SUMMARY
Ecological speciation with gene flow is widespread in nature [1], but presents a conundrum: how are associations between traits under divergent natural selection and traits that contribute to assortative mating maintained? Theoretical models suggest that genetic mechanisms inhibiting free recombination between loci underlying these two types of traits (hereafter, “genetic coupling”) can facilitate speciation [2–4]. Here, we perform a direct test for genetic coupling by mapping both divergent traits and female mate choice in a classic model of ecological speciation: sympatric benthic and limnetic threespine stickleback (Gasterosteus aculeatus). By measuring mate choice in F2 hybrid females, we allowed for recombination between loci underlying assortative mating and those under divergent ecological selection. In semi-natural mating arenas in which females had access to both benthic and limnetic males, we found that F2 females mated with males similar to themselves in body size and shape. In addition, we found two quantitative trait loci (QTL) associated with female mate choice that also predicted female morphology along the benthic-limnetic trait axis. Furthermore, a polygenic genetic model that explains adaptation to contrasting benthic and limnetic feeding niches [5] also predicted F2 female mate choice. Together, these results provide empirical evidence that genetic coupling of assortative mating with traits under divergent ecological selection helps maintain species in the face of gene flow, despite a polygenic basis for adaptation to divergent environments.
RESULTS
We tested for genetic coupling between loci underlying ecologically divergent traits and assortative mating by examining morphological and genomic determinants of female mate choice in a sympatric pair of benthic and limnetic threespine stickleback from Paxton Lake in British Columbia, Canada. Species pairs of stickleback have evolved repeatedly in multiple postglacial lakes in British Columbia [6,7]. Each lake contains a larger, deeper bodied benthic form that inhabits inshore habitats, and a smaller, shallow bodied limnetic form that inhabits open water [8,9]. These species are morphologically adapted to their contrasting food sources: benthic stickleback primarily feed on invertebrates inhabiting the substrate or attached to vegetation, whereas limnetics specialize on zooplankton [10–12]. Although hybrids exist in the wild [13–15] and there are no strong intrinsic incompatibilities [14,16], benthics and limnetics show nearly complete assortative mating in experimental trials [17]. Previous no-choice mating trials suggested that benthic and limnetic females prefer mates with similar body size [18–20] and shape [20]. In this study, we conducted female mate choice experiments in ponds that allowed females to access both benthic and limnetic males in habitats that closely mimic those found in the wild [5]. By examining whether recombinant F2 hybrid females that vary in phenotype mate with benthic or limnetic males, we tested whether females prefer to mate with individuals that have similar phenotypes to themselves. We also identified QTL for female mate choice and morphology to test whether genomic regions associated with mate choice correspond to regions determining phenotypic traits under divergent selection. These represent the first direct tests of genetic coupling in this vertebrate system.
F2 females prefer males with a similar body shape and size
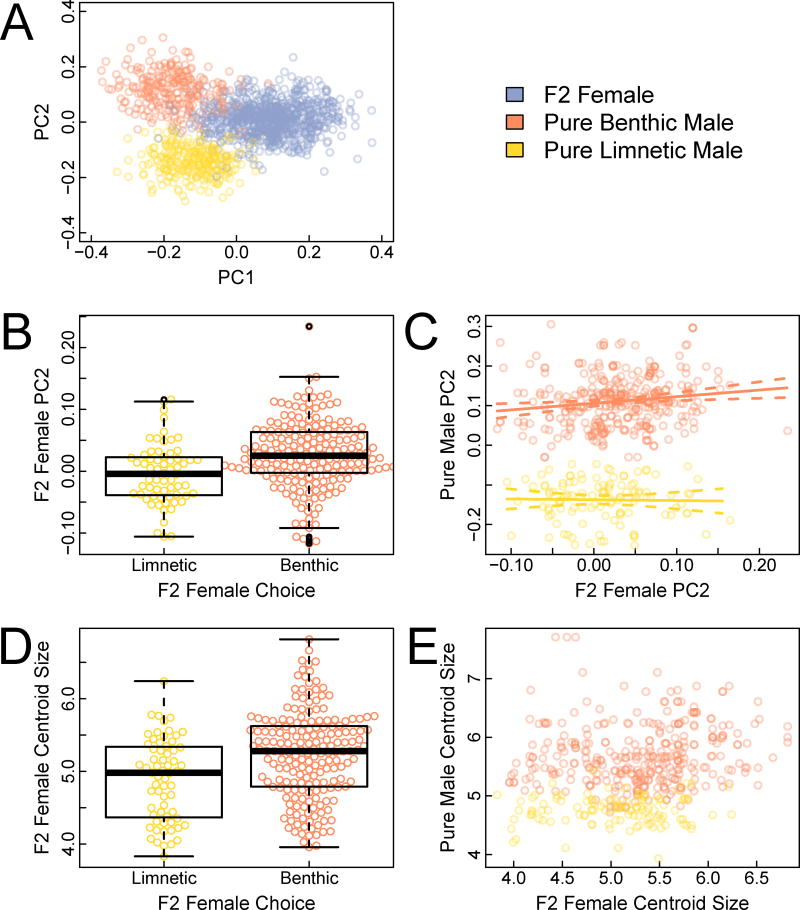
Body shape of F2 hybrid females was positively associated with the shape of chosen mates. We defined shape based on 17 external morphological landmarks (34 x- and y-coordinates; Figure S1), with landmarks for each fish rotated and scaled to the same centroid size. We used principal component (PC) analysis of the morphological landmark coordinates to summarize continuous variation in phenotypes of pure-species males and F2 females, which allowed us to examine shape variation associated with female choice within and between benthic and limnetic males. The first PC axis (PC1) separated male from female fish and was not analyzed further. PC2 separated benthic males from limnetic males, with F2 females intermediate (Figure 1A). Females that mated with limnetic males had lower (more limnetic-like) PC2 shape values than those that mated with benthic males (χ2=17.46; P=2.9×10−5: partial R2=0.072; Figure 1B). Remarkably, among F2 females that mated with benthic males, those most benthic-like in shape tended to mate with benthic males that were closer to the benthic extreme of the PC2 shape distribution (F1,444=6.65; P=0.01; partial R2=0.015; Figure 1C). We did not detect a similar trend in F2 females that mated with limnetic males (F1,444=0.33; P=0.56; partial R2=0.0008; Figure 1C). We also analyzed F2 female shape using a discriminant function that separates benthic from limnetic males based on the 34 external morphological landmark coordinates (Table S1). In accordance with the results above using PC2, females that mated with benthic males had a more benthic-like shape than those that mated with limnetic males (χ2=16.23; P=5.6×10−5; partial R2=0.065). When centroid size was used as a covariate in these analyses, the correlations between the shape of F2 hybrid females and the chosen males remained (data not shown), suggesting that body shape is an important component of female mate choice.
