ABSTRACT
Genetic and epigenetic alterations are required for carcinogenesis and the mutation burden across tumor types has been investigated. Here, we investigate epigenetic alterations with a novel measure of global DNA methylation dysregulation, the methylation dysregulation index (MDI), across 14 cancer types in The Cancer Genome Atlas (TCGA) database. DNA methylation data—obtained using Illumina HumanMethylation450 BeadChip—was accessed from TCGA. We calculated the MDI in 14 tumor types (n = 5,592 tumors), using adjacent normal tissues (n = 701) from each tumor site. Copy number alteration, and mutation burden were retrieved from cBioportal (n = 5,152). We tested the relation of subject MDI across tumors and with age, gender, tumor stage, estimated tumor purity, and copy number alterations for both overall MDI and genomic-context-specific MDI. We also investigated the top most dysregulated loci shared across tumor types. There was a broad range of extent in methylation dysregulation across tumor types (P < 2.2E-16). However, a consistent pattern of methylation dysregulation stratified by genomic context was observed across tumor types where the highest dysregulation occurred at non-CpG island regions. Considering other summary measures of somatic alteration, MDI was correlated with copy number alterations but not with mutation burden. Using the top dysregulated CpG sites in common across tumors, 4 classes of cancer types were observed, and the functional consequences of these alterations to gene expression were confirmed. This work identified the global DNA methylation dysregulation patterns across 14 cancer types showing a higher impact for the non-CpG island areas. The most dysregulated loci across cancer types identified common clusters across cancer types that may have implications for future treatment and prevention measures.
KEYWORDS: Copy number alterations, DNA methylation, epigenetics, pan-cancer
Abbreviations
- BLCA
bladder carcinoma
- BRCA
breast carcinoma
- COAD
colon adenocarcinoma
- ESCA
esophageal carcinoma
- HNSC
head and neck squamous cell carcinoma
- KIRC
renal clear cell carcinoma
- KIRP
renal papillary cell carcinoma
- LUSC
lung squamous cell carcinoma
- LIHC
hepatocellular carcinoma
- LUAD
lung adenocarcinoma
- PAAD
pancreatic adenocarcinoma
- PRAD
prostate adenocarcinoma
- THCA
thyroid carcinoma
- UCEC
endometrial carcinoma
Background
Cancer is a major source of morbidity and mortality; in 2015, 15 million new cases of malignant neoplasms were expected worldwide,1 and about 10% of these incident cases were expected in the USA.2 Typically, cancers include hundreds of somatic alterations to DNA, overcoming programs that control replication and apoptosis to allow survival and growth. There are more than 100 recognized types of cancer, usually named after the organ or tissue of origin and/or the cell type from which they are derived. Through a collaborative effort and integrative genomic profiling of somatic alterations, the Cancer Genome Atlas initiative (TCGA) has characterized the molecular alteration profiles of several cancer types with the goal of discovering new phenotypes and possibly new therapeutic molecular targets.
Interest in taking pan-cancer approaches to investigate molecular similarities and differences across tumors has grown as data from more tumor types is becoming available.3 Recently, pan-cancer analyses have reported functional mutations,4 immunogenomic signature,5 and copy number alterations.6,7 The global landscape of somatic tumor alterations is providing a new perspective relating the broad spectrum of cancer types through common molecular signatures and may offer opportunities for targeted treatments, off target therapeutic applications, and potential preventative measures with broader impact.
Epigenetic alterations are recognized to have consequences for gene expression patterns and chromosomal stability; and among epigenetic modifications, DNA methylation alterations are the best characterized. In cancer, initially two general patterns of DNA methylation alterations were observed: a global loss of methylation across the genome, and increased methylation at CpG rich promoter areas known as CpG islands.8 Loss of methylation across the genome at repetitive regions, gene bodies, centromeric DNA, and normally imprinted areas leads to genomic instability.9 Increased methylation of promoter CpG islands (CGI) and shore regions can result in gene repression and silencing, including tumor suppressor genes. Although this general paradigm has been observed across different tumor types, for several years an understanding of altered methylation in cancer was limited by the extent of measures being performed. More recently, genome-scale approaches have revealed that distinct methylomes are present within each tissue type,10,11 and the alteration profiles of methylation differ among tumor types as well. Diverse methylomes within tumor type have been associated with tumor characteristics and patient prognosis. For example, a CGI methylator phenotype (CIMP) has been described for several cancer types,12,13 and has been shown to associate with a differential prognosis dependent on the specific cancer type.
Current approaches to integrate DNA methylation analyses across tumor types have several limitations. Previous integration attempts relied on combining older microarray platforms (Illumina HumanMethylation27) with Illumina HumanMethylation450 (450K), limiting the coverage across gene regions to those shared between the two different generations of array platforms, and among those restricting the analyses only to those probes tracking to promoter CGI sites.14 One recently published approach used the information from the 450K microarray and integrated information from normal samples outside TCGA; however, the differentially methylated sites were based only on tumor/normal comparisons without taking into account subject covariates which may also affect the methylation signal.15 Importantly, incorporation of data on estimated tumor purity, which can affect tumor methylation signal, has only recently become practice.16
Here, we use an alternative to traditional bottom up approaches in which individual CpG loci are evaluated in relation to a particular tumor type. In addition, we include adjustment for potential confounders, including estimated tumor purity. Previously, we developed and applied a top down approach, in which the methylation dysregulation index (MDI) describes the cumulative absolute departure from normal DNA methylation between neoplastic and normal (disease-free) tissue.17 The MDI represents the average departure of DNA methylation in tumor cells from normal cells across all measured loci. In addition, this approach can be adapted to specific subsets of CpGs, such as those tracking to a specific genomic context (e.g., promoter CpG island), to evaluate the relative departure from normal among different genomic regions. To comprehensively catalog the similarities and differences of DNA methylation burden across solid tumor types, we used the MDI to investigate DNA methylation dysregulation across 14 cancer types in TCGA.
Results
In total, 14 tumor types (including 5,592 tumor samples and 701 normal adjacent samples) were analyzed; summary statistics are provided in Table 1. Across cancers, there were a similar total number of males and females, though this analysis includes several sex specific cancers—prostate adenocarcinoma (PRAD) and endometrial carcinomas (UCEC)—and some neoplasms with a marked gender predominance—breast carcinoma (BRCA). Overall, the mean age (and standard deviation, SD) of subjects was 61.7 (SD: 12.96) years, which was similar in all the cancer subtypes except thyroid carcinoma (THCA), which has a younger age at diagnosis (mean 47.8, SD: 15.77 years). Most tumors were early stage (62%). Early stage tumors (localized) were less prominently represented in bladder carcinoma (BLCA; 33%), because that study focused on muscle invasive tumors. This contrasts with the relative population incidence in the US, where the non-muscle invasive bladder tumors (localized plus in situ) are the most common stages diagnosed (86% of all the new cases).2 Similarly, 38% of PRAD were early stage tumors in TCGA, which is much lower than the relative population expected incidence in the US (approximately 80%).18 The pathologically estimated tumor cell percentage across samples was generally high with a median of 80% and an interquartile range (IQR) of 70 to 90%. Pancreatic adenocarcinoma (PAAD) was the only tumor with a median tumor cell percentage under this range (60%).
Table 1.
General characteristics of the study population.
