Abstract
Background
The Aurora kinase family is comprised of highly conserved serine/threonine protein kinases that are known to be crucial in the regulation of the cell cycle. Aberrant expression of Aurora kinases has been demonstrated in certain malignancies. We aimed to examine the expression of Aurora kinases in human breast cancer tissues and to investigate the cellular impact of Aurora kinases inhibitor on breast cancer cells.
Methods
The expression of Aurora kinase A/B/C was individually examined in tumor specimens (n = 106) and normal tissues (n = 29) from breast cancer patients using quantitative real-time PCR (Q-PCR) and immunohistochemistry. Cells were treated with the corresponding inhibitor, and then migration and adhesion were evaluated by electric cell impedance sensing assay. The proliferation of breast cancer cells treated with the inhibitor was examined using in vitro models.
Results
High levels of Aurora kinase B and C were found in the tumor tissues from breast cancer patients, but low levels of Aurora kinase A were seen in normal tissues at the mRNA level and immunohistochemistry. The mRNA expression level of Aurora kinase B and C had a negative correlation with grade staging, staging and survival rate in breast cancer patients, whilst Aurora kinase A exhibited a converse expression. The inhibitor ZM447439 promoted adhesion of the human breast cancer cell line MDA-MB-231 and inhibited the migration of MCF-7 human breast cancer cells.
Conclusion
Taken together, the expression of Aurora kinase B and C was down-regulated in breast tumor tissues but Aurora kinase A was not. Aurora kinase may have a key role in the progression and metastasis of breast cancer.
Keywords: Breast cancer; Metastasis; Aurora kinases A, B and C; Immunohistochemistry; Migration; Adhesion; Proliferation; ZM447439
Introduction
As reported, breast cancer is the most common carcinoma and a life-threatening disorder in females with high rates of morbidity and mortality due to distant metastasis. There are numerous metastatic sites for breast cancer, such as brain, bone, liver and lung. Amongst these, bone metastasis is regarded as the most common complication in breast cancer [1-4]. However, the metastatic mechanism regulating this complex process is little known. Therefore, understanding the mechanisms of metastatic disease is essential for the development of new diagnostic and therapeutic strategies.
The Aurora kinase family (Aurora A, B, and C) of highly conserved serine/threonine protein kinases are known to be crucial in the early stages of the mitotic cell cycle [5]. The Aurora kinase family consists of a regulatory N-terminal domain, a protein kinase domain and a C-terminal domain sharing high sequence homology and has a high degree of conserved functional motifs [6-8]. The functional role of Aurora family is associated with their localization and specific substrate proteins binding to them [9].
Aberrant expression of Aurora kinases may interfere with mitosis and cytokinesis, and then lead to genetic instability; this in turn may result in the development of tumors. The Aurora kinase action mechanism is based on different isoform and cancer type [6]. It is reported that Aurora kinase A is overexpressed during breast cancer cell transformation [10, 11]. Also, high expression of Aurora A is also found in ovarian, colon and lung cancer [11, 12]. It is also found that aberrant expression of Aurora B is associated with thyroid anaplastic carcinoma [13], colon cancer [14] and oral cancer [15]. Aurora C plays a role in meiosis [16] and its overexpression is also associated with certain somatic cancers [17]. Aurora kinases are validated as a therapeutic target for cancer and thus the development of Aurora kinase inhibitors has progressed from preclinical to clinical studies [17, 18]. After the discovery of the first Aurora kinase inhibitor ZM447494 as a potential drug in cancer treatment for targeted therapy, more than 30 Aurora kinase inhibitors have been identified in tumor treatment [19, 20]. Tavant et al indicated that Aurora kinases inhibitors VX-680 and ZM447439 were promising drug of potential clinical in connection with conventional osteosarcoma chemotherapeutic agents via the analysis of these two inhibitors [21]. AT9283 is one kind of Aurora kinases inhibitors, and it could inhibit Aurora kinases A and B, which has inhibition of proliferation and cellular division and the induction of apoptosis in cancer cells [22, 23]. ENMD-2076, another Aurora kinase inhibitor, has also been proved to inhibit cell growth and induce apoptosis in acute myeloid leukemia cells in vitro [24]. VX-680, also called MK0457, is a pan-Aurora kinase inhibitor that induces apoptosis and disrupts mitosis in cells cycling and plays an efficacious role in clinical like lymphomas and solid tumors. Recently, several studies showed VX-680 inhibition of tumor growth was associated with well-tolerated doses, and no signs of mechanism-independent toxicity were found [25-27]. Aurora kinases played key functions in the early phases of the mitotic cell cycle. Recent researches have demonstrated that the expression of Aurora kinases was valuable in targeted therapy development and disease stabilization. There are numerous preclinical studies and multiple phase I and II clinical trials taking place in both hematologic and solid malignancies about Aurora kinase inhibitors.
In this study, we investigated the possible clinical implication of Aurora kinases in human breast tumor tissues and whether the expression level is related to the clinical outcome of patients with breast cancer. We also detected the effect of Aurora kinase on human breast tumor cells with Aurora kinase inhibitor ZM447439 in vitro.
Materials and Methods
Human breast specimens and ethics, consent and permissions
Breast tissue samples were obtained immediately from breast cancer patients after surgery and snap frozen in liquid nitrogen for further use. The specimens included breast tumor tissues (n = 106) and normal background mammary tissues (n = 29) obtained from the same patients. The pathology of these samples was confirmed by a pathologist and the background tissues were free from tumor deposits. All protocols were approved by the local ethics committee. Ethics were under the Bro Taf Health Authority (01/4303 and 01/4046). Full patient consent was obtained. Full patient clinical data details are shown in Table 1.
