SUMMARY
The cytoplasmic tail of gp41 (gp41CT) remains the last HIV-1 domain with an unknown structure. It plays important roles in HIV-1 replication such as mediating envelope (Env) intracellular trafficking and incorporation into assembling virions, mechanisms of which are poorly understood. Herein, we present the solution structure of gp41CT in a micellar environment and characterize its interaction with the membrane. We show that the N-terminal 45 residues are unstructured and not associated with the membrane. However, the C-terminal 105 residues form three membrane-bound amphipathic a-helices with distinctive structural features such as variable degree of membrane penetration, hydrophobic and basic surfaces, clusters of aromatic residues, and a network of cation-p interactions. This work fills a major gap by providing the structure of the last segment of HIV-1 Env, which will provide insights into the mechanisms of Gag-mediated Env incorporation as well as the overall Env mobility and conformation on the virion surface.
eTOC blurb
Murphy et al. devised new approaches to prepare the HIV-1 gp41CT protein, solved the structure by NMR methods, characterized its interactions with the membrane, and provided a preferred topology of the protein when bound to the membrane.
INTRODUCTION
The envelope glycoprotein (Env) of human immunodeficiency virus type 1 (HIV-1) is synthesized as a 160-kDa precursor in the rough endoplasmic reticulum, where it is glycosylated and subsequently cleaved in the Golgi apparatus to the form of the surface (gp120) and transmembrane (gp41) subunits (reviewed in (Checkley et al., 2011)). The gp41 subunit comprises a fusogenic ectodomain, a transmembrane domain (TMD), and a C-terminal cytoplasmic tail (gp41CT). For the past two decades, research has mainly focused on elucidating the mechanisms of the gp120/gp41 trimer binding to host receptor, membrane fusion, and evasion of recognition by the immune system. It is established that binding of gp120 to the CD4 receptor and CCR5/CXCR4 co-receptors triggers large conformational changes in the gp41 ectodomain, allowing the viral membrane to fuse with the host membrane (reviewed in (Merk and Subramaniam, 2013)). Membrane-proximal external region (MPER) of the HIV-1 gp41 protein, which forms an α-helical trimer (Buzon et al., 2010), precedes the TMD and is a key target for broadly neutralizing monoclonal antibodies elicited during human infection (Alam et al., 2009). Current models suggest that receptor binding leads to the exposure of the gp41 fusion peptide, which anchors to the target cell membrane to produce an intermediate, pre-hairpin state bridging the two membranes.
One of the least understood phenomena in retroviral assembly is the mechanism by which the Env protein is recruited and incorporated into virus particles. There is mounting evidence that gp41CT plays a functional role in Env incorporation in physiologically relevant cell types (Akari et al., 2000; Murakami and Freed, 2000b). Deletion of gp41CT strongly reduces Env incorporation in permissive cell lines but has only minor effect in non-permissive cell lines (Freed and Martin, 1996; Freed and Martin, 1995; Murakami and Freed, 2000b). The CT domain is remarkably long (150 amino acids) for most lentiviruses but significantly shorter (~20–40 amino acids) for other retroviruses (Checkley et al., 2011). The biological implication of the variable length and its effect on Env incorporation are not well understood.
The assembly of HIV-1 particles is an orchestrated process that is driven by transport of Gag polyproteins to the plasma membrane (PM) (reviewed in (Freed, 2015)). Gag-membrane binding is mediated by the myristoylated N-terminal matrix (MA) domain. HIV-1 Gag targeting to the PM is dependent on phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2),(Chukkapalli and Ono, 2011; Ono et al., 2004) which binds directly to the MA domain (Anraku et al., 2010; Mercredi et al., 2016; Saad et al., 2006; Shkriabai et al., 2006; Vlach and Saad, 2013). Based on genetic, in vivo, and biochemical evidence, early studies suggested that Env incorporation is mediated by interactions between the MA domain of Gag and gp41CT (Cosson, 1996; Dorfman et al., 1994; Freed and Martin, 1996; Freed and Martin, 1995; Yu et al., 1992). Freed and coworkers have recently provided biochemical evidence that MA trimerization as an obligatory step in the assembly of infectious HIV-1 virions and demonstrated a correlation between loss of MA trimerization and loss of Env incorporation (Tedbury et al., 2016). It has also been shown that Gag assembly induced the aggregation of small Env clusters into larger domains that were completely immobile (Roy et al., 2013). Truncation of the CT domain abrogated Gag’s ability to induce Env clustering and restored Env mobility at assembly sites (Roy et al., 2013). Super-resolution microscopy data indicated that recruitment of HIV-1 Env to viral assembly sites is dependent on gp41CT (Muranyi et al., 2013). Spearman and co-workers also demonstrated that Rab11-family interacting protein 1C (FIP1C) is required for CT-dependent incorporation of Env into HIV-1 particles (Qi et al., 2015; Qi et al., 2013), and identified a tyrosine-based motif (Y795/W796) as critical in mediating cell-type–dependent Env incorporation (Qi et al., 2015). FIPs are effectors of Rab11 GTPases that mediate sorting of cargo from the endosomal recycling compartment to the PM (Hales et al., 2001). Altogether, these lines of evidence strongly suggest a role of gp41CT in Gag–Env co-localization and incorporation of Env into virus particles.
Despite the evidence for a Gag-mediated Env incorporation process, structural or biophysical evidence for direct gp41CT–MA interaction is still unavailable. A major barrier to characterizing a potential gp41CT–MA interaction by structural tools has been the unavailability of a recombinant gp41CT protein. A structural and functional topology of the gp41CT domain has been proposed based on sequence analysis and biophysical characterization of short peptide fragments (Boscia et al., 2013; Costin et al., 2007; Steckbeck et al., 2013; Steckbeck et al., 2010). A model of gp41CT has been proposed in which a portion of the protein appears to be associated with the membrane (Costin et al., 2007; Steckbeck et al., 2010). The membrane-associated domain contains three lentivirus lytic peptide motifs called LLP2, LLP3 and LLP1, which are highly conserved not only among HIV-1 strains but also among HIV-2, simian immunodeficiency virus (SIV), and equine infectious anemia virus (EIAV) (Srinivas et al., 1992; Steckbeck et al., 2011). However, an exact protein topology is still controversial. Conventional models based on primary sequence analysis and biochemical assays have placed the entirety of CT inside the cytoplasm of the cell/interior of the virus (Steckbeck et al., 2010). Alternative models based on the presence of an immunogenic phenotype (Kennedy epitope, KE) have been proposed, which postulate the existence of a transmembrane region within gp41CT allowing for the extracellular exposure of the immunogenic region (Cleveland et al., 2003; Hollier and Dimmock, 2005; Steckbeck et al., 2010). Structural characterization of the gp41CT protein is therefore critical for determining its precise topology and the functional role in Gag-mediated Env incorporation.
Herein, we devised new approaches that allowed for successful preparation of the gp41CT protein and determination of the NMR structure in micellar solution. We show that the N-terminal region (45 amino acids) of gp41CT lacks a regular secondary structure and is not associated with membrane. The C-terminal domain (105 amino acids), however, forms three consecutive amphipathic α-helices that are tightly associated to membrane. This study solves a longstanding challenge and provides new insights into the mechanisms of Gag-mediated Env incorporation as well as the overall Env mobility and conformation on the virion surface.
RESULTS
Two independent domains of gp41CT
We have devised a strategy to produce the gp41CT protein via recombinant techniques, which involved expression, purification and screening for the proper membrane mimetics to solubilize the protein and allow for detection of NMR signals. Details on gp41CT preparation, biochemical and biophysical properties are described in Supplemental Information. The gp41CT protein was, however, prone to proteolysis during expression and purification resulting in a soluble peptide comprising residues 707-751 (gp41CTN) and an insoluble fragment comprising residues 752-856 (gp41CTC) (Figure 1A). To facilitate their biochemical, biophysical and structural characterization, we have expressed and purified the two fragments independently. As described below, we show that the two domains adopt structures that are very similar to those within the intact protein.
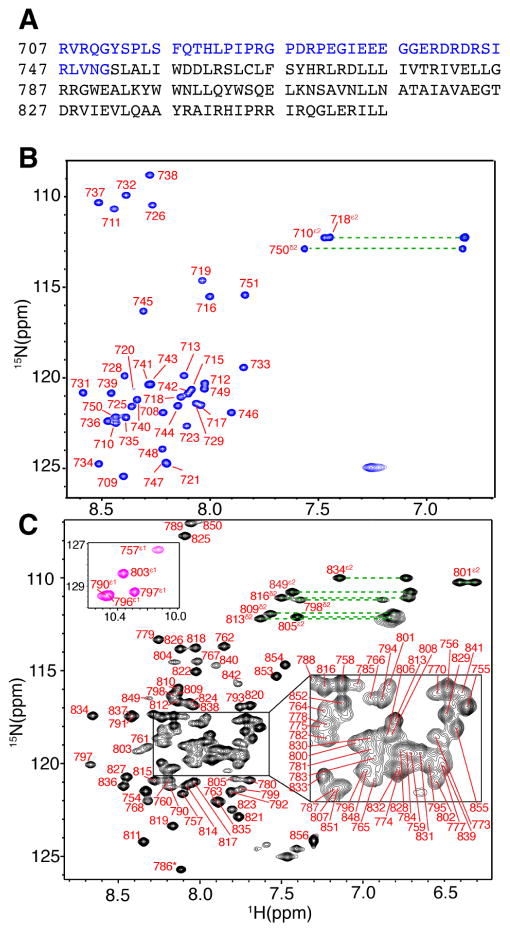
Figure 1. NMR Spectra of the Two Independent gp41CT Domains.