| AJCC tumor stagea |
|||||||
|---|---|---|---|---|---|---|---|
| Tumor | Normalb | Female | Male | Age | Stage I/II | Stage III/IV | |
| Cancer type | n | n | n (%) | n (%) | Mean (SD) | n (%) | n (%) |
| BLCA | 411 | 21 | 108 (26.3) | 303 (73.7) | 68.56 (10.60) | 133 (32.5) | 276 (67.5) |
| BRCA | 779 | 97 | 770 (98.8) | 9 (1.2) | 58.62 (13.10) | 563 (72.8) | 210 (27.2) |
| COAD | 283 | 38 | 131 (46.3) | 152 (53.7) | 65.37 (13.20) | 150 (54.5) | 125 (45.5) |
| ESCA | 185 | 16 | 27 (14.6) | 158 (85.4) | 62.95 (11.87) | 98 (60.1) | 65 (39.9) |
| HNSC | 527 | 50 | 142 (26.9) | 385 (73.1) | 61.40 (11.93) | 103 (22.6) | 353 (77.4) |
| KIRC | 319 | 160 | 114 (35.7) | 205 (64.3) | 61.85 (11.84) | 187 (58.6) | 132 (41.4) |
| KIRP | 275 | 45 | 73 (26.5) | 202 (73.5) | 62.18 (12.09) | 188 (74.3) | 65 (25.7) |
| LIHC | 374 | 50 | 120 (32.1) | 254 (67.9) | 59.83 (13.31) | 261 (73.9) | 92 (26.1) |
| LUAD | 457 | 32 | 244 (53.4) | 213 (46.6) | 65.53 (10.16) | 362 (79.4) | 94 (20.6) |
| LUSC | 369 | 42 | 96 (26.0) | 273 (74.0) | 68.05 (8.73) | 309 (84.0) | 59 (16.0) |
| PAAD | 183 | 10 | 82 (44.8) | 101 (55.2) | 65.28 (10.99) | 171 (94.5) | 10 (5.5) |
| PRAD | 497 | 50 | NA | 497 (100.0) | 61.56 (6.79) | 185 (37.8) | 305 (62.2) |
| THCA | 504 | 56 | 368 (73.0) | 136 (27.0) | 47.83 (15.77) | 334 (66.5) | 168 (33.5) |
| UCEC | 429 | 34 | 429 (100.0) | NA | 64.60 (11.18) | 284 (69.6) | 124 (30.4) |
| Overall | 5592 | 701 | 2704 (48.4) | 2888 (51.6) | 61.66 (12.96) | 3328 (61.6) | 2078 (38.4) |
| P-value | <2.2E-16c | <2.2E-16d | <2.2E-16c | ||||
General American Joint Committee on Cancer Classification,
Normal adjacent samples
P-values based on:
Fisher exact test,
ANOVA test NA: Not Available/Not Applicable
First, we compared the overall departure of methylation among all tumor types using the subject methylation dysregulation index (sMDI). The sMDI was significantly different among tumor types (Kruskal-Wallis rank sum test P < 2.2E-16, Table 2). The median sMDI across all tumors and all tumor types was 8.1 (the absolute difference between tumor and adjacent normal DNA methylation was, in average, 8.1% for all the CpGs measured) with an IQR between 6.24 and 10.16. Several tumor types showed lower median sMDI most markedly the group of THCA, PAAD, renal clear cell carcinoma (KIRC), and renal papillary cell carcinoma (KIRP), in which the median sMDI were 5.2, 6.1, 6.3, and 6.4, respectively. On the other hand, BLCA, hepatocellular carcinoma (LIHC), UCEC, and esophageal carcinoma (ESCA) showed the highest sMDI (all the medians above 10). We also observed large differences in levels of MDI across tumor types when stratifying MDI by the genomic context of CpG sites using the gcMDI (Kruskal-Wallis rank sum test P < 2.2E-16, Table 2). Despite large differences in the magnitude of sMDI and gcMDI among tumor types, a consistent pattern in the extent of gcMDI was observed across tumors. For example, CpG island regions had a consistently lower gcMDI compared with other genomic contexts. The CpG island shore and shelf regions that flank CpG islands had higher gcMDI levels than island regions and were consistently similar to each other. Farthest from CpG island regions, open sea gcMDI levels were the highest gcMDI across tumors types with only one exception (THCA, Table 2). Striking differences between genomic context MDI were observed and Wilcoxon-test pairwise false discovery rate (FDR) comparisons are provided in Supplementary Table S1.
Table 2.
Subject and genomic context methylation dysregulation indices, and fraction of genome altered by cancer types.
| gcMDI |
|||||||
|---|---|---|---|---|---|---|---|
| sMDI | CpG Island | Shores | Shelves | Open Sea | Fraction of genome altered | ||
| Cancer type | n | median (IQR) | median (IQR) | median (IQR) | median (IQR) | median (IQR) | median (IQR) |
| BLCA | 411 | 10.7 (8.5, 13.4) | 7 (5.6, 8.4) | 11.1 (8.6, 13.6) | 11.7 (9, 15.4) | 13.6 (10.3, 17.2) | 28.2 (13.9, 45.2) |
| LIHC | 374 | 10.6 (8.4, 13.4) | 7 (5.6, 8.1) | 10.6 (8.6, 12.8) | 11.3 (8.4, 15.4) | 13.2 (9.9, 17.9) | 25.2 (15.9, 37.8) |
| UCEC | 429 | 10.4 (9.2, 11.9) | 7.2 (6.2, 8.5) | 11.3 (10.1, 12.8) | 10.6 (9, 12.3) | 12.5 (10.7, 14.6) | 8.1 (0.2, 34.9) |
| ESCA | 185 | 10.1 (8.8, 11.8) | 7.2 (5.6, 9.6) | 10.2 (8.9, 11.9) | 10.6 (9, 12.5) | 11.9 (10.2, 14) | 35.8 (22.6, 50.1) |
| COAD | 283 | 9 (7.4, 10.4) | 7.2 (5.8, 9.1) | 8.9 (7.6, 10.5) | 8.7 (7, 10.2) | 10 (8.1, 11.7) | 20.7 (9, 32.6) |
| LUSC | 369 | 8.6 (7.3, 10.1) | 5.7 (4.9, 6.8) | 9.1 (7.6, 10.9) | 9.1 (7.6, 11.3) | 10.4 (8.7, 12.6) | 38.9 (3.8, 52.5) |
| HNSC | 527 | 8.5 (7.2, 10.1) | 6.3 (5.4, 7.4) | 8.8 (7.4, 10.4) | 8.8 (7.2, 11.2) | 9.7 (8.2, 12.3) | 22.9 (12.5, 33.4) |
| BRCA | 779 | 8.5 (7, 10.2) | 6.1 (4.9, 7.8) | 9 (7.6, 10.6) | 8.4 (6.9, 10.4) | 9.8 (8, 12.3) | 22.8 (11.2, 41.3) |
| LUAD | 457 | 7.5 (6.1, 8.9) | 5.7 (4.6, 7) | 8 (6.5, 9.5) | 7.2 (5.8, 8.8) | 8.4 (6.8, 10.3) | 22 (9.2, 38.7) |
| PRAD | 497 | 7.5 (5.5, 8.9) | 5.6 (4.4, 6.8) | 8 (6, 9.6) | 7.1 (5.3, 8.6) | 8.5 (6.2, 10.4) | 6.4 (1.8, 12.2) |
| KIRP | 275 | 6.4 (5.7, 7.7) | 3.7 (3.4, 4.7) | 7.4 (6.5, 8.8) | 6.7 (6.1, 7.8) | 8 (7.1, 9.2) | 14.9 (8.3, 22.8) |
| KIRC | 319 | 6.3 (5.7, 7.1) | 3.8 (3.3, 4.6) | 6.9 (6.2, 7.8) | 6.8 (6.2, 7.8) | 7.7 (7.1, 8.8) | 12.1 (6.3, 20) |
| PAAD | 183 | 6.1 (5.1, 7.8) | 4.9 (3.8, 6.2) | 6.5 (5.4, 8.3) | 6.3 (5.3, 7.8) | 7.1 (6, 8.8) | 9.4 (0.2, 22.8) |
| THCA | 504 | 5.2 (4.8, 5.7) | 3 (2.8, 3.4) | 6.3 (5.7, 6.9) | 5.5 (5, 6.1) | 6.2 (5.7, 6.9) | 0.01 (0.003, 1.1) |
| Overall | 5592 | 8.1 (6.2, 10.2) | 5.8 (4.3, 7.4) | 8.6 (6.8, 10.7) | 8.1 (6.4, 10.7) | 9.4 (7.4, 12.3) | 17.4 (4.8, 34.3) |
| P-valuea | <2.2E-16 | <2.2E-16 | <2.2E-16 | <2.2E-16 | <2.2E-16 | <2.2E-16 | |
Kruskal-Wallis rank sum test
Acronyms: Interquartile Range (IQR)
Extending our assessment to summary measures of genetic somatic alterations, overall copy number alterations—represented by the fraction of the genome altered (FGA)—median was 17.4%, though this had a wide IQR (4.75 to 34.3%). In addition, FGA was positively correlated with MDI when pooling all tumor types (Spearman correlation, ρ = 0.61, P < 2.2E-16). When stratifying by tumor type, the median THCA FGA was lower than the other tumor types (0.01%). MDI and FGA ρ ranged between 0.08 and 0.80 and was significant for all the tumors except UCEC. In addition, where available, the mutation count burden (MCB) was poorly to moderately correlated with MDI; ρ ranged between 0.05 (BLCA) and 0.38 (PRAD) (Supplementary Table S2).
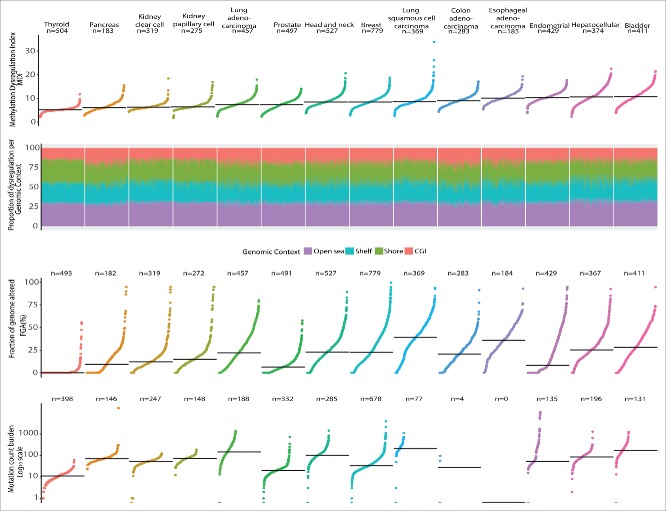
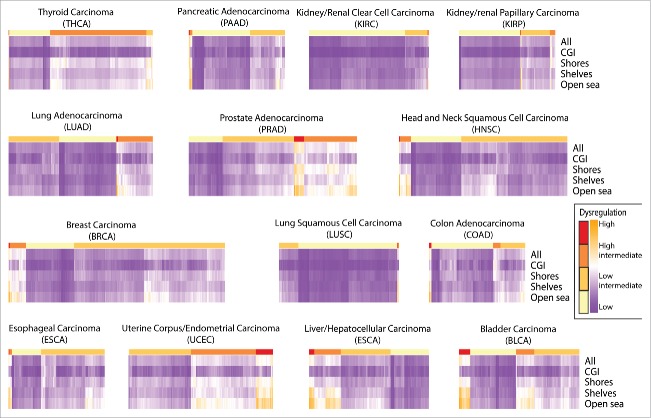
Figure 1 presents the patterns observed for DNA methylation dysregulation and compares tumor types sorted by MDI and the fraction contributed by genomic context to the global sMDI measure. In contrast, the summary measures of genetic somatic alterations FGA and MCB, also presented in the figure, showed a different order and magnitude across cancer types. In addition, Fig. 2 shows the proportion of the sMDI that is contributed by each genomic context and again shows larger gcMDI for shores, shelves, and open sea compared with CpG Islands. When adjusting for the number of probes measured in each genomic context the effect was reduced on the shelves, but remained high on the open sea and low for CpG islands. The magnitude and order of FGA and MCB differed compared with the MDI by cancer type. The ρ between sMDI and the gcMDI was >0.95, except for the CGI gcMDI which was only moderately correlated with the other scores (ρ range: 0.69–0.85, Supplementary Table S2). This also contrasted to the poor to moderate correlation observed with FGA (ρ range: 0.08–0.68) and MCB (ρ range: 0.05–0.38). In contrast, as observed in Fig. 1, the magnitude of dysregulation of both FGA and MCB differed to that observed for the sMDI. Next, we visualized the gcMDI for all samples using unsupervised clustering and for each cancer type set four classes: high dysregulation, high-intermediate dysregulation, low-intermediate dysregulation, and low dysregulation (Fig. 2). The variation across patients within each tumor type were moderate for most of the cancer types; however, the genomic context dysregulation was concentrated on the non-CpG island areas. When comparing clusters with the magnitude of FGA and MCB we did not observe consistent clustering (Supplementary Table S3).
