ABSTRACT
Abundant repetitive DNA sequences are an enigmatic part of the human genome. Despite increasing evidence on the functionality of DNA repeats, their biologic role is still elusive and under frequent debate. Macrosatellites are the largest of the tandem DNA repeats, located on one or multiple chromosomes. The contribution of macrosatellites to genome regulation and human health was demonstrated for the D4Z4 macrosatellite repeat array on chromosome 4q35. Reduced copy number of D4Z4 repeats is associated with local euchromatinization and the onset of facioscapulohumeral muscular dystrophy. Although the role other macrosatellite families may play remains rather obscure, their diverse functionalities within the genome are being gradually revealed. In this review, we will outline structural and functional features of coding and noncoding macrosatellite repeats, and highlight recent findings that bring these sequences into the spotlight of genome organization and disease development.
KEYWORDS: DNA repeats, epigenetics, genome organization, macrosatellite repeats, noncoding genome
Introduction
Recently, there has been substantial progress in understanding genome content and what is considered a functional DNA sequence, moving away from the classical dogma centered on protein coding genes. Even though repeats have been traditionally considered as junk DNA because their functionality was elusive, several repeat families have been recognized as important players in genome structure, evolution and diversity.1,2 Nevertheless, DNA repeats still remain one of the most puzzling components of the genome. As a constitutive part of the genome, these sequences are replicated and maintained through the individual's successive generations. They fulfill the concept of double helix “selfish” replicators, having their own survivability pressure during evolution.3,4,5 Leaving aside their own existence as individual “parasite” replicator entities, the scope of this review is to describe their structural and functional features in the context of their host human genome and their impact on disease development.
The human genome contains a large portion of repetitive DNA. While the current version of RepeatMasker identifies around 56% of the human genome as repetitive,6 recent studies propose even higher numbers, with estimates of up to 69%.7 The vast majority of repeats are still poorly investigated due to extensive computational and experimental limitations. Repeat-rich regions are difficult to align and assemble, thus they are frequently absent from the reference genome or not placed in their correct genomic context.8 Furthermore, high-throughput genome-wide studies, such as ChIP-seq and RNA-seq, which became essential tools in molecular biology research, are limited in analyzing repeat-derived reads as they present ambiguities in alignment to the reference genome.9 For all these reasons, the study of the genomic implications of repeat alterations at both DNA and RNA level is a difficult task. Although these drawbacks have significantly hindered the progress in understanding the role of repeats in genome stability and disease development, repeated DNA sequences are gaining attention as research on the noncoding genome is steadily growing.
Continuous work trying to illuminate the role of repeats in the genome has put forward new perspectives on the mechanisms by which they impact genome stability. Based on the pattern of their distribution, DNA repeats can be classified as interspersed repeats or tandem repeats10 (Fig. 1). Interspersed repeats are dispersed across the genome and include retro(pseudo)genes, tDNA, transposons, and local repeats. Tandem repeats are organized in a head-to-tail orientation and include ribosomal DNA (rDNA) and satellite repeats. Based on the size of each repeat unit, satellite repeats can be further divided in microsatellites with units of 1 to 6–10 bp, minisatellites with repeat units from 10 bp to few hundred bp, and macrosatellites with repeat units of several kb in length.10,11
Figure 1.
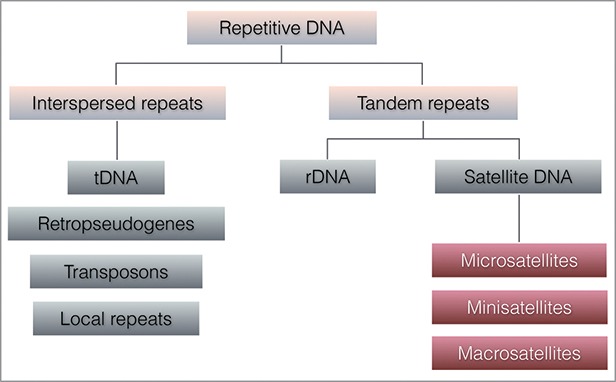
Repetitive DNA in the human genome. The diagram shows various classes of DNA repeats in the human genome, classified according to their pattern of occurrence.
It is estimated that satellite repeats cover around 3% of the human genome,9 with microsatellites being the most abundant. Both, micro- and mini-satellites, display notable instability and dynamics. Microsatellites are altered with a relative high frequency both ontogenetically, and especially phylogenetically, during DNA replication in mitosis due to slippage by strand misalignment.12
Minisatellites also exhibit extreme polymorphisms in the form of copy number, length, and sequence composition. Unlike microsatellites, minisatellites can undergo alterations during meiosis,13,14 which made them suitable for DNA fingerprinting and population studies.15,16 However, unlike the larger minisatellites, microsatellite-containing DNA fragments are usually small enough to be amplified by PCR, and hence microsatellites have almost completely replaced minisatellites as genetic markers.
Changes in mini- and micro-satellites correlate with various diseases including cancer, and have been extensively studied. For instance, microsatellite instability (MSI) is the landmark of hereditary non-polyposis colorectal cancer (HNPCC), and also accounts for around 10% of non-hereditary (sporadic) colorectal cancers.17-20 Another example of the involvement of microsatellites in pathogenesis are the expansions of triplet repeat motifs that are recognized as a cause of several neurologic and neuromuscular diseases.21,22 Triplet repeat expansion disorders include common inherited diseases, such as Huntington's disease, myotonic dystrophy, and fragile X syndrome. For example, expansion of the cytosine-adenine-guanine (CAG) repeat is the underlying cause of triplet repeat disorders collectively known as polyglutamine diseases, one of which is Huntington's disease. Extended CAG tracts are translated into a series of uninterrupted glutamine residues, which are prone to aggregation, thus causing cellular toxicity.
Accumulating evidence also reveals interesting aspects of repeats in gene regulation. For instance, changes in the length of GA and CA microsatellite dinucleotide repeats in gene promoters were associated with differences in gene expression.23-26 Recently, this phenomenon was attributed to dinucleotide repeat motifs having an effect on enhancer activity.27 Dinucleotide repeat motifs are highly enriched in enhancers, particularly in those that are broadly active across different cell types. The importance of these motifs in enhancer function was demonstrated by inserting these repeat motifs in an inactive sequence that became a de novo active enhancer. Moreover, repeats were shown to have a role in the regulation of long noncoding RNA (lncRNA) expression, protein interactions, and subcellular location. For instance, the X-inactive specific transcript lncRNA (Xist), which inactivates the female inactive X chromosome, was demonstrated to recruit Polycomb repressive complex 2 (PRC2) through the use of its repeat motifs located at the 5′ end of Xist RNA, known as Repeat A region.28 Another repeat motif in the first exon of Xist RNA, known as Repeat C, was shown to be necessary for Xist RNA loading on the inactive X chromosome by binding YY1, which bridges the interaction of Xist RNA to DNA.29 The recently discovered long intergenic noncoding RNA Firre (Functional Intergenic Repeating RNA Element) is a strictly nuclear RNA that plays a role in adipogenesis by mediating trans-chromosomal interactions.30,31 A local repeating RNA domain (named RRD) in the lincRNA Firre was shown to act as a ribonucleic nuclear retention signal, without which the Firre RNA location shifts from nuclear to cytoplasmic.32 Other examples demonstrate that transposable elements can regulate the expression of lncRNAs, dividing them into cell type-specific classes and possibly regulating their evolution.33,34 Collectively, these data suggest an important and unforeseen role of distinct repeat classes as RNA and DNA regulatory elements.
More recently, macrosatellite repeats (MSRs) are emerging as unique structures in the human genome. Although MSRs are sequence-unrelated, they share some features (Table 1). These include, spanning in tandem over hundreds of kilobases covering significant portions of the genome, being rich in CpGs, thus often regulated by DNA methylation, and frequently expressing noncoding and coding RNAs. Taken together, it is now well accepted that MSRs have a structural and regulatory role in the organization of the chromatin in the nucleus.
Table 1.
Main characteristics of some of the best-described macrosatellites in the human genome.
| Name | Repeat length (kb) | CNV | Location (hg38) | GC content (%) | Methylation changes in disease | Associated disease | Encoded product | ncRNA | Refs. |
|---|---|---|---|---|---|---|---|---|---|
| D4Z4 | 3.3 | 1–150 | 4q35 | 71% | DNA hypomethylation | FSHD, ICF syndrome | DUX4 | Long sense transcript (DBE-T), long sense and antisense transcripts originating within each repeat unit, siRNAs, miRNAs | 43-60,81 |
| DXZ4 | 3 | 12–100 | Xq23 | 62% | Long sense and antisense transcript and small antisense RNA | 61,65 | |||
| NBL2 | 1.4 | not determined | 21p11.2 | 62% | DNA hypomethylation* | Ovarian, colorectal, breast, gastrointestinal and hepatocelular cancer, neuroblastoma, ICF syndrome | 76-81,86 | ||
| RS447 | 4.7 | 20–103 | 4p16.1 | 50% | USP17 | Long antisense transcript | 89,91,92 | ||
| RNU2 | 6.1 | 5–82 | 17q21-q22 | 65% | U2 snRNA | 36,98,99 | |||
| TAF11-Like | 3.4 | 10–98 | 5p15.1 | 50% | Possible role in schizophrenia | TAF11 | 35,109 | ||
| CT47 | 4.8 | 4–17 | Xq24 | 48% | CT47 | 110,111 |
Besides their frequent hypomethylation in cancer, in ovarian cancer, and Wilms tumors, NBL2/SST1 repeats were reported to be more frequently hypermethylated at HhaI site, than hypomethylated at NotI site77.
