Abstract
The main approach of traditional information retrieval (IR) is to examine how many words from a query appear in a document. A drawback of this approach, however, is that it may fail to detect relevant documents where no or only few words from a query are found. The semantic analysis methods such as LSA (latent semantic analysis) and LDA (latent Dirichlet allocation) have been proposed to address the issue, but their performance is not superior compared to common IR approaches. Here we present a query-document similarity measure motivated by the Word Mover’s Distance. Unlike other similarity measures, the proposed method relies on neural word embeddings to compute the distance between words. This process helps identify related words when no direct matches are found between a query and a document. Our method is efficient and straightforward to implement. The experimental results on TREC Genomics data show that our approach outperforms the BM25 ranking function by an average of 12% in mean average precision. Furthermore, for a real-world dataset collected from the PubMed® search logs, we combine the semantic measure with BM25 using a learning to rank method, which leads to improved ranking scores by up to 25%. This experiment demonstrates that the proposed approach and BM25 nicely complement each other and together produce superior performance.
Keywords: PubMed literature search, Semantic similarity, Word Mover’s Distance, Word embeddings, Learning to rank
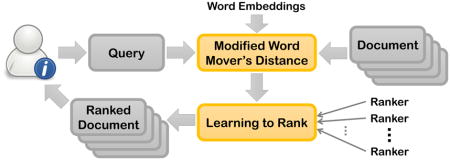
Graphical Abstract
1. Introduction
In information retrieval (IR), queries and documents are typically represented by term vectors where each term is a content word and weighted by tf-idf, i.e. the product of the term frequency and the inverse document frequency, or other weighting schemes [1]. The similarity of a query and a document is then determined as a dot product or cosine similarity. Although this works reasonably, the traditional IR scheme often fails to find relevant documents when synonymous or polysemous words are used in a dataset, e.g. a document including only “neoplasm” cannot be found when the word “cancer” is used in a query. One solution of this problem is to use query expansion [2, 3, 4, 5] or dictionaries, but these alternatives still depend on the same philosophy, i.e. queries and documents should share exactly the same words.
While the term vector model computes similarities in a sparse and high-dimensional space, the semantic analysis methods such as latent semantic analysis (LSA) [6, 7] and latent Dirichlet allocation (LDA) [8] learn dense vector representations in a low-dimensional space. These methods choose a vector embedding for each term and estimate a similarity between terms by taking an inner product of their corresponding embeddings [9]. Since the similarity is calculated in a latent (semantic) space based on context, the semantic analysis approaches do not require having common words between a query and documents. However, it has been shown that LSA and LDA methods do not produce superior results in various IR tasks [10, 11, 12] and the classic ranking method, BM25 [13], usually outperforms those methods in document ranking [14, 15].
Neural word embedding [16, 17] is similar to the semantic analysis methods described above. It learns low-dimensional word vectors from text, but while LSA and LDA utilize co-occurrences of words, neural word embedding learns word vectors to predict context words [11]. Moreover, training of semantic vectors is derived from neural networks. Both co-occurrence and neural word embedding approaches have been used for lexical semantic tasks such as semantic relatedness (e.g. king and queen), synonym detection (e.g. cancer and carcinoma) and concept categorization (e.g. banana and pineapple belong to fruits) [11, 18]. But, Baroni et al. [11] showed that neural word embedding approaches generally performed better on such tasks with less effort required for parameter optimization. The neural word embedding models have also gained popularity in recent years due to their high performance in NLP tasks [19].
Here we present a query-document similarity measure using a neural word embedding approach. This work is particularly motivated by the Word Mover’s Distance [20]. Unlike the common similarity measure taking query/document centroids of word embeddings, the proposed method evaluates a distance between individual words from a query and a document. Our first experiment was performed on the TREC 2006 and 2007 Genomics benchmark sets [21, 22], and the experimental results showed that our approach was better than BM25 ranking. This was solely based on matching queries and documents by the semantic measure and no other feature was used for ranking documents.
