Summary
Autophagy is required for benign hepatic tumors to progress into malignant hepatocellular carcinoma. However, the mechanism is unclear. Here we report that mitophagy, the selective removal of mitochondria by autophagy, positively regulates hepatic cancer stem cells (CSCs) by suppressing the tumor suppressor p53. When mitophagy is enhanced, p53 colocalizes with mitochondria and is removed by a mitophagy-dependent manner. However, when mitophagy is inhibited, p53 is phosphorylated at serine-392 by PINK1, a kinase associated with mitophagy, on mitochondria and translocated into the nucleus where it binds to the NANOG promoter to prevent OCT4 and SOX2 transcription factors from activating the expression of NANOG, a transcription factor critical for maintaining the stemness and the self-renewal ability of CSCs, resulting in the reduction of hepatic CSC populations. These results demonstrate that mitophagy controls the activities of p53 to maintain hepatic CSCs and provide an explanation to why autophagy is required to promote hepatocarcinogenesis.
eTOC Blurb
Autophagy is required for the malignant transformation of liver tumors. Liu et al. demonstrated that mitophagy, the selective removal of mitochondria by autophagy, was required to maintain the hepatic cancer stem cell population by removing mitochondria-associated p53, which otherwise would be activated by PINK1 to suppress the expression of NANOG.

Introduction
Autophagy (i.e., macroautophagy) is a catabolic process that removes protein aggregates and damaged organelles in cells. It is important for maintaining cellular homeostasis and has also been implicated in the development of cancers with two opposite functions (White, 2015). It may function as a tumor suppressor by preventing the accumulation of dysfunctional mitochondria, which can lead to increased oxidative stress and DNA damage (Tian et al., 2015), and the accumulation of the p62 sequestosome protein, which also promotes oxidative stress and tumor growth (Mathew et al., 2009). Autophagy may also function as a tumor promoter to alleviate metabolic stress during tumorigenesis and suppress the expression of tumor suppressors (Guo et al., 2013; Rosenfeldt et al., 2013; Tian et al., 2015), and impairing autophagy can impede hepatocarcinogenesis and prevent benign hepatic tumors from becoming malignant hepatocellular carcinoma (HCC) (Takamura et al., 2011; Tian et al., 2015), prevent low-grade pre-malignant pancreatic neoplastic lesions from progressing into high-grade pancreatic intraepithelial neoplasia and the pancreatic ductal adenocarcinoma (Rosenfeldt et al., 2013), and alter the fate of pulmonary tumors from adenomas and carcinomas to benign oncocytomas (Guo et al., 2013). Interestingly, in the above studies of hepatic, pancreatic and pulmonary tumors with impaired autophagy, tumor progression was restored or partially restored if the expression of the tumor suppressor p53 was suppressed, suggesting that autophagy may promote tumorigenesis via the control of p53 activities.
As an important tumor suppressor, p53 has many different activities. One of these activities is to act as a transcription factor to regulate the expression of its target genes via its response element in the promoters of those genes (Beckerman and Prives, 2010). The activities of p53 are regulated by a variety of post-translational modifications. For example, the phosphorylation of p53 at serine-392 (S392) can lead to its stabilization and tetramerization and the activation of its sequence-specific DNA binding activity (Dai and Gu, 2010). The role of p53 in the regulation of the homeostasis of stem cells has also been recognized. It can restrict the self-renewal of stem cells, inhibit symmetric division and block the reprogramming of somatic/progenitor cells into stem cells (Bonizzi et al., 2012). The loss of p53 will therefore facilitate the development of tumors due to the expansion of stem cells resulting from increased self-renewal and symmetric divisions and the reprogramming of somatic/progenitor cells (Bonizzi et al., 2012).
Cancer stem cells (CSCs), also known as tumor-initiating cells, are a subset of tumor cells that display the stem cell markers and, similar to stem cells, possess the ability to self-renew and produce heterogeneous progeny cells (Ailles and Weissman, 2007). They are found in solid tumors including HCC and may be derived from normal stem cells or differentiated cells (Ailles and Weissman, 2007; Ma et al., 2007; Yamashita et al., 2009). CSCs are highly tumorigenic and chemotherapy-resistant. They are thought to play important roles in the tumorigenesis of HCC (Yamashita and Wang, 2013).
In this report, we studied how autophagy might promote hepatocarcinogenesis. Our results indicated that autophagy was required to maintain the hepatic CSC population via the suppression of p53, which was removed by a pathway dependent on mitophagy, a selective autophagy that specifically removes mitochondria. We also found that p53 was phosphorylated at serine-392 and activated by Pten-induced putative kinase 1 (PINK1), a kinase associated with mitochondria and important for mitophagy, and when autophagy or mitophagy was impaired, the activated p53 was localized to the nucleus to suppress the expression of NANOG, a key transcription factor required for the self-renewal and the maintenance of the stemness of stem cells (Lin et al., 2005), resulting in the reduction of the hepatic CSC population. Our studies thus indicated that mitophagy positively regulated hepatic CSCs by suppressing p53, which otherwise would be activated by PINK1 to suppress the expression of NANOG and hepatic CSCs. These results provided an explanation to how autophagy and, more specifically, mitophagy promoted hepatocarcinogenesis.
Results
Autophagy positively regulates hepatic cancer stem cells
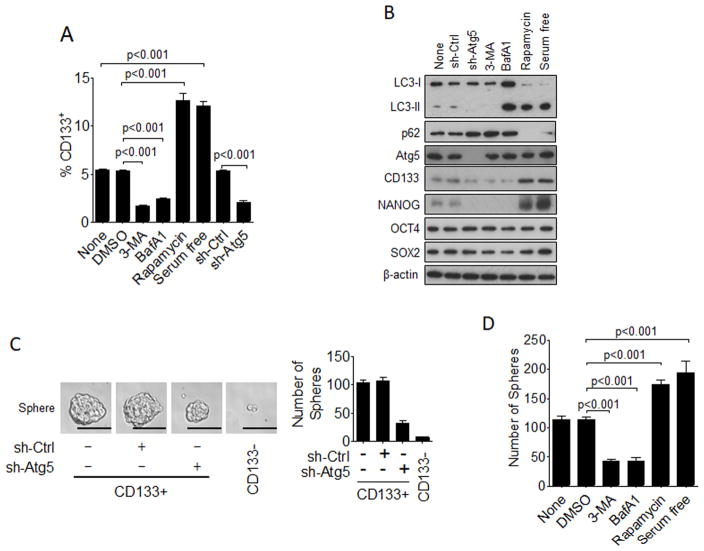
To understand why autophagy was required to promote hepatocarcinogenesis, we analyzed the effect of autophagy on hepatic CSCs using HepG2 cells, a human hepatoblastoma cell line. CD133 is a marker of CSCs, and CD133+ cells have tumor-initiating ability, including the initiation of liver tumors (Ailles and Weissman, 2007; Yamashita and Wang, 2013). As shown in Figure 1A, CD133+ cells comprised about 5% of the total HepG2 cell population. This percentage was reduced by 3-methyladenine (3-MA) and bafilomycin A1 (BafA1), which are autophagy inhibitors, and enhanced by rapamycin and serum deprivation, which are autophagy inducers. The silencing of ATG5, a gene essential for autophagy, also reduced the population of CD133+ cells (Figure 1A). Examples of the flow cytometry results for the analysis of CD133+ cells are shown in Figure S1A. These results indicated that autophagy played a positive role in the generation of CD133+ HepG2 cells. We had previously produced mice with hepatocyte-specific knockout of the ATG5 gene. These mice developed benign liver tumors with a high frequency with no detectable HCC, not even with the treatment of the carcinogen diethylnitrosamine (DEN), which induced HCC with a high frequency in wild-type mice (Tian et al., 2015). To ensure that the effect of autophagy on hepatic CSCs was not specific to HepG2 cell, we also analyzed the liver tumors isolated from ATG5-knockout mice and control mice that had been treated with DEN. As shown in Figure S1B, approximately 0.85% of liver tumor cells isolated from control mice were CD133+CD49f+ whereas only about 0.14% of liver tumor cells isolated from ATG5-knockout mice were CD133+CD49f+. CD49f is another liver stem cell marker (Tsukamoto et al., 2014). These results indicated that the effect of autophagy on CSC-like cells was not specific to HepG2 cells and could also be detected in vivo in mouse liver tumors.
Figure 1. Effect of autophagy on hepatic CSCs.
(a) HepG2 with or without various treatments for 24 hours and stable HepG2 cells that expressed a control shRNA (sh-Ctrl) or the Atg5 shRNA (sh-Atg5) were subjected to flow cytometry analysis for CD133+ cells. Results represent the mean ± SEM of three independent experiments. None, no treatment. (B) HepG2 cells with the treatments shown in (A) were lysed for immunoblot analysis. LC3-I, non-lipidated LC3; LC3-II, lipidated LC3. The β-actin protein was also analyzed to serve as the loading control. (C) Sphere-formation assay of CD133+ and CD133− HepG2 cells. The panels shown to the left are representative results of spheres formed by CD133+ and CD133− HepG2 cells with and without stable ATG5 knockdown. Scale bar=200 μm. The histogram shown to the right indicated the number of spheres larger than 100 μm in diameter when 500 CD133+ cells were seeded. The results represent the mean ± SEM of three independent experiments. (D) HepG2 cells with various treatments for 24 hours were incubated with MicroBeads (Miltenyi Biotec) for the isolation of CD133+ cells, which were then analyzed for their sphere-forming ability. 500 cells were seeded for the assay. Also see Figure S1.