(A) Amino acid sequence of the HIV-1 gp41CT protein (pNL4-3 isolate). Sequences of gp41CTN and gp41CTC are highlighted in blue and black, respectively. (B) 2D 1H-15N HSQC spectrum obtained for gp41CTN (200 μM) at 32 °C. (C) 2D 1H-15N HSQC spectrum obtained for gp41CTC (300 μM) at 50 °C. Tryptophan side chain signals are colored in magenta (inset). Dashed lines indicate side chain amide signals.
Characterization of gp41CTN
The gp41CTN fragment was soluble at concentrations > 1 mM in the absence of detergents, which enabled full structural and biophysical characterization. On a gel filtration column, gp41CTN elutes as a single peak at 17.9 mL (Figure S1), indicating a homogenous sample. When compared to known protein markers, the molecular mass of gp41CTN fragment corresponds to a ~13 kDa species (Figure S1). The 2D 1H-15N HSQC spectrum of 15N-labeled gp41CTN shows a narrow dispersion of the amide proton resonances, suggesting a lack of ordered structure (Figure 1B). The far-UV CD spectrum of gp41CTN displays a negative band at ~200 nm, consistent with a random coil conformation (Figure S2). No changes are observed in the CD spectrum of gp41CTN in the presence of DPC, demonstrating that protein does not associate with membrane. Proton, carbon and nitrogen chemical shifts of gp41CTN were used to predict order parameters and secondary structure content in TALOS+ (Shen et al., 2009), confirming that gp41CTN lacks any regular secondary structure (Figure S2). NOESY data also indicated that the protein lacks a regular secondary structure. No intermolecular NOEs indicative of a protein oligomer have been detected, confirming that gp41CTN is a monomer and that the smaller than expected elution volume obtained from gel filtration data is likely due to the shape and disordered conformation of the molecule.
Characterization of gp41CTC
Preparation and membrane reconstitution of the gp41CTC fragment was conducted in a manner that is essentially identical to that of the full-length gp41CT protein (Supplemental Information). The best results regarding protein reconstitution, stability, and quality of NMR data were achieved in DPC micellar solution. A gel filtration assay of gp41CTC incorporated in DPC micelle shows a single peak at 15.2 mL, which corresponds to ~65 kDa species (Figure S1). Similar to the gp41CT protein, the apparent molecular mass of gp41CTC in DPC micelles as determined by gel filtration is significantly larger. gp41CTC in DPC produced high-quality NMR spectra at 50 °C, which enabled a full structural characterization as described below. CD spectra obtained for gp41CTC in DPC micelles at 35 and 50 °C (Figure S2) are virtually identical, demonstrating that the protein structure was not compromised at 50 °C.
We also examined whether the gp41CTC protein can produce high-quality NMR data in dimyristoylphosphatidylcholine (DMPC)/dihexanoylphosphatidylcholine (DHPC) bicelles and whether the NMR spectra are similar to those obtained in DPC micelles. Although the 2D 1H-15N signals in the HSQC spectrum of gp41CTC in DMPC/DHPC bicelles were relatively broad compared to those observed in DPC micelles, their positions were remarkably similar (Figure S3) demonstrating that gp41CTC maintains a similar structure in both membrane environments. A question may arise whether the structural properties of the isolated gp41CTN and gp41CTC proteins are similar to the corresponding domains within the full-length gp41CT protein. To answer this question, we compared the 1H-15N HSQC spectrum of gp41CT to those of the isolated gp41CTN and gp41CTC domains under identical buffer conditions in DPC micelles. As shown in Figure S3, the 1H-15N resonances of gp41CTN and gp41CTC were at very similar positions as the corresponding signals in the spectrum obtained for the full-length gp41CT protein, which demonstrates that the two domains adopt structures that are very similar to those in the intact protein. Importantly, NMR data analysis of gp41CTC was facilitated by the absence of strong and overlapping gp41CTN signals allowing for full structural characterization of the protein.
NMR signal assignment of gp41CTC
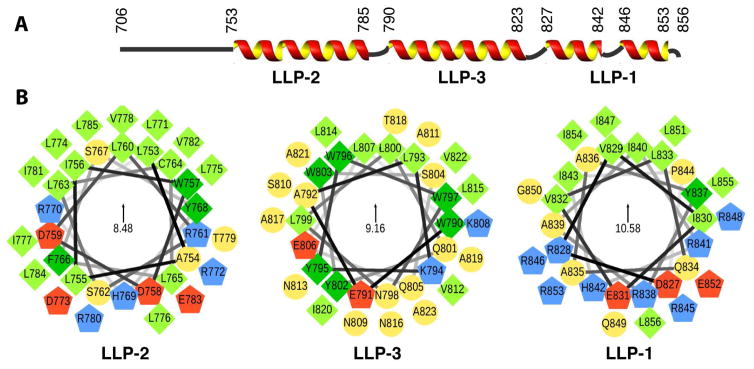
The majority of backbone amide NMR signals were assigned with the exception of L771, R772, L776, I843, R845, R846 and I847 (Figure 1C). Overall, more than 80% of gp41CTC 1H, 13C and 15N resonances were assigned, with the exception of L771, R845 and R846 side chains, which could not be resolved due to severe signal overlaps or signal broadening. For P844, only very broad signals of Cδ and Cγ groups were detected, suggesting that the lack of signals within this region (P844–R846) was due to an intermediate conformational exchange. The chemical shift assignments for gp41CTC were used to predict its order parameters and secondary structure content in TALOS+ (Figure S2). The TALOS+ data indicated that gp41CTC contains three α-helical motifs spanning residues 753-785, 790-823, and 827-853. These motifs were named LLP2, LLP3 and LLP1, respectively (Figure 2) (Steckbeck et al., 2010). In the two short linkers, the α-helical character appeared to be either reduced (residues 786–789 between LLP2 and LLP3) or not present (residues 824–826 between LLP3 and LLP1). Taken together, our data indicated that gp41CTN lacks an ordered structure and is not associated with membrane. gp41CTC, however, is tightly associated with membrane and formed three consecutive α-helices connected by short linkers. The structures and dynamics of the two domains appeared to be completely independent of each other.
Figure 2. Secondary Structure and Helical Wheel Diagrams of the gp41CTC Protein.
(A) Secondary structure representation of the gp41CT protein based on the NMR data. (B) Helical wheel diagrams of the LLP2, LLP3, and LLP1 motifs. Amino acid sequences are plotted clockwise. Hydrophobic and aromatic residues are represented by light and dark green squares, respectively. Polar residues are shown as yellow circles, while positively and negatively charged residues as blue and red pentagons, respectively. The helical wheels are oriented so that their hydrophobic moments (indicated in the centers) point upwards. Helical wheels were generated via a modified script obtained from http://rzlab.ucr.edu/scripts/wheel/.
Structure determination of gp41CTC
Sequential and medium-range NOEs obtained from 3D 15N-edited NOESY-HSQC and 13C-edited HMQC-NOESY spectra contained an extensive network of characteristic α-helical contacts. No long-range intra- or intermolecular NOEs indicative of a tertiary structural fold or formation of oligomer of the gp41CTC protein were detected. A total of 492 NOEs were used to calculate the structure (Table 1). The calculated ensemble of 20 lowest-energy structures showed a good convergence and correspondingly low positional RMSD values within the structured regions (Figures 3A,S3 and Table 1). The gp41CTC protein adopted an α-helical conformation with clearly defined α-helices spanning residues 753-785, 790-823, 827-841, and 847-853 (Figure 3). The linker regions (residues 786-789 and 824-826) were mostly unrestrained during structure calculations and the three α-helices in the resulting structures adopted pseudo-random orientations with respect to each other, limited only by steric factors (Figure S3). Therefore, the bending observed between the helical motifs is variable in the 20 calculated structures. Because signals corresponding to residues P844, R845 and R846 were undetectable, this region was also not restrained during calculation resulting in a small break in the LLP1 helical structure. Of note, previous NMR studies conducted in the presence of 2,2,2-trifluoroethanol on an LLP1 peptide spanning residues 827-853 have shown that the region involving residues 842-845 is highly flexible and adopts a type-II β-turn at P844 and R845 (Sham et al., 2008; Yuan et al., 1995). While we could not obtain any structural data for this region, the observed broadness of P844 methylene signals is consistent with a highly flexible region. Another structural feature of gp41CTC is the presence of strong NOE contacts between aromatic rings of tyrosine, tryptophan or phenylalanine that are in the i position and side chain groups of arginine (Hδ) or lysine (Hε) in the i+4 position, suggesting the presence of a network of cation-π interactions (Gallivan and Dougherty, 1999). Such NOEs were detected for F766–R770, Y768–R772, W790–K794, and Y737–R741, but not for the W757–R761 pair. Interestingly, a strong association also exists for W797–Q801 and W802–E806 pairs given the presence of strong NOEs between the respective side chains and significant upfield shifts of the Gln and Glu resonances.
Table 1.
