Staphylococcus aureus is a pathogen that can cause a wide range of infections in humans. Studies have suggested that CRISPR-Cas systems can drive the loss of integrated mobile genetic elements (MGEs) by chromosomal targeting. Here we demonstrate that CRISPR-mediated cleavage contributes to the partial deletion of integrated SCCmec in methicillin-resistant S. aureus (MRSA), which provides a strategy for the treatment of MRSA infections. The spacer within artificial CRISPR arrays should contain more than 25 nucleotides for immunity, and consecutive trinucleotide pairings between a selected target and the 5′ tag of crRNA can block targeting. These findings add to our understanding of the molecular mechanisms of the type III-A CRISPR-Cas system and provide a novel strategy for the exploitation of engineered CRISPR immunity against integrated MGEs in bacteria for clinical and industrial applications.
KEYWORDS: CRISPR-Cas system, Staphylococcus aureus, chromosomal targeting, mobile genetic element, staphylococcal cassette chromosome mec
ABSTRACT
CRISPR-Cas (clustered regularly interspaced short palindromic repeat [CRISPR]-CRISPR-associated protein [Cas]) systems can provide protection against invading genetic elements by using CRISPR RNAs (crRNAs) as a guide to locate and degrade the target DNA. CRISPR-Cas systems have been classified into two classes and five types according to the content of cas genes. Previous studies have indicated that CRISPR-Cas systems can avoid viral infection and block plasmid transfer. Here we show that chromosomal targeting by the Staphylococcus aureus type III-A CRISPR-Cas system can drive large-scale genome deletion and alteration within integrated staphylococcal cassette chromosome mec (SCCmec). The targeting activity of the CRISPR-Cas system is associated with the complementarity between crRNAs and protospacers, and 10- to 13-nucleotide truncations of spacers partially block CRISPR attack and more than 13-nucleotide truncation can fully abolish targeting, suggesting that a minimal length is required to license cleavage. Avoiding base pairings in the upstream region of protospacers is also necessary for CRISPR targeting. Successive trinucleotide complementarity between the 5′ tag of crRNAs and protospacers can disrupt targeting. Our findings reveal that type III-A CRISPR-Cas systems can modulate bacterial genome stability and may serve as a high-efficiency tool for deleting resistance or virulence genes in bacteria.
IMPORTANCE Staphylococcus aureus is a pathogen that can cause a wide range of infections in humans. Studies have suggested that CRISPR-Cas systems can drive the loss of integrated mobile genetic elements (MGEs) by chromosomal targeting. Here we demonstrate that CRISPR-mediated cleavage contributes to the partial deletion of integrated SCCmec in methicillin-resistant S. aureus (MRSA), which provides a strategy for the treatment of MRSA infections. The spacer within artificial CRISPR arrays should contain more than 25 nucleotides for immunity, and consecutive trinucleotide pairings between a selected target and the 5′ tag of crRNA can block targeting. These findings add to our understanding of the molecular mechanisms of the type III-A CRISPR-Cas system and provide a novel strategy for the exploitation of engineered CRISPR immunity against integrated MGEs in bacteria for clinical and industrial applications.
INTRODUCTION
Staphylococcus aureus is a bacterial pathogen that can cause infectious diseases in humans, ranging from skin or soft tissue infections to life-threatening illnesses (1). Recent studies have revealed that the emergence and resurgence of methicillin-resistant S. aureus (MRSA) are serious public health threats, especially the community-associated MRSA infections (2). Mechanisms of resistance to β-lactam antibiotics among MRSA strains are due to the acquisition of the mecA resistance gene, which is carried on staphylococcal cassette chromosome mec (SCCmec) and encodes an additional penicillin-binding protein PBP2a with low affinity for β-lactam antibiotics (3–5). The mobile genetic element SCCmec can conduct horizontal transfer among staphylococcal strains and accordingly lead to the prevalence of methicillin resistance (6).
Clustered regularly interspaced short palindromic repeats (CRISPRs) and CRISPR-associated proteins (Cas) constitute an adaptive immunity system that protects archaea and bacteria from threats of foreign mobile elements. According to the constitution and function of Cas proteins, CRISPR-Cas systems are currently classified into five distinctive types and diverse subtypes (7). Studies have mainly focused on types I, II, and III systems in the last decade. CRISPR loci, composed of conserved repeats and diverse spacers, are under the control of an AT-rich leader sequence. Repeats and spacers are first transcribed into precursor CRISPR RNAs (pre-crRNAs) and then are processed into small and mature crRNAs, which can guide the Cas complex for sequence-specific targeting (8–10). A recent study revealed that pre-crRNA processing is independent on its sequence, length, or secondary structure in Staphylococcus epidermidis type III-A CRISPR-Cas system (11). The protospacer adjacent motif (PAM) and seed sequence play a key role in recognition and targeting (12, 13), as well as new spacer acquisition (14) in type I and type II CRISPR-Cas systems. Type III systems do not require a PAM, and self/nonself discrimination relies on eight nucleotides of repeat sequence present at the 5′ handle of crRNA (crRNA 5′ tag). One early study has concluded that the 5′-tag noncomplementarity of protospacers and crRNAs at specific positions is responsible for interference, whereas extended pairing between the 5′ tag of crRNA and the target prevents autoimmunity in S. epidermidis (15). Similar results were observed but at different pairing positions in Sulfolobus solfataricus (16). However, until now, the role of a potential seed sequence for type III immunity has remained unknown. Intriguingly, a previous study implies that exact complementarity between crRNAs and protospacers in the 5′ end is necessary for antiplasmid immunity in S. aureus type III-A system (17).
Most of the spacers from multiple organisms are characterized to be homologous to the sequences derived from bacteriophages or conjugative plasmids, but a number of spacers are also found to match with archaeal or bacterial genomes. It has been reported that 59 of 330 CRISPR-positive organisms possess at least one spacer targeting endogenous genomic sequence (18), indicating that incorporation of a self-targeting spacer is not an accident. Another study suggests that among 4,500 spacers from various organisms, 35% have homologs to chromosomal sequences in the NCBI database (19). Some of these spacers target genes within integrated mobile genetic elements (MGEs), while others target nonmobile genes. For example, Pectobacterium atrosepticum contains a self-targeting spacer completely complementary to an endogenous gene within a horizontally acquired island named HAI2 (20). It has also been found that a spacer matches the sequence within hisS, which codes for the histidyl-tRNA synthetase in Pelobacter carbinolicus (21). These findings raise the question of what role self-targeting spacers may play. One controversial idea is that self-targeting spacers may participate in gene regulation and bacterial genome evolution (22, 23). A few authors proposed that chromosomal targeting has a deleterious effect, but bacteria can survive at the cost of the disruption of CRISPR arrays or Cas proteins (20, 21, 24). They were disposed to agree with the view that chromosomal targeting is a case of autoimmunity rather than a regulatory mechanism (18). Although incorporation of a self-targeting spacer is less common than spacers against MGEs, this phenomenon provides an insight into the biological application of CRISPR-Cas systems.
The interaction between the CRISPR-Cas system and prophage has been a subject of intense research in the last 10 years. Marraffini et al. pointed out numerous novel views about antibacteriophage immunity in S. epidermidis type III-A CRISPR-Cas system (25–27). Unfortunately, an active prophage in the CRISPR-positive S. aureus has not been found yet. A recent study concluded that CRISPR-negative strains contained significantly more prophages and larger genomes than the CRISPR-positive strains did (28). A possible reason is that the uptake of MGEs is prevented by CRISPR-Cas systems. A few studies actually supported this hypothesis. As the consequence of transforming an engineered plasmid with spacers targeting the chromosomal gene within HAI2, P. atrosepticum survived by excision of the entire HAI2 island or deletion of part of the pathogenicity island (20). A similar result has been observed in the Streptococcus thermophilus that carries a type II-A CRISPR-Cas system (29). When a plasmid with spacers targeting genomic islands was transformed, CRISPR-Cas systems can drive deletion of large genomic islands and genome evolution by insertion sequence (IS)-dependent recombination. Collectively, these observations indicate that CRISPR-Cas systems can direct bacterial genome rearrangement and evolution through deletion of the integrated MGEs. Spontaneous SCCmec excision events occur at a low frequency in the wild-type population (30, 31).
CRISPR-Cas systems have been found in several S. epidermidis and S. aureus strains, especially in SCCmec-positive strains (32–35). It has been demonstrated that CRISPR-Cas systems can limit plasmid conjugation and phage invasion in S. epidermidis strain RP62A (26, 36). In a previous study, we identified six clinical isolates of S. aureus that harbor type III-A CRISPR-Cas systems and demonstrated their immunity function (17). Here, we further performed experiments in S. aureus strain AH1, a methicillin-resistant clinical isolate containing type V SCCmec. To investigate the effect of CRISPR-mediated chromosomal targeting toward SCCmec, we constructed artificial CRISPR plasmids with spacers targeting the mecA gene within SCCmec. Our results demonstrate that spacers with a perfect match to the endogenous gene are actually detrimental, but bacteria can avoid this autoimmunity by various mutations. The most common mutation mechanism was reshaping the sequence within SCCmec instead of driving excision of the entire SCCmec. We further found that the appropriate length of crRNAs and successive mismatches between the 5′ tag of crRNAs and nucleotides adjacent to protospacers are required for type III-A CRISPR immunity. These findings provide novel insight into the molecular mechanisms of CRISPR targeting and clinical applications of CRISPR-Cas systems in the treatment of MRSA infection.