Figure 1.
Comparison of subject DNA methylation dysregulation index, fraction of genome altered, mutation count burden, and proportion of methylation dysregulation by genomic context by cancer type. Each dot corresponds to the sample specific methylation dysregulation index-MDI, fraction of genome altered, or mutation count burden. Tumor types are ordered by their median frequency of each alteration. Overall the median MDI was similar between the compared tumors. Individual MDI were disaggregated according to the unweighted proportion contributed by each genomic context. Thyroid cancers showed the lowest alterations in all the 3 measures, while bladder was consistently highly altered in all 3 measurements. For this specific data set there were only reported 4 mutations in colon adenocarcinoma, and there was not information available for esophageal carcinoma.
Figure 2.
MDI clustering by cancer and genomic context.
We next tested the association of sMDI with patient and tumor characteristics in each tumor type by fitting linear models for MDI with age, sex, cancer stage, estimated tumor purity, and FGA. In adjusted models age was related with increased sMDI in BRCA, LIHC, and THCA (P-values: 5.9E-10, 3.6E-06, and 6.4E-03, respectively, Table 3). Males had increased sMDI in lung adenocarcinoma (LUAD) and PAAD (P-values: 4.21E-04, 9.84E-03 respectively, Table 3). A consistent linear trend was observed with increased cancer stage in both KIRC and KIRP (P-values: 4.48E-06 and 7.14E-09). The coefficients for the relation of sMDI with estimated tumor purity were positive for all tumor types (Table 3). Similarly, the fraction of genome altered increased with most of the MDI scores, but the UCEC association was inconsistent. We extended our analysis of patient and tumor characteristics by fitting linear models for each tumor type with gcMDI values and these results are shown in Supplemental Table 3. Briefly, similar with sMDI results males had consistently increased gcMDI in LUAD for each genomic context except CpG islands (Supplementary Table S4). Due to the recent reclassification of some THCA tumors as benign tumors,19 as a sensitivity analysis, we excluded those THCA cases classified as follicular, without capsule involvement and/or extracapsular involvement (n = 75). The results of this sensitivity analysis were similar to those without the exclusion, thus we report the results using the complete set of cases. As a second sensitivity analysis, we replaced the histological tumor percentage for a DNA methylation estimate of cell purity.20 Both measurements were positively correlated (Pearson r = 0.27), being the estimation consistently lower than the reported percentage of histology. However, the estimates of cell purity for LUAD differed with those reported in the slide reading (Supplementary Figure S1), and the results of the models were severely deflated (R2 < 5%). Therefore, our approach to adjust for tumor purity used estimates from histopathological review.
Table 3.
Factors affecting the subject Methylation dysregulation index.
| Agea |
Female vs. Male |
Early vs. late stageb |
Estimated tumor purity |
Fraction of genome alteredc |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| β (95% CI) | P-value | β (95% CI) | P-value | β (95% CI) | P-value | β (95% CI) | P-value | β (95% CI) | P-value | R2 | |
| BLCA | −0.26 (−0.05,0.002) | 0.06 | 0.32 (−0.35,0.99) | 0.35 | −0.73 (−1.37,−0.1) | 0.02 | 0.49 (0.03,0.07) | 3.30E-07 | 0.63 (0.05,0.08) | 2.70E-15 | 25.1% |
| BRCA | 0.38 (0.03,0.05) | 5.87E-10 | 1.66 (0.24,3.09) | 0.02 | −0.03 (−0.37,0.32) | 0.87 | 0.12 (0.004,0.02) | 3.84E-03 | 0.42 (0.03,0.05) | 5.77E-26 | 20.2% |
| COAD | 0.16 (−0.004,0.04) | 0.11 | −0.13 (−0.64,0.38) | 0.61 | −0.47 (−1.01,0.06) | 0.08 | 0.42 (0.02,0.06) | 6.46E-06 | 0.27 (0.01,0.04) | 7.12E-04 | 12.2% |
| ESCA | 0.01 (−0.03,0.03) | 0.93 | −0.46 (−1.41,0.5) | 0.35 | −0.11 (−0.81,0.58) | 0.75 | 0.21 (0.004,0.04) | 0.01 | 0.43 (0.02,0.06) | 1.56E-05 | 14.8% |
| HNSC | −0.02 (−0.02,0.02) | 0.83 | −0.06 (−0.53,0.41) | 0.79 | 0.56 (0.06,1.05) | 0.03 | 0.12 (−0.001,0.02) | 0.07 | 0.6 (0.05,0.07) | 1.72E-19 | 19.4% |
| KIRC | 0.06 (−0.01,0.02) | 0.30 | 0.13 (−0.17,0.43) | 0.39 | 0.69 (0.4,0.98) | 4.48E-06 | 0.003 (−0.01,0.01) | 0.96 | 0.32 (0.02,0.04) | 2.42E-10 | 20.7% |
| KIRP | 0.01 (−0.02,0.02) | 0.90 | −0.17 (−0.69,0.35) | 0.52 | 1.6 (1.07,2.12) | 7.14E-09 | 0.05 (−0.01,0.02) | 0.57 | 0.15 (0.003,0.03) | 0.01 | 18.3% |
| LIHC | 0.68 (0.04,0.1) | 3.60E-06 | 0.39 (−0.4,1.19) | 0.33 | 0.68 (−0.18,1.54) | 0.12 | 0.35 (0.01,0.06) | 0.01 | 0.71 (0.05,0.09) | 6.07E-11 | 18.8% |
| LUAD | 0.04 (−0.01,0.02) | 0.58 | 0.57 (0.26,0.89) | 4.21E-04 | −0.05 (−0.44,0.34) | 0.80 | 0.04 (−0.01,0.01) | 0.39 | 0.77 (0.07,0.09) | 1.17E-53 | 46.8% |
| LUSC | 0.08 (−0.03,0.04) | 0.63 | 0.35 (−0.32,1.02) | 0.30 | 0.21 (−0.58,1.01) | 0.60 | 0.2 (0.003,0.04) | 0.02 | 0.7 (0.06,0.08) | 7.77E-20 | 25.8% |
| PAAD | −0.03 (−0.02,0.02) | 0.79 | 0.59 (0.14,1.04) | 9.84E-03 | −0.13 (−1.09,0.83) | 0.79 | 0.003 (−0.01,0.01) | 0.94 | 0.95 (0.08,0.11) | 1.81E-32 | 58.9% |
| PRAD | 0.28 (0.001,0.06) | 0.04 | NA | NA | −0.11 (−0.5,0.28) | 0.58 | 0.35 (0.02,0.05) | 3.26E-10 | 1.1 (0.09,0.13) | 5.31E-26 | 34.1% |
| THCA | 0.09 (0.002,0.01) | 6.44E-03 | 0.2 (0.01,0.38) | 0.04 | 0.14 (−0.07,0.35) | 0.18 | 0.1 (0.01,0.02) | 9.08E-05 | 0.29 (0.02,0.04) | 3.77E-05 | 11.7% |
| UCEC | 0.07 (−0.01,0.03) | 0.51 | NA | NA | −0.22 (−0.68,0.24) | 0.35 | 0.26 (0.01,0.04) | 3.43E-04 | 0.02 (−0.01,0.01) | 0.60 | 3.8% |
per each 10 y increase,
Early (I+II), and late (III+IV),
per each 10% increase
Note: In bold those P-values ≤ 0.01 (Bonferroni correction for 5 comparisons).
