Abstract
During early plant embryogenesis, precursors for all major tissues and stem cells are formed. While several components of the regulatory framework are known, how cell fates are instructed by genome-wide transcriptional activity remains unanswered - in part because of difficulties in capturing transcriptome changes at cellular resolution. Here, we have adapted a two-component transgenic labelling system to purify cell type-specific nuclear RNA and generate a transcriptome atlas of early Arabidopsis embryo development, with focus on root stem cell niche formation. We validated the dataset through gene expression analysis, and show that gene activity shifts in a spatio-temporal manner, likely signifying transcriptional reprogramming, to induce developmental processes reflecting cell states and state transitions. This atlas provides the most comprehensive tissue- and cell-specific description of genome-wide gene activity in the early plant embryo, and serves as a valuable resource for understanding the genetic control of early plant development.
Keywords: Cell type-specific, Nuclear isolation, Embryo, Transcriptome analysis, Root stem cell niche
Introduction
Land plants (Embryophytes) develop as a result of the continuous formation of tissue layers and organs from stem cells. This necessitates the coordination of oriented cell divisions, cell-cell communication and genetic regulatory mechanisms that together form a framework for cell fate determination and pattern formation. The framework is first established during early embryogenesis where it sets up the basic body pattern from tissue precursors and stem cells. Thus, the formation of all plant structures, regardless of complexity, is the results of iteration of a developmental process first established in the early embryo. As such, plant embryogenesis can be considered a miniature model for plant development. Consequently, a central question in plant biology is how the framework is established and regulated in the embryo to allow for cell and tissue specification, and, subsequently, an ordered 3D structure.
In seed plants, the mature embryo (seedling) displays the same basic body plan of the post-embryonic structure1: an overlapping apical-basal and radial pattern with a shoot and root apical meristem (SAM and RAM) along the apical-basal axis of polarity, and the basic tissues (epidermis, vascular tissue, ground tissue) forming concentric radial layers. One or more cotyledons flank the SAM. The meristems are tissues that contain specialized microenvironments called “niches” in which stem cells continuously divide to produce differentiated cells (for tissue/organ formation) and so-called organizer cells prevent stem cell differentiation2,3. The niche organizer is termed the organizer centre in shoots and the quiescent centre (QC) in roots. In the flowering plant Arabidopsis thaliana, a highly invariant cell division pattern makes it possible to track the origin of the basic body plan to specific cell types in the early embryo4–7. This, together with its small genome size and relative ease to genetically manipulate, has made Arabidopsis the de facto model organism to study the cell fate of stem cells, organizer cells and precursors of the major tissues. In past years, several important components of the regulatory framework underlying their establishment have been identified (reviewed previously8), however a key question of how these cell fates are instructed by cellular transcriptomes remains unanswered.
A major challenge has been to adapt cell-specific genome-wide approaches to the small plant embryo contained within seed and fruit tissues. Manual and laser capture microdissection (LCM)9 have been used to isolate whole embryos and larger embryonic tissue domains from various plant species for transcriptional analysis10–14. Notably, this has elucidated spatio-temporal gene expression patterns15–18,10, the dynamics of zygotic genome activation19,20, and microRNA function21. While important, these studies are limited when investigating developmental decisions since they, by definition, occur at a cellular scale. However, the recent development of several cell-specific approaches in other systems, such as fluorescence-activated cell/nuclei sorting (FACS/FANS)22,23 and transgenic labelling and affinity purification of nuclei/polysomes (INTACT/TRAP)24–26, have now made it possible to determine gene expression at the single cell type level in the plant embryo. Of these methods, INTACT holds the most promise for studying embryogenesis.
Although the use of both FACS/FANS and TRAP have provided important insights regarding cell fate during Arabidopsis development27–32 (reviewed previously33), a main concern is that promoters used to transgenically mark cells need to be exclusive to a specific cell or tissue type. This is an issue especially in embryos since many embryonically expressed genes are also found in equivalent tissues of the surrounding seed material. INTACT circumvents this by utilizing a two-component transgenic labelling system where biotin ligase (BirA) biotinylates a nuclear envelope-localised GFP protein (nuclear tagging factor or NTF) when co-expressed in the same cells26. The fast, specific and high affinity binding between biotin and streptavidin is then exploited to efficiently isolate biotin-tagged nuclei from crude nuclear preparations using streptavidin-coated beads. To date, INTACT has been used to isolate cell type-specific nuclei from Arabidopsis root26,34, female central cell35 and endosperm36, from tomato root37, and from several animal models38–40, for chromatin, proteomic and gene expression studies.
Here, we have optimized INTACT, both in regards to protocol setup and the two-component labelling system, to generate a transcriptome atlas of early Arabidopsis embryo development at cell type-specific resolution. Focus has been on the cell types essential for root stem cell niche formation. To accomplish this, we have established a wide array of INTACT-lines driven by promoters expressed in discrete cell types of the early embryo. By describing and integrating spatial and temporal genome-wide gene activity at the cellular level, our work provides a resource to explore the developmental processes and the genetic networks that shape the first tissues of the plant.
Results
Adaptation of INTACT for early embryo cell types
In order to adapt the INTACT method for use on early Arabidopsis embryos, a codon-optimized version of BirA was synthesized to replace the original E. coli-derived BirA fragment. In this mBirA, all codons were designed according to the Arabidopsis codon preference to facilitate translation (Supplementary Fig. 1). In addition, either a 3xMyc epitope tag (mBirA-3xMyc) or fluorescent mCherry (mBirA-mCherry) was fused to mBirA to allow determination of expression in transgenic plants.
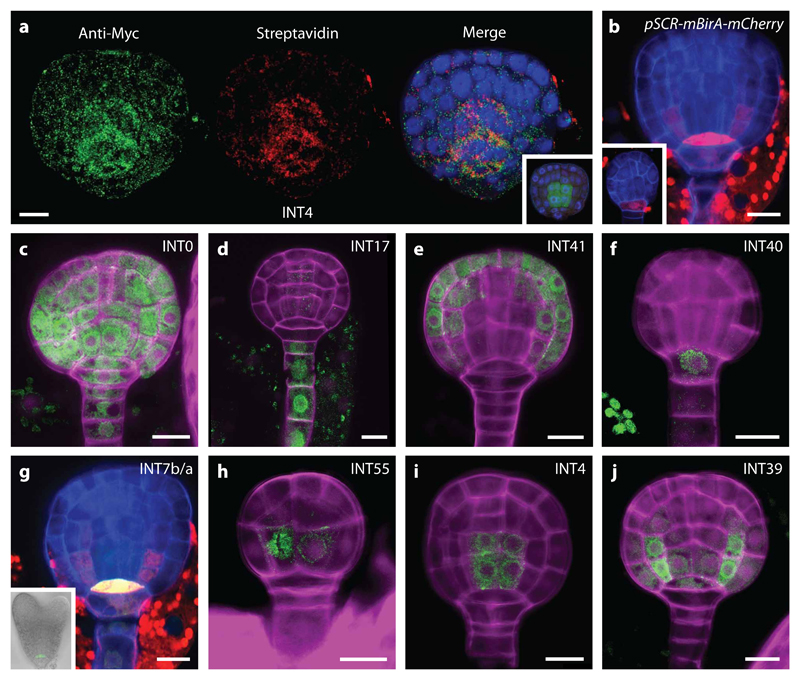
INTACT is an intrinsic two-component labelling system26. We exploited this capacity to isolate embryo-derived cell type-specific nuclei without the need to dissect embryos from seeds (Supplementary Fig. 2a). First, mBirA-3xMyc was uniformly expressed in the entire embryo by using the WOX2 promoter41 (pWOX2-mBirA-3xMyc; Fig. 1a, Supplementary Fig. 2b-e). A more restricted and cell-specific localisation of mBirA to the hypophysis and later to the QC and ground tissue precursors was achieved by expressing mCherry-tagged mBirA with the SCR promoter42 (pSCR-mBirA-mCherry; Fig. 1b). Second, a library of INTACT (INT) lines was generated as a resource for cell type-specific nucleus isolation in the embryo. 38 promoters were selected based on reported gene expression patterns (Supplementary Table 1), and used to express NTF in a genetic background containing either pWOX2-mBirA-3xMyc or pSCR-mBirA-mCherry. All INT lines were screened to obtain lines that show specific NTF expression during the early stages of embryo development (16-cell, early globular, late globular). Of the lines, 22 show cell type-specific expression at these stages (Supplementary Table 1, Fig. 3). From this collection, we selected “gold-standard” cell type-specific INT lines that mark the whole embryo, suspensor, protoderm, hypophysis, and the QC, vascular and ground tissue precursors (Fig. 1c-j, Supplementary Fig. 2f-l). BirA transfers biotin onto the BLRP moiety of NTF43. Thus, we used immunostaining to determine if NTF is efficiently biotinylated in vivo. Results confirmed local NTF biotinylation by mBirA in the embryo (Fig. 1a). Furthermore, since no visible phenotypes were observed in transgenic lines, neither NTF nor mBirA expression appears to affect early embryo development.
