Abstract
The alpha/beta hydrolase (ABH) superfamily is a widespread and functionally malleable protein fold recognized for its diverse biochemical activities across all three domains of life. ABH enzymes possess unexpected catalytic activity in the green plant lineage through selective alterations in active site architecture and chemistry. Furthermore, the ABH fold serves as the core structure for phytohormone and ligand receptors in the gibberellin, strigolactone, and karrikin signaling pathways in plants. Despite recent discoveries, the ABH family is sparsely characterized in plants, a sessile kingdom known to evolve complex and specialized chemical adaptations as survival responses to widely varying biotic and abiotic ecologies. This review calls attention to the ABH superfamily in the plant kingdom to highlight the functional adaptability of the ABH fold.
Introduction
Serine hydrolases are one of the most prevalent enzyme families constituting approximately 1% of the human genome [1]. The core catalytic machinery of these enzymes is composed of a conserved Ser/His/Asp(Asn) catalytic triad and a transition state stabilizing oxyanion hole provided by the peptide backbone [2]. The majority of these enzymes fall into the large α/β hydrolase (ABH) fold superfamily, first classified in 1992 [3]. This protein superfamily is found in all domains of life serving catalytic roles in primary and secondary metabolism as esterases, thioesterases, lipases, proteases, dehalogenases, haloperoxidases, and epoxide hydrolases [4–6]. Our understanding of the catalytic and non-catalytic versatility of this family continues to expand. As enzymes, ABHs are classically responsible for the hydrolysis of ester and peptide bonds. However, ABHs also participate in the breaking of carbon-carbon bonds[7], decarboxylation reactions [8–11], and the fascinating cofactor-independent dioxygenation of heteroaromatic rings [12].
The core fold of ABHs is an 8-stranded β-sheet surrounded by α-helices. Structural and functional variation is typically dependent on additional structural elements, often referred to as lid domains (Figure 1) [4]. The number of β-strands and α-helical segments vary, but the intervening loops carrying the catalytic Ser, His, and Asp/Asn residues are the most conserved features defining the ABH family. Nevertheless, across all three domains of life, ABH family members often share surprisingly low sequence identity (e.g. 6.2% identity/9.3% similarity between Pseudomonas sp. Dienelactone hydrolase and wheat serine caboxypeptidase II) while maintaining a highly conserved three-dimensional core architecture (Figure 1).
Figure 1. Structural overview of ABH proteins in green plant lineage.
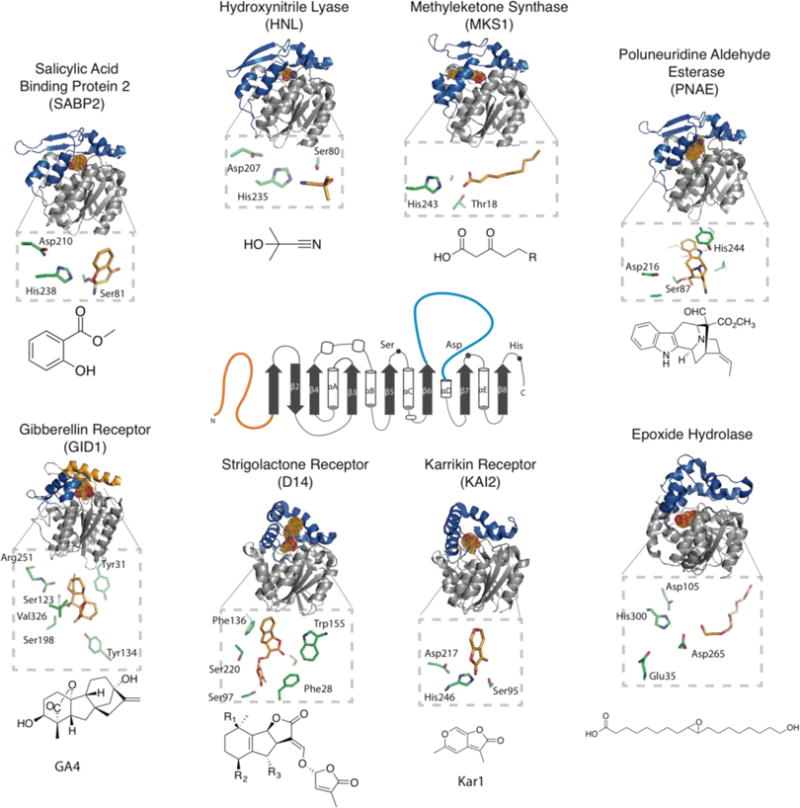
The 2D structure format of alpha/beta hydrolases is indicated in the center of the figure. N-terminal regions (orange) and inserted lid domains (blue) correspond to the 3D structures surrounding the 2D outline. The surrounding ABH structures are intended to demonstrate the diversity of currently solved structures from the green plant lineage. Some ABHs may not contain complete catalytic triads, therefore their active site region along with their catalytic residues are shown with their co-crystalized substrates. The ligand in each structure is represented as dots and colored by atom with Carbon - bright orange, Oxygen - red, and Nitrogen - blue. A close-up of the active site is shown below each, oriented to best view the ligand and catalytic/relevant residues, rather than to remain in the same orientation as the cartoon full view. Chemical structures of the biologically relevant ligands/substrates are also shown. PDB codes are as follows: SABP2 (1XKL), HNL(1SCQ), MKS1(3STU), PNAE(3GZJ), Epoxide Hydrolase (2CJP), KAI2 (3JKM), D14 (5DJ5), GID1(3EBL).
Due to their sessile nature, plants rely on a diverse repertoire of specialized, often taxon-specific metabolites and sophisticated signaling systems to communicate and survive in ecosystems challenged by a myriad of biotic and abiotic factors. The evolutionary adaptability of the ABH fold has allowed it to serve as an exemplary scaffold for the evolutionary ‘design’ of additional enzyme chemistries and biological functions in the green plant lineage. Recent discoveries include hydroxy nitrile lyases [7,13], polyneuridine aldehyde esterase [11], and methyl ketone synthase[8–10]. Notably, the α/β hydrolase fold also functions as bona fide hormone receptors in the strigolactone, karrikin-smoke receptor, and gibberellin response pathways [14–19]. Despite mounting evidence for the importance of these enzymes in plant physiology and specialized metabolism, the Arabidopsis thaliana genome alone contains hundreds of uncharacterized ABH-like genes (Figure 2). Notably, only a small number of ABH structures from all green plants have been experimentally determined. This review seeks to provide a representative overview of the diverse functions of the ABH superfamily in the green plant lineage, with a focus on unique structural elements recently uncovered in plant ABHs related to unanticipated catalytic and signaling functions. Given the large number of ABHs encoded in plant genomes, future work will undoubtedly discover additional roles for ABHs in plant metabolism, growth, development, survival and overall fitness.
Figure 2. Maximum Parsimony Tree showing all identifiable ABH-domain containing genes from A. thaliana.
