Abstract
Animal models of the rhesus macaque (Macaca mulatta), the most widely used nonhuman primate, have been irreplaceable in neurobiological studies. However, a population-averaged macaque brain diffusion-tensor-imaging (DTI) atlas, including comprehensive gray and white matter labeling as well as bony and facial landmarks guiding invasive experimental procedures, is not available. The macaque white matter tract pathways and microstructures have been rarely recorded. Here, we established a population-averaged macaque brain atlas with high-resolution ex vivo DTI integrated into in vivo space incorporating bony and facial landmarks, and delineated microstructures and three-dimensional pathways of major white matter tracts. In vivo MRI/DTI and ex vivo (postmortem) DTI of 10 rhesus macaque brains were acquired. Single-subject macaque brain DTI template was obtained by transforming the postmortem high resolution DTI data into in vivo space. Ex vivo DTI of 10 macaque brains was then averaged in the in vivo single-subject template space to generate population-averaged macaque brain DTI atlas. The white matter tracts were traced with DTI-based tractography. 118 neural structures including all cortical gyri, white matter tracts and subcortical nuclei, were labeled manually on population-averaged DTI-derived maps. The in vivo microstructural metrics of fractional anisotropy, axial, radial and mean diffusivity of the traced white matter tracts were measured. Population-averaged digital atlas integrated into in vivo space can be used to label the experimental macaque brain automatically. Bony and facial landmarks will be available for guiding invasive procedures. The DTI-metric measurements offer unique insights into heterogeneous microstructural profiles of different white matter tracts.
Keywords: Macaque brain, atlas, population-averaged, high resolution DTI, white matter tracts, invasive procedures
Introduction
Animal models of rhesus macaque (Macaca mulatta), the most widely used nonhuman primate, have been irreplaceable in neurobiological studies. Invasive experiments with macaque models such as electrophysiological recording (e.g. Hendry and Yoshioka, 1994; Steimnetz et al., 2000) and chemical tracing (e.g. Schmanhmann and Pandya, 2006) answer fundamental questions on brain functional systems. More complex cognitive functions and social behavior can also be examined in this intelligent animal. Macaque brain atlases are invaluable for neurobiological investigations. They offer neuroanatomical knowledge of various brain structures, serve as a reference for mapping the functional information gained by other techniques and provide anatomical guidance for invasive experiments.
Previous neuroanatomical atlases were established with histological staining (e.g. Martin and Bowden, 1996; Martin and Bowden, 2000; Saleen and Logothetis, 2007; Paxinous et al, 2009). They usually include the two-dimensional (2D) histological slides with annotation and contours of neural structures and have been a great anatomical reference for primate studies. However, the macaque histological atlases have their limitations. These 2D atlases only provide limited slices and orientations of images. It is difficult to three-dimensionally (3D) reconstruct the neural structures. It is also difficult to make the probabilistic atlas with histological images. Relaxation-based structural magnetic resonance imaging (MRI) macaque brain atlases and templates (Black et al., 2001; McLaren et al., 2009; Frey et al., 2011; Rohlfing et al. 2012) are 3D and digital, making establishment of the probabilistic atlas possible. They have excellent contrasts for segmenting gray matter and ventricle. The digital format is interactive, searchable and extensible with many advantages over conventional histology print atlases. The anatomical information from the atlases can be transferred to imaging data of the experimental subjects through image registration, with their digital format. In addition, parcellation and representation based on cortical surface (Van Essen and Dierker, 2007) have become available with structural MRI. However, these atlases usually lack the detailed anatomical information of white matter (WM) tracts with the limitation of T1- or T2-weighted contrasts.
Diffusion tensor imaging (DTI) (Basser et al., 1994), a modality of MRI, characterizes the water diffusion properties in the brain voxels with a tensor model. With the high contrasts of images derived from DTI, both white and gray matter neural structures can be delineated. DTI-based human brain atlases (e.g. Mori et al., 2008) have been widely used for normal and neuropathological brain research. DTI-based macaque brain atlases and templates from in vivo (Adluru et al., 2012; Zakszewski et al., 2014) and ex vivo (Calabrese et al., 2015) macaque brains have become available only recently. With the DTI image registration technique (e.g. Xu et al., 2003), the probabilistic DTI macaque atlases can also be established with a population average. The atlases with in vivo DTI usually have the limitation of low spatial resolution. On the other hand, the sample distortion due to fixation and confinement of the container during lengthy scans is unavoidable and affects the neuroanatomical accuracy of the atlases based on DTI of postmortem macaque brains. In addition, the skulls were usually stripped for ex vivo DTI to reduce field of view, resulting in removal of the bony and facial landmarks essential for the invasive procedures that count on the guidance of the atlases. In vivo T1 weighted data is usually considered the image with high anatomical fidelity, as it has relatively higher resolution than in vivo DTI, no geometric distortion (e.g. Huang et al., 2008) related to echo planar imaging (EPI) used in DTI, and no distortion related to fixation or container confinement in postmortem brain MRI. Therefore, a digital 3D probabilistic high-resolution DTI macaque brain atlas characterized with comprehensive white and gray matter labeling, averaged from a population, and integrated into in vivo space with bony and facial landmarks incorporated is needed. In vivo space here specifically refers to an anatomical space of in vivo T1 weighted image.