NMR refinement statistics of the 20 model ensemble of gp41CTC.
| NMR-derived restraints | |
| 1H-1H distance restraints | 492 |
| Intraresidue | 115 |
| Sequential (|i − j| = 1) | 156 |
| Medium range (1 < |i − j| = 4) | 221 |
| Long range (4 < |i − j|) | 0 |
| Backbone H-bonds (4/H-bond) | 77 |
| Distance restraints per refined residue | 8.9 |
| Torsion angles | 198 |
| Restraint violations (mean ± standard deviation) | |
| Ave. max. upper dist. viol. (Å) | 0.20 ± 0.02 |
| Ave. max. van der Waals viol. (Å) | 0.19 ± 0.04 |
| Ave. max. torsion angle viol. (Å) | 0.87 ± 0.52 |
| Cyana target function (Å2) | 0.65 ± 0.14 |
| Residue distribution in Ramachandran plot (%) | |
| Most favored regions | 97.0 |
| Additional allowed regions | 2.6 |
| Generously allowed regions | 0.4 |
| Disallowed regions | 0.0 |
| Structure convergence (Å)1 (mean ± standard deviation) | |
| Backbone N, Cα, C′ atoms RMSD | |
| residues 754–785 | 0.62 ± 0.24 |
| residues 791–820 | 0.26 ± 0.10 |
| residues 827–842 | 0.22 ± 0.07 |
| residues 848–855 | 0.15 ± 0.03 |
| All heavy atoms RMSD | |
| residues 754–785 | 1.14 ± 0.16 |
| residues 791–820 | 0.77 ± 0.10 |
| residues 827–842 | 1.09 ± 0.13 |
| residues 848–855 | 1.26 ± 0.15 |
Structures were superimposed with the representative model and RMSDs against an average structure calculated for the indicated residue ranges.
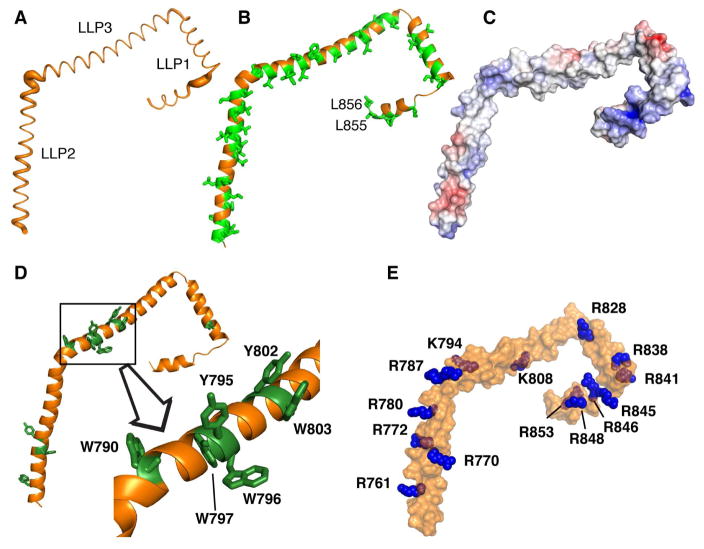
Figure 3. Structural Features of the gp41CTC Protein.
(A) The Cα positional RMSDs between the 20 low-energy structures of gp41CTC are represented as a sausage plot of the representative gp41CTC structure (line thickness is proportional to the RMSD values). The RMSDs from an average structure were calculated after aligning each α-helical segment separately. For residues located at the overlapping boundaries of individual segments, an average of the two RMSD values was calculated. Bending between the helical fragments is variable in the 20 calculated structures. (B) Cartoon representation of the gp41CTC protein showing the extensive hydrophobic interface formed by Leu, Ile, Val, Ala, Trp and Phe residues (green sticks). (C) Surface representation of the gp41CTC colored according to electrostatic surface potential. A basic patch is formed in the arginine-rich LLP1 motif. (D) Cartoon representation of the gp41CTC protein showing Tyr and Trp residues as green sticks. Notice the cluster of six aromatic residues at the beginning of LLP3. The majority of aromatic residues are deeply buried in the interior of DPC micelle. (E) Surface representation of the gp41CTC protein showing Arg and Lys residues (blue spheres).
A striking feature gleaned from the gp41CTC structure was the extensive distribution of hydrophobic surface (Figure 3B,C). Hydrophobic and aromatic residues comprise > 50% of the total gp41CTC protein sequence. The hydrophobic surface is a characteristic feature of the amphipathic gp41CTC and indicates that the interaction interface with the membrane is extensive (see below). An unusual feature of the gp41CTC sequence was the clustering of six aromatic residues at the beginning of helix LLP3 (Figure 3D). Several of these aromatic residues were implicated in the functional role of gp41CT in Env incorporation and infectivity (Lambele et al., 2007; Murakami and Freed, 2000a; Qi et al., 2015; Qi et al., 2013). As shown in Figure 2B, these residues form three pairs with differential localization in the amphipathic structure. Residues W790 and W797 are located on the hydrophobic–hydrophilic interface and, as described above, are engaged in the cation-π and anion-π interactions, whereas W796 and W803 are located on the hydrophobic side of the α-helix. On the other hand, Y795 and Y802 are both localized on the polar side. Interestingly, Y795/W796 has been shown to be a critical motif mediating cell-type–dependent Env incorporation (Qi et al., 2015).
Another characteristic feature of the gp41CTC protein was the unusually high concentration of arginine residues in LLP2 and LLP1 (Figure 3C,E). Recent studies have shown that while conservative substitution of LLP2 Arg residues displayed wild-type phenotypes, similar substitution of LLP1 Arg residues resulted in a significant impairment of Env expression, fusogenicity and incorporation, as well as virus replication (Kuhlmann et al., 2014). As shown in Figures 2B and 3E, six arginine residues were concentrated in the polar side of LLP1 which generated a basic surface in the C-terminus of gp41CTC (Figure 3C,E).
Taken together, our structural data revealed that gp41CTC adopted an α-helical conformation with unique characteristics such as the extensive hydrophobic surface, clustering of aromatic residues in LLP3, unusually high number of cation-π interactions, and formation of a basic patch on the polar side of LLP1. Of note, our structure is among the largest membrane-associated systems studied by NMR spectroscopy, second only to the reported structure and dynamics of micelle–bound α-synuclein, a 140-amino acid protein implicated in Parkinson’s disease (Ulmer and Bax, 2005; Ulmer et al., 2005).
Protein–micelle interactions
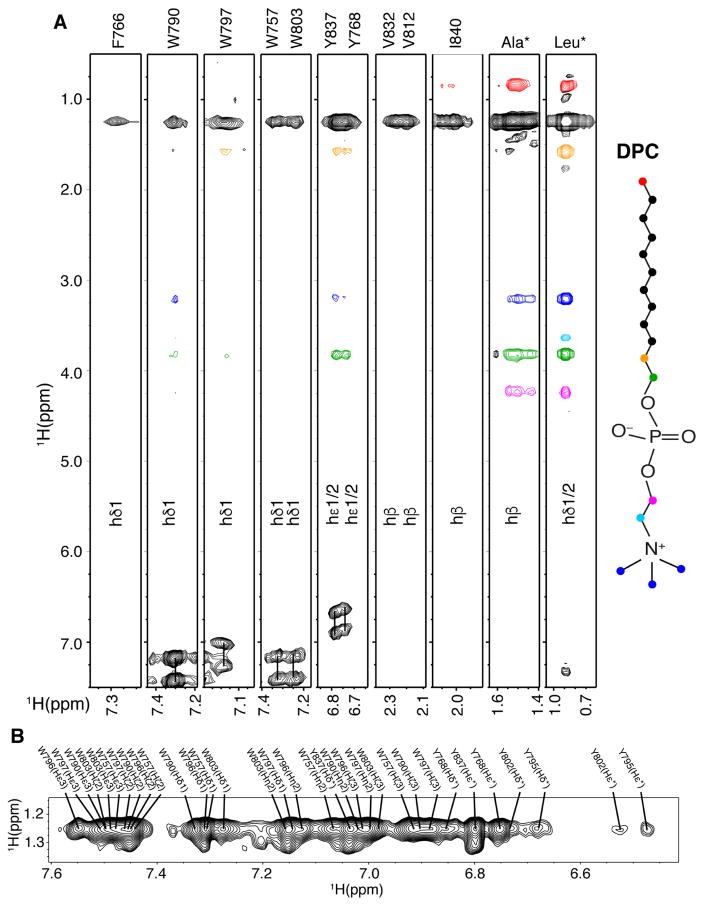
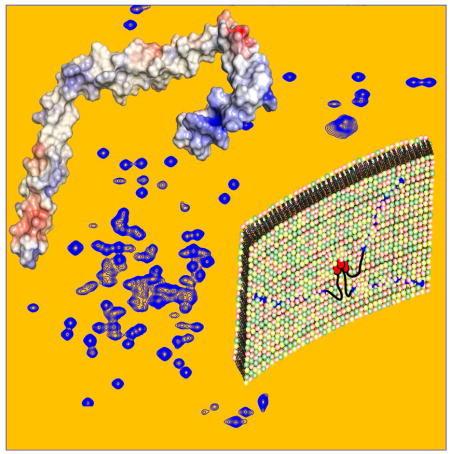
The structure of the micelle-bound gp41CTC protein revealed a large hydrophobic surface that is predisposed to interact with membrane (Figures 2B and 3B). We employed three complementary approaches to probe the depth of membrane insertion by gp41CTC. First, we obtained 2D NOESY, 3D 13C-edited HMQC-NOESY and 13C-half-filtered/13C-edited NOESY-HSQC data and identified numerous intermolecular NOE cross-peaks between DPC and the side chains of hydrophobic and aromatic residues (Figures 4). The methylene resonances of DPC chain exhibited strong intermolecular NOE correlations to the side chains of W757, F766, Y768, W790, W797, W803, V812, V832, Y837, and I840. Residues Y795 and Y802 exhibited only weak NOEs with the DPC micelle interior groups but relatively strong NOEs with the DPC polar head (Figure 4), confirming their exposure on the micelle surface. As expected, strong NOEs were also observed with the side chains of the bulk of Leu, Ile and Ala residues (Figure 4). In general, NOEs between the side chains of most hydrophobic/aromatic residues and the polar head of DPC were virtually nonexistent or very weak, indicating a significant penetration of the protein in the interior of DPC micelles.