Figure 1. mBirA expression and selected gold-standard INTACT (INT) lines.
(a) Immunostaining of a late globular INT line (INT4; pIQD15-NTF, pWOX2-mBirA-3xMyc) embryo showing whole embryo mBirA expression (Anti-Myc Tag and Alexa Fluor 488 antibody; green) and NTF biotinylation in the vascular tissue precursors (Strepdavidin conjugated with Alexa Fluor 647; red). Counterstaining by DAPI (blue). Insert: negative control (INT4). GFP signal (green) from the pIQD15-NTF is visible in the vascular tissue precursors after fixation. (b) Expression of pSCR-mBirA-mCherry in hypophysis at early globular stage (insert) and in QC and ground tissue precursors at late globular stage. (c-j) INT lines where NTF is expressed in the whole embryo (INT0; c), suspensor (INT17; d), protoderm (INT41; e), hypophysis (INT40; f), QC precursor (INT7b/a; g), and precursor cells (INT55; h) of the vascular (INT4; i) and ground tissue (INT39; j) initials. Scale bar represent 10 μm in all panels.
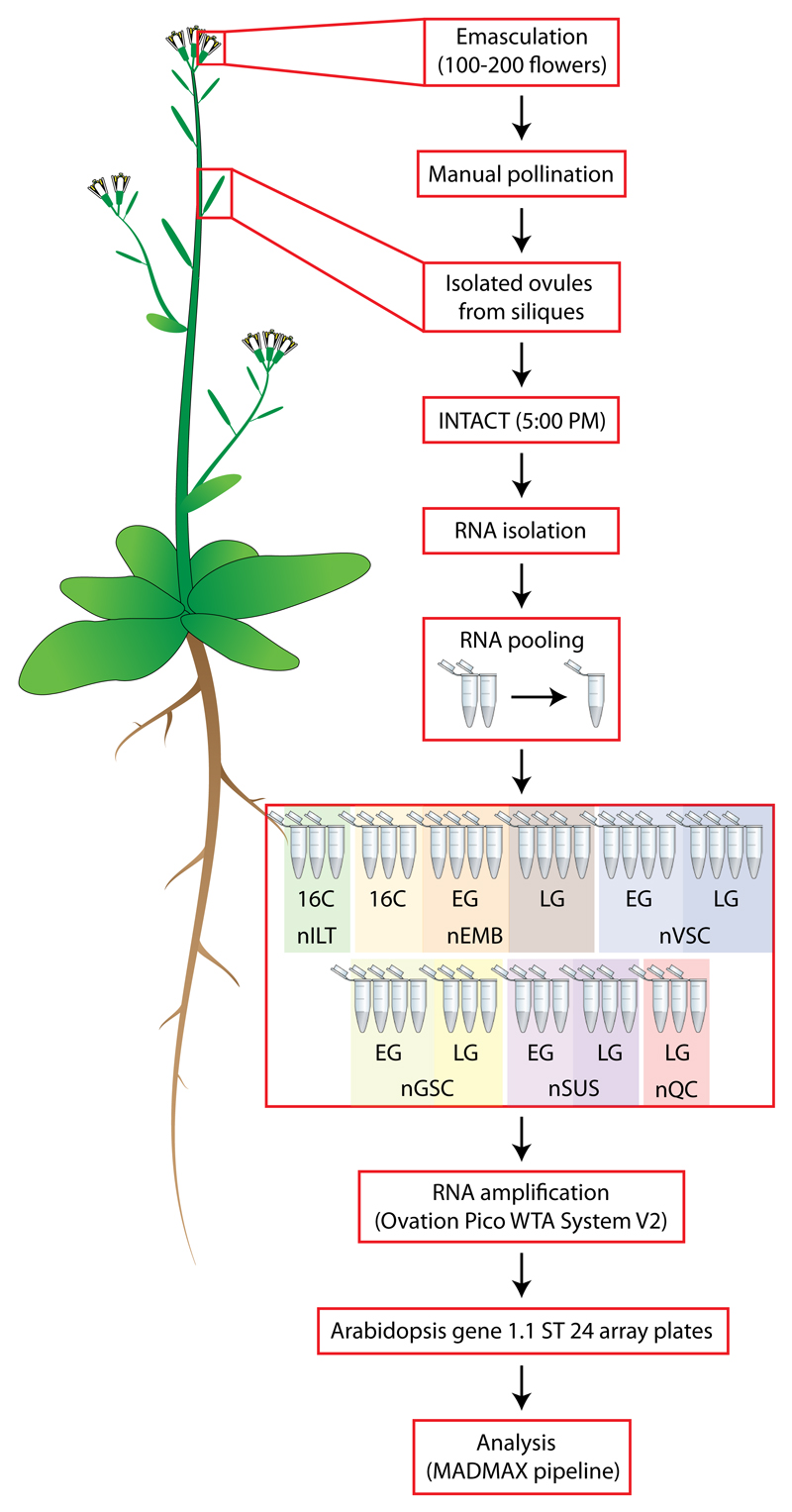
Figure 3. Schematic diagram of experimental setup.
Abbreviations are described in Table 1.
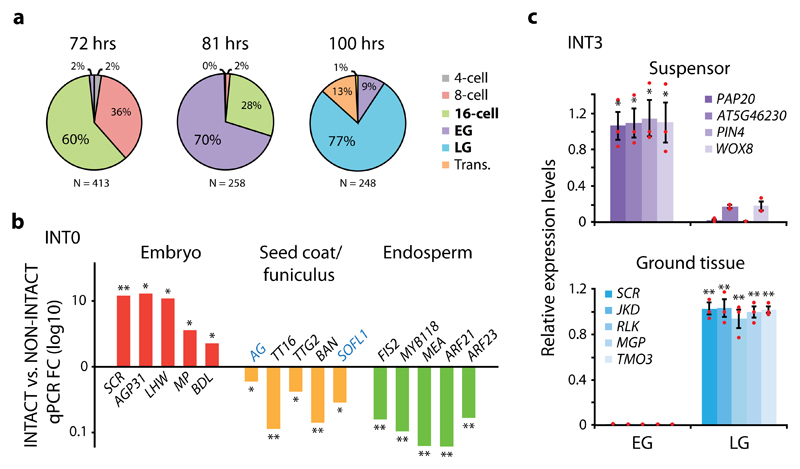
Arabidopsis embryos are small, contain few cells and are enclosed by a seed coat and surrounded by the micropylar endosperm4. This affected final yield and purity (data not shown) when INTACT was performed according to the original protocol, which was established for roots44, a larger and more accessible tissue. Therefore, several steps in the INTACT protocol were optimized (see Materials and Methods) to obtain a recovery efficiency of 20-50% with a purity (±SD) of 86.2% ± 6.6% for all nuclei of interest, as based on 50 experiments (Supplementary Table 2). Since embryo development is a progressive process, with several cell specification events happening at specific moments for the first time in plant life, it is important to isolate embryos from defined stages to isolate nuclei from exact cell types. We performed manual pollination of emasculated pWOX2-mBirA-3xMyc flowers to control timing of fertilization, and harvested embryos at varying time intervals. A reasonable degree of temporal homogeneity in embryonic stages within each fruit were found, and we determined 72, 81 and 100 hours after pollination as optimal intervals for 16-cell, early globular and late globular stages, respectively (Fig. 2a). This was subsequently verified in each INT line selected for transcriptomic profiling.
Figure 2. INTACT efficiently isolate cell type- and stage-specific nuclei.
(a) Optimal time intervals in hours (hrs) for the isolation of 16-cell (72 hrs), early globular (EG; 81 hrs) and late globular (LG; 100 hrs) embryonic stages following manual pollination of pWOX2-mBirA-3xMyc plants. (b) Quantitative RT-PCR (qPCR) analysis showing enrichment (FC; fold change) of genes predominantly expressed in embryo, seed coat, seed coat/funiculus (blue text) or endosperm when comparing INT0 (whole) embryo-derived nuclei (INTACT) to total seed nuclei (NON-INTACT). (c) qPCR relative expression levels of genes predominantly expressed in the suspensor or ground tissue in nuclei derived from the INT3 line, where pOFP8-NTF is expressed first in the suspensor at EG and then primarily in the ground tissue at LG. Suspensor- and ground tissue-expressed genes show enrichment in EG and LG, respectively. Results are based on mean values and SEM of 3 biological replicates in both b and c. In c, individual data points are plotted as red dots. Asterisks denote significance levels (*p <0.05, **p <0.01) as determined by one-tailed Welch’s t-test. Normal distribution is assumed.