DTI-based tractography has been an effective means to delineate WM tracts (Catani et al., 2002; Wakana et al., 2004; Benhrens et al., 2007). The major WM tracts connecting different brain regions are often categorized into different tract groups based on their distinct functions. Despite the significance of the macaque WM tracts in functions and connectivity, their in vivo microstructural profiles have not been quantitatively characterized. For human brains, the major cerebral WM tracts are roughly categorized into five tract groups, limbic, projection, commissural, association and thalamic (e.g. Wakana et al., 2004; Huang et al., 2012a; Huang et al., 2012b). The WM tracts within a tract group perform similar functions. For example, limbic tracts underlie the connectivity in the limbic system and association tracts connect between cerebral cortical areas. With the close relationship of the macaque brain to the human brain, it is possible to categorize the macaque WM tract groups following the same categorization in the human brains. For each WM tract or tract group, besides widely used DTI-derived fractional anisotropy (FA) (Pierpaoli and Basser, 1996; Beaulieu 2002) and mean diffusivity (MD) for microstructural characterization, radial diffusivity (RD) and axial diffusivity (AxD) are linked to microstructural properties of myelination and axonal integrity, respectively (Song et al., 2002).
In this study, we presented a population-averaged, high-resolution, digital macaque brain DTI atlas that is in in vivo space with bony and facial landmarks preserved. Both in vivo MRI/DTI and ex vivo (postmortem) DTI of 10 rhesus macaque brains were acquired. All major WM tracts were traced with DTI tractography. By seamlessly warping high resolution ex vivo DTI into in vivo space using diffeomorphic correspondences, we built up the comprehensive 2D and 3D digital macaque brain atlas with comprehensive grey and white matter labels. 3D morphology of the cortical gyri, subcortical nuclei and WM tracts of the macaque brain was also delineated comprehensively in this digital atlas. The presented atlas will be freely downloadable at www.brainmrimap.org. Furthermore, the FA, MD, AxD and RD of all major macaque cerebral WM tracts categorized into 5 functionally distinctive tract groups were measured with in vivo DTI.
Materials and Methods
Macaques
Ten young adult macaques (age: 5.3±2.8 years; body weight=5.67±2.34kg; 6 male and 4 female) obtained from the rhesus macaque colony in the Mind and Brain Institute of Johns Hopkins University underwent in vivo MRI scan and then sacrificed for ex vivo MRI scan. As the macaque life span is about 25 years, the recruited macaques are at the stage comparable to young human adults. All studies were done with great care to ensure the wellbeing of the macaques and were approved by Institutional Animal Care and Use Committee at Johns Hopkins University. After in vivo MRI/DTI, all these 10 macaques were sacrificed by perfusion fixation with 4% paraformaldehyde after anesthesia with intramuscular injection of ketamine hydrochloride. Then the postmortem macaque brains were kept in 10% formalin for at least two months before ex vivo high resolution diffusion tensor microimaging (DTMI).
Acquisition of DTI and T1-weighted images of In vivo macaque brains
High resolution DTI and T1-weighted images were acquired in vivo for 10 macaques. A Philips 3T Achieva MR system at Kennedy Krieger Institute was used for in vivo MRI of macaques. After induction by intramuscular injection of ketamine hydrochloride (5-10mg/kg), general anesthesia during scanning was maintained by either sodium pentobarbital (25mg/kg IV) or a mixture of ketamine (7mg/kg) and xylazine (0.5-2mg/kg), with additional doses given if necessary. The macaque’s head was placed in an MRI-compatible stereotactic apparatus to stabilize its position during the scan. The macaque’s head in the stereotactic apparatus was placed inside the MRI head coil. For DTI of macaque brains, a single-shot EPI sequence with SENSE parallel imaging scheme (SENSitivity Encoding, reduction factor =2.5) was used. The diffusion weighting was encoded along 30 independent orientations (Jones et al., 1999) and the b value was 1000s/mm2. The imaging matrix was 80×80 or 100×100 with a field of view (FOV) of 120×120mm or 150×150mm (imaging resolution of 1.5mm) depending on the head size, which was zero-filled to a 160×160 matrix. To reduce the effects of distortion caused by B0-6 inhomogeneity on the brain anatomy of the acquired DTI data, coronal slices of 1.5mm thickness were acquired perpendicular to the anterior-posterior commissure line (AC-PC). A total of 60 to 70 slices covered the entire cerebrum and brainstem without gap. The echo time (TE) and repetition time (TR) were 71ms and 5.41s without cardiac gating. To increase the signal-to-noise ratio (SNR), three repetitions were performed, with a total imaging time of 10 minutes and 30 seconds. Co-registered magnetization-prepared rapid gradient-echo (MPRAGE) images at a resolution of 0.75×0.75×0.75mm3 were also acquired with scan time 6 minutes and 22 seconds.
Acquisition of high resolution DTI data of postmortem macaque brains
A Bruker 4.7 T scanner was used for ultra-high resolution DTI data acquisition. Before ex vivo DTI, the macaque brains were placed in 10% phosphate-buffered saline (PBS) for at least 120 hours to allow the exchange of fixation solution and PBS. The macaque brains were then transferred into a custom-made MRI-compatible container and bathed with fomblin (Fomblin Profludropolyether; Ausimont, Thorofare, NJ). A 3D multiple spin echo diffusion tensor sequence with eight echoes was used for DTI imaging. From the eight echoes, eight individual 3D volume images were obtained, which were averaged to enhance the SNR (Zhang et al., 2003; Huang et al., 2008). A set of diffusion-weighted images (DWI) were acquired in 8 linearly independent directions with b value 1000s/mm2. DWI parameters were: TE=32.5ms, TR=0.7s, FOV=78mm/56mm/58mm, imaging matrix=200×108×108 for a nominal resolution of 0.39×0.52×0.54mm3 (this was zero filled to data matrix 256×128×128 yielding 0.30×0.44×0.45mm3 interpolated resolution). Two repetitions were performed to increase SNR, with a total imaging time of 45 hours for acquiring DTI data of one postmortem brain.
