Abstract
Purpose
Osteosarcoma is the most prevalent primary bone tumor in children, adolescents, and older adults, typically presenting with poor survival outcomes. In recent years, ample evidence has shown that many long noncoding RNAs (lncRNAs) have been aberrantly expressed in osteosarcoma, demonstrating their potential to serve as prognostic markers. In this study, we performed a meta-analysis on four lncRNAs (TUG1, UCA1, BCAR4, and HULC) to systematically evaluate their prognostic value in osteosarcoma.
Materials and methods
The eligible articles were systematically searched in PubMed, Web of Science, Embase, and Elsevier ScienceDirect (up to September 22, 2017), and one meta-analysis concerning the association between lncRNA expression and the overall survival (OS) of osteosarcoma patients was performed. Survival outcomes were analyzed by OS. Subgroup analyses were performed.
Results
A total of 1,361 patients with osteosarcoma and 12 lncRNAs from 16 articles were included in the study. Of the listed lncRNAs, the high expression of 10 lncRNAs indicated worse survival outcomes, while only two lncRNAs were shown to positively affect patients’ OS.
Conclusion
This meta-analysis indicated that the abnormally expressed lncRNAs might significantly affect the survival of osteosarcoma patients. Combined use of these lncRNAs may serve as potential novel biomarkers for the indication of clinical outcomes of osteosarcoma patients as well as the selection of adjuvant chemotherapy strategies for clinical treatment of this disease.
Keywords: lncRNAs, osteosarcoma, prognosis, meta-analysis
Introduction
Osteosarcoma is an aggressive malignancy primarily occurring in the bone of both children and adults. Although primary bone cancers account for less than 0.2% of all cancers,1 their frequency has kept increasing by 0.3% annually over the past decade according to the National Cancer Institute SEER program.2 The clinical treatment of osteosarcoma involves chemotherapy in combination with surgery. Depending on the success of therapy, the 5-year survival rates of patients with localized tumors have increased ranging from 60% to 80%, while patients with detectable metastases at the time of diagnosis or recurrences usually have poor prognoses, with reports of 5-year survival rates as low as 19%.3–6 In addition, many targeted therapies have been tried over the past 20 years.7 However, these therapeutic strategies have had minimal beneficial effects on the improvement of survival outcomes.7–10 This is primarily due to the fact that the genetic etiology of osteosarcoma is extremely heterogeneous and has not been fully understood.11 Thus, the identification of new molecule targets is urgently needed for the improvement of clinical approaches and outcomes of osteosarcoma patients.
Insights gained from the latest research on cancer-related molecules may lead to better clinical prognostic evaluations for patients with cancer. Long noncoding RNAs (lncRNAs), previously viewed as transcriptional “noise,”12 have been currently recognized as functional molecules that play essential roles in various biological processes and diseases including cancers.13,14 Accumulating evidence has confirmed that abnormal expression of multiple lncRNAs confers cell proliferation, metastasis, and drug resistance in osteosarcoma.15–18
Hence, we conducted a meta-analysis to identify the exact role of lncRNAs in the prognosis of osteosarcoma patients. Moreover, we summed up the correlation of distinct lncRNAs and the prognosis of osteosarcoma patients in the current study. The pooled results from meta-analysis indicated that deregulated expression of lncRNAs might considerably affect the overall survival (OS) of osteosarcoma patients and may serve as useful biomarkers of osteosarcoma prognosis. This may provide us with new insights into the clinical value of application of joint detection of some of these potential biomarkers in predicting prognosis and the determination of therapeutic strategies of osteosarcoma.
Materials and methods
Literature search strategy
We primarily conducted a systematic selection of literature published in English from the databases of PubMed, Web of Science, and Embase. A comprehensive search was performed based on the varying combination of the following subject terms: osteosarcoma, prognosis, long non-coding RNA, or lncRNA. The last update of searching time was May 19, 2017. A second search was performed on September 22, 2017, using the same keywords in the databases including PubMed, Web of Science, Embase, and Elsevier ScienceDirect. A total of 116 articles were returned in the second search.