INTACT allows efficient isolation of targeted nuclei
Isolation of embryo nuclei through INTACT on whole seeds requires that the overlap of BirA and NTF expression is restricted to embryo cells. Due to chlorophyll autofluorescence in the seed coat and funiculus, we could not fully exclude the possibility of pWOX2 expression in this region, even though no difference in red fluorescence (outside of the embryo) was detected between pWOX2-nDsRed and wild-type seeds (Supplementary Fig. 2d,e). Several INT lines did not show any NTF expression outside the embryo, but some others did (e.g. INT55, INT39; Supplementary Fig. 2j,l). Therefore, to ascertain that INTACT can be used on our INT lines to enrich cell type-specific nuclei exclusive to the embryo, we investigated the expression of a panel of genes predominantly expressed in embryo, endosperm, or seed coat/funiculus in nuclei derived from the gold-standard whole embryo INT0 line (Fig. 1c, 2b and Supplementary Fig. 4, Table 3). This line contains pWOX2-mBirA-3xMyc and NTF driven by the RPS5A promoter, which is expressed in all seed tissues45. When compared to total seed (NON-INTACT) nuclei we found strong enrichment of embryo-expressed genes in the embryo-derived (INTACT) nuclei. In contrast, expression levels of endosperm genes were strongly depleted in INTACT-purified nuclei. For seed coat/funiculus genes, we found depletion in INTACT-derived nuclei, but to a lesser degree to that of endosperm-expressed genes. Thus, while contamination with seed coat/funiculus genes is observed, INTACT strongly enriched for embryo-expressed genes, and thus, potential expression of the WOX2 and NTF promoters outside the embryo is not expected to impact expression profiles.
We next verified stage-specific nuclei isolation by using the INT3 line (Supplementary Fig. 3) to investigate the expression of genes predominantly expressed in the suspensor or ground tissue (Fig. 2c and Supplementary Table 3). In INT3, pOFP8-NTF expression is detected in the early globular suspensor after which it shifts to the ground tissue in late globular stage. Reflecting the distribution of embryo stages at optimal time points (Fig. 2a), early globular-derived nuclei showed strong enrichment of suspensor-expressed genes but next to no expression of ground tissue genes (Fig. 2c). In contrast, these genes were strongly enriched in late globular-derived nuclei while suspensor genes were lowly expressed. Together, these results demonstrate the efficient isolation of both cell type- and stage-specific nuclei.
A transcriptome atlas of the early Arabidopsis embryo
With a collection of gold-standard INT lines and an optimized INTACT workflow, we set out to generate a transcriptome atlas of the early Arabidopsis embryo containing cell types important for root stem cell niche formation. INTACT relies on profiling of nuclear RNA, which consists of both nascent transcripts and mature mRNA not yet exported to the cytosol. To determine if nuclear RNA is representative of total cellular mRNA, we compared whole genome expression data from nuclear (INTACT) and cellular (manual whole embryo isolation) RNA of the entire INT0 embryo at 16-cell stage. We found a high correlation coefficient (r = 0.82) in the composition of these RNA pools (Supplementary Table 4a), demonstrating that RNA isolated from INTACT-derived nuclei is a reasonable proxy for steady-state transcript levels and can thus be used for gene expression profiling. Interestingly, differences between nuclear and total mRNA do exist at the individual gene level. In addition to contribution of seed tissue contamination (Supplementary Table 4b), this may reflect the regulation, transient state and stability of mRNA (reviewed previously46,47), as well as selective enrichment associated with cellular functions and nuclear retention48,49.
As a first step in generating the atlas, we isolated between 1000-5000 cell type-specific nuclei from 100-200 manually pollinated siliques for each of 6-8 biological replicates of a selected INT line and embryonic stage (listed in Table 1). After RNA extraction, multiple biological replicates were first pooled and then all amplified simultaneously to reduce technical variance between samples and to avoid batch-to-batch effects50,51 (Fig. 3). Transcriptomic profiling was carried out using Affymetrix Arabidopsis Gene 1.1 ST arrays, which covers 84.2% (28,501) of the current total of annotated genes (33,602) (TAIR10)52. Using this approach, a spatio-temporal differential gene expression dataset was generated (Supplementary Table 5a,b). Similar to what was shown in the qPCR analysis (Fig. 2b), manual and software-assisted53 comparison to previous seed tissue transcriptome datasets10,54 revealed that seed coat and funiculus contamination is present in the dataset (Supplementary Table 5c), but given that its level is comparable across all samples, we do not expect this generic contamination to significantly affect the interpretation of differential gene expression between stages or cell types.
Table 1. Abbreviations and representations of selected INTACT (INT) lines and developmental stages used for transcriptomic profiling.
| INT line | Cell type | Development stage | Abbreviation | Representation |
|---|---|---|---|---|
| INT0 (pRPS5A-NTF, pWOX2-mBirA-3xMyc) |
Whole embryo | 16-cell | nEMB_16C | 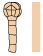 |
| Early globular | nEMB_EG |  |
||
| Late globular | nEMB_LG |  |
||
| INT4 (pIQD15-NTF, pWOX2-mBirA-3xMyc) |
Vascular tissue precursor | Early globular | nVSC_EG | 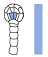 |
| Late globular | nVSC_LG |  |
||
| INT7b (pWOX5-NTF, pSCR-mBirA-mCherry) |
Quiescent centre precursor | Late globular | nQC_LG |  |
| INT17 (pKMD2-NTF, pWOX2-mBirA-3xMyc) |
Suspensor | Early globular | nSUS_EG |  |
| Late globular | nSUS_LG |  |
||
| INT39 (pMGP-NTF, pWOX2-mBirA-3xMyc) |
Ground tissue precursor | Early globular | nGSC_EG |  |
| Late globular | nGSC_LG |  |
||
| INT55 (pSHR-NTF, pWOX2-mBirA-3xMyc) |
Inner lower tier | 16-cell | nILT_16C |  |
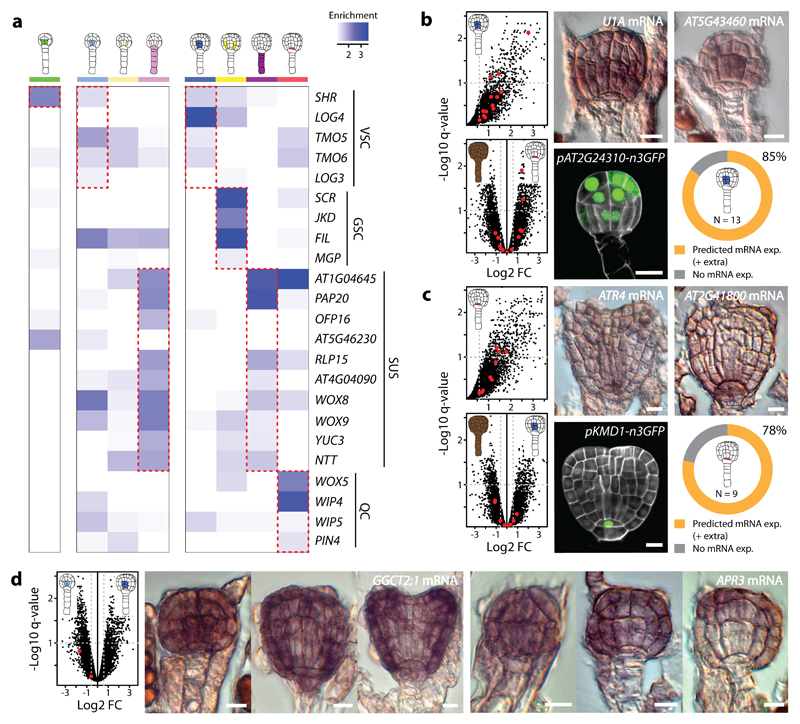
To validate the cell-specific profiles, we first surveyed genes with well-known expression patterns and found that a majority of the genes showed enrichment above fold change (FC) 1.5 in the expected cell type when compared to the whole embryo (Fig. 4a and Supplementary Table 3). Based on this result, we then investigated the expression pattern of genes for which no independent expression pattern had been reported. We selected genes that were increasingly enriched (FC >1.5, regardless of q-value) in (1) nVSC and (2) nQC compared to late globular nEMB, or in early compared to late globular nVSC (Fig. 4b-d and Supplementary Fig. 5). RNA in situ results were largely in agreement with INTACT-based predictions. Most (≥78%) of the 22 spatially enriched genes were detected in, and concentrated to, the expected cell type (Fig. 4b,c and Supplementary Fig. 5a,b). No expression could be detected for the remaining genes. Likewise, both temporally enriched genes disappeared from the vascular tissue precursors after early globular stage (Fig. 4d and Supplementary Fig. 5c). Promoter-reporter fusion analysis of a selected subset of these genes supported these findings. No clear correlation was observed between expression patterns and FC of expression enrichment, nor with the statistical q-values. Several genes were more broadly expressed than only in the predicted cell type, which is a logical consequence of our (spatial) selection strategy in which enrichment in a single cell type relative to the entire embryo was considered. Such enrichment does not exclude expression in other cell types. This was especially evident for nVSC selected genes since they, in general, were also enriched in other cell types (Fig. 4b and Supplementary Fig. 5a). Also, in situ hybridization mostly detects cytosolic mRNA, while INTACT captures nuclear RNA, and some investigated genes showed divergent expression in the nuclear vs. whole cellular comparison (Supplementary Table 4c). Nevertheless, cell-specific enrichment is consistent with observed expression patterns, validating the applicability of the atlas.