Figure 4. Intermolecular Contacts Between gp41CTC and Micelles.
(A) 3D 13C-half-filtered/13C-edited NOESY spectrum for 13C-labeled gp41CTC showing intermolecular NOEs between DPC and gp41CTC residues. Colored intermolecular NOE cross-peaks correspond to colored atoms on the DPC structure. (B) A selected region of 2D 1H-1H NOESY spectrum for gp41CTC showing NOEs between side chains of aromatic residues and DPC acyl chain methylene groups.
In the second approach, we assessed the depth of membrane insertion by utilizing two paramagnetic spin-labeled “depth” probes, 5-doxyl stearic acid (5-DSA) and 16-doxyl stearic acid (16-DSA). Both molecules are incorporated into DPC micelles and cause severe broadening of the NMR signals of nuclei that are within ~10–12 Å of the paramagnetic center (Jarvet et al., 1997; Papavoine et al., 1994). For 16-DSA, only the intensities of resonances buried in the micellar core are dampened. Addition of 16-DSA to 15N- or 13C-labeled gp41CTC samples in DPC micelles caused a substantial reduction of intensities of numerous signals in the 2D 1H-15N and 1H-13C HSQC spectra, respectively. Quantification of the line-broadening of the amide resonances due to the paramagnetic relaxation enhancement clearly showed that the effect is widespread throughout the entire gp41CTC protein (Figures S4). Likewise, significant reduction of signal intensities was observed upon addition of 5-DSA to gp41CTC in DPC micelles (Figures S4). Overall, signal dampening caused by 16-DSA was larger for the majority of residues compared to the 5-DSA analog indicating the that the gp41CTC protein was significantly inserted in the interior of the micelle core.
In the third approach, we probed the regions that are most exposed to solvent by titrating gadodiamide, which is excluded from the hydrophobic interior of the micelle. Paramagnetic gadodiamide induces distance-dependent broadening of NMR signals of residues that are solvent-exposed or near the aqueous environment. We monitored the gadodiamide-induced line-broadening in the 1H-15N HSQC spectra obtained for gp41CTC. Although there was a moderate widespread reduction in signal intensity at high concentration of gadodiamide, a substantial decrease in the intensity of 1H-15N resonances was observed for residues 825-827 (Figure S4), indicating the exposure of the linker between LLP3 and LLP1 to solvent. Other residues such as R788 (located in the linker between LLP2 and LLP3), E791, Y802, S810, A817, Q831, and G850 were also affected but to a lesser extent, suggesting that these regions are transiently exposed or less buried in micelles. Taken together, our NMR data have shown that gp41CTC interacted extensively with the micelle and that numerous hydrophobic residues were buried in the interior of the membrane.
DISCUSSION
Several points have emerged from this work. Most importantly, this study fills a major gap by providing the structure of the last remaining segment of HIV-1 Env, which will likely provide insights into the mechanisms of Env incorporation as well as the overall Env mobility and conformation on the virion surface. We show that gp41CT consists of an unstructured N-terminal domain (residues 707–751) and a membrane-bound C-terminal domain (residues 752–856). A recent NMR structure of the transmembrane segment of gp41 revealed that residues 707–710 (RVRQ) located at the C-terminus of the transmembrane α-helix play a role in stabilizing the trimer (Dev et al., 2016). In gp41CTN, we have not observed any propensity for residues 707–710 to form an α-helix, most likely because of the missing context of the transmembrane region. The C-terminal domain of gp41CT consists of three consecutive amphipathic α-helices (LLP2, LLP3 and LLP1). By defining the boundaries of individual α-helices, we are able to remove the long-standing uncertainty about the exact topology of the LLP segments. Our data revealed variable degree of membrane penetration among the three helices with the N-terminal LLP2 helix penetrating deeper than LLP3 and LLP1 (Figure 5). We have also shown that the helical structures of LLP2 and LLP3 contain several cation-π interactions between aromatic and basic residues in the i and i+4 positions, respectively. Cation-π interacting pairs tend not to be buried while at the same time they are not as exposed as cationic residues alone (Gallivan and Dougherty, 2000). As pointed out previously,(Steckbeck et al., 2011) HIV-1 gp41CTC contains a number of highly conserved arginines and lysines within the LLP1 and LLP2 motifs. Our results show that several of those residues are involved in cation-π interactions. Analysis of over 5000 Env sequences of HIV-1 M group isolates in the Los Alamos database revealed that those pairs are highly conserved (Figure S5). The unusually high number and conservation of these interactions suggests that they play a role in the stabilization of gp41CTC α-helical structure. We speculate that they may also contribute to bilayer destabilization by keeping the charged side chain in the hydrophobic interior of PM or vice versa exposing the bulky aromatic residues to the polar membrane interface. LLP peptides derived from the gp41CT protein were shown to disrupt membranes at very low concentrations (Costin et al., 2007). Interestingly, protein sequence alignment suggests that cation-π interactions are less prominent in HIV-2 and SIV strains (Figure S6). Several arginine residues are also highly conserved across the gp41CT sequence in 128 HIV-2 strains, but much less so in 206 SIV strains. However, an arginine-rich segment (IPRRIRQG850) in LLP1 appears to be almost strictly conserved among all HIV-1, HIV-2 and SIV isolates (Figures S5 and S6). We found that this motif is structurally flexible, which suggests that it may have a functional role in gp41CT-mediated processes. Interestingly, sequence analysis of EIAV gp41CT (GenBank: M16575.1), the longest among retroviruses (200 amino acids), revealed almost no sequence identity to HIV-1, HIV-2 or SIV, and appears to lack the LLP1 Arg-rich domain and frequent cation-π interactions. In summary, the structure of HIV-1 gp41CT revealed novel features that may be important for its function.
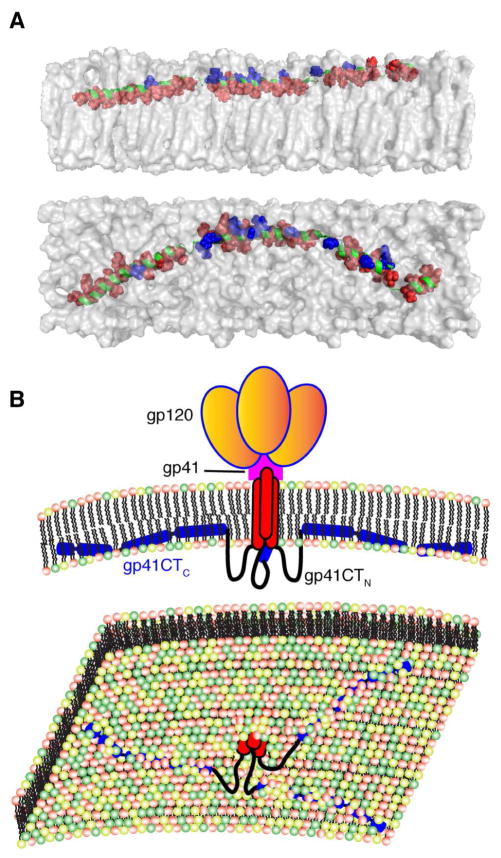
Figure 5. Membrane Interaction of gp41CTC and Overall Env Organization on the Virion Surface.
(A) A model of gp41CTC bound to a membrane bilayer constructed based on the NMR data using the representative structure of gp41CTC with only minor modifications of the dihedral angles in the hinge regions to create an extended molecule. Length of the extended gp41CTC domain shown here is 160 Å. Top and bottom panels show side and top views of the protein, respectively. Residues indicated as red spheres interact extensively with the interior of the membrane while those in blue are mostly exposed and interact with the polar head. Membrane bilayer was generated in VMD membrane builder plug-in (Humphrey et al., 1996). (B) Top panel: A model depicting the gp120 and gp41 proteins on the surface of HIV-1 particles. The gp41CTC domain is penetrating deeply in the inner leaflet of the membrane. Lower panel: An expanded view of the inner leaflet of the PM showing gp41CT penetrating the bilayer.
Early studies reported that antiserum produced against a synthetic peptide from gp41CT (residues 728–745) bound to HIV-1 Env, and that serum from HIV-1 infected humans also recognized this synthetic peptide (Kennedy et al., 1986). This observation led to the suggestion that this sequence is exposed on the virion surface to allow antibody binding and neutralization. Similarly, studies by Lu et al. supported the transient exposure of LLP2 on the cell surface during cell-cell fusion (Lu et al., 2008). In contrast, it was reported that antibodies that bind to Kennedy sequence do not bind Env on intact virions (Steckbeck et al., 2010). In an attempt to explain these observations, Dimmock and colleagues proposed an alternative model by which the KE is exposed (Cleveland et al., 2003; Hollier and Dimmock, 2005). In this model, a three β–sheet membrane-spanning domain (MSD) was proposed based on theoretical sequence analysis. Our data revealed that the recombinant gp41CTN protein does not contain a MSD or any secondary structure either in the absence or in the presence of membrane. Therefore, at least in vitro, our structural findings support a topology by which gp41CTN is unstructured and not membrane-associated (Figure 5).