The effects of autophagy inhibitors and inducers as well as the effect of ATG5 knockdown on autophagy and the expression of CD133 in HepG2 cells were also confirmed by immunoblot. As shown in Figure 1B, ATG5 knockdown and 3-MA, which both inhibited the early stage of autophagy, suppressed the lipidation of LC3 (i.e., LC3-II), an important step for the formation of autophagosomes (Klionsky et al., 2016). BafA1, which inhibited the maturation of autophagosomes and hence the recycling of lipidated LC3, increased the level of LC3-II. These treatments all led to the increase of p62, a protein removed by autophagy, and the reduction of the protein levels of CD133 and NANOG. In contrast, rapamycin and serum deprivation both led to the increase of LC3-II, the reduction of p62, and the increase of CD133 and NANOG. None of the treatments affected OCT4 and SOX2, two upstream transcription factors that activate the NANOG gene and are also important for the self-renewal of stem cells (Rodda et al., 2005). These results indicated that autophagy increased the level of CD133+ HepG2 cells via a step downstream or independent of OCT4 and SOX2 factors but upstream of NANOG. We had also conducted the sphere-formation assay, which is an assay frequently used to determine the self-renewal ability of CSCs (Pastrana et al., 2011), to test the effect of autophagy on the self-renewal ability of CD133+ HepG2 cells. As shown in Figure 1C, ATG5 knockdown reduced the sphere-forming ability (i.e., self-renewal ability) of CD133+ HepG2 cells. Few spheres could be detected when CD133− HepG2 cells were used for the assay. Similarly, both 3-MA and BafA1 reduced the sphere-forming ability of CD133+ HepG2 cells, but rapamycin and serum deprivation increased it (Figure 1D). The silencing of ATG7, another gene essential for autophagy, led to the loss of LC3-II and increase of p62. This silencing of ATG7 also suppressed the expression of CD133 and NANOG and increased the CD133+ cell population and their sphere-forming ability (Figure S1C–E). These results together indicated that CD133+ HepG2 cells possessed the property of CSCs and that autophagy played an important role in maintaining their population.
The effect of autophagy on hepatic cancer stem cells is p53-dependent
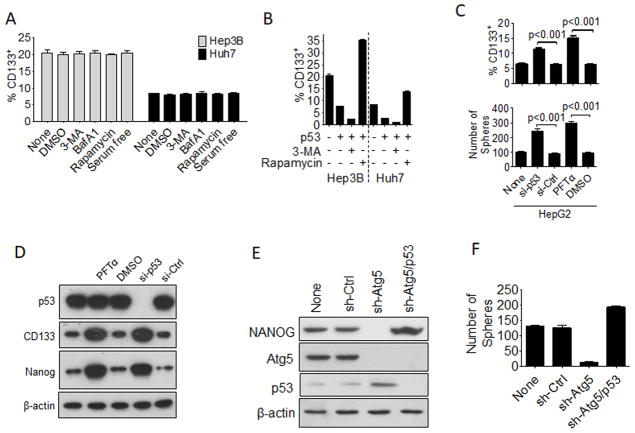
In contrast to HepG2 cells and mouse liver tumors, autophagy inhibitors and inducers had no effect on the CD133+ cell population of Hep3B and Huh7 cells (Figure 2A), two different human hepatoma cell lines. HepG2 cells expressed a wild-type p53 whereas Hep3B cells were p53-negative and Huh7 cells expressed a p53 mutant with a tyrosine-to-cysteine mutation at amino acid 220 (i.e., the Y220C mutation) that had impaired DNA binding ability (Hsu et al., 1993; Kubicka et al., 1997). To determine whether the lack of effect of autophagy on the CD133+ cells of these two cell lines was due to the defect in p53 in these two cell lines, we transfected these two cell lines with a plasmid that expressed the wild-type p53. As shown in Figure 2B, the expression of p53 in Hep3B cells reduced the CD133+ cell population from 20% to 8%, which could be further reduced by the autophagy inhibitor 3-MA to approximately 2% and restored by the autophagy inducer rapamycin to over 30%. Similarly, as shown in the same figure, the expression of wild-type p53 in Huh7 cells reduced the CD133+ CSCs from 9% to 3%, which could be further reduced by 3-MA and restored by rapamycin. These results indicated that p53 could reduce the CD133+ cell population of these two cell lines, and this activity of p53 on CD133+ cells was antagonized by autophagy. To determine whether p53 also negatively regulated the CD133+ population of HepG2 cells, we transfected HepG2 cells with the p53 siRNA or treated the cells with Pifithrin-α (PFTα), which inhibits the nuclear transport of p53 (Lei et al., 2013) (Figure S2A). PFTα did not affect the viability of cells (Figure S2B). As shown in Figure 2C, the suppression of p53 expression or the inhibition of its nuclear localization increased the population of CD133+ HepG2 cells and their self-renewal ability. Both p53 siRNA and PFTα also increased the expression levels of CD133 and NANOG in HepG2 cells as revealed by the immunoblot analysis (Figure 2D). We also analyzed whether the suppression of p53 expression could restore the expression of NANOG and sphere-forming ability of HepG2 cells with ATG5 knockdown. As shown in Figure 2E and 2F, this was indeed the case. These results demonstrated that p53 negatively regulated CD133+ HepG2 cells and their self-renewal ability.
Figure 2. Role of p53 in the regulation of hepatic CSCs by autophagy.
(A) Hep3B and Huh7 cells with various treatments for 24 hours were subjected to flow cytometry analysis for CD133+ cells. (B) HepG2 and Huh7 cells were transfected with the p53-expressing plasmid for two days, treated with 3-MA or rapamycin for another 24 hours and then subjected to flow cytometry analysis for CD133+ cells. (C) HepG2 cells transfected with the control siRNA (si-Ctrl) or the p53 siRNA (si-p53) for two days or treated with PFTα or DMSO for one day were analyzed for their CD133+ cells by flow cytometry (top panel) or sphere-forming ability of their CD133+ cells (bottom panel). The results shown in (A), (B) and (C) represent the mean ± SEM of three independent experiments. (D) HepG2 cells treated with DMSO or PFTα for one day or with siRNA for two days were lysed for immunoblot analysis. (E) Stable HepG2 cells that expressed control shRNA (sh-Ctrl), sh-Atg5, or both sh-Atg5 and sh-p53 were lysed for immunoblot analysis. (F) Cells mentioned in (E) were used for the sphere-formation assay. Also see Figure S2.
Phosphorylation of p53 at serine-392 reduces the hepatic CSC population
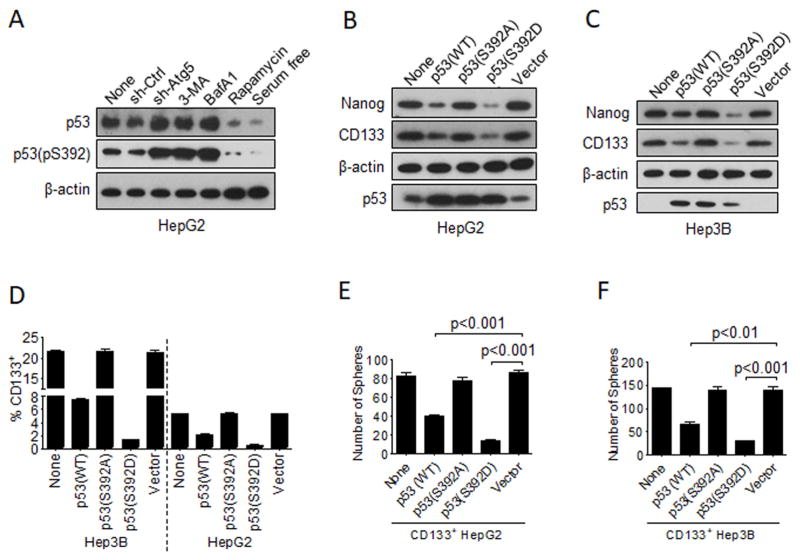
To further understand the relationship between autophagy and p53 and their effects on hepatic CSCs, we analyzed the effects of autophagy inhibitors and inducers as well as the effect of ATG5 knockdown on the expression of p53 in HepG2 cells. As shown in Figure 3A, the suppression of autophagy with Atg5 shRNA, 3-MA or BafA1 all led to the increase of p53 and its phosphorylation at serine-392 (i.e., p53(pS392)). Similar results were obtained when the mouse liver tumors were analyzed. As shown in Figure S3A, liver tumors isolated from control mice expressed higher levels of CD133 and NANOG, in agreement with the results shown in Figure S1B, and lower levels of p53 and p53(pS392) than those isolated from mice with hepatocyte-specific knockout of ATG5. In contrast, rapamycin or serum deprivation reduced p53 and p53(pS392) protein levels in HepG2 cells (Figure 3A). None of these treatments affected the p53 mRNA level (Figure S3B), indicating that their effects on p53 were most likely post-translational. The phosphorylation of p53 at S392 can activate p53 and lead to its stabilization and nuclear localization to bind to its target genes (Dai and Gu, 2010). To test the possible importance of this S392 phosphorylation of p53 on hepatic CSCs, we transfected HepG2 cells and Hep3B cells with an expression plasmid that expressed the wild-type p53 (p53(WT)), the non-phosphorylated analog of p53(S392A) or the phosphorylated analog of (S392D). As shown in Figure 3B, 3C and 3D, the expression of either p53(WT) or p53(S392D) reduced the expression levels of CD133 and NANOG in HepG2 and Hep3B cells and the population of CD133+ cells. In contrast, the expression of p53(S392A) or the control vector had no effect on the expression of these two proteins and the population of CD133+ cells. Similarly, although p53(WT) and p53(S392D) reduced the sphere-forming ability of CD133+ HepG2 and Hep3B cells, p53(S392A) and the control vector had no effect (Figure 3E and 3F). These results indicated that the phosphorylation of p53 at S392 likely played an important role in suppressing the expression of NANOG and reducing the CSC population of HepG2 and Hep3B cells.