Figure 4. Expression patterns of selected genes validate atlas transcriptomic profiles.
(a) Heat map show enrichment (fold change; FC) of genes with known expression pattern in the embryo in all cell-specific samples when compared to corresponding nEMB stages, regardless of overall expression values. Enrichment is in blue (>1). The enrichment correlate well with published expression data for a majority of the genes, however, TMO6, LOG3, MGP, AT5G46230, WIP5 and PIN4 show enrichment below FC 1.5 in the expected cell type. Abbreviations: VSC = vascular tissue precursors, GSC = ground tissue precursors, QC = quiescent centre precursor, SUS = suspensor. (b-d) Scatter plots (left upper panel [b,c], left panel [d]) and mRNA (RNA in situ hybridization) and promoter-n3GFP expression of genes enriched in nVSC (b) and nQC (c) when compared to nEMB at late globular stage, and in early compared to late globular nVSC (d). Red dots mark the genes selected for gene expression analysis (here and Supplementary Fig. 5). Volcano plots in b and c (lower left panel) show enrichment of the selected nVSC and nQC genes in nQC (b) and nVSC (c), respectively. Log2 fold FC is plotted against the negative log10 q-value, where vertical dotted line = FC 1.5 and horizontal dotted line = q-value 0.1. Doughnut graphs in b and c (lower right panel) summarizes mRNA expression patterns of all spatially enriched genes. In total, 11 of 13 (85%) nVSC genes and 7 of 9 (78%) nQC genes showed expression in the correct cell type while the rest showed no expression. Plus (+) extra denotes broader expression pattern than exclusively in the predicted cell type. For both temporally nVSC enriched genes (d), expression gradually disappeared from the vascular tissue precursors after the early globular stage. Scale bar represent 10 μm in all panels. Representations are described in Table 1.
Developmental stage has a dominant impact on transcriptome dynamics
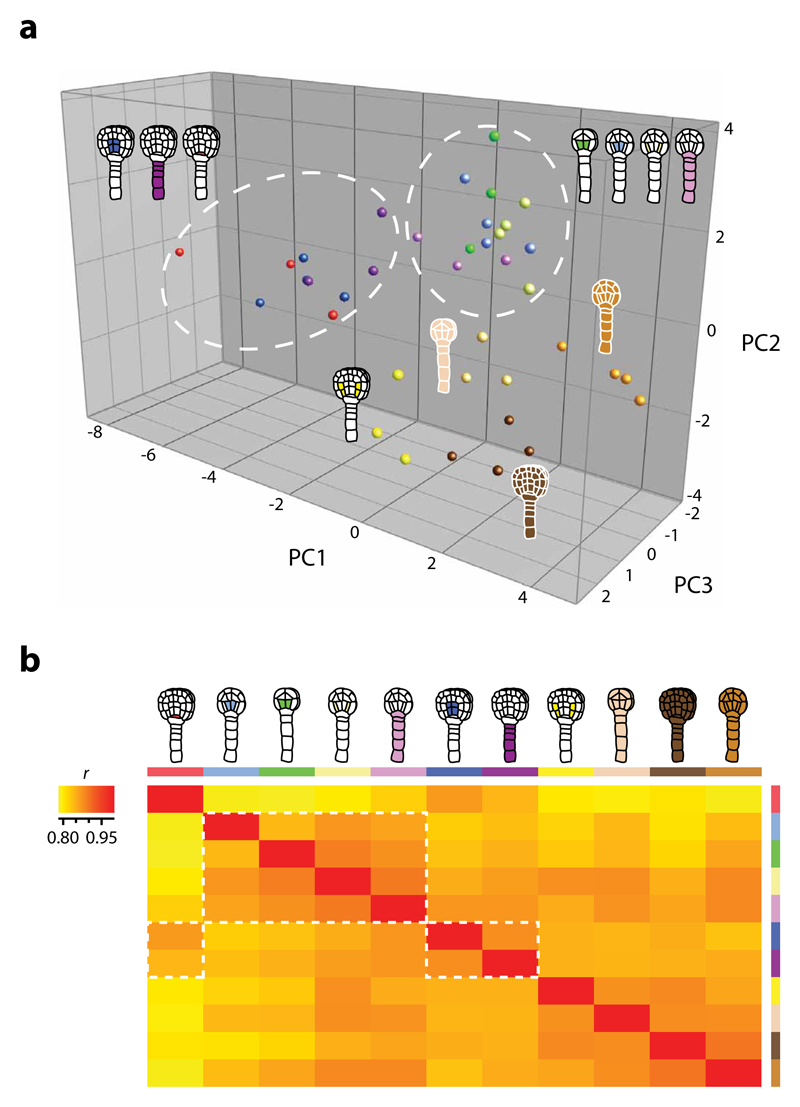
Since the atlas provides spatial and temporal distinct expression values for individual genes, we next estimated overall cell type- and stage-specific diversity by comparing mRNA populations using principal component analysis (PCA) on the top 2000 variable genes (Fig. 5a and Supplementary Fig. 6), and pairwise expression profile correlation on mean replicate values of the entire populations (Fig. 5b and Supplementary Table 5d). The cell-specific samples, excluding late globular nGSC, formed stage-specific subgroups that clustered separately from (stage-separated) nEMB. Surprisingly, late globular nGSC associated closer with nEMB of the same stage than with any other samples. A closer study of the ground tissue line (INT39; Fig. 1j) revealed weak NTF expression also in neighbouring tissues (vascular, QC, protoderm) at this stage. This likely resulted in broader nuclei isolation and, thus, could explain the similarity between these populations. Also, the low overall variability seen in the PCA analysis (Fig. 5a) is a probable consequence of the pooling of RNA samples and the simultaneous RNA amplification. Nevertheless, all replicates except for nQC clustered according to cell type, though in some cases (e.g. nSUS, nILT) not very distinctly. For nQC, the result reflects the low correlation observed between the nQC replicates (Supplementary Table 5a). Hierarchical clustering confirmed the PCA results (Supplementary Fig. 7).
Figure 5. Expression diversity is cell type- and stage-specific.
(a) PCA comparing the top 2000 variable genes of the atlas, selected based on the interquartile range of all samples. The PCA distinguish spatial and (more distinct) temporal patterns where the cell-specific transcriptomes (except late globular nGSC) cluster separately from nEMB and according to developmental stage (white circles). Principal components 1 through 3 collectively represent 34.5% of the total variance in the dataset. Log2 normalised values were used. (b) Heat map of Pearson′s correlation coefficients (r) in pairwise comparisons of the whole atlas dataset where low correlation is in yellow and high correlation in red. Samples are assorted according to amount of correlation (low to high: left to right, top to bottom), which show a pattern that reflect the PCA results (white rectangles). Representations are described in Table 1.
The analyses demonstrate that INTACT-derived transcriptomes are reproducible, as most biological replicates cluster together. Secondly, transcriptomes are different both between cell types, and between stages. However, embryo stage has a more profound impact on transcriptome differences than cell type does, as is particularly evident from pairwise correlation analysis (Fig. 5; white outline). This suggests that developmental stage greatly influence cell state.