Coding, noncoding, and architectural roles of macrosatellite repeats in genome organization and disease development
MSRs epigenetic and/or genetic alterations are associated with several human diseases, including cancer. The mechanistic contribution of MSRs to disease development has been analyzed in detail for some MSRs, such as D4Z4, while for others it is unclear to which extent they may contribute to disease development and genome stability. The number of macrosatellite repeats in a tandem array is known to vary between different individuals, from several to hundreds of copies, contributing to significant copy number variation (CNV) that may be related to disease.8,35-37 The true copy number of many macrosatellites is probably underestimated.38
It has been shown that repetition of a sequence in tandem triggers automatic heterochromatization in cis in a copy number dependent manner.39-41 In 1998 Garrick et al. demonstrated that higher copy number of a transgene is associated with its hypermethylation and adaptation of a repressive local chromatin configuration, resulting in transcriptional silencing of the transgene.40 On the contrary, reduction of transgene number to just a few copies resulted in high transgene expression and more accessible local chromatin structure. This repeat feature has been proposed to serve as protection against the consequences of parasitic sequence elements integrated in the genome in high copy number, namely viruses and transposons.42 It has also been proposed that during evolution this putative general feature of repeat elements was adapted to regulate expression of adjacent genes, mostly to induce silencing.38
D4Z4 macrosatellite regulates local chromatin structure in a copy number dependent manner
In somatic cells, many MSRs display features of heterochromatin, with high DNA methylation and repressive histone marks, such as trimethylation of the lysine 9 residue of histone H3 (H3K9me3) and in some cases trimethylation of lysine 27 of histone H3 (H3K27me3). Accordingly, it has been shown that high copy number of some MSRs can have an effect on the chromatin structure in cis and on the regions immediately proximal, and thus contribute to genome stability by triggering gene silencing. D4Z4 is a 3.3 kb repeat located at the subtelomeric regions of chromosomes 4q35 and 10q26. It has been a major research focus due to the link of the D4Z4 array from chromosome 4q35 with the development of facioscapulohumeral muscular dystrophy (FSHD). FSHD is characterized by progressive wasting of muscles in the face, shoulders, and upper arms. The most common form of FSHD is FSHD1, accounting for 95% of cases. FSHD1 is an autosomal dominant disease, with the only detectable genetic defect being the reduction in the copy number of D4Z4 repeats to less than 11 units within the 4q35 subtelomeric repeat array, with the presence of at least 1 repeat unit necessary for FSHD1 development.43 Healthy individuals carry between 11 to 150 copies of D4Z4 that are characterized by highly methylated DNA and organized in heterochromatic structure with H3K9me3 and H3K27me3. In FSHD1 patients, reduced copy number of D4Z4 repeats is accompanied by local loss of repressive marks and overexpression of surrounding genes.44-47
In rare FSHD cases, D4Z4 copy number is not altered and is within the normal range. However, mutations in proteins SMCHD148 and DNMT3B,49 which are involved in heterochromatin formation at D4Z4 locus, can lead to occurrence of this disease, further reinforcing the importance of heterochromatinization of the tandem repeat. This type of FSHD occurs in approximately 5% of patients and is referred to as FSHD2. FSHD2 is clinically identical to FSHD1 and both are characterized by a loss of heterochromatin at D4Z4 locus and thus a de-repression of the region.
D4Z4 repeats contain an open reading frame (ORF) coding for a double homeobox 4 gene (DUX4), usually silenced in normal tissues except for testis. The DUX4 protein has been shown to be pro-apototic and thus could explain muscle weakness observed in FSHD patients.50,51 Aberrant production of DUX4 protein requires an open chromatin conformation in addition to specific polymorphisms involved in RNA stabilization and processing. Expression of DUX4 in FSHD contributes to upregulation of germline genes, endogenous retrotransposons [long-terminal repeat (LTR) elements from MaLR class], RNA splicing and processing genes, atrophy-ubiquitin ligases, noncoding RNAs, and skeletal muscle suppressors of differentiation.46,52,53 All together, this DUX4-induced gene expression signature in FSHD is a major contributor to the disease's pathophysiology.54
There are several models for FSHD1 pathogenesis. D4Z4 is GC rich and displays features of a CpG island. Hence, it has been hypothesized that contraction of the array induces changes in chromatin structure leading to inappropriate transcriptional regulation of several FSHD candidate genes: either the DUX4 gene in D4Z4 unit, genes adjacent to D4Z4 tandem array, or genes that might be regulated by D4Z4 region in trans.55 In this context, 3C analysis of this region revealed specific chromatin contacts between DUX4 and adjacent genes in normal conditions, whereas in FSHD other interactions take place, probably as a result of global reorganization of the 4q35 region upon D4Z4 contraction.56 Although there is a disagreement on which of the FSHD candidate genes is causing FSHD1, it is clear that the presence of D4Z4 contracted alleles is essential for disease development, indicating an important role of D4Z4.
Recently, in efforts to explain the mechanism underlying the epigenetic changes at the contracted D4Z4 array in FSHD1, Cabianca et al. demonstrated that upon D4Z4 copy number reduction, the effect of Polycomb silencing is reduced, resulting in expression of long sense RNAs originating in a distal region of D4Z4 array and extending through multiple repeats. Although D4Z4 repeats contain DUX4 ORFs, these long sense transcripts were nuclear and associated to the chromatin in cis, and thus they were considered noncoding and accordingly named D4Z4 binding element transcript (DBE-T).57 DBE-T recruits the Tritorax group protein Ash1L to D4Z4 repeats in cis, resulting in H3 lysine 36 dimethylation, and long-range gene upregulation (Fig. 2). This study was the first to show how a macrosatellite repeat-derived long noncoding RNA can alter chromatin composition in cis, an important step in understanding macrosatellite biology and its causative role in disease. In addition, it demonstrated a clear association of repeat number reduction and production of a long noncoding RNA.
Figure 2.
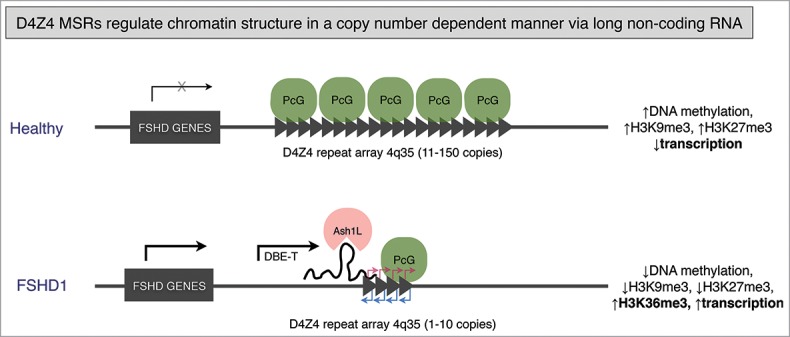
D4Z4 regulates local chromatin structure and expression of surrounding genes via long noncoding RNA. Healthy individuals carry between 11 and 150 copies of D4Z4 macrosatellite in the subtelomeric regions of chromosome 4 (4q35). D4Z4 repeats are highly methylated and enriched in H3K9me3. D4Z4 repeats are targets of Polycomb group proteins (PcG), with a resulting repressive chromatin structure and surrounding genes in transcriptionally silent state. In patients with facioscapulohumeral dystrophy 1 (FSHD1) there is a reduction of D4Z4 copy number to between 1 and 10 copies. The contracted allele loses heterochromatin features (DNA methylation, H3K9me3, H3K27me3) and expresses a long noncoding RNA DBE-T, from a promoter distal to the repeat array, that binds and recruits Tritorax group protein Ash1L in cis, resulting in H3 lysine 36 dimethylation and long-range gene up regulation. In addition, transcription within each repeat unit was reported to occur bidirectionally, indicated by red (sense transcription) and blue (antisense transcription) arrows. Sense and antisense transcripts originate from promoters mapped upstream and downstream of DUX4 ORF, respectively, and are transcribed through multiple D4Z4 repeat units. Those transcripts are suggested to give rise to small ncRNAs. Model design based on,ref. 57, 59 and 60.
Upon D4Z4 epigenetic de-repression, long transcripts through multiple D4Z4 repeat monomers in both sense and antisense directions have also been detected.58 In contrast to DBE-T, these transcripts originate at each repeat unit either near DUX4 promoter (for sense transcription) or at a distal region of DUX4 ORF (for antisense transcripts). These sense and antisense transcriptions modulate DUX4 expression,59 and give rise to siRNA and miRNA which contribute to epigenetic silencing of the locus, opening new avenues to potential therapeutic approaches.60
Considering that many MSRs show significant CNV between individuals, D4Z4 study encourages further research toward unveiling whether similar mechanisms may occur with other MSRs types. Brachmachary et al. provided further evidence for a strong correlation between macrosatellite copy number and epigenetic modifications, and in several cases with nearby gene expression,38 supporting the hypothesis of repeat-induced gene silencing as a mechanism of gene regulation in humans. Their research included MSat10, a relatively unknown GC-rich 5.4 kb macrosatellite repeat, located several kb distal to the ZFP37 gene on chromosome 9q32. Similar to D4Z4, high copy number of Msat10 associates with high local DNA methylation and H3K9me3. On the other hand, reduction in copy number leads to the loss of heterochromatic features and de-repression of the adjacent ZFP37 gene, although in this case generation of a lncRNA was not reported.