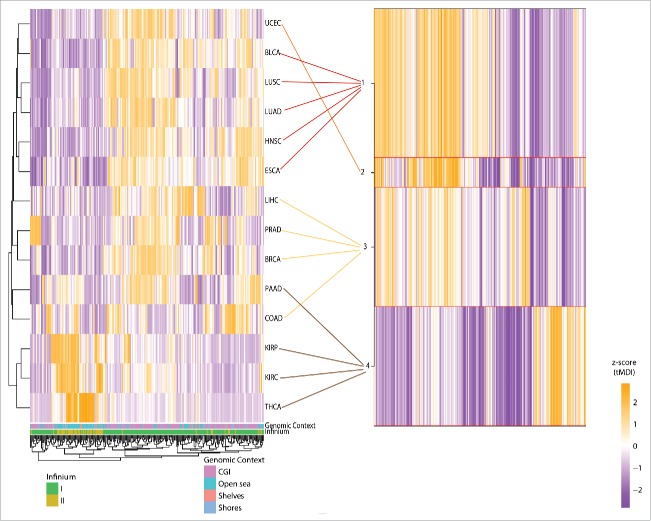
We next sought to identify the specific CpG loci with the largest DNA methylation alteration level in cancers. In each tumor type we derived the mean departure of DNA methylation for all subjects in each tumor (tMDI); 14 probe lists including 399,294 loci were analyzed separately for each cancer type and the resulting tMDI lists were ranked individually from the highest to the lowest most deregulated CpGs. Probes with missing information due to filtering during quality control were kept as NAs in the lists. We used a moderate-deviation algorithm and identified 346 probes as commonly dysregulated among the different tumor types (Supplementary Table S5). A subset of 275 probes without missing values was analyzed for all tumor types using two clustering approaches: an unsupervised hierarchical clustering on Euclidean distance and a semi-supervised recursively partitioned mixture model (RPMM) (Fig. 3).21 Unsupervised clustering showed two major branches separating the less dysregulated tumors (THCA, KIRP, and KIRC) from the others cancer types. Using RPMM to cluster tumor types based on the 275 CpG loci commonly dysregulated across tumors resulted in four methylation classes (Fig. 3B). The methylation class 1 included four tumor types: bladder, lung cancers, esophageal, and head and neck. Endometrial cancer was exclusive to RPMM class 2; class 3 included cancers included prostate, breast, colorectal and liver. Finally, RPMM methylation class 4 grouped the remainder tumors: pancreatic, thyroid, renal clear cell, and renal papillary cancers. A bias corrected enrichment analysis of the 346 probes displayed a significant enrichment of several pathways using the curated GSEA gene sets (Supplementary Table S6). In the curated GSEA gene set 2, 14 pathways were significantly enriched to CpG promoters and histone trimethylation (H3K27me3) in several tissues, Polycomb-group proteins, and related to esophageal, bladder, thyroid, and hepatocellular carcinomas (FDR<0.05). When looking for specific oncogenic signatures (Supplementary Table S6), 11 pathways were enriched to genes related to KRAS, SUZ12, WNT1, and EED (FDR<0.05). As a sensitivity analysis, we calculated the standard deviation of the β-values of the adjacent normal samples (SDN), and the CpG density of the probes used for the tMDI calculation. Using all the cancer types we observed a positive correlation between the tMDI and the SDN (Pearson r = 0.72), and a negative correlation with the CpG density (Pearson r = −0.25). The observed distributions of the tMDI and SDN per cancer type are available as the Supplementary Figure S2, and the heatmaps showing the top tMDI compared with SDN and CpG density as Supplementary Figure S3.
Figure 3.
Unsupervised clustering of tumors using the top most commonly dysregulated probes (n = 275). Using 275/384 most dysregulated loci several patterns were explored using 2 clustering approaches among cancer types. On the left an unsupervised hierarchical Euclidean distance. On the right, the 4 groups derived from a semi-supervised recursively partitioned mixture model (RPMM) classification.
To illuminate potential common pathways derived from the tMDI loci, 34 CpGs were selected from among the 346 probes top loci using the Cross Entropy Monte Carlo (CEMC) algorithm (Supplementary Table S5). These 34 CpG sites tracked to genes related to different cell developmental and proliferation pathways including several Homeobox related genes (OTX2-AS1, HOXD9, OTX1, SIX6, PRRX1, LHX5-AS1, TLX1 and IRX4) in BLCA, BRCA, colon adenocarcinoma (COAD), ESCA, head and neck (HNSC), LUAD, lung squamous cell carcinomas (LUSC), PAAD, PRAD, and UCEC. The group of THCA, KIRC, and KIRP showed dysregulation of loci associated to inflammation (ANXA1), growth hormone regulation (GABRR2), and metabolic genes (AHR). Several CpG sites were related to long noncoding RNA or antisense genes (OTX2-AS1, LINC00466, TFAP2A-AS1, LHX5-AS1, LOC100507443, LIN28B-AS1, PAUPAR, DKFZp686K1684), transcription and regulatory factors (TFAP2B, USP44), and genes encoding tumor suppressor proteins (TRIM15, CASZ1). In addition, several commonly altered CpG sites were located in enhancer elements (cg10903903, cg17754510, cg22797031, cg13539545, cg08443563) and a few were spatially near to the histone related cluster of chromosome 6 (cg10903903-HIST1H2BL, and cg01518607-PRSS16). Lastly, we investigated the functional implication of DNA methylation at the 34 CpGs identified by the CEMC algorithm by testing the association of methylation with gene expression. The β-values of the samples were adjusted for the subject specific covariates, and their linear relationship was tested against the gene expression of the associated gene, or the nearest gene available or sense gene if the CpG probe was tracked to the antisense gene. For those probes associated with the promoter genomic context [CpG located 200 bp upstream of the transcriptional start site (TSS200), CpG located in 1500 bp upstream of the transcriptional start site, or CpG located in 5’untranslated regions (UTR)] we observed a consistent reduction in gene expression related with increased DNA methylation. In contrast, for CpG probes in intragenic regions (gene body, first exon or 3’UTR), increased DNA methylation was related with increased gene expression, though in a few cases gene expression was reduced. A less consistent pattern of methylation related with gene expression was observed for intergenic regions as the tested transcripts corresponded to genes not tracking to the probe. The association here could be related to other regulatory elements as enhancers (present in 11 of those probes), which may differ by tissue/cell of origin. Cancer specific results are summarized in Table 4 and detailed results are provided in Supplementary Table S7.
Table 4.
Relation between the top ranked dysregulated loci and the gene expression of the closest gene.
| BLCA | BRCA | COAD | HNSC | KIRC | KIRP | LIHC | LUAD | LUSC | PAAD | PRAD | THCA | UCEC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CGID(Gene) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) | % Δ(P-value) |
| 1) Promoter related probes | |||||||||||||
| a) CpG located 200 bp upstream of the transcriptional start site (TSS200) | |||||||||||||
| cg15811515 (CSDAP1) | −3.1 (0.04) |
−0.2 (0.91) |
−8 (0.01) |
5.4 (0.03) |
4.5 (3.20E-04) |
0.3 (0.87) |
−0.2 (0.93) |
−1.8 (0.20) |
0.1 (0.95) |
4.6 (0.06) |
−0.1 (0.94) |
−0.2 (0.93) |
−7.1 (0.05) |
| cg02772121 (TRIM15) | −4 (0.28) |
−8.7 (1.18E-19) |
−17.5 (2.98E-05) |
−5.4 (0.21) |
−40.9 (5.68E-25) |
−42.7 (4.97E-44) |
−30.8 (6.35E-07) |
−28.9 (1.86E-16) |
−12.6 (0.01) |
−35.2 (3.15E-19) |
−2.5 (2.41E-05) |
−4.8 (1.40E-06) |
−23.9 (1.59E-03) |
| cg13356896 (BOLL) | −0.1 (0.95) |
−2.3 (4.68E-07) |
−2.5 (1.25E-03) |
−2.9 (0.02) |
−0.1 (0.90) |
−0.2 (0.78) |
−0.7 (0.26) |
−4.2 (1.98E-06) |
−3.7 (1.32E-03) |
−0.3 (0.67) |
−0.6 (0.28) |
−0.5 (0.31) |
−7.3 (1.55E-04) |
| cg11201447 (PVT1) | −23.5 (1.89E-23) |
−23 (1.04E-58) |
−10.3 (4.77E-06) |
−22.2 (2.91E-16) |
−28.2 (2.21E-29) |
−20.7 (1.35E-33) |
−25.9 (1.03E-42) |
−24.2 (2.37E-41) |
−27.9 (1.85E-25) |
−29.1 (8.68E-40) |
−22.9 (4.92E-22) |
−22.4 (1.04E-46) |
−16.8 (1.00E-13) |
| b) CpG located 1500 bp upstream of the transcriptional start site (TSS1500) | |||||||||||||
| cg13879483 (USP44) | −19.2 (6.69E-30) |
−13.9 (5.85E-43) |
−6.6 (2.09E-03) |
−20 (1.55E-51) |
−5.7 (2.20E-03) |
−10.6 (2.73E-04) |
−17.7 (6.73E-22) |
−26.1 (1.40E-29) |
−18.8 (3.40E-24) |
−15 (2.36E-10) |
−9.4 (1.46E-07) |
−15.6 (3.88E-07) |
NA |
| cg13591783 (ANXA1) | −32.4 (6.25E-60) |
−15.1 (1.13E-40) |
−17.2 (3.62E-15) |