Several models have been proposed to explain Env incorporation into virus particles (Checkley et al., 2011). The “direct Gag–gp41CT interaction” model is based on genetic and limited biochemical data (Cosson, 1996; Dorfman et al., 1994; Freed and Martin, 1996; Freed and Martin, 1995; Murakami and Freed, 2000a; Yu et al., 1992). It was shown that mutations in MA (L13E and L31E) block the incorporation of Env into HIV-1 particles (Freed and Martin, 1995). This phenotype, however, is reversed by truncating the gp41CT by 104 or 144 amino acids (Freed and Martin, 1995). Subsequent studies revealed that incorporation into virions of Env lacking the last 23, 30, 51, or 56 residues from the CT domain is specially blocked by MA L13E mutation, whereas truncations greater than 93 amino acids reverse this defect (Freed and Martin, 1996). Recently, a model has been proposed whereby MA hexamers of trimers form a lattice capable of accommodating gp41CT (Tedbury et al., 2016). According to our structural data, the region spanning LLP2-LLP3 which has been implicated in MA interaction (Murakami and Freed, 2000a) is the most membrane-sequestered region of gp41CT (Figure 5). Therefore, we propose a potential link between the degree of membrane association of gp41CT and its function in mediating Env incorporation. Because MA binding to the inner leaflet of the membrane is also required for Gag assembly, docking of the two proteins on a specific PM compartment is perhaps needed for a potential interaction to occur. This model, which is called “Gag-Env cotargeting” was among those proposed previously (Checkley et al., 2011). Based on previous studies and the current work, other scenarios for MA-gp41CT interaction are still possible: the gp41CTN loop may play a role in MA interaction, and the gp41CT can adopt multiple conformations depending on whether Gag is present and/or whether the underlying Gag lattice is in the immature or mature state. The advancement achieved here will allow for the development of new approaches to characterize the interplay between gp41CT protein and Gag.
In the “indirect interaction model”, it was proposed that gp41CT–Gag interaction is mediated by cellular factor(s) (Checkley et al., 2011). This model is supported by the finding that the requirement for gp41CT in Env incorporation is cell-type dependent (Akari et al., 2000; Murakami and Freed, 2000b). Numerous cellular factors have been described to interact with gp41CT but their exact functional role is not well understood (reviewed in (Santos da Silva et al., 2013)). Nonetheless, in most of the cases these interactions enhance virus replication. Among the cellular proteins most implicated in Env trafficking and incorporation are the adaptor protein complexes, AP-1 and AP-2 (Berlioz-Torrent et al., 1999; Boge et al., 1998; Ohno et al., 1997). These complexes are known to direct the sorting and trafficking of proteins in the secretory and endocytic pathway. AP-1 has been shown to regulate the subcellular localization of Env via binding to a dileucine motif (L855-L856) in gp41CT (Berlioz-Torrent et al., 1999; Byland et al., 2007; Wyss et al., 2011). AP-2, on the other hand, has been shown to drive clathrin-mediated endocytosis of Env from the PM by binding to gp41CT via a conserved Yxxϕ (ϕ indicates a hydrophobic residue; Y712SPL in our sequence) motif located at the N-terminus of gp41CT (Boge et al., 1998; Ohno et al., 1997). Interestingly, AP-1 and AP-2 have also been shown to interact with the MA domain of Gag (Batonick et al., 2005; Camus et al., 2007). Our structural data show that the YSPL motif is unstructured and not membrane-bound; the dileucine motif, which is located at the end of the gp41CT protein (Figure 3B), interacts only weakly with the membrane. Therefore, both motifs are likely accessible to interact with cellular factors. A recent structural study revealed that even in the presence of the transmembrane segment, the YSPL motif is disordered (Dev et al., 2016). Whereas the YSPL motif and L855 are strictly conserved in HIV-1 isolates, L856 is more variable (Figure S5). In addition to those two motifs, it has been shown Y795/W796 is critical in mediating cell-type–dependent Env incorporation by interacting with the FIP1C protein (Qi et al., 2015; Qi et al., 2013). Our structural data revealed that while W796 is buried and interacts with the membrane, Y795 is exposed to the cytoplasm and accessible for interactions with cellular factors (Figures 2B and 3D). It has yet to be determined how these motifs are involved in Env incorporation.
It has been suggested that part of MPER helices is inserted in the viral membrane, which would induce membrane curvature required for fusion (Buzon et al., 2010). Our finding that the majority of gp41CT protein is embedded in the membrane, and the fact that gp41CT LLP peptides are able to disrupt membranes (Costin et al., 2007) support the hypothesis that gp41CT may contribute additional free energy to the fusion reaction. Indeed, it has been previously shown that during HIV-1 fusion LLP2 may be transiently exposed on the surface of the effector cell in the presence of the target cell (Lu et al., 2008). A synthetic peptide derived from the LLP2 sequence was shown to interact with the gp41 core, suggesting that gp41CT may regulate the fusogenicity of viral Env through the interaction of the transiently exposed LLP2 region with the gp41 core on virions or HIV-1-infected cells (Lu et al., 2008).
This work may also help advance the current understanding of the structure, function, mobility, and lateral movement of Env on the virion surface. The mobility of Env spikes on the virion surface has been of high interest, as motion between these spikes might allow for Env clustering, receptor binding, and membrane fusion, and even IgG avidity (Sougrat et al., 2007). By employing electron tomographic methods, it has been shown that HIV-1 and SIV viruses make contact with T cells via a unique structure called viral “entry claw”, which is typically composed of about six clustered rods of density that span the contact region (Sougrat et al., 2007). The finding that spacing of these rods is slightly closer than the average spacing of Env spikes observed in free viruses led to the suggestion that rearrangement of spikes within the viral membrane may be involved in formation of the structure. Interestingly, Env clustering has been recently recapitulated upon murine leukemia virus binding to the host cell (Riedel et al., 2017). Based on these observations, it is reasonable to hypothesize that the largely membrane-embedded HIV-1 gp41CT domain may facilitate or, at least does not constrain, clustering and lateral movement of Env spikes on the virion surface upon binding to host receptor.
Recent studies have shown that unliganded HIV-1 Env is intrinsically dynamic, transitioning between three distinct pre-fusion conformations, whose relative occupancies were remodeled by CD4 and antibody binding (Munro et al., 2014). CT truncation affected the antigenic surface of the ectodomain of HIV-1 Env on the opposite side of the PM (Chen et al., 2015). It was suggested that a physical coupling (conformation and/or dynamics) between the CT and the ectodomain is mediated by the transmembrane domain (Dev et al., 2016). In the absence of a detailed molecular model of the intact Env including gp41CT, it is difficult to understand this coupling relationship and how the CT domain can influence Env conformation and behavior to play a role in antibody recognition. We believe that our findings may help in development of a platform to produce the intact Env protein, which may ultimately aid in developing new approaches for antiviral drug design, broadly neutralizing antibodies, and vaccine candidates.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Jamil S. Saad (saad@uab.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Bacterial Strains
His6-SUMO-gp41CT, His6-SUMO-gp41CTN, and His6-SUMO-gp41CTC proteins were overexpressed in Escherichia coli BL21 (DE3) codon plus RIL cells with Luria-Bertani (LB) broth or M9 minimal media supplemented with 15NH4Cl and D-glucose-13C6 and containing 50 μg/mL Kanamycin.
METHOD DETAILS
Plasmid construction
To construct the gp41CT clones, their coding sequences were PCR-amplified from the pNL4-3 isolate (NCBI accession code: AF324493) and ligated to the 3′-end of Small Ubiquitin-like Modifier (SUMO) gene via BamHI and XhoI sites within a pET28 vector. The resulting plasmid has a His6 tag encoded on the N-terminus of SUMO. Plasmids were verified by DNA sequencing at the Heflin Center for Genomic Sciences at the University of Alabama at Birmingham.
Preparation of the gp41CT protein
The gp41CT protein was prone to proteolysis even during expression and purification. The proteolytic cleavage resulted in two fragments as detected by chromatography and SDS PAGE. Purification of the cleaved products yielded soluble (~6 kDa) and insoluble (~12 kDa) fragments. The small soluble fragment was isolated and identified by mass spectrometry as the N-terminal 45 amino acids (residues 707-751; gp41CTN). The remaining insoluble C-terminal 105-amino acid region (residues 752-856) is referred to as gp41CTC. We constructed two plasmids to express and purify the gp41CTN and gp41CTC proteins independently. Cleavage of SUMO tag leaves a non-native N-terminal serine residue on all three proteins (gp41CT, gp41CTN and gp41CTC).