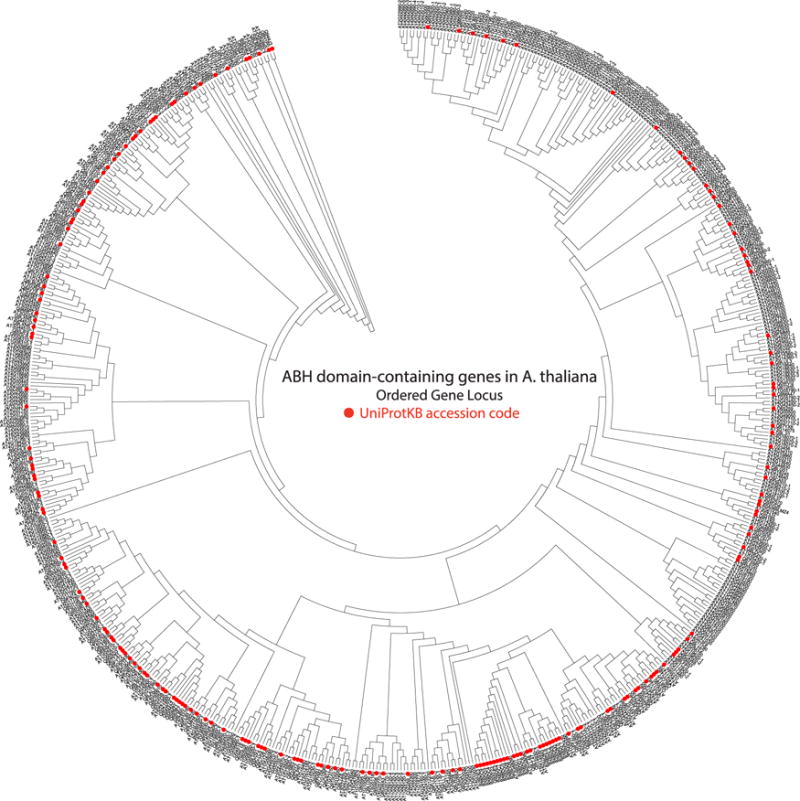
These sequences were collected from the Interpro database (Alpha/Beta hydrolase fold: IPR029058, [80]). Phylogeny inferred using the Jones-Taylor-Thornton model and all sites with gaps were included. Protein sequences that have been reviewed and entered into the Swiss-Prot have been assigned an Ordered Gene Locus name, while proteins that have not been verified remain in the TrEMBL, have not been assigned an Ordered Gene Locus, and are labeled in the tree based on their UniProtKB accession code (Red dots). Proteins that are splice forms are identified with -1,-2,-3 and so on, after the ordered gene locus. Phylogenetic and molecular evolutionary analyses were conducted using MEGA version 5. [20]
ABHs and their role in primary and specialized metabolism
ABHs are commonly associated with housekeeping roles that participate in the breakdown and recycling of cellular metabolites, processing of external nutrients and detoxification of xenobiotics [2,20–22]. In addition, ABHs play key regulatory roles in metabolism. C-terminal peptidases, such as carboxypeptidase II, are essential for modulating protein lifetime, function, and turnover [23,24]. Notably, carboxypeptidase II from wheat served as one of the founding members of the plant ABH family [3,24]. In addition to peptidases, lipases play central roles in lipid metabolism and signaling. Phospholipases, many belonging to the plant ABH family, are key to generating chemical signals at the cell membrane [21]. Phospholipases produce second messengers, such as Diacyl Glycerol (DAG) and Phosphatidic Acid (PA) to regulate cell function and respond to environmental cues [21]. Plants also depend on lipases as catalytic hubs in the biosynthesis of the volatile and phytohormone jasmonic acid [21]. The A. thaliana phospholipase, A. thaliana DAD1-like Seedling Establishment-related Lipase (AtDSEL), whose three dimensional structure has been elucidated, likely plays a critical role in mediating lipid mobilization during seed germination and growth [25].
Diversity of catalytic ABHs in the green plant lineage
In addition to serving roles in core metabolic processes, ABHs also support a variety of unique catalytic functions for defense and hormone regulation. Salicylic acid binding protein (SABP2) is an ABH esterase that regulates responses to Salicylic Acid (SA), a key hormone for plant immune responses [26–28]. SA signaling activity can be modified through carboxyl-directed methylation by a Salicylic Acid Methyl Transferase (SAMT), an S-Adenosyl-L-Methionine (SAM)-dependent Methyl Transferase (MT) in the SABATH family MTs [29–31]. Methylated SA (MeSA) curtails SA signaling and renders the core SA molecule more volatile and lipophilic. These properties afford the plant with a store of inactive SA to ensure rapid response to challenges by immune elicitors and possibly furnish enhanced transport of SA metabolites within and between plants. SABP2, initially described as a SA binding protein, was later shown to act as a MeSA esterase, hydrolyzing MeSA to produce active SA [32,33]. SABP2’s catalytic mechanism follows the canonical ABH mechanism, outlined in Figure 3. Interestingly, the reaction product SA possesses high affinity for SABP2’s binding pocket. This facile binding to the SABP active site is thought to participate in feedback inhibition to control the strength and duration of the SA response [33].
Figure 3. Proposed mechanism of alpha-beta hydrolases in four different plant alpha/beta hydrolases.
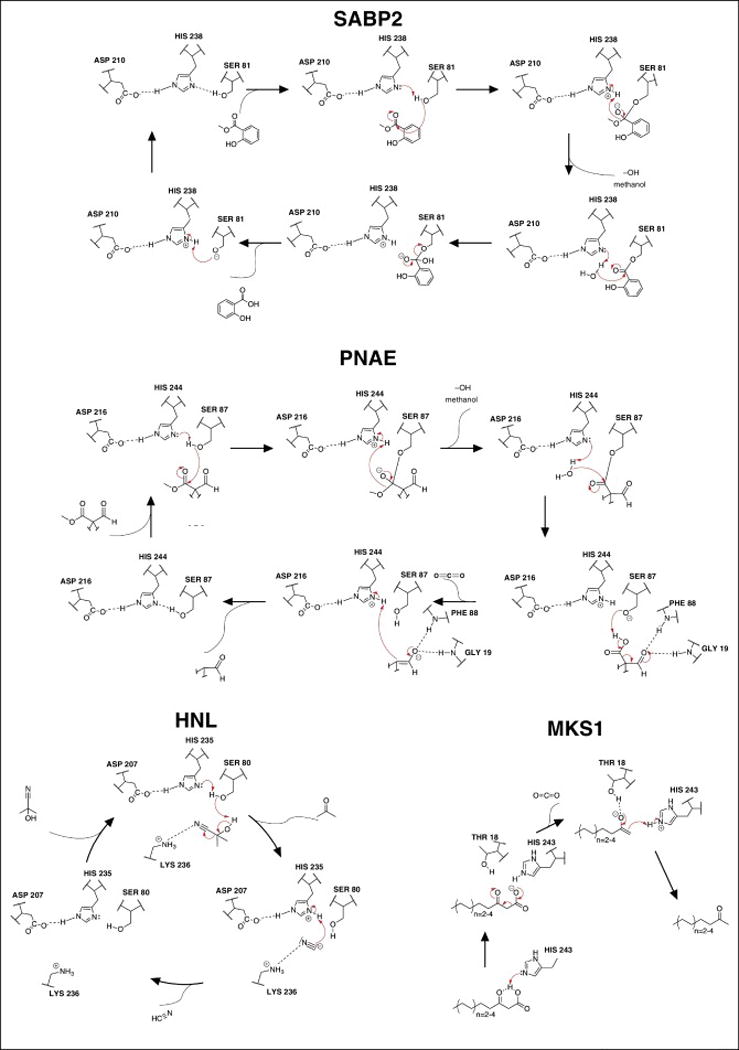
Figure highlights the catalytic diversity present in plant alpha/beta hydrolases. SABP2 serves as the canonical esterase mechanism, while hyroxynitrile lyase (HNL), polyneuridine aldehyde esterase (PNAE), and methyl ketone synthase (MKS1) serve as non-canonical alpha/beta hydrolase reaction mechanisms that have evolved in plants. The full PNAE molecule is not represented in the figure and can be found in figur
More unexpected deviations from the canonical ABH reaction scheme in green plants are typified by hydroxynitrile lyase (HNL), methyl ketone synthase 1 (MKS1), and polyneuridine aldehyde esterase (PNAE) (Figure 1). HNLs from Manihot esculenta and Hevea brasiliensis, have been studied structurally and enzymatically [7,13,34–36]. HNLs produce protective cyanide from plant phytoanticipins, cyanohydrin glucosides, during defense against herbivory [37–39]. HNLs catalyze the breaking of cyanohydrin C-C bonds to yield lethal doses of cyanide that kill chewing insects by inhibiting key enzymes of the insect electron transport chain [38]. HNL retains the typical ABH catalytic triad (Ser80/His235/Asp207), but also depends upon an emergent active site lysine residue, Lys236, that modulates the pKa of His235 and stabilizes charged reaction intermediates during substrate turnover [35,36]. The mechanism deviates significantly from standard ABHs, whereby the active site serine residue acts as a catalytic base instead of a nucleophile (Figure 3). Additionally, HNL organizes a non-canonical oxyanion hole provided by the backbone amide of Cys81 and β-hydroxyl group of Thr11. These evolutionary adaptations illustrate the propensity of the ABH fold to rapidly adapt to catalyze non-canonical reactions essential to organismal survival in challenging ecosystems [7,13,34]. Commercially, HNLs are valued industrially for their ability to catalyze the biosynthesis of cyanohydrins with high stereoselectivity [40].
The methyl ketone (MK) 2-tridecanone, initially isolated from wild tomato (Lycopersicon hirsutum glabratum), serves as a chemical defense against herbivory [41]. MK biosynthesis occurs by a two-step enzymatic process that interrupts normal fatty acid elongation. The first step, catalyzed by MKS2, a member of the hotdog fold thioesterase (TE) family, intercepts C12 and C14 β-keto acyl-ACP intermediates during fatty acid biosynthesis to release free β-keto fatty acids [42]. Structures for this TE class of plant and algal enzymes are lacking, but homologous structures from bacterial systems (PDB IDs: 2OWN, 2ESS, and 4GAK) provide reasonable structural models for plant systems.