−17.4 (2.21E-09) |
−16.6 (4.65E-35) |
−20.4 (8.02E-36) |
−12.8 (2.87E-11) |
−30.9 (1.83E-60) |
−24 (2.20E-22) |
−23.1 (5.26E-20) |
−27.1 (7.54E-22) |
−20.5 (4.43E-104) |
−21.2 (7.45E-13) |
| cg21039708 (OTX2)b | −0.3 (0.81) |
0.9 (0.16) |
NA | −0.5 (0.83) |
1.1 (0.17) |
−0.1 (0.74) |
0.2 (0.70) |
−7.6 (1.43E-03) |
−12.4 (0.02) |
−1.4 (0.03) |
−0.3 (0.47) |
0.3 (0.09) |
15.2 (5.72E-04) |
| c) CpG located in 5’ untranslated region (5’UTR) | |||||||||||||
| cg22399133 (CRYGD) | NA | NA | −0.1 (0.61) |
−5.3 (1.57E-07) |
−0.6 (0.17) |
NA | −0.2 (0.66) |
NA | −1.3 (0.09) |
−1.9 (0.50) |
−3.1 (9.78E-03) |
−0.8 (0.56) |
−16.2 (1.29E-16) |
| 2) Intragenic CpG related probes | |||||||||||||
| d) 1st Exon | |||||||||||||
| cg22674699 (HOXD9) | −6.1 (2.23E-05) |
−2.8 (6.75E-03) |
−5.8 (0.03) |
−1.6 (0.61) |
−10.3 (0.05) |
−12.5 (0.05) |
−0.6 (0.82) |
−14.3 (1.04E-06) |
−1 (0.79) |
−6.2 (0.01) |
−30.6 (1.21E-13) |
21.4 (2.10E-05) |
−4.7 (0.02) |
| e) Gene body | |||||||||||||
| cg27260772 (TFAP2B) | 7.3 (0.05) |
−39.5 (9.07E-77) |
−6.2 (5.75E-03) |
−5.8 (0.10) |
−7.2 (0.04) |
7.1 (0.07) |
2.9 (1.64E-03) |
−4.3 (0.17) |
−17.4 (6.65E-04) |
−0.1 (0.96) |
−0.4 (0.70) |
0.4 (0.41) |
−12.1 (0.05) |
| cg06766860 (GALNT9) | −1 (0.52) |
−0.1 (0.85) |
−3.6 (0.23) |
1.9 (0.32) |
−10.5 (5.03E-04) |
−16.2 (2.97E-05) |
−0.6 (0.68) |
0.8 (0.67) |
−0.5 (0.82) |
−6.4 (0.02) |
7.4 (1.60E-05) |
−3.2 (0.35) |
15.8 (0.06) |
| cg07974511 (OTX1) | 18.6 (4.74E-10) |
33 (3.32E-39) |
34.7 (9.50E-13) |
36.5 (2.75E-11) |
15.7 (3.47E-12) |
24.6 (4.10E-27) |
23.3 (2.09E-10) |
19.8 (1.62E-07) |
43.2 (7.66E-11) |
39 (2.99E-16) |
17.3 (2.80E-05) |
13.1 (1.98E-25) |
43 (1.81E-12) |
| cg00817367 (GRASP) | −3.4 (6.46E-03) |
−3.7 (1.61E-04) |
−10.5 (3.46E-07) |
−4.6 (1.08E-03) |
−6 (8.14E-04) |
−6.1 (1.05E-03) |
−8 (1.87E-06) |
−4.4 (8.57E-04) |
−7.8 (7.66E-04) |
−8.3 (1.69E-05) |
−19.5 (2.25E-29) |
−3.8 (0.46) |
−6.5 (2.25E-03) |
| cg18236877 (CASZ1) | NA | NA | 5.1 (0.10) |
5.2 (0.29) |
−14 (7.80E-17) |
−8.6 (6.50E-04) |
NA | −3.2 (0.47) |
NA | NA | −7 (3.05E-14) |
−11.2 (2.98E-21) |
NA |
| cg17774559 (IRX4) | 28.1 (3.10E-10) |
20.6 (1.48E-04) |
−3 (0.20) |
11.4 (0.25) |
2.2 (0.12) |
0.6 (0.43) |
0.3 (0.51) |
9.1 (5.32E-04) |
95.2 (1.56E-15) |
−0.8 (0.76) |
40.4 (9.23E-06) |
1.3 (0.04) |
6.6 (0.19) |
| cg14861089 (TLX1) | 16.6 (9.55E-09) |
−2.2 (0.32) |
24.7 (1.15E-06) |
−6.1 (0.09) |
9.7 (1.37E-07) |
8.5 (4.54E-07) |
26.1 (2.20E-22) |
10.4 (8.30E-07) |
11.9 (8.13E-03) |
20.1 (4.07E-07) |
4.4 (2.46E-03) |
−3.7 (0.14) |
NA |
| cg03301058 (GABRR2) | −1.7 (0.47) |
0.4 (0.73) |
−3.7 (0.47) |
−2.4 (0.58) |
−3.2 (0.42) |
6.7 (0.02) |
−2.2 (0.08) |
1.4 (0.62) |
2.1 (0.48) |
−2.5 (0.32) |
−0.2 (0.89) |
6 (1.72E-14) |
−2.2 (0.17) |
| cg09296001 (SND1) | −1.3 (0.05) |
0.1 (0.79) |
2.9 (0.07) |
2.2 (3.51E-05) |
3.1 (1.94E-04) |
0.7 (0.39) |
1.6 (0.25) |
2 (1.29E-04) |
3.8 (3.64E-06) |
0.1 (0.90) |
8.4 (1.68E-24) |
5.1 (6.50E-03) |
2.3 (5.51E-03) |
| 3) Intergenic CpG related probes | |||||||||||||
| cg10903903 (HIST1H2BL)a | 0.6 (0.75) |
5.4 (2.83E-07) |
6.4 (0.01) |
5.5 (1.72E-03) |
1.8 (0.31) |
3.2 (0.03) |
−2.4 (0.02) |
2.2 (0.15) |
5.4 (0.01) |
3.2 (0.02) |
NA | NA | 6.2 (0.20) |
| cg06962177 (FOXD3) a | −6.2 (0.05) |
23.9 (5.68E-13) |
−5.2 (0.05) |
24.8 (2.25E-04) |
0.6 (0.59) |
3.4 (2.75E-03) |
2.5 (0.03) |
24.7 (1.72E-08) |
45.5 (6.14E-08) |
−6.6 (0.06) |
30.1 (1.14E-17) |
−1.4 (0.10) |
17.8 (6.92E-06) |
| cg11536474 (OTX1) a | 12.4 (1.52E-07) |
23.7 (3.24E-25) |
26.7 (7.22E-31) |
36.9 (7.93E-11) |
17.2 (1.03E-13) |
26.1 (2.07E-15) |
17.3 (5.67E-12) |
21.4 (3.63E-11) |
41.2 (4.80E-10) |
23.9 (7.69E-13) |
17.2 (6.66E-06) |
21.5 (5.59E-17) |
37.6 (1.47E-17) |
| cg08364561 (RCN1)a | 8 (8.53E-04) |
3.6 (7.93E-03) |
3 (0.32) |
13.7 (4.47E-05) |
3 (0.04) |
3.2 (0.08) |
6.1 (5.19E-07) |
−1.5 (0.32) |
26.1 (2.82E-10) |
−0.6 (0.92) |
−0.7 (0.34) |
−3.8 (2.20E-05) |
14.6 (2.12E-07) |
| cg22797031 (PRRX1) a | −0.6 (0.86) |
−3.4 (0.04) |
−26.1 (6.80E-07) |
−12 (4.06E-03) |
7.5 (0.02) |
NA | 4.3 (0.03) |
1.9 (0.41) |
7.2 (0.09) |
6.7 (0.06) |
−3.7 (0.02) |
3.8 (0.08) |
8.5 (0.41) |
| cg13539545 (SIX6) a | 0.1 (0.60) |
1.3 (0.01) |
0 (0.76) |
−0.6 (0.71) |
0.2 (0.67) |
−0.1 (0.62) |
−0.1 (0.49) |
1.1 (0.40) |
1.2 (0.50) |
−1.6 (9.76E-03) |
3.5 (0.21) |
−0.1 (0.68) |
3.7 (0.07) |
| cg17754510 (TFAP2A) b | 17.4 (2.77E-10) |
23.9 (2.70E-36) |
10.1 (0.03) |
8.7 (1.20E-14) |
59.7 (1.02E-33) |
88.1 (3.05E-55) |
37.2 (3.45E-22) |
29.8 (1.19E-29) |
2.1 (0.20) |
31.4 (4.34E-16) |
12.3 (1.55E-05) |
33.8 (4.09E-04) |
45 (6.02E-10) |
| cg01518607 (PRSS16) a | 8 (1.03E-04) |
16.4 (4.06E-08) |
6.1 (7.62E-06) |
8.2 (7.77E-04) |
8.3 (0.02) |
−1.2 (0.76) |
5.2 (0.13) |
11.4 (3.08E-09) |
13.7 (4.34E-09) |
17.6 (1.52E-07) |
−4.9 (0.08) |
2.1 (0.02) |
22.1 (1.02E-07) |
| cg09887059 (LHX5) a | 7.8 (0.03) |
−0.5 (0.59) |
−2.4 (0.52) |
−18.1 (2.50E-03) |
0.1 (0.69) |
2.1 (0.06) |
0.2 (0.62) |
−5.5 (0.14) |
0.1 (0.98) |
−7.7 (0.05) |
1.4 (0.10) |
−0.2 (0.80) |
−0.2 (0.96) |
| cg18177414 (KRBA1) a | 6.2 (2.65E-06) |
2.7 (2.69E-03) |
NA | 4.2 (6.23E-03) |
5.5 (0.01) |
−2.3 (0.09) |
6.6 (9.96E-04) |
2.1 (0.06) |
3.3 (0.05) |
−14.9 (2.19E-13) |
1.6 (0.23) |
2.4 (0.13) |
0.2 (0.92) |
| cg22620090 (LIN28B)b | 7.6 (0.16) |
1.5 (0.34) |
1.1 (0.75) |
−1.2 (0.80) |
2.5 (0.09) |
1.5 (0.14) |
NA | 5.6 (0.21) |
8.5 (0.16) |
NA | 4.2 (1.03E-04) |
0.5 (0.03) |
36.2 (2.41E-03) |
| cg04456219 (AHR) a | −26.2 (6.27E-12) |
−2 (0.08) |
3 (0.56) |
−6.5 (5.17E-03) |
−16.2 (5.20E-13) |
−14.3 (2.22E-07) |
1.4 (0.38) |
−3.8 (0.02) |
1 (0.46) |
−15.3 (5.68E-10) |
4.8 (0.04) |
−11 (1.51E-17) |
−2.9 (0.21) |
| cg08443563 (MERTK) a | 19.8 (7.35E-04) |
3.3 (0.05) |
41.4 (5.10E-04) |
48.1 (5.29E-07) |
11.1 (4.94E-04) |
3.4 (0.13) |
NA | 11.9 (4.48E-05) |
20.9 (5.19E-06) |
15.2 (1.33E-04) |
1.6 (0.21) |
0.7 (0.44) |
5.9 (0.06) |
| cg18103859 (MNAT1) a | 4.7 (2.68E-04) |
3.3 (3.84E-08) |
NA | 6.6 (1.22E-07) |
2.4 (2.69E-03) |
0.6 (0.40) |
1.5 (0.03) |
5.3 (1.34E-05) |
11.6 (5.91E-11) |
4.9 (1.43E-07) |
3.5 (1.85E-03) |
1.7 (0.04) |
4.1 (2.77E-03) |
| cg03498081 (DYNLRB2) a | −3.4 (0.56) |
13.2 (1.27E-10) |
−1.3 (0.53) |
4.9 (0.35) |
−15.2 (3.87E-07) |
−12.5 (5.17E-08) |
4.6 (0.01) |
4.7 (0.48) |
6 (0.44) |
28.2 (7.32E-09) |
7.1 (0.03) |
8.8 (2.18E-08) |
14.5 (9.93E-03) |
| cg02389317 (ZDHHC17)a | −0.2 (0.92) |
2.3 (6.77E-04) |
2 (0.26) |
−4.7 (0.02) |
−2.4 (0.04) |
−0.5 (0.71) |
0.4 (0.72) |
0.1 (0.90) |
1.8 (0.13) |
4.4 (0.09) |
0.9 (0.34) |
−2.4 (4.82E-07) |
−0.2 (0.91) |
% Δ: Percent change in gene expression per 10% increase of methylation. Both gene expression and methylation were adjusted for age, sex, cancer stage, percent of tumor cells on the sample, and fraction of genome altered. The residuals of both linear models were used for the calculations.
Nearest gene.
CpG is annotated to the antisense transcript
Discussion