Selection criteria
Titles and abstracts of the retrieved articles were first checked by two authors (Ying Wang and Yuelong Huang), and 42 duplicate articles were removed. Afterward, we came up with inclusion criteria for eligible articles that were analyzed for our full-text evaluation: 1) studies about the correlation between lncRNA expression in tumor tissues or blood and prognosis of patients with osteosarcoma; 2) the survival outcomes that were measured with OS; 3) the detection method of lncRNAs which was restricted to quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) or fluorescence in situ hybridization (FISH). The inclusion criteria are shown in Table 1. Studies that are not fitting the aforementioned inclusion criteria, laboratory studies, reviews, and articles of other tumors were excluded.
Table 1.
Inclusion criteria for studies on the prognosis of osteo-sarcoma based on lncRNA expression levels
| Inclusion items | Selection criteria |
|---|---|
| Tumor type | Osteosarcoma |
| Sample type | Tumor tissue or blood |
| Assay method | qRT-PCR or FISH |
| Time of study | January 2003 to September 2017 |
| Follow-up (months) | ≥60 |
| Included results | Multivariate analysis of OS and Kaplan–Meier survival curves |
Abbreviations: FISH, fluorescence in situ hybridization; lncRNA, long noncoding RNA; OS, overall survival; qRT-PCR, quantitative reverse transcriptase polymerase chain reaction.
Data extraction
All data were retrieved from eligible studies independently by two authors (Ying Wang and Yuelong Huang). Data extracted from eligible studies were shown as follows: first author’s name; study publication year; investigated lncRNA; sample size; survival outcome parameter; follow-up time; hazard ratios (HR) with 95% confidence interval (95% CI); P-value. If HR and 95% CI were not directly given in the original literature, data in tables or figures were extracted. Differences of opinion were discussed with a third investigator (Peng Xiang).
Statistical analysis
Some individual HRs with 95% CI were calculated based upon the extracted log-rank P-value and Kaplan–Meier survival curves of survival outcomes with the number of patients using Engauge Digitizer version 4.1 and Tierney’s method.19 The log HR and standard error (SE) were used for aggregation of the survival outcomes of different lncRNAs. In our meta-analysis, we evaluated the heterogeneity using a fixed-effect model without significant heterogeneity (P>0.1 and I2<50%); otherwise, the random-effect model was applied in RevMan 5.2 software. All P-values were two sided, and a P-value of <0.05 was considered significant.
Results
Characteristics of included studies
As shown in Figure 1, 116 studies were returned after we searched for the second time through databases, such as PubMed, Web of Science, Embase, and Elsevier Science-Direct. A total of 42 duplicated records were eliminated. After two authors carefully reviewed the titles and abstracts independently, we further removed 50 unqualified publications, including 38 laboratory studies, four review articles, seven other types of cancer study, and one retracted study. Moreover, studies that did not use OS as survival parameters (n=8) were excluded as well. Thus, 16 eligible articles were finally included.
Figure 1.
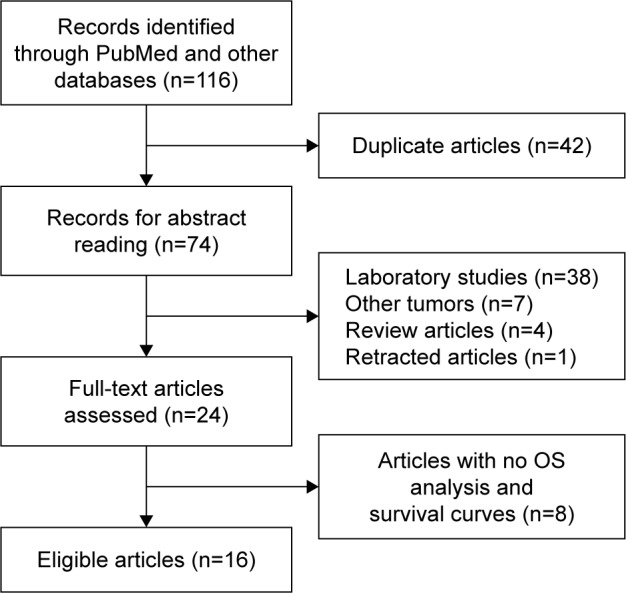
The flowchart of the selection process.
Abbreviation: OS, overall survival.