Figure 3. Effect of autophagy on serine-392 phosphorylation of p53 and hepatic CSCs.
(A) HepG2 cells with various treatments for 24 hours or stably expressing the control shRNA or the Atg5 shRNA were lysed for immunoblot analysis. (B) Immunoblot analysis of HepG2 cells transfected with either the control vector or the expression vector for various p53 proteins. Cells were lysed two days after DNA transfection for immunoblot analysis. None, control cells with no DNA transfection. (C) The experiments were conducted the same way as in (B), with the exception that Hep3B cells were used for the expression studies. (D) Hep3B or HepG2 cells were transfected with various p53-expressing plasmids or the control vector as indicated for two days followed by flow cytometry analysis for CD133+ cells. (E) HepG2 cells were transfected with the p53-expressing plasmids for two days, and CD133+ cells were then isolated for the sphere-formation assay. (F) The experiments were conducted the same way as in (E), with the exception that Hep3B cells were used for the studies. The results in (D–F) represented the mean ± SEM of three independent experiments. Also see Figure S3.
p53 antagonizes the activities of OCT4-SOX2 to suppress the expression of NANOG
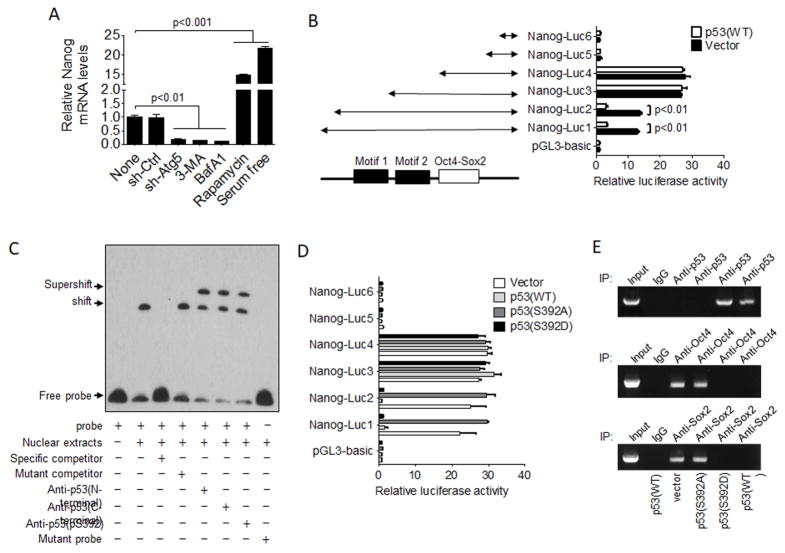
As mentioned above, NANOG is a key transcription factor that regulates the self-renewal of stem cells. To understand how p53 might suppress the expression of NANOG, we first examined the expression levels of the NANOG mRNA. As shown in Figure 4A, Atg5 shRNA, 3-MA and BafA1 all reduced the NANOG mRNA level in HepG2 cells and, in contrast, rapamycin and serum deprivation both increased the NANOG mRNA level. These results suggested a transcriptional regulation of NANOG by autophagy. An examination of the NANOG promoter led to the identification of two possible p53-binding motifs, which are separated by two base-pairs and located in the upstream region of the OCT4-SOX2 binding site that was previously reported (Rodda et al., 2005) (Figure 4B and Figure S4A). These two putative p53 binding motifs resemble the p53 response element, which consists of two tandem p53 recognition sequences that are separated by 0–13 base pairs (Beckerman and Prives, 2010; Lin et al., 2005). To test whether these two putative p53 binding motifs were indeed important for p53 to suppress the expression of NANOG, we conducted a deletion-mapping experiment on the NANOG promoter using firefly luciferase as the reporter, and co-transfected the reporter constructs with either the control vector or the p53-expressing plasmid into HepG2 cells. As shown in Figure 4B, the co-expression of p53 could lead to the suppression of luc1 and luc2 reporter constructs, which contained the two putative p53 binding motifs. However, the deletion of motif 1 (i.e., the luc3 construct) or both motifs 1 and 2 (i.e., the luc4 construct) led to a significant increase of the reporter activity, which could no longer be suppressed by the p53-expressing plasmid. Further deletion of the OCT4-SOX2 binding site (i.e., luc5 and luc6 reporters) led to the significant reduction of the luciferase activity, confirming the previous report that this site was important for OCT4-SOX2 to activate the NANOG promoter. The ability of p53 to bind to motifs 1 and 2 was further confirmed by the electrophoretic mobility-shift assay (EMSA). As shown in Figure 4C, nuclear extracts of HepG2 cells could cause the bandshift of the probe containing the sequence of motifs 1 and 2 but not the probe containing mutations in these two motifs. This binding could be abolished by the specific competitor but not by the competitor containing mutations in the two putative p53 binding motifs. The bandshift could be supershifted by three different anti-p53 antibodies that recognized the N-terminus, the C-terminus and phosphoserine-392, confirming the binding of p53. The binding of p53 to the DNA probe apparently required the phosphorylation at S392, as the anti-p53(pS392) antibody could supershift the p53-probe complex in a dose-dependent manner to completion (Figure S4B). These results confirmed that p53 could indeed bind to motifs 1 and 2, which together constituted the p53 response element.
Figure 4. Analysis of the effect of p53 on the NANOG promoter.
(A) HepG2 cells with various treatments for 24 hours or stably expressing a control shRNA or Atg5 shRNA were lysed for quantification of the NANOG mRNA. The NANOG mRNA level in non-treated cells was arbitrarily defined as 1. (B) HepG2 cells were transfected with the Nanog-luc reporter constructs illustrated together with the p53-expressing plasmid or the control vector. The plasmid pSV-RL, which expressed the renila luciferase under the control of the SV40 promoter was used an as internal control to monitor the transfection efficiency. Cells were lysed two days later for the analysis of the luciferase activities using the Promega dual luciferase assay kit. The firefly luciferase activity expressed by the pGL3-basic control vector was arbitrarily defined as 1. The Nanog-luc reporter constructs are illustrated to the right. The black boxes indicate motifs 1 and 2 of the putative p53 response element. The empty box denotes the OCT4-SOX2 binding site. (C) EMSA analysis. Details of the analysis are described in Experimental Procedures. The specific competitor used was the non-labeled probe. The mutated competitor contained nucleotide mutations in motifs 1 and 2. This mutated probe was also used as the control for EMSA. Three different anti-p53 antibodies that recognize the N-terminus, the C-terminus and the phosphoserine-392 were used for the supershift assay. (D) Relative luciferase activities of various Nanog-luc constructs in Hep3B cells. The experiments were conducted the same way as in (B) with the exceptions that Hep3B cells were used and that the expression plasmids for p53(S392A) and p53(S392D) were also included in the studies. (E) ChIP analysis of the endogenous Nanog promoter of Hep3B cells. Hep3B cells transfected with various p53-expressing plasmids or the control vector were subjected to ChIP analysis for the binding of p53, OCT4 and SOX2 to the Nanog promoter. Top panel, the anti-p53 antibody was used; middle panel, the anti-OCT4 antibody was used; and the bottom panel, the anti-SOX2 antibody was used. See also Figure S4.
The same reporter studies were also repeated using Hep3B cells. As our EMSA results indicated that p53(pS392) could bind to motifs 1 and 2, we also included the expression plasmid for p53(S392A) and p53(S392D) in the studies. As shown in Figure 4D, both p53(WT) and p53(S392D) could suppress the expression of the luciferase reporter from luc1 and luc2, which contained the putative p53 response element, but not from luc3–6 constructs, which did not contain the putative p53 response element. p53(S392A) had no effect on the expression of any of the reporter constructs. These results were consistent with the results shown in Figure 4B and further supported the finding of Figure S4B that the suppressive effect of p53 on the NANOG promoter was mediated by p53(pS392). The repeat of the studies using Huh7 cells generated similar results (Figure S4C). To further understand how p53 suppressed the NANOG promoter activity, we conducted a ChIP assay to determine the effect of p53(WT), p53(S392A) and p53(S392D) on the binding of OCT4-SOX2 to the NANOG promoter in Hep3B cells. As shown in Figure 4E, top panel, in agreement with the EMSA result, both p53(WT) and p53(S392D) could bind to the NANOG promoter, but p53(S392A) could not. Interestingly, the expression of p53(WT) and p53(S392D), but not p53(S392A), inhibited the binding of OCT4 and SOX2 to the NANOG promoter. These results indicated that the binding of p53 to the NANOG promoter could prevent the binding of OCT4-SOX2 to this promoter and was likely the reason why p53 suppressed the expression of NANOG.