DTI processing
After acquisition, all data were transferred to an off-line workstation where tensor fitting and postprocessing was performed. Automated image registration (AIR) (Woods et al., 1998) was conducted on raw DWIs to correct distortion caused by eddy currents. Six elements of the 3×3 7 diffusion tensor were determined by multivariate least-square fitting of DWIs. The tensor was diagonalized to obtain three eigenvalues (λ1-3) and eigenvectors (V1-3). Anisotropy was measured by calculating fractional anisotropy (FA) (Pierpaoli and Basser, 1996). The tensor fitting was conducted with DtiStudio (Jiang et al., 2006). Based on fitted tensor, maps of FA, mean diffusivity (MD), axial diffusivity (AxD) and radial diffusivity (RD) were obtained. These procedures of tensor fitting and generation of DTI-metric maps were repeated for both in vivo and ex vivo DTI data. The in vivo DTI metric maps were used for microstructural measurement of WM tracts, while ex vivo DTI maps were used for the labeling of neural structures.
Population-averaged macaque brain template with ex vivo imaging resolution integrated into in vivo space
Two steps were conducted to generate population-averaged macaque brain in in vivo space while keeping ex vivo high resolution. The large deformation diffeomorphic metric mapping (LDDMM) (Miller et al., 2002) has been used for both steps. First, a middle-sized brain was selected as the single-subject (SS) template. LDDMM was used to align the averaged diffusion weighted image (aDWI) of ex vivo brain to the skull-stripped T1 weighted image of in vivo brain of the same SS-template macaque subject, after AIR (Woods et al., 1998) for linear affine alignment. Geometric deformations caused by brain tissue shrinkage in the fixation and confinement of the container in lengthy ex vivo scanning are clear on the left panel of Fig. 1. Of the note, aDWI is the average of all non-b0 image in the diffusion MRI. These geometric deformations incorporate both low-dimensional and high-dimensional geometric differences. LDDMM transformation was then used to “fine tune” the high frequency geometric differences by providing diffeomorphic correspondences between in vivo and ex vivo space and generalizing the low dimensional rotations and translation used for rigid alignment to the infinite dimensional case appropriate for correspondences between the two spaces. On the right panel of Fig. 1, the identical red contour that was delineated with skull-stripped in vivo T1 weighted imaging fits well with the ex vivo brain after LDDMM transformation. In this way, an SS-template was generated in in vivo space while keeping ex vivo high resolution. Secondly, affine and LDDMM transformations to the SS-template in in vivo space were applied to high-resolution ex vivo DTI data of other 9 macaque brains to generate the population-averaged macaque brain template. For tensor transformation, the affine transformation matrix and LDDMM transformation matrix obtained from the scalar images were applied to the tensor field to create normalized tensor fields with details described in previous literature (Xu et al., 2003). Such tensor transformations were conducted with DiffeoMap software (mristudio.org). After co-registration, the cropped brain from T1 weighted images were replaced by high resolution population-averaged or single-subject DTI data in in vivo space. In this way, the population-averaged or single-subject macaque brain template was generated with ex vivo imaging resolution integrated into in vivo space, with bony and facial landmarks kept.
Figure 1.

Illustration of integrating the high resolution ex vivo macaque brain DTI data (left) into in vivo space (right) using elastic warping with LDDMM transformation T(•). The identical red contour, characterizing the tissue boundary of in vivo space, was delineated from in vivo T1 weighted images and overlaid on each image to demonstrate the warping effects. aDWI and DTI orientation-encoded colormap were acquired ex vivo and T1 weighted image was acquired in vivo. aDWI and ss-T1w stand for averaged diffusion weighted image and skull-stripped T1 weighted image, respectively.
DTI-based tractography of macaque white matter tracts
DTI-based tractography was used to segment WM tracts, which were subsequently used as regions of interests (ROI) for measuring tract-specific DTI-derived microstructural metrics. A streamline propagation tractography method (Mori et al., 1999) was used for DTI fiber tracking of all ex and in vivo macaque brain DTI data. All major macaque WM tracts were categorized into limbic, commissural, thalamic, projection and association tracts, similar to the categorization used in the human brain (Wakana et al., 2004; Huang et al., 2012a, b). The tractography protocol for tracing cerebral and brain stem tracts in human brain (Stieltjes et al., 2001; Wakana et al., 2007) was also used to trace these tracts in the macaque brains. Most of major WM tracts including left and right cingulum bundles in the cingulate gyrus (cgc-L and cgc-R) and cingulum to the hippocampus (cgh-L and cgh-R) of the limbic system tracts, left and right cortico-spinal tract (cst-L and cst-R) of the projection tracts, forceps major (fmajor) and forceps minor (fminor) of the commissural tracts, left and right anterior thalamic radiation (atr-L and atr-R) of thalamic tracts and left and right inferior longitudinal fasciculus (ilf-L and ilf-R) and uncinate fasciculus (unc-L and unc-R) of the associate tracts could be reproducibly and reliably traced with both in vivo and ex vivo macaque DTI data. Due to MR susceptibility distortions at the brain stem area and resolution limitations of in vivo DTI data, inferior fronto-occipital fasciculus, fornix and the brain stem tracts including middle cerebellar peduncle, left and right inferior and superior cerebellar peduncle could only be reproducibly traced with ex vivo macaque DTI, but not in vivo macaque DTI. To demonstrate the 3D pathways of macaque WM, all the tracts described above were reconstructed with Amira (FEI software, Hillsboro, OR). The inter-subject transformation matrices were then applied to transfer the tracts into the population-averaged brain template in in vivo template space and generate the probabilistic tracts.