Figure 1. Mate choice of F2 females is associated with shape and centroid size of males and females.
Shape is summarized using principal component analysis of 17 landmarks. (A) PC2 separates benthic and limnetic males, with F2 females intermediate. In F2 females, PC2 is significantly associated with the male species chosen (B) and with variation in male PC2 scores when benthic males were chosen (C). In F2 females, centroid size is significantly associated with mate choice (D), but not with variation in male centroid size when benthic males were chosen or when limnetic males were chosen (E). See also Figure S1, Table S1, Table S3.
Body size of hybrid F2 females also predicted mate choice. Females with larger centroid sizes preferentially mated with males of the larger, benthic species (χ2=17.79; P=2.5×10−5; partial R2=0.103; Figure 1D). Among F2 females that mated with benthic males, there was a non-significant tendency for the largest of them to mate with the largest benthic males (F1,444=1.89; P=0.17; partial R2=0.004; Figure 1E). There was a similar positive tendency among F2 females that mated with limnetic males, though again this pattern was not significant (F1,444=1.06; P=0.30; partial R2=0.002; Figure 1E).
Eight of 34 shape traits (x- and y-coordinates of 17 morphological landmarks; Figure S1), found mainly in the head and the caudal region of F2 females, were significantly associated with mate choice when tested one at a time: y1, x2, y2, x5, y6, y9, y15, and y16 (FDR-adjusted P<0.05). The importance of some of these traits to mate choice is also indicated by their contribution to scaling on the benthic-limnetic discriminant function and their loading on PC2 (Table S1). The scaling values of two jaw coordinates (y1 and y2) are within the top five scaling values on the first linear discriminant axis. Both of those coordinates, along with coordinates at the insertion of the dorsal (y15) and anal (y16) fins, were also within the top five loadings on PC2.
Genetic coupling of mate choice and ecological traits
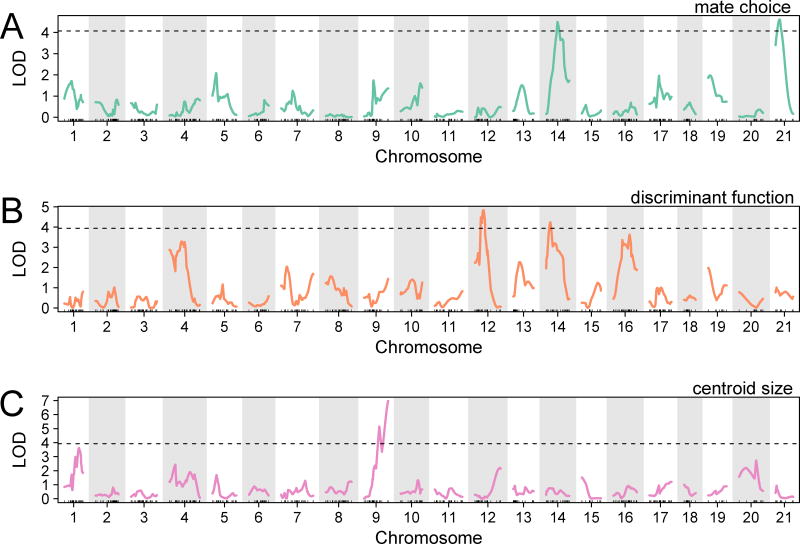
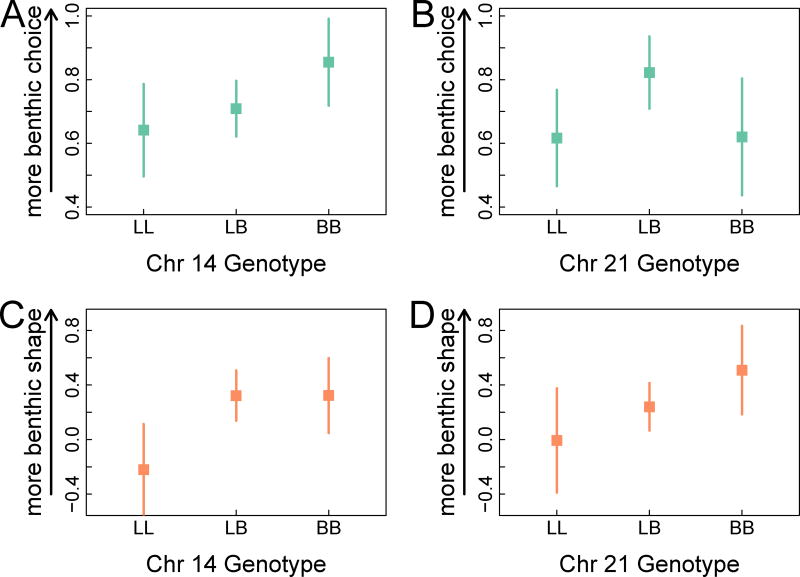
We found two QTL peaks for F2 female mate choice (Figure 2A; Table S2), on chromosomes 14 (LOD= 4.5, PVE=7.52) and 21 (LOD=4.61, PVE=10.07). For the QTL on chromosome 14, F2 females homozygous for the benthic allele (BB) were more likely to choose a benthic mate than either the limnetic homozygotes (LL) or heterozygotes (LB) (Figure 3; means: LL=0.64, LB=0.71, BB=0.86, where 0 and 1 indicate limnetic and benthic mate choice, respectively). The QTL on chromosome 21 showed a different pattern, where the heterozygote was more likely to choose a benthic mate than either homozygote (means: LL=0.58, LB=0.83, BB=0.61).