In general, conventional ranking models (e.g. BM25) rely on a manually designed ranking function and require heuristic optimization for parameters [23, 24]. In the age of information explosion, this one-size-fits-all solution is no longer adequate. For instance, it is well known that links to a web page are an important source of information in web document search [25], hence using the link information as well as the relevance between a query and a document is crucial for better ranking. In this regard, learning to rank [23] has drawn much attention as a scheme to learn how to combine diverse features. Given feature vectors of documents and their relevance levels, a learning to rank approach learns an optimal way of weighting and combining multiple features.
We argue that the single scores (or features) produced by BM25 and our proposed semantic measure complement each other, thus merging these two has a synergistic effect. To confirm this, we measured the impact on document ranking by combining BM25 and semantic scores using the learning to rank approach, LamdaMART [26, 27]. Trained on PubMed user queries and their click-through data, we evaluated the search performance based on the most highly ranked 20 documents. As a result, we found that using our semantic measure further improved the performance of BM25.
Taken together, we make the following important contributions in this work. First, to the best of our knowledge, this work represents the first investigation of query-document similarity for information retrieval using the recently proposed Word Mover’s Distance. Second, we modify the original Word Mover’s Distance algorithm so that it is computationally less expensive and thus more practical and scalable for real-world search scenarios (e.g. biomedical literature search). Third, we measure the actual impact of neural word embeddings in PubMed by utilizing user queries and relevance information derived from click-through data. Finally, on TREC and PubMed datasets, our proposed method achieves stronger performance than BM25.
2. Methods
A common approach to computing similarity between texts (e.g. phrases, sentences or documents) is to take a centroid of word embeddings, and evaluate an inner product or cosine similarity between centroids1 [15, 28]. This has found use in classification and clustering because they seek an overall topic of each document. However, taking a simple centroid is not a good approximator for calculating a distance between a query and a document [20]. This is mostly because queries tend to be short and finding the actual query words in documents is feasible and more accurate than comparing lossy centroids. Consistent with this, our approach here is to measure the distance between individual words, not the average distance between a query and a document.
2.1. Word Mover’s Distance
Our work is based on the Word Mover’s Distance between text documents [20], which calculates the minimum cumulative distance that words from a document need to travel to match words from a second document. In this subsection, we outline the original Word Mover’s Distance algorithm, and our adapted model is described in Section 2.2.
First, following Kusner et al. [20], documents are represented by normalized bag-of-words (BOW) vectors, i.e. if a word wi appears tfi times in a document, the weight is
| (1) |
where n is number of words in the document. The higher the weight, the more important the word. They assume a word embedding so that each word wi has an associated vector xi. The dissimilarity c between wi and wj is then calculated by
| (2) |
The Word Mover’s Distance makes use of word importance and the relatedness of words as we now describe.
Let D and D′ be BOW representations of two documents D and D′. Let T ∈ ℛn×n be a flow matrix, where Tij ≥ 0 denotes how much it costs to travel from wi in D to wj in D′, and n is the number of unique words appearing in D and/or D′. To entirely transform D to D′, we ensure that the entire outgoing flow from wi equals di and the incoming flow to wj equals . The Word Mover’s Distance between D and D′ is then defined as the minimum cumulative cost required to move all words from D to D′ or vice versa, i.e.
| (3) |
The solution is attained by finding Tij that minimizes the expression in Eq. (3). Kusner et al. [20] applied this to obtain nearest neighbors for document classification, i.e. k-NN classification and it produced outstanding performance among other state-of-the-art approaches. What we have just described is the approach given in Kusner et al. We will modify the word weights and the measure of the relatedness of words to better suit our application.
2.2. Our query-document similarity measure
While the prior work gives a hint that the Word Mover’s Distance is a reasonable choice for evaluating a similarity between documents, it is uncertain how the same measure could be used for searching documents to satisfy a query. First, it is expensive to compute the Word Mover’s Distance. The time complexity of solving the distance problem is O(n3 log n) [29]. Second, the semantic space of queries is not the same as those of documents. A query consists of a small number of words in general, hence words in a query tend to be more ambiguous because of the restricted context. On the contrary, a text document is longer and more informational. Having this in mind, we realize that ideally two distinctive components could be employed for query-document search: 1) mapping queries to documents using a word embedding model trained on a document set and 2) mapping documents to queries using a word embedding model obtained from a query set. In this work, however, we aim to address the former, and the mapping of documents to queries remains as future work.