DXZ4 macrosatellite regulates higher order nuclear architecture
DXZ4 is a 3 kb, CpG-rich macrosatellite present between 12 and 100 tandem copies on chromosome Xq23. Gialcone et al. discovered DXZ4 in 1992 as a novel X-linked variable number tandem repeat (VNTR) harboring different DNA methylation levels on the active and inactive X chromosome.61 In mammalian females, one of the two X chromosomes is subjected to a process known as X chromosome inactivation to ensure similar levels of expression of X-linked genes compared to males.62 Thus, females have one active X chromosome (Xa), and one inactive X chromosome (Xi). Xi is transcriptionally silenced, characterized by facultative heterochromatin and organized in a 3D configuration within the nucleus, known as the Barr body.63,64 Since its discovery, DXZ4 drew attention because it adopts alternative chromatin states on Xa and Xi chromosome, which differ from the surrounding chromatin.65 On the Xa in males and females, DXZ4 is organized in constitutive heterochromatin, characterized by the presence of H3K9me3 and DNA hypermethylation. On the contrary, DXZ4 harbors opposite chromatin structure on the Xi: DNA hypomethylation, H3K4me2 and H3K9Ac, hallmarks of euchromatin.61-64 Due to the lack of active histone marks on the Xi chromosome, DXZ4 can be visualized on the metaphase Xi chromosome by immunofluorescence as an intensive H3K4me2 signal, surrounded by heterochromatin.66-68 On Xi, but not on Xa chromosome, DXZ4 is bound by CTCF,65,69,70 a highly conserved multifunctional DNA-binding protein implicated in multiple processes throughout the genome, including chromatin insulation and interchromosomal interactions.69
In 2008 Chadwick demonstrated that the CTCF-binding region of DXZ4 unidirectionally interferes with promoter-enhancer communication, supporting the hypothesis that DXZ4 repeat array might act as an insulator.65 On a transcriptional level, the authors demonstrated that DXZ4 is expressed from a bi-directional promoter located within each repeat unit. Their analysis revealed long sense transcripts originating from Xa and Xi arrays, and a long antisense transcript specific to the Xi. Moreover, small antisense RNAs originate from four specific regions of DXZ4, and the site of origin of three of them overlaps precisely with the H3K9me3 and H3K4me2 peaks. This led them to speculate that the small RNAs are involved in heterochromatin formation and maintenance at the DXZ4 locus (Fig. 3).
Figure 3.
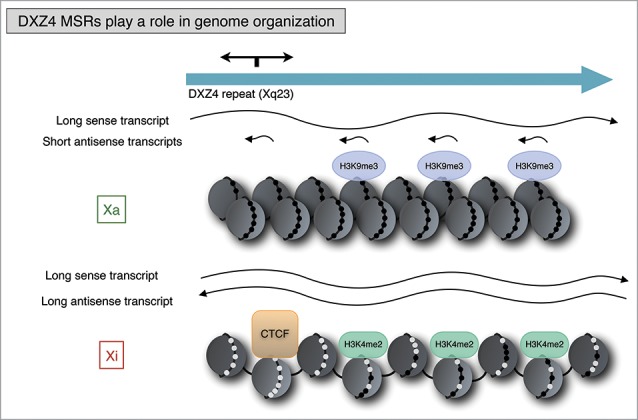
DXZ4 plays a role in genome organization and Xi chromosome higher order structure. DXZ4 harbors opposite chromatin structures on Xa and Xi chromosome, which differ from the surrounding chromatin. On Xi, DXZ4 displays features of euchromatin (hypomethylated CpGs and H3K4me2) and is bound by CTCF. Arrays on Xa and Xi chromosome are transcriptionally active; however, on Xa DXZ4 is transcribed in a long sense transcript and four small antisense RNAs, 3 of which overlap H3K9me3 peaks. On Xi chromosome, DXZ4 is transcribed into long sense and antisense transcripts. DXZ4 on Xi chromosome is necessary for regulating Xi higher order structures. Black dots represent methylated CpGs, white dots unmethylated CpGs. Model design based on ref. 65.
For a long time it has been hypothesized that DXZ4 might have a role in X chromosome inactivation and/or chromatin organization, especially considering it is bound by CTCF on the Xi chromosome. The first observation that DXZ4 indeed does participate in higher order structure organization on the Xi chromosome was made by Horakova et al.70 By applying DNA FISH and 3C analysis, the authors demonstrated Xi-specific long-range interactions between DXZ4 and two newly described tandem repeats, named X56 and X130, in a CTCF-dependent manner. Recent studies using chromosome conformation capture approaches confirmed those results and revealed that human and mouse Xi chromosomes are split into two large superdomains separated by a region containing DXZ4 repeats.71-73 Rao et al. also reported that Xi chromosome forms very large chromatin loops called superloops, with some of them anchored at the DXZ4 macrosatellite.71 Two recent studies provide strong evidence of DXZ4 having an essential role in the regulation of Xi higher order structure and nuclear organization.74,75 By applying genome-wide chromosome conformation capture analysis, authors found that deletion of DXZ4 from Xi chromosome led to the loss of bipartite structure of Xi, disrupted superloops anchored at DXZ4, and induced changes in compartmentalization of the nucleus and in chromatin marks.
NBL2 macrosatellite is hypomethylated in many types of cancer
Some MSRs, such as NBL2, have been shown to be frequently hypomethylated in various types of cancer. NBL2 is a 1.4 kb macrosatellite repeat found mostly on the short arm of acrocentric chromosomes 13, 14, 15 and 2176,77 (intriguingly, not 22), and belongs to a family of macrosatellite repeats known as SST1. NBL2 is CpG-rich and highly methylated in somatic cells. Thoraval et al. discovered NBL2 in 1996 during genome-wide screening for DNA methylation differences in neuroblastoma tumors compared with normal cells by 2D separations of human genomic restriction fragments. They digested DNA from neuroblastoma cells and peripheral blood lymphocytes with methylation sensitive NotI restriction enzyme and two additional cutters, and labeled NotI-derived 5’ ends with 32P. Among fragments appearing hypomethylated at the NotI site they found a previously unreported repeat family, which they named NBL2 and whose sequence was submitted to EMBL database under accession number U59100.76 By applying the same approach, in 1999 Nagai et al. independently found the same sequence hypomethylated in 75% of hepatocellular carcinomas, especially in those with hepatitis B virus infection, which they named NotI repeat (submitted to EMBL database under the accession number Y10751).78
The majority of NotI sites of the human genome lie within CpG islands. Because NBL2 is CpG rich and contains a NotI site, it was suitable for detection with 2D separations of human genomic fragments with NotI restriction enzyme. Thus far, in addition to neuroblastoma and hepatocellular carcinoma, NBL2 was found to be hypomethylated at NotI sites in high risk gastrointestinal tumors,79 bladder cancer,80 immunodeficiency, centromeric instability, and facial abnormalities syndrome patients,81 which have DNMT3B gene mutated. Additionally, it was also found strongly hypomethylated in sperm.78 In ovarian cancer and Wilms tumors, NBL2 was reported to be more frequently hypermethylated at HhaI site than hypomethylated at NotI site.77
Previous work in our group also identified a prominent hypomethylated sequence in colon cancers by the methylation sensitive amplified fragment polymorphism (MS-AFLP) DNA fingerprinting technique.82-85 The sequence was later on identified as SST1/NBL286. In-depth analysis of NBL2 hypomethylation by bisulfite sequencing of an internal 317 bp region, containing a NotI site, showed that SST1/NBL2 was hypomethylated in 22% of colorectal cancers (CRCs), in 15% of gastric cancers, in 20% of ovarian cancers, and in 20% of breast cancers.86
Thus, alterations in NBL2 methylation are characteristic of many cancer types. Nevertheless, the advancement in understanding either the cause or the consequence of this hypomethylation has been slowed down, mostly because NBL2 is not assembled in the reference genome. With the release of genome version hg38, a group of NBL2 repeats were mapped to chromosome 21p11.2, although some other genomic NBL2 loci still remain unassembled. Consequently, the genomic context of NBL2, such as distance to protein coding genes and other regulatory features, are not known. Despite all these challenges, we could determine that in CRC, somatic demethylation of NBL2 was associated with genomic damage assessed by arbitrary primed PCR (AP-PCR),87 especially in tumors with wild type TP5386. Furthermore, in CRC cell lines and primary tumor samples NBL2 hypomethylation is accompanied by local changes in chromatin structure (Fig. 4). In normal somatic cells, NBL2 displays features of constitutive heterochromatin, with high levels of DNA methylation and H3K9me3. However, upon hypomethylation, H3K9me3 levels are decreased, accompanied by a gain in Polycomb repressive mark H3K27me3, typical for facultative heterochromatin.86 Both chromatin states observed at NBL2 region are considered to form stable chromatin; however, there are important differences in the plasticity of both states.
Figure 4.
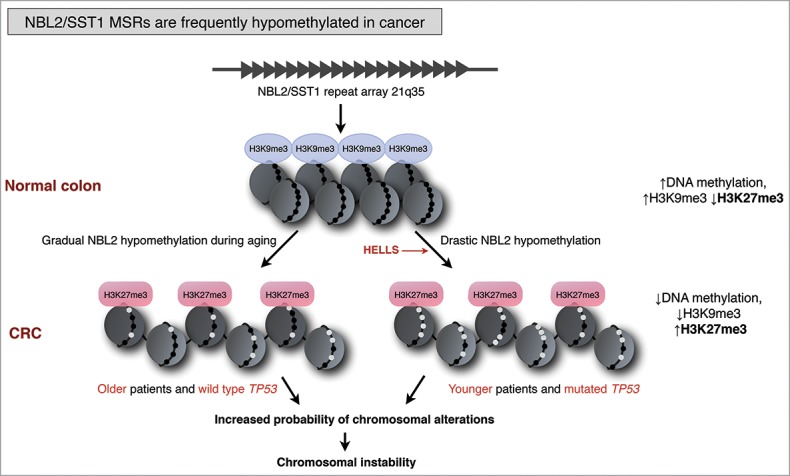
NBL2 macrosatellites are frequently hypomethylated in colorectal cancer (CRC). In normal colon epithelium, NBL2/SST1 repeats display features of constitutive heterochromatin, with high levels of DNA methylation and H3K9me3. In CRC, NBL2/SST1 repeats undergo gradual hypomethylation during aging associated with wild type TP53. Some CRC patients harbor strongly hypomethylated NBL2/SST1 repeats that implicates mechanisms other than aging, and preferentially occurs in mutated TP53 tumors. Hypomethylation of NBL2 results in reprogramming of NBL2 chromatin state from constitutive heterochromatin to facultative heterochromatin characterized by a gain in H3K27me3. NBL2 DNA hypomethylation is linked to increased genomic damage in cases with wild-type TP53. Black dots represent methylated CpGs, white dots unmethylated CpGs. Model design based on ref. 86.