Establishment of the comprehensive white and gray matter macaque brain atlas
The annotation and labeling of the major white and gray matter structures were conducted on the population-averaged high-resolution aDWI images and DTI orientation-encoded color maps warped to in vivo template space. ROIEditor (www.mristudio.org) was used for manual labeling. With the integration of ex vivo high resolution DTI data into in vivo space, the underlying ex vivo high resolution orientation-encoded DTI colormaps were used for annotation of the major WM tracts while the underlying ex vivo aDWI images were used for annotation of cortical gyri and subcortical nuclei. A total of 118 comprehensive white and gray matter neural structures were labeled, as shown in the table 1. Both WM tracts and cortical surface was reconstructed in 3D. The contrasts of DTI orientation-encoded color maps and aDWI images and available atlases (Martin and Bowden, 1996; Martin and Bowden, 2000; Saleem and Logothetis, 2007) were used as guidance for labeling. In addition, the WM tractography information were used to manually delineate WM tracts on the orientation-encoded DTI color maps in in vivo template space, similar to the WM labeling of human brain (Wakana et al., 2004). The cerebral cortical gyri, the subcortical gray matter and ventricles were manually delineated with the contrast of the high resolution aDWI images in in vivo template space. All the labeling was conducted on the 2D coronal slices. The cross-section of the 3D reconstructed WM tracts and parcellated cortical gyral surfaces in the 2D slices and two other views (axial and sagittal) were used to adjust and 10 refine the labeling. 3D visualizations of the parcellated cortex and subcortical gray matter were generated with Amira (FEI software, Hillsboro, OR).
Table 1.
Anatomical labels of 118 brain neural structures and their abbreviations.
| Abbreviations | Structures | Left/right | Level | |
|---|---|---|---|---|
| 1 | RG | Gyrus rectus | Left | Frontal Lobe |
| 2 | RG | Gyrus rectus | Right | Frontal Lobe |
| 3 | MOG | Medial orbital gyrus | Left | Frontal Lobe |
| 4 | MOG | Medial orbital gyrus | Right | Frontal Lobe |
| 5 | LOG | Lateral orbital gyrus | Left | Frontal Lobe |
| 6 | LOG | Lateral orbital gyrus | Right | Frontal Lobe |
| 7 | FOG | Fronto-orbital gyrus | Left | Frontal Lobe |
| 8 | FOG | Fronto-orbital gyrus | Right | Frontal Lobe |
| 9 | OT | Olfactory tubercle | Left | Frontal Lobe |
| 10 | OT | Olfactory tubercle | Right | Frontal Lobe |
| 11 | SFG | Superior frontal gyrus | Left | Frontal Lobe |
| 12 | SFG | Superior frontal gyrus | Right | Frontal Lobe |
| 13 | MFG | Middle frontal gyrus | Left | Frontal Lobe |
| 14 | MFG | Middle frontal gyrus | Right | Frontal Lobe |
| 15 | IFG | Inferior frontal gyrus | Left | Frontal Lobe |
| 16 | IFG | Inferior frontal gyrus | Right | Frontal Lobe |
| 17 | PrCG | Precentral gyrus | Left | Frontal Lobe |
| 18 | PrCG | Precentral gyrus | Right | Frontal Lobe |
| 19 | PoCG | Postcentral gyrus | Left | Parietal Lobe |
| 20 | PoCG | Postcentral gyrus | Right | Parietal Lobe |
| 21 | SPL | Superior parietal lobule | Left | Parietal Lobe |
| 22 | SPL | Superior parietal lobule | Right | Parietal Lobe |
| 23 | SMG | Superior marginal gyrus | Left | Parietal Lobe |
| 24 | SMG | Superior marginal gyrus | Right | Parietal Lobe |
| 25 | AnG | Angular gyrus | Left | Parietal Lobe |
| 26 | AnG | Angular gyrus | Right | Parietal Lobe |
| 27 | Ins | Insular cortex | Left | Insula Lobe |
| 28 | Ins | Insular cortex | Right | Insula Lobe |
| 29 | OG | Occipital gyrus | Left | Occipital Lobe |
| 30 | OG | Occipital gyrus | Right | Occipital Lobe |
| 31 | IOG | Inferior occipital gyrus | Left | Occipital Lobe |
| 32 | IOG | Inferior occipital gyrus | Right | Occipital Lobe |
| 33 | AG | Annectant gyrus | Left | Occipital Lobe |
| 34 | AG | Annectant gyrus | Right | Occipital Lobe |
| 35 | PrCu | Precuneus | Left | Occipital Lobe |
| 36 | PrCu | Precuneus | Right | Occipital Lobe |
| 37 | Cun | Cuneus | Left | Occipital Lobe |
| 38 | Cun | Cuneus | Right | Occipital Lobe |
| 39 | LiG | Lingual gyrus | Left | Occipital Lobe |
| 40 | LiG | Lingual gyrus | Right | Occipital Lobe |
| 41 | STG | Superior temporal gyrus | Left | Temporal Lobe |
| 42 | STG | Superior temporal gyrus | Right | Temporal Lobe |
| 43 | MTG | Middle temporal gyrus | Left | Temporal Lobe |
| 44 | MTG | Middle temporal gyrus | Right | Temporal Lobe |
| 45 | ITG | Inferior temporal gyrus | Left | Temporal Lobe |
| 46 | ITG | Inferior temporal gyrus | Right | Temporal Lobe |
| 47 | PA | Prepyriform area | Left | Temporal Lobe |
| 48 | PA | Prepyriform area | Right | Temporal Lobe |
| 49 | Hippo | Hippocampus | Left | Temporal Lobe |
| 50 | Hippo | Hippocampus | Right | Temporal Lobe |
| 51 | PHG | Parahippocampal gyrus | Left | Temporal Lobe |
| 52 | PHG | Parahippocampal gyrus | Right | Temporal Lobe |