RESULTS
Determination of the functional CRISPR promoter region.
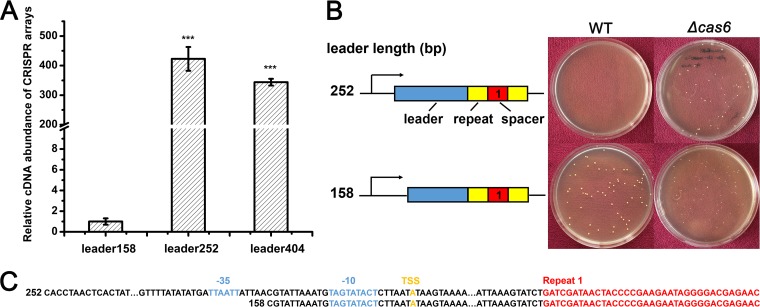
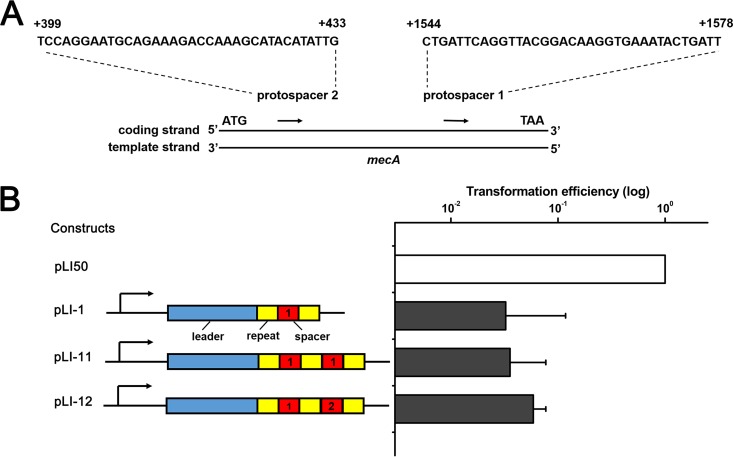
To investigate the effect of chromosomal targeting by the type III-A CRISPR-Cas system in S. aureus strain AH1, we constructed artificial CRISPR plasmids containing chromosome-targeting spacers. We first identified the functional promoter region of the CRISPR array by constructing a series of plasmids with truncated leader sequences of 404, 252, and 158 bp of the first repeat and native CRISPR arrays. These plasmids were transformed into the CRISPR knockout strain, and the transcription efficiencies of different leader sequences were detected by real-time quantitative reverse transcription-PCR (qRT-PCR). The transcriptional level of native crRNAs driven by the 158-bp leader sequence decreased more than 300-fold (Fig. 1A). Then, we constructed artificial CRISPR plasmids with 252-bp or 158-bp leader sequence and a mini-CRISPR array generating crRNAs targeting mecA, yielding plasmids pLI-252 and pLI-158. The mecA gene is located on SCCmec and encodes an alternative penicillin-binding protein PBP2a, which exhibits a much lower affinity to β-lactam antibiotics than PBP2 does (4). These two plasmids were transformed into the wild-type (WT) and cas6 knockout strains. Transformation results showed that only the 252-bp leader sequence exhibited obvious transcriptional activity, which was detrimental to bacterial cell growth (Fig. 1B). The low transcription efficiency of the 158-bp leader may be due to its position that is too close to the predicted −35 and −10 promoter regions and the putative transcription start site of the CRISPR array (Fig. 1C). As a result, the 252-bp leader was chosen as the promoter of the artificial CRISPR array in our research. The targeting activity of artificial CRISPR plasmids was assessed by the transformation efficiency relative to the transformation efficiency of the empty plasmid pLI50. There was no apparent additional effect with the mecA-targeting constructs containing one spacer (pLI-1), two identical spacers (pLI-11), or two individual spacers (pLI-12) (Fig. 2), suggesting that a single spacer is sufficient for targeting.
FIG 1 .
Identification of the functional CRISPR promoter region. (A) Relative transcription level of the native CRISPR array under the control of the truncated leader in the CRISPR knockout strain. The lengths of truncated leader were 404, 252, and 158 bp. Values that are significantly different from the value for the leader158 (P < 0.001) are indicated by three asterisks. (B) Artificial mini-CRISPR arrays with truncated leaders of 158 and 252 bp were constructed and transformed into the WT and cas6 knockout strains. At least three independent transformation experiments were performed, and representative plates are shown. (C) The sequences of the truncated 252-bp and 158-bp leaders, the predicted −35 and −10 promoter regions (blue), and the transcription start site (TSS) (in orange) relative to the first CRISPR repeat (red) are shown.
FIG 2 .
One chromosome-targeting spacer is sufficient for CRISPR targeting. (A) Schematic of two sequence regions selected as an artificial CRISPR array-targeting site. Sequences of the coding strand from 1544 to 1578 nt and from 399 to 433 nt relative to the start codon (ATG) of mecA constituted protospacer 1 and protospacer 2, respectively. (B) mecA-targeting constructs pLI-1 (one spacer), pLI-11 (two identical spacers), and pLI-12 (two different spacers) displayed similar toxicity. The transformation efficiency of the empty plasmid pLI50 (no spacer) was set at 100%. Transformations were performed three times, and average relative transformation efficiencies plus standard deviations (error bars) are shown in the graph.
Chromosomal targeting by the type III-A CRISPR-Cas system in S. aureus.
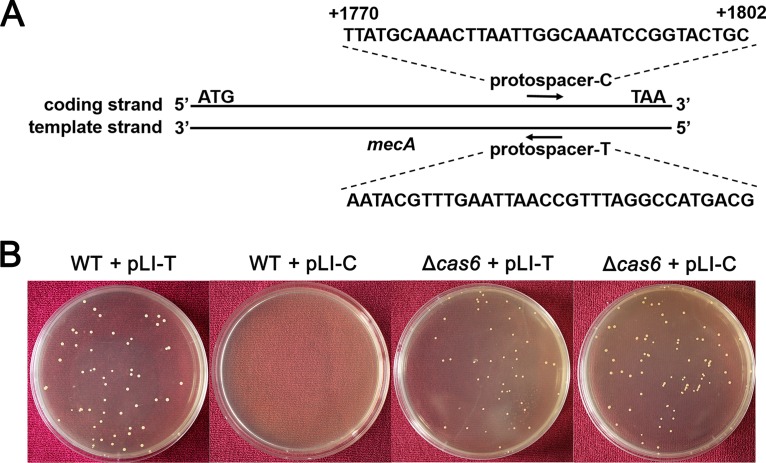
To further investigate whether the effect of chromosomal targeting by the type III-A CRISPR-Cas system is dependent on the transcription of the target gene, we constructed artificial CRISPR plasmids with spacers targeting the coding strand (pLI-C) and the template strand (pLI-T) of mecA and transformed them into the WT and cas6 knockout strains (Fig. 3). The results indicated that nearly no transformant was obtained in the WT strain with spacers targeting the coding strand of mecA, whereas many transformants were obtained with spacers targeting the template strand of mecA, and many transformants were obtained when CRISPR immunity was abolished in the cas6 knockout strain (Fig. 3B) (17). In addition, we detected the oxacillin MIC level of transformants generated from the cas6 knockout strain. The transformants exhibited the same MIC level with the WT and cas6 knockout strains (Table 1), indicating again that Cas6 is essential for immunity function. These results demonstrate that chromosomal targeting by the type III-A CRISPR-Cas system is dependent on the transcription of the target gene.
FIG 3 .
An artificial CRISPR plasmid with spacers targeting mecA displays Cas-dependent toxicity. (A) Schematic of sequence regions selected as artificial CRISPR plasmid targeting sites. The sequence of protospacer-C is in the coding strand of mecA, whereas protospacer-T is the complementary sequence of protospacer-C in the template strand. (B) Transformation plates of the WT strain and the Δcas6 mutant strain after growth for 36 h on TSB containing chloromycetin (Chl). CRISPR plasmids contained spacers targeting the coding strand and template strand of mecA.
TABLE 1 .
Oxacillin susceptibility of S. aureus strains
| Strain and relevant characteristic(s) | Oxacillin MIC (mg/liter)a |
|---|---|
| AH1 strains | |
| WT | 2 |
| Containing CRISPR plasmid; mecA deletion | <0.5 |
| Containing CRISPR plasmid; cas mutation | 2 |
| Containing destroyed CRISPR plasmid | 2 |
| Δcas6 strains | |
| S. aureus AH1; cas6-deleted strain | 2 |
| S. aureus AH1, cas6-deleted strain; containing CRISPR plasmid | 2 |
Oxacillin MIC in Mueller-Hinton broth.