The second step in MK production was unexpected given the assumed reactivity of β-keto acids to undergo spontaneous decarboxylation. Instead, the released β-keto fatty acids are generally stable, and decarboxylation is catalyzed by the ABH family member, MKS1 [8–10]. Surprisingly, MKS1’s active site lacks a canonical catalytic triad, possessing only a conserved histidine residue, His243, while the established active site serine and aspartic acid residues are substituted by Ala87 and Asn215, respectively. In fact, an emergent and catalytically essential threonine residue, Thr18, is juxtaposed next to His243 and trapped substrate analogs, providing the necessary hydrogen bonding architecture for the efficient decarboxylation of bound β-keto acids (Figure 3) [8]. Alanine substitutions of Thr18 or His243 severely reduce MKS1 catalytic activity. Attempts at converting MKS1 to an active thioesterase via homology-based mutations (Ala87Ser and Asn215Asp mutations) proved unsuccessful, indicating that additional features of MKS1 topological evolution drove its positive selection as a key catalytic component in defensive MK production in plants [8].
Polyneuridine aldehyde esterase (PNAE) plays a key role in the biosynthesis and chemical diversification of monoterpene indole alkaloids of the sarpagine/ajmaline family of plant alkaloids [43]. Well-known compounds from this class of molecules include sarpagine, ajmaline (anti-arrhythmia) and raumacline [43,44]. Additionally, these compounds serve in plant defense largely as insecticides [45]. PNAE resides at a key branch point in the production of downstream ajmaline/sarpagine C9 terpene indole alkaloids by converting polyneuridine aldehyde into 16-epivellosimine, CO2 and methanol [11,43,46]. PNAE retains the canonical catalytic triad, but has diverged mechanistically from traditional ABHs to catalyze a bifunctional reaction. It first utilizes the conventional esterase mechanism, releasing methanol, but then upon water-mediated hydrolysis of the covalent Ser87-substrate ester adduct catalyzes decarboxylation of the released intermediate, forming the final product 16-epivellosimine (Figure 3) [11].
The above examples indicate that the plant ABH enzyme family, including HNL, MKS1 and PNAE, catalyze a wide variety of chemical reactions to create structurally diverse classes of primary and specialized metabolites often in restricted plant taxa. Given their key roles in primary and secondary metabolism, hormone regulation and signaling, and defense, ABHs have evolved wide-ranging functional versatility during land plant evolution.
ABH fold recruitment as ligand receptors in plants
Plant hormone metabolism, temporal and spatial regulation of biosynthesis and transport, and signaling through receptors, are all critical topics of research in plant biology. In the past 10 years, there has been substantial progress in identifying and structurally characterizing the receptors for the currently identified plant hormones [19,47–50]. Additionally, plant hormones of the strigolactone family and hormone-like karrikins derived from burnt plant tissue, have been extensively characterized for their biosynthesis and signaling capacities [17,51]. Many of these newly elucidated hormone signaling pathways regulate gene expression by forming SCF E3 ubiquitin ligase complexes that degrade downstream transcriptional repressors through the 26s proteasome pathway (Figures 4 and 5), thereby activating hormone-specific suites of genes. An interesting facet to this rapidly developing field is the evolutionary selection and adaptation of the ABH fold as hormone–small molecule receptors in the gibberellin, strigolactone, and karrikin signaling pathways. The gibberellin receptor in Oryza stativa, gibberellin insensitive dwarf 1 (GID1), is an ABH that exhibits structural similarity to the hormone sensitive lipases and is a member of the plant carboxylesterase family in the ESTHER database [39,52]. Additionally, the karrikin and strigolactone receptors, KAI2 and DAD2/D14, respectively, are ABH folds belonging to the RsbQ-like family of αβ-hydrolase folds [14,15,39,53]. These three receptor families provide an evolutionary glimpse into the selection of a wide-spread protein fold re-appropriated in the green plant lineage to recognize and transduce signals from small molecules to regulate growth and development in a myriad of challenging plant ecosystems.
Figure 4. Signal transduction by the Gibberellin receptor (GID1).
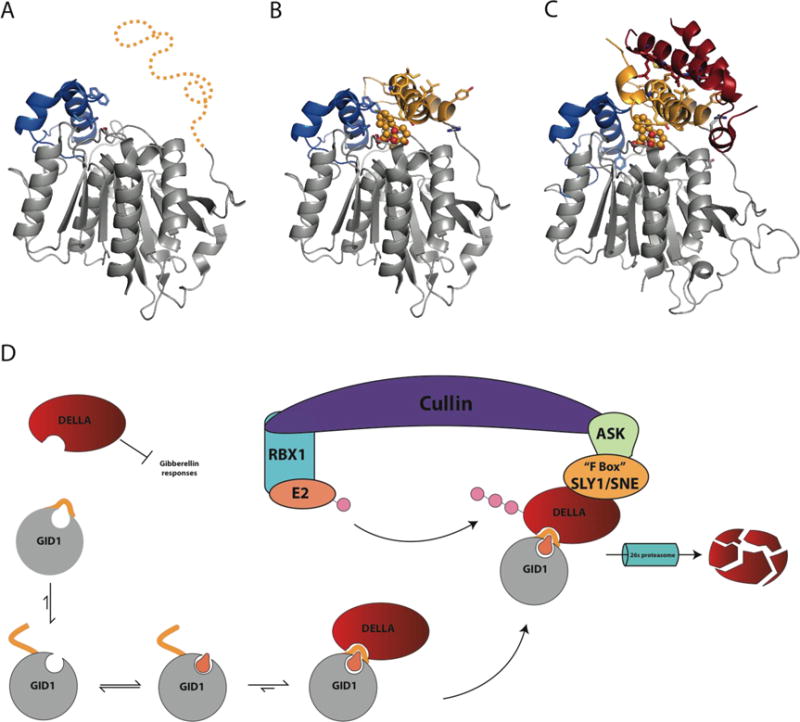
A) Proposed Apo GID1 structure (3EBL with ligand deleted) of flexible N-terminal lid region (Gold) from OsGID1 before becoming ordered due to substrate binding (PDB 3EBL). B) Crystal structure of OsGID1A in complex with gibberellin (GA4) covered by N-terminal lid region. Solvent exposed hydrophobic residues on the N-terminal lid are shown in stick (PDB 3EBL). C) Complex formed between AtGID1 and the helical N-terminal DELLA domain of GAI (red). Residues involved with the recognition and interaction between GID1 and GAI are shown as stick and colors correspond to their respective structures (red, DELLA; Gray/Gold, GID1) (PDB 2ZSI). D) The regulation of gibberellin hormone signaling by DELLA proteins and the GID1 receptor. Upon binding GA, the N-terminal region orders itself over the GA binding site, providing an interaction site for DELLA proteins. GID1-DELLA complex is then recruited to an SCF(SKP1-like Cul1 F-box) E3 ubiquitin-ligase complex, which is comprised of Cullin, the RING-H2 protein RBX1, a ubiquitin conjugating enzyme E2, and an SKP1-like protein that acts as a substrate adaptor to recruit specific FBOX proteins. FBOX proteins are named for their 50 amino acid FBOX domain and are responsible for recruiting E3 ligase substrates destined for 26s proteasomal degradation through polyubiquitinylation.[61] After recruitment of the GID1-DELLA complex to the SCF complex by SLY1/SNE, DELLA proteins are polyubiquitinylated and destroyed by the 26s proteasome, releasing GA signaling inhibition, allowing transcription of genes under GA control.
Figure 5. Structural and functional homology of Strigolactone and Karrikin receptors.