Table 1. Breast Cancer Patient Clinical Data Details.
| Clinical data | Sample no. |
|---|---|
| Tissue sample | |
| Normal | 34 |
| Tumor | 118 |
| NPI | |
| 1 | 63 |
| 2 | 36 |
| 3 | 15 |
| Tumor grade | |
| 1 | 21 |
| 2 | 41 |
| 3 | 7 |
| TNM staging | |
| I | 2 |
| II | 36 |
| III | 7 |
| IV | 4 |
| Survival status | |
| 1 | 84 |
| 2 | 6 |
| 3 | 5 |
| 4 | 16 |
Cell culture
Breast cancer cell lines BT549, MCF-7, MCF-10A and MDA-MB-231 were purchased from the European Collection of Animal Cell Cultures (ECACC; Salisbury, UK). Invasions of MCF-7 and BT-549 are low, while MDA-MB-231 represents high invasiveness, and MCF-10A is an immortalized mammary epithelial cell. Of these cell lines used, MCF-10A and MCF-7 are ER positive, while MDA-MB-231 and BT-549 are ER negative. MCF-10A cells were cultured in mammary epithelial cell medium (Lonza Biologics Inc., Portsmouth, NH, USA), the remaining cells were routinely cultured in DMEM/F12 HAM with L-Glutamine (Dulbecco’s modified Eagle’s medium; Sigma-Aldrich, Dorset, UK) supplemented with 10% fetal calf serum (FCS; PAA Laboratories, Somerset, UK), streptomycin (Sigma-Aldrich) and penicillin, in an incubator at 37 °C, 5% CO2 and 95% humidity.
Processing of tissues and extraction of RNA and generation of cDNA
Tissue specimens were homogenized in RNA extraction solution with hand held homogenizer to extract total RNA. RNA concentration was quantified using UV spectrophotometer. Total cDNA was generated from 1 µg RNA using commercially available RT kit (AbGene Laboratories, Essex, England).
Immunohistochemical staining of Aurora kinase A, B and C in breast specimens
Frozen sections of breast tumor and background tissues were cut at a thickness of 6 µm using a cryostat (Leica Microsystems (UK) Ltd, Bucks, UK). Briefly, immunohistochemistry staining followed the protocol as described previously [28].
The dilution chosen of primary antibodies and secondary antibody was based on evaluation test run, during which the antibodies were tested over a range from 1:10 to 1:1,000. Primary antibodies were omitted in negative controls. Following cultured with antibodies, Avidin Biotin Complex (Vector Laboratories) was applied to the sections followed by extensive washing. Diaminobenzidine chromogen (Vector Labs) was then added to the sections, which were kept in the dark for 5 min. Independent observers carried out the microvessels counts as we have reported in a study before [29].
Real-time reverse transcription quantitative PCR
Real-time quantitative PCR, based on the Amplifluor™ technology, was to quantify the mRNA expression level of Aurora kinases from above cDNA samples of breast tissues following the method previous reported [30]. The pairs of Q-PCR primers were designed with Beacon Designer software which includes complementary sequence to universal Z probe (Intergen Inc., Oxford, UK). The pair Q-PCR primers sequences are shown in Table 2. Real-time PCR was carried out using an IcyclerIQ™ (Bio-Rad, Hemel Hempstead, UK) kit with the following cycling conditions: 94 °C for 12 min, 60 cycles of 94 °C for 15 s, 55 °C for 40 s (the data capture step) and 72 °C for 20 s. The transcripts levels were generated from an internal standard which was simultaneously amplified with the samples.
Table 2. Primers Sequences (Q-PCR).
| Molecular | Sense primers (5′-3′) | Antisense primers (5′-3′) |
|---|---|---|
| Aurora kinase A | GGTCAGTACATGCTCCATCT | ACTGAACCTGACCGTACACAGAGATCCACCTTCTCATC |
| Aurora kinase B | GCAGAGAGATCGAAATCCA | ACTGAACCTGACCGTACAGGGGCATACTCTAGGAATCAA |
| Aurora kinase C | TGGAGTGCTCTGCTATGAG | ACTGAACCTGACCGTACCGTACAGCATTGATAGTGGAAACCTC |
| GAPDH (Q-PCR) | CTGAGTACGTCGTGGAGTC | ACTGAACCTGACCGTACAGAGATGATGACCCTTTTG |
Electric cell-substrate impedance sensing (ECIS) based analyses on cell adhesion and cell migration
The ECIS Ztheta system (Applied Biophysics Ltd, Troy, NJ, USA) evaluated cells adhesion and migration as described by Jiang et al before [31]. ECIS needed special 96-well W96E1 microarrays. Following 100 µL DMEM medium with different concentration inhibitor (ZM447439: IC50 = 50 nM) added in a 96-well W96E1 microarray, BT-549, MCF-7, MCF-10A and MDA-MB-231 cells (40,000 per well) were separately seeded into wells of array. Live tracking of cell adhesion was carried out over a range of frequencies from 1,000 to 64,000 Hz using automated modules for 24 h. Confluent breast cancer cell monolayers seeded in the arrays were electrically wounded (2,000 mA for 20 s each). Following the track, migration of the cells was tracked over a range of frequencies for 5 h. All the experiments were conducted at least three times.
In vitro cell growth assay
Cells suspension treated with different concentration inhibitor (ZM447439) was seeded into a 96-well plate (3,000 cells/200 µL/well). Cell growth was assessed after a period of incubation in quadruplicate (overnight, day 3 and day 5). Four percent formalin was used to fix cells and 0.5% crystal violet was used for stained cells. Crystal violet was extracted with 100 µL 10% acetic acid and the absorbance of the dissolved dye determined using an ELx800 spectrophotometer (BIO-TEK, ELx800) at a wavelength of 540 nm.