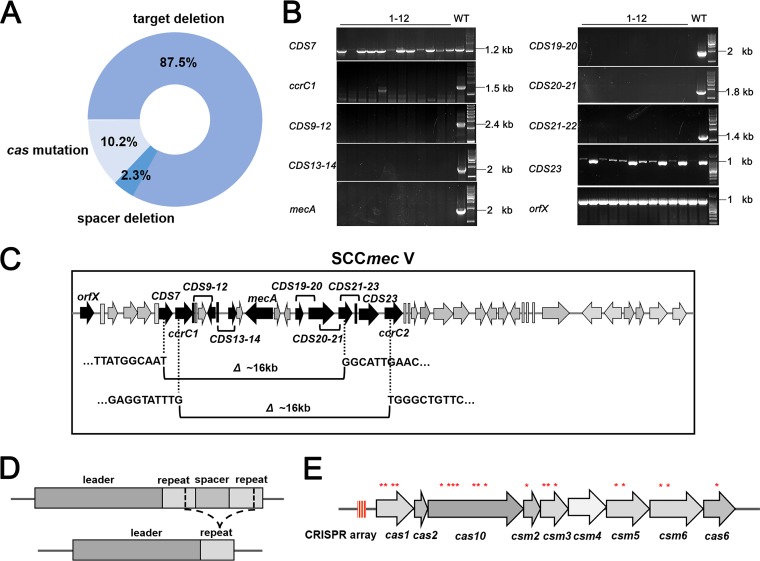
The chromosome-targeting spacers displayed extremely high chromosomal targeting capacity, leading to the death of more than 95% of the transformed bacterial cells. The surviving clones evaded CRISPR attack by various mutations. To distinguish the mutations, we analyzed 128 transformants that had been obtained in several transformation experiments. Mutation analysis was implemented by determining the presence of any mutation in the target, CRISPR plasmid, or cas genes. We extracted genomic DNA from all transformants and amplified mecA as well as its surrounding regions by PCR. Surprisingly, large fragment deletions of similar sizes across the targeted region occurred in more than 87% of the transformants (Fig. 4A and B). To map the accurate deletion region, we randomly chose two transformants to perform whole-genome sequencing, and reads were mapped to the reference genome sequence using software. Sequence analysis revealed the deletion of fragments (~16 kb) within SCCmec (Fig. 4C). The deleted fragments contain 15 to 17 coding sequences (CDS) and constitute ~0.55% of the 2,900-kb genome of S. aureus.
FIG 4 .
Transformants evade CRISPR targeting by different mutations. (A) Summary of different mutation types and corresponding proportions of 128 surviving clones. (B) PCR amplification for identification of large fragment deletions across SCCmec. Deletions occurred between CDS7 and CDS23. The weaker PCR bands reflected gene breaking regions. 1-12, lanes 1 to 12. (C) Schematic of two representative transformants contained about 16-kb deletion within SCCmec. The deletion regions were between CDS7 and CDS21 and between ccrC1 and ccrC2. (D) Schematic of transformants avoiding CRISPR attack by removal of the spacer repeat unit. (E) Distribution of mutations within different cas genes. Red asterisks indicate the mutation sites of single-nucleotide insertions, deletions, or substitutions.
We further sequenced the mecA PCR products from the transformants harboring mecA and found that no nucleotide mutation occurred in the matching region. The remaining transformants survived due to the deletion of the anti-mecA spacers or mutations in cas genes required for targeting. We found three transformants with deletion of anti-mecA spacer repeat unit within the impaired CRISPR constructs, which presumably occurred via recombination of repeat sequences (Fig. 4A and D). To assay inactivating mutations, we amplified the full CRISPR-Cas loci of the remaining 13 transformants and found 10 amplicons containing mutations (Fig. 4A). Sequencing results of the PCR products identified the loss-of-function mutations in different cas genes, including cas1, cas10, csm2, csm3, csm5, csm6, and cas6 (Fig. 4E and Table 2). Intriguingly, we obtained three transformants with the chromosome-targeting spacer and corresponding protospacer, but no mutation was observed in mecA, the CRISPR array, or cas genes. In addition, we detected the oxacillin MIC level of all transformants. Transformants in which mecA was deleted were all sensitive to oxacillin, and transformants with mutations in cas genes or CRISPR plasmids were still resistant to oxacillin and displayed the same MIC level as the WT strain (Table 1).
TABLE 2 .
Characteristics of cas mutations in S. aureus AH1 transformants
| Mutation site | Mutation type(s) | No. of transformants |
|---|---|---|
| cas1 | Nucleotide substitution | 3 |
| Nucleotide insertion, frameshift | 1 | |
| cas6 | Nucleotide insertion, frameshift | 1 |
| cas10 | Nucleotide substitution | 1 |
| Nucleotide insertion, frameshift | 2 | |
| Nucleotide deletion, frameshift | 1 | |
| csm2 | Nucleotide substitution | 1 |
| csm3 | Nucleotide substitution | 2 |
| Nucleotide insertion, frameshift | 1 | |
| csm5 | Nucleotide insertion, frameshift | 1 |
| Nucleotide deletion, frameshift | 1 | |
| csm6 | Nucleotide insertion, frameshift | 1 |
| Nucleotide deletion, frameshift | 1 |
The lengths of mature crRNAs were constant.
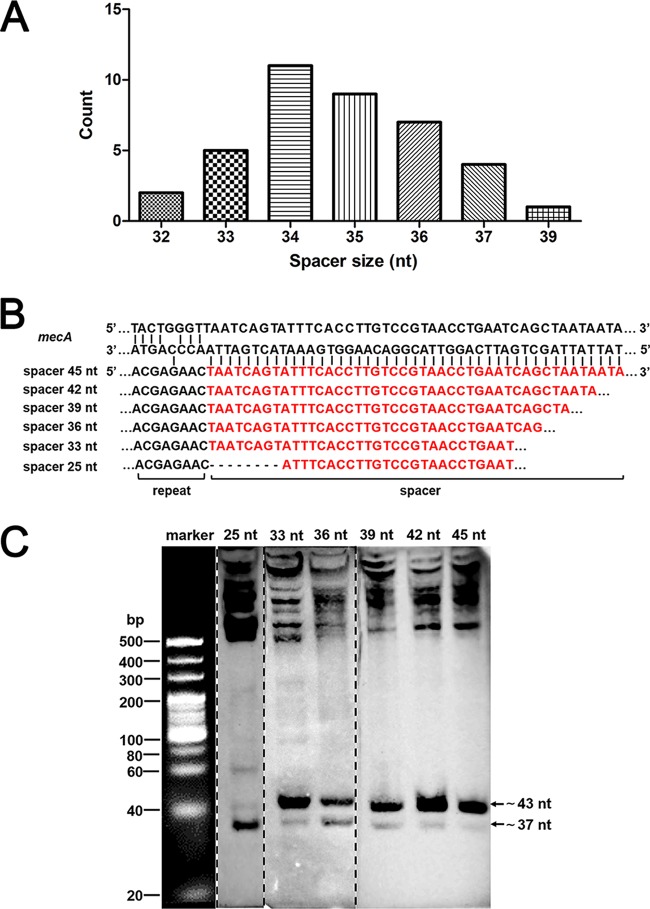
S. aureus strain AH1 harbors three distinct spacers, one of which was 35 nucleotides (nt) long and two were 37 nt long (17). Characterization and comparison of 39 spacers from six CRISPR-positive S. aureus strains (AH1, AH2, AH3, SH1, SH2, and SH3) indicate that the size of the spacer was not constant, with the longest spacer being 39 nt, the shortest spacer being 32 nt, and the most common sizes being 34 and 35 nt (Fig. 5A). The range of spacer size was variable among different species. The longer spacers were observed in Methanopyrus kandleri, which possesses 51- to 72-nt spacers. In some bacteria, the spacer size is even less than 30 nt (37). To determine whether spacer size can affect crRNA processing, we introduced a series of mecA-targeting CRISPR arrays with spacers of different lengths and distinguished the lengths of crRNAs by Northern blotting. We found that the transcripts of artificial CRISPR arrays of different sizes were all processed into two mature crRNAs that were comparable in size (Fig. 5B and C). These results indicate that the plasmid-borne CRISPR array can be successfully transcribed and processed into mature crRNAs and that the size of the spacer is not the critical factor in crRNA processing. More interestingly, the primary CRISPR transcript with a spacer length of less than 30 nt showed a stronger hybridization signal than the 37-nt band did (Fig. 5C).
FIG 5 .
Determination of the intermediate products and mature crRNAs generated from mecA-targeting CRISPR arrays by Northern blotting. (A) Size distribution of 39 spacers from six CRISPR-positive S. aureus strains. (B) Schematic of sequences with mecA-targeting spacers of different lengths (red). (C) The processed crRNAs generated from spacers of different lengths showed similar sizes. The arrows indicate the positions of mature crRNAs with sizes of ~37 nt and ~43 nt. The higher bands indicate the intermediate products. Portions of the gel were taken from three different gels and joined together. The broken lines show the spliced portions.
To precisely determine the sizes and sequences of mature crRNAs, we performed 5′ and 3′ rapid amplification of cDNA ends (RACE). Our RACE data indicated that all primary CRISPR transcripts were reduced to mature crRNAs with sizes of 43 and 37 nt (Table 3). The sequence of the first 8 nt (ACGAGAAC) of mature crRNAs was constant, and this crRNA 5′ tag was conservative in staphylococci (11). The 3′ end of crRNAs differed and maturation followed a rule that primary CRISPR transcripts were trimmed on the 3′ end and retained the 35 or 29 nt following the 5′ tag in vivo (Table 3). These data suggested that maturation of crRNAs is independent of the sequence and length of intermediate crRNAs and that the crRNA 3′ end maintained a constant distance from its 5′ tag (11).