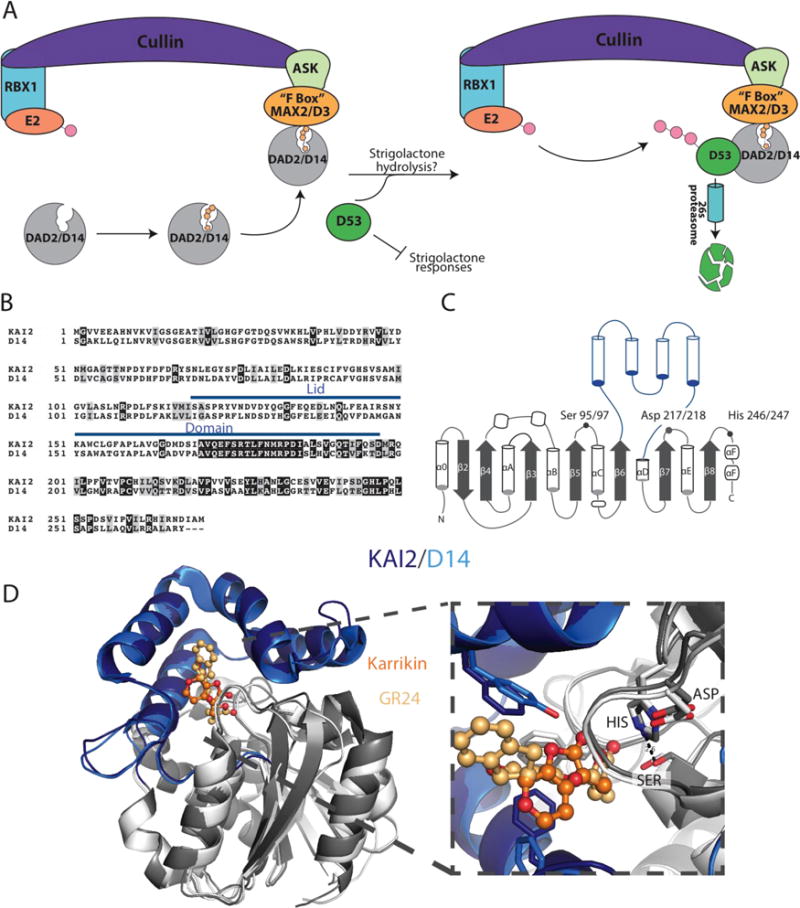
A) Proposed signaling pathway for D14/DAD2. D14/DAD2 binds strigolactone and is recruited to the SCF E3 ligase associated FBOX protein, MAX2, in order to form a complex between DAD2/D14, MAX2, and D53. SCF(SKP1-like Cul1 F-box) E3 ubiquitin-ligase complex are comprised of Cullin, the RING-H2 protein RBX1, a ubiquitin conjugating enzyme E2, and an SKP1-like protein that acts as a substrate adaptor to recruit specific FBOX proteins. FBOX proteins are named for their 50 amino acid FBOX domain, originally identified in the mammalian cyclin F protein, and are responsible for recruiting E3 ligase substrates that undergo 26s proteasomal degradation through polyubiquitinylation.[61] Upon recruitment by MAX2, D53 is then polyubiquitinylated and destroyed through the 26s proteasome pathway, thereby removing inhibition of strigolactone responses. The underlying importance of strigolactone hydrolysis and signal transduction is still not well understood and is therefore left open to interpretation. The above figure does not take into account that D14/DAD2 can independently bind with D53 or MAX2/D3 in the presence of strigolactone, which may indicate that D14/DAD2 interacts with D53 before MAX2/D3. B) Sequence alignment of A. thaliana KAI2(PDB: 4jym) and O. sativa D14 (PDB: 5dj5), with the lid domains indicated with a blue line. Identical and similar positions are indicated in black and grey boxes, respectively. C) Structural topology of KAI2 and D14 with the lid domains indicated in blue helices and the core domain in grey. The catalytic triad residues are labeled; the positions are numbered in the order, Kai2/D14. D) X-ray crystal structures of Kai2 and D14 are shown as cartoon diagrams and superimposed. Lid domains are colored in dark blue (KAI2) and light blue (D14), core domains in dark grey (KAI2) and light grey (D14). Zoom in picture of the active site shows the catalytic triads and ligands Karrikin (bright orange) and the strigolactone analog, G24 (light orange).
Gibberellin receptor, a catalytically inactive hormone receptor
Gibberellins (GAs) are an extraordinarily diverse class of diterpenoid-derived molecules that play key roles in the regulation of seed germination, plant growth, and flowering [18,54]. GA biosynthesis and regulation have been previously reviewed [18,54,55]. GA activated gene pathways are repressed by the GRAS family transcriptional repressors known as DELLA proteins, named for the conserved DELLA sequence residing on their N-terminal domains. This DELLA sequence is essential for productive DELLA protein/GID1 interactions upon binding of GA to GID1. Recent structural studies have shed light on the substrate recognition and signal transduction mechanisms of the GID1 receptor [19,56–58]. Structure and sequence comparisons demonstrate that GID1 is an ABH family member with structural similarity to the hormone sensitive lipase ABH subfamily [39,52]. Interestingly, GID1 lacks a catalytic histidine residue that is part of the canonical ABH family catalytic triad. Moreover, GID1 does not appear to possess catalytic activity, suggesting that it likely acts only as a receptor with no general biosynthetic or metabolic function [19,52,59].
The GID1 structure from Oryza sativa in complex with GA4, a physiologically active GA (Figure 1), and the ternary complex of GID1-GA3-GAI (DELLA protein) from A. thaliana provide atomic resolution insights into the GA-mediated mechanism of hormone recognition and signal transduction (Figure 4) [19,56,59]. Based on difficulties in crystallizing the GID1 apo structure and proteolysis studies, researchers concluded that the N-terminal region is disordered until the receptor binds its GA substrate [58,60]. Unfortunately, no structures are available for apo GID1, as attempts to crystallize the protein in the absence of its ligand were unsuccessful [19]. Upon GA binding, the apparently disordered N-terminal domain undergoes a conformational transition to an ordered state, wrapping over the GA molecule completing the top and back wall of the GA binding pocket (Figure 4B). Interestingly, GA3/GA4 binding to GID1 is enhanced by the coordination of a GA carboxylate moiety by GID1’s predicted oxyanion hole. Notably, mutations to these residues conforming to a canonical ABH oxyanion hole significantly decrease GA binding affinity [59]. Solvent exposed hydrophobic residues on the GID1 N-terminal domain serve as the recognition site for DELLA proteins. The interface between GID1 and DELLA buries ~2,600 Å2 and is largely composed of hydrophobic interactions between GID1’s N-terminal region and the conserved DELLA and LExLE motifs of GAI. Upon complex formation, GID1-GAI recruits the SCF E3 ubiquitin ligase complex through the FBOX protein SLY1. SCF E3 then polyubiquitinylates DELLA, marking it for degradation by the 26S proteasome (Figure 5) [55,61]. Destruction of DELLA GRAS family transcriptional repressors then relieves the negative regulation of GA-controlled gene expression.
Strigolactone and karrikin receptors KAI2 and D14
Strigolactones and karrikins are recently discovered butenolide-containing plant signaling molecules that have garnered much attention in the plant community [62–64]. Strigolactones, initially discovered for their ability to induce germination of the parasitic plant Striga hermonthica, serve as endogenous plant hormones that control shoot branching and as excreted signaling molecules to mediate symbiosis with arbuscular mycorrhizal fungi [51,53,65]. Karrikins are smoke derived compounds shown to stimulate seed germination after fires [15,53,64,66]. Interestingly, karrikins do not appear to be synthesized in plants, but form during the burning of plant tissue. Most notably, they are assumed to mimic a yet to be identified endogenous plant signaling molecule [67,68]. The biosynthesis, formation, and signaling pathways of strigolactones and karrikins have been reviewed previously [62–64,69].
Early studies in Petunia hybrid (Ph), Arabidopsis thaliana (At), and Oryza sativa (Os) identified mutations that resulted in dwarfed plants with highly branched phenotypes classified as decreased apical dominance (DAD), more axillary growth (MAX), dwarf (D), or high tillering dwarf (htd) mutants, respectively. Similarly, mutations that resulted in karrikin insensitivity (KAI) and light hyposensitivity in Arabidopsis led to the discovery of the karrikin receptor KAI2 [62,64]. With regard to the strigolactone and karrikin receptors, D14/DAD2 and KAI2, respectively, they exhibit high sequence identity (~50%, see sequence alignment in Figure 5B), and appear to converge on the same FBOX protein signaling partner, MAX2/D3. However, they are differentially regulated by distinct small molecules resulting in non-overlapping plant growth responses. The latter roles are most clearly discernable by disparate phenotypic effects in plants lacking the respective D14/DAD2 or KAI2 receptors. [70–72]
While KAI2 responds to the exogenous presence of smoke-derived karrikins, the unique phenotypes of KAI2 knock-out plants suggests KAI2 recognizes an endogenous plant molecule possibly emergent during early plant evolution [71,73]. Recently, structures of these ABHs provided ligand-dependent architectural transitions and catalytic hypotheses for these ABH folds possessing canonical Ser/His/Asp catalytic triads (Figure 5C). Notably, this includes a conserved four-helical, V-shaped lid domain important for hormone binding/recognition and signal transduction mediated by protein-protein interactions (Figure 5D) [14–16,74]. Comparison of the two receptors shows a smaller binding pocket for KAI2 than D14, but overall the structures superimpose with an RMSD of ~1.3 Å.