Moreover, a detailed bisulfite analysis revealed two types of NBL2 hypomethylation: moderate hypomethylation, with 5–10% average hypomethylation in tumor compared with adjacent normal tissue, and severe hypomethylation with equal or more than 10% of NBL2 average hypomethylation in tumors compared with adjacent normal tissue. While moderate hypomethylation of NBL2 in CRC patients appeared age-dependent, the severe cases tended to occur in younger patients. Therefore, in those severe cases, NBL2 hypomethylation could be caused by mechanisms other than gradual, stochastic erasure of methylation patterns during aging. The precise mechanism that may causally link NBL2 somatic demethylation and chromosome instability remains to be established.
In this context, a chromatin remodeler enzyme, called helicase lymphoid specific (HELLS), which is known for its role as the “epigenetic guardian of repetitive elements”88 was found to associate with methylated NBL2 in cell lines.86 Furthermore, downregulation of HELLS resulted in NBL2 hypomethylation. The mechanism by which HELLS altered function or impaired recruitment to NBL2 loci could contribute to the somatic demethylation of NBL2 before and/or during CRC development is under investigation.
RS447 macrosatellite codes for ubiquitin-specific protease 17
In addition to their noncoding functions, some macrosatellites have been shown to encode for functional proteins. RS447 is a 4.7 kb macrosatellite present on chromosome 4p1589 and several copies on chromosome 8p.90 RS447 repeat units display promoter activity and contain USP17 (ubiquitin-specific protease 17) gene, which codes for a functional deubiquitinating enzyme.91,92 USP17 removes ubiquitin from target proteins, and it has been shown to play important roles in tumor pathology. Several reports have proved that USP17 acts as a critical regulator of cell proliferation, migration and survival through regulating Ras pathway.93 In 2011 de Vega et al. demonstrated that USP17 depletion blocks chemokine-induced subcellular relocalization of GTPases Cdc42, Rac and RhoA, which are GTPases essential for cell motility, thus demonstrating that USP17 has a critical role in cell migration.94
Okada et al. performed a pedigree analysis of RS447 transmission, and detected that RS447 copy number is highly variable, ranging between 20–103 copies.95 Furthermore, they showed a high frequency (8.3%) of meiotic instability and somatic mosaicism. Because USP17 forms a part of RS447 repeats, the difference in the copy number of RS447 could result in altered USP17 expression levels and thus possibly affect several cellular processes. However, using cosmid vectors containing different numbers of RS447 repeat, Saitoh et al. demonstrated that the level of RS447 sense transcripts and USP17 protein was independent to the integrated copy number of RS447, while abundance of a high molecular weight RS447 antisense transcript was proportional.91 The process of antisense transcripts regulating the expression of their complementary sense transcripts on a transcriptional or post-transcriptional level has been recognized as a mechanism of antisense-mediated gene regulation.96,97 This opens the possibility that the copy number dependent large RS447 antisense transcript may act as a suppressor of the sense transcripts, thus buffering the difference in the repeat copy number. Okada et al. also reported that the RS447 allele can be partially methylated.95 It is probable that a combination of antisense transcripts and DNA methylation regulate the levels of USP17 protein in a copy number dependent manner.
RNU2 macrosatellite encodes for a housekeeping small noncoding RNA
RNU2 is a 6.1 kb macrosatellite present from 5 to 82 tandem copies on chromosome 17q21-q22.36,98,99 Although RNU2 arrays differ in repeat copy number from individual to individual, the arrays are stably inherited.100 Each RNU2 unit encodes for a housekeeping noncoding RNA, U2 small nuclear RNA (U2 snRNA). U2 snRNA is ubiquitously expressed and is an essential component of RNA splicing machinery (spliceosome). Every repeat unit contains snRNA transcriptional control elements (TATA-less promoter/enhancer and 3’ end formation signal), 5 Alu, one LTR retrotransposon and a polymorphic tract of a CT microsatellite, being an example of a microsatellite repeat embedded within a macrosatellite.
Repeat units within an individual RNU2 tandem array appear to be identical, except for the CT microsatellite, which exhibits minor length and sequence polymorphisms. Various roles for the CT microsatellite were proposed: required in DNA recombination for the concerted evolution of RNU2 repeats101; establishment, and/or maintenance of U2 tandem arrays;102 and maintenance of an open chromatin structure.103 Importantly, the nearest gene to the RNU2 tandem array is BRCA1, located 124 kb away.104 Both loci lie within the same linkage disequilibrium block, which allowed to calculate RNU2 macrosatellite mutation rate by tracing BRCA1 mutations in different families. This gives an estimation by maximum likelihood of 5 × 10−3 mutations per generation, which is close to that of microsatellites.105 RNU2 macrosatellite is evolutionarily conserved through speciation in baboon, orangutan, gorilla and chimpanzee.106 Mutations in one copy of the U2 snRNAs have been shown to cause splicing alterations that lead to neurodegeneration in mice; however, whether mutations in human RNU2 genes or CNV in this array may contribute to disease is not known. Importantly, a 3´ fragment of U2 small nuclear RNA, miR-U2–1, could be a marker for non-small cell lung carcinoma.107
Due to the difference in copy number, RNU2 genes must be subjected to some form of dosage compensation, although the mechanism is still not clear. There is evidence for RNU2 bimodal pattern of methylation: the 1.5 kb region covering the U2 transcriptional control elements, the U2 snRNA gene sequence and the CT microsatellite is completely unmethylated, whereas the rest of the U2 repeat (approximately 4.6 kb) is heavily methylated.108 The authors propose that this type of bimodal methylation may permit both efficient expression of U2 snRNA and stable maintenance of U2 tandem arrays in somatic cells.
TAF11-Like macrosatellite codes for a TAF family factor
This tandem array is located in the short arm of human chromosome 5p15.137. Each repeat unit is approximately 3.4 kb in length and contains a short open reading frame of 594 bp encoding for a predicted TATA binding associated factor 11 like protein, which gives the name to the macrosatellite. The repeat unit also contains LTR retrotransposon (MLT1E3), a disrupted DNA transposon (Charlie 2a) and a partial Alu repeat. Pulsed field gel electrophoresis (PFGE) and Southern blot analysis with a probe specific to the TAF11-Like macrosatellite revealed that the size of the TAF11-Like array is very polymorphic in the population, ranging from 34 to 335 kb, thus indicating that the copy number of TAF11-Like macrosatellite can range between 10 and 98 tandem copies.
The closest genes to TAF11-Like macrosatellite are brain abundant, membrane attached signal protein (BASP1) at 210 kb and cadherin 18 type 2 preproprotein (CDH18) at over 1.8 Mb. However, whether TAF11-Like macrosatellite length or its epigenetic status could influence the expression of these genes or contribute to disease is not clear. One study showed that alleles contracted to less than 21 tandem repeats associate to schizophrenia,109 which could support a contributory role to the disease, perhaps in a regulatory manner similar to what has been shown for contracted D4Z4 macrosatellites and facioscapulohumeral dystrophy. However, other schizophrenia families did not show contracted TAF11-Like macrosatellite array according to quantitative PCR (qPCR) analysis, and therefore the results on TAF11-Like possible contribution to schizophrenia were inconclusive. Nevertheless, the authors hypothesize that the low monomer numbers in one 5p15.1 allele may be masked by the uncontracted allele when analyzed by qPCR, since the average number of both alleles may be higher than 21 repeats.37,109 These results highlight that qPCR, which represents the sum of the repeat number, may not be sensitive enough to measure differences in allele size of tandemly repeated DNA.
Expression of TAF11-Like RNA has been detected in testes, brain and fetal tissues from brain, liver and prostate; however, the biologic significance of TAF11-Like expression remains elusive. Nevertheless, the TAF11-Like ORF sequence is conserved in primates, revealing a translated 198 aminoacid sequence with 90.9 to 96% identity in great apes and 86.4% in Macaque (199 aminoacids).37 Furthermore, several peptides from the putative protein [accession A6NLC8 in the PRoteomics IDEntifications (PRIDE) database] can be detected by mass spectrometry in different analyses. These data point toward a functional TAF11-Like protein, but more research is required to fully understand its functionality.
CT47, a macrosatellite with testis-specific expression
The cancer/testis gene CT47 is located on Xq24, arranged in 4 to 17 tandem repeats of 4.8 kb each.110 CT47 RNA is putatively coding, 1,286 bp in length [excluding the poly(A) tail] with the coding region of 867 bp encoding a protein of 288 aminoacids. Chimpanzee is the only species other than human with a gene homologous to CT47, which is also located on chromosome X. The predicted protein is approximately 80% identical in its carboxy terminal region between the two species. CT47 is highly expressed only in testis, and low levels are detectable in placenta and brain, while silenced during early development and in other normal somatic tissues tested.110 CT47 expression was detected in 14% of lung cancer, in 15% of esophageal cancer and in 11% of endometrial cancer specimens, but not in colorectal, breast and bladder tumors tested.110,111 In normal somatic cells, CT47 is organized in heterochromatin, characterized by high levels of H3K9me3, H3K27me3, methylated CpG sites around CT47 promoter and silenced CT47. Balog et al. encouraged with the clear correlation between D4Z4 copy number and local heterochromatin formation, studied whether reduced CT47 copy number would result in a loss of heterochromatin features, and consequently CT47 expression. Their results indicate that within the tested copy number range (4 to 17 CT47 copies), CT47 copy number does not correlate with local H3K9me3 and H3K27me3 levels,111 thus arguing against a direct link between repeat copy number and heterochromatization. However, since the lowest CT47 copy number analyzed was four, authors hypothesize that each MSR might have a minimal number of repeat units that would ensure proper heterochromatin formation. Small cell lung cancer (SCLC) cell lines that express CT47 (albeit at much lower levels than detected in testis) showed H3K9me3 decrease and demethylation of CpG sites near the transcription start site, which suggests that loss of heterochromatin features at CT47 array may result in CT47 expression. However, the causes of CT47 array heterochromatin loosening were not studied, nor whether CT47 RNA or protein contributes to SCLC disease.