Mitophagy suppresses p53 and regulates its subcellular localization
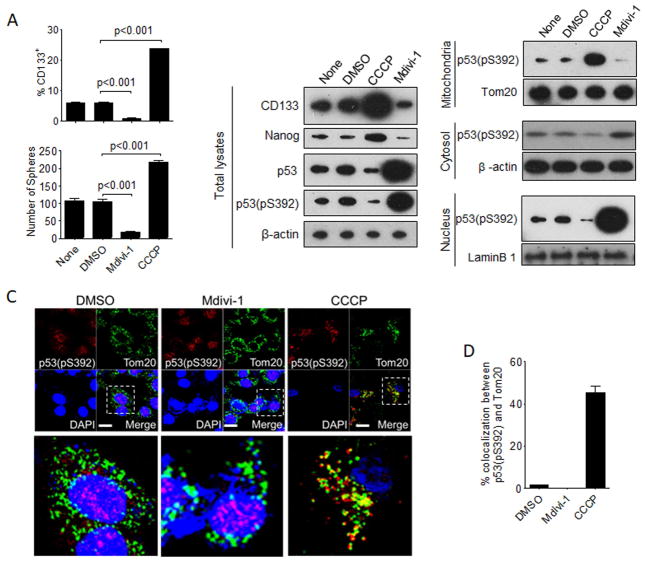
Mitophagy is the selective removal of mitochondria by autophagy (Youle and Narendra, 2011). To determine whether, similar to autophagy, mitophagy also affects CSCs, we treated HepG2 cells with Mdivi-1, a mitophagy inhibitor (Cui et al., 2010), and carbonyl cyanide m-chlorophenylhydrazone (CCCP), a mitophagy inducer (Ding et al., 2012). As shown in Figure S5A, Mdivi-1 increased TOM20 and TIM23 protein levels, which are translocases associated with mitochondrial outer and inner membranes, respectively (Aguileta et al., 2015). It also decreased the levels of mitochondrion-associated LC3-II and PINK1, a protein kinase important for the initiation of mitophagy (Youle and Narendra, 2011), confirming its ability to suppress mitophagy. In contrast, CCCP reduced the levels of TOM20 and TIM23 and increased the levels of mitochondrion-associated LC3-II and PINK1, also confirming the role of CCCP in the induction of mitophagy. The effects of CCCP1 and Mdivi on mitophagy were also confirmed by the analysis of mitochondrial DNA levels (Figure S5B) and the mito-Keima reporter assay (Yamashita and Kanki, 2017) (Figure S5C). We then tested the possible effect of Mdivi-1 and CCCP on CSCs of HepG2 cells. As shown in Figure 5A, Mdivi-1 reduced the CD133+ cell population of HepG2 cells and their self-renewal ability whereas CCCP had the opposite effects. The immunoblot analysis also confirmed that Mdivi-1 could reduce the expression levels of CD133 and NANOG, and CCCP could increase them (Figure 5B). These results indicated that, the same as autophagy, mitophagy could also positively regulate the CSC population of HepG2 cells, raising the possibility that the effect of autophagy on CSCs was mediated by mitophagy. Interestingly, while Mdivi-1 significantly increased the levels of p53 and its activated form p53(pS392), CCCP reduced them, indicating that mitophagy could reduce the levels of p53 and p53(pS392). The treatment of HepG2 cells with deferiperone (DFP), which induces mitophagy (Figure S5B and S5C) (also see (Allen et al., 2013)), also increased the levels of CD133 and NANOG, slightly reduced the level of p53 and significantly reduced the level of p53(pS392) (Figure S5D). The subcellular fractionation analysis indicated that p53(S392) was present in mitochondrial, cytosolic and nuclear fractions of control cells and cells treated with dimethyl sulfoxide (DMSO) and associated primarily with the mitochondrial fraction in cells treated with CCCP. In contrast, p53(pS392) was associated primarily with the nuclear fraction in cells treated with Mdivi-1. The subcellular localization of p53(pS392) was also confirmed by confocal microscopy. As shown in Figure 5C, in HepG2 cells treated with DMSO, p53(pS392) was detected in both the cytoplasm and the nucleus, and in HepG2 cells treated with Mdivi-1, p53(pS392) was localized primarily to the nucleus. In contrast, in CCCP-treated cells, p53(pS392) was localized almost exclusively to the cytoplasm, with a significant fraction of it colocalized with TOM20 (Figure 5C and 5D). These results indicated that the induction of mitophagy could lead to the association of p53(pS392) with mitochondria and its loss from the nucleus, and the inhibition of mitophagy could increase the level of p53(pS392) and its nuclear localization. The need of mitophagy to suppress p53 and subsequently induce NANOG explained how mitophagy positively regulated CSCs.
Figure 5. Effects of mitophagy on hepatic CSCs and p53.
(A) Top panel, HepG2 cells were treated with DMSO, Mdivi-1 or CCCP for one day and then subjected to flow cytometry analysis for CD133+ cells; bottom panel, CD133+ HepG2 cells were isolated and treated with Mdivi-1 or CCCP for two days and then analyzed for their sphere-forming ability. (B) HepG2 cells without treatment or with the treatment of DMSO, CCCP or Mdivi-1 for one day were lysed for immunoblot analysis. Cells were also subjected to subcellular fractionation for the isolation of mitochondria, cytosol, and nuclei for immunoblot analysis. Tom20, β-actin and lamin B1 were used as the loading controls for mitochondria, cytosol and nucleus, respectively, to ensure equal amount of proteins were loaded on the gel. (C) Confocal microscopy for the analysis of the subcellular localization of p53(pS392) in HepG2 cells treated with DMSO, Mdivi-1 or CCCP. TOM20 was used as the marker for mitochondria. The areas boxed are enlarged at the bottom. Scale bar, 10 μm. (D) The results shown in (C) were quantified with a Leica TCS SP8 fluorescent confocal microscope. The results indicated the percentages of p53(pS392) that colocalized with TOM20. The results represent the mean ± SEM of at least 30 cells that were analyzed. See also Figure S5.
PINK1 suppressed hepatic CSCs by phosphorylating serine-392 of p53
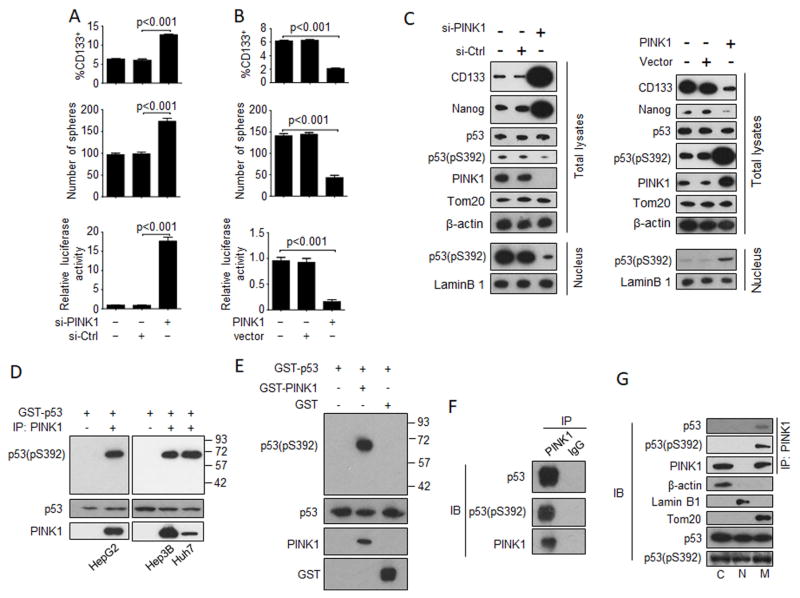
To further investigate the relationship between mitophagy and CSCs, we conducted the siRNA knockdown experiment to suppress the expression of PINK1, a serine/threonine kinase that plays an important role in the initiation of mitophagy (Youle and Narendra, 2011). Surprisingly, as shown in Figure 6A, the suppression of PINK1 expression increased the CD133+ HepG2 cell population, their sphere-forming ability, and the NANOG promoter activity, suggesting that PINK1 negatively regulated hepatic CSCs. To confirm the role of PINK1 in the suppression of hepatic CSCs, we transfected HepG2 cells with the PINK1-expressing plasmid. As shown in Figure 6B, the over-expression of PINK1 reduced the CD133+ cell population, their sphere-forming ability, and the NANOG promoter activity. The immunoblot analysis also confirmed that the knockdown of PINK1 could increase the expression levels of CD133 and NANOG and the over-expression of PINK1 had the opposite effects (Figure 6C). Interestingly, the knockdown and the over-expression of PINK1 also reduced and increased, respectively, the p53(pS392) level in cells and in the nuclei (Figure 6C). Neither the knockdown nor the over-expression of PINK1 had any effect on total p53 and TOM20, indicating a specific effect of PINK1 on the S392 phosphorylation of p53. The lack of effect of PINK1 knockdown or over-expression on TOM20 indicated that PINK1 did not affect the basal mitophagy in cells, which was likely mediated by a PINK1-independent mechanism (Youle and Narendra, 2011). The over-expressed PINK1 was apparently biologically active, as its over-expression increased the phosphorylation of ubiquitin and PARKIN, two of its substrates, in HepG2 cells (Figure S6A).
Figure 6. Phosphorylation of S392 of p53 by PINK1.