All the enrolled studies analyzed the prognosis of 1,361 patients diagnosed with osteosarcoma with or without initial evidence of metastasis and the association between the expression level of 12 lncRNAs and survival outcomes. All the necessary information of included studies is displayed in Tables 1 and 2. As shown in Table 2, the number of patients included in each study ranged from 33 to 168, and the duration of follow-up varied from 60 months to a maximum of 120 months. Of the enrolled studies, 15 included participants from China15,17,20–32 and one involved patients from Brazil.33 All studies investigated patients with osteosarcoma and qRT-PCR was applied for the evaluation of the expression level of each lncRNA.
Table 2.
Basic information of included articles
| Reference | Year | LncRNA | Total patients (n) | Survival analysis | Follow-up (months) |
|---|---|---|---|---|---|
| Wang et al21 | 2017 | TUG1 | 44 | OS | 120 |
| Wen et al23 | 2017 | UCA1 | 151 | OS | 60 |
| Zhou et al30 | 2017 | CCAL | 46 | OS | 60 |
| Chen et al24 | 2016 | BCAR4 | 60 | OS | 60 |
| Cong et al32 | 2016 | TUSC7 | 82 | OS | 120 |
| Gao and Lian27 | 2016 | MALAT1 | 162 | OS | 65 |
| Ju et al25 | 2016 | BCAR4 | 168 | OS | 68 |
| Li et al26 | 2016 | HIF2PUT | 82 | OS | 60 |
| Li et al22 | 2016 | UCA1 | 135 | OS | 60 |
| Ma et al20 | 2016 | TUG1 | 76 | OS | 60 |
| Uzan et al33 | 2016 | HULC | 33 | OS | 96 |
| Xia et al29 | 2016 | 91H | 67 | OS | 60 |
| Zhao et al15 | 2016 | HNF1A-AS1 | 43 | OS | 60 |
| Li et al28 | 2015 | HOTTIP | 68 | OS | 60 |
| Sun et al17 | 2015 | HULC | 78 | OS | 60 |
| Tian et al31 | 2015 | MEG3 | 64 | OS | 60 |
Abbreviations: lncRNA, long noncoding RNA; OS, overall survival.
Overall analysis
A total of 10 lncRNAs were indicated to negatively affect the OS of patients, while two lncRNAs were associated with favorable OS outcomes. All HRs, 95% CI, and P-values of included studies are listed in Table 3.
Table 3.
HRs of lncRNA expression in osteosarcoma
| LncRNA | HR (OS) | 95% CI
|
P-value | lnHR | SE | Related to poor prognosis | |
|---|---|---|---|---|---|---|---|
| Lower | Upper | ||||||
| TUG1 | 2.78 | 1.29 | 6.00 | 0.009 | 1.02 | 0.39 | High |
| TUG1 | 3.6 | 1.01 | 12.8 | 0.035 | 1.28 | 0.65 | High |
| UCA1 | 2.52 | 1.32 | 4.83 | 0.011 | 0.92 | 0.33 | High |
| UCA1 | 3.13 | 1.29 | 7.55 | 0.015 | 1.14 | 0.45 | High |
| BCAR4 | 3.22 | 0.89 | 7.88 | 0.014 | 1.17 | 0.55 | High |
| BCAR4 | 2.32 | 1.24 | 5.62 | 0.018 | 0.84 | 0.40 | High |
| HULC | 2.27 | 0.61 | 8.44 | 0.016 | 0.82 | 0.67 | High |
| HULC | 2.28 | 1.48 | 5.43 | 0.009 | 0.83 | 0.33 | High |
| HIF2PUT | 5.48 | 1.99 | 12.29 | 0.01 | 1.70 | 0.46 | High |
| MALAT1 | 3.16 | 1.56 | 6.88 | 0.003 | 1.15 | 0.38 | High |
| HNF1A-AS1 | 2.64 | 1.39 | 7.42 | 0.005 | 0.97 | 0.43 | High |
| HOTTIP | 2.89 | 1.37 | 7.06 | 0.007 | 1.06 | 0.42 | High |
| 91H | 3.13 | 1.32 | 7.49 | 0.010 | 1.04 | 0.44 | High |
| CCAL | 3.18 | 1.64 | 9.94 | 0.021 | 1.16 | 0.46 | High |
| MEG3 | 0.81 | 0.27 | 0.97 | 0.006 | −0.21 | 0.32 | Low |
| TUSC7 | 0.31 | 0.09 | 0.87 | 0.020 | −1.17 | 0.58 | Low |
Abbreviations: CI, confidence interval; HR, hazard ratio; lncRNA, long noncoding RNA; lnHR, log hazard ratio; OS, overall survival; SE, standard error.