Conclusion and future perspectives
Macrosatellite repeats, along with many other repeats, remain poorly investigated and thus are considered the “dark matter” of the human genome. They span through relatively large stretches of the genome; however, technical limitations together with the view that those sequences were functionally irrelevant (junk or garbage DNA) led to a considerable neglect in analyzing repeats as valuable components of human genetic material. With massive sequencing technologies, many noncoding regions of the genome have been discovered to be transcribed and to play distinct roles in cell biology. These discoveries have revolutionized the field, and the previous dogma of what is considered a functional genomic region has changed, leading to the acceptance of repeats as important architectural DNA building blocks and functional components of the transcriptome. Nevertheless, we are still largely unaware of many macrosatellite features such as their precise location, sequence composition, epigenetic regulation, copy number variation, transcription, and function. Substantial efforts are being placed to develop strategies that would overcome the obstacles in aligning next generation sequencing data and in de novo genome assembly of these regions. Longer read-lengths will reduce difficulties related to repeat alignment and thus fuel a more thorough analysis of those regions, which may lead to breakthrough discoveries.
Increasing evidence suggests that macrosatellites are unique regulatory sequences, each of them with distinct functions. Macrosatellites encompass coding, noncoding and structural roles in the genome and seem to undergo frequent epigenetic and genetic alterations in disease. Sequence complexity and long repeat nature may allow macrosatellites to play significant roles in genome architecture, organization and regulation, representing an additional layer to fine-tune complex and dynamic regulatory networks of the genome. Mechanisms by which they accomplish those roles may be by anchoring chromatin remodeling complexes and transcription factors to form loops that regulate higher order genome architecture, as was demonstrated with DXZ4 repeats. Other mechanisms include generation of noncoding RNAs that modulate local transcription and chromatin formation, as was shown for D4Z4 macrosatellite. Furthermore, some parallels exist between several macrosatellite families. Those commonalities are most notable between D4Z4 and DXZ4, since both are well-studied, and include presence of internal promoters, bidirectional transcription, and generation of long and short ncRNAs. This poses interesting questions whether these noncoding transcripts play similar roles in both macrosatellites or even if those similarities may be extended to other macrosatellite families.
What appears to be a general rule is that greater numbers of repeats associate to heterochromatic features, and this is also the case for other repeats such as some microsatellites. For instance, CTG triplet repeat expansion found in myotonic dystrophy 1 results in acquisition of heterochromatin. This “heterochromatinization” mechanism involves CTCF loss, bidirectional transcription and generation of siRNAs.112 Again, this highlights the fact that noncoding transcription may contribute to opposite chromatin statuses that can mediate either silencing (as in CTG microsatellite expansion) or activation (such as in D4Z4 contraction). All these facts expose the complexity of DNA repeat regulation and transcription of noncoding regions.
Todate, D4Z4 remains one of the best described macrosatellite repeats and the only one reported to be transcribed into a regulatory, chromatin-associated, long noncoding RNA. Recently, lncRNAs have become a major research focus due to their versatile functions and key roles in cell physiology. They can regulate chromatin structure in cis by targeting protein complexes to a specific chromatin loci, or in trans by anchoring chromosomal interactions. They can also interact with mRNAs and regulate their metabolism, or interact with proteins and regulate protein complex assembly.113 Since many macrosatellite repeats remain poorly investigated, especially at the transcriptional level, it might be plausible that they contain a hoard of regulatory ncRNAs or even coding RNAs that have yet to be discovered. Exciting years of research in the field of repetitive DNA are ahead of us, which will shed more light on these still veiled regions of the genome and determine their possible relevance in genome organization, cellular biology, and disease pathogenesis.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgements
The authors thank Michael Maher for revising the manuscript for English corrections.
Funding
This work was supported in part by the Spanish Ministry of Health Plan Nacional de I+D+I, ISCIII, FEDER, FIS PI09/02444, PI12/00511 and PI15/01763 grants, and 2014-SGR-1269 from the Agència de Gestió d'Ajuts Universitaris i de Recerca (AGAUR). GD is supported by FPU fellowship “beca de formación de profesorado universitario” (FPU12/06605). SVF is supported by grants from Fundación Olga Torres and MINECO (PIE16–00011).
References
- 1.Bowen NJ, Jordan IK. Transposable elements and the evolution of eukaryotic complexity. Curr Issues Mol Biol 2002; 4:65-76; PMID:12074196; https://doi.org/ 10.21775/cimb.004.065 [DOI] [PubMed] [Google Scholar]
- 2.Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P. The struggle for life of the genome's selfish architects. Biol Direct 2011; 6(1):19; PMID:21414203; https://doi.org/ 10.1186/1745-6150-6-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawkins R. The Selfish Gene. Oxford University Press; 1976 [Google Scholar]
- 4.Doolittle F, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature 1980; 284:601-3; PMID:6245369; https://doi.org/ 10.1038/284601a0 [DOI] [PubMed] [Google Scholar]
- 5.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature 1980; 284:604-7; PMID:7366731; https://doi.org/ 10.1038/284604a0 [DOI] [PubMed] [Google Scholar]
- 6.Smit AFA, Hubley R, Green P. http://www.repeatmasker.org RepeatMasker Open-4.0. 2013-2015. [Google Scholar]
- 7.de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7(12):e1002384; PMID:22144907; https://doi.org/ 10.1371/journal.pgen.1002384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warburton PE, Hasson D, Guillem F, Lescale C. Analysis of the largest tandemly repeated DNA families in the human genome. BMC Genomics 2008; 18:533; PMID:18992157; https://doi.org/ 10.1186/1471-2164-9-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing : computational challenges and solutions. Nat Rev Genet 2012; 13:36-46; PMID:22124482; https://doi.org/ 10.1038/nrg3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richard GF, Kerrest A, Dujon B. Comparative genomics and molecular dynamics of DNA repeats in eukaryotes. Microbiol Mol Biol Rev 2008; 72(4):686-727; PMID:19052325; https://doi.org/ 10.1128/MMBR.00011-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rich J, Ogryzko VV, Pirozhkova IV. Satellite DNA and related diseases. Biopolymers Cell 2014; 30(4):249-59; https://doi.org/ 10.7124/bc.00089E [DOI] [Google Scholar]
- 12.Streisinger G, Okada J, Emrich J, Newton J, Tsugita A, Terzaghi E, et al.. Frameshift mutations and the genetic code. Cold Spring Harbor Symp Quant Biol 1967; 31:77-84; PMID:5237214; https://doi.org/ 10.1101/SQB.1966.031.01.014 [DOI] [PubMed] [Google Scholar]
- 13.Jarman AP, Wells RA. Hypervariable minisatellites: recombinators or innocent bystanders. Trends Genet 1989; 5(11):367-71; PMID:2692244; http://dx.doi.org/9670029 10.1016/0168-9525(89)90171-6 [DOI] [PubMed] [Google Scholar]
- 14.Jeffreys AJ, Neil DL, Neumann R. Repeat instability at human minisatellites arising from meiotic recombination. EMBO J 1998; 17(14):4147-57; PMID:9670029; https://doi.org/ 10.1093/emboj/17.14.4147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyman AR, White R. A highly polymorphic locus in human DNA. Proc Natl Acad Sci USA 1980; 77(11):6754-8; PMID:6935681; https://doi.org/ 10.1073/pnas.77.11.6754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffreys A, Wilson V, Thein SL. Hypervariable ´minisatellite´ regions in human DNA. Nature 1985; 314:67-73; PMID:3856104; https://doi.org/ 10.1038/314067a0 [DOI] [PubMed] [Google Scholar]
- 17.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 1993; 363:558-61; PMID:8505985; https://doi.org/ 10.1038/363558a0 [DOI] [PubMed] [Google Scholar]
- 18.Aaltonen LA, Peltomäki P, Leach FS, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Powell SM, Jen J, Hamilton SR, et al.. Clues to the pathogenesis of familial colorectal cancer. Science 1993; 260(5109):816–20; PMID:8484122; https://doi.org/ 10.1126/science.8484121 [DOI] [PubMed] [Google Scholar]
- 19.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science 1993; 90:816-9; https://doi.org/ 10.1126/science.8484122 [DOI] [PubMed] [Google Scholar]
- 20.Marra G, Boland CR. Hereditary nonpolyposis colorectal cancer: the syndrome, the genes, and historical perspectives. J Natl Cancer Inst 1995; 87(15):1114-25; PMID:7674315; https://doi.org/ 10.1093/jnci/87.15.1114 [DOI] [PubMed] [Google Scholar]