(A) Effects of PINK1 knockdown on CD133+ HepG2 cells (top panel), their sphere-forming ability (middle panel) and their effects on the Nanog promoter using the Nanog-luc1 reporter (bottom panel). HepG2 cells transfected with either the control siRNA or the PINK1 siRNA for two days were analyzed. In the bottom panel, HepG2 cells were also transfected with the Nanog-luc1 reporter (see Figure 3B) for the analysis of luciferase activity. The luciferase activity of cells without the transfection of siRNA was arbitrarily defined as 1. The results represented the mean ± SEM of three independent experiments. (B) Effects of PINK1 over-expression on CD133+ HepG2 cells (top panel), their sphere-forming ability (middle panel) and their effects on the Nanog promoter (bottom panel). The experiments were conducted the same way as in (A), except that instead of using siRNA, cells were transfected with either the control vector or the PINK1-expressing plasmid. (C) Immunoblot analysis of HepG2 cells with PINK1 knockdown (left panels) or PINK1 over-expression (right panels) were lysed for immunoblot analysis. Total cell lysates as well as the nuclear lysates (bottom two panels) were analyzed. (D) PINK1 in HepG2, Hep3B or Huh7 cells was immunoprecipitated with a control antibody (−) or the anti-PINK1 antibody (+) and then incubated with GST-p53 in the presence of ATP. The GST-p53 phosphorylated at S392 was analyzed using the anti-p53 antibody that recognized phosphoserine-392. GST-p53 added in the reaction and PINK1 immunoprecipitated were also analyzed by immunoblot (bottom two panels). Numbers to the left of the top panel indicate protein molecular weight markers. (E) GST-p53 was mixed with GST-PINK1 or GST and incubated in the presence of ATP. The phosphorylation of p53 at S392 was then analyzed with the antibody that recognized phosphoserine-392. GST-p53, GST-PINK1 and GST used for the reaction was also analyzed by anti-p53, anti-PINK1 and anti-GST antibodies, respectively (bottom three panels). Numbers to the left indicate protein molecular weight markers. (F) Co-immunoprecipitation of p53 and p53(pS392) with PINK1. HepG2 cells were lysed and immunoprecipitated using the anti-PINK1 antibody or the control antibody followed by immunoblot analysis for p53, p53(pS392) and PINK1. (G) Co-immunoprecipitation of p53 and p53(pS392) with PINK1 using the anti-PINK1 antibody in different subcellular fractions (top 3 panels). β-actin, lamin B1 and Tom20 were used as the markers for cytosolic (C), nuclear (N) and mitochondrial (M) fractions. Equal amounts of p53 were used for the co-immunoprecipitation experiment (bottom 2 panels). See also Figure S6.
The correlation between the expression level of PINK1 and the level of p53(pS392) prompted us to examine the relationship between PINK1 and the S392 phosphorylation of p53. Several kinases including CK2α, p38MAPK, PKR and CDK9 had been proposed to phosphorylate p53 at S392 (Cox and Meek, 2010). As shown in Figure S6B, the suppression of expression of any of these kinases in HepG2 cells had no effect on the S392 phosphorylation of p53. However, in agreement with the results shown in Figure 6C, the suppression of expression of PINK1 reduced the 53(pS392) signal to an almost undetectable level. The suppression of expression of PINK1 in Huh7 cells also prevented the S392 phosphorylation of the p53(Y220C) mutant (Figure S6C), indicating that PINK1 was also required for the phosphorylation of this defective p53 mutant at S392 in Huh7 cells. PINK1 is a serine/threonine kinase. To determine whether this kinase activity is important for the phosphorylation of p53, we produced a stable HepG2 cell line with PINK1 knockdown using shRNA (sh-PINK1) (Figure S6D). As shown in Figure S6E, the expression of sh-PINK1 resistant PINK1 increased the p53(pS392) level, but the expression of the kinase-dead PINK1 mutant did not, indicating an important role of the kinase activity of PINK1 in the phosphorylation of S392 of p53. To determine whether PINK1 could directly phosphorylate p53, we immunoprecipitated PINK1 from HepG2, Hep3B and Huh7 cells and incubated the immunoprecipitated PINK1 with the recombinant GST-p53 fusion protein. As shown in Figure 6D, this incubation led to the phosphorylation of p53 at S392, which could be detected by the anti-p53(S392) antibody. The ability of PINK1 to phosphorylate p53 was further confirmed in vitro using the GST-PINK1 fusion protein expressed in E. coli. This recombinant protein could phosphorylate GST-PARKIN and tetraubiquitin in vitro (Figure S7A–C). As shown in Figure 6E, GST-p53 could also be phosphorylated at S392 by GST-PINK1, but not by the control GST, in the presence of ATP, confirming that PINK1 could indeed directly phosphorylate p53 at S392.
To further determine the relationship between PINK1 and p53, we conducted the co-immunoprecipitation experiment. As shown in Figure 6F, both p53 and p53(pS392) could be co-immunoprecipitated with PINK1 by the anti-PINK1 antibody but not by the control antibody. The ability of PINK1 to bind to p53 was also confirmed by the single-molecule pull-down (SiMPull) assay (Jain et al., 2012), which also determined that the dissociation constant (Kd) between PINK1 and p53 was approximately 5 nM (Figure S7D). Interestingly, the subcellular fractionation experiment indicated that p53 and p53(pS392) that were associated with mitochondria could be co-immunoprecipitated with PINK1, but not those in the cytosolic fraction. PINK1 was not detected in the nuclear fraction (Figure 6G). These results indicated that the phosphorylation of p53 at S392 by PINK1 likely took place on mitochondria.
As the inhibition of autophagy enhanced the S392 phosphorylation of p53, we analyzed the effect of autophagy on PINK1. As shown in Figure S7E, the suppression of autophagy with 3-MA, shATG5 or shATG7 led to an increase of PINK1. We had also investigated the relationship between autophagy, PINK1 and p53 on CSCs by producing HepG2 cells with ATG5 knockdown, PINK1 knockdown, and ATG5 and PINK1 double knockdown (Figure S7F). As shown in Figure S7G, although ATG5 knockdown increased p53(pS392) and decreased NANOG, these increase and decrease were abolished when PINK1 was also knocked down. CD133+ cells were then isolated from these stable cells and grafted into nude mice for tumorigenesis analysis. As shown in Figure S6G, the knockdown of ATG5 suppressed the tumorigenesis of HepG2 cells, which was reversed if the expression of PINK1 was also suppressed. Interestingly, cells with PINK1 single knockdown were the most tumorigenic, suggesting a tumor suppressor role of PINK1 in hepatocarcinogenesis.
Discussion
Previous studies indicated that autophagy was essential for benign hepatic tumors to progress into malignant HCC (Takamura et al., 2011; Tian et al., 2015). However, the mechanism for this requirement was unclear. In this report, we demonstrated that autophagy was required to maintain the hepatic CSC population and their self-renewal ability (Figure 1). This effect of autophagy on hepatic CSCs was due to its ability to suppress the activity of p53 and, as such, Hep3B and Huh7 CSCs, which lacked functional p53, were not affected by autophagy (Figure 2). These results indicated that the use of autophagy inhibitors to treat HCC patients will need to take into consideration the p53 status in cancer cells, as autophagy had no effect on hepatic CSCs that express defective or no p53.
Our further studies indicated that p53 suppressed CSCs by binding to the promoter of the NANOG gene, which plays a critical role in maintaining the stemness and the self-renewal of CSCs (Lin et al., 2005). This binding prevented the binding of OCT4-SOX2 to the NANOG promoter and resulted in the suppression of NANOG expression (Figure 3). How the binding of p53 to the NANOG promoter inhibited the binding of OCT4-SOX2 is unclear. Due to the proximity of the p53 binding site to the OCT4-SOX2 biding site (Figure S4), it is conceivable that the binding of the tetrameric p53 and the subsequent recruitment of its coactivators/repressors might create a steric hindrance to occlude the binding of OC4-SOX2. An interesting finding of ours is that autophagy, and more specifically mitophagy, could reduce the levels of p53 and its activated form p53(pS392) in cells. As p53(pS392) was associated primarily with mitochondria when mitophagy was stimulated (Figure 4), it is likely that p53(pS392) was removed together with mitochondria by mitophagy. This could explain why the levels of p53 and p53(pS392) significantly increased when mitophagy was inhibited (Figure 4).