Transcriptional reprogramming during embryogenesis
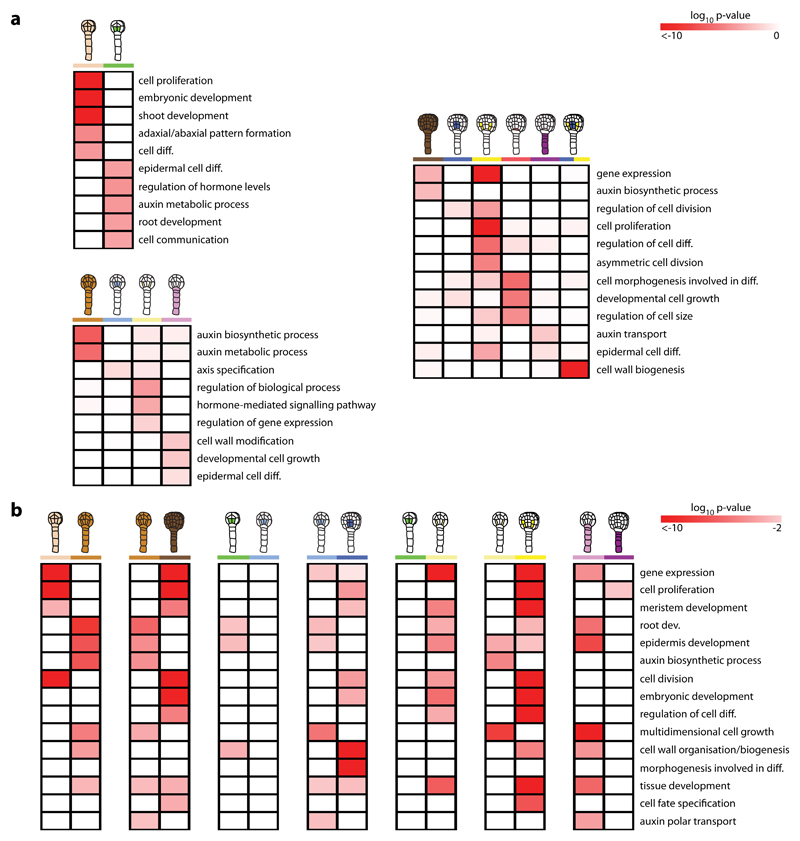
In order to explore how the transcriptomes of the atlas define cell states and state transitions during development, we investigated gene ontogeny (GO) terms and transcription factor (TF) families in dominant spatial and temporal gene expression patterns of unique and shared genes (Supplementary Fig. 8). Here, “spatial” refers to comparison to all other cell types of the same stage and “temporal” to comparison to the same cell type of a different stage. Of note, GO-term enrichment analysis revealed that terms such as “cell wall biogenesis”, “cell growth”, “epidermal cell differentiation” and “auxin polar transport” were spatially enriched in early globular nSUS (Fig. 6a and Supplementary Table 5e), but not or to a lesser degree in late globular stage. This developmental shift was also seen in the temporal analysis (Fig. 6b and Supplementary Table 5f), since the same or related GO-terms were only enriched in early globular nSUS. These results corroborate previously published data demonstrating the critical developmental importance of the pre- but not (post-) globular suspensor7,55. In addition, the temporal analysis revealed that several developmental GO-terms, including “tissue and meristem development” were enriched in early globular nGSC but not nVSC when compared to nILT. Since the vascular and ground tissue precursors are established following the periclinal division of the four inner lower tier cells during the 16-cell to early globular stage transition6, the lack of developmental progression in nVSC suggest that vascular identity is already established in these cells at 16-cell stage, while ground tissue cell fate is acquired through transcriptional reprogramming after their division. Supporting this, the vascular markers SHR and ZLL are both expressed in the inner lower tier cells at 16-cell stage56 (INT55 [SHR], INT56 [ZLL]; Fig. 1h, Supplementary Fig. 3).
Figure 6. Spatio-temporal shifts in developmental processes defines early embryonic cell fate.
Heat maps show enriched biological gene ontology (GO)-terms in dominant spatial (a) and temporal (b) gene expression patterns of the atlas. High p-value is in white, and low p-value is in red. Enrichment patterns generally reflect cell state and state transitions in the early embryo. Representations are described in Table 1.
During the transition from early to late globular stage, asymmetric and anticlinal division of the hypophysis and the vascular and ground tissue precursors generate small apical cells that differentiate into the QC and the vascular and ground tissues, respectively6. Results from the GO-term enrichment analysis reflect this development. Terms associated with cell division, differentiation and specification were spatially and/or temporally enriched in late globular nVSC and nGSC (Fig. 6). As previously noted, transcripts from the vascular tissue precursors are likely also present in the late globular nGSC samples. For late globular nQC, terms associated with cell morphology and its involvement in differentiation were enriched (Fig. 6a).
These results suggest that, as a reflection of the shifts in cell-specific gene expression (Fig. 5), developmental processes change in a corresponding manner to define cell fates during root stem cell niche formation in the early embryo. This conclusion is supported by TF family enrichment analysis that showed highly dynamic expression patterns of TF families crucial for early embryo patterning8, including auxin response factor (ARF), basic-helix-loop-helix (bHLH) and Homeobox (HOX) (Supplementary Fig. 9, Table 5e,f).
Discussion
In this study, we adapted, optimized and validated a two-component transgenic labelling system (INTACT) to efficiently isolate cell type-specific nuclei from the early seed-enclosed Arabidopsis embryo. With this approach, a transcriptome atlas was generated as a resource to explore how genome-wide gene activity shape fates at the cellular level during the formation of the root stem cell niche.
We identified major shifts in unique cell type-specific gene sets in the early embryo that, while associated with clonal origin, was more dependent on the developmental stage of the embryo. Dominant expression patterns, likely signifying transcriptional reprogramming, revealed enrichment of transcription factor families (ARF, bHLH, HOX) and biological processes important for cell fate determination that correspond to known activities of the cell types they encompass and reflect developmental transitions. Indeed, our results verify earlier data showing that the pre- but not (post-) globular suspensor is necessary for development, and reveal that vascular identity is likely established prior to ground tissue cell fate specification. A substantial coordination between dynamic gene activity, reprogramming and biological processes in defining developmental transitions has earlier been shown in seed tissues (including the embryo)10,17,54 and during stomatal lineage progression30. This, together with the expression of known tissue markers, provides support for previously published results suggesting that cell types that will form the root apical stem cell niche acquire their fates already at this early stage of development, and that this is, at least partially, mediated by auxin signalling through ARFs and their downstream targets, including bHLHs and WOX557–61. From this, it is tempting to speculate that self-regulating auxin response provides the spatio-temporal cues that initiate the observed shifts in cell type-specific gene expression. Future investigations using our atlas together with other published datasets may help to dissect the source and functional outcome of these shifting patterns.
To explore the atlas, it is important to have confidence that the microarray results represent true in vivo gene expression differences. By surveying expression profiles of well-known genes and using in vivo expression analysis of selected genes, we could show that atlas predictions correlate with observed expression patterns, even for nQC where, presumably, small cell numbers caused high variation between replicates. Seed tissue contamination is common in the atlas, however, this is expected since the INTACT-method, as applied here, isolates between 10-20% of non-biotinylated and, thus, total seed nuclei. Nevertheless, this is not expected to greatly impact overall expression profile comparisons since contamination levels are similar across all samples. Residual values from the nuclear vs. whole cellular comparison can be consulted to identify putative contamination or other nuclear/cellular-derived RNA differences that may affect predictions for specific genes. When compared, our atlas performs favourably to and, by providing more pattern information, supplements already available cellular10 and nuclear28 transcriptomes (Supplementary Fig. 10, Table 5g), even though the datasets are different (Supplementary Fig. 11). It also appears to contain similar, or even less seed tissue contamination, than the FANS-generated nuclear28 transcriptomes of comparable cell types and developmental stages (Supplementary Table 6). It should be noted, that unlike the other datasets, the atlas contains both pre-processed and mature mRNA as a consequence of the RNA amplification kit used. Furthermore, the higher differential gene expression observed for the cellular transcriptome suggests that small initial differences in transcription level between cells are amplified by post-transcriptional processes.
Much remains to be learned about cell fate in the early embryo. We anticipate that this atlas, in providing the most comprehensive description of plant embryonic tissue- and cell-specific gene activity to date, will be a valuable resource. Not only to investigate patterning events involved in cell fate specification and differentiation during root apical stem cell formation, but also together with other genetic resources to understand the regulatory mechanisms underlying plant development. For this purpose, an interactive graphical representation of the atlas has been established at http://www.albertodb.org (AlBERTO).
Materials and Methods
Plant material
Plants used in all experiments were Columbia (Col-0) ecotype. The pWOX2-nDsRed reporter line was kindly provided by Thomas Laux and has been described previously41.
After seed sterilization, transgene selection was performed on solid Murashige and Skoog medium supplemented with 50 mg/L kanamycin or 15 mg/L phosphinothricin. Resistant seedlings were transferred to soil and further grown at a constant temperature of 22°C under long-day conditions (16 hours light/8 hours dark). Floral dip was used for plant transformations62.
Generation of transgenic constructs
All transgenic constructs were generated using the LIC cloning system and vectors described by De Rybel et al.63 and Wendrich et al.64. To generate promoter fusions for nuclei purification, a nuclear targeting factor (NTF) fragment were cloned into pPLV1 (LIC-NTF). The E. coli biotin ligase (mBirA) was synthesized with codon usage according to Arabidopsis preference (Mr. Gene, Regensburg; codon choice according to manufacturer’s specifications) and cloned into pPLV2 tagged with either 3xMyc (LIC-mBirA-3xMyc) or mCherry (LIC-mBirA-mCherry). The original NTF and BirA fragments were kindly provided by Roger B. Deal and have been previously described26. For promoter-NTF, pWOX2-mBirA-3xMyc and pSCR-BirA-mCherry constructs, promoter fragments corresponding to a region approximately 3 kbp upstream from the start codon of the genes were cloned into the corresponding LIC vectors. To generate promoter-n3GFP reporter constructs, promoter fragments were cloned into pPLV4. Amplification primers of each promoter fragment are listed in Supplementary Table 7.