Statistical analysis
Statistical analysis was carried out using the Minitab statistical software package (version 14) and Mann-Whitney test, whilst normally distributed data used t-test. Significant differences were taken at P<0.05.
Results
Expression of Aurora kinases in human breast cancer tissues and the histopathological/clinical characteristics of the disease
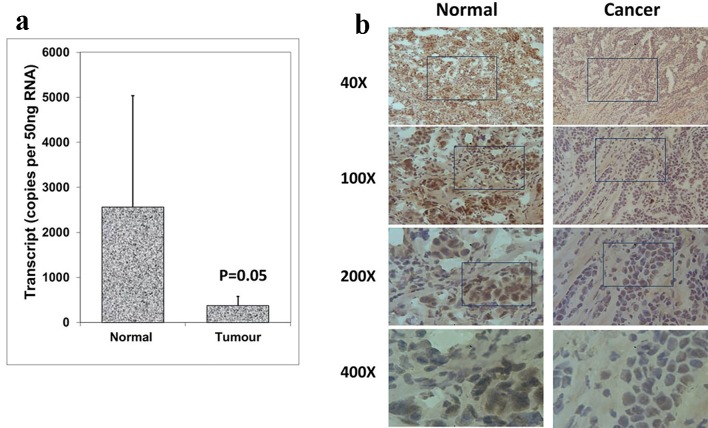
The expression patterns of Aurora kinase A, B and C were analyzed in breast cancer tissues and adjacent normal tissues (tumor, n = 118; background, n = 34) using real-time quantitative PCR (all values are displayed as mean Aurora kinase transcript copies/µL of cDNA from 50 ng total RNA). In comparison with normal background tissues, a significantly decreased expression of Aurora kinase A was found in breast tumor tissues (P = 0.05) (Fig. 1a). Aurora kinase A protein was observed in the cytoplasmic region of normal mammary epithelial cells, while stromal cells had very little staining. Immunochemical staining was conducted on a portion of paired normal background tissues and tumor tissues (n = 34 pairs). Intense expression of Aurora kinase A was primarily observed in normal background tissues compared with breast tumor tissues (Fig. 1b). These results together suggested that Aurora kinase A is lowly expressed in breast cancer and may act a role in the progression of breast cancer.
Figure 1.
Expression of Aurora kinase A in breast cancer tissues. (a) Aurora kinase A transcripts level was decreased in human breast cancer. (b) Immunohistochemical staining of Aurora kinase A in normal breast tissue (left panel) and tumor tissue (right panel).
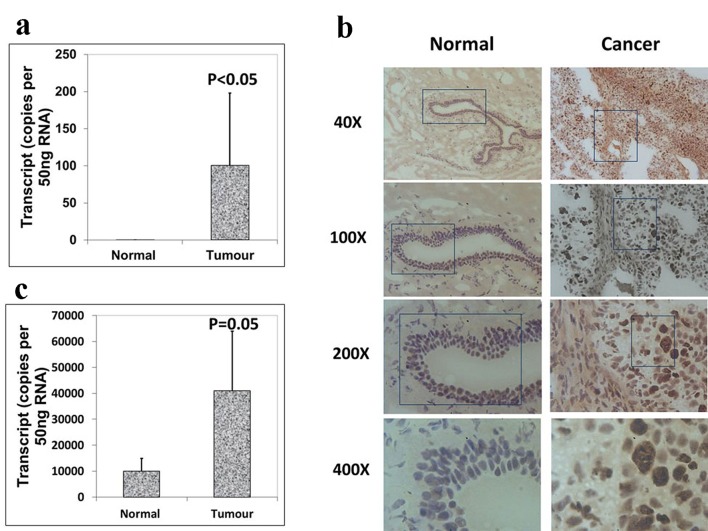
We also analyzed the expression levels of Aurora B and C in paired normal tissues and breast tumor tissues within the same specimen. However, increased levels of Aurora B demonstrated a higher level in breast tissues than in normal background tissues (Fig. 2a), which were consistent with the immunohistochemistry results of Aurora B RNA transcript expression (Fig. 2b), and the difference was statistically significant (P < 0.05). Also there was a significantly increased expression of Aurora kinase C in breast tumor tissues compared with adjacent normal tissues (P = 0.05) (Fig. 2c). These results together suggested that Aurora kinase B and C are highly expressed in breast cancer. Taken together, the expression levels of Aurora kinase A, B and C are different in the same tissues though they belong to the same family. Thus, they may play different roles in the progression of breast cancer.
Figure 2.
Expression of Aurora kinase B and C in breast cancer tissues. (a) The expression of Aurora kinase B is higher in breast cancer tissues. (b) Immunohistochemical staining of Aurora kinase B in normal background tissue (left panel) and breast tumor tissue (right panel). (c) Aurora kinase C transcripts level was increased in human breast cancer.