Quantifying the extent of DNA methylation alterations allows the comparison of different data sets from a broader perspective. Our analysis revealed different levels of methylation dysregulation in different cancer types; however, a similar pattern of methylation dysregulation in the different genomic contexts was observed. Globally, the amount of dysregulation was lower in CpG island regions compared with other genomic regions, such as CpG island shores and shelves, and those in less CpG dense regions across tumor types. A group of four tumor types (thyroid, kidney papillary and clear cell carcinomas, and pancreatic adenocarcinoma), all from organs with primary or secondary endocrine function, showed a lower level dysregulation at the subject level than the other cancer types. This finding was further supported when analyzing the specific loci dysregulation, in which these cancer types cluster together; three of the four showed a pattern of loci dysregulation associated to specific metabolic genes, proto-oncogenes and oncogenes. The most dysregulated areas for these tumors were located on open sea, shores, and shelves areas. In contrast, age was consistently associated with MDI in breast cancer and hepatocellular carcinoma globally and across the different genomic contexts. Age was also associated to dysregulation in the CpG islands for colon, prostate, and thyroid cancers. Again, BRCA, PRAD, COAD, and LIHC top loci (mainly CpG island probes) clustered together. Among those tumors clustered in class 1 of RPMM (BLCA, ESCA, HNSC, LUAD and LUSC) the subject specific covariates did not provide common patterns, but the loci specific analyses showed a common cluster related to dysregulation of both CpG islands and open sea areas. Endometrial cancer was the only tumor not associated with the fraction of genome altered, while at the loci level showed a pattern similar to the tumors of class 1.
An alternative index of altered DNA methylation from Yang et al used a DNA methylation “instability” index to quantify aberrant DNA methylation in cancer.22 This approach summarized two different z-scores using the probes according to those located on the CGI promoters (hyperZ score) and the other using the probes on the open sea (hypoZ score). However, this index does not take into account shore regions, areas where we also observe high levels of dysregulation. In particular, shore dysregulation have been previously associated with increased variability in gene expression in cancer more than CGI.23 In our approach, we investigated both the overall methylation dysregulation level across tumors and performed a stratified analysis of DNA methylation dysregulation by genomic context which revealed striking differences between genomic context.
Our results are consistent with previous reports of several DNA methylation phenotypes for the different cancer types. Specific reported mutations associated with differential methylation (CIMP phenotypes) differ by cancer subtype, which precludes an integrative analysis of specific mutation signatures for all the samples.13,24-33 In addition, depending of the cancer type, DNA methylation is related to cancer stage,30 lifestyle/environmental factors,29,31 germline/somatic mutations,13,25,27,31 microsatellite instability (MSI phenotypes),24,33 miRNA risk phenotypes,26 synchronic copy number alterations changes,28 and has partial overlapping with other molecular signatures (e.g., tumor hormone receptor status).32 The use of the estimated cell purity was previously related to altered transcriptomic signals,16 although this could be also true for the DNA methylation signal these estimates have not been used in previous pan-cancer analyses. We observed an improvement using the estimated cell purity (based on the slide readings) in several of our models. We explored using inference-based purity estimations with DNA methylation data though observed some inconsistent results compared with histopathologic slide readings. We therefore used histopathologic estimates in our models, though in cases where such data are not available, inference-based approaches are expected to have utility.
Our work adds the integration of an agnostic approach to evaluate the non-CGI methylation status using information derived from the Illumina 450K platform. In the previous integrative reports, only a few integrated non-CGI information,25,26 and several used the older 27K platform or integrated only the common probes between 27K and 450K platforms.28,33 Moreover, as most of the previous integrative analyses were focused on specific pathways some of them only relied on promoter DNA methylation. One limitation for the generalizability of our findings is the lack of some common epidemiological information between the data sets (i.e., smoking status), this precludes an integrative analysis of some exposures as they cannot be adjusted for in all the data sets. On the other hand, the absence of these tumor specific covariates is a caveat that could produce some uncontrolled confounding in the common models.
The use of a summary dysregulation index by loci has not been previously reported. The ranked algorithm selected the common probes across several cancers, this approach was intended initially to perform gene expression meta-analyses in which technical variability might preclude direct comparisons or pooled analyses.34 The main disadvantage of this approach is that the cut-offs for the selection process rely on the user. For smaller databases the use of graphic approaches to select an appropriate delta is advised, but in longer lists, as those derived from microarrays, this process is burdensome and not completely reproducible. On the other hand, a mathematical limitation is given by the distance selected, and the number of lists that are compared which is computationally intensive. However, using a different range of deltas the top list of the CEMC list was quite robust, the delta only affected the inclusion of some additional probes under the cut-off. Biologically, the genes associated on the CEMC list are consistent with common pathways among the cancer types, and some areas have also been previously reported in relation to DNA methylation changes. When using the complete list for the GSEA enrichment analysis, the pathways were associated with multiple cancer pathways including epigenetic histones modifications, Polycomb repression complexes. The finding of a consistent association between methylation levels and gene expression for those CpGs located on the gene coding areas is also encouraging for future analyses. The dysregulation of enhancer related areas in non-coding areas of the genome, and the cis and trans regulation of distant genes for those CpGs located on the open sea should be explored further in the future and goes beyond the scope of this paper.
The use of a summary index instead of the original β-values has several strengths and limitations. The use of summary indices reduces the number of tested comparisons and outperforms the delta β in terms of global information provided on the genomic context. The MDI follow a Berkson error modeling35 in which the cumulative deviation from the mean in a genomic context increases the sample power to find differences in terms of the genomic context. Furthermore, if a few outliers were present in the samples, the median constrains the differences and the MDI might attenuate the effect of these outliers. This is an advantage and increases the comparability for a pan-cancer approach. Nevertheless, with the use of summary measures the granularity of the data (locus specific methylation by subject) is lost and therefore the results lose precision. When the aim is to obtain specific CpG site changes using the β-values or M-values might provide better answers, but, as a trade-off, the use of individual CpG increases the probability of estimates biased to the null (classical measurement error).
Conclusions
This work contributes to the understanding of the impact of epigenetic alterations from a pan-cancer perspective. The pattern of DNA methylation dysregulation showed a higher impact of CpG-poor areas of the genome across tumor types. In addition, the most dysregulated loci across cancer types identified common clusters across cancer types that may have implications for future treatment and prevention measures. The full extent of functional implications of the integrative pan-cancer somatic alteration portraits presented here will require additional investigation.