Subgroup analysis
Among the 12 listed lncRNAs, four (TUG1, UCA1, BCAR4, and HULC) have been studied in more than one article. We therefore carried out meta-analysis and obtained the combined HRs.
TUG1
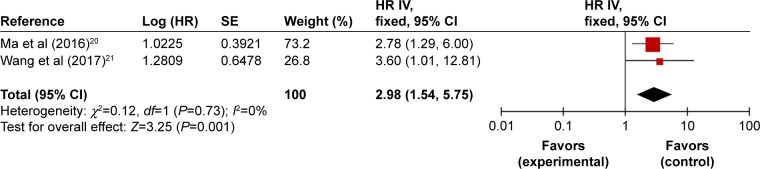
We included two studies concerning the correlation of TUG1 expression with OS of osteosarcoma patients. Ma et al20 studied a cohort of 76 pairs of osteosarcoma tumor tissues. Multivariate analysis using Cox’s proportional hazards model was performed, and HR for high TUG1 expression was 2.78 (95% CI: 1.29–6.00, P=0.009). In Wang et al’s21 study, 44 patients diagnosed with osteosarcoma were enrolled. This study presented log-rank test P-value and Kaplan–Meier survival curves of survival outcomes in high and low expression group, instead of multivariate Cox hazard regression analysis. The calculated HR (HR =3.6, 95% CI: 1.01–12.8, P=0.035) is shown in Table 3.
We then carried out meta-analysis with these two articles. No heterogeneity between studies was observed, and therefore fixed-effect model was applied. A combined HR of 2.98 (95% CI: 1.54–5.75, P=0.001) for those patients with high expression of TUG1 was observed (Figure 2). These results indicate that overexpressed TUG1 could predict worse prognosis of osteosarcoma patients concerning OS and could be used as an independent prognostic marker.
Figure 2.
Forest plots of studies evaluating HRs of high TUG1 expression as compared to low expression.
Abbreviations: CI, confidence interval; HR, hazard ratio; IV, inverse variance; SE, standard error.
UCA1
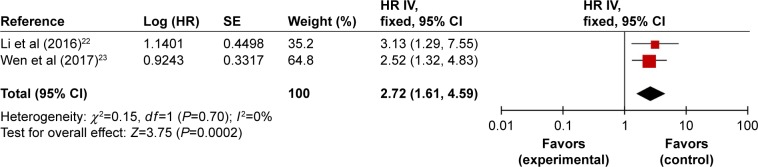
Two studies described the overexpressed UCA1 as a significant predictor of poor OS in osteosarcoma. Li et al22 reported the relevance of UCA1 expression to prognosis of osteosarcoma patients in a cohort of 135 patients. This article presented Kaplan–Meier survival curves, HR, and log-rank P-value, without information about 95% CI; as such, HR with 95% CI was calculated and is shown in Table 3 (HR =3.13, 95% CI: 1.29–7.55, P=0.015). Wen et al23 enrolled 151 patients diagnosed with osteosarcoma who had never received radiotherapy and chemotherapy prior to surgical resection. Multivariate Cox hazard regression analysis was carried out, and HR for high tumoral UCA1 expression was 2.52 (95% CI: 1.32–4.83, P=0.011). To clarify the impact of UCA1 on patients’ OS, we performed a meta-analysis. We observed no heterogeneity between studies (I2=0%, P=0.70), and hence the fixed-effect model was used to calculate the association between high UCA1 expression and OS (HR =2.72, 95% CI: 1.61–4.59, P=0.0002; Figure 3). These results suggest that high expression of UCA1 might develop as an independent predictor of survival for osteosarcoma patients.
Figure 3.
Forest plots of studies evaluating HRs of high UCA1 expression as compared to low expression.
Abbreviations: CI, confidence interval; HR, hazard ratio; IV, inverse variance; SE, standard error.