TABLE 3 .
Sequences and sizes of mature crRNAs with different length spacers
| CRISPR plasmid spacer length (nt) |
Mature crRNA sequencea | crRNA size (nt) |
|---|---|---|
| 36 | ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCUGAAUCA | 43 |
| ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCU | 37 | |
| 39 | ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCUGAAUCA | 43 |
| ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCU | 37 | |
| 42 | ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCUGAAUCA | 43 |
| ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCU | 37 | |
| 45 | ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCUGAAUCA | 43 |
| ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCU | 37 | |
| 33 | ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCUGAAUGA | 43 |
| ACGAGAACUAATCAGUAUUUCACCUUGUCCGUAACCU | 37 | |
| 25 | ACGAGAACAUUUCACCUUGUCCGUAACCUGAAUGAUCGAUAAC | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAACCUGAAUGAUC | 37 | |
| 23 | ACGAGAACAUUUCACCUUGUCCGUAACCUGAGAUCGAUAACUA | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAACCUGAGAUCGA | 37 | |
| 22 | ACGAGAACAUUUCACCUUGUCCGUAACCUGGAUCGAUAACUAC | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAACCUGGAUCGAU | 37 | |
| 21 | ACGAGAACAUUUCACCUUGUCCGUAACCUGAUCGAUAACUACC | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAACCUGAUCGAUA | 37 | |
| 20 | ACGAGAACAUUUCACCUUGUCCGUAACCGAUCGAUAACUACCC | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAACCGAUCGAUAA | 37 | |
| 17 | ACGAGAACAUUUCACCUUGUCCGUAGAUCGAUAACUACCCCGA | 43 |
| ACGAGAACAUUUCACCUUGUCCGUAGAUCGAUAACUA | 37 |
Spacer sequences are underlined.
Spacer size played an important role in CRISPR targeting.
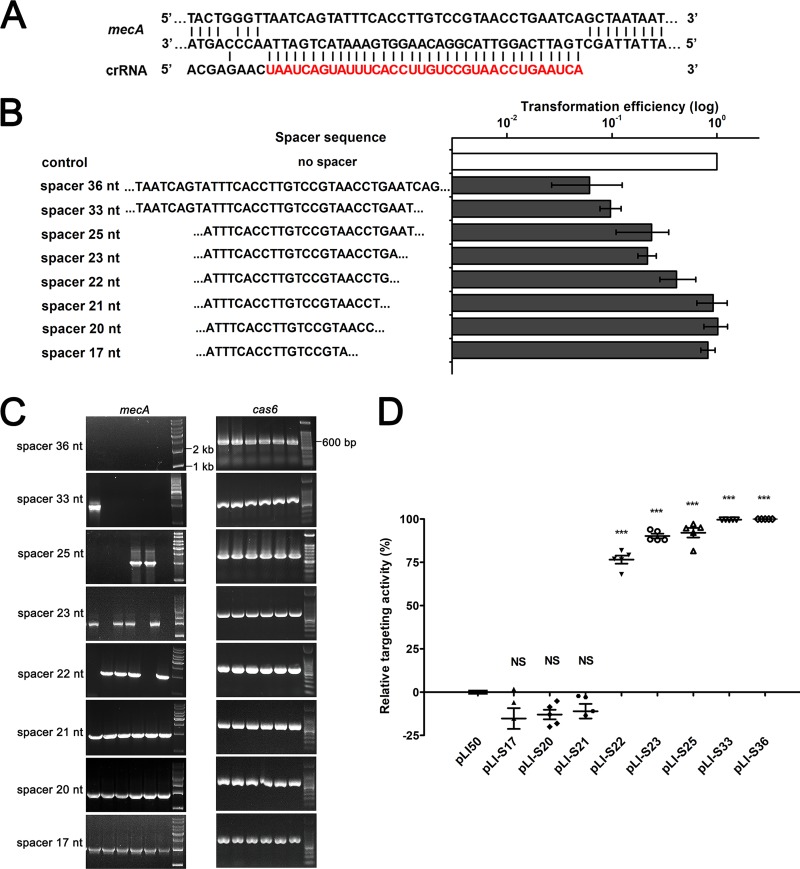
While the variation in spacer size had no influence on crRNA processing, the mature crRNAs had multiple mismatches with the protospacer sequence of mecA, especially when the length of the spacer was reduced (Fig. 6A and B). To investigate whether these mismatches abolish CRISPR-mediated immunity, we performed transformation experiments and detected the transformation efficiencies of each CRISPR construct (17). The results indicated that CRISPR plasmids with 33-nt (pLI-S33) and 36-nt spacers (pLI-S36) exhibited obvious targeting capacity. The relative transformation efficiencies of pLI-S36 and pLI-S33 were only about 5% (Fig. 6B). CRISPR plasmids with spacers ranging in size from 22 to 25 nt (pLI-S22, pLI-S23, and pLI-S25) displayed strong reductions in targeting capacity. The relative transformation efficiencies of pLI-S22, pLI-S23, and pLI-S25 were about 20% to 40% (Fig. 6B). CRISPR plasmids with spacer lengths of less than 21 nt had no effect on targeting. The transformation efficiencies of pLI-S17, pLI-S20, and pLI-S21 were comparable to that of the control pLI50 (Fig. 6B). To further determine the targeting capacity of these CRISPR plasmids, the presence of mecA for each transformant was detected by PCR amplification, and cas6 was amplified as a control (Fig. 6C). Unexpectedly, crRNAs and mecA coexisted in the daughter clones of the transformants containing mecA-targeting construct pLI-S17, pLI-S20, or pLI-S21, suggesting that the truncation of spacers may cause the loss of targeting activity (Fig. 6C). In the daughter clones of the transformants containing pLI-S22, pLI-S23, or pLI-S25, some lost the target gene mecA, while others did not (Fig. 6C). In contrast, mecA was deleted in all the daughter clones of the transformants containing pLI-S33 and pLI-S36 (Fig. 6C). We assumed that CRISPR targeting was partially impaired due to the truncation of spacers. Therefore, we detected the positive ratio of mecA in each transformant population, and the result was consistent with our hypothesis. CRISPR plasmids with spacer lengths of less than 21 nt (pLI-S17, pLI-S20, and pLI-S21) showed no targeting activity, and the transformant populations were all mecA-positive clones (Fig. 6D). CRISPR plasmids with 22-nt, 23-nt, and 25-nt spacers displayed higher targeting activities. The average targeting activities of pLI-S22, pLI-S22, and pLI-S25 were about 75%, 85%, and 90%, respectively (Fig. 6D). CRISPR plasmids with 33-nt and 36-nt spacers exhibited strong targeting activities. The average targeting activities of pLI-S33 and pLI-S36 were more than 99% (Fig. 6D). Altogether, these data suggest that appropriate spacer size is required for CRISPR targeting and that targeting capacity is positively associated with the spacer length within a certain range.
FIG 6 .
The spacer length has influence on targeting activity. (A) Schematic of base pairing between mature crRNA generated from pLI-S36 and its target sequence. Nucleotides from the spacer are highlighted in red. (B) The relative transformation efficiencies of S. aureus strain AH1 with artificial CRISPR plasmids containing spacers of different lengths. Transformations were performed at least three times. The transformation efficiency of the control (pLI50) was set at 100%. (C) PCR amplification for the detection of the mecA target gene. PCR performed for cas6 was shown as a control. (D) Relative activity of mecA-targeting constructs containing spacers of different lengths. The targeting activity of empty plasmid pLI50 was set at zero. Five independent transformants were analyzed for each construct. The values are means ± standard deviations (error bars). Values that are significantly different (P < 0.001) from the value for pLI50 are indicated by three asterisks. Values that are not significantly different (NS) from the value for pLI50 are also indicated.
Mutations in the 5′ tag of crRNAs can partially block CRISPR targeting.