- 21.Budworth H, Mcmurray CT. A brief history of triplet repeat diseases. Methods Mol Biol 2013; 1010:3-17; PMID:23754215; https://doi.org/ 10.1007/978-1-62703-411-1_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.La Spada AR, Taylor JP. Repeat expansion disease: Progress and puzzles in disease pathogenesis. Nat Rev Genet 2010; 11(4):247-58; PMID:20177426; https://doi.org/ 10.1038/nrg2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada H, Seidman M, Howard BH, Gorman CM. Enhanced gene expression by the poly (dT-dG) poly (dC-dA) sequence. Mol Cell Biol 1984; 4(12):2622-30; PMID:6098815; https://doi.org/ 10.1128/MCB.4.12.2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B, Ren J, Ooi LL, Chong SS, Lee CG. Dinucleotide repeats negatively modulate the promoter activity of Cyr61 and is unstable in hepatocellular carcinoma patients. Oncogene 2005; 24:3999-4008; PMID:15782120; https://doi.org/ 10.1038/sj.onc.1208550 [DOI] [PubMed] [Google Scholar]
- 25.Baranovskaya S, Martin Y, Alonso S, Pisarchuk KL, Dai Y, Khaldoyanidi S, Krajewski S, Novikova I, Sidorenko YS, Perucho M, et al.. Down-regulation of epidermal growth factor receptor by selective expansion of a 5′-end regulatory dinucleotide repeat in colon cancer with microsatellite instabillity. Clin Cancr Res 2009; 15(14):4531-7; PMID:19584170; https://doi.org/ 10.1158/1078-0432.CCR-08-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris EE, Amria MY, Kistner-griffin E, Svenson JL, Kamen DL, Gilkeson GS, Nowling TK. A GA microsatellite in the Fli1 promoter modulates gene expression and is associated with systemic lupus erythematosus patients without nephritis. Arthritis Res Ther 2010; 12:1-9; PMID:21087477; https://doi.org/ 10.1186/ar3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yáñez-Cuna O, Arnold CD, Stampfel G, Boryn M, Ya JO, Gerlach D, Rath M, Stark A. Dissection of thousands of cell type-specific enhancers identifies dinucleotide repeat motifs as general enhancer features. Genome Res 2014; 24:1147-56; PMID:24714811; https://doi.org/ 10.1101/gr.169243.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Sun BK, Erwin JA, J-j Song, Jeannie T. Polycomb proteins targeted by a short repeat RNA to the mouse X-chromosome. Science 2009; 322(5902):750-6; PMID:18974356; https://doi.org/ 10.1126/science.1163045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jeon Y, Lee JT. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2012; 146(1):119-33; PMID:21729784; https://doi.org/ 10.1016/j.cell.2011.06.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun L, Goff LA, Trapnell C, Alexander R, Alice K, Hacisuleyman E. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci USA 2013; 110(9):3387-92; PMID:23401553; https://doi.org/ 10.1073/pnas.1222643110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al.. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 2014; 21(2):198-206; PMID:24463464; https://doi.org/ 10.1038/nsmb.2764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hacisuleyman E, Shukla CJ, Weiner CL, Rinn JL. Function and evolution of local repeats in the Firre locus. Nat Commun 2016; 6:1-12; PMID:27009974; https://doi.org/ 10.1038/ncomms11021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley D, Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol 2012; 13:R107; PMID:23181609; https://doi.org/ 10.1186/gb-2012-13-11-r107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kapusta A, Kronenberg Z, Lynch VJ, Zhuo X, Ramsay L, Bourque G, Yandell M, Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet 2013; 9(4):e1003470; PMID:23637635; https://doi.org/ 10.1371/journal.pgen.1003470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tremblay DC, Moseley S, Chadwick BP. Variation in array size, monomer composition and expression of the macrosatellite DXZ4. PLoS One 2011; 6(4):e18969; PMID:21544201; https://doi.org/ 10.1371/journal.pone.0018969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schaap M, Lemmers RJ, Maassen R, van der Vliet PJ, Hoogerheide LF, van Dijk HK, Baştürk N, de Knijff P, van der Maarel SM. Genome-wide analysis of macrosatellite repeat copy number variation in worldwide populations : evidence for differences and commonalities in size distributions and size restrictions. BMC Genomics 2013; 14:143; PMID:23496858; https://doi.org/ 10.1186/1471-2164-14-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tremblay DC, Alexander G Jr, Moseley S, Chadwick BP. Expression, tandem repeat copy number variation and stability of four macrosatellite arrays in the human genome. BMC Genomics 2010; 11(1):632; PMID:21078170; https://doi.org/ 10.1186/1471-2164-11-632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brahmachary M, Guilmatre A, Quilez J, Hasson D, Borel C, Warburton P, Sharp AJ. Digital genotyping of macrosatellites and multicopy genes reveals novel biological functions associated with copy number variation of large tandem repeats. PLoS Genet 2014; 10(6):e1004418; PMID:24945355; https://doi.org/ 10.1371/journal.pgen.1004418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Assaad FF, Tucker KL, Signer ER. Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol Biol 1993; 22:1067-85; PMID:8400126; https://doi.org/ 10.1007/BF00028978 [DOI] [PubMed] [Google Scholar]
- 40.Garrick D, Fiering S, Martin DI, Whitelaw E. Repeat induced gene silencing in mammals. Nat Genet 1998; 18:56-9; PMID:9425901; https://doi.org/ 10.1038/ng0198-56 [DOI] [PubMed] [Google Scholar]
- 41.Ye F, Signer ER. RIGS (repeat-induced gene silencing) in Arabidopsis is transcriptional and alters chromatin configuration. Proc Natl Acad Sci USA 1996; 93:10881-6; PMID:8855276; https://doi.org/ 10.1073/pnas.93.20.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henikoff S. Conspiracy of silence among repeated transgenes. Bioessays 1998; 20:532-5; PMID:9723001; https://doi.org/ 10.1002/(SICI)1521-1878(199807)20:7%3c532::AID-BIES3%3e3.0.CO;2-M [DOI] [PubMed] [Google Scholar]
- 43.van Deutekom JC, Wljmenga C, van Tienhoven EA, Grater AM, Hewitt JE, Padberg GW, van Ommen GJ, Hofker MH, Frants RR. FSHD associated DNA rearrangements are due to deletions of integral copies of a 3.2 kb tandemly repeated unit. Hum Mol Genet 1993; 2(12):2037-42; PMID:8111371; https://doi.org/ 10.1093/hmg/2.12.2037 [DOI] [PubMed] [Google Scholar]
- 44.Gabellini D, Green MR, Tupler R. Inappropriate gene activation in FSHD: a repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 2002; 110:339-48; PMID:12176321; https://doi.org/ 10.1016/S0092-8674(02)00826-7 [DOI] [PubMed] [Google Scholar]
- 45.Rijkers T, Deidda G, van Koningsbruggen S, van Geel M, Lemmers RJ, van Deutekom JC, Figlewicz D, Hewitt JE, Padberg GW, Frants RR, et al.. FRG2, an FSHD candidate gene, is transcriptionally upregulated in differentiating primary myoblast cultures of FSHD patients. J Med Genet 2004; 41:826-36; PMID:15520407; https://doi.org/ 10.1136/jmg.2004.019364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dixit M, Tassin A, Winokur S, Shi R, Qian H, Acker AM, Leo O, Figlewicz D, Barro M, Laoudj-Chenivesse D, et al.. DUX4, a candidate gene of facioscapulohumeral muscular dystrophy, encodes a transcriptional activator of PITX1. Proc Natl Acad Sci USA 2007; 104(46):18157-62; PMID:17984056; https://doi.org/ 10.1073/pnas.0708659104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bodega B, Di G, Ramirez C, Grasser F, Cheli S, Brunelli S, et al.. Remodeling of the chromatin structure of the facioscapulohumeral muscular dystrophy (FSHD) locus and upregulation of FSHD-related gene 1 (FRG1) expression during human myogenic differentiation. BMC Biol 2009; 16:7–41; PMID:19607661; https://doi.org/23143600 10.1186/1741-7007-7-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lemmers RJ, Tawil R, Petek LM, Balog J, Block GJ, Santen GW, Amell AM, van der Vliet PJ, Almomani R, Straasheijm KR, et al.. Digenic inheritance of an SMCHD1 mutation and an FSHD- permissive D4Z4 allele causes facioscapulohumeral muscular dystrophy type 2. Nat Genet 2012; 44(12):1370-4; PMID:23143600; https://doi.org/ 10.1038/ng.2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van den Boogaards ML, Lemmers RJ, Balog J, Wohlgemuth M, Auranen M, Mitsuhashi S, van der Vliet PJ, Straasheijm KR, van den Akker RF, Kriek M, et al.. Mutations in DNMT3B modify epigenetic repression of the D4Z4 repeat and the penetrance of facioscapulohumeral dystrophy. Am J Hum Genet 2016; 98(5):1020-9; PMID:27153398; https://doi.org/ 10.1016/j.ajhg.2016.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bosnakovski D, Xu Z, Ji Gang E, Galindo CL, Liu M, Simsek T, Garner HR, Agha-Mohammadi S, Tassin A, Coppée F, et al.. An isogenetic myoblast expression screen identifies DUX4-mediated FSHD-associated molecular pathologies. EMBO J 2008; 27(20):2766-79; PMID:18833193; https://doi.org/ 10.1038/emboj.2008.201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kowaljow V, Marcowycz A, Ansseau E, Conde C, Sauvage S, Mattéotti C, Arias C, Corona ED, Nuñez NG, Leo O, et al.. The DUX4 gene at the FSHD1A locus encodes a pro-apoptotic protein. Neuromuscul Disord 2017; 17(8):611-23; PMID:17588759; https://doi.org/ 10.1016/j.nmd.2007.04.002 [DOI] [PubMed] [Google Scholar]