Western blotting and immunostaining
Protein was extracted by grinding siliques in liquid nitrogen, adding 200 μl Lysis buffer (50 mM Tris-HCl [pH 7.4]; 150 mM NaCl; 1% Triton X-100; 1% [v/v]) protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) and then centrifugation (18.000 g). The supernatant was incubated with 50 μl 1 x sample buffer (62.5 mM Tris-HCl [pH 6.8]; 10% [v/v] glycerol, 2% [w/v] SDS, 1% [v/v] β-mercaptoethanol, 0.0005% [w/v] bromophenol blue) for 5 min (100°C) before proteins were separated by SDS/PAGE 10% and transferred onto a PVDF membrane (Bio-Rad Laboratories, CA, USA) by electroblotting. The membrane was blocked for 2 hours (3% [w/v] dried milk, 0.05% [v/v] Tween in PBS [pH 7.4]) after which it was incubated for 1 hour with 0.5 μg/ml Anti-Myc Tag antibody (clone 9E10) (RRID:AB_309725) (Merck Millipore, CA, USA). After washing (3 x 15 min, 0.1% [v/v] Tween in PBS), the membrane was incubated with a secondary Rabbit Anti-Mouse HRP-conjugated antibody (RRID:AB_92531) (Merck Millipore) at 1:5000 dilution for 1 hour and then washed again. Blots were developed using Lumigen ECL (GE Healthcare, Buckinghamshire, UK) and analysed on the ChemiDoc™ XRS+ system (Bio-Rad Laboratories).
Immunostaining was performed as previously described65. The Anti-Myc Tag antibody (clone 9E10) (Merck Millipore), and a secondary Anti-Mouse Alexa Fluor 488 (RRID:AB_141514) and/or Strepdavidin conjugated with Alexa Fluor 647 antibody (RRID:AB_2336066) (Thermo Fisher Scientific, OR, USA) were used (at 1:400 dilution). DAPI (3 ng/ml, Thermo Fisher Scientific) was used for counterstaining.
Western blotting and immunostaining experiments were successfully repeated 3 times.
Purification of biotin-tagged nuclei
Biotin-tagged nuclei from Arabidopsis embryos were isolated and purified as previously described44 with several modifications. For each purification, pistils (exposed by emasculation 1 day earlier) from an NTF/BirA transgenic plant line were manually pollinated at a specific time point for no longer than 1 hour to synchronize embryo development. After a determined amount of time, siliques containing seeds with embryos of the appropriate stage were collected at 5:00 PM (corresponding to 10 hours after subjective dawn; ZT10) to minimize variation due to circadian rhythm effects and placed on double-sided adhesive tape-covered microscopy slides. Using a Microlance 3 needle (Becton Dickinson, Dublin, Ireland), the valves of the siliques were opened along the replum to expose the septum and seeds. The slides were transferred to a square petri dish (120x120 mm; Gosselin, Borre, France) under a stereo microscope (SMZ-140; Motic Deutschland GmbH, Wetzlar, Germany) and covered with 1xPBS (pH 7). The seeds were then isolated by suctioning them up onto a 20 μm nylon net filter (NY20; Merck Millipore, Cork, Ireland) with a Pasteur pipette attached by rubber tubing to a filter holder (25 mm In-Line; Pall Life Sciences, MI, USA) connected to a pump generating up to 40 mbar of vacuum. A continuous flow of new 1xPBS (pH 7) was provided to facilitate suctioning and to prevent the seeds on the filter of drying out. The filter was dipped several times in two or three 2 ml eppendorf tubes (Eppendorf AG, Hamburg, Germany) containing 1xPBS (pH 7) to wash of the seeds. The PBS was carefully removed using a pipette and the exposed seeds were then frozen in liquid nitrogen and homogenized using 4.8 mm ball bearings in a mixer mill (MM400; Retsch, Düsseldorf, Germany) at a vibrational frequency of 30 Hz for 2x30 sec. All subsequent steps were performed at 4°C.
The homogenized sample was resuspended in 10 ml nuclear purification buffer (NPB)44 and 200 U of Protector RNase Inhibitor (Roche Diagnostics GmbH) was added. The suspension was filtered through a 40 μm nylon mesh (Becton Dickinson, NJ, USA) and the nuclei were pelleted at 1,200 x g for 7 min. The nuclei were then resuspended in 1 ml of NPB and incubated with 10 μl of washed M-280 streptavidin-coated Dynabeads (Invitrogen, Oslo, Norway) for 30 min, with end-over-end rotation. During incubation, 1 ml Rainin LTS pipette tips (Labcon, CA, USA) were treated with NPBc (NPB with 1% [v/v] casein) for 20 min after which a tip was inserted firmly (without cracking) in the groove of an OctoMACS separator magnet (Miltenyl Biotec, Bergisch Gladback, Germany). A two-way stopcock was attached to narrow tip end and the tip was pre-loaded with NPBt (NPB with 0.1% [v/v] Triton X-100). After 30 min of incubation, the beads and nuclei mixture was diluted to 10 ml with NPBt, mixed gently for 30 sec and drawn into a plastic 10 ml serological pipette (ALP, Chorges, France) with 40x20 mm parafilm wrapped around its tip. The serological pipette was inserted and fastened to the broad opening of the pre-loaded 1 ml micropipette. The mixture was drained past the magnet at a rate of approximately 0.75 ml/min allowing the capture of bead-bound nuclei to the sides of the micropipette tip. Bubbles were removed using a Pasteur pipette before passing the magnet. Unbound nuclei were washed away from the outer tip wall by drying it off with moist paper tissue, and from the inner wall (up until the bead-bound nuclei) by carefully drawing NBPt in and out 5 times without disturbing the beads attached to the wall. The bead-bound nuclei were rinsed in 1 ml NPBt, diluted to 10 ml with NPBt and the magnetic purification was repeated again. After rinsing, the bead-bound nuclei were collected by centrifugation (1000 x g for 5 min), decanted and resuspended in 25 μl NPB.
Purity and yield of nuclei after purification were determining by counting DAPI-stained (2μg/ml) bead-bound (biotinylated) and unbound (non-biotinylated) nuclei in the purified sample using a hemacytometer (Neubauer improved; Brand GmbH, Wertheim, Germany).
Isolation of embryos
Embryo isolation was performed according to Raissig et al.66, using seeds collected and isolated, as described above, from ~60 siliques after manual pollination. A Zeiss Confocor 1 inverted microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) together with an Eppendorf Transferman 4r micromanipulator (Eppendorf AG) were used to isolate about 60 washed embryos in 50 μl isolation buffer.
RNA extraction
For RNA extraction, (1) bead-bound nuclei and (2) isolated embryos were immediately incubated with 500 μl of TRIzol reagent (Ambion, CA, USA) for (1) 5 min at RT (vortexing) and (2) 30 min at 60°C, and then purified according to the TRIzol reagent protocol for small sample quantities. GlycoBlue (2 μl of 15 mg/ml; Ambion, TX, USA) was used as a co-precipitant. The RNA was treated with DNase I (~14 Kunitz units; QIAGEN GmbH, Hilden, Germany) for 5 min at RT and then purified (RNeasy Micro Kit; QIAGEN GmbH) and concentrated to 5 μl (Concentrator plus; Eppendorf AG) before being stored at -80°C until further processing.
Microarray analysis
RNA was amplified using the Ovation Pico WTA System V2 (NuGEN, CA, USA), labelled with the ENCORE Biotin Module (NuGEN) and hybridized to Arabidopsis Gene 1.1 ST 24-array plates (atlas) or single array strips (nuclear vs. cellular RNA) (Affymetrix, CA, USA) according to the manufacturers protocol. Microarray analysis was performed using the MADMAX pipeline67 and a custom CDF file (MBNI CustomCDF version 19.0.0)68. Here, all expression values were (quantile) normalised by the Robust multi-array average algorithm (RMA)69 using the median polish (nuclear vs. cellular RNA) or M-estimator (atlas) algorithm for probes to probe set summarisation. Probe sets were redefined using current genome information68 and re-organized according to TAIR10 gene definitions. Linear models and an intensity-based moderated t statistic approach70,71 were used to identify differentially expressed genes (probe sets). P-values were corrected for multiple testing using an optimized false discovery rate (FDR) approach72.