BCAR4
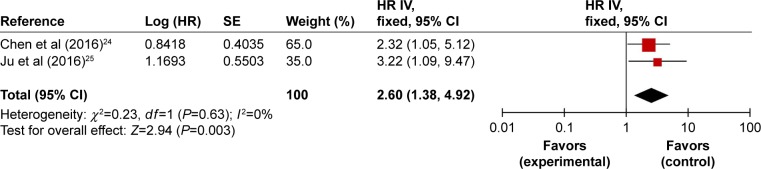
We carried out meta-analysis with two studies demonstrating the elevated expression of BCAR4 as predictive of poor OS in osteosarcoma. Chen et al24 and Ju et al25 enrolled, respectively, 60 patients and 168 patients diagnosed with osteosarcoma who had never received any therapy prior to surgery. Both studies performed Kaplan–Meier survival analysis. We observed no significant heterogeneity between these two studies (I2=0%, P=0.63). Further meta-analysis using fixed-effect model revealed that high expression of BCAR4 could be used as an independent predictor of inferior prognosis of osteosarcoma patients (HR =2.60, 95% CI: 1.38–4.92, P=0.003; Figure 4).
Figure 4.
Forest plots of studies evaluating HRs of high BCAR4 expression as compared to low expression.
Abbreviations: CI, confidence interval; HR, hazard ratio; IV, inverse variance; SE, standard error.
HULC
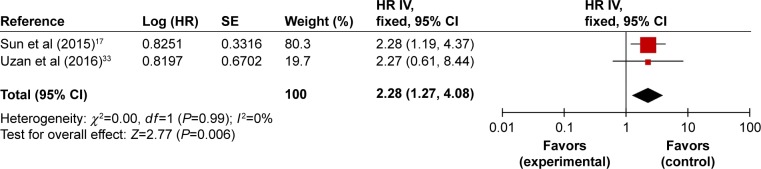
We performed meta-analysis on articles choosing lncRNA HULC as a prognostic marker. There were two studies included in this meta-analysis. In Sun et al’s17 study, multivariate Cox regression analysis was conducted, the data such as HR and 95% CI were subsequently directly extracted and put into pooled analysis. Uzan et al33 enrolled 33 patients in the study, which presented Kaplan–Meier OS curves with corresponding log-rank P-value, in place of multivariate Cox regression analysis result, the HR (HR =2.27, 95% CI: 0.61–8.44, P=0.016) for death was therefore calculated and shown in Table 3. Since there was no evidence of considerable heterogeneity in these two studies (I2=0%, P=0.99), the fixed-effect model was selected. A combined HR of 2.28 (95% CI: 1.27–4.08, P=0.006) for patients with high expression of HULC was found (Figure 5), from which we drew the conclusion that high expression of HULC is an independent predictor of poorer survival in patients diagnosed with osteosarcoma.
Figure 5.
Forest plots of studies evaluating HRs of high HULC expression as compared to low expression.
Abbreviations: CI, confidence interval; HR, hazard ratio; IV, inverse variance; SE, standard error.
Discussion
Up to date, the clinical treatment for osteosarcoma is still limited and little prognostic improvement has been achieved from the past 2 decades’ research on either traditional or novel therapeutic approaches. Hence, novel biomarkers that may facilitate the prediction of prognosis of patients with osteosarcoma are still urgently needed. Recently, increasing evidence has demonstrated that lncRNAs can help assist or hinder tumor growth and metastasis. Even more important, multiple aberrantly expressed lncRNAs have been found to be involved in the development of osteosarcoma and may have prognostic potential in this disease.
In our study, a total of 16 recently published articles comprising 12 lncRNAs that were potential biomarkers for survival prognosis and 1,361 patients were included. In this study, we performed a meta-analysis to investigate the potential role of these lncRNAs as novel prognostic biomarkers of survival in osteosarcoma. All qualified studies used qRT-PCR to detect lncRNAs expression. In most of the selected studies, the patients with osteosarcoma were classified into high and low expression groups according to the median expression level of each examined lncRNA, while in studies by Ma et al20 and Uzan et al,33 the lncRNA expression cutoff value was determined based upon the receiver operating characteristic analysis. In terms of the prognostic value, the high expression level of 10 lncRNAs was associated with poor prognosis, as was the low expression level of two lncRNAs.