- 52.Hewitt JE, Lyle R, Clark LN, Valleley EM, Wright TJ, Wijmenga C, van Deutekom JC, Francis F, Sharpe PT, Hofker M, et al.. Analysis of the tandem repeat locus D4Z4 associated with facioscapulohumeral muscular dystrophy. Hum Mol Genet 1994; 3(8):1287-95; PMID:7987304; https://doi.org/ 10.1093/hmg/3.8.1287 [DOI] [PubMed] [Google Scholar]
- 53.Geng Linda N, Yao Z, Snider L, Fong Abraham P, Cech Jennifer N, Young Janet M, van der Maarel SM, Ruzzo WL, Gentleman RC, Tawil R, et al.. DUX4 activates germline genes, retroelements, and immune mediators: implications for facioscapulohumeral dystrophy. Dev Cell 2012; 22(1):38-51; PMID:22209328; https://doi.org/ 10.1016/j.devcel.2011.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao Z, Snider L, Balog J, Lemmers RJLF, Van Der Maarel SM, Tawil R, Tapscott SJ. DUX4-induced gene expression is the major molecular signature in FSHD skeletal muscle. Hum Mol Genet 2014; 23(20):5342-52; PMID:24861551; https://doi.org/ 10.1093/hmg/ddu251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Greef JC, Frants RR, van der Maarel SM. Epigenetic mechanisms of facioscapulohumeral muscular dystrophy. Mutat Res 2008; 647:94-102; PMID:18723032; https://doi.org/ 10.1016/j.mrfmmm.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pirozhkova I, Petrov A, Dmitriev P, Laoudj D, Lipinski M, Vassetzky Y. A functional role for 4qA/B in the structural rearrangement of the 4q35 region and in the regulation of FRG1 and ANT1 in facioscapulohumeral dystrophy. PLoS One 2008; 3(10):1-9; PMID:18852887; https://doi.org/ 10.1371/journal.pone.0003389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cabianca DS, Casa V, Bodega B, Xynos A, Ginelli E, Tanaka Y, Gabellini D. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell 2012; 149(4):819-31; PMID:22541069; https://doi.org/ 10.1016/j.cell.2012.03.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snider L, Asawachaicharn A, Tyler AE, Geng LN, Petek LM, Maves L, Miller DG, Lemmers RJ, Winokur ST, Tawil R, et al.. RNA transcripts, miRNA-sized fragments and proteins produced from D4Z4 units: new candidates for the pathophysiology of facioscapulohumeral dystrophy. Hum Mol Genet 2009; 18(13):2414-30; PMID:19359275; https://doi.org/ 10.1093/hmg/ddp180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Block GJ, Petek LM, Narayanan D, Amell AM, Moore JM, Rabaia NA, Tyler A, van der Maarel SM, Tawil R, Filippova GN, et al.. Asymmetric bidirectional transcription from the FSHD- causing D4Z4 array modulates DUX4 production. PLoS One 2012; 7(4):e35532; PMID:22536400; https://doi.org/ 10.1371/journal.pone.0035532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lim J-W, Snider L, Yao Z, Tawil R, van der Maarel SM, Rigo F, Bennett CF, Filippova GN, Tapscott SJ. DICER/AGO-dependent epigenetic silencing of D4Z4 repeats enhanced by exogenous siRNA suggests mechanisms and therapies for FSHD. Hum Mol Genet 2015; 24(17):4817-28; PMID:26041815; https://doi.org/ 10.1093/hmg/ddv206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gialcone J, Friedes J, Francke U. A novel GC-rich human macrosatellite VNTR in Xq24 is differentially methylated on active and inactive X chromosomes. Nature Genet 1992; 1:137-43; PMID:1302007; https://doi.org/ 10.1038/ng0592-137 [DOI] [PubMed] [Google Scholar]
- 62.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961; 190:372-3; PMID:13764598; https://doi.org/ 10.1038/190372a0 [DOI] [PubMed] [Google Scholar]
- 63.Barr M, Bertram E. Morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 1949; 163:676-7; PMID:18120749; https://doi.org/ 10.1038/163676a0 [DOI] [PubMed] [Google Scholar]
- 64.Naughton C, Sproul D, Hamilton C, Gilbert N. Analysis of active and inactive X chromosome architecture reveals the independent organization of 30 nm and large-scale chromatin structures. Mol Cell 2010; 40(3):397-409; PMID:21070966; https://doi.org/ 10.1016/j.molcel.2010.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chadwick BP. DXZ4 chromatin adopts an opposing conformation to that of the surrounding chromosome and acquires a novel inactive X-specific role involving CTCF and antisense transcripts. Genome Res 2008; 18:1259-69; PMID:18456864; https://doi.org/ 10.1101/gr.075713.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chadwick BP, Willard HF. Cell cycle – dependent localization of macroH2A in chromatin of the inactive X chromosome. J Cell Biol 2002; 157(7):1113-23; PMID:12082075; https://doi.org/ 10.1083/jcb.200112074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chadwick BP, Willard HF. Multiple spatially distinct types of facultative heterochromatin on the human inactive X chromosome. Proc Natl Acad Sci USA 2004; 101(50):17450-5; PMID:15574503; https://doi.org/ 10.1073/pnas.0408021101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boggs BA, Cheung P, Heard E, Spector DL, Chinault AC, Allis CD. Differentially methylated forms of histone H3 show unique association patterns with inactive human X chromosomes. Nature Genet 2002; 30:73-6; PMID:11740495; https://doi.org/ 10.1038/ng787 [DOI] [PubMed] [Google Scholar]
- 69.Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Curr Topics Dev Biol 2008; 80(07):337-60; PMID:17950379; https://doi.org/ 10.1016/S0070-2153(07)80009-3 [DOI] [PubMed] [Google Scholar]
- 70.Horakova AH, Moseley SC, Mclaughlin CR, Tremblay DC, Chadwick BP. The macrosatellite DXZ4 mediates CTCF-dependent long-range intrachromosomal interactions on the human inactive X chromosome. Hum Mol Genet 2012; 21(20):4367-77; PMID:22791747; https://doi.org/ 10.1093/hmg/dds270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rao SSP, Huntley MH, Durand NC, Stamenova EK. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014; 159(7):1665-80; PMID:25497547; https://doi.org/ 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deng X, Ma W, Ramani V, Hill A, Yang F, Ay F, Berletch JB, Blau CA, Shendure J, Duan Z, et al.. Bipartite structure of the inactive mouse X chromosome. Genome Biol 2015; 16:1-21; PMID:25583448; https://doi.org/ 10.1186/s13059-015-0728-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Minajigi A, Froberg J, Wei C, Sunwoo H, Kesner B, Lessing D, Payer B, Boukhali M, Haas W, Lee JT. A comperhensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 2016; 349(6245):aab2276; PMID:26089354; https://doi.org/ 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Darrow EM, Huntley MH, Dudchenko O, Stamenova EK, Durand NC. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc Natl Acad Sci USA 2016; 113:E4504-12; PMID:27432957; https://doi.org/ 10.1073/pnas.1609643113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, Chen CJ, Kaplan N, Chang HY, Heard E, et al.. Structural organization of the inactive X in the mouse. Nature 2016; 535(7613):575-9; PMID:27437574; https://doi.org/ 10.1038/nature18589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Thoraval D, J Asakawa, Wimmer K, Kuick R, Lamb B, Richardson B. Demethylation of repetitive DNA sequences in neuroblastoma. Genes Chromosomes Cancer 1996; 17(4):234-44; PMID:8946205; https://doi.org/ [DOI] [PubMed] [Google Scholar]
- 77.Nishiyama R, Qi L, Tsumagari K, Weissbecker K, Dubeau L, Champagne M, Sikka S, Nagai H, Ehrlich M. A DNA repeat, NBL2, is hypermethylated in some cancers but hypomethylated in others. Cancer Biol Ther 2005; 4:440-8; PMID:15846090; https://doi.org/ 10.4161/cbt.4.4.1622 [DOI] [PubMed] [Google Scholar]
- 78.Nagai H, Sung Y, Yasuda T, Ohmachi Y, Yokouchi H, Monden M, Emi M, Konishi N, Nogami M, Okumura K, et al.. A novel sperm-specific hypomethylation sequence is a demethylation hotspot in human hepatocellular carcinomas. Gene 1999; 237:15-20; PMID:10524231; https://doi.org/ 10.1016/S0378-1119(99)00322-4 [DOI] [PubMed] [Google Scholar]
- 79.Igarashi S, Suzuki H, Niinuma T, Shimizu H, Nojima M, Iwaki H, Nobuoka T, Nishida T, Miyazaki Y, Takamaru H, et al.. A Novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors a novel correlation between LINE-1 hypomethylation and the malignancy of gastrointestinal stromal tumors. Clin Cancer Res 2010; 16:5114-23; PMID:20978145; https://doi.org/ 10.1158/1078-0432.CCR-10-0581 [DOI] [PubMed] [Google Scholar]
- 80.Choi SH, Worswick S, Byun H-M, Shear T, Soussa JC, Wolff EM, Douer D, Garcia-Manero G, Liang G, Yang AS. Changes in DNA methylation of tandem DNA repeats are different from interspersed repeats in cancer. Int J Cancer 2009; 125:723-9; PMID:19437537; https://doi.org/ 10.1002/ijc.24384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kondo T, Bobek MP, Kuick R, Lamb B, Zhu X, Narayan A, Bourc'his D, Viegas-Péquignot E, Ehrlich M, Hanash SM. Whole-genome methylation scan in ICF syndrome : hypomethylation of non-satellite DNA repeats D4Z4 and NBL2. Hum Mol Genet 2000; 9(4):597-604; PMID:10699183; https://doi.org/ 10.1093/hmg/9.4.597 [DOI] [PubMed] [Google Scholar]