While both receptors retain catalytic activity for the hydrolysis of reactive and non-physiological substrates, namely p-nitrophenylphosphate (PNP) esters, a common hydrolase assay, only D14/DAD2 catalyzes hydrolysis of the synthetic strigolactone GR24, albeit with significantly compromised turnover [17]. Neither KAI2 nor D14 appear to modify karrikins, although a futile cycle of catalytic ring opening and closing is possible [70]. It is interesting to note that AtKAI2 responded to racemic mixtures of GR24 but not to an enantiomer of GR24, specifically GR245DS. GR245DS mimics the naturally biosynthesized enantiomer of strigolactones and is specifically turned over by D14/DAD2 ABHs [70]. Biochemically, AtKAI2 catalyzes hydrolysis of the non-natural enantiomeric analog GR24ent5DS, suggesting the endogenous ligand of KAI2 is likely to possess a butenolide-like moiety susceptible to turnover by the catalytic triad of KAI2 [72].
Yeast two-hybrid studies indicate that Petunia hybrida DAD2 binds PhMAX2, an SCF E3 ligase FBOX protein, in the presence of strigolactones. Interestingly, mutation of the active site Ser96 to Ala in DAD2 abolishes detectable interactions between DAD2 and PhMAX2A, indicating that the coupling of hormone recognition and hydrolysis likely plays a central role in mediating strigolactone-dependent signal transduction in the D14/DAD2 ABH fold subfamily [14]. It is important to note that strigolactone hydrolysis products themselves do not appear to possess in vivo activity [14]. Despite the noted lack of in vivo activity, Nakumara et al. obtained a complex structure of D14 with a hydrolyzed product of strigolactone, D-OH. They also found extremely high concentrations of D-OH (at 50 μM) could slightly rescue the strigolactone biosynthetic mutant phenotype. They proposed D-OH bound D14 is important for signal transduction, suggesting that hydrolysis of full length strigolactones is important for D14’s signal transducing role [75].
The strigolactone and karrikin signaling pathways are reminiscent of those of auxin, jasmonic acid (JA), and GAs, where receptors are FBOX proteins (auxin and JA) or bind to FBOX proteins (GA) in SCF E3 ubiquitin ligase complexes (Figure 5A) to regulate the degradation of downstream transcriptional repressors. Interestingly, D14/DAD2 and KAI2 appear to share the same E3 ligase target FBOX protein, D3/MAX2. Nevertheless, ongoing studies are needed to mechanistically elucidate how these two receptors independently regulate signals downstream of D3/MAX2. Recent work has identified D53 in Oryza sativa as a negative regulator of the strigolactone response, and published results indicate that activation of D14 by strigolactones induce the degradation of D53, relieving repression in the absence of strigolactones [76]. Similarly, work by Stanga et al. identified SMAX1, a D53 homologue in Arabidopsis, as a KAI2 signal regulator [77].
Work elucidating the underlying mechanism of strigolactone signaling indicates that D14 can interact with MAX2/D3 or D53 independently of one another in the presence of strigolactones. While the exact sequence of molecular events accompanying ligand recognition, catalysis, protein conformational changes, and formation of protein-protein complexes is still a subject of debate, D14 appears to become more thermally unstable in the presence of strigolactones and its signaling partners MAX2/D15 or D53 resulting in ligand-dependent target degradation [14,74]. To the best of our knowledge, regulation of D14/DAD2/KAI2 catalytic activity through protein-protein interactions remains unresolved and would represent an unprecedented function of ABH fold enzymes. Additionally, how the timing and chemical structures accompanying the complex cycle of ABH hydrolytic mechanisms modulates signaling and signal termination through D14/DAD2/KAI2s are a subject of intense current debate. The integration of structural biology with genetics, chemistry and biochemistry will very likely be key to unraveling the events accompanying ligand recognition, catalysis, signaling, and evolution in the strigolactone/karrikin family of ABH receptors.
Conclusion
The evolution and continuing expansion of protein folds possessing diverse chemistries associated with ligand recognition and catalysis has been an essential feature of the success of sessile plant colonization of the terrestrial earth. This is particularly notable with regard to the diversity of forms and mechanisms of chemical adaptation to some of the most challenging biotic and abiotic features of global ecosystems. In addition to the already broad spectrum of ABH catalytic reactions, recent work on two soil dwelling bacteria in contact with plant roots uncovered cofactor independent oxygenases unexpectedly possessing the ABH fold, namely Arthrobacter nitroguajacolicus Rü61a 1-H-3-hydroxy-4-oxoquinaldine 2,4-dioxygenase (HOD) and Pseudomonas putida 33/1 1-H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (QDO). Both catalyze the dioxygenation of heteroaromatic rings, however, they employ a nucleophile/histidine/acid triad in which the aspartic acid is transmigrated to the end of strand β6 from its standard position at the end of strand β7 (Figure 6) [12]. Research on ABH fold proteins in green plants has so far uncovered notable examples of the kind of catalytic diversity represented by HOD and QDO and highly conserved three dimensional structures (Figures 1 and 6).
Figure 6. Superposition of KAI2 and QDO shows structural conservation and functional diversity.
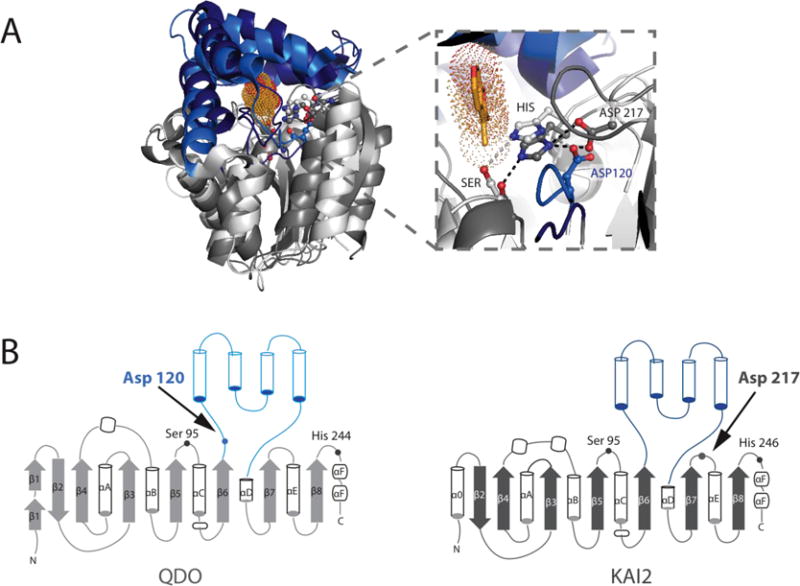
A) Sequence-independent structural superposition of A. thaliana KAI2 (PDB: 3JKM) and Pseudomonas putida 1H-3-hydroxy-4-oxoquinoline 2,4-dioxygenase (QDO, PDB: 3IBT), with core domains shown in dark and light grey, and the lid domains shown in dark and light blue, respectively. Karrikin is in bright orange sticks with dots in the KAI2 active site. Zoom in of the active site highlights that despite the distance in primary sequence, the two enzymes have their catalytic triad Aspartates in largely the same location, enabling efficient H-bonding with their respective catalytic Histidines. B) Topology diagrams showing various differences in the core domains of both enzymes and the differences in Catalytic Aspartate placement within the structure.
A cursory survey of the Arabidopsis genome alone reveals the presence of well-over 638 genes likely encoding ABH folds, representing ~2.3% of the ~27,379 annotated protein coding genes, nearly all of which have yet to be studied [78,79]. As in other organisms, the ABHs will continue to provide a treasure trove of proteins likely to encode unanticipated catalytic and/or receptor functions. Much work remains in order to characterize the full breadth of plant ABH biochemical activities and biological functions across widely divergent taxa of plants. Given the sessile nature of plants and their dependence on specialized metabolism and small molecule signaling for survival and overall fitness in extreme environments, plant ABH research will provide a greater understanding of the adaptive evolution of the ABH fold with likely translational applications to sustainable agriculture, ecosystem revitalization, and global health.