Figure 2. QTL mapping of female mate choice, body shape, and body size.
The graphs show LOD scores across the 21 stickleback chromosomes for: (A) female mate choice, (B) benthic-limnetic discriminant function, and (C) centroid size. Dotted lines: α=0.1 genome-wide significance cutoff based on 10,000 permutations. See also Table S2, Table S3, Table S4.
Figure 3. Effects of two QTL on female mate choice and body shape.
The effects of the mate choice QTL on chromosome 14 (A,C) and 21 (B,D) are shown for mate choice (A,B) and shape, represented by discriminant function score (C, D). QTL for mate choice is based on a binary response variable with 0=limnetic and 1=benthic. Points represent mean for each female genotype and error bars indicate 95% confidence intervals. See also Table S2.
F2 female mate choice is associated with her own shape and size, despite the opportunity for recombination between loci underlying the traits, suggesting either pleiotropy or physical linkage between morphology and mate choice loci. For this reason, we also investigated the genetic architecture of F2 female morphology. The results suggest that the genetic basis of morphological traits correlated with mate choice is more widely distributed across the genome than implied by the two QTL we identified for mate choice. Of the QTL for body size and the three measures of shape variation predicting mate choice (i.e. eight x- and y-landmark coordinates, PC2, and the benthic-limnetic linear discriminant function), a single QTL for body shape overlaps with a mate choice QTL (Table S2). We found a single QTL on chromosome 9 for centroid size (Figure 2C; LOD=6.97, PVE=10.08) and two QTL for PC2: one on chromosome 4 (LOD=4.04; PVE=5.97) and one on chromosome 7 (LOD=6.67; PVE=9.71). Of the eight landmark traits correlated with mate choice, five were influenced by QTL distributed across five chromosomes (Table S2). One of these QTL, for a jaw landmark coordinate (y2), overlapped with the QTL for PC2 on chromosome 4. Finally, two QTL were associated with the discriminant function separating benthic and limnetic morphology (Figure 2B). One of the QTL overlapped with the mate choice QTL on chromosome 14 (LOD=4.23; PVE=6.24), and the other mapped to chromosome 12 (LOD=4.83; PVE=7.10). At both QTL, the benthic allele was associated with a higher (more benthic-like) value of the morphological trait (Table S2).
Despite this distributed genetic architecture for F2 female body size and shape, two lines of evidence suggest genetic coupling between the QTL detected for mate choice and those detected for ecologically divergent traits. First, a linear model containing the two QTL detected for mate choice on chromosomes 14 and 21 explained a significant amount of variation in the benthic-limnetic discriminant function (Figure 3; P=0.015; LOD=2.73; PVE=4.08). Second, an additive, polygenic QTL model that predicted F2 hybrid position along the benthic-limnetic ecological niche axis provided by an earlier study of the same species pair [5] also accounted for a significant proportion of the variance in mate choice in the current study (P=0.001; LOD=10.48; PVE=21.4). The linear model based on these QTL genotypes also explained a significant proportion of the variance in the benthic-limnetic discriminant function in our experiment (P=0.00005; LOD=13.24; PVE=18.29).
DISCUSSION
The genetic basis of mate choice has consequences for the efficacy of ecological speciation with gene flow. We used data on associations between morphology, genetics, and mate choice to test predictions of the “genetic coupling” model for the evolution of mate choice. We investigated the genetic basis of interspecific mate choice in a sympatric species pair of stickleback that continue to undergo a low level of hybridization in the wild [13–15]. By measuring mate choice in F2 hybrids, which allowed the opportunity for some recombination between loci encoding mate choice and those encoding traits under divergent selection, we found strong evidence for genetic coupling. First, we found that F2 hybrid females mated with males that were more similar to themselves in shape and size. This result implies that assortative mating between like phenotypes was not eliminated by recombination in this hybrid population. Second, we found two QTL for mate choice that also explained variation in body shape. Finally, we found that a QTL model that explained variation in F2 hybrid niche use along the benthic-limnetic axis in a previous study [5] also explained variation in both F2 female shape and mate choice in our study. Together, these results are consistent with genetic coupling for the evolution and maintenance of assortative mating in this stickleback species pair.