- 82.Yamamoto F, Yamamoto M, Soto JL, Kojima E, Wang EN, Perucho M, Sekiya T, Yamanaka H. NotI-MseI methylation-sensitive amplified fragment length polymorphism for DNA methylation analysis of human cancers. Electrophoresis 2001; 22(10):1946-56; PMID:11465493; https://doi.org/ 10.1002/1522-2683(200106)22:10%3c1946::AID-ELPS1946%3e3.0.CO;2-Y [DOI] [PubMed] [Google Scholar]
- 83.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell 2003; 4:121-31; PMID:12957287; https://doi.org/ 10.1016/S1535-6108(03)00190-9 [DOI] [PubMed] [Google Scholar]
- 84.Samuelsson JK, Alonso S, Yamamoto F, Perucho M. DNA fingerprinting techniques for the analysis of genetic and epigenetic alterations in colorectal cancer. Mutat Res 2010; 693(1-2):61-76; PMID:20851135; https://doi.org/ 10.1016/j.mrfmmm.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell 2006; 9:199-207; PMID:16530704; https://doi.org/ 10.1016/j.ccr.2006.02.016 [DOI] [PubMed] [Google Scholar]
- 86.Samuelsson JK, Dumbovic G, Polo C, Moreta C, Alibés A, Ruiz-Larroya T, et al.. Helicase lymphoid-specific enzyme contributes to the maintenance of methylation of SST1 pericentromeric repeats that are frequently demethylated in colon cancer and associate with genomic damage. Epigenomes 2017; 1(1):2; https://doi.org/ 10.3390/epigenomes1010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peinado MA, Malkhosyan S, Velazquez A, Perucho M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proc Natl Acad Sci USA 1992; 89:10065-9; PMID:1359533; https://doi.org/ 10.1073/pnas.89.21.10065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Huang J, Fan T, Yan Q, Zhu H, Fox S, Issaq HJ, Best L, Gangi L, Munroe D, Muegge K. Lsh, an epigenetic guardian of repetitive elements. Nucleic Acids Res 2004; 32(17):5019-28; PMID:15448183; https://doi.org/ 10.1093/nar/gkh821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kogi M, Fukushige S, Lefevre C, Hadano S, Ikeda JE. A novel tandem repeat sequence located on human chromosome 4p: isolation and characterization. Genomics 1997; 42(2):278-83; PMID:9192848; https://doi.org/ 10.1006/geno.1997.4746 [DOI] [PubMed] [Google Scholar]
- 90.Gondo Y, Okada T, Matsuyama N, Saitoh Y, Yanagisawa Y, Ikeda JE. Human megasatellite DNA RS447: copy-number polymorphisms and interspecies conservation. Genomics 1998; 54(1):39-49; PMID:9806828; https://doi.org/ 10.1006/geno.1998.5545 [DOI] [PubMed] [Google Scholar]
- 91.Saitoh Y, Miyamoto N, Okada T, Gondo Y, Showguchi-Miyata J, Hadano S, Ikeda JE. The RS447 human megasatellite tandem repetitive sequence encodes a novel deubiquitinating enzyme with a functional promoter. Genomics 2000; 67(3):291-300; PMID:10936051; https://doi.org/ 10.1006/geno.2000.6261 [DOI] [PubMed] [Google Scholar]
- 92.Burrows JF, Mcgrattan MJ, Johnston JA. The DUB/USP17 deubiquitinating enzymes, a multigene family within a tandemly repeated sequence. Genomics 2005; 85:524-9; PMID:15780755; https://doi.org/ 10.1016/j.ygeno.2004.11.013 [DOI] [PubMed] [Google Scholar]
- 93.Burrows JF, Scott CJ, Johnston JA. The DUB/USP17 deubiquitinating enzymes : A gene family within a tandemly repeated sequence, is also embedded within the copy number variable Beta-defensin cluster. BMC Genomics 2010; 11(1):250; PMID:20044946; https://doi. org/ 10.1186/1471-2164-11-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.de la Vega M, Kelvin AA, Dunican DJ, McFarlane C, Burrows JF, Jaworski J, et al.. The deubiquitinating enzyme USP17 is essential for GTPase subcellular localization and cell motility. Nat Commun 2011; 2:257-9; PMID:21448156; https://doi.org/ 10.1038/ncomms1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Okada T, Gondo Y, Goto J, Kanazawa I, Hadano S, Ikeda JE. Unstable transmission of the RS447 human megasatellite tandem repetitive sequence that contains the USP17 deubiquitinating enzyme gene. Hum Genet 2002; 110(4):302-13; PMID:11941478; https://doi.org/ 10.1007/s00439-002-0698-2 [DOI] [PubMed] [Google Scholar]
- 96.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 2009; 10:637-43; PMID:19638999; https://doi.org/ 10.1038/nrm2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vanhée-Brossollet C, Vaquero C. Do natural antisense transcripts make sense in eukaryotes? Gene 1998; 211(1):1–9; PMID:9573333; https://doi.org/6201719 10.1016/S0378-1119(98)00093-6 [DOI] [PubMed] [Google Scholar]
- 98.van Arsdell SW, Weiner AM. Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol 1984; 4(3):492-9; PMID:6201719; https://doi.org/ 10.1128/MCB.4.3.492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hammarstrom K, Santesson B, Westin G, Pettersson U. The gene cluster for human U2 RNA is located on chromosome 17q21. Exp Cell Res 1985; 159:473-8; PMID:2411580; https://doi.org/ 10.1016/S0014-4827(85)80020-3 [DOI] [PubMed] [Google Scholar]
- 100.Liao D, Pavelitz T, Kidd JR, Kidd KK, Weiner AM. Concerted evolution of the tandemly repeated genes encoding human U2 snRNA (the RNU2 Locus) involves rapid intrachromosomal homogenization and rare interchromosomal gene conversion. EMBO J 1997; 16(3):588-98; PMID:9034341; https://doi.org/ 10.1093/emboj/16.3.588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liao D, Weiner AM. Concerted evolution of the tandemly repeated genes encoding primate U2 small nuclear RNA (the RNU2 Locus ) does not prevent rapid diversification of the (CT)n (GA)n microsatellite embedded withing the U2 repeat unit. Genomics 1995; 30:583-93; PMID:8825646; https://doi.org/ 10.1006/geno.1995.1280 [DOI] [PubMed] [Google Scholar]
- 102.Bailey AD, Pavelitz T, Weiner AM. The microsatellite sequence (CT)n (GA)n promotes stable chromosomal integration of large tandem arrays of functional human U2 small nuclear RNA genes. Mol Cell Biol 1998; 18(4):2262-71; PMID:9528797; https://doi.org/ 10.1128/MCB.18.4.2262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Htun H, Lund E, Westin G, Pettersson U, Dahlberg JE. Nuclease Si-sensitive sites in multigene families : human U2 small nuclear RNA genes. EMBO J 1985; 4(7):1839-45; PMID:2411549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tessereau C, Buisson M, Monnet N, Imbert M, Barjhoux L, Schluth-Bolard C, Sanlaville D, Conseiller E, Ceppi M, Sinilnikova OM, et al.. Direct visualization of the highly polymorphic RNU2 locus in proximity to the BRCA1 gene. PLoS One 2013; 8(10):e76054; PMID:24146815; https://doi.org/ 10.1371/journal.pone.0076054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tessereau C, Lesecque Y, Monnet N, Buisson M, Barjhoux L, Léoné M, Feng B, Goldgar DE, Sinilnikova OM, Mousset S, et al.. Estimation of the RNU2 macrosatellite mutation rate by BRCA1 mutation tracing. Proc Natl Acad Sci USA 2014; 42(14):9121-30; PMID:25034697; https://doi.org/ 10.1093/nar/gku639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pavelitz T, Rusche L, Matera AG, Scharf JM, Weiner AM. Concerted evolution of the tandem array encoding primate U2 snRNA occurs in situ, without changing the cytological context of the RNU2 locus. EMBO J 1995; 14(1):169-77; PMID:7828589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mazières J, Catherinne C, Delfour O, Gouin S, Rouquette I, Delisle MB, Prévot G, Escamilla R, Didier A, Persing DH, et al.. Alternative processing of the U2 small nuclear RNA produces a 19 – 22nt fragment with relevance for the detection of non-small cell lung cancer in human serum. PLoS One 2013; 8(3):e60134; PMID:23527303; https://doi.org/ 10.1371/journal.pone.0060134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang C, Liao D. Striking bimodal methylation of the repeat unit of the tandem array encoding human U2 snRNA (the RNU2 Locus). Genomics 1999; 62(3):508–18; PMID:10644450; https://doi.org/ 10.1006/geno.1999.6052 [DOI] [PubMed] [Google Scholar]
- 109.Bruce HA, Sachs N, Rudnicki DD, Lin SG, Willour VL, Cowell JK, Conroy J, McQuaid DE, Rossi M, Gaile DP, et al.. Long tandem repeats as a form of genomic copy number variation : structure and length polymorphism of a chromosome 5p repeat in control and schizophrenia populations. Psychiatr Genet 2009; 19:64-71; PMID:19672138; https://doi.org/ 10.1097/YPG.0b013e3283207ff6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen YT, Iseli C, Venditti CA, Old LJ, Simpson AJ, Jongeneel CV. Identification of a new cancer/testis gene family, CT47, among expressed multicopy genes on the human X chromosome. Genes Chromosomes Cancer 2005; 45(4):392-400; PMID:16382448; https://doi.org/ 10.1002/gcc.20298 [DOI] [PubMed] [Google Scholar]
- 111.Balog J, Miller D, Sanchez-Curtailles E, Carbo-Marques J, Block G, Potman M, de Knijff P, Lemmers RJ, Tapscott SJ, van der Maarel SM. Epigenetic regulation of the X-chromosomal macrosatellite repeat encoding for the cancer/testis gene CT47. Euro J Hum Genet 2011; 20(2):185-91; PMID:21811308; https://doi.org/ 10.1038/ejhg.2011.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 2005; 20(3):483-9; PMID:16285929; https://doi.org/ 10.1016/j.molcel.2005.09.002 [DOI] [PubMed] [Google Scholar]
- 113.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell 2016; 29(4):452-63; PMID:27070700; https://doi.org/ 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]