| 53 | FuG | Fusiform gyrus | Left | Temporal Lobe |
| 54 | RFu | Fusiform gyrus | Right | Temporal Lobe |
| 55 | Ent | Entorhinal area | Left | Temporal Lobe |
| 56 | Ent | Entorhinal area | Right | Temporal Lobe |
| 57 | ACgG | Anterior cingulate gyrus | Left | Temporal Lobe |
| 58 | ACgG | Anterior cingulate gyrus | Right | Temporal Lobe |
| 59 | PCgG | Posterior cingulate gyrus | Left | Temporal Lobe |
| 60 | PCgG | Posterior cingulate gyrus | Right | Temporal Lobe |
| 61 | cst | Corticospinal tract | Left | Projection tracts |
| 62 | cst | Corticospinal tract | Right | Projection tracts |
| 63 | atr | Anterior thalamic radiation | Left | Thalamic tracts |
| 64 | atr | Anterior thalamic radiation | Right | Thalamic tracts |
| 65 | str | Superior thalamic radiation | Left | Thalamic tracts |
| 66 | str | Superior thalamic radiation | Right | Thalamic tracts |
| 67 | ilf | Inferior longitudinal fasciculus | Left | Association tracts |
| 68 | ilf | Inferior longitudinal fasciculus | Right | Association tracts |
| 69 | ifo | Inferior fronto-occipital fasciculus | Left | Association tracts |
| 70 | ifo | Inferior fronto-occipital fasciculus | Right | Association tracts |
| 71 | unc | Uncinate fasciculus | Left | Association tracts |
| 72 | unc | Uncinate fasciculus | Right | Association tracts |
| 73 | slf III | Superior longitudinal fasciulus III | Left | Association tracts |
| 74 | slf III | Superior longitudinal fasciulus III | Right | Association tracts |
| 75 | cgh | Cingulum hippocampal part | Left | Limbic tracts |
| 76 | cgh | Cingulum hippocampal part | Right | Limbic tracts |
| 77 | cgc | Cingulum cingulate gyrus part | Left | Limbic tracts |
| 78 | cgc | Cingulum cingulate gyrus part | Right | Limbic tracts |
| 79 | cc-body | Corpus callosum (body) | Commissural tracts | |
| 80 | Fminor | Forceps minor | Commissural tracts | |
| 81 | Fmajor | Forceps major | Commissural tracts | |
| 82 | Tap | Tapetum | Left | Commissural tracts |
| 83 | Tap | Tapetum | Right | Commissural tracts |
| 84 | fx | Fornix | Left | Limbic tracts |
| 85 | fx | Fornix | Right | Limbic tracts |
| 86 | ac | Anterior commissure | Commissural tracts | |
| 87 | mcp | Middle cerebellar peduncle | Cerebellar tracts | |
| 88 | scp | Superior cerebellar peduncle | Left | Cerebellar tracts |
| 89 | scp | Superior cerebellar peduncle | Right | Cerebellar tracts |
| 90 | icp | Inferior cerebellar peduncle | Left | Cerebellar tracts |
| 91 | icp | Inferior cerebellar peduncle | Right | Cerebellar tracts |
| 92 | Caud | Caudate nucleus | Left | Basal Nuclei |
| 93 | Caud | Caudate nucleus | Right | Basal Nuclei |
| 94 | Put | Putamen | Left | Basal Nuclei |
| 95 | Put | Putamen | Right | Basal Nuclei |
| 96 | GP | Globus pallidus | Left | Basal Nuclei |
| 97 | GP | Globus pallidus | Right | Basal Nuclei |
| 98 | Cl | Claustrum | Left | Basal Nuclei |
| 99 | Cl | Claustrum | Right | Basal Nuclei |
| 100 | Amyg | Amygdala | Left | Basal Nuclei |
| 101 | Amyg | Amygdala | Right | Basal Nuclei |
| 102 | HT | Hypothalamus | Left | Diencephalon |
| 103 | HT | Hypothalamus | Right | Diencephalon |
| 104 | opt | Optic tract | Left | Diencephalon |
| 105 | opt | Optic tract | Right | Diencephalon |
| 106 | SN | Septal nucleus | Left | Diencephalon |
| 107 | SN | Septal nucleus | Right | Diencephalon |
| 108 | NA | Nucleus accumbens | Left | Diencephalon |
| 109 | NA | Nucleus accumbens | Right | Diencephalon |
| 110 | Tha | Thalamus | Left | Diencephalon |
| 111 | Tha | Thalamus | Right | Diencephalon |
| 112 | LGN | Lateral geniculate nucleus | Left | Diencephalon |
| 113 | LGN | Lateral geniculate nucleus | Right | Diencephalon |
| 114 | PaR | Pars reticulata | Left | Diencephalon |
| 115 | PaR | Pars reticulata | Right | Diencephalon |
| 116 | Lat Ven | Lateral ventricle | Left | Ventricle |
| 117 | Lat Ven | Lateral ventricle | Right | Ventricle |
| 118 | 3rd Ven | Third ventricle | Ventricle |
Microstructural measurements of macaque brain white matter tracts
To avoid postmortem or fixation effects on WM microstructures, in vivo DTI data of the 10 macaque brains were used for the microstructural DTI metric measurements of WM tracts. All the tracts for DTI metric measurements were reproducibly traced with all 10 in vivo macaque DTI data. Note that the brain stem tracts can be reproducibly traced with ex vivo high resolution DTI, but not in vivo DTI. Hence brain stem tracts were not included for in vivo microstructural measurements. Tract-level FA, MD, AxD and RD of individual WM tracts were measured with the traced tracts as the ROIs and these metric measurements averaged over the traced tract ROIs. All four DTI metrics were measured to reveal the comprehensive WM microstructural profiles. To demonstrate the distinctive microstructural properties of categorized WM tract groups, the plots of MD versus FA, AxD versus FA and RD versus FA were generated.