The absence of free recombination between loci for mate choice and loci for traits under divergent selection (i.e. genetic coupling) could be due to either pleiotropy or close linkage. Felsenstein [2] showed that both mechanisms increase the likelihood of speciation and species persistence in the face of gene flow. Pleiotropy can result from phenotype matching, whereby individuals in both species (and their hybrids) prefer to mate with individuals having a similar phenotype to their own. This corresponds to Felsenstein’s “one-allele” model for the evolution of mate choice, because at a given mating locus the same allele encodes conspecific preference in both species (e.g., it encodes a phenotype matching behavior “mate with like”). When the phenotype matching alleles are fixed in both species, the observed genetic determinants of variation in mate choice are the allelic variants at the loci underlying traits upon which matching is based. This contrasts with Felsenstein’s “two-allele” model with linkage, in which distinct alleles controlling assortative mating between alternative phenotypes are physically linked to genes for traits under divergent natural selection.
By themselves, our results do not allow us to distinguish between genetic coupling caused by phenotype matching (one-allele model) and genetic coupling caused by physical linkage between alleles for mate choice and traits (two-allele model with linkage), because in both cases mate choice in recombinant hybrids should map to the regions of the genome responsible for variation in phenotypic traits. However, previous studies in this system are most consistent with a one-allele mechanism. In no-choice mating trials between heterospecifics, females mate with males that are similar in size and shape to themselves [18–20]. Importantly, non-genetic manipulation of the sizes of females changes the size of males with which they prefer to mate [19]. This result is strong evidence for the one-allele phenotype matching mechanism, at least for body size, because this non-genetic phenotypic manipulation of female body size yields no change in genes for body size preference, even if linked to genes for body size [19]. However, longer-term studies with more advanced generation hybrids to break down potential linkage between the loci that underlie body size and shape and the loci that underlie mate preferences are needed to provide more direct evidence that a one-allele mechanism contributes to genetic coupling of traits under divergent selection and mate choice in this system.
The proximate mechanism for phenotype matching suggested by our data and demonstrated by other studies is not clear [19]. How do female fish perceive and match subtle variations in their own shape and size to that of their mate? Proposed mechanisms often include sexual imprinting or social learning. A few studies have found evidence for sexual imprinting in mate preference between stickleback species [21,22]. Yet, all F2 females used in our study were produced by natural mating between F1 hybrid parents, which possess a much lower amount of size and shape variation than is seen between the two parent species, thus reducing the opportunity for imprinting or learning. It is possible to imagine that during courtship a female would be capable of evaluating her own body size relative to that of a male, but it seems far less plausible that she would be able to compare subtle differences in their body shapes. Instead, phenotype matching might occur not by direct comparison of morphology but rather by a shared feeding habitat preference between individuals that are similar in morphology. In threespine stickleback, size and shape is strongly associated with niche use both among species and among F2 hybrid individuals varying in morphology [5,10–12]. For example, the most benthic-like F2 females might feed preferentially in the same pond regions as do male benthics, and this higher encounter rate between like individuals might then lead to a higher probability of mating.
Regardless of the underlying mechanism, our results provide empirical evidence that genetic coupling is important for the persistence of species in the face of gene flow. Although genetic coupling, either via a one-allele mechanism [23] or a two-allele mechanism with linkage, has now been shown in a few other systems, in all of these cases the divergent traits are encoded by one or a few loci of relatively large effect [24–30]. However, such a simple genetic architecture for traits under divergent selection might be relatively rare. Our previous studies in stickleback have indeed shown that the genetic architecture of adaptation in this system is highly polygenic [5,31,32]. This diffuse genetic architecture of adaptation makes a two-allele model with tight linkage seem less plausible, because this would require a large number of mate choice alleles to be distributed across the genome, all in tight linkage with alleles for traits under divergent selection. Under either model, our results suggest that even when the underlying genetic architecture of phenotypes under divergent selection is polygenic and distributed across the genome, genetic coupling with assortative mating will contribute to the persistence of species in the face of gene flow.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Catherine Peichel (catherine.peichel@iee.unibe.ch).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All animal experiments involved threespine stickleback (Gasterosteus aculeatus) fish and were approved by the University of British Columbia Animal Care Committee (protocols A07-0293, A11-0402) and the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1797).
Generation of F2 females
In 2007, we used wild-caught adult fish to make six in vitro interspecific crosses. Three crosses involved a limnetic female and three crosses involved a benthic female. We stored their bodies in 95% ethanol for DNA analysis. We reared the resulting F1 hybrids in the laboratory. In March 2008, F1 hybrids were introduced to two outdoor experimental ponds on the campus of University of British Columbia (described in [5]). For the first, we randomly selected 24 F1 hybrid adults from a cross involving a limnetic female, and 24 F1 hybrid adults from a cross involving a benthic female. We took a sample of caudal fin tissue from each individual F1 hybrid for DNA analysis and then released them into two separate mesh enclosures within a pond. The enclosures were designed to allow only full-sib matings between F1s and to allow F2 hybrid offspring to escape the enclosure into the pond. However, we realized that the enclosures were limiting the number of F2 hybrids that were produced. Thus, we established a second rearing pond, for which we randomly selected five F1 hybrids of each sex from the remaining four crosses. We took a sample of caudal fin tissue from each individual for DNA analysis and then released them into the pond. This design allowed interbreeding between F1s from different crosses. In 2009, we used wild-caught adult fish to make two additional in vitro interspecific crosses. One cross involved a limnetic female and the other a benthic female. We stored their bodies in 95% ethanol for DNA analysis. We reared the resulting F1 hybrids in the laboratory. In May 2010, we initiated two F2 rearing ponds to increase the number of F2 hybrids generated and to allow only full-sib matings between F1 hybrids. We randomly selected 35 F1 hybrid adults from the cross involving a limnetic female and 35 F1 hybrid adults from the cross involving a benthic female. We took a sample of caudal fin tissue from each individual F1 hybrid for DNA analysis and then released them into their respective ponds. After release, the F1 hybrids were allowed to mate freely with their full-siblings in the same pond throughout the breeding season. For an overview of the source and numbers of the F2 females used in these experiments in both years, see Figure S2.