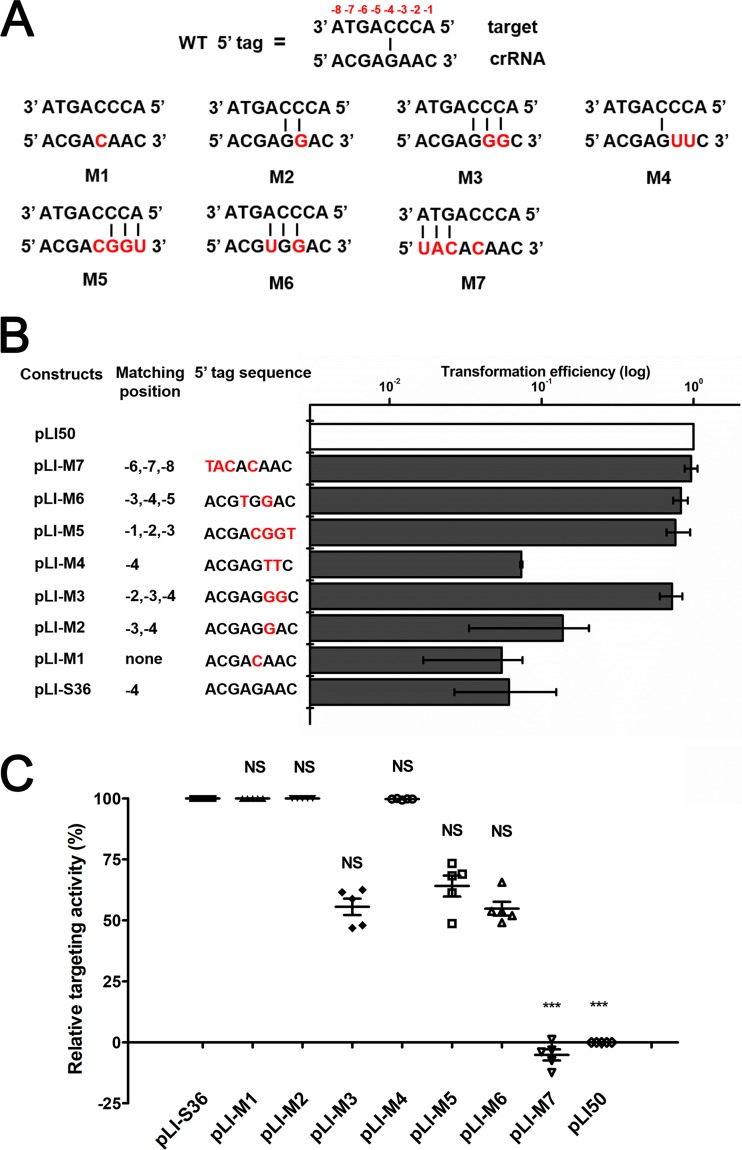
In S. epidermidis, CRISPR immunity against nonself targets is enabled by mismatches between the 5′ upstream sequence of target DNA and crRNAs. Formation of at least three base pairings at positions −4, −3, and −2 eliminates targeting. Self-recognition and protection are achieved by complementarity between the CRISPR locus and the crRNAs. Disruption of base pairings at positions −4 and −3 or −3 and −2 abolishes protection (15).
To further verify this hypothesis in the S. aureus type III-A CRISPR-Cas system, some mutations were introduced into the upstream repeat sequence of pLI-S36, yielding a variety of complementary sequences between the crRNAs and associated protospacers (Fig. 7A). The 5′ tag of crRNAs generated from the mecA-targeting CRISPR construct pLI-S36 exhibited pairing with protospacers at position −4, but it did not influence CRISPR targeting (Fig. 7B). It was possible that a single nucleotide mutation was not sufficient to completely block CRISPR targeting. We then introduced some mutations at positions −2 to −4 within the 5′ tag of crRNAs. The transformation results indicated that noncomplementarity (G-4C) between the crRNA and the upstream flanking sequences of protospacer can absolutely ensure targeting (Fig. 7B). Base pairings at positions −4 and −3 (A-3G) did not significantly disrupt CRISPR targeting, whereas three consecutive matches at positions −2 to −4 (A-2G and A-3G) almost eliminated targeting (Fig. 7B). To confirm that the decisive requirement for targeting is noncomplementarity with the crRNA 5′ tag rather than nucleotide identity, we introduced mutations at the same positions (−2 and −3) but with nucleotide T, not G, and the result was consistent with our hypothesis. In contrast to mutation M3 (A-2G and A-3G), mutation M4 (A-2T and A-3T) yielded base pairing only at position −4 and could not eliminate CRISPR targeting (Fig. 7B).
FIG 7 .
Mutations in the crRNA 5′ tag eliminate CRISPR attack. (A) Schematic of the complementarity between the flanking sequences (positions −1 to −8) of crRNAs (bottom) and target DNA (top). The mutated nucleotides are shown in red. (B) Effects of progressive sequence mutations in the 5′-tag sequence on transformation efficiency. The transformation efficiency of the empty plasmid pLI50 was set at 100%. The construct pLI-S36, which contains the native repeat, was used as a positive control. The mutated nucleotides are shown in red. (C) The relative targeting activity of mecA-targeting constructs contained a series of mutations in the 5′-tag sequence. The targeting activity of empty plasmid pLI50 was set at zero. The construct pLI-S36 was taken as a positive control. Five independent transformants were analyzed for each construct with bars indicating standard deviations.
To identify the positions important for protection, we introduced three consecutive nucleotide pairings at different positions in the 5′ tag (M5, M6, and M7) (Fig. 7A). Contrary to a previous report (15), mutations M5 and M6 exhibited the same transformation efficiencies as the mutation M3 did, suggesting that any three consecutive matches at positions −1 to −5 could protect the target from degradation (Fig. 7B). Mutation M7 showed nearly the same transformation efficiency as the negative control did (Fig. 7B). It was possible that mutations at positions −6, −7, and −8 eliminated crRNA maturation and targeting (15). To investigate how spacer sequence may affect CRISPR attack in the presence of a 5′-tag mutation, we detected the relative targeting activity of crRNAs with mutations in the 5′ tag. The crRNAs generated from constructs pLI-M1, pLI-M2, and pLI-M4 showed the similar targeting capacities as pLI-S36 did (Fig. 7C), which could fully degrade protospacers. In contrast, crRNAs generated from constructs pLI-M3, pLI-M5, and pLI-M6 exhibited significantly reduced targeting activities, and only ~40% to 50% protospacers were cleaved (Fig. 7C), revealing that at least three consecutive matches at positions −1 to −5 could partially disturb CRISPR targeting and protect protospacers from degradation. As expected, the pLI-M7 construct displayed no targeting activity, as did the empty vector pLI50 (Fig. 7C).
Taken together, these results demonstrate that the 5′-tag sequence can play an important role in the recognition of self/nonself. In addition, three consecutive base pairings between the 5′ tag of crRNAs and protospacer-adjacent sequences have a negative effect on CRISPR targeting.
DISCUSSION
The CRISPR-Cas system is a typical immune system that can protect bacteria and archaea against invading foreign DNA. As an important element in the evolution process of prokaryotic organisms, how does a host distinguish between the advantages and disadvantages of a CRISPR-Cas system? Recently, the origin of diverse spacers and the mechanism of spacer acquisition have become the focus of attention. Bioinformatic analysis shows that in addition to attacking conjugative plasmid and bacteriophage, a small number of spacers match with archaeal or bacterial genomes (18, 19, 38, 39). Remarkably, although only a minority of spacers share homology with prokaryotic genomes, they present at a high frequency. About one in every 5.5 CRISPR-positive organisms contains at least one spacer matching with archaeal or its own bacterial genome (18). However, a reasonable and convincing explanation for the existence of chromosome-targeting spacers has not been provided yet. One theory is that chromosomal targeting is detrimental and bacteria escape from autoimmunity at a severe fitness cost of CRISPR-Cas system inactivation (18). A few studies have provided experimental evidence to support this hypothesis. In P. carbinolicus type I-E CRISPR-Cas system, the CRISPR locus contains a spacer against the housekeeping gene hisS. Transformation of the artificial plasmid with spacers targeting hisS into a Geobacter sulfurreducens strain could inhibit its growth (21). Introduction of an artificial mini-CRISPR locus with a spacer against the beta-galactosidase gene in S. solfataricus by transfection caused growth inhibition, and the host cells can survive by eliminating the corresponding CRISPR locus (40). In addition, spacers against integrated MGEs exhibited unexpected effects. Although the type I-F CRISPR-positive P. atrosepticum contained a spacer completely complementary to an endogenous gene within genomic island HAI2, CRISPR lethality was abolished due to a single nucleotide mutation in the PAM. Engineering a CRISPR locus with a correct PAM could recover the deleterious effect and promote bacterial genome evolution (20). A similar result was observed in the S. thermophilus type II-A system. When an artificial spacer targeting lacZ located in the integrated genomic island was introduced, most of the transformants were killed. Lac survivors showed large-scale genome deletion via IS-dependent recombination (29). However, in the type III-B system, chromosome-targeting spacers could be used as a tool to silence endogenous genes instead of killing cells due to the fact that the target is RNA, not DNA (41).
In this study, we have demonstrated that chromosomal targeting by the type III-A CRISPR-Cas system is significantly deleterious. Chromosomal targeting was achieved by transforming plasmids containing engineered CRISPR arrays with chromosome-targeting spacers. Importantly, the resistance gene mecA within SCCmec is chosen as the target. Neither the activity of a CRISPR-Cas system against integrated SCCmec nor its consequence for genome-scale evolution has been detected before. We have revealed that the most common fitness cost corresponding to chromosomal targeting is deletion of the target sequence. It seems that chromosomal targeting can provide a great selective pressure for bacterial genome evolution. Other types of negative fitness cost were also observed, such as loss-of-function mutations in cas genes and deletion of responsible spacers (Fig. 4D and E). Nevertheless, we did not observe any transposon insertion mutation or the deletion of the entire CRISPR-Cas locus among all 128 transformants. In a very small proportion of survivors, no mutation was found in protospacers, cas genes, or plasmids carrying a mini-CRISPR array. It seems reasonable to assume that CRISPR-Cas immunity is not absolutely abolished in these strains and that partial immunity leads to tolerance of self-targeting, which is in agreement with the results reported in S. epidermidis (22). Also, the proportion of different types of mutations in our experiments (Fig. 4A) differed from those observed by others in S. epidermidis and in Sulfolobus islandicus (22, 42). These results suggest that bacteria deal with the evolution downside of selective pressure through different mechanisms and produce preference according to differential conditions (targeting conjugative plasmid or chromosome). Moreover, bacteria can escape from chromosomal targeting at the negative cost of loss-of-function mutations in diverse cas genes. In addition, multiple point mutations were identified within the cas1 gene (Table 2), which is not responsible for CRISPR immunity.