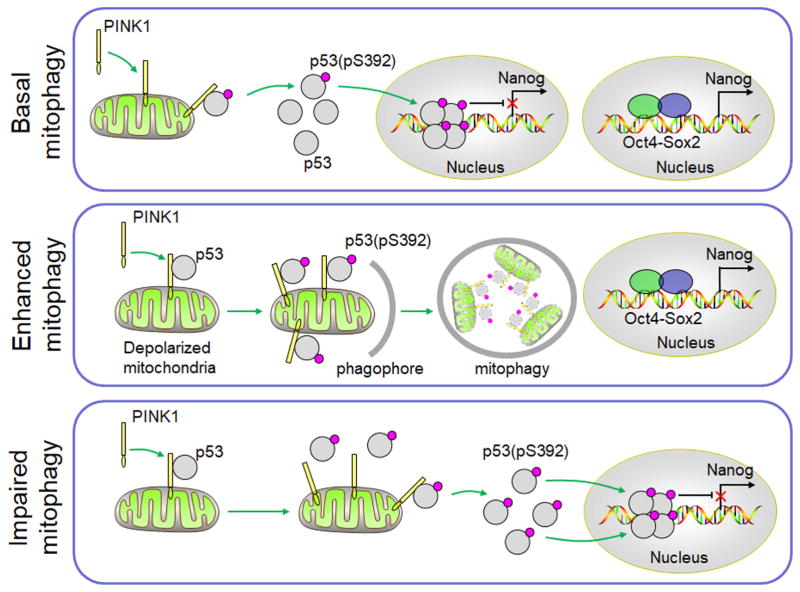
The phosphorylation of p53 at S392 is important for the activation of the DNA binding activity of p53. Several kinases including CK2α, p38MAPK, PKR and CDK9 had been proposed to phosphorylate p53 at S392 (Cox and Meek, 2010). Curiously, the suppression of none of these kinases was able to suppress the phosphorylation of S392 of p53 in HepG2 cells. Instead, our results shown in Figure 5 provided a strong argument that PINK1 could phosphorylate p53 at this serine residue. This argument was supported by the observations that first, the knockdown of PINK1 expression could significantly reduce the level of p53(pS392); second, PINK1 isolated from HepG2, Hep3B and Huh7 cells could phosphorylate p53 at S392; and third, recombinant PINK1 could phosphorylate p53 at S392 in vitro. An interesting question is how the activity of PINK1 on p53 is regulated. As illustrated in Figure 7, it is conceivable that when the basal autophagy is not perturbed, a low level of PINK1 that is associated with mitochondria recruits and phosphorylates a low level of p53 at S392, which may dislodge from mitochondria and localize to the nucleus to partially control the expression of NANOG and the population of CSCs. When autophagy/mitophagy is enhanced such as that induced by CCCP, p53 recruited by PINK1 to mitochondria is entrapped by the elongating membranes of phagophores and subsequently removed by mitophagy or by a protein degradation mechanism that is dependent on mitophagy, resulting in the increase of the NANOG expression and the CSC population. However, when mitophagy is impaired, p53 phosphorylated by PINK1 can no longer be removed by mitophagy and is rapidly translocated into the nucleus where it suppresses the expression of NANOG to reduce the stemness and the self-renewal ability of hepatic CSCs, resulting in the suppression of hepatocarcinogenesis. Note that, although our results indicated that p53 could only interact with PINK1 in association with mitochondria (Figure 6G), we cannot rule out the possibility that when PINK1 is over-expressed, it may also directly phosphorylate p53 without the association with mitochondria due to its high concentration in the cytosol. Our studies thus demonstrated a critical role of mitophagy in the maintenance of the hepatic CSC population and the control of p53 activities, which otherwise would be activated by PINK1. Although our studies were focused on hepatic tumor cells, it is likely that mitophagy and PINK1 may also play similar roles in the control of CSCs of other tumor types.
Figure 7. A model for the role of mitophagy and PINK1 in the regulation of p53 and the expression of NANOG.
See the Discussion section for details.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, J.-H. James Ou (jamesou@hsc.usc.edu).
Experimental Model and Subject Details
Cell cultures
HepG2, Hep3B and Huh7 cells were human liver tumor cell lines. They were maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) in a humidified incubator at 37°C with 5% CO2. Stable HepG2 cells with ATG5, ATG7, PINK1 or p53 knockdown were produced by the transfection of an expression plasmid that expressed their respective shRNAs.
Mouse models
The production of ATG5-KO mice and the induction of liver tumors using diethylnitrosamine (DEN) had been described before (Tian et al., 2015). Briefly, male C57BL/6 mice with or without hepatocyte-specific knockout of ATG5 were intraperitoneally injected with DEN on day 16 of age and once a week for 4 weeks thereafter. The liver tumor tissues were homogenized in the RIPA buffer and, after a brief centrifugation to remove cell debris, the protein samples were stored at −80°C for immunoblot analysis. For tumorigenesis analysis, CD133+ HepG2 cells were isolated from stable cell lines that expressed control shRNA, ATG5 shRNA, PINK1 shRNA, or both ATG5 and PINK1 shRNAs. 8000 CD133+ cells were subcutaneously injected into 8-week old athymic nude mice (both male and female). Four mice were used for each cell line. After 3 months, bioluminescence was used to analyze the tumor volumes. Prior to imaging, mice were anesthetized with isoflurane. Approximately 10 minutes prior to imaging using Kodak In-vivo Imaging System FX Pro, animals were injected with 100 mg/kg D-Luciferin sodium salt in 0.1 ml phosphate-buffered saline (PBS). The pseudoimages were obtained by superimposing the emitted light over the gray-scale photographs of the animal. Our mouse studies were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and approved by the Institutional Animal Care and Use Committee of the University of Southern California.
Methods Details
DNA Plasmids
The kinase-dead PINK1 mutant, which contained the K219A/D362A/D384A mutations (Beilina et al., 2005), were generated by site-directed mutagenesis using the QuikChange kit. Plasmids encoding p53 mutant S392A and S392D were generated using the QuikChange® II XL Site-Directed Mutagenesis kit. The construction of Nanog-Luc reporter plasmids was conducted using PCR to isolate NANOG promoter DNA fragments, which were then digested with KpnI and XhoI and cloned into the pGL3 luciferase vector. The PCR primers for the construction of p53 mutants and Nanog-Luc reporter DNA plasmids are shown in Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD133 | Cell Signaling | 3663 |
| NANOG | Santa Cruz | sc-374001 |
| OCT4 | Cell Signaling | 2840 |
| SOX2 | Cell Signaling | 3579 |
| LC3 | Sigma-Aldrich | L7543 |
| p62 | Cell Signaling | 5114 |
| β-ACTIN | Cell Signaling | 3700 |
| ATG5 | Sigma-Aldrich | 0731 |
| p53(pS392) | Santa Cruz | sc-7997 |
| p53 | Calbiochem | OP09, OP03 |
| LAMIN-B1 | Abcam | ab16048 |
| TOM20 | Abcam | ab56783 |
| TIM23 | Santa Cruz | sc-13298 |
| PINK1 | Cell Signaling | 6946 |
| CK2α | Cell Signaling | 2656 |
| p38(MAPK) | Cell Signaling | 9212 |
| PKR | Abcam | ab28943 |
| CDK9 | Cell Signaling | 2316 |
| Bacterial and Virus Strains | ||
| E. coli BL21 | ThermoFisher | C601003 |
| E. coli BL21, BirA-transformed | BPS Bioscience | 27462 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-Methyladenine | Sigma-Aldrich | M9281 |
| Pifithrin-α | Sigma-Aldrich | P4359 |
| Bafilomycin A1 | Sigma-Aldrich | B1793 |
| Rapamycin | Sigma-Aldrich | R0395 |
| Mdivi-1 | Sigma-Aldrich | M0199 |
| Carbonyl cyanide m-chlorophenylhydrazone | Sigma-Aldrich | 215911 |
| Deferiprone | Sigma-Aldrich | 379409 |
| Lipofectamine 2000 | Invitrogen | 11668019 |
| D-Luciferin sodium salt | Sigma-Aldrich | L6882 |
| Human EFG | Life Technologies | PHG0311 |
| Human FGF | Life Technologies | PHG0266 |
| B27 supplement without vitamin A | Life Technologies | 12587-010 |
| N2 supplement | Life Technologies | 17502-048 |
| Trizol | ThermoFisher | 15596026 |
| VectaShield with DAPI | Vector Lab | H-1200 |
| 1X Kinase buffer | Cell Signaling | 9802 |
| Critical Commercial Assays | ||
| Pierce BCA protein assay | ThermoFisher | 23225 |
| LightShift Chemiluminescent EMSA Kit | ThermoFisher | 20148 |
| MTT Colorimetric Kit | Promega | G4000 |
| Experimental Models: Cell Lines | ||
| HepG2 | ATCC | HB-8065 |
| Hep3B | ATCC | HB-8064 |
| Huh7 | This paper | N/A |
| Experimental Models: Organisms/Strains | ||
| ATG5f/f mice | (Tian et al., 2015) | N/A |
| Oligonucleotides | ||
| PCR reverse primer for Nanog-Luc reporters: CCGCTCGAGTTAAAATCCTGGAGTCTCTAGATTT | This paper | N/A |
| PCR forward primer for Nanog-Luc1 reporter: CGGGGTACCATTCCTGATTTAAAAGTTGGAAACG | This paper | N/A |
| PCR forward primer for Nanog-Luc2 reporter: CGGGGTACCGCTCGGTTTTCTAGTTCCCCACCTA | This paper | N/A |
| PCR forward primer for Nanog-Luc3 reporter: CGGGGTACCCCCCACCTAGTCTGGGTTACTCTGC | This paper | N/A |
| PCR forward primer for Nanog-Luc4 reporter: CGGGGTACCGGGTTACTCTGCAGCTACTTTTGCA | This paper | N/A |
| PCR forward primer for Nanog-Luc5 reporter: CGGGGTACCGGCCTTGGTGAGACTGGTAGACGGG | This paper | N/A |
| PCR forward primer for Nanog-Luc6 reporter: CGGGGTACCAGACTGGTAGACGGGATTAACTGAG | This paper | N/A |
| PCR forward primer for constructing p53 mutants: CATGTTCAAGACAGAAGGGCCTGACGCAAGTCTAGAGGGCCCGTT | This paper | N/A |
| PCR reverse primer for p53(S392A) mutant: AACGGGCCCTCTAGACTTGCGTCAGGCCCTTCTGTCTTGAACATG | This paper | N/A |
| PCR reverse primer for p53(S392D) mutant: AACGGGCCCTCTAGACTGTCGTCAGGCCCTTCTGTCTTGAACATG | This paper | N/A |
| Recombinant DNA | ||
| pcDNA-DEST53 PINK1-N-GFP | (Beilina et al., 2005) | Addgene |
| pGEX5X.1-PINK1 WT | (Beilina et al., 2005) | Addgene |
| pGEX-human p53-(1-393) | (Ayed et al., 2001) | Addgene |
| pGEX5X.1-PINK1 (K219A/D362A/D384A) | This paper | N/A |
| pGL3-basic | Promega | E1751 |
| pNanog-Luc1 | This paper | N/A |
| pNanog-Luc2 | This paper | N/A |
| pNanog-Luc3 | This paper | N/A |
| pNanog-Luc4 | This paper | N/A |
| pNanog-Luc5 | This paper | N/A |
| pNanog-Luc6 | This papaer | N/A |
| pMT-mKeima-Red | MBL International | AM-V0251M |
| Software and Algorithms | ||
| Carestream Molecular Imaging 5.0 | Bruker Corp. | N/A |
| FlowJo Software | FLOWJO | N/A |
| Other | ||
| QuikChange Kit | Agilent Technologies | 200515 |
| QuikChange® II XL Site-Directed Mutagenesis Kit | Agilent Technologies | 200521 |
| Nuclear Extraction Kit | Abcam | ab113474 |
| Mitochondria Isolation Kit | ThermoFisher | 89874 |
| Human CD133 MicroBead Kit | Miltenyi Biotech | 130-050-801 |
| Ultra-low attachment plate | Corning Inc. | 3261 |
| SuperScript II First-strand Synthesis System | ThermoFisher | N/A |
| Biotin 3′-end DNA Labeling Kit | Pierce | 89818 |
| PureLink DNA Mini Kit | ThermoFisher | K182001 |
Subcellular fractionation and immunoblot analysis
For subcellular fractionation, the Nuclear Extraction Kit (Abcam) and the Mitochondria Isolation Kit (ThermoFisher) were used to isolate nuclear extracts and mitochondria, respectively. For the preparation of whole cell lysates, cells were homogenized in the RIPA buffer (10 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate and 0.1% sodium dodecyl sulfate (SDS)). Protein concentrations were determined using the Pierce BCA protein assay, and an equal amount of proteins (15–100 μg/sample, depending on the experiments) were electrophoresed on 10% or 15% polyacrylamide gels for immunoblot analysis.