For software-assisted53 comparison of the atlas dataset to Belmonte et al.10 seed tissue transcriptomic data, a minimum fold of 2 and p-value of 0.05 was considered “enriched” and temporal changes were not buffered. Genes not considered “expressed” (mean expression value <9.85; see Supplementary Fig. 12) in each tested sample were removed before analysis. Dominant expression patterns were determined after first filtering the dataset by (1) removing non-“expressed” genes in any of the compared samples, and (2) selecting the top 50% (4796 to 6368) varying genes across the dataset. Then, either manual selection (2 datasets; fold change >1.5) or fuzzy k-means and hierarchical clustering (>2 datasets)73, with K = 1.05 and a cluster membership probability (m-values) at or above 0.4, were performed. Biological significance of co-expressed genes was explored using agriGO74, where significance of gene ontology (GO) term enrichment was relative to TAIR locus IDs covered by the Gene 1.1 ST array. P-values from fisher distributions where adjusted via the Benjamini-Yekutieli method to assess the FDR. GO terms with p ≤0.001 or FDR <0.05 were considered significantly enriched. Transcription factors (TFs) belonging to a family were identified using the Arabidopsis TF database at AGRIS75.
RNA in situ hybridization
In situ hybridisation was performed as described in Saiga et al.76 and successfully repeated at least 2 times for each gene. Target gene primers are listed in Supplementary Table 7.
Microscopy
Differential interference contrast (DIC) microscopy, fluorescence microscopy and confocal microscopy were performed as previously described65 with the following modifications: confocal imaging was performed on a Leica SP5-II system (HyD detector) and 0.1% (v/v) SR2200 (Renaissance Chemicals, Selby, UK) was used in the mounting solution (0.4% [v/v] DMSO, 5% [v/v] glycerol, 4% [w/v] paraformaldehyde in PBS [pH 8.0]) for counterstaining of transgenic plant embryos carrying a reporter construct.
Quantative RT-PCR analysis
Poly(dT) cDNA, prepared with an iScript cDNA Synthesis Kit (Bio-Rad Laboratories) from nuclear RNA, was analysed on a Bio-Rad CFX384 Real-Time PCR detection system with iQ SYBR Green Supermix (Bio-Rad Laboratories) according to the manufacturer´s instructions. Targets were quantified with primers designed with Beacon Designer 7.0 (PREMIER Biosoft, CA, USA) and listed in Supplementary Table 7. All individual reactions where done in triplicate. Data were analysed with qBase77. Expression levels were normalised to those of the reference genes Elongation Factor 1-aplha (EF1A) and Glyceraldehyde-3-phosphate dehydrogenase C subunit (GAPC).
Data availability
Raw microarray expression data are available at the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) as series GSE89101.
Supplementary Material
Acknowledgements
The authors thank Thomas Laux, Roger Deal and Steven Henikoff for sharing materials and Tatyana Radoeva for supplying the image of INT56 (ZLL) at 16-cell stage. Further, the authors thank students Maartje Geerlings, Diana van der Plaat, Evans Asamoah Gyimah, Marieta Goranova, André Kuhn, Leontien van der Bent, Ioanna Papakosta, Sofia Thomaidou, Stina Hedžet and Katrin Heidemeyer for their contributions. This study was supported by the Federation of European Biochemical Societies (FEBS) (J.P.), ERA-CAPS project EURO-PEC (Grant number 849.13.006) and the European Research Council (ERC Starting Grant “CELLPATTERN”; Contact number 281573 to D.W.).
Footnotes
Author contributions: D.W. conceived the study; J.P., S.S., J.R.W. and D.W. designed research; S.S. generated INTACT lines; J.P. and N.W.H. optimized and adapted experimental setup; J.P., S.S. and J.R.W. performed INTACT; F.H. and J.P. performed nuclear vs. cellular RNA comparison; M.B., G.J.H. and J.P. performed transcriptomic profiling and analysed data with support from S.M.; J.P., S.S., J.R.W., J.P.S. and J.S. validated expression patterns; J.P.S. and J.P. designed and developed the AlBERTO browser; J.P. and D.W. wrote the paper with input from all other authors.
Competing interests
The authors declare no competing interests.
References
- 1.Esua K. Anatomy of seed plants 2nd edition. Wiley; 1977. pp. 475–495. [Google Scholar]
- 2.van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature. 1997;390:287–289. doi: 10.1038/36856. [DOI] [PubMed] [Google Scholar]
- 3.Weigel D, Jürgens G. Stem cells that make stems. Nature. 2002;14:751–754. doi: 10.1038/415751a. [DOI] [PubMed] [Google Scholar]
- 4.Mansfield SG, Briarty LG. Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Can J Bot. 1991;69:461–476. [Google Scholar]
- 5.Jürgens G, Mayer U. In: A colour atlas of developing embryos. Bard JBL, editor. Wolfe Publishing; 1994. pp. 7–21. [Google Scholar]
- 6.Yoshida S, et al. Genetic control of plant development by overriding a geometric division rule. Dev Cell. 2014;29:75–87. doi: 10.1016/j.devcel.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Gooh K, et al. Live-cell imaging and optical manipulation of Arabidopsis early embryogenesis. Dev Cell. 2015;34:242–251. doi: 10.1016/j.devcel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Palovaara J, de Zeeuw T, Weijers D. Tissue and organ initiation in the plant embryo: a first time for everything. Annu Rev Cell Dev Biol. 2016;32:47–75. doi: 10.1146/annurev-cellbio-111315-124929. [DOI] [PubMed] [Google Scholar]
- 9.Kerk NM, Ceserani T, Tausta SL, Sussex IM, Nelson TM. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003;132:27–35. doi: 10.1104/pp.102.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belmonte MF, et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. PNAS. 2013;110:E435–E444. doi: 10.1073/pnas.1222061110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vega-Bartol JJ, et al. Transcriptomic analysis highlights epigenetic and transcriptional regulation during zygotic embryo development of Pinus pinaster. BMC Plant Biol. 2013;13:123. doi: 10.1186/1471-2229-13-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venglat P, et al. Gene expression profiles during embryo development in Brassica napus. Plant Breeding. 2013;132:514–522. [Google Scholar]
- 13.Chen J, et al. Dynamic transcriptome landscape of maize embryo and endosperm development. Plant Physiol. 2014;166:252–264. doi: 10.1104/pp.114.240689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Itoh J-I, et al. Genome-wide analysis of spatiotemporal gene expression patterns during early embryogenesis in rice. Development. 2016;143:1217–1227. doi: 10.1242/dev.123661. [DOI] [PubMed] [Google Scholar]
- 15.Casson S, Spencer M, Walker K, Lindsey K. Laser capture microdissection for the analysis of gene expression during embryogenesis of Arabidopsis. Plant J. 2005;42:111–123. doi: 10.1111/j.1365-313X.2005.02355.x. [DOI] [PubMed] [Google Scholar]
- 16.Spencer MWB, Casson SA, Lindsey K. Transcriptional profiling of the Arabidopsis embryo. Plant Physiol. 2007;143:924–940. doi: 10.1104/pp.106.087668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le BH, et al. Global analysis of gene activity during Arabidopsis seed development and identification of seed-specific transcription factors. PNAS. 2010;107:8063–8070. doi: 10.1073/pnas.1003530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang D, et al. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiol. 2011;156:346–356. doi: 10.1104/pp.110.171702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Autran D, et al. Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell. 2011;145:707–719. doi: 10.1016/j.cell.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Nodine MD, Bartel DP. Maternal and paternal genomes contribute equally to the transcriptome of early plant embryos. Nature. 2012;482:94–97. doi: 10.1038/nature10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nodine MD, Bartel DP. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010;24:2678–2692. doi: 10.1101/gad.1986710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Birnbaum K, et al. A gene expression map of the Arabidopsis root. Science. 2003;302:1956–1960. doi: 10.1126/science.1090022. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C, Barthelson RA, Lambert GM, Galbraith DW. Global characterization of cell-specific gene expression through fluorescence-activated sorting of nuclei. Plant Physiol. 2008;147:30–40. doi: 10.1104/pp.107.115246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zanetti ME, Chang I-F, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005;138:624–635. doi: 10.1104/pp.105.059477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mustroph A, et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia in Arabidopsis. PNAS. 2009;106:18843–18848. doi: 10.1073/pnas.0906131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deal RB, Henikoff S. A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev Cell. 2010;18:1030–1040. doi: 10.1016/j.devcel.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin S-Y, et al. Profiling of translatomes of in vivo–grown pollen tubes reveals genes with roles in micropylar guidance during pollination in Arabidopsis. Plant Cell. 2014;26:602–618. doi: 10.1105/tpc.113.121335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slane D. Cell type-specific transcriptome analysis in the early Arabidopsis thaliana embryo. Development. 2014;141:4831–4840. doi: 10.1242/dev.116459. [DOI] [PubMed] [Google Scholar]