Finally, throughout this review we have described ABH superfamily members assuming that, since they all share highly similar core three dimensional architectures and fold topologies, they also trace their ancestry back to a common aboriginal ABH gene. While it is convenient to infer that all differences among the ABH family are ultimately the result of divergent evolution from a single ABH ancestral fold, this is not necessarily the only explanation, given the extreme breadth of sequence space sampled by ABH family members across all three domains of life. Indeed, the vast degree of sequence, biochemical and functional diversity encoded by the ABH fold superfamily is a testament to the relative disconnection between fold/biological function/biochemical activity and nucleotide sequence. Put another way, commonality of fold is neither a proxy for related primary sequences nor shared ancestry. This being the case, it is an intriguing possibility that the modern members of the ABH fold family trace their genetic ancestry back to several, independent ancestral starting points. In support of this notion, remarkable advances in computational protein design approaches, including the Rosetta software suite, have established that protein folds can be accurately designed ‘from scratch’ without reliance on a natural protein sequence to serve as their common ancestor [80,81]. Whether we will be able to infer with statistical confidence the likelihood of such events, and, if so, what would be the number of such starting points are provocative questions to consider in contemporary studies of protein/enzyme design and evolution.
Acknowledgments
Research in our laboratories is supported by NSF EEC-0813570 and MCB- 0645794 to JPN and NIH GM095970 and NSF IOS-1516156 to MDB. JTM acknowledges training support from the NIH Molecular Biophysics Training Grant T32GM008326. YS is a JSPS Postdoctoral Fellow for Research Abroad. JPN is an investigator with the Howard Hughes Medical Institute.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
● of special interest
●● of outstanding interest
- 1●.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: Serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. This review describes the prevalence of ABH proteins and presents the application of reactive probes to evaluate their activity and determine their physiological role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Long JZ, Cravatt BF. The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem Rev. 2012;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David L, Cheah E, Cygler M, Dijkstra B, Frolow F, Sybille M, Harel M, Remington SJ, Silman I, Schrag J, et al. The alpha/beta hydrolase fold. Protein Eng Des Sel. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 4.Nardini M, Dijkstra BW. Chemistry B, Groningen AG: α / β Hydrolase fold enzymes : the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 5.De Simone G, Menchise V, Manco G, Mandrich L, Sorrentino N, Lang D, Rossi M, Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus. J Mol Biol. 2001;314:507–18. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- 6.Klenk H-P, Clayton RA, Tomb J-F, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 7.Gruber K, Gugganig M, Wagner UG, Kratky C. Atomic resolution crystal structure of hydroxynitrile lyase from Hevea brasiliensis. Biol Chem. 1996;380:993–1000. doi: 10.1515/BC.1999.123. [DOI] [PubMed] [Google Scholar]
- 8●.Auldridge ME, Guo Y, Austin MB, Ramsey J, Fridman E, Pichersky E, Noel JP. Emergent Decarboxylase Activity and Attenuation of / -Hydrolase Activity during the Evolution of Methylketone Biosynthesis in Tomato. Plant Cell. 2012;24:1596–1607. doi: 10.1105/tpc.111.093997. This paper provides the structural characterization of MKS1, verifying the absence of a canonical alpha/beta hydrolase triad as well the elucidation of key residues in involved in MKS1 decarboxylation reaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E. Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 2009;151:1952–1964. doi: 10.1104/pp.109.146415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridman E, Wang J, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E. Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell. 2005;17:1252–67. doi: 10.1105/tpc.104.029736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang L, Hill M, Wang M, Panjikar S, Stockigt J. Structural basis and enzymatic mechanism of the biosynthesis of C 9from C10-monoterpenoid indole alkaloids. Angew Chemie - Int Ed. 2009;48:5211–5213. doi: 10.1002/anie.200900150. [DOI] [PubMed] [Google Scholar]
- 12●.Steiner RA, Janssen HJ, Roversi P, Oakley AJ, Fetzner S. Structural basis for cofactor-independent dioxygenation of N-heteroaromatic compounds at the alpha/beta-hydrolase fold. Proc Natl Acad Sci U S A. 2010;107:657–62. doi: 10.1073/pnas.0909033107. Paper provides structural and mechanistic insights into the unprecedented cofactor independent dioxygenation of aromatic N-heterocyclic compounds. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gruber K, Gugganig M, Wagner UG, Kratky C. Atomic resolution crystal structure of hydroxynitrile lyase from Hevea brasiliensis. Biol Chem. 1999;380:993–1000. doi: 10.1515/BC.1999.123. [DOI] [PubMed] [Google Scholar]
- 14●.Hamiaux C, Drummond RSM, Janssen BJ, Ledger SE, Cooney JM, Newcomb RD, Snowden KC. DAD2 is an a/b hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr Biol. 2012;22:2032–2036. doi: 10.1016/j.cub.2012.08.007. The structural elucidation of the strigolactone receptor from petunia, DAD2, and the role strigolactones play in mediating DAD2 interactions with the FBOX protein PtMAX2. [DOI] [PubMed] [Google Scholar]
- 15●.Guo Y, Zheng Z, La Clair JJ, Chory J, Noel JP. Smoke-derived karrikin perception by the / -hydrolase KAI2 from Arabidopsis. Proc Natl Acad Sci. 2013;110:8284–8289. doi: 10.1073/pnas.1306265110. Co-crystal structure of the karrikin receptor, KAI2, in complex with the karrikin, KAR1. This work highlights the important structural characteristics and features for karrikin binding and KAI2 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16●.Kagiyama M, Hirano Y, Mori T, Kim SY, Kyozuka J, Seto Y, Yamaguchi S, Hakoshima T. Structures of D14 and D14L in the strigolactone and karrikin signaling pathways. Genes to Cells. 2013;18:147–160. doi: 10.1111/gtc.12025. Structural elucidation of D14 (strigolactone receptor) and D14L (karrikin receptor) from Oryza sativa. Additionally, work provides direct evidence of strigolactone binding to D14 using isothermal titration calorimetry. [DOI] [PubMed] [Google Scholar]
- 17●.Zhao L-H, Zhou XE, Wu Z-S, Yi W, Xu Y, Li S, Xu T-H, Liu Y, Chen R-Z, Kovach A, et al. Crystal structures of two phytohormone signal-transducing α/β hydrolases: karrikin-signaling KAI2 and strigolactone-signaling DWARF14. Cell Res. 2013;23:436–439. doi: 10.1038/cr.2013.19. Elucidation of strigolactone and karrikin receptor structures from Arabidopsis thaliana (AtD14 and AtKAI2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hedden P, Thomas SG. Gibberellin biosynthesis and its regulation. Biochem J. 2012;444:11–25. doi: 10.1042/BJ20120245. [DOI] [PubMed] [Google Scholar]
- 19.Shimada A, Ueguchi-Tanaka M, Nakatsu T, Nakajima M, Naoe Y, Ohmiya H, Kato H, Matsuoka M. Structural basis for gibberellin recognition by its receptor GID1. Nature. 2008;456:520–523. doi: 10.1038/nature07546. [DOI] [PubMed] [Google Scholar]
- 20.Gershater MC, Edwards R. Regulating biological activity in plants with carboxylesterases. Plant Sci. 2007;173:579–588. [Google Scholar]
- 21.Canonne J, Froidure-Nicolas S, Rivas S. Phospholipases in action during plant defense signaling. Plant Signal Behav. 2011;6:13–8. doi: 10.4161/psb.6.1.14037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satoh T, Hosokawa M. The mammalian carboxylesterases: from molecules to functions. Annu Rev Pharmacol Toxicol. 1998;38:257–288. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- 23.van der Hoorn RaL. Plant proteases: from phenotypes to molecular mechanisms. Annu Rev Plant Biol. 2008;59:191–223. doi: 10.1146/annurev.arplant.59.032607.092835. [DOI] [PubMed] [Google Scholar]