The ponds (25 × 15 m surface area) contained a sloping shallow zone and a deep open-water zone (6 m deep), thereby providing feeding and nesting habitat for both species [5]. In each spring of 2007 – 2010, we inoculated the ponds with macrophytes, sediments and water full of aquatic insects, mollusks and plankton from Paxton Lake. Each time we added 1.25kg of a 25.5:1 mix of 50% pure KNO3 : KH2PO4 to stimulate primary production.
METHOD DETAILS
F2 female mate choice experiment in ponds
We established three ‘mating arena’ ponds during the study (Figure S2), one in the summer of 2009 and two in the summer of 2011 to increase the area available for males to establish territories. On April 20 and 21, 2009, we added 122 wild-caught limnetic males and 117 wild-caught benthic males to the mating arena pond. From April 22 to June 1, 2009, we used minnow traps to catch 331 gravid F2 females from the two rearing ponds initiated the previous year and transferred them to the mating arena. On April 28 and 29 2011, we added 64 wild-caught limnetic males and 61 wild-caught benthic males to mating arena 1, and 64 wild-caught limnetic males and 62 wild-caught benthic males to mating arena 2. From May 2 to June 23, 2011, we used minnow traps to catch gravid F2 females from the rearing ponds initiated the previous year and transferred 219 F2 females to mating arena 1 and 218 F2 females to mating arena 2. We photographed all fish on their left side and took a sample of caudal fin tissue for DNA analysis before releasing fish into mating arenas.
From April 30 to July 17, 2009 and May 17 to July 14, 2011, we used snorkeling and SNUBA (Surface Nexus Underwater Breathing Apparatus) gear in each mating arena pond once every 3–4 days (2009) or once per week (2011) to collect fertilized eggs from male’s nests. Upon collection, eggs were inspected for their extent of development. If eyes were visible, the entire clutch was stored directly in 95% ethanol for DNA parentage analysis. If eyes were not yet visible, the clutch was split approximately in half. One half was stored directly in 95% ethanol and the other half was incubated in an aquarium to allow further development to ensure enough DNA for parentage analysis before being stored in 95% ethanol. When multiple clutches were found within the same nest (determined visually via different egg clumps and extent of egg development), each clutch was treated separately.
Parentage assignment
For the mate choice experiment conducted in 2009, we genotyped 331 F2 females, 117 benthic males, 122 limnetic males, and 245 fertilized eggs (1 per clutch) or free-swimming juveniles with 18 microsatellite markers (Table S3) following [33]. For the mate choice experiment conducted in 2011, we genotyped 437 F2 females (219 in arena 1, 218 in arena 2), 123 benthic males (61 in arena 1, 62 in arena 2), 128 limnetic males (64 in arena 1, 64 in arena 2), and 328 fertilized eggs (1 per clutch) or free-swimming juveniles (186 in arena 1, 142 in arena 2) with 19 microsatellite markers (Table S3). Parentage was assigned using the R package ‘MasterBayes’ [34] with the following parameters: E1=0.01, E2=0.01, mm.tol=10 (DRYAD data file ‘pedigree.all.csv’).
In total, 383 unique F2 females were identified in the parentage analyses (Figure S2). However, for further analyses, we only considered the 467 unique mating events for which the probability of parentage assignment of a fertilized egg or free-swimming juvenile was greater than 0.75. Using these assignments, we assessed mate choice for 291 unique F2 females, of which 255 mated exclusively with a single male species while the remaining 36 chose males of the two species for separate clutches (DRYAD data file ‘choice.all.csv’). Of the 255 F2 females that mated with only one species, 191 mated once, while 64 mated multiple times including one F2 female that mated with benthic males ten times.
Morphological analysis
We used 17 morphological landmarks to summarize morphology in wild-caught benthic and limnetic males and F2 females (Figure S1; DRYAD data file ‘phenotypes.all.csv’). Using digital images taken of live fish alongside a ruler for scale, we recorded the x- and y-coordinates of each landmark and scaled the values using ‘tpsDig’ v2.12 [35]. Coordinates were superimposed, and scaled values as well as centroid sizes were calculated using Generalized Procrustes Analysis in the R package ‘shapes’ [36]. We summarized these landmarks using principal component analysis with the ‘prcomp’ function in R [37]. Custom R scripts (‘Morphology.R’ and ‘landmarks.R’) for these analyses are provided on DRYAD (http://dx.doi.org/10.5061/dryad.bs7sg).
Association of F2 mate choice and morphology
We tested for associations between mate choice and morphology of the 255 F2 females that mated with only a single male species using centroid size as a measure of body size and three measures of F2 female shape based on landmarks: (1) principal component analysis, (2) discriminant function analysis; (3) individual × and y coordinates of landmarks. For centroid size, we tested associations between F2 female size and female mate choice using a binomial generalized linear model with experimental pond as a covariate.
Principal component analysis
We used principal component analysis to examine morphological variation within F2 females as well as within and between benthic and limnetic males. A single principal component axis (PC2) separated benthic and limnetic males, with F2 females intermediate. We used this benthic-limnetic PC axis to test associations between female morphology and female mate choice. Parents of each egg clutch, as determined from the parentage analysis, were used to determine the species of male chosen by each F2 female. We used a binomial generalized linear model to test associations between female mate choice (benthic or limnetic) and her score along the benthic-limnetic PC axis, with experimental pond as a covariate and significance assessed using the drop1 function in R. We also used linear models to compare female PC scores with the PC scores of the chosen males, with experimental pond as a covariate and mother as a random effect. Coefficients of partial determination (partial R2) were calculated using the ‘rsq’ package in R [37]. We repeated these analyses using centroid size as a measure of body size in place of the benthic-limnetic PC axis.