Methods
Cancer data sets
Fourteen cancers: bladder (BLCA), breast (BRCA), renal clear cell (KIRC), renal papillary cell (KIRP), esophageal (ESCA), thyroid (THCA) and endometrial carcinomas (UCEC); colon (COAD), lung (LUAD), pancreatic (PAAD), and prostate (PRAD) adenocarcinomas; head and neck (HNSC) and lung (LUSC) squamous cell carcinomas; and hepatocellular carcinomas (LIHC), were downloaded from The Cancer Genome Atlas (TCGA) public database and are available from the TCGA legacy repository (https://gdac.broadinstitute.org/). Only those tumor types in which there were samples representing the primary tumor and at least 10 normal adjacent samples were included. For cases where several samples from the same tumor were processed and included on the database, only the first listed was included in subsequent statistical analyses.
Clinical, histological, and additional molecular information
For every case, we obtained the clinical information from TCGA [age, gender, and pathological stage according the latest available American Joint Committee on Cancer (AJCC) classification]. Tumor stage was recoded as early (stages I and II) and late (stages III and IV). For comparability, we excluded the cancer specific sub-classification. Although copy number alterations do not bias DNA methylation signals,36 biologically they are drivers of other cancer alterations which may alter DNA methylation levels, and were therefore included in our models. Information on the fraction of the genome with copy number alterations (FGA) and mutation count burden (MCB) were retrieved from cBioportal.37,38 FGA was measured using Affymetrix SNP arrays and corresponds to the fraction of the genome affected by copy number alterations, which is equivalent to the number of bases in segments with mean log2 greater than 0.2 or smaller than −0.2 divided by the number of bases in all segments profiled by the array. MCB, defined as the total number of non-synonymous substitutions in exome sequencing was excluded from multivariate analyses because not all the data sets included this information for the DNA methylation samples (COAD, ESCA). As the sample purity may alter the DNA methylation landscape16 we retrieved the estimated tumor cell percentage from the histologic database in TCGA. Given that some of the slide samples were read per duplicate (bottom and top), the mean reported percentage of tumor cells was used for the analyses. As a sensitivity analysis, we compared our results using a DNA methylation estimate of cell purity (InfiniumPurify) instead of the slide reported percentage.20 Finally, for some selected loci, we retrieved level 3 data for gene expression from cBioportal to be analyzed with loci specific methylation changes. In total, clinical data from 5592 tumors and 701 adjacent normal tissues were considered in our analyses.
Preprocessing and quality control of DNA methylation data sets
The Level 1 intensity data files (i.e., .idat) derived from the Infinium HumanMethylation450 BeadChip (Illumina, Inc., San Diego, CA) were imported and preprocessed using the RnBeads package in R.39 DNA methylation β-values were estimated based on the measured intensities of the 2-paired channels (i.e., red and green) and computed as the ratio of the methylated probe intensity, divided by the sum of the unmethylated plus the methylated intensities signals plus an offset (usually 100).40 β-values range between 0 and 1, and can be interpreted as the proportion of methylated alleles at a specific CpG site. β-values were background corrected using methylumi-noob41 and normalized with a functional normalization procedure42 to increase comparability across data sets and to reduce potential batch effects. Probes marked as Non-CpG, CpG loci on the X and Y chromosomes, and those previously documented as polymorphic or cross-reactive, were excluded from subsequent analyses.43 The Greedycut hierarchical algorithm was applied to exclude unreliable probes and samples.39,44 Briefly, the Greedycut is an iterative algorithm that filters out probes or samples with the highest fraction of unreliable measurements removing/entering one sample/probe per iteration (P detection >0.05). After every iteration, the matrices with and without the unreliable probes or samples were compared using the expression sensitivity +1 – false positive rate. Those further away from the diagonal (highest area under the curve) were retained.
Genome-wide DNA methylation dysregulation index
To evaluate dysregulation of tumor DNA methylation compared with normal tissue we calculated the methylation dysregulation index (MDI) as described in O'Sullivan et al.17 Briefly, the MDI measure represents the cumulative departure from normal DNA methylation in a CpG locus-specific manner calculated by summing the absolute difference in DNA methylation β-values at each CpG between each tumor sample and the median β-value for each CpG across all normal-adjacent samples specific to each tumor tissue type. MDI was calculated by subject, and was summed across all the CpGs interrogated in a subject and then divided by the total number of CpGs (subject-specific MDI, sMDI). To better reflect scale MDI was multiplied by 100. The MDI represents the average change in β-value per CpG in the tumor sample compared with adjacent-normal tissue. Therefore, a MDI value close to 0 suggests a similar methylation profile to its component normal while increasing levels of MDI indicates that the DNA methylome has been deregulated to a greater extent. To assess context-specific relationships between methylation dysregulation across different cancer types, we calculated specific methylation dysregulation indices for each genomic context (gcMDI) in the subject (i.e., CGI, CGI shores, CGI shelves, and open sea). Lastly, to summarize the average locus specific DNA methylation change across all the subjects with the same tumor type we calculated a tumor specific MDI, tMDI. The tMDI represents the average departure from normal of a specific CpG site within a tumor type. The tMDI was calculated for each tumor type and then ranked, to determine if there were specific CpGs that were commonly altered across cancer types.
Statistical analysis
Descriptive statistics (mean t-test, Mann-Whitney, χ2, and Fisher tests) were used to summarize the subject level information. FGA was analyzed as a proportion and modeled per 10% increase in multivariable models. MCB was Log10 transformed for exploratory analyses. Spearman rank test was used to evaluate correlations between continuous covariates. Clustering analyses of the different relationships between the sMDI, gcMDI and the top loci of the tMDI were performed using unsupervised hierarchical clustering (Euclidean distance and complete linkage) by each cancer type. In addition, for the tMDI a semi-supervised recursively partitioned mixture model (RPMM),21 a hierarchical model-based clustering methodology that assumes an underlying mixture of β-distributions was used to confirm the results of the unsupervised clustering. Linear regression models were used to model the relationship between sMDI and gcMDI, and the clinical data of each subject stratified by each tumor type. Multivariable linear regression models were adjusted for age, gender (except UCEC and PRAD), cancer stage, FGA, and tumor cell percentage.
The CpG/methylation probes per tumors were ranked according to their tMDI value. During the Greedycut preprocessing different probes were excluded due to quality issues in different cancers, those missing probes were kept as missing on the common list. Posteriorly, the cancer ranked lists were merged and a global rank was calculated using the TopKLists R package. The k top ranked CpG sites (those in which the slope was less steep) were analyzed using the moderate-deviation-based inference for random degeneration in paired rank lists, and from them a subset of highly concordant probes were derived from the Cross Entropy Monte Carlo (CEMC) algorithm.34,45 For the ranking, a maximal distance on the ranking (delta) of 50, a pilot subset of 1000, a threshold of 21% (at least present in 3 lists within the delta) were used to determine the top k sites to be evaluated. The gene symbols associated with the probes of the tMDI were annotated using the most recent Illumina files; the annotated genes were updated using mygene.46 Those probes of the open sea were annotated to the nearest gene. For the CEMC highly concordant probes obtained using the tMDI we explored the relationship between DNA methylation and gene expression: the residuals of the Log2 transcripts of the gene expressed and the residuals of the β-values were obtained from multivariable linear regression adjusted for the covariates described above for the sMDI models. Finally, the complete top ranked list was enriched using the collections 2 (curated database) and 6 (oncogenic signatures) of GSEA (http://software.broadinstitute.org/gsea/msigdb/collection_details.jsp) applying a test adapted for the 450K microarray based on Wallenius’ noncentral hypergeometric distribution included on missMethyl.47 Statistical significance was considered as P-values < 0.05, or its equivalent after Bonferroni correction for multiple comparisons (e.g., ≤ 0.01 for the multivariable linear models), or FDR<0.05 (Benjamini-Hochberg) for the enrichment analyses.
Ethics approval and consent to participate
The current analyses were based on public information from The Cancer Genome Atlas. The prospective participants provided an informed consent about the purpose of the project, samples, and medical information collected and coding to preserve the anonymity of the participants on the public databases. They received information regarding the potential benefits and risks for their participation plus specific information according to the IRB of the participating centers. The TCGA project is framed on the genetic information nondiscrimination act (GINA), and all the databases that could contain sensible personal information are protected from general access.