Volumetric measurements of subcortical gray matter structure, ventricle, cerebrum, cerebellum and brain stem
The volumes were measured with in vivo T1-weighted images in the native space of each macaque brain. Semi-automated tissue segmentation with region growing and manual adjustment was conducted by three operators (LF, TJ and QY) to ensure accuracy. Differences between the three operators were less than 5% for all volumes and the average was used for final measurement results. The individual absolute volumes of ventricle, caudate nucleus, putamen, thalamus, amygdala and hippocampus were measured for all 10 macaques. In addition, the volumes of cerebrum, cerebellum and brain stem were measured.
Results
Population-averaged macaque brain template in in vivo space
DTI of a population-averaged macaque brain template is shown in the Fig 2. The population-averaged DTI template was created based on an average of ex vivo DTI data of 10 macaque brains and integrated into in vivo space. The orientation-encoded colormap, FA map and aDWI of the population-averaged template are shown in the Fig 2. High spatial resolution and superior signal-to-noise ratio are clear. As shown in the Fig 2, the contrasts of population-averaged DTI images remain sharp, suggesting high quality of inter-subject registration. Due to these factors, the white and gray matter neural structures were readily delineable on population-averaged template. For example, various WM fibers including small WM fibers (e.g. fornix or fx) can be clearly appreciated from the orientation-encoded colormap of population-averaged macaque template, as shown in the enlarged white circle on the right panel of the Fig 2.
Figure 2.
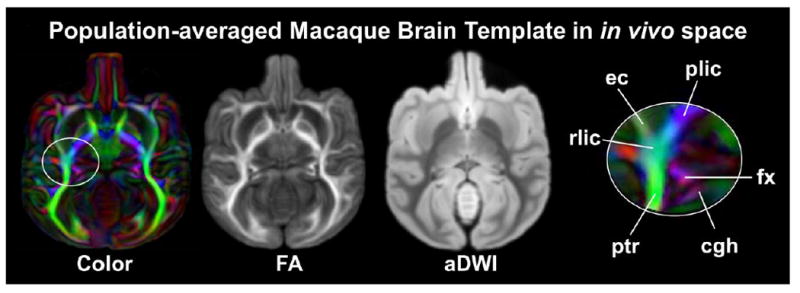
Orientation-encoded colormap, FA map and aDWI averaged from ex vivo DTI data of 10 macaque brains and integrated into in vivo space were generated as the population-averaged template. The white circle highlights the sharp contrasts of different WM tracts in the population-averaged orientation-encoded colormap. Abbreviations: cgh: cingulum in the hippocampal part; ec: external capsule; fx: fornix; plic/rlic: posterior limb / retrolenticular part of the internal capsule; ptr: posterior thalamic radiation.
3D reconstructed and probabilistic white matter tracts
The 3D reconstructed pathways of the major macaque WM tracts are illustrated in Fig. 3. Similar to human WM tracts, major macaque WM tracts are categorized into six distinctive functional tract groups: limbic tracts (Fig 3a), projection tracts (Fig 3b), commissural tracts (Fig 3c), association tracts (Fig 3d), thalamic tracts (Fig 3e) and brain stem tracts (Fig 3f). All these tracts were reproducibly traced with ex vivo DTI data of macaque brains. The anatomy of these major macaque WM tracts from a population of macaque brains can be further illustrated with the probabilistic tract maps overlaid on averaged FA map in the in vivo template space in Fig 4. Individual differences on tract anatomical pathways are found around the boundaries of the WM bundles, indicated by blue colors with low probability. The core of the anatomical pathways of the WM tracts are consistent, indicated by red and yellow color with high probability (Fig 4).
Figure 3.