Discriminant function analysis
We used discriminant function analysis to summarize F2 female shape morphology along a benthic-limnetic axis. We used morphological landmarks from wild-caught benthic and limnetic males to build a discriminant function with the R package ‘MASS’ [38]. This model had 99.8% classification accuracy using 10-fold cross-validation; only a single individual male was incorrectly classified. The model was used to predict discriminant function values for F2 females based on the same morphological landmarks. We then tested for association between this benthic-limnetic discriminant function value and mate choice in F2 females using a binomial generalized linear model with experimental pond as a covariate.
Individual x- and y-coordinates
To identify specific morphological landmarks that are most strongly correlated with female mate choice, we also tested for associations between female mate choice and body shape landmarks of F2 female phenotype. For this, we used the scaled x- and y-landmark coordinates for F2 females and tested associations with female mate choice using a binomial generalized linear model with experimental pond as a covariate.
Genotyping F2 females
We isolated genomic DNA from caudal fin tissue of the 16 F0 progenitors, 158 F1 hybrids, and the 383 F2 hybrid females identified in the parentage analyses using Proteinase K digestion, phenol-chloroform extraction, ethanol precipitation and re-suspension of the precipitated DNA in 30 µL of TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0). We genotyped all F0, F1, and F2 individuals using Illumina’s GoldenGate assay and a custom multiplex oligonucleotide pool developed for a previously published collection of single nucleotide polymorphisms (SNPs; [7]; Table S4). We found 494 of these SNPs to be polymorphic in at least one of our crosses. The Illumina Sentrix Array Matrices used for genotyping were processed at the Genomics Shared Resource of the Fred Hutchinson Cancer Research Center (Seattle, WA, USA). We scored genotypes from the raw data using GenomeStudio software (Illumina Inc.).
Linkage map construction
To build a linkage map, we started with the 383 genotyped F2 females in this experiment (Figure S2), along with 1,348 F2 individuals from the same crosses but used in another experiment [39]. Following [5], we only used F2 individuals that could be assigned to an F1 × F1 family having at least 10 full-siblings for linkage map construction and subsequent QTL analyses, resulting in the inclusion of 302 F2 females from this experiment. We first calculated pairwise recombination frequencies for each F1 × F1 family using JoinMap ver 3.0 [40]; recombination frequencies were concatenated and imported into JoinMap to produce a single linkage map. We found 21 linkage groups, which were assigned to the 21 chromosomes from the stickleback genome assembly using known SNP locations.
QTL analysis
All QTL analysis was performed in the ‘R/qtl’ package [41], and a custom R script ‘QTL.R’ is provided on DRYAD (http://dx.doi.org/10.5061/dryad.bs7sg). Although power to detect QTL of small effect is increased by having more individuals, power to detect QTL at all is reduced if the phenotypic analysis is not robust. To map mate choice, we therefore conservatively used the 200 F2 females that were: (1) included in the linkage map construction; (2) had a parentage assignment probability greater than 0.75; and (3) mated with only a single species of male (DRYAD data files: ‘purechoice.gen.csv’ and ‘purechoice.pheno.csv’). We used the ‘scanone’ command with Haley-Knott regression and a binary response variable (1 = chose benthic; 0 = chose limnetic), with both family and experimental pond as covariates. To determine significance, we used 10,000 permutations and a genome-wide cutoff of α=0.1. We used this lenient threshold because our main goal was to determine whether QTL for mate choice and morphology lie in the same regions, so false positives were less of a concern than missing QTL.
To increase our power to detect QTL for morphological traits associated with mate choice, we included all 302 F2 females used in the linkage map construction (DRYAD data files: ‘all.gen.csv’ and ‘all.pheno.csv’). We conducted a similar analyses as above to find QTL for centroid size as well as for our three shape measurements: 1) the PC axis that differentiated benthic and limnetic shapes; 2) the benthic-limnetic discriminant function; and 3)×and y coordinates of morphological landmarks. For these, we assumed a Gaussian distribution for the response variable. For each significant QTL, we calculated percent variance explained (PVE) under a single QTL model using the function PVE=1–10(−2*LOD/n) [41]. All shape QTL remained significant even after using centroid size as a covariate in the analyses (data not shown).
Additionally, we used ‘fitqtl’ to investigate whether QTL peaks for mate choice could also explain the predicted benthic-limnetic discriminant function values of the 200 F2 females used to map mate choice. We calculated significance (χ2 test), log odds ratio (LOD), and PVE as above.
Arnegard et al. [5] defined an additive model of 11 QTL loci and significant interactions that predicted F2 phenotype along the benthic-limnetic niche axis. Because the same SNP assay was used here as in Arnegard et al. [5], we were able to use the same markers to test whether this model could explain both morphology and mate choice in our experiment. We used ‘fitqtl’ to compare the sum of squares of a model with pond and family covariates only to a model that also included genotypes at the 11 markers identified by Arnegard et al. [5] to explain the predicted benthic-limnetic discriminant function value as well as mate choice in the 200 F2 females used to map mate choice.
QUANTIFICATION AND STATISTICAL ANALYSIS
All analysis was conducted in R [37]. Statistical tests and software used are described in Method Details (above).
DATA AND SOFTWARE AVAILABILITY
All data files and custom R scripts required to recreate these analyses are available on DRYAD: http://dx.doi.org/10.5061/dryad.bs7sg.
SUPPLEMENTAL INFORMATION
Supplemental Information PDF contains 2 figures and 3 tables.