Our meta-analysis focused on four lncRNAs (TUG1, UCA1, BCAR4, HULC), the putative prognostic value of which in osteosarcoma has been clearly demonstrated in more than one study. The lncRNA TUG1, short for Taurine upregulated gene 1, was originally reported to be upregulated in response to taurine treatment of developing mouse retinal cells and has been frequently described as an oncogene in various human cancers.34–36 UCA1 (urothelial carcinoma associated 1) was initially identified in human bladder carcinoma and has been shown to exert oncogenic activity in other human malignant tumors such as pancreatic cancer and lung cancer.37–39 BCAR4 (breast cancer antiestrogen resistance 4) was first identified in screening for tamoxifen resistance-associated genes in breast cancer and has been recently implicated in the positive regulation of cell proliferation, invasion, and metastasis,40,41 demonstrating this molecule as a critical oncogene. HULC (highly upregulated in liver cancer) is one of the first strongly overexpressed lncRNAs to be identified in primary tumors of the liver.42 Since it was discovered, the aberrant upregulation of HULC has been frequently observed in other types of tumors and has been described in various cellular processes such as cell proliferation, metastasis, and angiogenesis.43,44 To enhance the statistic power to confirm the prognostic efficacy of these lncRNAs in osteosarcoma, subgroup analysis with a fixed- or random-effect-model was carried out in the current study. The combined HRs indicated that elevated expression of TUG1, UCA1, BCAR4, and HULC was significantly correlated with poor prognosis of osteosarcoma patients (Figures 2–5), suggesting their potential role as prognostic biomarkers. It is noteworthy in this study that the qRT-PCR technique was applied in all enrolled studies to detect the expression level of lncRNAs that made pooled data from different independent studies more convincing in the case of consistent measurement background, yet it was also noticed that the detailed procedure of this technique used across the studies was similar but not identical. To be used as a prognostic biomarker, the qRT-PCR detection method should be routinely implemented to detect and quantify the expression of specific lncRNA, and this calls for the establishment of the standardization of the implementation processes across different clinical laboratories and appropriate quality controls that directly relate to the feasibility and reproducibility of the application of qRT-PCR, as well as the evaluation of the sensitivity and specificity of this technique. In consideration of the complicated experimental processes and influencing factors, it is essential that large-scale multicenter prospective studies need to be conducted after the standardization of the experimental scheme to investigate the feasibility and reproducibility of this technique in the future.
It should be highlighted that there are some limitations in the current study where further refinement is needed. First, as studies involved were published in English only, the bias of selection could occur. Second, due to the reason that the size of the eligible literatures was relatively small, the meta-analysis was carried out on very few numbers of studies and there was only one study for most of the investigated lncRNAs, which will probably lead to an overestimate of the prognostic value of these lncRNAs. Third, even though these involved lncR-NAs could independently predict the prognostic outcomes, there is still lack of lncRNAs that are specific biomarkers for clinical evaluation of patients with osteosarcoma. Finally, the dominant ethnic group of patients in our study was Chinese; therefore, our results should be interpreted with great caution when applying our results to other populations.
Conclusion
In summary, our results confirmed the relatively strong prognostic value of the transcription level of lncRNAs that have been detected in different population-based cohort studies in osteosarcoma. Nevertheless, in consideration of the abovementioned limitations, these findings need to be further verified by larger numbers of clinical trials in the future to confirm the clinical utility of these lncRNAs in osteosarcoma prognosis evaluation.
Acknowledgments
This work was largely supported by the National Natural Science Foundation of China grant 81402219 to Ying Wang. This work was also supported, in part, by the National Natural Science Foundation of China grant 81330043 to Wei Tian.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Biermann JS, Adkins DR, Agulnik M, et al. Bone cancer. J Natl Compr Canc Netw. 2013;11(6):688–723. doi: 10.6004/jnccn.2013.0088. [DOI] [PubMed] [Google Scholar]
- 2.National Institutes of Health SEER; Cancer Stat Facts: Bone and Joint Cancer. [Accessed November 2, 2017]. Available from: https://seer.cancer.gov/statfacts/html/bones.html.