For our purpose, we will change the word weight di to incorporate inverse document frequency (idf), i.e.
| (4) |
where . K is the size of a document set and ki is the number of documents that include the ith term. The rationale behind this is to weight words in such a way that common terms are given less importance. It is the idf factor normally used in tf-idf and BM25 [30, 31]. In addition, our word embedding is a neural word embedding trained on the 25 million PubMed titles and abstracts.
Let Q and D be BOW representations of a query Q and a document D. D and D′ in Section 2.1 are now replaced by Q and D, respectively. Since we want to have a higher score for documents relevant to Q, c(i, j) is redefined as a cosine similarity, i.e.
| (5) |
In addition, the problem we try to solve is the flow Q → D. Hence, Eq. (3) is rewritten as follows.
| (6) |
where di represents the word wi in Q. idf(i) in Eq. (4) is unknown for queries, therefore we compute idf(i) based on the document collection. The optimal solution of the expression in Eq. (6) is to map each word in Q to the most similar word in D based on word embeddings. The time complexity for getting the optimal solution is O(mn), where m is the number of unique query words and n is the number of unique document words. In general, m ≪ n and evaluating the similarity between a query and a document can be implemented in parallel computation. Thus, the document ranking process can be quite efficient.
2.3. Learning to rank
In our study, we use learning to rank to merge two distinctive features, BM25 scores and our semantic measures. This approach is trained and evaluated on real-world PubMed user queries and their responses based on click-through data [32]. While it is not common to use only two features for learning to rank, this approach is scalable and versatile. Adding more features subsequently should be straightforward and easy to implement. The performance result we obtain demonstrates the semantic measure is useful to rank documents according to users’ interests.
We briefly outline learning to rank approaches [33, 34] in this subsection. For a list of retrieved documents, i.e. for a query Q and a set of candidate documents, D = {D1, D2, …, Dm}, we are given their relevancy judgements y = {y1, y2, …, ym}, where yi is a positive integer when the document Di is relevant and 0 otherwise. The goal of learning to rank is to build a model h that can rank relevant documents near or at the top of the ranked retrieval list. To accomplish this, it is common to learn a function h(w, ψ(Q,D)), where w is a weight vector applied to the feature vector ψ(Q,D). A part of learning involves learning the weight vector but the form of h may also require learning. For example, h may involve learned decision trees as in our application.
In particular, we use LambdaMART [26, 27] for our experiments. LambdaMART is a pairwise learning to rank approach and is being used for PubMed relevance search. While the simplest approach (pointwise learning) is to train the function h directly, pairwise approaches seek to train the model to place correct pairs higher than incorrect pairs, i.e. h(w, ψ(Q,Di)) ≥ h(w, ψ(Q,Dj))+ε, where the document Di is relevant and Dj is irrelevant. ε indicates a margin. LambdaMART is a boosted tree version of LambdaRank [27]. An ensemble of LambdaMART, LambdaRank and logistic regression models won the Yahoo! learning to rank challenge [24].
3. Results and discusssion
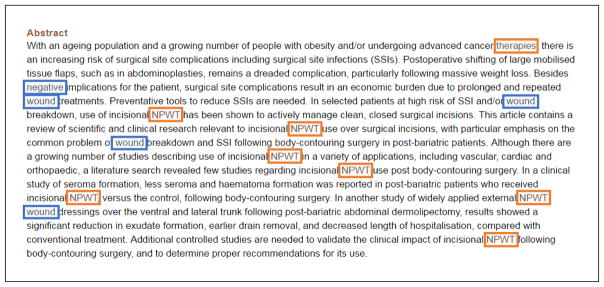
Our resulting formula from the Word Mover’s Distance seeks to find the closest terms for each query word. Figure 1 depicts an example with and without using our semantic matching. For the query, “negative pressure wound therapy”, a traditional way of searching documents is to find those documents which include the words “negative”, “pressure”, “wound” and “therapy”. As shown in the figure, the words, “pressure” and “therapy”, cannot be found by perfect string match. On the other hand, within the same context, the semantic measure finds the closest words “NPWT” and “therapies” for “pressure” and “therapy”, respectively. Identifying abbreviations and singular/plural would help match the same words, but this example is to give a general idea about the semantic matching process. Also note that using dictionaries such as synonyms and abbreviations requires an additional effort for manual annotation.