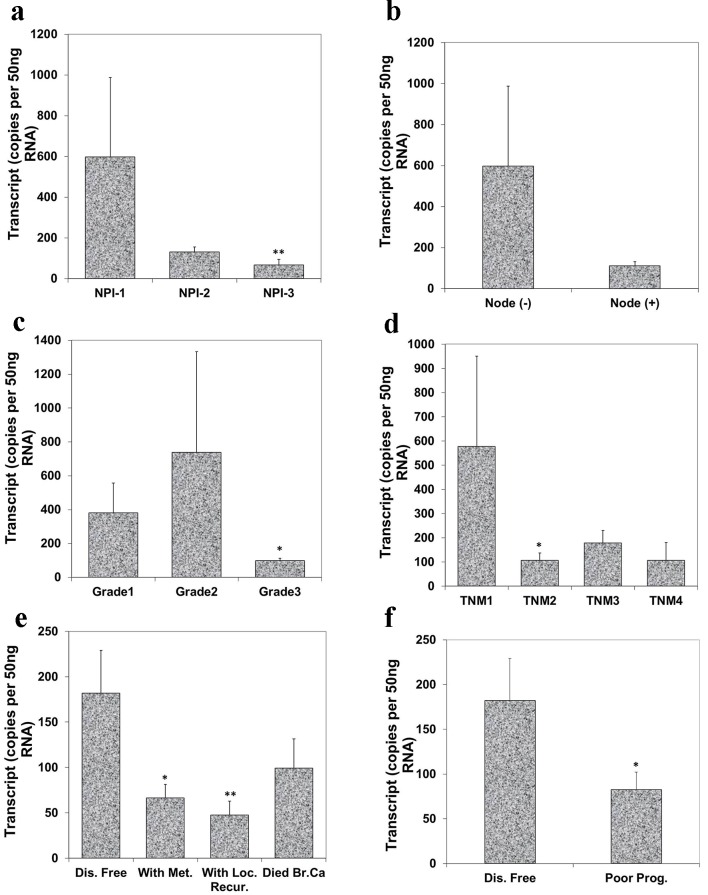
Correlation of Aurora kinase A expression with Nottingham prognostic index (NPI), grade, TNM staging and prognosis
To evaluate the relationship of Aurora kinase A expression with disease progression, we analyzed the level of Aurora kinase A transcripts in connection with NPI, grade and TNM staging of breast tumors. NPI was used as an indicator to assess the relationship between Aurora kinase A transcript and a predicted prognosis. Based on the NPI value, breast cancer patients were divided into three groups: good (NPI-1), moderate (NPI-2) and poor prognosis (NPI-3). It was suggested that the lowest level of Aurora kinase A was observed in patients with poor prognostic index (67.0 ± 28 copies/µL) among these three groups. The difference between patients with good prognosis and poor prognosis was highly significant (P = 0.0092) (Fig. 3a). Although node negative tumors had higher levels of Aurora kinase A transcript (597.0 ± 391 copies/µL) when compared with node positive tumors (111.0 ± 20 copies/µL), the difference was nonetheless insignificant (Fig. 3b).
Figure 3.
Correlation of Aurora kinase A expression with Nottingham prognostic index (NPI), grade, TNM staging and prognosis. (a) Lowest level of Aurora kinase A was observed in patients with poor prognostic index among these three groups (**P < 0.01 NPI-3 vs. NPI-1). (b) Node negative tumors had higher levels of Aurora kinase A transcript than node positive tumor. (c) Levels of expression of Aurora kinase A in breast tumor tissues in connection with grade and (d) with TNM status (*P < 0.05 grade 3 vs. grade 1; *P < 0.05 TNM 2 vs. TNM 1). (e) Levels of Aurora kinase A transcript and clinical outcomes. Aurora kinase A expression progressively decreased in the patients with metastasis disease, with local recurrence and die of breast cancer, compared with patient with disease free (*P < 0.05 metastasis vs. disease free; **P < 0.01 local recurrence vs. disease free). (f) Lower level of Aurora kinase A expression in patients with poor prognosis.
We further evaluated the levels of Aurora kinase A transcript in connection with breast tumor grade. The expression level of Aurora kinase A was lowest in grade 3 (97.0 ± 15.7 copies/µL) and grade 3 tumor had significantly lower levels compared with grade 1 (380.0 ± 177 copies/µL) breast tumors (P = 0.0401) (Fig. 3c). The relationship between Aurora kinase A and TNM status was also analyzed. Although the expression level of Aurora kinase A in TNM2, TNM3 and TNM4 was all lower than TNM1, statistical significant difference only disappeared in TNM2 tumor when compared with TNM1 tumor (P = 0.0292) (Fig. 3d).
Regarding the clinical outcomes at the final follow-up, the patients were divided into remaining disease free, with metastasis disease, with local recurrence and died of breast tumor after a median 120 months follow-up. Patients who developed tumor metastasis and local recurrence, and who died of breast cancer all had lower levels of Aurora kinase A. Patients with metastatic disease and with local recurrence showed significantly lower level in comparison with those who remained disease free (P = 0.021, P = 0.008) (Fig. 3e). Decreased transcript levels of Aurora kinase A was found in patients with poor prognosis and reached statistical significance (P = 0.05) (Fig. 3f).