Flow cytometry
Cells were fixed with 1% paraformaldehyde, washed with PBS three times and then incubated with the anti-CD133-PE antibody at room temperature for 30 minutes. Cells were then washed with PBS three more times for flow cytometry. The FlowJo software was used to analyze the data.
Sphere-formation assay
The Human CD133 MicroBead Kit (Miltenyi Biotec) was used to isolate CD133+ cells from HepG2, Hep3B and Huh7 cells. CD133+ cells were resuspended in serum-free DMEM/F12 (1:1 ratio) supplemented with 100 IU/ml penicillin, 100 μg/ml streptomycin, 20 ng/ml human EGF, 10 ng/ml human FGF, 2% B27 supplement without vitamin A, and 1% N2 supplement as previously described (Cao et al., 2011). Cells (500 cells/well) were subsequently cultured in the ultra-low attachment plate for one week for the analysis of their sphere-forming ability.
Real-time PCR for RNA quantification
Cells were homogenized in Trizol (Invitrogen) and total RNA was isolated following the manufacturer’s protocol. The reverse transcription for the synthesis of cDNA was performed using the SuperScript II First-Strand Synthesis System. The PCR primers for β-actin, Nanog and p53 mRNAs for the Taqman assay were purchased from ThermoFisher. The real-time PCR was conducted using the Applied Biosystem 7500 Fast PCR System. The mRNA levels determined were normalized against the housekeeping gene β-actin mRNA.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from HepG2 or Huh7 cells were prepared using the Nuclear Extraction Kit (Abcam) according to the manufacturer’s instructions. Oligonucleotides for the EMSA were labeled with the Biotin 3′-End DNA Labeling Kit (Pierce). The nucleotide sequence of the probe was 5′-GTTTTCTAGTTCCCCACCTAGTCTGG-3′ and the sequence of the oligonucleotide mutant was 5′-GTTTTATCGCTCCCCACATCGCCTGG-3′, which contained six nucleotide mutations (underlined) in putative p53-binding motifs 1 and 2. For the supershift assay, the antibody was added into the nuclear extracts and incubated on ice for 10 minutes prior to the addition of the probe. The LightShift™ Chemiluminescent EMSA Kit (ThermoFisher) was used to detect the signals of the probe and the bandshifts.
Chromatin Immunoprecipitation (ChIP) assay
The ChIP assay was performed as described (Li et al., 2012). The resulting PCR products (20 μl) were analyzed on a 1.5% agarose gel. 1% of starting chromatin DNA was used for PCR and served as the input control. The primers used for amplifying the Nanog promoter (−147~−60) were: forward primer, 5′-GCTCGGTTTTCTAGTTCCCCACCTA-3′; and reverse primer, 5′-CTTGTGAATTCTCAGTTAATCCCGT-3′.
Confocal microscopy
Cells were fixed with 4% paraformaldehyde in PBS for 5 minutes at room temperature and then permeabilized with the immunofluorescence buffer (1X PBS containing 0.1% saponin, 1% bovine serum albumin (BSA) and 0.05% sodium azide) for another 5 minutes. Cells were then incubated with the primary antibody overnight at 4°C and the fluorescein/rhodamine-conjugated secondary antibody for one hour at room temperature. Cells were then washed three times with the immunofluorescence buffer, mounted in VectaShield (Vector) containing DAPI, and imaged with a Leica TCS SP8 fluorescent confocal microscope.
Co-immunoprecipitation assay (CO-IP)
CO-IP was performed as previously described (Liu et al., 2015). Cell lysates (1 mg of protein in 500 μl of SDS-free RIPA buffer) were pre-cleared using protein A/G PLUS-agarose beads (Santa Cruz Biotechnology) and then incubated at 4°C overnight with the anti-PINK1 antibody. The immunocomplexes were precipitated with protein A/G agarose beads and then subjected to immunoblot analysis using the anti-p53 antibodies.
Protein expression in Escherichia coli
Detailed procedures for the expression of proteins in E. coli and their purification had been described before (Beilina et al., 2005). Briefly, the E. coli BL21 strain was transformed with pGEX5X.1-PINK1 for the expression of the GST-PINK1 fusion protein or pGEX-human p53 for the expression of GST-p53. For the expression of biotinylated and 6xHis-tagged PINK1 for the SiMPull analysis, E. coli BL21 strain containing an IPTG-inducible BirA biotin ligase plasmid was used. Cells were grown at 37°C until the OD600 reached 0.7–0.9 and then treated with isopropyl β-D-thiogalactopyranoside (0.2 mM) at 25°C for 4 hou rs for the induction of protein expression. Cells were harvested and lysed by sonication on ice in the lysis buffer (200 mM NaCl, 50 mM Tris-HCl, 1 mM EDTA, 1 mM DTT and 0.2% Triton X-100) containing protease inhibitors. The Pierce GST Spin Purification Kit (Thermo Scientific) was then used to purify the recombinant proteins. The 6xHis-tagged PINK1 was purified using a nickel column.
In vitro kinase assay
The nonradioactive kinase assay was used to analyze the role of PINK1 on the phosphorylation of S392 of p53. PINK1 immunoprecipitated from HepG2, Hep3B and Huh7 cells or recombinant GST-PINK1 (10 μg) was mixed with 50 μg recombinant GST-p53 in 10 μl 1X Kinase Buffer (Cell Signaling) containing 200 μM ATP. After the incubation at 30°C for 30 minutes, the reaction was stopped by the addition of 10 μl 3X SDS sample buffer. The protein samples were then subjected to immunoblot analysis using the anti-p53(pS392) antibody.
MTT assay
The cell viability was analyzed using the MTT colorimetric assay kit (Promega) as previously described (Liu et al., 2014). Briefly, cells were incubated in the incubation medium containing MTT (0.5 mg/ml) at 37°C for 4 hours. After the remo val of the culture medium, the blue precipitates were solubilized with DMSO and the absorbance at 570 nm was measured.
In vitro phosphorylation assay using γ-32P-ATP
Recombinant GST-PINK1 (10μg) was incubated with recombinant GST-p53 (50μg) in the kinase buffer (20 mM Tris-HCl pH7.4, 5 mM EGTA and 20 mM β-glycerol phosphate). The reaction was initiated by the addition to final concentrations of 0.1 mM [32P]-γ-ATP (0.2 μCi/reaction) and 20 mM MgCl2. The reaction was incubated at 30°C with shaking f or 30 minutes and terminated by placing the tube on ice. The protein was then analyzed on an SDS-PAGE gel and exposed to a phosphor-imager screen for the detection of 32P-labeled protein.
Detection of phosphorylation of ubiquitin and PARKIN by PINK1
HepG2 cells were transfected with a control vector or the plasmid encoding PINK1 for 48 hours, followed by isolation of total cell lysates for immunoblot analysis using anti-p-ubiquitin(S65) and anti-p-PARKIN(S65) antibodies.
Mito-Keima analysis of mitophagy
HepG2 cells were transfected with the plasmid that expressed the mitochondria-targeted Keima-Red (pMT-mKeima-Red, MBL International) for 24 hours and then treated with the drugs for another 24 hours. The fluorescence of Keima-Red, which displays the green color under normal conditions and red color when mitophagy is activated, was then analyzed using the Keyence All-in-One fluorescence microscope.
Mitochondria DNA assay
Mitochondrial DNA was quantified by real-time PCR as previously described (Okatsu et al., 2010). Briefly, total DNA containing genomic DNA and mitochondrial DNA was purified from cells using PureLink DNA Mini Kit (ThermoFisher). Mitochondrial DNA was then quantified by real-time PCR using primers that targets the mitochondrial 12S rRNA: 5″-AAC TCA AAG GAC TTG GCG GTA CTT TAT ATC-3′ and 5′-GAT GGC GGT ATA TAG GCT GAA TTA GCA AGA G-3′. The level of mitochondrial DNA was normalized against nuclear GAPDH DNA, which served as the internal control.