Figure 1.
Matching the query, “negative pressure wound therapy” with the words in the abstract of PMID 25853645 using the exact string match vs. the semantic string match. The blue boxes indicate that the words appear in the query. Additional words (orange boxes) are found using our semantic measure.
In the following subsections, we describe the datasets and experiments, and discuss our results.
3.1. Datasets
To evaluate our word embedding approach, we used two scientific literature datasets: TREC Genomics data and PubMed. Table 1 shows the number of queries and documents in each dataset. TREC represents the benchmark sets created for the TREC 2006 and 2007 Genomics Tracks [21, 22]. The original task is to retrieve passages relevant to topics (i.e. queries) from full-text articles, but the same set can be utilized for searching relevant PubMed documents. We consider a PubMed document relevant to a TREC query if and only if the full-text of the document contains a passage judged relevant to that query by the TREC judges. Our setup is more challenging because we only use PubMed abstracts, not full-text articles, to find evidence.
Table 1.
Number of queries and documents for TREC and PubMed experiments. TREC 2006 includes 28 queries originally but two were removed because there were no relevant documents.
| Dataset | # Queries | # Documents |
|---|---|---|
| TREC 2006 | 26 | 162,259 |
| TREC 2007 | 36 | 162,259 |
| PubMed | 27,870 | 27,098,6292 |
Machine learning approaches, especially supervised ones such as learning to rank, are promising and popular nowadays. Nonetheless, they usually require a large set of training examples, and such datasets are particularly difficult to find in the biomedical domain. For this reason, we created a gold standard set based on real (anonymized) user queries and the actions users subsequently took, and named this the PubMed set.
To build the PubMed set, we collected one year’s worth of search logs and restricted the set of queries to those where users requested the relevance order and which yielded at least 20 retrieved documents. This set contained many popular but duplicate queries. Therefore, we merged queries and summed up user actions for each of them. That is, for each document stored for each query, we counted the number of times it was clicked in the retrieved set (i.e. abstract click) and the number of times users requested full-text articles (i.e. full-text click). We considered the queries that appeared less than 10 times to be less informative because they were usually very specific, and we could not collect enough user actions for training. After this step, we further filtered out non-informational queries (e.g. author and journal names). As the result, 27,870 queries remained for the final set.
The last step for producing the PubMed set was to assign relevance scores to documents for each query. We will do this based on user clicks. It is known that click-through data is a useful proxy for relevance judgments [35, 36, 37]. Let a(D,Q) be the number of clicks to the abstract of a document D from the results page for the query Q. Let f(D,Q) be the number of clicks from D’s abstract page to its full-text, which result from the query Q. Let λ ∈ ℝ+ be the boost factor for documents without links to full-text articles. FT(D) is the indicator function such that FT(D) = 1 if the document D includes a link to full-text articles and FT(D) = 0 otherwise. We can then calculate the relevance, y, of a document for a given query:
| (7) |
μ is the trade-off between the importance of abstract clicks and full-text clicks. The last term of the relevance function gives a slight boost to documents without full-text links, so that they get a better relevance (thus rank) than those for which full-text is available but never clicked, assuming they all have the same amount of abstract clicks. We manually tuned the parameters based on user behavior and log analyses, and used the settings, μ = 0.33 and λ = 15.
Compared to the TREC Genomics set, the full PubMed set is much larger, including all 27 million documents in PubMed. While the TREC and PubMed sets share essentially the same type of documents, the tested queries are quite different. The queries in TREC are a question type, e.g. “what is the role of MMS2 in cancer?” However, the PubMed set uses actual queries from PubMed users.
In our experiments, the TREC set was used for evaluating BM25 and the semantic measure separately and the PubMed set was used for evaluating the learning to rank approach. We did not use the TREC set for learning to rank due to the small number of queries. Only 62 queries and 162,259 documents are available in TREC, whereas the PubMed set consists of many more queries and documents.