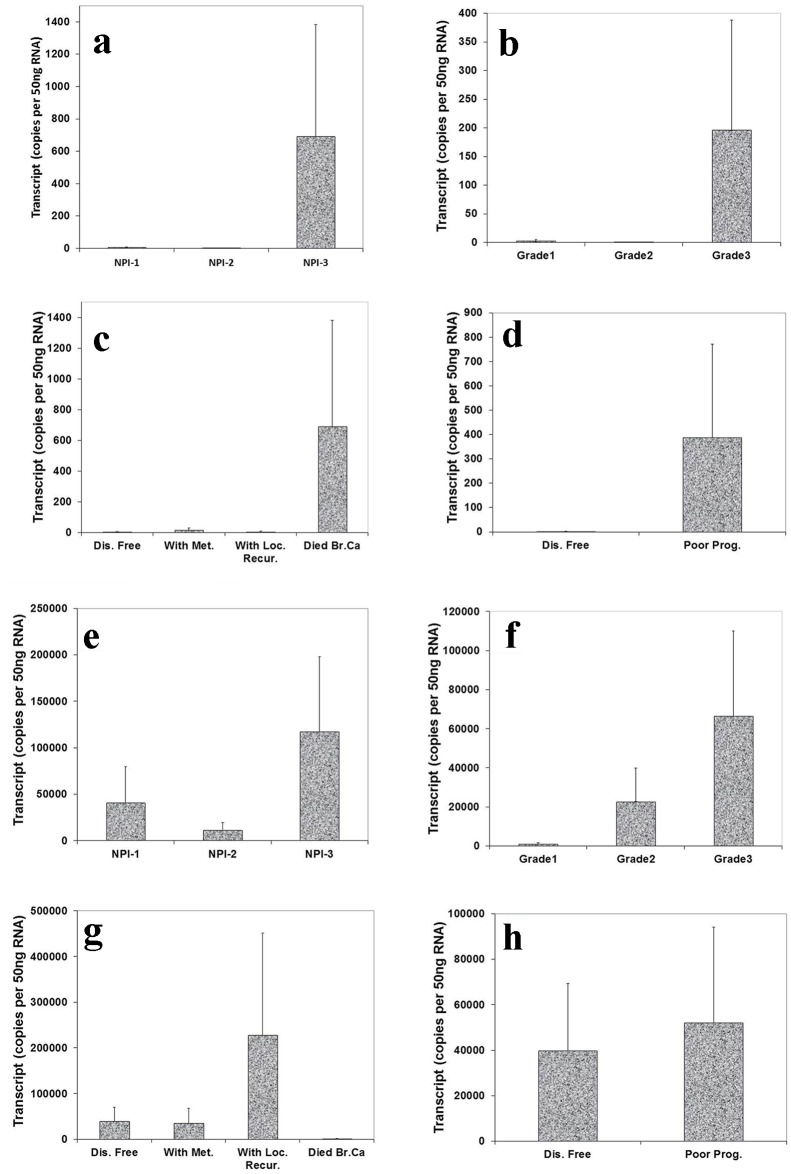
Correlation of Aurora kinase B and C expression with NPI, grade staging and prognosis
We also analyzed the level of Aurora kinase B and C transcripts in connection with NPI, grade and TNM staging of breast tumors. This showed that the highest level of Aurora kinase B transcript was seen in NPI-3 compared with NPI-1 and NPI-2 (Fig. 4a). We also found that the expression level of Aurora kinase B was higher in grade 3 than in grade 1 and grade 2 (Fig. 4b). Patients who died of breast cancer had the highest levels of Aurora kinase B (Fig. 4c). Increased transcript levels of Aurora kinase B were found in patients with poor prognosis (Fig. 4d). Although these tests did not reach statistical significance, the increased tendency of Aurora kinase B expression level was clear in breast tumor tissues.
Figure 4.
Correlation of Aurora kinase B and C expression with Nottingham prognostic index (NPI), grade staging and prognosis. Top panel (Aurora kinase B): (a) Highest level of Aurora kinase B in patients with poor prognostic index among these three groups. (b): Grade 3 tumor had higher level of Aurora kinase B transcript than grade 1 and 2. (c) Levels of Aurora kinase B transcript and clinical outcomes. Aurora kinase B expression progressively increased in the patients with local recurrence and die of breast cancer, compared with patient with disease free. (d) Higher level of Aurora kinase B expression in patients with poor prognosis. Bottom panel (Aurora kinase C): (e) Higher level of Aurora kinase C in patients with poor prognostic index than with moderate and poor prognosis. (f) A stepwise increase of levels of Aurora kinase C was found from grade 1 to grade 3 staging. (g) Lever of Aurora kinase C expression was higher in the patients with local recurrence than with metastasis disease and disease free. (h) Lower level of Aurora kinase C expression in patients with good prognosis.
The expression level of Aurora kinase C was highest in poor prognostic index compared with good and moderate prognosis (Fig. 4e). There appeared to be a stepwise increase of levels of Aurora kinase C from grade 1 to grade 3 staging (Fig. 4f). Patients who developed local recurrence had higher Aurora kinase C expression level than those remaining disease free and with metastasis disease (Fig. 4g). Higher levels of Aurora kinase C were seen in patients with poor prognosis compared with remaining disease free (Fig. 4h). The increased tendency of Aurora kinase C expression level was clear in breast tumor tissues, though differences did not reach significant.