Protein Expression and Purification
gp41CT and gp41CTC proteins
To make uniformly 15N- and 15N-,13C-labelled proteins, transformed cells were grown overnight in 25 mL LB medium (50 mg/L kanamycin) at 37 °C. Cells were then spun down and transferred into 1 L of modified M9 minimal medium containing 15NH4Cl and D-glucose-13C6 as the sole nitrogen and carbon sources to produce 15N- and 13C-labelled proteins, respectively. The M9 minimal medium contained 6.8 g/L Na2HPO4, 3 g/L KH2PO4 (pH 7.4), 0.5 g/L NaCl, 5 mM MgSO4, 0.2 mM CaCl2, 0.25 mL/L trace metal solution (1000x), 2.5 mL/L minimum essential medium (MEM) vitamin solution, 1 g/L 15NH4Cl, 2 g/L 13C D-glucose, and 50 mg/L kanamycin. Cells were grown at 37 °C until A600 reached 1.6–1.8, then induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) and grown at 37 °C for 2 h. Cells were then spun down and stored overnight at −80 °C. Next day, the cell pellet was resuspended in lysis buffer containing 100 mM sodium phosphates (pH 8.0), 500 mM NaCl, 0.5% Triton X-100, 10% glycerol, 20 mM imidazole, 0.5 mg/mL lysozyme, 2 mM benzamidine and 0.1 mM phenylmethanesulfonyl fluoride. Cells were sonicated and cell lysate was spun down at 35,000 g for 40 min at 4 °C. The protein-co ntaining supernatant was applied to cobalt affinity resin. The resin was washed with 10 column volumes of buffer A (20 mM sodium phosphates (pH 8.0), 500 mM NaCl, 10% glycerol, and 0.1% Triton X-100), buffer B (20 mM sodium phosphates (pH 8.0), 500 mM NaCl and 10% glycerol), and buffer C (20 mM sodium phosphates (pH 8.0), 500 mM NaCl, 10% glycerol and 500 mM imidazole). To remove imidazole, the resin was washed with 20 mM sodium phosphates (pH 8.0), 500 mM NaCl and 10% glycerol. His6-SUMO protease was then added to the resin and left to rock overnight at room temperature.
A detergent screen kit containing 96 unique conditions was used to elute gp41CT and gp41CTC proteins from the cobalt resin, yielding twenty-three detergent conditions in which the proteins were partially or completely soluble. Among those detergents were dodecylphosphocholine (DPC), dihexanoylphosphatidylcholine (DHPC) and sodium dodecyl sulfate (SDS, Sigma-Aldrich) micelles. In addition, we were able to solubilize the protein in dimyristoylphosphatidylcholine (DMPC)/DHPC bicelles (prepared as described below). DPC was chosen for further work. The protein was eluted off the resin using 20 mM sodium phosphates (pH 8.0), 500 mM NaCl, 10% glycerol and 5.7 mM DPC. The fractions considered as judged by Coomassie-stained SDS–polyacrylamide gel electrophoresis (SDS-PAGE) were pooled, concentrated, and buffer was exchanged to 50 mM sodium phosphates (pH 6.0), 50 mM NaCl, 1 mM TCEP, and 5% D2O (NMR buffer) by using Amicon Ultra Centrifugal Filters (3 kDa membrane cut-off). Final concentration of DPC in NMR samples was ~25 mM.
gp41CTN protein
To make uniformly 15N- and 15N-,13C-labelled samples, transformed cells were grown in 25 mL LB medium (50 mg/L kanamycin) at 37 °C overnight. Cells were then spun down and transferred into 1 L M9 minimal medium containing 15NH4Cl and D-glucose-13C6 as the sole nitrogen and carbon sources to produce 15N- and 13C-labelled proteins, respectively. Cells were grown at 37 °C until A600 was 0.8, induced with 1 mM IPTG and grown at 37 °C overnight. Cells were spun down and stored overnight at −80 °C. Next day, the cell pellet was resuspended in lysis buffer containing 100 mM sodium phosphates (pH 8.0), 500 mM NaCl, 2 mM benzamidine and 0.1 mM phenylmethanesulfonyl fluoride. Cells were sonicated and cell lysate was spun down at 35,000 g for 40 min at 4 °C. The protein-containing supernatant was applied to cobalt affinity resin that was subsequently washed with 5 column volumes of a buffer containing 20 mM sodium phosphates (pH 8.0) and 300 mM NaCl. The His6-SUMO-gp41CTN protein was eluted using an imidazole gradient (0–300 mM). Fractions containing the protein were pooled and His6-SUMO protease was added. Protein was dialyzed overnight at 4 °C against a buffer containing 20 mM sodium phosphates (pH 8.0) and 300 mM NaCl. His6-SUMO and His6-SUMO protease were removed by cobalt affinity resin. The gp41CTN fractions were pooled, concentrated and further purified by gel filtration chromatography. Fractions containing the gp41CTN protein were then pooled and buffer was exchanged to NMR buffer by using Amicon Ultra Centrifugal Filters (3 kDa membrane cut-off).
Preparation of DMPC/DHPC bicelles
Bicelles consisting of DMPC/DHPC in molar ratio (q) of 0.5 were prepared at 2 % (w/v) total lipid concentration by mixing appropriate amounts of solid DMPC with 336 mM DHPC in 20 mM sodium phosphates (pH 8.0), 500 mM NaCl and 10% glycerol buffer, followed by three freeze-thaw cycles. 2 mL of the bicelle solution was added to 1 mL cobalt resin containing His6-SUMO-gp41CT (or His6-SUMO-gp41CTC) after imidazole removal (see above). His6-SUMO protease was then added and the resin was rocked overnight at room temperature. After separating the resin by filtration, gp41CT (or gp41CTC) in bicelles was concentrated and buffer exchanged to NMR buffer using Amicon Ultra centrifugal filters. DHPC was added to the final sample to compensate for any loss during concentration. Final DHPC/DMPC ratio was verified by 1H NMR.
Biochemical and biophysical characterization of the gp41CT protein
NMR and CD spectroscopic methods were employed to identify the proper buffer conditions that maintained protein solubility, secondary structure, and most importantly, allowed for detection of NMR signals. Our initial attempts focused on properly reconstituting the protein in DMPC/DHPC bicelles. Unfortunately, the overall quality of NMR data was poor. Only a very small subset of 1H-15N resonances were observed in the HSQC spectrum. The best results in terms of high solubility and protein stability were achieved with DPC micelles.
The oligomerization properties of gp41CT in DPC micelles were analyzed by a gel filtration assay and verified by SDS-PAGE (Figure S1). Protein elutes as a single peak at 14.8 mL (Figure S1), indicating a homogenous DPC-reconstituted protein. Interestingly, compared to known protein standards the apparent molecular mass of gp41CT protein in DPC micelle corresponds to a ~75 kDa species (Figure S1). Previous studies have shown that the size of a DPC micelle can range from 50 to 90 molecules which corresponds to 18–32 kDa (Kim et al., 2010; Lazaridis et al., 2005). Therefore, the larger than expected estimated molecular weight is either due to the elongated shape of gp41CT and formation of a larger micelle to accommodate the protein or due to formation of a trimer in a smaller micelle. The latter has been ruled out as described below. Next, we examined the secondary structure of DPC-associated gp41CT protein by CD spectroscopy. The far-UV CD spectrum of gp41CT in DPC micelles shows two minima at 208 and 222 nm, features distinctive of an α-helical secondary structure (Figure S2). Our data are in agreement with previous CD studies conducted on short peptides derived from gp41CT that indicated the presence of α-helical structures in membranes consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG) (Costin et al., 2007) or SDS micelles (Steckbeck et al., 2011). For DPC-solubilized gp41CT, NMR spectra were collected at different temperatures with best data obtained at 50 °C. However, numerous signals were relatively weak and severely overlapping which rendered signal assignments impractical (Figure S2). The CD spectra obtained for gp41CT in DPC micelles at 20 and 50 °C (Figure S2) are virtually identical, demonstrating that the protein structure was not compromised at 50 °C. In summary, none of the conditions applied to the gp41CT protein produced sufficient quality of NMR data to enable full structural characterization. This result is likely attributed to the presence of two independent domains (free and membrane-bound) with very different tumbling and relaxation rates.
Gel filtration assay
The mobility of gp41CTC was analyzed by a gel filtration assay. Briefly, 0.5 mL of ~200 μM sample was loaded onto a HiLoad Superdex 75 (10/300) column in a buffer containing 50 mM sodium phosphates (pH 6), 50 mM NaCl, 1 mM TCEP, and 3 mM DPC. Protein fractions were analyzed by SDS-PAGE and stained by Coomassie brilliant blue. The approximate molecular weight of gp41CTC was determined by a molecular weight gel filtration calibration kit.
Circular dichroism (CD) spectroscopy
CD spectra were acquired on a Jasco J815 spectropolarimeter at 20, 35 and 50 °C from 260 to 185 nm. Scanning rate was set to 50 nm/min. Loading concentrations were 20 μM for gp41CTN in a buffer containing 10 mM potassium phosphates (pH 6) and 100 mM potassium fluoride. The gp41CTC was in a buffer containing 10 mM potassium phosphates (pH 6), 100 mM potassium fluoride and 3 mM DPC. The background signal from the buffer solution was subtracted from each spectrum. Protein samples were run on gel filtration column (as described above) to ensure high purity and homogeneity prior to collection of the CD spectra.