3.2. Word embeddings and other experimental setup
We used the skip-gram model of word2vec [17] to obtain word embeddings. The alternative models such as GloVe [12] and Fast Text [38] are available, but their performance varies depending on tasks and is comparable to word2vec overall [39, 40]. word2vec was trained on titles and abstracts from over 25 million PubMed documents. Word vector size and window size were set to 100 and 10, respectively. These parameters were optimized to produce high recall for synonyms [41]. Note that an independent set (i.e. synonyms) was used for tuning word2vec parameters, and the trained model is available online (https://www.ncbi.nlm.nih.gov/IRET/DATASET).
For experiments, we removed stop words from queries and documents. BM25 was chosen for performance comparison and the parameters were set to k = 1.9 and b = 1.0 [42]. Among document ranking functions, BM25 shows a competitive performance [43]. It also outperforms co-occurrence based word embedding models [14, 15]. For learning to rank approaches, 70% of the PubMed set was used for training and the rest for testing. The RankLib library (https://sourceforge.net/p/lemur/wiki/RankLib/) was used for implementing LambdaMART and the PubMed experiments.
3.3. TREC experiments
Table 2 presents the average precision of tf-idf (TFIDF), BM25, word vector centroid (CENTROID) and our embedding approach on the TREC dataset. Average precision [44] is the average of the precisions at the ranks where relevant documents appear. Relevance judgements in TREC are based on the pooling method [45], i.e. relevance is manually assessed for top ranking documents returned by participating systems. Therefore, we only used the documents that annotators reviewed for our evaluation [2].
Table 2.
Mean average precision of tf-idf (TFIDF), BM25, word vector centroid (CENTROID) and our semantic approach (SEM) on the TREC set.
| Method | TREC 2006 | TREC 2007 |
|---|---|---|
| TFIDF | 0.3018 | 0.2375 |
| BM25 | 0.3136 | 0.2463 |
| CENTROID | 0.2363 | 0.2459 |
| SEM | 0.3732 | 0.2601 |
As shown in Table 2, BM25 performs better than TFIDF and CENTROID. CENTROID maps each query and document to a vector by taking a centroid of word embedding vectors, and the cosine similarity between two vectors is used for scoring and ranking documents. As mentioned earlier, this approach is not effective when multiple topics exist in a document. From the table, the embedding approach boosts the average precision of BM25 by 19% and 6% on TREC 2006 and 2007, respectively. However, CENTROID provides scores lower than BM25 and SEM approaches.
Although our approach outperforms BM25 on TREC, we do not claim that BM25 and other traditional approaches can be completely replaced with the semantic method. We see the semantic approach as a means to narrow the gap between words in documents and those in queries (or users’ intentions). This leads to the next experiment using our semantic measure as a feature for ranking in learning to rank.
3.4. PubMed experiments
For the PubMed dataset, we used learning to rank to combine BM25 and our semantic measure. An advantage of using learning to rank is its flexibility to add more features and optimize performance by learning their importance. PubMed documents are semi-structured, consisting of title, abstract and many more fields. Since our interest lies in text, we only used titles and abstracts, and applied learning to rank in two different ways: 1) to find semantically closest words in titles (BM25 + SEMTitle) and 2) to find semantically closest words in abstracts (BM25 + SEMAbstract). Although our semantic measure alone produces better ranking scores on the TREC set, this does not apply to user queries in PubMed. It is because user queries are often short, including around three words on average, and the semantic measure cannot differentiate documents when they include all query words.
Table 3 shows normalized discounted cumulative gain (NDCG) scores for top 5, 10 and 20 ranked documents for each approach. NDCG [46] is a measure for ranking quality and it penalizes relevant documents appearing in lower ranks by adding a rank-based discount factor. In the table, reranking documents by learning to rank performs better than BM25 overall, however the larger gain is obtained from using titles (BM25 + SEMTitle) by increasing NDCG@20 by 23%. NDCG@5 and NDCG@10 also perform better than BM25 by 23% and 25%, respectively. It is not surprising that SEMTitle produces better performance than SEMAbstract. The current PubMed search interface does not allow users to see abstracts on the results page, hence users click documents mostly based on titles. Nevertheless, it is clear that the abstract-based semantic distance helps achieve better performance.