- 29.Yadav RK, Tavakkoli M, Xie M, Girke T, Reddy GV. A high-resolution gene expression map of the Arabidopsis shoot meristem stem cell niche. Development. 2014;141:2735–2744. doi: 10.1242/dev.106104. [DOI] [PubMed] [Google Scholar]
- 30.Adrian J, et al. Transcriptome dynamics of the stomatal lineage: Birth, amplification, and termination of a self-renewing population. Dev Cell. 2015;33:107–118. doi: 10.1016/j.devcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Antoniadi I, et al. Cell-type-specific cytokinin distribution within the Arabidopsis primary root apex. Plant Cell. 2015;27:1955–1967. doi: 10.1105/tpc.15.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vragović K, et al. Translatome analyses capture of opposing tissue-specific brassinosteroid signals orchestrating root meristem differentiation. PNAS. 2015;112:923–928. doi: 10.1073/pnas.1417947112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palovaara J, Saiga S, Weijers D. Transcriptomics approaches in the early Arabidopsis embryo. Trends Plant Sci. 2013;18:514–521. doi: 10.1016/j.tplants.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 34.Foley SW, et al. A global view of RNA-protein interactions identifies post-transcriptional regulators of root hair cell fate. Dev Cell. 2017;41:204–220. doi: 10.1016/j.devcel.2017.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park K, et al. DNA demethylation is initiated in the central cells of Arabidopsis and rice. PNAS. 2016;113:15138–15143. doi: 10.1073/pnas.1619047114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moreno-Romero J, Santos-González J, Hennig L, Köhler C. Applying the INTACT method to purify endosperm nuclei and to generate parental-specific epigenome profiles. Nat Protoc. 2017;12:238–254. doi: 10.1038/nprot.2016.167. [DOI] [PubMed] [Google Scholar]
- 37.Ron M, et al. Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 2014;166:455–469. doi: 10.1104/pp.114.239392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henry GL, Davis FP, Picard S, Eddy SR. Cell type–specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40:9691–9704. doi: 10.1093/nar/gks671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner FA, Talbert PB, Kasinathan S, Deal RB, Henikoff S. Cell-type-specific nuclei purification from whole animals for genome-wide expression and chromatin profiling. Genome Res. 2012;22:766–777. doi: 10.1101/gr.131748.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amin NM, et al. Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (INTACT) Development. 2014;141:962–973. doi: 10.1242/dev.098327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ueda M, Zhang Z, Laux T. Transcriptional activation of Arabidopsis axis patterning genes WOX8/9 links zygote polarity to embryo development. Dev Cell. 2011;20:264–270. doi: 10.1016/j.devcel.2011.01.009. [DOI] [PubMed] [Google Scholar]
- 42.Wysocka-Diller JW, Helariutta Y, Fukaki H, Malamy JE, Benfey PN. Molecular analysis of SCARECROW function reveals a radial patterning mechanism common to root and shoot. Development. 2000;127:595–603. doi: 10.1242/dev.127.3.595. [DOI] [PubMed] [Google Scholar]
- 43.Beckett D, Kovaleva E, Schatz PJ. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deal RB, Henikoff S. The intact method for cell type-specific gene expression and chromatin profiling in Arabidopsis thaliana. Nat Protoc. 2011;6:56–68. doi: 10.1038/nprot.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weijers D, et al. An Arabidopsis Minute-like phenotype caused by a semi-dominant mutation in a RIBOSOMAL PROTEIN S5 gene. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 46.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 47.Hocine S, Singer RH, Grünwald D. RNA processing and export. Cold Spring Harb Perspect Biol. 2010;2:a000752. doi: 10.1101/cshperspect.a000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barthelson RA, Lambert GM, Vainer C, Lynch RM, Gailbraith DW. Comparison of the contributions of the nuclear and cytosplasmic compartments to global gene expression in human cells. BMC Genomics. 2007;8:340. doi: 10.1186/1471-2164-8-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grindberg RV, et al. RNA-sequencing from single nuclei. PNAS. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clément-Ziza M, et al. Evaluation of methods for amplification of picogram amounts of total rna for whole genome expression profiling. BMC Genomics. 2009;10:246. doi: 10.1186/1471-2164-10-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morse AM, Carballo V, Baldwin DA, Taylor CG, McIntyre LM. Comparison between NuGEN's WT-OVATION PICO and One-Direct Amplification Systems. J Biomol Tech. 2010;21:141–147. [PMC free article] [PubMed] [Google Scholar]
- 52.Lamesch P, et al. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr Protoc Bioinformatics. 2002;30 doi: 10.1002/0471250953.bi0111s30. 1.11:1.11.1–1.11.51. [DOI] [PubMed] [Google Scholar]
- 53.Schon MA, Nodine MD. Widespread contamination of Arabidopsis embryo and endosperm transcriptome data sets. Plant Cell. 2017;29:608–617. doi: 10.1105/tpc.16.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Khan D, et al. Transcriptome atlas of the Arabidopsis funiculus – a study of maternal seed subregions. Plant J. 2015;82:41–53. doi: 10.1111/tpj.12790. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, et al. Direct evidence that suspensor cells have embryogenic potential that is suppressed by the embryo proper during normal embryogenesis. PNAS. 2015;112:12432–12437. doi: 10.1073/pnas.1508651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Radoeva T, ten Hove CA, Saiga S, Weijers D. Molecular characterization of Arabidopsis GAL4/UAS enhancer trap lines identifies novel cell type-specific promoters. Plant Physiol. 2016;171:1169–1181. doi: 10.1104/pp.16.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sarkar AK, et al. Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature. 2007;446:811–814. doi: 10.1038/nature05703. [DOI] [PubMed] [Google Scholar]
- 58.Schlereth A, et al. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature. 2010;464:913–916. doi: 10.1038/nature08836. [DOI] [PubMed] [Google Scholar]
- 59.De Rybel B, et al. A BHLH complex controls embryonic vascular tissue establishment and indeterminate growth in Arabidopsis. Dev Cell. 2013;24:426–437. doi: 10.1016/j.devcel.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 60.De Rybel B, et al. Integration of growth and patterning during vascular tissue formation in Arabidopsis. Science. 2014;345:1255215. doi: 10.1126/science.1255215. [DOI] [PubMed] [Google Scholar]
- 61.Crawford BCW, et al. Genetic control of distal stem cell fate within root and embryonic meristems. Science. 2015;347:655–659. doi: 10.1126/science.aaa0196. [DOI] [PubMed] [Google Scholar]
- 62.Clough SJ, Bent AF. Floral dip: A simplified method foragrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 63.De Rybel B, et al. A versatile set of ligation-independent cloning vectors for functional studies in plants. Plant Physiol. 2011;156:1292–1299. doi: 10.1104/pp.111.177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wendrich JR, Liao CY, van den Berg WA, De Rybel R, Weijers D. Ligation-independent cloning for plant research. Methods Mol Biol. 2015;1284:421–431. doi: 10.1007/978-1-4939-2444-8_21. [DOI] [PubMed] [Google Scholar]
- 65.Llavata-Peris C, Lokerse A, Möller B, De Rybel B, Weijers D. In: Plant organogenesis: Methods and protocols. De Smet I, editor. Humana Press; 2013. pp. 137–148. [Google Scholar]
- 66.Raissig MT, Gagliardini V, Jaenisch J, Grossniklaus U, Baroux C. Efficient and rapid isolation of early-stage embryos from Arabidopsis thaliana seeds. J Vis Exp. 2013;76:e50371. doi: 10.3791/50371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lin K, et al. MADMAX – Management and analysis database for multiple ~omics experiments. J Integr Bioinform. 2011;8:160. doi: 10.2390/biecoll-jib-2011-160. [DOI] [PubMed] [Google Scholar]
- 68.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of genechip data. Nucleic Acids Res. 2005;33:e175–e75. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 70.Sartor MA, et al. Intensity-based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics. 2006;7:1–17. doi: 10.1186/1471-2105-7-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Phipson B, Lee S, Majewski IJ, Alexander WS, Smyth GK. Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann App Stat. 2016;10:946–963. doi: 10.1214/16-AOAS920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Storey JD, Tibshirani R. Statistical significance for genomewide studies. PNAS. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orlando DA, Brady SM, Koch JD, Dinneny JR, Benfey PN. In: Plant systems biology. Belostotsky AD, editor. Humana Press; 2009. pp. 57–77. [Google Scholar]
- 74.Du Z, Zhou X, Ling Y, Zhang Z, Su Z. agrigo: a GO analysis toolkit for the agricultural community. Nucleic Acids Res. 2010;38:W64–W70. doi: 10.1093/nar/gkq310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Palaniswamy SK, et al. AGRIS and AtRegNet. a platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Saiga S, et al. The Arabidopsis OBERON1 and OBERON2 genes encode plant homeodomain finger proteins and are required for apical meristem maintenance. Development. 2008;135:1751–1759. doi: 10.1242/dev.014993. [DOI] [PubMed] [Google Scholar]
- 77.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:1–14. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw microarray expression data are available at the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) as series GSE89101.