ECIS based cell adhesion and cell migration assay
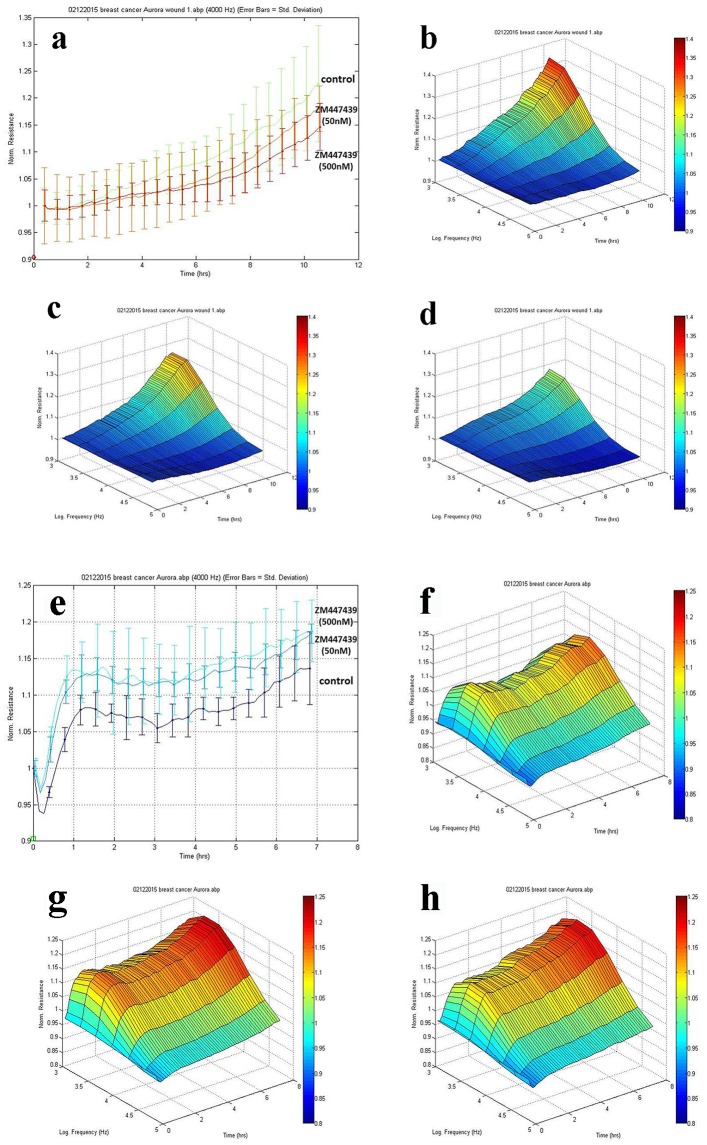
To investigate the effects of Aurora kinases inhibitor ZM447439 on breast cell migration and adhesion, we adopted the ECIS method in tracking cell motility. As is shown in Figure 5a, MCF-7 cells treated with inhibitor ZM447439 showed a slowdown in recovery after electric wounding. 3D imaging confirmed the migration of MCF-7 cells. The migration ability of MCF-7 treated with ZM447439 (Fig. 5c, d) decreased compared with control group (no treatment) (Fig. 5b). We also analyzed other breast cancer cells such as MDA-MB-231, BT549 and MCF-10A, but the difference was not clear (data not shown).
Figure 5.
Effects of Aurora kinases inhibitor on the adhesion of breast cancer cells. Top panel: The response of MCF-7 to ZM447439. (a) Migration traces of MCF-7 cell’s response to ZM447439 at different concentrations. (b) 3D image of MCF-7 (control); (c and d) Response of MCF-7 to ZM447439 at 50 and 500 nM, respectively. Bottom panel: The response of MDA-MB-231 to ZM447439. (e) Traces of MDA-MB-231 cell’s response to ZM447439 at different concentrations. (f) 3D image of MDA-MB-231 (control); (g and h) Response of MDA-MB-231 to ZM447439 at 50 and 500 nM, respectively.
There appeared some difference in adherence of MDA-MB-231 cells when treated with Zm447439. The adherence of MDA-MB-231 cells treated with inhibitor ZM447439 increased compared with the control group (no treatment) (Fig. 5e). In 3D imaging, it was confirmed (Fig. 5f, g, h). In comparison with control group (no treatment), higher level of adhesion was found in MDA-MB-231 cell group treated with inhibitor ZM447439. We also analyzed the effect of ZM447439 on proliferation and invasion of breast cancer cells, but this did not reach statistical significance (data not shown).
Discussion
In this study, we have demonstrated that Aurora kinase A had lower expression levels in human breast tumor tissues compared with normal background tissues. Aurora kinase B and C were also aberrantly expressed in clinical breast tumor tissues; however, both of them showed higher expression in breast tumor tissues than in adjacent normal tissues. The abnormal expression of Aurora kinases in breast tumor tissues may act a role in the progression of breast cancer.
The relationship between the level of Aurora kinase expression and clinical outcome is an important observation. An inverse correlation was found between the level of the Aurora kinase A transcript and tumor grade and TNM staging. However, the relationship between Aurora kinase B and C expression level had a positive correlation. The levels of Aurora kinase B and C were correlated with the clinical outcomes and long term survival of breast cancer patients, but Aurora kinase A was opposite. According to analysis of the clinical data, Aurora kinases may act as a potential prognostic indicator in patients with breast tumor.
Recently, some reports have shown that the role of Aurora kinases is to act in some cancer and signaling pathways. A new study showed KRAS could induce knockdown of Aurora kinase A and B in the KRAS-positive lung cancer cell lines H358 and A549. Furthermore, the growth, proliferation, transformation and viability levels of these two cell lines decreased when treated with Aurora kinase A and B inhibitors in vitro. This suggested that Aurora kinases were important KRAS targets in lung cancer and Aurora kinases inhibition therapy may specially target KRAS-transformed cells [32]. However, another study found overexpression in 73% of the examined specimens. Furthermore, the expression of Aurora kinase A decreased taxane sensitivity [33]. Aurora kinase A has also been shown to play a novel non-mitotic role in promoting tumor progression via activation of epithelial-mesenchymal transition reprograming. This resulted in the genesis of tumor-initiating cells [34]. In our research, however, the expression level of Aurora kinase A decreased in breast cancer tissues samples. Thus, the role of Aurora kinase A playing in breast cancer is unclear. In our current study, we detected the effect of Aurora kinase inhibitor ZN447439 on the motility of breast cancer cells. We found that the adherence of MDA-MB-231 cells decreased when treated with ZM447439. ZM447439 promoted the migration of MCF-7 cells. We also analyzed the effect of ZM447439 on proliferation and invasion of breast cancer cells, but the difference was not significant. Interestingly, the results in vitro are associated with clinical outcome of human breast cancer.
In conclusion, Aurora kinases act as regulators of cellular motility of breast cancer cells. With cells treated with inhibitor ZM447439, we predicted that Aurora kinases inhibitor could inhibit tumor metastasis by reducing migration of breast cancer cells. Together with the clinical relevance as demonstrated above, Aurora kinases play an important role as prognostic indicators and may be an important target when considering therapies.