Table 3.
NDCG scores for BM25 and learning to rank (BM25 + SEM) search results. We used two fields from PubMed documents for the learning to rank approach. “Title” and “Abstract” mean only words from titles and abstracts were used to compute semantic scores, respectively. The scores in parentheses show the improved ratios of BM25 + SEM to BM25 ranking.
| Method | NDCG@5 | NDCG@10 | NDCG@20 |
|---|---|---|---|
| BM25 | 0.0854 | 0.1145 | 0.1495 |
| BM25 + SEMTitle | 0.1048 (22.72%) | 0.1427 (24.59%) | 0.1839 (23.03%) |
| BM25 + SEMAbstract | 0.0917 (7.38%) | 0.1232 (7.57%) | 0.1592 (6.51%) |
After our experiments for Table 3, we also assessed the efficiency of learning to rank (BM25 + SEMTitle) by measuring query processing speed in PubMed relevance search. Using 100 computing threads, 900 queries are processed per second, and for each query, the average processing time is 100 milliseconds, which is fast enough to be used in the production system.
4. Conclusion
We presented a word embedding approach for measuring similarity between a query and a document. Starting from the Word Mover’s Distance, we reinterpreted the model for a query-document search problem. Even with the Q → D flow only, the word embedding approach is already efficient and effective. In this setup, the proposed approach cannot distinguish documents when they include all query words, but surprisingly, the word embedding approach shows remarkable performance on the TREC Genomics datasets. Moreover, applied to PubMed user queries and click-through data, our semantic measure allows to further improves BM25 ranking performance. This demonstrates that the semantic measure is an important feature for IR and is closely related to user clicks.
While many deep learning solutions have been proposed recently, their slow training and lack of flexibility to adopt various features limit real-world use. However, our approach is more straightforward and can be easily added as a feature in the current PubMed relevance search framework. Proven by our PubMed search results, our semantic measure improves ranking performance without adding much overhead to the system.
This work investigates query-document similarity for information retrieval using the Word Mover’s Distance.
We modify the original Word Mover’s Distance algorithm so that it is computationally less expensive and thus more practical and scalable for real-world search scenarios.
We measure the actual impact of neural word embeddings in PubMed by utilizing user queries and click-through based relevance information.
Applied to TREC Genomics and PubMed datasets, our approach achieves stronger performance than traditional ranking functions.
Acknowledgments
Funding
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine.
Footnotes
The implementation of word2vec also uses centroids of word vectors for calculating similarities (https://code.google.com/archive/p/word2vec).
This is the number of PubMed documents as of Apr. 6, 2017. This number and the actual number of documents used for our experiments may differ slightly.
Conflicts of interest: none
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Salton G, Buckley C. Term-weighting approaches in automatic text retrieval. Information Processing & Management. 1988;24(5):513–523. [Google Scholar]
- 2.Lu Z, Kim W, Wilbur WJ. Evaluation of query expansion using MeSH in PubMed. Information Retrieval. 2009;12(1):69–80. doi: 10.1007/s10791-008-9074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carpineto C, Romano G. A survey of automatic query expansion in information retrieval. ACM Computing Surveys. 2012;44(1):1:1–1:50. [Google Scholar]
- 4.Diaz F, Mitra B, Craswell N. Query expansion with locally-trained word embeddings. Proc. Annual Meeting of the Association for Computational Linguistics (ACL 2016); 2016. pp. 367–377. [Google Scholar]