- 3.Mialou V, Philip T, Kalifa C, et al. Metastatic osteosarcoma at diagnosis: prognostic factors and long-term outcome – the French pediatric experience. Cancer. 2005;104(5):1100–1109. doi: 10.1002/cncr.21263. [DOI] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kager L, Zoubek A, Potschger U, et al. Primary metastatic osteosarcoma: presentation and outcome of patients treated on neoadjuvant Cooperative Osteosarcoma Study Group protocols. J Clin Oncol. 2003;21(10):2011–2018. doi: 10.1200/JCO.2003.08.132. [DOI] [PubMed] [Google Scholar]
- 6.Gorlick R, Anderson P, Andrulis I, et al. Biology of childhood osteogenic sarcoma and potential targets for therapeutic development: meeting summary. Clin Cancer Res. 2003;9(15):5442–5453. [PubMed] [Google Scholar]
- 7.Omer N, Le Deley MC, Piperno-Neumann S, et al. Phase-II trials in osteosarcoma recurrences: a systematic review of past experience. Eur J Cancer. 2017;75:98–108. doi: 10.1016/j.ejca.2017.01.005. [DOI] [PubMed] [Google Scholar]
- 8.Ebb D, Meyers P, Grier H, et al. Phase II trial of trastuzumab in combination with cytotoxic chemotherapy for treatment of metastatic osteosarcoma with human epidermal growth factor receptor 2 overexpression: a report from the children’s oncology group. J Clin Oncol. 2012;30(20):2545–2551. doi: 10.1200/JCO.2011.37.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pappo AS, Vassal G, Crowley JJ, et al. A phase 2 trial of R1507, a monoclonal antibody to the insulin-like growth factor-1 receptor (IGF-1R), in patients with recurrent or refractory rhabdomyosarcoma, osteosarcoma, synovial sarcoma, and other soft tissue sarcomas: results of a Sarcoma Alliance for Research Through Collaboration study. Cancer. 2014;120(16):2448–2456. doi: 10.1002/cncr.28728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zalupski MM, Rankin C, Ryan JR, et al. Adjuvant therapy of osteosarcoma – a phase II trial: Southwest Oncology Group study 9139. Cancer. 2004;100(4):818–825. doi: 10.1002/cncr.20021. [DOI] [PubMed] [Google Scholar]
- 11.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36:5661–5667. doi: 10.1038/onc.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathieu EL, Belhocine M, Dao LT, Puthier D, Spicuglia S. Rôle des longs ARN non codants dans le développement normal et pathologique [Functions of lncRNA in development and diseases] Med Sci (Paris) 2014;30(8–9):790–796. doi: 10.1051/medsci/20143008018. French. [DOI] [PubMed] [Google Scholar]
- 15.Zhao H, Hou W, Tao J, et al. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/beta-catenin signaling pathway. Am J Transl Res. 2016;8(8):3503–3512. [PMC free article] [PubMed] [Google Scholar]
- 16.Yun-Bo F, Xiao-Po L, Xiao-Li L, Guo-Long C, Pei Z, Fa-Ming T. LncRNA TUG1 is upregulated and promotes cell proliferation in osteosarcoma. Open Med (Wars) 2016;11(1):163–167. doi: 10.1515/med-2016-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun XH, Yang LB, Geng XL, Wang R, Zhang ZC. Increased expression of lncRNA HULC indicates a poor prognosis and promotes cell metastasis in osteosarcoma. Int J Clin Exp Pathol. 2015;8(3):2994–3000. [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. doi: 10.1016/j.canlet.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 19.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17(24):2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 20.Ma B, Li M, Zhang L, et al. Upregulation of long non-coding RNA TUG1 correlates with poor prognosis and disease status in osteosar-coma. Tumour Biol. 2016;37(4):4445–4455. doi: 10.1007/s13277-015-4301-6. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Yang T, Zhang Z, et al. Long non-coding RNA TUG1 promotes migration and invasion by acting as a ceRNA of miR-335-5p in osteosarcoma cells. Cancer Sci. 2017;108(5):859–867. doi: 10.1111/cas.13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Xie P, Ruan WH. Overexpression of lncRNA UCA1 promotes osteosarcoma progression and correlates with poor prognosis. J Bone Oncol. 2016;5(2):80–85. doi: 10.1016/j.jbo.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen JJ, Ma YD, Yang GS, Wang GM. Analysis of circulating long non-coding RNA UCA1 as potential biomarkers for diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2017;21(3):498–503. [PubMed] [Google Scholar]