Among staphylococcal strains with type III-A CRISPR-Cas systems, most strains contain two CRISPR arrays with 14 or 15 spacers upstream and downstream of the cas locus, respectively (17). However, S. aureus strain AH1 has only one CRISPR array with three spacers. Similarly, S. epidermidis strain RP62A has only five spacers, three spacers located upstream of the cas locus and two spacers located downstream of cas (35). The number of CRISPR arrays and spacers may be associated with the background, environment, and evolution process of different strains. However, it does not influence the immunity function of the CRISPR-Cas system in different strains (17, 36). The sizes of the three native spacers were 35 or 37 nt in S. aureus strain AH1 (17). By changing the length of chromosome-targeting spacers in our experiments, we found that it had no influence on the size of mature crRNAs. Northern blot results showed two clear bands with sizes of about 43 and 37 nt as previously described (11). RACE assays further confirmed Northern blot results, indicating that the sizes of mature crRNAs are constant. We further demonstrated that spacer length has an effect on the targeting activity. The artificial spacers with the sizes of 36 or 33 nt exhibited high targeting capacity and triggered more than 99% of DNA degradation (Fig. 6D). Introduction of 13-nt mismatches between the target gene and the 3′ ends of crRNAs by truncating the spacer length to 22 nt could still result in more than 75% of DNA degradation (Fig. 6D). Further truncation (17 to 21 nt) completely abrogated CRISPR attack (Fig. 6D), indicating that more than 13 consecutive mutations in the 3′ ends of crRNAs can fully abolish CRISPR targeting activity. Similar conclusions were proposed in previous studies. For example, Cao et al. demonstrated that 12 consecutive nucleotide mutations resulted in a decreased immunity activity in S. aureus and that 13 consecutive nucleotide mutations completely disrupted CRISPR antiplasmid immunity (17). Manica et al. reported that more than 15 nucleotide mutations fully blocked CRISPR interference in S. solfataricus (16). These observations imply that mutations are highly tolerated between crRNAs and their protospacers and that the number of paired nucleotides between the crRNAs and protospacers is the decisive characteristic for CRISPR targeting.
The CRISPR-Cas system is a simple but ingenious defense system, and it can precisely discriminate self/nonself to prevent autoimmunity. In type I and II systems, host distinguishes self from nonself via the recognition of specific nucleotides in the PAM region. The type III CRISPR-Cas system is independent of the PAM and identifies targets by a distinctive mechanism. In S. epidermidis, three or more successive base parings between the 5′ tags of crRNAs and targets are necessary for self-recognition (15). One previous study has indicated that base pairing at positions −2, −3, and −4 is crucial and that this recognition process is independent of the nucleotide sequence (15). In S. solfataricus, similar conclusions are proposed but for positions −3, −4, and −5 (16). To figure out the key nucleotides for self/nonself discrimination in our strain, we constructed a chromosome-targeting spacer with multiple mutations in the first 8 nt of the repeat and performed the transformation experiments. Significantly higher transformation efficiencies were observed, suggesting that any consecutive three-nucleotide complementarity between the 5′ tag of crRNAs and the adjacent region of protospacers can block attack (Fig. 7B). This self-recognition was independent of position or sequence (Fig. 7). Interestingly, most of these transformants exhibited small and rough colonies, and further experiments confirmed that only ~50% chromosome degradation was realized in these clones, implying that CRISPR attack was not completely abolished (Fig. 7C). These data imply that the mechanism of self/nonself recognition in the type III CRISPR-Cas system is more complicated than we thought.
In conclusion, we use engineered chromosomal targeting as an alternative strategy to investigate the immunity function and molecular mechanisms of the type III-A CRISPR-Cas system in S. aureus. Our findings indicate that chromosomal targeting can drive large-scale deletion within integrated SCCmec and contribute to bacterial genome reshaping. In addition, this study may provide a promising tool to delete resistance and virulence genes in bacterial pathogens by CRISPR-Cas systems.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 4. Escherichia coli was grown (220 rpm) in lysogeny broth medium (Franklin Lakes) or on lysogeny broth agar (LA) at 37°C. Staphylococcus aureus strains were grown (220 rpm) in tryptic soy broth (TSB) (Difco) or on tryptic soy agar plates (Difco) at 37°C. When needed, 150 μg/ml ampicillin sodium salt or 50 μg/ml kanamycin sulfate for E. coli or 15 μg/ml chloromycetin for S. aureus strains was added to the bacterial cultures.
TABLE 4 .
Bacterial strains and plasmids used in this study
| Strain or plasmid(s) | Characteristicsa | Source or referenceb |
|---|---|---|
| Strains | ||
| E. coli TransT1 | Clone host strain; F− φ80(lacZ) ΔM15 ΔlacX74 hsdR (rK − mK+) ΔrecA1398 endA1 tonA | TransGen |
| S. aureus | ||
| RN4220 | 8325-4; restriction-negative strain | NARSA |
| AH1 | CA-MRSA; SCCmec type V | Hospital |
| Δcas6 | AH1; cas6-deleted strain | |
| Plasmids | ||
| pLI50 | Shuttle vector; Ampr Chlr | 46 |
| pLIC-404 | pLI50 derivative with 404 bp of leader sequence and native CRISPR locus from S. aureus strain AH1 |
This study |
| pLIC-252 | pLI50 derivative with 252 bp of leader sequence and native CRISPR locus from S. aureus strain AH1 |
This study |
| pLIC-158 | pLI50 derivative with 158 bp of leader sequence and a native CRISPR array from S. aureus strain AH1 |
This study |
| pLI-252 | pLI50 derivative with 252 bp of leader sequence and an artificial CRISPR array targeting mecA | This study |
| pLI-C | pLI50 derivative with an artificial CRISPR array targeting the coding strand of mecA | This study |
| pLI-T | pLI50 derivative with an artificial CRISPR array targeting the template strand of mecA | This study |
| pLI-1 | pLI50 derivative with an artificial CRISPR array containing one spacer targeting mecA | This study |
| pLI-11 | pLI50 derivative with artificial CRISPR arrays containing two identical spacers targeting mecA | This study |
| pLI-12 | pLI50 derivative with artificial CRISPR arrays containing two different spacers targeting mecA | This study |
| pLI-S17, pLI-S20, pLI-S21,pLI-S22, pLI-S23, pLI-S25, pLI-S33, pLI-S36, pLI-S39, pLI-S42, pLI-S45 |
LI50 derivative containing mecA-targeting spacers with the spacer length of 17, 20, 21, 22, 23, 25, 33, 36, 39, 42, or 45 nt |
This study |
| pLI-M1, pLI-M2, pLI-M3, pLI-M4, pLI-M5, pLI-M6, pLI-M7 |
pLI-S36 derivative with different mutations in the first repeat sequence | This study |
| pEASY blunt simple | Commercial cloning vector; Ampr Kanr | TransGen |
CA-MRSA, community-associated MRSA; Ampr, ampicillin resistant; Chlr, chloramphenicol resistant; Kanr, kanamycin resistant.
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
Construction of artificial CRISPR arrays.
To construct CRISPR plasmids that can be used further for cloning and expression of any spacer and repeat sequence, 404, 252, or 158 bp of the native CRISPR leader and CRISPR arrays were amplified with forward primers leader404-f (f stands for forward), leader252-f, or leader158-f and the reverse primer CRISPR-r (r stands for reverse). The products were then digested with KpnI and SacI and ligated to pLI50 previously digested with the same enzymes, generating plasmids pLIC-404, pLIC-252, and pLIC-158. These plasmids were then digested with ClaI and ligated with engineered spacer repeat units, yielding artificial CRISPR plasmids pLI-252 and pLI-158. The repeat and target-specific spacer regions were amplified by PCR with the primer pairs that contained engineered spacer repeat units. The repeats were digested with the enzyme Cla, which resulted in the introduction of subsequent spacer repeat units, and this procedure could be performed to construct any artificial CRISPR array. These plasmids were first introduced into S. aureus strain RN4220 for modification and subsequently transformed into S. aureus strain AH1 and its mutant strains. All plasmids extracted from S. aureus strain RN4220 were sequenced to confirm that no mutation occurred during the modification process. The sequences of the primers used in plasmid construction are shown in Table 5.
TABLE 5 .