- 5.Roy D, Paul D, Mitra M, Garain U. Using word embeddings for automatic query expansion. Proc. SIGIR 2016 Workshop on Neural Information Retrieval; 2016. [Google Scholar]
- 6.Deerwester S, Dumais ST, Furnas GW, Landauer TK, Harshman R. Indexing by latent semantic analysis. Journal of the American Society for Information Science. 1990;41(6):391–407. [Google Scholar]
- 7.Hofmann T. Probabilistic latent semantic indexing. Proc. Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; 1999. pp. 50–57. [Google Scholar]
- 8.Blei DM, Ng AY, Jordan MI. Latent Dirichlet allocation. Journal of Machine Learning Research. 2003;3:993–1022. [Google Scholar]
- 9.Sordoni A, Bengio Y, Nie J-Y. Learning concept embeddings for query expansion by quantum entropy minimization. Proc. AAAI Conference on Artificial Intelligence; 2014. pp. 1586–1592. [Google Scholar]
- 10.Maas AL, Daly RE, Pham PT, Huang D, Ng AY, Potts C. Learning word vectors for sentiment analysis. Proc. Annual Meeting of the Association for Computational Linguistics (ACL 2011); 2011. pp. 142–150. [Google Scholar]
- 11.Baroni M, Dinu G, Kruszewski G. Don’t count, predict! a systematic comparison of context-counting vs. context-predicting semantic vectors. Proc. Annual Meeting of the Association for Computational Linguistics (ACL 2014); 2014. pp. 238–247. [Google Scholar]
- 12.Pennington J, Socher R, Manning C. GloVe: Global vectors for word representation. Proc. Conference on Empirical Methods in Natural Language Processing (EMNLP 2014); 2014. pp. 1532–1543. [Google Scholar]
- 13.Robertson S, Zaragoza H. The probabilistic relevance framework: BM25 and beyond. Foundations and Trends in Information Retrieval. 2009;3(4):333–389. [Google Scholar]
- 14.Atreya A, Elkan C. Latent semantic indexing (LSI) fails for TREC collections. ACM SIGKDD Explorations Newsletter. 2011;12(2):5–10. [Google Scholar]
- 15.Nalisnick E, Mitra B, Craswell N, Caruana R. Improving document ranking with dual word embeddings. Proc. International World Wide Web Conference (WWW 2016); 2016. pp. 83–84. [Google Scholar]
- 16.Bengio Y, Ducharme R, Vincent P, Janvin C. A neural probabilistic language model. Journal of Machine Learning Research. 2003;3:1137–1155. [Google Scholar]
- 17.Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J. Distributed representations of words and phrases and their compositionality. Advances in Neural Information Processing Systems (NIPS 2013) 2013:3111–3119. [Google Scholar]
- 18.Schnabel T, Labutov I, Mimno D, Joachims T. Evaluation methods for unsupervised word embeddings. Proc. Conference on Empirical Methods in Natural Language Processing (EMNLP 2015); 2015. pp. 298–307. [Google Scholar]
- 19.Levy O, Goldberg Y. Neural word embedding as implicit matrix factorization. Advances in Neural Information Processing Systems (NIPS 2014) 2014:2177–2185. [Google Scholar]
- 20.Kusner MJ, Sun Y, Kolkin NI, Weinberger KQ. From word embeddings to document distances. Proc. International Conference on Machine Learning (ICML 2015); 2015. pp. 957–966. [Google Scholar]
- 21.Hersh W, Cohen AM, Roberts P, Rekapalli HK. TREC 2006 Genomics Track overview. Proc. Text REtrieval Conference; 2006; 2006. [Google Scholar]
- 22.Hersh W, Cohen AM, Ruslen L, Roberts P. TREC 2007 Genomics Track overview. Proc. Text REtrieval Conference; 2007; 2007. [Google Scholar]
- 23.Liu TY. Learning to rank for information retrieval. Foundations and Trends in Information Retrieval. 2009;3(3):225–331. [Google Scholar]
- 24.Chapelle O, Chang Y. Yahoo! learning to rank challenge overview. Proc. Learning to Rank Challenge; 2011. pp. 1–24. [Google Scholar]
- 25.Brin S, Page L. The anatomy of a large-scale hypertextual web search engine. Proc. International Conference on World Wide Web; 1998. pp. 07–117. [Google Scholar]
- 26.Burges CJC, Svore KM, Wu Q, Gao J. Ranking, boosting, and model adaptation. Tech rep. 2008 [Google Scholar]