Single-molecule pull-down (SiMPull) assay
The SiMPull assay was conducted as described (Jain et al., 2012). Briefly, flow chambers were constructed by compartmentalizing and sealing the sandwich of methoxypolyethylene glycol (mPEG)-coated microscope slides and coverslips. The chambers were washed once with T50-BSA (10 mM Tris-HCl, pH8, 0.1 mg/ml bovine serum albumin) and then incubated with NeutrAvidin (0.2 mg/ml) at room temperature for 5 minutes. After the removal of unbound NeutrAvidin, the slides were blocked with FBS and, after which, biotinylated PINK1 protein bait (40 nM) was added. After further washing to remove unbound PINK1, GST or GST-p53 at concentrations of 20, 40 and 80 nM was added. The bait-prey complex was stained by sequential incubations with the anti-GST antibody and the FITC-conjugated secondary antibody. The fluorescent images were captured with a prism-type total internal reflection fluorescence (TIRF) microscope. The dissociation constant (Kd) was calculated based on 50% binding at equilibrium.
Quantification and Statistical Analysis
All of the data shown in the histograms were the results of at least three independent experiments and are presented as the mean ± SEM. The differences between groups were compared using Student’s t-test. Quantitative analysis of tumor sizes in mice was done with Carestream molecular imaging software, version 5.0 (Rastegar et al., 2010).
Supplementary Material
Highlights.
Autophagy positively regulates hepatic cancer stem cells via the suppression of p53.
p53 down-regulates NANOG and is removed with mitochondria by mitophagy.
PINK1 binds to p53 on mitochondria and phosphorylates p53 at serine-392.
PINK1-activated p53 is localized to the nucleus when mitophagy is impaired.
Acknowledgments
We would like to thank the Confocal Imaging Core of the USC Research Center for Liver Diseases for help with confocal microscopy and Dr. Jae Jung’s laboratory at USC for help with the SiMPull assay. This work was supported by NIH grants DK100257, DK094652, AI129540 and CA177337.
Footnotes
Author Contributions
Conceptualization, K.L., J.L., and J.H.J.O.; Methodology, K.L., J.L., and J.H.J.O.; Investigation, K.L., J.L., J.Y.K., L.W., S.T.C., C.C., and Y.T.; Writing – Original Draft, K.L.; Writing – Review and Editing, K.L., J.L., and J.H.J.O.; Visualization, K.L., J.L., J.Y.K., L.W., and Y.T.; Supervision, K.M., D.C., and J.H.J.O.; Project Administration, J.H.J.O.; Funding Acquisition, J.H.J.O.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguileta MA, Korac J, Durcan TM, Trempe JF, Haber M, Gehring K, Elsasser S, Waidmann O, Fon EA, Husnjak K. The E3 ubiquitin ligase parkin is recruited to the 26 S proteasome via the proteasomal ubiquitin receptor Rpn13. J Biol Chem. 2015;290:7492–7505. doi: 10.1074/jbc.M114.614925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ailles LE, Weissman IL. Cancer stem cells in solid tumors. Curr Opin Biotechnol. 2007;18:460–466. doi: 10.1016/j.copbio.2007.10.007. [DOI] [PubMed] [Google Scholar]
- Allen GF, Toth R, James J, Ganley IG. Loss of iron triggers PINK1/Parkin-independent mitophagy. EMBO Rep. 2013;14:1127–1135. doi: 10.1038/embor.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayed A, Mulder FA, Yi GS, Lu Y, Kay LE, Arrowsmith CH. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- Beckerman R, Prives C. Transcriptional regulation by p53. Cold Spring Harb Perspect Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, Cookson MR. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A. 2005;102:5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonizzi G, Cicalese A, Insinga A, Pelicci PG. The emerging role of p53 in stem cells. Trends Mol Med. 2012;18:6–12. doi: 10.1016/j.molmed.2011.08.002. [DOI] [PubMed] [Google Scholar]
- Cao L, Zhou Y, Zhai B, Liao J, Xu W, Zhang R, Li J, Zhang Y, Chen L, Qian H, et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011;11:71. doi: 10.1186/1471-230X-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox ML, Meek DW. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal. 2010;22:564–571. doi: 10.1016/j.cellsig.2009.11.014. [DOI] [PubMed] [Google Scholar]
- Cui M, Tang X, Christian WV, Yoon Y, Tieu K. Perturbations in mitochondrial dynamics induced by human mutant PINK1 can be rescued by the mitochondrial division inhibitor mdivi-1. J Biol Chem. 2010;285:11740–11752. doi: 10.1074/jbc.M109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–536. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding WX, Guo F, Ni HM, Bockus A, Manley S, Stolz DB, Eskelinen EL, Jaeschke H, Yin XM. Parkin and mitofusins reciprocally regulate mitophagy and mitochondrial spheroid formation. J Biol Chem. 2012;287:42379–42388. doi: 10.1074/jbc.M112.413682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, Chen G, Price S, Lu W, Teng X, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu IC, Tokiwa T, Bennett W, Metcalf RA, Welsh JA, Sun T, Harris CC. p53 gene mutation and integrated hepatitis B viral DNA sequences in human liver cancer cell lines. Carcinogenesis. 1993;14:987–992. doi: 10.1093/carcin/14.5.987. [DOI] [PubMed] [Google Scholar]
- Jain A, Liu R, Xiang YK, Ha T. Single-molecule pull-down for studying protein interactions. Nat Protoc. 2012;7:445–452. doi: 10.1038/nprot.2011.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubicka S, Trautwein C, Niehof M, Manns M. Target gene modulation in hepatocellular carcinomas by decreased DNA-binding of p53 mutations. Hepatology. 1997;25:867–873. doi: 10.1002/hep.510250414. [DOI] [PubMed] [Google Scholar]
- Lei XH, Zhao D, Li YL, Li XF, Sun X, Du WZ, Sun Y, Hao ZF, Xin SY, Liu C, et al. Pifithrin-alpha enhances the survival of transplanted neural stem cells in stroke rats by inhibiting p53 nuclear translocation. CNS Neurosci Ther. 2013;19:109–116. doi: 10.1111/cns.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Jin R, Zhang X, Lv F, Liu L, Liu D, Liu K, Li N, Chen D. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology. 2012;56:1380–1390. doi: 10.1002/hep.25891. [DOI] [PubMed] [Google Scholar]
- Lin T, Chao C, Saito S, Mazur SJ, Murphy ME, Appella E, Xu Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- Liu K, Jiang T, Ouyang Y, Shi Y, Zang Y, Li N, Lu S, Chen D. Nuclear EGFR impairs ASPP2-p53 complex-induced apoptosis by inducing SOS1 expression in hepatocellular carcinoma. Oncotarget. 2015;6:16507–16516. doi: 10.18632/oncotarget.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Shi Y, Guo X, Wang S, Ouyang Y, Hao M, Liu D, Qiao L, Li N, Zheng J, et al. CHOP mediates ASPP2-induced autophagic apoptosis in hepatoma cells by releasing Beclin-1 from Bcl-2 and inducing nuclear translocation of Bcl-2. Cell Death Dis. 2014;5:e1323. doi: 10.1038/cddis.2014.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO, Zheng BJ, Guan XY. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology. 2007;132:2542–2556. doi: 10.1053/j.gastro.2007.04.025. [DOI] [PubMed] [Google Scholar]
- Mathew R, Karp CM, Beaudoin B, Vuong N, Chen G, Chen HY, Bray K, Reddy A, Bhanot G, Gelinas C, et al. Autophagy suppresses tumorigenesis through elimination of p62. Cell. 2009;137:1062–1075. doi: 10.1016/j.cell.2009.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okatsu K, Saisho K, Shimanuki M, Nakada K, Shitara H, Sou YS, Kimura M, Sato S, Hattori N, Komatsu M, et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells. 2010;15:887–900. doi: 10.1111/j.1365-2443.2010.01426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, Doetsch F. Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell. 2011;8:486–498. doi: 10.1016/j.stem.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastegar F, Gao JL, Shenaq D, Luo Q, Shi Q, Kim SH, Jiang W, Wagner ER, Huang E, Gao Y, et al. Lysophosphatidic acid acyltransferase beta (LPAATbeta) promotes the tumor growth of human osteosarcoma. PLoS One. 2010;5:e14182. doi: 10.1371/journal.pone.0014182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, Eishi Y, Hino O, Tanaka K, Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y, Kuo CF, Sir D, Wang L, Govindarajan S, Petrovic LM, Ou JH. Autophagy inhibits oxidative stress and tumor suppressors to exert its dual effect on hepatocarcinogenesis. Cell Death Differ. 2015;22:1025–1034. doi: 10.1038/cdd.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto H, Mishra L, Machida K. Alcohol, TLR4-TGF-beta antagonism, and liver cancer. Hepatol Int. 2014;8(Suppl 2):408–412. doi: 10.1007/s12072-013-9489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. The role for autophagy in cancer. J Clin Invest. 2015;125:42–46. doi: 10.1172/JCI73941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita SI, Kanki T. Detection of Hypoxia-Induced and Iron Depletion-Induced Mitophagy in Mammalian Cells. Methods Mol Biol. 2017 doi: 10.1007/7651_2017_19. [DOI] [PubMed] [Google Scholar]
- Yamashita T, Ji J, Budhu A, Forgues M, Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136:1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Wang XW. Cancer stem cells in the development of liver cancer. J Clin Invest. 2013;123:1911–1918. doi: 10.1172/JCI66024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.