NMR spectroscopy
NMR data were collected at 32 or 50 °C on a Bruker Avance II (700 MHz 1H) spectrometer equipped with a cryogenic triple-resonance probe, processed with NMRPipe (Delaglio et al., 1995) and analyzed with NMRVIEW (Johnson and Blevins, 1994) or CCPN Analysis (Vranken et al., 2005). Isotopically unlabeled and uniformly 13C-, 15N-, or 13C-/15N-labelled gp41CTN and gp41CTC protein samples were prepared at ~400–500 μM concentrations. The backbone and side-chain atom resonances were assigned using HNCA, HN(CO)CA, HNCACB, HN(CO)CACB, HNCO, HN(CA)CO, 15N-edited NOESY- and TOCSY-HSQC, and (H)CCH-TOCSY experiments. Assignments of aromatic signals for gp41CTC protein were confirmed by the 1H-13C TROSY, TROSY-(H)CCH-COSY and (HB)CB(CGCD)HD experiments. Intermolecular NOE contacts between DPC and gp41CTC were observed in 2D NOESY, 3D 1H-15N NOESY-HSQC, and 13C-half-filtered/13C-edited NOESY-HSQC spectra (all mixing times 120 ms). Chemical shifts were referenced to 2,2-dimethyl-2-silapentanesulfonic acid (Markley et al., 1998).
Paramagnetic Relaxation Enhancement NMR
Doxyl stearic acid titrations
The additions of solid aliquots of 5- or 16-doxyl stearic acid (5-DSA and 16-DSA, respectively) to 150 μM 15N-labelled gp41CTC in NMR buffer and 25 mM DPC to final concentrations of 0.1, 0.2, 0.4, 0.8, and 1.6 mM. 1H-15N HSQC spectra were collected at 50 °C after each addition. The spin la bels did not alter the protein structure given the absence of chemical shift perturbations.
Gadodiamide titration
A stock solution of 100 mM gadodiamide in H2O was titrated to 90 μM 15N-labelled gp41CTC in NMR buffer to final concentrations of 0.05, 0.1, 0.2, 0.4, 0.8, 1.6, 3.2, 6.4 and 9.6 mM. 1H-15N HSQC spectra were collected at 50 °C after each a ddition.
Data analysis
For each titration, peak heights were determined for non-overlapping signals and relative reductions (RR) of the heights calculated according to RR = (I0–In)/I0, where I0 is peak height in the control experiment and In is peak height at titration point n.
Structure calculations
Phi and psi dihedral angle restraints were generated in TALOS+ (Shen et al., 2009) based on 1HN, 15N, 13C′, 1Hα, 13Cα and 13Cβ chemical shifts and used with an uncertainty of ±2 × standard deviation or ± 20 deg, whichever was larger, only for residues with a “Good” score (Shen et al., 2009). Structure of the gp41CTC was calculated in CYANA (Güntert, 2004) using ϕ/ψ dihedral angles as predicted with TALOS+ and assigned NOE restraints from 1H-15N NOESY-HSQC and 1H-13C HMQC-NOESY. Upper distance limits were set to 2.7, 3.3, 4.0, 4.5 or 5.0 Å based on the intensities of NOE cross-peaks and standard pseudo atom distance corrections were applied to groups of degenerate hydrogen atoms during calculations. Hydrogen bond restraints with O–H and O–N distances of 1.8–2.0 and 2.7–3.0 Å, respectively, were used in α-helical regions identified by characteristic α-helical NOEs and TALOS prediction. Side chain rotamers were treated with low-weight torsion angle restraints during the initial stages of simulated annealing. Structure calculations were initiated with random torsion angle values and standard protocols for simulated annealing and subsequent molecular dynamics minimization were used. In the final calculation cycle, 100 random input structures were minimized and 20 structures with the best agreement with the final restraint set were selected as a representative ensemble. Statistical information is provided in Table 1. The quality of structures was assessed using PROCHECK-NMR (Laskowski et al., 1996), and visualization was performed via PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4 Schrödinger, LLC.). Electrostatic potential maps were generated using PDB2PQR and APBS software (Baker et al., 2001; Dolinsky et al., 2004).
DATA AND SOFTWARE AVAILABILITY
The atomic coordinates and restraints have been deposited in the Protein Data Bank, (PDB ID code 5VWL), and chemical shifts have been deposited in the Biological Magnetic Resonance Data Bank (accession codes 27110 and 30297 for gp41CTN and gp41CTC, respectively).
Supplementary Material
Highlights.
Devised new approaches to generate HIV-1 gp41CT protein via recombinant techniques.
Determined the NMR structure of gp41CT in micellar solution.
Characterized gp41CT interaction with the membrane.
Provided a preferred topology of gp41CT bound to the membrane.
Acknowledgments
This work was supported by the National Institute of Health (R01 GM117837) and the Center for AIDS Research at the University of Alabama at Birmingham.
Footnotes
Supplemental Information includes 6 figures can be found with this article online.
AUTHOR CONTRIBUTIONS
Conceptualization, R.E.M., A.B.S., J.V., and J.S.S.; Methodology, R.E.M., A.B.S., J.V., and J.S.S.; Investigation, R.E.M., A.B.S., and J.V.; Writing – Original Draft, R.E.M., J.V., and J.S.S.; Writing – Review & Editing, R.E.M., J.V., and J.S.S.; Funding Acquisition, J.S.S.; Resources, J.S.S.; Supervision, J.S.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akari H, Fukumori T, Adachi A. Cell-dependent requirement of human immunodeficiency virus type 1 gp41 cytoplasmic tail for Env incorporation into virions. J Virol. 2000;74:4891–4893. doi: 10.1128/jvi.74.10.4891-4893.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, et al. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proc Natl Acad Sci U S A. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anraku K, Fukuda R, Takamune N, Misumi S, Okamoto Y, Otsuka M, Fujita M. Highly sensitive analysis of the interaction between HIV-1 Gag and phosphoinositide derivatives based on surface plasmon resonance. Biochemistry. 2010;49:5109–5116. doi: 10.1021/bi9019274. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batonick M, Favre M, Boge M, Spearman P, Höning S, Thali M. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology. 2005;342:190–200. doi: 10.1016/j.virol.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Berlioz-Torrent C, Shacklett BL, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar MC, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boge M, Wyss S, Bonifacino JS, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- Boscia AL, Akabori K, Benamram Z, Michel JA, Jablin MS, Steckbeck JD, Montelaro RC, Nagle JF, Tristram-Nagle S. Membrane structure correlates to function of LLP2 on the cytoplasmic tail of HIV-1 gp41 protein. Biophys J. 2013;105:657–666. doi: 10.1016/j.bpj.2013.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzon V, Natrajan G, Schibli D, Campelo F, Kozlov MM, Weissenhorn W. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6:e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byland R, Vance PJ, Hoxie JA, Marsh M. A conserved dileucine motif mediates clathrin and AP-2-dependent endocytosis of the HIV-1 envelope protein. Mol Biol Cell. 2007;18:414–425. doi: 10.1091/mbc.E06-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camus G, Segura-Morales C, Molle D, Lopez-Vergès S, Begon-Pescia C, Cazevieille C, Schu P, Bertrand E, Berlioz-Torrent C, Basyuk E. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol Biol Cell. 2007;18:3193–3203. doi: 10.1091/mbc.E06-12-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Checkley MA, Luttge BG, Freed EO. HIV-1 Envelope Glycoprotein Biosynthesis, Trafficking, and Incorporation. J Mol Biol. 2011;410:582–608. doi: 10.1016/j.jmb.2011.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. HIV-1 ENVELOPE. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science. 2015;349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chukkapalli V, Ono A. Molecular Determinants that Regulate Plasma Membrane Association of HIV-1 Gag. J Mol Biol. 2011;410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland SM, McLain L, Cheung L, Jones TD, Hollier M, Dimmock NJ. A region of the C-terminal tail of the gp41 envelope glycoprotein of human immunodeficiency virus type 1 contains a neutralizing epitope: evidence for its exposure on the surface of the virion. J Gen Virol. 2003;84:591–602. doi: 10.1099/vir.0.18630-0. [DOI] [PubMed] [Google Scholar]
- Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- Costin JM, Rausch JM, Garry RF, Wimley WC. Viroporin potential of the lentivirus lytic peptide (LLP) domains of the HIV-1 gp41 protein. Virol J. 2007;4:123. doi: 10.1186/1743-422X-4-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dev J, Park D, Fu Q, Chen J, Ha HJ, Ghantous F, Herrmann T, Chang W, Liu Z, Frey G, et al. Structural basis for membrane anchoring of HIV-1 envelope spike. Science. 2016;353:172–175. doi: 10.1126/science.aaf7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA. PDB2PQR: an automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004;32:W665–667. doi: 10.1093/nar/gkh381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfman T, Mammano F, Haseltine WA, Göttlinger HG. Role of the Matrix Protein in the Virion Association of the Human Immunodeficiency Virus Type 1 Envelope Glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. HIV-1 assembly, release and maturation. Nat Rev Microbiol. 2015;13:484–496. doi: 10.1038/nrmicro3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO, Martin AM. Domains of the Human Immonodeficiency Virus Type 1 Matrix and gp41 Cytoplasmic Tail Required for Envelope Incorporation into Virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed OE, Martin AM. Virion Incorporation of Envelope Glycoproteins with Long but Not Short Cytoplasmic Tails Is Blocked by Specific, Single Amino Acid Substitutions in the Human Immunodeficiency Virus Type 1 Matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. A Computational Study of Cation–π Interactions vs Salt Bridges in Aqueous Media: Implications for Protein Engineering. J Am Chem Soc. 2000;122:870–874. [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- Hales CM, Griner R, Hobdy-Henderson KC, Dorn MC, Hardy D, Kumar R, Navarre J, Chan EK, Lapierre LA, Goldenring JR. Identification and characterization of a family of Rab11-interacting proteins. J Biol Chem. 2001;276:39067–39075. doi: 10.1074/jbc.M104831200. [DOI] [PubMed] [Google Scholar]