- 27.Burges CJ. From RankNet to LambdaRank to LambdaMART: An overview. Tech rep. 2010 [Google Scholar]
- 28.Furnas GW, Deerwester S, Dumais ST, Landauer TK, Harshman RA, Streeter LA, Lochbaum KE. Information retrieval using a singular value decomposition model of latent semantic structure. Proc. Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; 1988. pp. 465–480. [Google Scholar]
- 29.Pele O, Werman M. Fast and robust earth mover’s distances. Proc. IEEE International Conference on Computer Vision (ICCV 2009); 2009. pp. 460–467. [Google Scholar]
- 30.Witten IH, Moffat A, Bell TC. Managing Gigabytes. Morgan-Kaufmann; San Francisco, California, USA: 1999. [Google Scholar]
- 31.Wilbur WJ. Global term weights for document retrieval learned from trec data. Journal of Information Science. 2001;27(5):303–310. [Google Scholar]
- 32.Joachims T. Optimizing search engines using clickthrough data. Proc. ACMSIGKDD International Conference on Knowledge Discovery and Data Mining; 2002. pp. 133–142. [Google Scholar]
- 33.Severyn A, Moschitti A. Learning to rank short text pairs with convolutional deep neural networks. Proc. International ACM SIGIR Conference on Research and Development in Information Retrieval; 2015. pp. 373–382. [Google Scholar]
- 34.Freund Y, Iyer R, Schapire RE, Singer Y. An efficient boosting algorithm for combining preferences. Journal of Machine Learning Research. 2003;4:933–969. [Google Scholar]
- 35.Joachims T. Evaluating retrieval performance using clickthrough data. Proc. SIGIR Workshop on Mathematical/Formal Methods in Information Retrieval; 2002. [Google Scholar]
- 36.Agrawal R, Halverson A, Kenthapadi K, Mishra N, Tsaparas P. Generating labels from clicks. Proc. ACM International Conference on Web Search and Data Mining (WSDM 2009); 2009. pp. 172–181. [Google Scholar]
- 37.Xu J, Chen C, Xu G, Li H, Abib ERT. Improving quality of training data for learning to rank using click-through data. Proc. ACM International Conference on Web Search and Data Mining (WSDM 2010); 2010. pp. 171–180. [Google Scholar]
- 38.Bojanowski P, Grave E, Joulin A, Mikolov T. Enriching word vectors with subword information. Transactions of the Association for Computational Linguistics. 2017;5:135–146. [Google Scholar]
- 39.MTH, Sahu SK, Anand A. Evaluating distributed word representations for capturing semantics of biomedical concepts. Proc. ACL Workshop on Biomedical Natural Language Processing (BioNLP 2015); 2015. pp. 158–163. [Google Scholar]
- 40.Cao S, Lu W. Improving word embeddings with convolutional feature learning and subword information. Proc. AAAI Conference on Artificial Intelligence (AAAI 2017); 2017. pp. 3144–3151. [Google Scholar]
- 41.Yeganova L, Kim W, Kim S, Dögan RI, Liu W, Comeau DC, Lu Z, Wilbur WJ. Pubtermvariants: Biomedical term variants and their use for pubmed search. Proc. ACL Workshop on Biomedical Natural Language Processing (BioNLP 2016); 2016. pp. 141–145. [Google Scholar]
- 42.Lin J, Wilbur WJ. PubMed related articles: a probabilistic topic-based model for content similarity. BMC Bioinformatics. 2007;8:423. doi: 10.1186/1471-2105-8-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trotman A, Puurula A, Burgess B. Improvements to BM25 and language models examined. Proc. 2014 Australasian Document Computing Symposium (ADCS 2014); 2014. pp. 58:58–58:65. [Google Scholar]
- 44.Turpin A, Scholer F. User performance versus precision measures for simple search tasks. Proc. Annual International ACM SIGIR Conference on Research and Development in Information Retrieval; 2006. pp. 11–18. [Google Scholar]
- 45.Manning CD, Raghavan P, Schütze H. Introduction to Information Retrieval. Cambridge University Press; New York, New York, USA: 2008. [Google Scholar]
- 46.Burges C, Shaked T, Renshaw E, Lazier A, Deeds M, Hamilton N, Hullender G. Learning to rank using gradient descent. Proc. International Conference on Machine Learning; 2005. pp. 89–96. [Google Scholar]