A) Tau or (B) rigor kinesin (RK) obstacles are used. Tau and rigor kinesin obstacle ratios are relative to tubulin dimer concentration. L = left protofilament switches and R = right protofilament switches. Median lateral displacements with the interquartile range displayed. Kinesin-1 values are in blue and kinesin-2 values are in red. A nonparametric ANOVA followed by a Dunn’s Multiple Comparisons Test was used to determine significance (p-value < 0.05). No statistical differences between lateral displacements of different experimental conditions were found. (Note: lateral distances were slightly larger than the predicted (≤ 2 fold change). This is likely due to additional distance not accounted for between the neck and coil-1 of the rod domain. See section above.)
TABLE S1 Summary of rigor kinesin (RK) density along microtubules (MT)