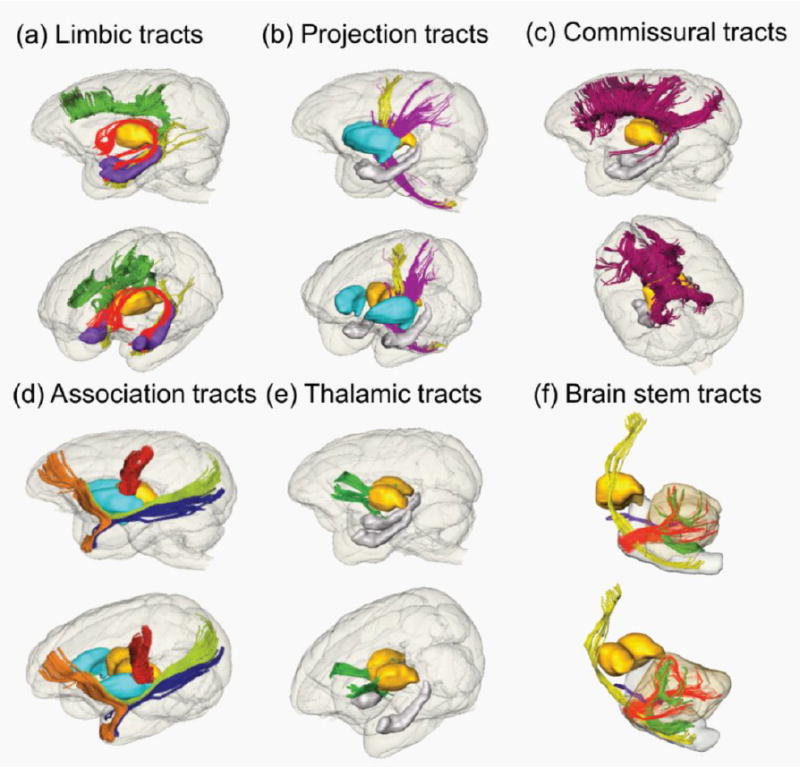
3D reconstructed limbic (a), projection (b), commissural (c), association (d), thalamic (e) and brain stem (f) tracts of a macaque brain. Lateral and oblique views are displayed for each panel. Reconstructed tracts are cingulum in the cingulate gyrus (green), cingulum to the hippocampus (yellow) and fornix (red) in (a); corticospinal tract (yellow) and cerebral peduncle (purple) in (b); corpus callosum including genu, body, splenium and tapetum projecting to the temporal lobe (crimson) in (c); uncinate fasciculus (orange), fronto-parietal short tract (red), inferior longitudinal fasciculus (blue) and inferior fronto-occipital fasciculus (green) in (d); thalamic tract (green) in (e); and corticospinal tract (yellow), middle cerebellar peduncle (red), inferior cerebellar peduncle (green) and superior cerebral peduncle (purple). For anatomical guidance, thalamus (yellow), hippocampus (purple in (a) and gray in (e)) and putamen (cyan in (d)) are also displayed.
Figure 4.
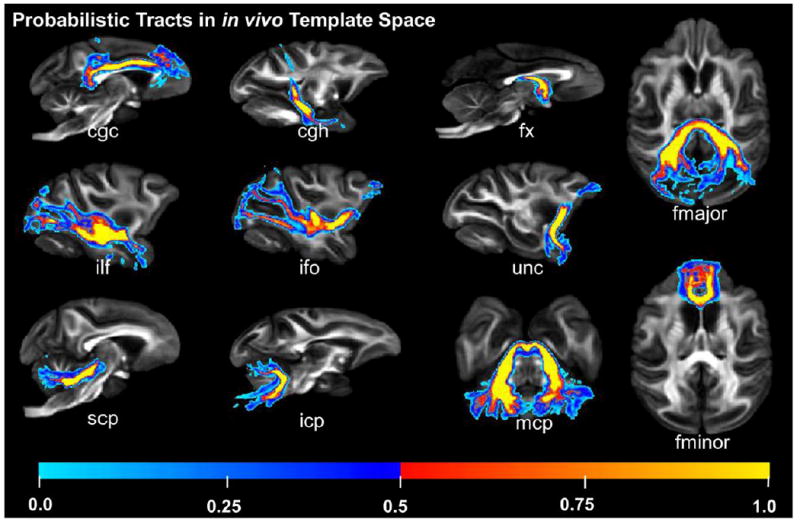
Probabilistic tracts in in vivo template space. The underlying FA map is from the averaged macaque brain template in in vivo space. Individual differences around the boundaries of WM bundles, indicated by cold color with low probability, and consistency at the core of the WM bundles, indicated by warm color with high probability, can be observed. Abbreviations: atr: anterior thalamic radiation; cgc: cingulum in the cingulate cortex; cgh: cingulum in the hippocampal area; cst: corticospinal tract; fx: fornix; ilf: inferior longitudinal fasciculus; ifo: inferior fronto-occipital fasciculus; unc: uncinate fasciculus; scp: superior cerebellar peduncle; icp: inferior cerebellar peduncle; mcp: middle cerebellar peduncle; fmajor/fminor: forceps major/minor of corpus callosum.
Digital macaque brain DTI atlas with comprehensive neuroanatomical labels and bony landmarks included
All atlas labels were established on the population-averaged template in in vivo space. The labels have been refined several rounds to ensure the naturally smooth boundaries of all delineated neural structures appear smooth not only in coronal planes, but also in axial and sagittal planes (Figs 5-8). Fig. 5 shows an overview of comprehensive labels of white and gray matter. The labels of both white and gray matter are displayed in Fig 5a. Using the same colors for the identical gray or white matter neural structures on the left and right hemispheres in Fig 5a, the symmetry of the white and gray matter labels is clear. The labels of all major WM tracts are demonstrated in Fig 5b, while gray matter cortical gyri and subcortical nuclei are demonstrated in Fig 5c. The orientation-encoded colormaps (Fig 5b) and aDWIs (Fig 5c) are shown on the left side of the coronal and axial planes for reference. In addition, the anterior, lateral and superior view of the 3D reconstructed cortical surface with labeled gyri are displayed in Fig 5d. From Figs 5b-5d, it is evident that both white and gray matter labels match their corresponding anatomical structures.