Acknowledgments
The Authors wish to thank Cancer Research Wales, Life Sciences Research Network Wales and the Albert Hung Foundation for supporting this study. Huishan Zhao is a recipient of China Medical Scholarship awarded by Cardiff University.
Grant Support
Cancer Research Wales and Life Sciences Research Network Wales. The Albert Hung Foundation and the China Medical Scholarship.
Financial Disclosure
The authors have no financial disclosures to make.
Competing Interest
None of the authors have any competing interests.
Author Contributions
HZ carried out most of the experiments and drafted the paper. SO participated in the experimental data. TAM participated in the study design and completed the manuscript. ED collated the tissue samples. WGJ conceived the study and carried out the statistical analysis. All authors read and approved the final manuscript.
Abbreviations
- Q-PCR
quantitative polymerase chain reaction
- ECACC
European Collection of Animal Cell Culture
- DMEM
Dulbecco’s modified Eagle’s medium
- FCS
fetal calf serum
- NPI
Nottingham prognostic indicator
- TNM
tumor-nodal involvement
- ECIS
electric cell impedance sensing
References
- 1.Porter PL. Global trends in breast cancer incidence and mortality. Salud Publica Mex. 2009;51(Suppl 2):s141–146. doi: 10.1590/S0036-36342009000800003. [DOI] [PubMed] [Google Scholar]
- 2.Weil RJ, Palmieri DC, Bronder JL, Stark AM, Steeg PS. Breast cancer metastasis to the central nervous system. Am J Pathol. 2005;167(4):913–920. doi: 10.1016/S0002-9440(10)61180-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans AJ, James JJ, Cornford EJ, Chan SY, Burrell HC, Pinder SE, Gutteridge E. et al. Brain metastases from breast cancer: identification of a high-risk group. Clin Oncol (R Coll Radiol) 2004;16(5):345–349. doi: 10.1016/j.clon.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 4.Lorincz T, Toth J, Badalian G, Timar J, Szendroi M. HER-2/neu genotype of breast cancer may change in bone metastasis. Pathol Oncol Res. 2006;12(3):149–152. doi: 10.1007/BF02893361. [DOI] [PubMed] [Google Scholar]
- 5.Crane R, Gadea B, Littlepage L, Wu H, Ruderman JV. Aurora A, meiosis and mitosis. Biol Cell. 2004;96(3):215–229. doi: 10.1016/j.biolcel.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Kollareddy M, Dzubak P, Zheleva D, Hajduch M. Aurora kinases: structure, functions and their association with cancer. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2008;152(1):27–33. doi: 10.5507/bp.2008.004. [DOI] [PubMed] [Google Scholar]
- 7.Fu J, Bian M, Liu J, Jiang Q, Zhang C. A single amino acid change converts Aurora-A into Aurora-B-like kinase in terms of partner specificity and cellular function. Proc Natl Acad Sci U S A. 2009;106(17):6939–6944. doi: 10.1073/pnas.0900833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hans F, Skoufias DA, Dimitrov S, Margolis RL. Molecular distinctions between Aurora A and B: a single residue change transforms Aurora A into correctly localized and functional Aurora B. Mol Biol Cell. 2009;20(15):3491–3502. doi: 10.1091/mbc.E09-05-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung CH, Sarvagalla S, Lee JY, Huang YC, Coumar MS. Aurora kinase inhibitor patents and agents in clinical testing: an update (2011 - 2013) Expert Opin Ther Pat. 2014;24(9):1021–1038. doi: 10.1517/13543776.2014.931374. [DOI] [PubMed] [Google Scholar]
- 10.Sen S, Zhou H, White RA. A putative serine/threonine kinase encoding gene BTAK on chromosome 20q13 is amplified and overexpressed in human breast cancer cell lines. Oncogene. 1997;14(18):2195–2200. doi: 10.1038/sj.onc.1201065. [DOI] [PubMed] [Google Scholar]
- 11.Zhou H, Kuang J, Zhong L, Kuo WL, Gray JW, Sahin A, Brinkley BR. et al. Tumour amplified kinase STK15/BTAK induces centrosome amplification, aneuploidy and transformation. Nat Genet. 1998;20(2):189–193. doi: 10.1038/2496. [DOI] [PubMed] [Google Scholar]
- 12.Gu J, Gong Y, Huang M, Lu C, Spitz MR, Wu X. Polymorphisms of STK15 (Aurora-A) gene and lung cancer risk in Caucasians. Carcinogenesis. 2007;28(2):350–355. doi: 10.1093/carcin/bgl149. [DOI] [PubMed] [Google Scholar]
- 13.Sorrentino R, Libertini S, Pallante PL, Troncone G, Palombini L, Bavetsias V, Spalletti-Cernia D. et al. Aurora B overexpression associates with the thyroid carcinoma undifferentiated phenotype and is required for thyroid carcinoma cell proliferation. J Clin Endocrinol Metab. 2005;90(2):928–935. doi: 10.1210/jc.2004-1518. [DOI] [PubMed] [Google Scholar]
- 14.Katayama H, Ota T, Jisaki F, Ueda Y, Tanaka T, Odashima S, Suzuki F. et al. Mitotic kinase expression and colorectal cancer progression. J Natl Cancer Inst. 1999;91(13):1160–1162. doi: 10.1093/jnci/91.13.1160. [DOI] [PubMed] [Google Scholar]
- 15.Qi G, Ogawa I, Kudo Y, Miyauchi M, Siriwardena BS, Shimamoto F, Tatsuka M. et al. Aurora-B expression and its correlation with cell proliferation and metastasis in oral cancer. Virchows Arch. 2007;450(3):297–302. doi: 10.1007/s00428-006-0360-9. [DOI] [PubMed] [Google Scholar]