Supplementary Material
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by the National Institutes of Health (NIH) grant number R01 DE022772 to B.C.C. The research reported in this publication was also supported by the Center for Molecular Epidemiology COBRE program with grant funds from the National Institute of General Medical Sciences (NIGMS) of the NIH under award number P20 GM104416. The NIH grant 1KL2TR000119 supported DCK.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11. Lyon, France: International Agency for Research on Cancer, 2013 [accessed 2017 February 02] http://globocan.iarc.fr [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin 2016; 66:7-30; PMID:26742998; https://doi.org/ 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network, Weinstein JN, Collisson Ea, Mills GB, Shaw KRM, Ozenberger Ba, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet 2013; 45:1113-20; PMID:24071849; https://doi.org/ 10.1038/ng.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghazanfar S, Yang JYH. Characterizing mutation-expression network relationships in multiple cancers. Comput Biol Chem 2016; 63:73-82; PMID:26935398; https://doi.org/ 10.1016/j.compbiolchem.2016.02.009 [DOI] [PubMed] [Google Scholar]
- 5.Ock C-Y, Keam B, Kim S, Lee J-S, Kim M, Kim TM, Jeon YK, Kim D-W, Chung DH, Heo DS. Pan-cancer immunogenomic perspective on the tumor microenvironment based on PD-L1 and CD8 T cell infiltration. Clin Cancer Res 2016; 22:2261-70; PMID:26819449; https://doi.org/ 10.1158/1078-0432.CCR-15-2834 [DOI] [PubMed] [Google Scholar]
- 6.Andor N, Graham TA, Jansen M, Xia LC, Aktipis CA, Petritsch C, Ji HP, Maley CC. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat Med 2015; 22:105-13; PMID:26618723; https://doi.org/ 10.1038/nm.3984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao M, Liu Y, Qu H. Expression of epithelial-mesenchymal transition-related genes increases with copy number in multiple cancer types. Oncotarget 2016; 7:24688-99; PMID:27029057; https://doi.org/ 10.18632/oncotarget.8371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Witte T, Plass C, Gerhauser C. Pan-cancer patterns of DNA methylation. Genome Med 2014; 6:66; PMID:25473433; https://doi.org/ 10.1186/s13073-014-0066-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luczak MW, Jagodziński PP. The role of DNA methylation in cancer development. Folia Histochem Cytobiol 2006; 44:143-54; PMID:16977793 [PubMed] [Google Scholar]
- 10.Christensen BC, Houseman EA, Marsit CJ, Zheng S, Wrensch MR, Wiemels JL, Nelson HH, Karagas MR, Padbury JF, Bueno R, et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS Genet 2009; 5:e1000602; PMID:19680444; https://doi.org/ 10.1371/journal.pgen.1000602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rakyan VK, Down Ta, Maslau S, Andrew T, Yang T-P, Beyan H, Whittaker P, McCann OT, Finer S, Valdes AM, et al. Human aging-associated DNA hypermethylation occurs preferentially at bivalent chromatin domains. Genome Res 2010; 20:434-9; PMID:20219945; https://doi.org/ 10.1101/gr.103101.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, Berman BP, Pan F, Pelloski CE, Sulman EP, Bhat KP, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 2010; 17:510-22; PMID:20399149; https://doi.org/ 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network, Linehan WM, Spellman PT, Ricketts CJ, Creighton CJ, Fei SS, Davis C, Wheeler DA, Murray BA, Schmidt L, et al. Comprehensive molecular characterization of papillary renal-cell carcinoma. N Engl J Med 2016; 374:135-45; PMID:26536169; https://doi.org/ 10.1056/NEJMoa1505917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gevaert O, Tibshirani R, Plevritis SK. Pancancer analysis of DNA methylation-driven genes using MethylMix. Genome Biol 2015; 16:17; PMID:25631659; https://doi.org/ 10.1186/s13059-014-0579-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang X, Gao L, Zhang S. Comparative pan-cancer DNA methylation analysis reveals cancer common and specific patterns. Brief Bioinform 2016; pii:bbw063; PMID:27436122; https://doi.org/ 10.1093/bib/bbw063 [DOI] [PubMed] [Google Scholar]
- 16.Aran D, Sirota M, Butte AJ. Systematic pan-cancer analysis of tumour purity. Nat Commun 2015; 6:8971; PMID:26634437; https://doi.org/ 10.1038/ncomms9971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Sullivan DE, Johnson KC, Skinner L, Koestler DC, Christensen BC. Epigenetic and genetic burden measures are associated with tumor characteristics in invasive breast carcinoma. Epigenetics 2016; 11(5):344-53; PMID:27070496; https://doi.org/ 10.1080/15592294.2016.1168673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Cancer Institute SEER cancer statistics review, 1975-2013. Bethesda, MD: National Cancer Institute, 2013. PMID:27078145; https://doi.org/ 10.1001/jamaoncol.2016.0386 [DOI] [Google Scholar]
- 19.Nikiforov YE, Seethala RR, Tallini G, Baloch ZW, Basolo F, Thompson LDR, Barletta JA, Wenig BM, Al Ghuzlan A, Kakudo K, et al. Nomenclature revision for encapsulated follicular variant of papillary thyroid carcinoma: A paradigm shift to reduce overtreatment of indolent tumors. JAMA Oncol 2016; 15213:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng X, Zhang N, Wu H-J, Wu H. Estimating and accounting for tumor purity in the analysis of DNA methylation data from cancer studies. Genome Biol 2017; 18:17; PMID:28122605; https://doi.org/ 10.1186/s13059-016-1143-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koestler DC, Christensen BC, Marsit CJ, Kelsey KT, Houseman EA. Recursively partitioned mixture model clustering of DNA methylation data using biologically informed correlation structures. Stat Appl Genet Mol Biol 2013; 12:225-40; PMID:23468465; https://doi.org/ 10.1515/sagmb-2012-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Z, Jones A, Widschwendter M, Teschendorff AE. An integrative pan-cancer-wide analysis of epigenetic enzymes reveals universal patterns of epigenomic deregulation in cancer. Genome Biol 2015; 16:140; PMID:26169266; https://doi.org/ 10.1186/s13059-015-0699-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer 2013; 13:497-510; PMID:23760024; https://doi.org/ 10.1038/nrc3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas Research Network, Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson aG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013; 497:67-73; PMID:23636398; https://doi.org/ 10.1038/nature12113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cancer Genome Atlas Research Network The molecular taxonomy of primary prostate cancer. Cell 2015; 163:1011-25; PMID:26544944; https://doi.org/ 10.1016/j.cell.2015.10.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Research Network Integrated genomic characterization of papillary thyroid carcinoma. Cell 2014; 159:676-90; PMID:25417114; https://doi.org/ 10.1016/j.cell.2014.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511:543-50; PMID:25079552; https://doi.org/ 10.1038/nature13385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489:519-25; PMID:22960745; https://doi.org/ 10.1038/nature11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cancer Genome Atlas Research Network Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014; 507:315-22; PMID:24476821; https://doi.org/ 10.1038/nature12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cancer Genome Atlas Research Network Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013; 499:43-9; PMID:23792563; https://doi.org/ 10.1038/nature12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cancer Genome Atlas Network Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015; 517:576-82; PMID:25631445; https://doi.org/ 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature 2012; 490:61-70; PMID:23000897; https://doi.org/ 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012; 487:330-7; PMID:22810696; https://doi.org/ 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall P, Schimek MG. Moderate-deviation-based inference for random degeneration in paired rank lists. J Am Stat Assoc 2012; 107:661-72; https://doi.org/ 10.1080/01621459.2012.682539 [DOI] [Google Scholar]
- 35.Berkson J. Are there two regressions? J Am Stat Assoc 1950; 45:164-80; https://doi.org/ 10.1080/01621459.1950.10483349 [DOI] [Google Scholar]
- 36.Houseman EA, Christensen BC, Karagas MR, Wrensch MR, Nelson HH, Wiemels JL, Zheng S, Wiencke JK, Kelsey KT, Marsit CJ. Copy number variation has little impact on bead-array-based measures of DNA methylation. Bioinformatics 2009; 25:1999-2005; PMID:19542153; https://doi.org/ 10.1093/bioinformatics/btp364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2:401-4; PMID:22588877; https://doi.org/ 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6:pl1; PMID:23550210; https://doi.org/ 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assenov Y, Müller F, Lutsik P, Walter J, Lengauer T, Bock C. Comprehensive analysis of DNA methylation data with RnBeads. Nat Methods 2014; 11:1138-40; PMID:25262207; https://doi.org/ 10.1038/nmeth.3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, Lin SM. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 2010; 11:587; PMID:21118553; https://doi.org/ 10.1186/1471-2105-11-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Triche TJ, Weisenberger DJ, Van Den Berg D, Laird PW, Siegmund KD. Low-level processing of Illumina Infinium DNA methylation beadarrays. Nucleic Acids Res 2013; 41:e90; PMID:23476028; https://doi.org/ 10.1093/nar/gkt090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fortin J, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, Greenwood C, Hansen KD. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 2014; 15:503; PMID:25599564; https://doi.org/ 10.1186/s13059-014-0503-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, Gallinger S, Hudson TJ, Weksberg R. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013; 8:203-9; PMID:23314698; https://doi.org/ 10.4161/epi.23470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hazewinkel M. Greedy algorithm. Encyclopedia of Mathematics 2014; Available from: http://www.encyclopediaofmath.org/index.php?title = Greedy_algorithm&oldid = 34496 [Google Scholar]
- 45.Lin S, Ding J. Integration of ranked lists via cross entropy Monte Carlo with applications to mRNA and microRNA studies. Biometrics 2009; 65:9-18; PMID:18479487; https://doi.org/ 10.1111/j.1541-0420.2008.01044.x [DOI] [PubMed] [Google Scholar]
- 46.Xin J, Mark A, Afrasiabi C, Tsueng G, Juchler M, Gopal N, Stupp GS, Putman TE, Ainscough BJ, Griffith OL, et al. High-performance web services for querying gene and variant annotation. Genome Biol 2016; 17:91; PMID:27154141; https://doi.org/ 10.1186/s13059-016-0953-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina's HumanMethylation450 platform. Bioinformatics 2016; 32:286-8; PMID:26424855; https://doi.org/ 10.1093/bioinformatics/btv560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.