Supplementary Material
Related to Figure 2. The positions in bp refer to the original threespine stickleback genome assembly (Broad S1, Feb. 2006; http://www.ensembl.org/Gasterosteus_aculeatus/Info/Index).
Highlights.
Mate choice of benthic-limnetic stickleback F2 hybrid females was tested in ponds
F2 hybrid females choose males with a similar body size and shape to their own
QTL associated with F2 female mate choice also predict F2 female morphology
Speciation with gene flow in sticklebacks is facilitated by genetic coupling
Acknowledgments
We thank Cassie Sather and Elizabeth Jensen for assistance with SNP and microsatellite genotyping, and Joey Courchesne, Travis Ingram, Sahriar Kabir, Kerry Marchinko, and Patrick Tamkee for assistance with fieldwork. This research was funded by grants from the National Institutes of Health (F32 GM086125 to M.E.A., P50 HG002568 to D.M.K. and C.L.P., R01 GM089733 to D.S. and C.L.P.) and a Discovery grant from the Natural Sciences and Engineering Research Council to D.S. The ponds were built using an infrastructure grant from the Canada Foundation for Innovation to D.S., with matching funds from the Province of British Columbia and the University of British Columbia. D.M.K. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization: M.E.A., G.L.C., D.S., and C.L.P.; Methodology: M.E.A., G.L.C., D.S., and C.L.P.; Formal Analysis: R.A.B., M.E.A., G.L.C., D.S., and C.L.P.; Investigation: M.E.A., G.L.C., J.B., N.L.B., S.R.M., and M.E.D.; Resources: Y.F.C., F.C.J., and D.M.K.; Writing – Original Draft: R.A.B., G.L.C., D.S., and C.L.P.; Writing – Review & Editing: – M.E.A., N.L.B., Y.F.C., and D.M.K.; Supervision: M.E.A., D.S., and C.L.P.; Funding Acquisition: M.E.A., D.S., and C.L.P.
References
- 1.Schluter D. Evidence for ecological speciation and its alternative. Science. 2009;323:737–741. doi: 10.1126/science.1160006. [DOI] [PubMed] [Google Scholar]
- 2.Felsenstein J. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution. 1981;35:124–38. doi: 10.1111/j.1558-5646.1981.tb04864.x. [DOI] [PubMed] [Google Scholar]
- 3.Gavrilets S. Fitness Landscapes and the Origin of Species. Princeton: Princeton University Press; 2004. [Google Scholar]
- 4.Smadja CM, Butlin RK. A framework for comparing processes of speciation in the presence of gene flow. Mol. Ecol. 2011;20:5123–5140. doi: 10.1111/j.1365-294X.2011.05350.x. [DOI] [PubMed] [Google Scholar]
- 5.Arnegard ME, McGee MD, Matthews B, Marchinko KB, Conte GL, Kabir S, Bedford N, Bergek S, Chan YF, Jones FC, et al. Genetics of ecological divergence during speciation. Nature. 2014;511:307–311. doi: 10.1038/nature13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Taylor EB, McPhail JD. Historical contingency and ecological determinism interact to prime speciation in sticklebacks, Gasterosteus. Proc. Roy. Soc. Lond. B. 2000;267:2375–2384. doi: 10.1098/rspb.2000.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones FC, Chan YF, Schmutz J, Grimwood J, Brady SD, Southwick AM, Absher DM, Myers RM, Reimchen TE, Deagle BE, et al. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr. Biol. 2012;22:83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schluter D, McPhail JD. Ecological character displacement and speciation in sticklebacks. Am. Nat. 1992;140:85–108. doi: 10.1086/285404. [DOI] [PubMed] [Google Scholar]
- 9.McPhail JD. Speciation and the evolution of reproductive isolation in the sticklebacks (Gasterosteus) of south-western British Columbia. In: Bell MA, Foster SA, editors. The Evolutionary Biology of the Threespine Stickleback. New York: Oxford University Press; 1994. pp. 399–437. [Google Scholar]
- 10.Schluter D. Adaptive radiation in sticklebacks: trade-offs in feeding performance and growth. Ecology. 1995;76:82–90. [Google Scholar]
- 11.McGee MD, Schluter D, Wainwright PC. Functional basis of ecological divergence in sympatric stickleback. BMC Evol. Biol. 2013;13:277. doi: 10.1186/1471-2148-13-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews B, Marchinko KB, Bolnick DI, Mazumder A. Specialization of trophic position and habitat use by sticklebacks in an adaptive radiation. Ecology. 2010;91:1025–1034. doi: 10.1890/09-0235.1. [DOI] [PubMed] [Google Scholar]
- 13.McPhail JD. Ecology and evolution of sympatric sticklebacks (Gasterosteus): evidence for a species-pair in Paxton Lake, Texada Island, British Columbia. Can. J. Zool. 1992;70:361–369. [Google Scholar]
- 14.Gow JL, Peichel CL, Taylor EB. Contrasting hybridization rates between sympatric three-spined sticklebacks highlight the fragility of reproductive barriers between evolutionarily young species. Mol. Ecol. 2006;15:739–752. doi: 10.1111/j.1365-294X.2006.02825.x. [DOI] [PubMed] [Google Scholar]
- 15.Gow JL, Peichel CL, Taylor EB. Ecological selection against hybrids in natural populations of sympatric threespine sticklebacks. J. Evol. Biol. 2007;20:2173–2180. doi: 10.1111/j.1420-9101.2007.01427.x. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield T, Schluter D. Ecological speciation in sticklebacks: environment-dependent hybrid fitness. Evolution. 1999;53:866–873. doi: 10.1111/j.1558-5646.1999.tb05380.x. [DOI] [PubMed] [Google Scholar]