- 24.Liao DI, Breddam K, Sweet RM, Bullock T, Remington SJ. Refined atomic model of wheat serine carboxypeptidase II at 2.2-A resolution. Biochemistry. 1992;31:9796–9812. doi: 10.1021/bi00155a037. [DOI] [PubMed] [Google Scholar]
- 25.Kim EY, Seo YS, Kim WT. AtDSEL, an Arabidopsis cytosolic DAD1-like acylhydrolase, is involved in negative regulation of storage oil mobilization during seedling establishment. J Plant Physiol. 2011;168:1705–1709. doi: 10.1016/j.jplph.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Park S-W, Kaimoyo E, Kumar D, Mosher S, Klessig DF. Methyl Salicylate Is a Critical Mobile Signal for Plant Systemic Acquired Resistance. Science (80-) 2007;318:113–116. doi: 10.1126/science.1147113. [DOI] [PubMed] [Google Scholar]
- 27.Tripathi D, Jiang YL, Kumar D. SABP2, a methyl salicylate esterase is required for the systemic acquired resistance induced by acibenzolar-S-methyl in plants. FEBS Lett. 2010;584:3458–3463. doi: 10.1016/j.febslet.2010.06.046. [DOI] [PubMed] [Google Scholar]
- 28●●.Kumar D. Salicylic acid signaling in disease resistance. Plant Sci. 2014;228:127–134. doi: 10.1016/j.plantsci.2014.04.014. A concise review on the biosynthesis, signaling, and function of salicylic acid in the plant disease resistance. The authors discuss classic signaling pathways and binding proteins as well as newly discovered salicylic acid receptors and effectors such as nonexpressor of pathogensis related proteins (NPRs). [DOI] [PubMed] [Google Scholar]
- 29.Noel JP, Dixon RA, Pichersky E, Zubieta C, Ferrer J-L. Chapter two Structural, functional, and evolutionary basis for methylation of plant small molecules. Recent Adv Phytochem. 2003;37:37–58. [Google Scholar]
- 30.Tieman D, Zeigler M, Schmelz E, Taylor MG, Rushing S, Jones JB, Klee HJ. Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 2010;62:113–123. doi: 10.1111/j.1365-313X.2010.04128.x. [DOI] [PubMed] [Google Scholar]
- 31.Zubieta C, Ross JJR, Koscheski P, Yang Y, Pichersky E, Noel JP. Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell. 2003;15:1704–1716. doi: 10.1105/tpc.014548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar D, Klessig DF. High-affinity salicylic acid-binding protein 2 is required for plant innate immunity and has salicylic acid-stimulated lipase activity. Proc Natl Acad Sci. 2003;100:16101–16106. doi: 10.1073/pnas.0307162100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forouhar F, Yang Y, Kumar D, Chen Y, Fridman E, Park SW, Chiang Y, Acton TB, Montelione GT, Pichersky E, et al. Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc Natl Acad Sci U S A. 2005;102:1773–8. doi: 10.1073/pnas.0409227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lauble H, Miehlich B, Förster S, Kobler C, Wajant H, Effenberger F. Structure determinants of substrate specificity of hydroxynitrile lyase from Manihot esculenta. Protein Sci. 2002;11:65–71. doi: 10.1110/ps.33702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gruber K, Gartler G, Krammer B, Schwab H, Kratky C. Reaction mechanism of hydroxynitrile lyases of the α/ β-hydrolase superfamily: The three-dimensional structure of the transient enzyme-substrate complex certifies the crucial role of Lys236. J Biol Chem. 2004;279:20501–20510. doi: 10.1074/jbc.M401575200. [DOI] [PubMed] [Google Scholar]
- 36.Gartler G, Kratky C, Gruber K. Structural determinants of the enantioselectivity of the hydroxynitrile lyase from Hevea brasiliensis. J Biotechnol. 2007;129:87–97. doi: 10.1016/j.jbiotec.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 37.Gregory RJH. Cyanohydrins in Nature and the Laboratory: Biology, Preparations, and Synthetic Applications. Chem Rev. 1999;99:3649–3682. doi: 10.1021/cr9902906. [DOI] [PubMed] [Google Scholar]
- 38.Zagrobelny M, Bak S, Rasmussen AV, Jorgensen B, Naumann CM, Muller BL. Cyanogenic glucosides and plant-insect interactions. Phytochemistry. 2004;65:293–306. doi: 10.1016/j.phytochem.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 39●●.Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A. ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins. Nucleic Acids Res. 2004;32:D145–D147. doi: 10.1093/nar/gkh141. The ESTHER database holds all 45990 non-redundant alpha/beta hydrolase protein/gene sequences. These protein sequences have been allocated into 187 subfamilies that can be accessed and searched for based on a variety of characteristics on the ESTHER website ( http://bioweb.ensam.inra.fr/ESTHER/general?what=index). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bracco P, Busch H, von Langermann J, Hanefeld U. Enantioselective synthesis of cyanohydrins catalysed by hydroxynitrile lyases – a review. Org Biomol Chem. 2016;14:6375–6389. doi: 10.1039/c6ob00934d. [DOI] [PubMed] [Google Scholar]
- 41.Williams WG, Kennedy GG, Yamamoto RT, Thacker JD, Bordner J. 2-Tridecanone : A Naturally Occurring Insecticide from the Wild Tomato Lycopersicon hirsutum f. glabratum Science. 1980;207:888–889. doi: 10.1126/science.207.4433.888. [DOI] [PubMed] [Google Scholar]
- 42.Yu G, Nguyen TTH, Guo Y, Schauvinhold I, Auldridge ME, Bhuiyan N, Ben-Israel I, Iijima Y, Fridman E, Noel JP, et al. Enzymatic functions of wild tomato methylketone synthases 1 and 2. Plant Physiol. 2010;154:67–77. doi: 10.1104/pp.110.157073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Connor SE, Maresh JJ. Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep. 2006;23:532–547. doi: 10.1039/b512615k. [DOI] [PubMed] [Google Scholar]
- 44.Rolf S, Bruns HJ, Wichter T, Kirchhof P, Ribbing M, Wasmer K, Paul M, Breithardt G, Haverkamp W, Eckardt L. The ajmaline challenge in Brugada syndrome: Diagnostic impact, safety, and recommended protocol. Eur Heart J. 2003;24:1104–1112. doi: 10.1016/s0195-668x(03)00195-7. [DOI] [PubMed] [Google Scholar]
- 45.Menke F, Parchmann S, Mueller M, Kijne J, Memelink J. Involvement of the octadecanoid pathway and protein phosphorylation in fungal elicitor-induced expression of terpenoid indole alkaloid biosynthetic genes in catharanthus roseus. Plant Physiol. 1999;119:1289–96. doi: 10.1104/pp.119.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pfitzner A, Stockigt J. a Key Enzyme in the Biosynthesis of Sarpagine / Ajmaline Type Alkaloids. 1983;48:221–227. doi: 10.1055/s-2007-969924. [DOI] [PubMed] [Google Scholar]
- 47.Dharmasiri N, Dharmasiri S, Estelle M. The F-box protein TIR1 is an auxin receptor. Nature. 2005;435:441–5. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 48.Miyazono K-I, Miyakawa T, Sawano Y, Kubota K, Kang H-J, Asano A, Miyauchi Y, Takahashi M, Zhi Y, Fujita Y, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–14. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 49.Yin P, Fan H, Hao Q, Yuan X, Wu D, Pang Y, Yan C, Li W, Wang J, Yan N. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–6. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 50.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Umehara M, Hanada A, Yoshida S, Akiyama K, Arite T, Takeda-Kamiya N, Magome H, Kamiya Y, Shirasu K, Yoneyama K, et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature. 2008;455:195–200. doi: 10.1038/nature07272. [DOI] [PubMed] [Google Scholar]
- 52.Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow TY, Hsing YI, Kitano H, Yamaguchi I, et al. GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature. 2005;437:693–698. doi: 10.1038/nature04028. [DOI] [PubMed] [Google Scholar]
- 53.Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun E a, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, et al. Strigolactone inhibition of shoot branching. Nature. 2008;455:189–194. doi: 10.1038/nature07271. [DOI] [PubMed] [Google Scholar]
- 54●.Davière J-M, Achard P. Gibberellin signaling in plants. Development. 2013;140:1147–51. doi: 10.1242/dev.087650. A short overview of the recent advances in understanding the molecular mechanisms of Gibberellin signaling in plants. Review covers the role of GID1 and DELLA proteins in mediating hormone signaling. [DOI] [PubMed] [Google Scholar]
- 55●●.Nelson SK, Steber CM. Gibberellin hormone signal perception: down-regulating DELLA repressors of plant growth and development. Annu Plant Rev. 2016;49:153–188. A more recent and in depth review of the gibberellin signaling pathway in plants. Review includes discussions and recent work on GID1 and DELLA proteins as well as post translational modifications and subtle signaling caveats that may attenuate these signals. [Google Scholar]