- 16.Hu HM, Chuang CK, Lee MJ, Tseng TC, Tang TK. Genomic organization, expression, and chromosome localization of a third aurora-related kinase gene, Aie1. DNA Cell Biol. 2000;19(11):679–688. doi: 10.1089/10445490050199063. [DOI] [PubMed] [Google Scholar]
- 17.Falchook GS, Bastida CC, Kurzrock R. Aurora Kinase Inhibitors in Oncology Clinical Trials: Current State of the Progress. Semin Oncol. 2015;42(6):832–848. doi: 10.1053/j.seminoncol.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 18.Choudary I, Barr PM, Friedberg J. Recent advances in the development of Aurora kinases inhibitors in hematological malignancies. Ther Adv Hematol. 2015;6(6):282–294. doi: 10.1177/2040620715607415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cai J, Li L, Hong KH, Wu X, Chen J, Wang P, Cao M. et al. Discovery of 4-aminoquinazoline—urea derivatives as Aurora kinase inhibitors with antiproliferative activity. Bioorg Med Chem. 2014;22(21):5813–5823. doi: 10.1016/j.bmc.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 20.Gavriilidis P, Giakoustidis A, Giakoustidis D. Aurora kinases and potential medical applications of aurora kinase inhibitors: a review. J Clin Med Res. 2015;7(10):742–751. doi: 10.14740/jocmr2295w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavanti E, Sero V, Vella S, Fanelli M, Michelacci F, Landuzzi L, Magagnoli G. et al. Preclinical validation of Aurora kinases-targeting drugs in osteosarcoma. Br J Cancer. 2013;109(10):2607–2618. doi: 10.1038/bjc.2013.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dawson MA, Curry JE, Barber K, Beer PA, Graham B, Lyons JF, Richardson CJ. et al. AT9283, a potent inhibitor of the Aurora kinases and Jak2, has therapeutic potential in myeloproliferative disorders. Br J Haematol. 2010;150(1):46–57. doi: 10.1111/j.1365-2141.2010.08175.x. [DOI] [PubMed] [Google Scholar]
- 23.Santo L, Hideshima T, Cirstea D, Bandi M, Nelson EA, Gorgun G, Rodig S. et al. Antimyeloma activity of a multitargeted kinase inhibitor, AT9283, via potent Aurora kinase and STAT3 inhibition either alone or in combination with lenalidomide. Clin Cancer Res. 2011;17(10):3259–3271. doi: 10.1158/1078-0432.CCR-10-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao H, Li M, Qian WB. [Killing effect of aurora kinase inhibitor ENMD-2076 on acute myelogenous leukemia cells] Zhejiang Da Xue Xue Bao Yi Xue Ban. 2012;41(5):479–484. [PubMed] [Google Scholar]
- 25.Bebbington D, Binch H, Charrier JD, Everitt S, Fraysse D, Golec J, Kay D. et al. The discovery of the potent aurora inhibitor MK-0457 (VX-680) Bioorg Med Chem Lett. 2009;19(13):3586–3592. doi: 10.1016/j.bmcl.2009.04.136. [DOI] [PubMed] [Google Scholar]
- 26.Harrington EA, Bebbington D, Moore J, Rasmussen RK, Ajose-Adeogun AO, Nakayama T, Graham JA. et al. VX-680, a potent and selective small-molecule inhibitor of the Aurora kinases, suppresses tumor growth in vivo. Nat Med. 2004;10(3):262–267. doi: 10.1038/nm1003. [DOI] [PubMed] [Google Scholar]
- 27.Karthigeyan D, Prasad SB, Shandilya J, Agrawal S, Kundu TK. Biology of Aurora A kinase: implications in cancer manifestation and therapy. Med Res Rev. 2011;31(5):757–793. doi: 10.1002/med.20203. [DOI] [PubMed] [Google Scholar]
- 28.Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, Mansel RE. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res. 2003;9(17):6432–6440. [PubMed] [Google Scholar]
- 29.Martin TA, Parr C, Davies G, Watkins G, Lane J, Matsumoto K, Nakamura T. et al. Growth and angiogenesis of human breast cancer in a nude mouse tumour model is reduced by NK4, a HGF/SF antagonist. Carcinogenesis. 2003;24(8):1317–1323. doi: 10.1093/carcin/bgg072. [DOI] [PubMed] [Google Scholar]
- 30.Jiang WG, Watkins G, Fodstad O, Douglas-Jones A, Mokbel K, Mansel RE. Differential expression of the CCN family members Cyr61, CTGF and Nov in human breast cancer. Endocr Relat Cancer. 2004;11(4):781–791. doi: 10.1677/erc.1.00825. [DOI] [PubMed] [Google Scholar]
- 31.Jiang WG, Martin TA, Lewis-Russell JM, Douglas-Jones A, Ye L, Mansel RE. Eplin-alpha expression in human breast cancer, the impact on cellular migration and clinical outcome. Mol Cancer. 2008;7:71. doi: 10.1186/1476-4598-7-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dos Santos EO, Carneiro-Lobo TC, Aoki MN, Levantini E, Basseres DS. Aurora kinase targeting in lung cancer reduces KRAS-induced transformation. Mol Cancer. 2016;15:12. doi: 10.1186/s12943-016-0494-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cirak Y, Furuncuoglu Y, Yapicier O, Aksu A, Cubukcu E. Aurora A overexpression in breast cancer patients induces taxane resistance and results in worse prognosis. J BUON. 2015;20(6):1414–1419. [PubMed] [Google Scholar]
- 34.D’Assoro AB, Haddad T, Galanis E. Aurora-a kinase as a promising therapeutic target in cancer. Front Oncol. 2015;5:295. doi: 10.3389/fonc.2015.00295. [DOI] [PMC free article] [PubMed] [Google Scholar]