- 24.Chen F, Mo J, Zhang L. Long noncoding RNA BCAR4 promotes osteosarcoma progression through activating GLI2-dependent gene transcription. Tumour Biol. 2016;37(10):13403–13412. doi: 10.1007/s13277-016-5256-y. [DOI] [PubMed] [Google Scholar]
- 25.Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. 2016;20(21):4445–4451. [PubMed] [Google Scholar]
- 26.Li W, He X, Xue R, et al. Combined over-expression of the hypoxia-inducible factor 2alpha gene and its long non-coding RNA predicts unfavorable prognosis of patients with osteosarcoma. Pathol Res Pract. 2016;212(10):861–866. doi: 10.1016/j.prp.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Gao KT, Lian D. Long non-coding RNA MALAT1 is an independent prognostic factor of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(17):3561–3565. [PubMed] [Google Scholar]
- 28.Li F, Cao L, Hang D, Wang F, Wang Q. Long non-coding RNA HOTTIP is up-regulated and associated with poor prognosis in patients with osteosarcoma. Int J Clin Exp Pathol. 2015;8(9):11414–11420. [PMC free article] [PubMed] [Google Scholar]
- 29.Xia WK, Lin QF, Shen D, Liu ZL, Su J, Mao WD. Clinical implication of long noncoding RNA 91H expression profile in osteosarcoma patients. Onco Targets Ther. 2016;9:4645–4652. doi: 10.2147/OTT.S103376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou DK, Yang XW, Li H, Yang Y, Zhu ZJ, Wu N. Up-regulation of long noncoding RNA CCAL predicts poor patient prognosis and promotes tumor metastasis in osteosarcoma. Int J Biol Markers. 2017;32(1):e108–e112. doi: 10.5301/jbm.5000240. [DOI] [PubMed] [Google Scholar]
- 31.Tian ZZ, Guo XJ, Zhao YM, Fang Y. Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma. Int J Clin Exp Pathol. 2015;8(11):15138–15142. [PMC free article] [PubMed] [Google Scholar]
- 32.Cong M, Li J, Jing R, Li Z. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016;37(7):9441–9450. doi: 10.1007/s13277-015-4414-y. [DOI] [PubMed] [Google Scholar]
- 33.Uzan VR, Lengert A, Boldrini E, et al. High expression of HULC is associated with poor prognosis in osteosarcoma patients. PLoS One. 2016;11(6):e0156774. doi: 10.1371/journal.pone.0156774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregu-lated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15(6):501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- 35.Qin CF, Zhao FL. Long non-coding RNA TUG1 can promote proliferation and migration of pancreatic cancer via EMT pathway. Eur Rev Med Pharmacol Sci. 2017;21(10):2377–2384. [PubMed] [Google Scholar]
- 36.Li T, Liu Y, Xiao H, Xu G. Long non-coding RNA TUG1 promotes cell proliferation and metastasis in human breast cancer. Breast Cancer. 2017;24(4):535–543. doi: 10.1007/s12282-016-0736-x. [DOI] [PubMed] [Google Scholar]
- 37.Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi: 10.1016/j.febslet.2008.05.012. [DOI] [PubMed] [Google Scholar]
- 38.Chen P, Wan D, Zheng D, Zheng Q, Wu F, Zhi Q. Long non-coding RNA UCA1 promotes the tumorigenesis in pancreatic cancer. Biomed Pharmacother. 2016;83:1220–1226. doi: 10.1016/j.biopha.2016.08.041. [DOI] [PubMed] [Google Scholar]
- 39.Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi: 10.1016/j.canlet.2015.11.024. [DOI] [PubMed] [Google Scholar]
- 40.Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res. 2006;4(6):379–386. doi: 10.1158/1541-7786.MCR-05-0156. [DOI] [PubMed] [Google Scholar]
- 41.Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21(9):2075–2086. [PubMed] [Google Scholar]
- 42.Panzitt K, Tschernatsch MM, Guelly C, et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology. 2007;132(1):330–342. doi: 10.1053/j.gastro.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 43.Zhao Y, Guo Q, Chen J, Hu J, Wang S, Sun Y. Role of long non-coding RNA HULC in cell proliferation, apoptosis and tumor metastasis of gastric cancer: a clinical and in vitro investigation. Oncol Rep. 2014;31(1):358–364. doi: 10.3892/or.2013.2850. [DOI] [PubMed] [Google Scholar]
- 44.Wang YF, Zhang S, Li XQ, Wang Y. Expression of lncRNA HULC in cervical cancer and its correlation with tumor progression and patient survival. Eur Rev Med Pharmacol Sci. 2016;20(19):3987–3991. [PubMed] [Google Scholar]