- 56.Murase K, Hirano Y, Sun T, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. [DOI] [PubMed] [Google Scholar]
- 57.Hirano K, Asano K, Tsuji H, Kawamura M, Mori H, Kitano H, Ueguchi-Tanaka M, Matsuoka M. Characterization of the molecular mechanism underlying gibberellin perception complex formation in rice. Plant Cell. 2010;22:2680–2696. doi: 10.1105/tpc.110.075549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ueguchi-Tanaka M, Matsuoka M. The perception of gibberellins: clues from receptor structure. Curr Opin Plant Biol. 2010;13:503–508. doi: 10.1016/j.pbi.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 59.Ueguchi-Tanaka M, Nakajima M, Katoh E, Ohmiya H, Asano K, Saji S, Hongyu X, Ashikari M, Kitano H, Yamaguchi I, et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell. 2007;19:2140–55. doi: 10.1105/tpc.106.043729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Achard P, Genschik P. Releasing the brakes of plant growth: How GAs shutdown della proteins. J Exp Bot. 2009;60:1085–1092. doi: 10.1093/jxb/ern301. [DOI] [PubMed] [Google Scholar]
- 61.Chen L, Hellman H. Plant E3 Ligases: Flexible Enzymes in a Sessile World. Mol Plant. 2013;6:1388–1404. doi: 10.1093/mp/sst005. [DOI] [PubMed] [Google Scholar]
- 62●.Seto Y, Yamaguchi S. Strigolactone biosynthesis and perception. Curr Opin Plant Biol. 2014;21:1–6. doi: 10.1016/j.pbi.2014.06.001. A cogent review on karrikin/strigolactone biosynthesis and signaling. The authors outline the discovery and biosynthetic pathway of carlactone as a strigolactone precursor as well as a short summary of the importance of strigolactone hydrolysis and binding in signal transduction. [DOI] [PubMed] [Google Scholar]
- 63●.Al-Babili S, Bouwmeester HJ. Strigolactones, a novel carotenoid-derived plant hormone. Annu Rev Plant Biol. 2015;66:161–86. doi: 10.1146/annurev-arplant-043014-114759. A more in depth review of strigolactone biosynthesis and function in plants. In addition to the recent work elucidating strigolactone biosynthesis, this review also provides a more focused analysis on environmental factors that influence strigolactone pathways as well as their interactions with other hormones, such as auxin and abscisic acid. [DOI] [PubMed] [Google Scholar]
- 64●●.Morffy N, Faure L, Nelson DC. Smoke and Hormone Mirrors: Action and Evolution of Karrikin and Strigolactone Signaling. Trends Genet. 2016;32:176–88. doi: 10.1016/j.tig.2016.01.002. A recent and up to date review on the discovery, function, and perception of karrikins in plants. Review compares the signaling pathways of strigolactones and karrikins. Additionally, the authors provide a detailed discussion of the evolution of karrikin and strigolactone signaling pathways in parasitic plants and their hosts. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Akiyama K, Matsuzaki K, Hayashi H. Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature. 2005;435:824–827. doi: 10.1038/nature03608. [DOI] [PubMed] [Google Scholar]
- 66.Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006;4:1239–1247. doi: 10.1371/journal.pbio.0040226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Flematti GR, Waters MT, Scaffidi A, Merritt DJ, Ghisalberti EL, Dixon KW, Smith SM. Karrikin and cyanohydrin smoke signals provide clues to new endogenous plant signaling compounds. Mol Plant. 2013;6:29–37. doi: 10.1093/mp/sss132. [DOI] [PubMed] [Google Scholar]
- 68●●.Conn CE, Nelson DC. Evidence that KARRIKININSENSITIVE2 (KAI2) Receptors may Perceive an Unknown Signal that is not Karrikin or Strigolactone. Front Plant Sci. 2016;6:1–7. doi: 10.3389/fpls.2015.01219. Paper reviews recent work that provide support for an unkown endogenous hormone responsible for KAI2 signaling in plants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xie X, Yoneyama K. Strigolactone Story. Annu Rev Phytopathol. 2010;48:119–139. doi: 10.1146/annurev-phyto-073009-114453. [DOI] [PubMed] [Google Scholar]
- 70.Scaffidi A, Waters MT, Bond CS, Dixon KW, Smith SM, Ghisalberti EL, Flematti GR. Exploring the molecular mechanism of karrikins and strigolactones. Bioorganic Med Chem Lett. 2012;22:3743–3746. doi: 10.1016/j.bmcl.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 71.Waters MT, Nelson DC, Scaffidi A, Flematti GR, Sun YK, Dixon KW, Smith SM. Specialisation within the DWARF14 protein family confers distinct responses to karrikins and strigolactones in Arabidopsis. Development. 2012;139:1285–95. doi: 10.1242/dev.074567. [DOI] [PubMed] [Google Scholar]
- 72●●.Waters MT, Scaffidi A, Moulin SLY, Sun YK, Flematti GR, Smith SM. A Selaginella moellendorffii Ortholog of KARRIKIN INSENSITIVE2 Functions in Arabidopsis Development but Cannot Mediate Responses to Karrikins or Strigolactones. Plant Cell. 2015;27:1–21. doi: 10.1105/tpc.15.00146. Study provides analysis of the independence of KAI2 and D14 signalling by analyzing differnces in promoters as well as demonstrating that KAI2 catalytic triad is important for signaling. Additionally, the group performed complementation assays with parasitic plant KAI2 homologues, which in one case rescued the leaf development phenotypes of Arabodopsis KAI2 mutants. This KAI2 homolog showed no activity with karrikins, strigolactones, or carlactones, providing evidence for a yet to be discovered endogenous KAI2 signaling hormone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lopez-obando M, Conn CE, Hoffmann B, Bythell-Douglas R, Nelson DC, Rameau C, Bonhomme S. Structural modelling and transcriptional responses highlight a clade of PpKAI2- LIKE genes as candidate receptors for strigolactones in Physcomitrella patens. Planta. 2016 doi: 10.1007/s00425-016-2481-y. [DOI] [PubMed] [Google Scholar]
- 74●●.Zhao L-H, Zhou XE, Yi W, Wu Z, Liu Y, Kang Y, Hou L, de Waal PW, Li S, Jiang Y, et al. Destabilization of strigolactone receptor DWARF14 by binding of ligand and E3-ligase signaling effector DWARF3. Cell Res. 2015;25:1219–1236. doi: 10.1038/cr.2015.122. This paper provides the first Cocrystal structure of a strigolactone receptor in complex with the synthetic strigolactone GR24. Additionally, this work shows that D14 is more unstable upon binding GR24 and interestingly hydrogen deuterium exchange experiments indicate that D14 has increased exchange upon forming a complex with the FBOX protein D53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nakamura H, Xue Y-L, Miyakawa T, Hou F, Qin H-M, Fukui K, Shi X, Ito E, Ito S, Park S-H, et al. Molecular mechanism of strigolactone perception by DWARF14. Nat Commun. 2013;4:2613. doi: 10.1038/ncomms3613. [DOI] [PubMed] [Google Scholar]
- 76●.Jiang L, Liu X, Xiong G, Liu H, Chen F, Wang L, Meng X, Liu G, Yu H, Yuan Y, et al. DWARF 53 acts as a repressor of strigolactone signalling in rice. Nature. 2013;504:401–5. doi: 10.1038/nature12870. Work provides the basis for D53 as a strigolactone signaling repressor downstream of MAX2 signaling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77●.Stanga JP, Smith SM, Briggs WR, Nelson DC. SUPPRESSOR OF MORE AXILLARY GROWTH2 1 controls seed germination and seedling development in Arabidopsis. Plant Physiol. 2013;163:318–330. doi: 10.1104/pp.113.221259. Study that identifies SMAX1 as a strigolactone independent MAX2 downstream repressor of the KAI2 signaling pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lamesch P, Berardini TZ, Li D, Swarbreck D, Wilks C, Sasidharan R, Muller R, Dreher K, Alexander DL, Garcia-Hernandez M, et al. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012;40:1202–1210. doi: 10.1093/nar/gkr1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mitchell A, Chang HY, Daugherty L, Fraser M, Hunter S, Lopez R, McAnulla C, McMenamin C, Nuka G, Pesseat S, et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–7. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nagarajan D, Deka G, Rao M. Design of symmetric TIM barrel proteins from first principles. BMC Biochem. 2015;16:18. doi: 10.1186/s12858-015-0047-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