Primers used in this study
| Primer | Sequence (5′–3′)a | Application |
|---|---|---|
| Leader404-f | CGGggtaccCATCTCAATTAAGCAGCTA | Amplification for 404-bp leader |
| Leader252-f | CGGggtaccCACCTAACTCACTATCAAT | Amplification for 252-bp leader |
| Leader158-f | CGGggtaccCGTATTAAATGTAGTATACT | Amplification for 158-bp leader |
| CRISPR-r | CCGgagctcCCATCCCCTAAAAATTAATCC | Amplification for a native CRISPR array |
| CRISPR-Cas-f1 | TAACTCACTATCAATCATTTCTCCAC | Amplification for CRISPR-Cas locus |
| CRISPR-Cas-r1 | GCATAATCCATCATCATTAATATCTATG | Amplification for CRISPR-Cas locus |
| CRISPR-Cas-f2 | TATAGAACTATTTGGCGTAATG | Amplification for CRISPR-Cas locus |
| CRISPR-Cas-r2 | GTAATCTTGCTTCTTCATAACT | Amplification for CRISPR-Cas locus |
| CRISPR-Cas-f3 | TTTATGGTTGGAGGTATAAGTATGAC | Amplification for CRISPR-Cas locus |
| CRISPR-Cas-r3 | TATATTATACTATATTTCCCCATGCC | Amplification for CRISPR-Cas locus |
| R1-S1-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-1, pLI-11, pLI-12 |
| S1-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCCTGATTCAGGTTACGGACAAGGTGAAATACTGATT | pLI-1,pLI-11, pLI-12 |
| R2-S2-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACCAATATGTATGCTTTGGTCTTTCTGCATTCCTGGA | pLI-12 |
| S2-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTCCAGGAATGCAGAAAGACCAAAGCATACATATTG | pLI-12 |
| R1-mecAC-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACGCAGTACCGGATTTGCCAATTAAGTTTGCATAA | pLI-C |
| mecAC-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTTATGCAAACTTAATTGGCAAATCCGGTACTGC | pLI-C |
| R1-mecAT-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTTATGCAAACTTAATTGGCAAATCCGGTACTGC | pLI-T |
| mecAT-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCGCAGTACCGGATTTGCCAATTAAGTTTGCATAA | pLI-T |
| R1-S17-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTA | pLI-S17 |
| S17-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTACGGACAAGGTGAAAT | pLI-S17 |
| R1-S20-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTAACC | pLI-S20 |
| S20-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCGGTTACGGACAAGGTGAAAT | pLI-S20 |
| R1-S21-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTAACCT | pLI-S21 |
| S21-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCAGGTTACGGACAAGGTGAAAT | pLI-S21 |
| R1-S22-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTAACCTG | pLI-S22 |
| S22-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCCAGGTTACGGACAAGGTGAAAT | pLI-S22 |
| R1-S23-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTAACCTGA | pLI-S23 |
| S23-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTCAGGTTACGGACAAGGTGAAAT | pLI-S23 |
| R1-S25-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACATTTCACCTTGTCCGTAACCTGAAT | pLI-S25 |
| S25-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCATTCAGGTTACGGACAAGGTGAAAT | pLI-S25 |
| R1-S33-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAAT | pLI-S33 |
| S33-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCATTCAGGTTACGGACAAGGTGAAATACTGATTA | pLI-S33 |
| R1-S36-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-S36 |
| S36-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCCTGATTCAGGTTACGGACAAGGTGAAATACTGATTA | pLI-S36 |
| R1-S39-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAGCTA | pLI-S39 |
| S39-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTAGCTGATTCAGGTTACGGACAAGGTGAAATACTGATTA | pLI-S39 |
| R1-S42-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAGCTAATA | pLI-S42 |
| S42-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTATTAGCTGATTCAGGTTACGGACAAGGTGAAATACTGATTA | pLI-S42 |
| R1-S45-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAGCTAATAATA | pLI-S45 |
| S45-R2-r | CACTCTGTCCCCTATTCTTCGGGGTAGTTATCGATCTATTATTAGCTGATTCAGGTTACGGACAAGGTGAAATACTGATTA | pLI-S45 |
| R1-S36m1-f | GATCGATAACTACCCCGAAGAATAGGGGACGACAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M1 |
| R1-S36m2-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGGACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M2 |
| R1-S36m3-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGGGCTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M3 |
| R1-S36m4-f | GATCGATAACTACCCCGAAGAATAGGGGACGAGTTCTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M4 |
| R1-S36m5-f | GATCGATAACTACCCCGAAGAATAGGGGACGACGGTTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M5 |
| R1-S36m6-f | GATCGATAACTACCCCGAAGAATAGGGGACGTGGACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M6 |
| R1-S36m7-f | GATCGATAACTACCCCGAAGAATAGGGGTACAGAACTAATCAGTATTTCACCTTGTCCGTAACCTGAATCAG | pLI-M7 |
| mecA-f | TAATAGTTGTAGTTGTCGGGTTTGG | mecA detection |
| mecA-r | CATCGTTACGGATTGCTTCACTGTT | mecA detection |
| cas6-f | TTTAGGAAGTATTTTACATGGCGTG | cas6 detection |
| cas6-r | CCAGAAAATTCACCAAACTTCAATA | cas6 detection |
| CRISPR-RT-f | GGGACGAGAACTTCAAAT | qRT-PCR |
| CRISPR-RT-r | CAGTATGAAACAAATCAAGGT | qRT-PCR |
| mecA-r-biotin | ATTCAGGTTACGGACAAGGTGAAATACTGATTA | Northern blotting |
Nucleotides in the restriction sites are indicated by lowercase letters.
Preparation of electrocompetent S. aureus cells.
S. aureus cells from 15% glycerol stock were streaked on a TSB agar plate and incubated at 37°C. A single colony was selected and incubated in 5 ml TSB at 37°C overnight. One-milliliter portions of the overnight culture were added to 100 ml TSB in a 500-ml flask and shaken at 37°C until an optical density at 600 nm (OD600) of 0.4 was reached. The culture was put on ice for 5 min and then transferred to a sterile, round-bottom centrifuge tube. The cells were collected by centrifugation at 2,500 × g at 4°C for 10 min, and the supernatant was discarded. The cells were gently resuspended in 10 ml of ice-cold 0.5 M sucrose, and the suspension was kept on ice for 5 min. The centrifugation and resuspension steps were repeated twice. The cells were then resuspended in 1 ml of ice-cold 0.5 M sucrose, and the suspension was kept on ice for 15 min. Finally, 100-μl aliquots were prepared in sterile microcentrifuge tubes and frozen in liquid nitrogen. The competent cells were stored at −80°C.
Plasmid extraction and transformation in S. aureus.
Plasmids from all S. aureus strains were isolated using a plasmid purification kit (Sangon Biotech) according to the manufacturer’s instructions, except that the cells were pretreated with digestion buffer containing 40 U/ml lysostaphin, 10 mg/ml lysozyme, and 10% (vol/vol) glycerol for 30 to 60 min. Plasmids were transformed into all S. aureus strains by electroporation. Plasmid DNA (100 to 500 ng) and electrocompetent S. aureus cells (100 μl) were mixed and placed in a Gene Pulser cuvette with a 0.2-cm electrode gap. The settings for electroporation are as follows: voltage, 2.5 kV; capacitor, 50 μF; resistance, 200 Ω. After electroporation, 400 μl TSB was immediately added to the cuvette, and the cuvette was put on ice for 15 min. The cells were then transferred into a 1.5-ml Eppendorf tube and incubated with shaking (220 rpm, 37°C) for 1 h before being spread on a TSB plate.
Oxacillin susceptibility assay.
The oxacillin susceptibility of the WT strain and transformants was evaluated by detecting the microbroth MIC of oxacillin according to Clinical and Laboratory Standards Institute (CLSI) criteria (43). The cultures of all strains were diluted to a final test concentration of approximately 5 × 104 CFU/well and incubated at 37°C for 24 h.
Total RNA extraction and qRT-PCR.
Total RNA was extracted by RNAiso plus according to the manufacturer’s instructions (TaKaRa). Residual DNA was digested with RNase-free DNase I (TaKaRa). Reverse transcription was carried out with the PrimeScript first-strand cDNA synthesis kit (TaKaRa), and real-time PCR was performed with SYBR Premix Ex Taq (TaKaRa) using a StepOne real-time system (Applied Biosystems). The quantity of cDNA was normalized to the abundance of pta cDNA (44). All the qRT-PCR assays were repeated at least three times.
Evaluation of DNA targeting efficiency by real-time PCR.
To analyze the ratio of mecA-positive clones in the S. aureus population, strains carrying mecA-targeting constructs were cultivated in TSB with chloromycetin (15 μg/ml) at 37°C for 24 h, then cells were collected, and genomic DNA was extracted. A final concentration of 200 ng/ml genomic DNA was used as the template. The real-time PCR was performed with SYBR Premix Ex Taq (TaKaRa) using the StepOne real-time PCR system (Applied Biosystems). The quantity of mecA measured by real-time PCR was normalized to the abundance of pta DNA (44). All the real-time PCR assays were repeated at least three times. The relative targeting activity of mecA-targeting spacer was equal to one minus the value of the relative quantity of mecA.
Northern blot analysis.
Total RNA (30 mg) was denatured at 95°C for 5 min and then separated with a 12% denatured polyacrylamide–7 M urea gel (100 V, 1.5 h) in 1× Tris-borate-EDTA (TBE) and transferred onto a nylon membrane in 0.5× TBE. The product was then immobilized by UV cross-linking and blotted with the biotin-labeled oligonucleotide probes. RNA-DNA hybridization detection using a North2South chemiluminescence hybridization and detection kit (Thermo Scientific) was performed to detect crRNAs.