- 17.Rundle HD, Nagel L, Boughman JW, Schluter D. Natural selection and parallel speciation in sympatric sticklebacks. Science. 2000;287:306–308. doi: 10.1126/science.287.5451.306. [DOI] [PubMed] [Google Scholar]
- 18.Nagel L, Schluter D. Body size, natural selection, and speciation in sticklebacks. Evolution. 1998;52:209–218. doi: 10.1111/j.1558-5646.1998.tb05154.x. [DOI] [PubMed] [Google Scholar]
- 19.Conte GL, Schluter D. Experimental confirmation that body size determines mate preference via phenotype matching in a stickleback species pair. Evolution. 2013;67:1477–1484. doi: 10.1111/evo.12041. [DOI] [PubMed] [Google Scholar]
- 20.Head ML, Kozak GM, Boughman JW. Female mate preferences for male body size and shape promote sexual isolation in threespine sticklebacks. Ecol. Evol. 2013;3:2183–2196. doi: 10.1002/ece3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albert AYK. Mate choice, sexual imprinting, and speciation: a test of a one-allele isolating mechanism in sympatric sticklebacks. Evolution. 2005;59:927–6. [PubMed] [Google Scholar]
- 22.Kozak GM, Head ML, Boughman JW. Sexual imprinting on ecologically divergent traits leads to sexual isolation in sticklebacks. Proc. Roy. Soc. Lond B. 2011;278:2604–2610. doi: 10.1098/rspb.2010.2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortiz-Barrientos D, Noor MAF. Evidence for a one-allele assortative mating locus. Science. 2005;310:1467–1477. doi: 10.1126/science.1121260. [DOI] [PubMed] [Google Scholar]
- 24.Hawthorne DJ, Via S. Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature. 2001;412:904–907. doi: 10.1038/35091062. [DOI] [PubMed] [Google Scholar]
- 25.Shaw KL, Lesnick SC. Genomic linkage of male song and female acoustic preference QTL underlying a rapid species radiation. Proc. Natl. Acad. Sci. USA. 2009;106:9737–9742. doi: 10.1073/pnas.0900229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wiley C, Shaw KL. Multiple genetic linkages between female preference and male signal in rapidly speciating Hawaiian crickets. Evolution. 2010;64:2238–2245. doi: 10.1111/j.1558-5646.2010.01007.x. [DOI] [PubMed] [Google Scholar]
- 27.Wiley C, Ellison CK, Shaw KL. Widespread genetic linkage of mating signals and preferences in the Hawaiian cricket Laupala. Proc. R. Soc. Lond B. 2012;279:1203–1209. doi: 10.1098/rspb.2011.1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kronforst MR, Young LG, Kapan DD, McNeely C, ONeill RJ, Gilbert LE. Linkage of butterfly mate preference and wing color preference cue at the genomic location of wingless. Proc. Natl. Acad. Sci. USA. 2006;103:6575–6580. doi: 10.1073/pnas.0509685103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrill RM, Van Schooten B, Scott JA, Jiggins CD. Pervasive genetic associations between traits causing reproductive isolation in Heliconius butterflies. Proc. R. Soc. Lond B. 2011;278:511–518. doi: 10.1098/rspb.2010.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung H, Loehlin DW, Dufour HD, Vaccarro K, Millar JG, Carroll SB. A single gene affects both ecological divergence and mate choice in Drosophila. Science. 2014;343:1148–1151. doi: 10.1126/science.1249998. [DOI] [PubMed] [Google Scholar]
- 31.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peichel CL, Marques DA. The genetic and molecular architecture of phenotypic diversity in sticklebacks. Phil. Trans. R. Soc. B. 2017;372:20150486. doi: 10.1098/rstb.2015.0486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peichel CL, Nereng KS, Ohgi KA, Cole BLE, Colosimo PF, Buerkle CA, Schluter D, Kingsley DM. The genetic architecture of divergence between threespine stickleback species. Nature. 2001;414:901–905. doi: 10.1038/414901a. [DOI] [PubMed] [Google Scholar]
- 34.Hadfield JD, Richardson DS, Burke T. Towards unbiased parentage assignment:combining genetic, behavioural and spatial data in a Bayesian framework. Mol. Ecol. 2006;15:3715–3730. doi: 10.1111/j.1365-294X.2006.03050.x. [DOI] [PubMed] [Google Scholar]
- 35.Rohlf FJ. tpsDig v 2.12. Department of Ecology and Evolution; State University of New York at Stony Brook: 2010. [Google Scholar]
- 36.Dryden IL. R Foundation for Statistical Computing. Vienna, Austria: 2017. Shapes package. [Google Scholar]
- 37.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: 2014. [Google Scholar]
- 38.Venables WN, Ripley BD. Modern Applied Statistics with S. New York: Springer; 2002. [Google Scholar]
- 39.Conte GL, Arnegard ME, Best J, Chan YF, Jones FC, Kingsley DM, Schluter D, Peichel CL. Extent of QTL reuse during repeated phenotypic divergence of sympatric threespine stickleback. Genetics. 2015;201:1189–1200. doi: 10.1534/genetics.115.182550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Ooijen JW, Voorrips RE. JoinMap® 3.0, Software for the calculation of genetic linkage maps. Wageningen, the Netherlands: Plant Research International; 2001. [Google Scholar]
- 41.Broman KW, Sen S. A Guide to QTL Mapping with R/qtl. New York: Springer; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 2. The positions in bp refer to the original threespine stickleback genome assembly (Broad S1, Feb. 2006; http://www.ensembl.org/Gasterosteus_aculeatus/Info/Index).