- Hollier MJ, Dimmock NJ. The C-terminal tail of the gp41 transmembrane envelope glycoprotein of HIV-1 clades A, B, C, and D may exist in two conformations: an analysis of sequence, structure, and function. Virology. 2005;337:284–296. doi: 10.1016/j.virol.2005.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jarvet J, Zdunek J, Damberg P, Graslund A. Three-dimensional structure and position of porcine motilin in sodium dodecyl sulfate micelles determined by 1H NMR. Biochemistry. 1997;36:8153–8163. doi: 10.1021/bi970193b. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Kennedy RC, Henkel RD, Pauletti D, Allan JS, Lee TH, Essex M, Dreesman GR. Antiserum to a synthetic peptide recognizes the HTLV-III envelope glycoprotein. Science. 1986;231:1556–1559. doi: 10.1126/science.3006246. [DOI] [PubMed] [Google Scholar]
- Kim YP, Yeo KJ, Kim MH, Kim YC, Jeon YH. Structural characterization of the intra-membrane histidine kinase YbdK from Bacillus subtilis in DPC micelles. Biochem Biophys Res Commun. 2010;391:1506–1511. doi: 10.1016/j.bbrc.2009.12.106. [DOI] [PubMed] [Google Scholar]
- Kuhlmann AS, Steckbeck JD, Sturgeon TJ, Craigo JK, Montelaro RC. Unique functional properties of conserved arginine residues in the lentivirus lytic peptide domains of the C-terminal tail of HIV-1 gp41. J Biol Chem. 2014;289:7630–7640. doi: 10.1074/jbc.M113.529339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambele M, Labrosse B, Roch E, Moreau A, Verrier B, Barin F, Roingeard P, Mammano F, Brand D. Impact of natural polymorphism within the gp41 cytoplasmic tail of human immunodeficiency virus type 1 on the intracellular distribution of envelope glycoproteins and viral assembly. J Virol. 2007;81:125–140. doi: 10.1128/JVI.01659-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. AQUA and Procheck NMR: Programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Lazaridis T, Mallik B, Chen Y. Implicit Solvent Simulations of DPC Micelle Formation. J Phys Chem B. 2005;109:15098–15106. doi: 10.1021/jp0516801. [DOI] [PubMed] [Google Scholar]
- Lu L, Zhu Y, Huang J, Chen X, Yang H, Jiang S, Chen YH. Surface exposure of the HIV-1 env cytoplasmic tail LLP2 domain during the membrane fusion process: interaction with gp41 fusion core. J Biol Chem. 2008;283:16723–16731. doi: 10.1074/jbc.M801083200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. J Mol Biol. 1998;280:933–952. doi: 10.1006/jmbi.1998.1852. [DOI] [PubMed] [Google Scholar]
- Mercredi PY, Bucca N, Loeliger B, Gaines CR, Mehta M, Bhargava P, Tedbury PR, Charlier L, Floquet N, Muriaux D, et al. Structural and Molecular Determinants of Membrane Binding by the HIV-1 Matrix Protein. J Mol Biol. 2016;428:1637–1655. doi: 10.1016/j.jmb.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk A, Subramaniam S. HIV-1 envelope glycoprotein structure. Curr Opin Struct Biol. 2013;23:268–276. doi: 10.1016/j.sbi.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro JB, Gorman J, Ma X, Zhou Z, Arthos J, Burton DR, Koff WC, Courter JR, Smith AB, 3rd, Kwong PD, et al. Conformational dynamics of single HIV-1 envelope trimers on the surface of native virions. Science. 2014;346:759–763. doi: 10.1126/science.1254426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Freed EO. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and α-helix 2 of the gp41 cytoplasmic tail. J Virol. 2000a;74:3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T, Freed EO. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci. 2000b;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muranyi W, Malkusch S, Müller B, Heilemann M, Kräusslich HG. Super-Resolution Microscopy Reveals Specific Recruitment of HIV-1 Envelope Proteins to Viral Assembly Sites Dependent on the Envelope C-Terminal Tail. PLoS Pathog. 2013;9:e1103198. doi: 10.1371/journal.ppat.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H, Aguilar RC, Fournier MC, Hennecke S, Cosson P, Bonifacino JS. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci. 2004;101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papavoine CH, Konings RN, Hilbers CW, van de Ven FJ. Location of M13 coat protein in sodium dodecyl sulfate micelles as determined by NMR. Biochemistry. 1994;33:12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- Qi M, Chu H, Chen X, Choi J, Wen X, Hammonds J, Ding L, Hunter E, Spearman P. A tyrosine-based motif in the HIV-1 envelope glycoprotein tail mediates cell-type- and Rab11-FIP1C-dependent incorporation into virions. Proc Natl Acad Sci U S A. 2015;112:7575–7580. doi: 10.1073/pnas.1504174112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi M, Williams JA, Chu H, Chen X, Wang JJ, Ding L, Akhirome E, Wen X, Lapierre LA, Goldenring JR, et al. Rab11-FIP1C and Rab14 direct plasma membrane sorting and particle incorporation of the HIV-1 envelope glycoprotein complex. PLoS Pathog. 2013;9:e1003278. doi: 10.1371/journal.ppat.1003278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel C, Vasishtan D, Siebert CA, Whittle C, Lehmann MJ, Mothes W, Grunewald K. Native structure of a retroviral envelope protein and its conformational change upon interaction with the target cell. J Struct Biol. 2017;197:172–180. doi: 10.1016/j.jsb.2016.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy NH, Chan J, Lambele M, Thali M. Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion. J Virol. 2013;87:7516–7525. doi: 10.1128/JVI.00790-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. Structural basis for targeting HIV-1 Gag to virus assembly sites on the plasma membrane. Proc Natl Acad Sci U S A. 2006;103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal AB, Ghanam RH, Fernandez TF, Monroe EB, Saad JS. NMR, Biophysical and Biochemical Studies Reveal the Minimal Calmodulin-Binding Domain of the HIV-1 Matrix Protein. J Biol Chem. 2011;286:33533–33543. doi: 10.1074/jbc.M111.273623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos da Silva E, Mulinge M, Perez Bercoff D. The frantic play of the concealed HIV envelope cytoplasmic tail. Retrovirology. 2013;10:54. doi: 10.1186/1742-4690-10-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sham SW, McDonald JM, Micoli KJ, NRK Solution structure of a calmodulin-binding domain in the carboxy-terminal region of HIV type 1 gp160. AIDS Res Hum Retroviruses. 2008;24:607–616. doi: 10.1089/aid.2007.0202. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry. 2006;45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog. 2007;3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivas SK, Srinivas RV, Anantharamaiah GM, Segrest JP, Compans RW. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- Steckbeck JD, Craigo JK, Barnes CO, Montelaro RC. Highly conserved structural properties of the C-terminal tail of HIV-1 gp41 protein despite substantial sequence variation among diverse clades: implications for functions in viral replication. J Biol Chem. 2011;286:27156–27166. doi: 10.1074/jbc.M111.258855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckbeck JD, Kuhlmann AS, Montelaro RC. C-terminal tail of human immunodeficiency virus gp41: functionally rich and structurally enigmatic. J Gen Virol. 2013;94:1–19. doi: 10.1099/vir.0.046508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steckbeck JD, Sun C, Sturgeon TJ, Montelaro RC. Topology of the C-terminal tail of HIV-1 gp41: differential exposure of the Kennedy epitope on cell and viral membranes. PLoS One. 2010;5:e15261. doi: 10.1371/journal.pone.0015261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedbury PR, Novikova M, Ablan SD, Freed EO. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc Natl Acad Sci U S A. 2016;113:E182–190. doi: 10.1073/pnas.1516618113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmer TS, Bax A. Comparison of structure and dynamics of micelle-bound human alpha-synuclein and Parkinson disease variants. J Biol Chem. 2005;280:43179–43187. doi: 10.1074/jbc.M507624200. [DOI] [PubMed] [Google Scholar]
- Ulmer TS, Bax A, Cole NB, Nussbaum RL. Structure and dynamics of micelle-bound human alpha-synuclein. J Biol Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- Vlach J, Saad JS. Trio engagement via plasma membrane phospholipids and the myristoyl moiety governs HIV-1 matrix binding to bilayers. Proc Natl Acad Sci USA. 2013;110:3525–3530. doi: 10.1073/pnas.1216655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wyss S, Berlioz-Torrent C, Boge M, Blot G, Höning S, Benarous R, Thali M. The highly conserved C-terminal dileucine motif in the cytosolic domain of the human immunodeficiency virus type 1 envelope glycoprotein is critical for its association with the AP-1 clathrin adaptor [correction of adapter] J Virol. 2011;75:2982–2992. doi: 10.1128/JVI.75.6.2982-2992.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yuan X, Matsuda Z, Lee T-H, Essex M. The Matrix Protein of Human Immunodeficiency Virus Type I is Required for Incorporation of Viral Envelope Protein into Mature Virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Mietzner TA, Montelaro RC, Vogel HJ. Characterization of the calmodulin binding domain of SIV transmembrane glycoprotein by NMR and CD spectroscopy. Biochemistry. 1995;34:10690–10696. doi: 10.1021/bi00033a045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.