Determination of mature crRNA sequences by RACE.
The 5′ and 3′ ends of crRNAs were determined by RACE using the full 3′ RACE core set version 2.0 and the full 5′ RACE kit (TaKaRa) as previously described (45). PrimeSTAR HS DNA polymerase (TaKaRa) was used for PCR amplification, and the amplified RACE products were ligated with pEASY-Blunt Simple Cloning vector (pEASY-Blunt Simple Cloning kit; TransGen Biotech). The ligation was transformed into E. coli TransT1, and transformants were characterized by colony PCR to amplify the RACE products. The positive colonies were sequenced using the M13 forward sequencing primer (Sangon Biotech).
Genomic DNA extraction and sequencing.
Genomic DNA was extracted and sequenced using an Illumina Hiseq 2000 platform (Institute of Microbiology, Chinese Academy of Sciences, Beijing, China). About 1.8 GB of high-quality sequence data of each genome was then mapped using SOAP (short oligonucleotide alignment program; BGI) software.
Statistical analysis.
First, F test for two samples was performed for variances. Unpaired two-tailed t test for equal or unequal variances was then performed to calculate the significant differences (P value). All the tests were performed by the data analysis tool in Microsoft Excel.
ACKNOWLEDGMENTS
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (XDPB03) and the National Natural Science Foundation of China (31670133).
We thank Linyan Cao for technical assistance.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Grundmann H, Aires-De-Sousa M, Boyce J, Tiemersma E. 2006. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public-health threat. Lancet 368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 3.Song MD, Wachi M, Doi M, Ishino F, Matsuhashi M. 1987. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett 221:167–171. [DOI] [PubMed] [Google Scholar]
- 4.Chambers HF, Deleo FR. 2009. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malachowa N, DeLeo FR. 2010. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci 67:3057–3071. doi: 10.1007/s00018-010-0389-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Katayama Y, Ito T, Hiramatsu K. 2000. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother 44:1549–1555. doi: 10.1128/AAC.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJ, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. 2015. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol 13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. 2008. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. 2010. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 10.Haurwitz RE, Jinek M, Wiedenheft B, Zhou KH, Doudna JA. 2010. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatoum-Aslan A, Maniv I, Marraffini LA. 2011. Mature clustered, regularly interspaced, short palindromic repeats RNA (crRNA) length is measured by a ruler mechanism anchored at the precursor processing site. Proc Natl Acad Sci U S A 108:21218–21222. doi: 10.1073/pnas.1112832108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. 2011. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wiedenheft B, van Duijn E, Bultema JB, Bultema J, Waghmare SP, Waghmare S, Zhou K, Barendregt A, Westphal W, Heck AJ, Heck A, Boekema EJ, Boekema E, Dickman MJ, Dickman M, Doudna JA. 2011. RNA-guided complex from a bacterial immune system enhances target recognition through seed sequence interactions. Proc Natl Acad Sci U S A 108:10092–10097. doi: 10.1073/pnas.1102716108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C. 2009. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 15.Marraffini LA, Sontheimer EJ. 2010. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature 463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manica A, Zebec Z, Steinkellner J, Schleper C. 2013. Unexpectedly broad target recognition of the CRISPR-mediated virus defence system in the archaeon Sulfolobus solfataricus. Nucleic Acids Res 41:10509–10517. doi: 10.1093/nar/gkt767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao L, Gao CH, Zhu J, Zhao L, Wu Q, Li M, Sun B. 2016. Identification and functional study of type III-A CRISPR-Cas systems in clinical isolates of Staphylococcus aureus. Int J Med Microbiol 306:686–696. doi: 10.1016/j.ijmm.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 18.Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. 2010. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. 2005. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 20.Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. 2013. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aklujkar M, Lovley DR. 2010. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol Biol 10:230. doi: 10.1186/1471-2148-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. 2013. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet 9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R, Fang L, Tan S, Yu M, Li X, He S, Wei Y, Li G, Jiang J, Wu M. 2016. Type I CRISPR-Cas targets endogenous genes and regulates virulence to evade mammalian host immunity. Cell Res 26:1273–1287. doi: 10.1038/cr.2016.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath P, Coûté-Monvoisin AC, Romero DA, Boyaval P, Fremaux C, Barrangou R. 2009. Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131:62–70. doi: 10.1016/j.ijfoodmicro.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 25.Bikard D, Hatoum-Aslan A, Mucida D, Marraffini LA. 2012. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12:177–186. doi: 10.1016/j.chom.2012.06.003. [DOI] [PubMed] [Google Scholar]
- 26.Goldberg GW, Jiang W, Bikard D, Marraffini LA. 2014. Conditional tolerance of temperate phages via transcription-dependent CRISPR-Cas targeting. Nature 514:633–637. doi: 10.1038/nature13637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiang W, Samai P, Marraffini LA. 2016. Degradation of phage transcripts by CRISPR-associated RNases enables type III CRISPR-Cas immunity. Cell 164:710–721. doi: 10.1016/j.cell.2015.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heussler GE, O’Toole GA. 2016. Friendly fire: biological functions and consequences of chromosomal targeting by CRISPR-Cas systems. J Bacteriol 198:1481–1486. doi: 10.1128/JB.00086-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Selle K, Klaenhammer TR, Barrangou R. 2015. CRISPR-based screening of genomic island excision events in bacteria. Proc Natl Acad Sci U S A 112:8076–8081. doi: 10.1073/pnas.1508525112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bloemendaal AL, Brouwer EC, Fluit AC. 2010. Methicillin resistance transfer from Staphylocccus epidermidis to methicillin-susceptible Staphylococcus aureus in a patient during antibiotic therapy. PLoS One 5:e11841. doi: 10.1371/journal.pone.0011841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boundy S, Zhao Q, Fairbanks C, Folgosa L, Climo M, Archer GL. 2012. Spontaneous staphylococcal cassette chromosome mec element excision in Staphylococcus aureus nasal carriers. J Clin Microbiol 50:469–471. doi: 10.1128/JCM.01063-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golding GR, Bryden L, Levett PN, McDonald RR, Wong A, Wylie J, Graham MR, Tyler S, Van Domselaar G, Simor AE, Gravel D, Mulvey MR. 2010. Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg Infect Dis 16:587–594. doi: 10.3201/eid1604.091435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holt DC, Holden MT, Tong SY, Castillo-Ramirez S, Clarke L, Quail MA, Currie BJ, Parkhill J, Bentley SD, Feil EJ, Giffard PM. 2011. A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3:881–895. doi: 10.1093/gbe/evr078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinnevey PM, Shore AC, Brennan GI, Sullivan DJ, Ehricht R, Monecke S, Slickers P, Coleman DC. 2013. Emergence of sequence type 779 methicillin-resistant Staphylococcus aureus harboring a novel pseudo staphylococcal cassette chromosome mec (SCCmec)-SCC-SCCCRISPR composite element in Irish hospitals. Antimicrob Agents Chemother 57:524–531. doi: 10.1128/AAC.01689-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill SR, Fouts DE, Archer GL, Mongodin EF, Deboy RT, Ravel J, Paulsen IT, Kolonay JF, Brinkac L, Beanan M, Dodson RJ, Daugherty SC, Madupu R, Angiuoli SV, Durkin AS, Haft DH, Vamathevan J, Khouri H, Utterback T, Lee C, Dimitrov G, Jiang L, Qin H, Weidman J, Tran K, Kang K, Hance IR, Nelson KE, Fraser CM. 2005. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187:2426–2438. doi: 10.1128/JB.187.7.2426-2438.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marraffini LA, Sontheimer EJ. 2008. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grissa I, Vergnaud G, Pourcel C. 2007. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pourcel C, Salvignol G, Vergnaud G. 2005. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 39.Briner AE, Lugli GA, Milani C, Duranti S, Turroni F, Gueimonde M, Margolles A, van Sinderen D, Ventura M, Barrangou R. 2015. Occurrence and diversity of CRISPR-Cas systems in the genus Bifidobacterium. PLoS One 10:e0133661. doi: 10.1371/journal.pone.0133661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manica A, Zebec Z, Teichmann D, Schleper C. 2011. In vivo activity of CRISPR-mediated virus defence in a hyperthermophilic archaeon. Mol Microbiol 80:481–491. doi: 10.1111/j.1365-2958.2011.07586.x. [DOI] [PubMed] [Google Scholar]
- 41.Peng W, Feng M, Feng X, Liang YX, She Q. 2015. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res 43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L, Garrett RA, Shah SA, Peng X, She Q. 2013. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol Microbiol 87:1088–1099. doi: 10.1111/mmi.12152. [DOI] [PubMed] [Google Scholar]
- 43.Clinical and Laboratory Standards Institute 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 9th ed. Approved standard. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 44.Valihrach L, Demnerova K. 2012. Impact of normalization method on experimental outcome using RT-qPCR in Staphylococcus aureus. J Microbiol Methods 90:214–216. doi: 10.1016/j.mimet.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 45.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol 11:941–950. doi: 10.1016/S0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 46.Lee CY, Buranen SL, Ye ZH. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]