Figure 5.
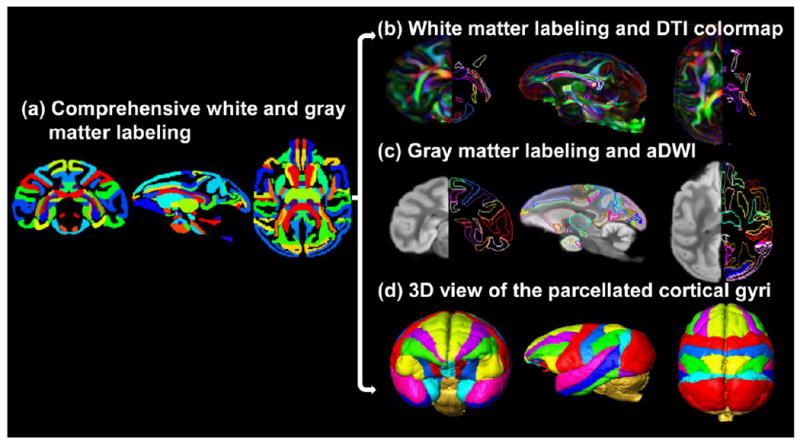
Comprehensive white and gray matter labels in the axial, parasagittal and coronal orientation. The labels of both white and gray matter are displayed in a typical axial, parasagittal and coronal plane in (a). In (b), the left and right sides of the coronal and axial planes show the population-averaged orientation-encoded colormaps and their corresponding WM tract labels, respectively. In (c), the left and right sides of the coronal and axial planes show the population-averaged aDWI images and their corresponding labels of cortical gyri and subcortical nuclei, respectively. The WM and gray matter labels are directly overlaid on a parasagittal plane of DTI colormap and aDWI image in (b) and (c), respectively. The anterior, lateral and superior view of the 3D reconstructed labeled cortical gyri are displayed in (d).
Figure 8.
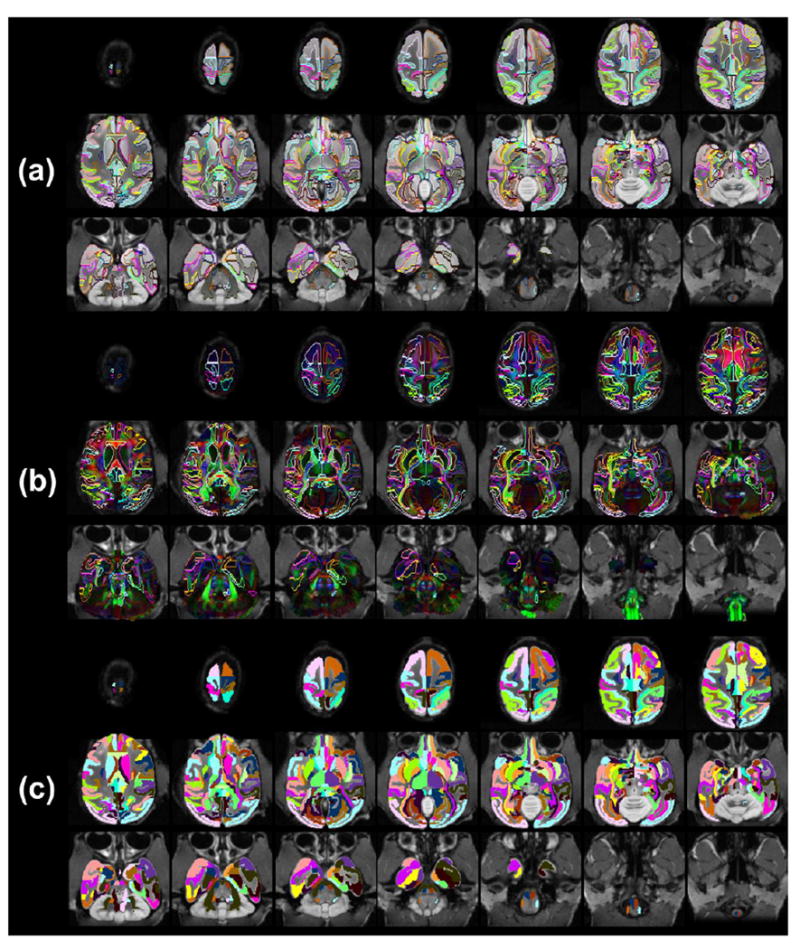
All atlas labels on the population-averaged aDWIs (a), population-averaged orientation-encoded colormaps (b) and single-subject T1 weighted image (c) in the evenly sampled axial planes from superior to inferior brain. Population-averaged aDWIs and orientation-encoded colormaps were integrated into in vivo space with bony and facial landmarks kept from in vivo T1 weighted images.
Fig 6 shows the contours of white and gray matter neural structures overlaid on ex vivo high resolution population-averaged aDWI (upper two rows of Fig 6a and Fig 6b) and DTI orientation-encoded colormaps (lower two rows in Fig 6a and Fig 6b), both integrated into in vivo space with bony and facial landmarks included. These contours are based on comprehensive labels of all cortical gyri, subcortical nuclei and WM tracts. The bony, facial and other outside landmarks are from in vivo T1-weighted images. The high degree of alignment between the ex vivo high resolution brain DTI image and the in vivo skull and tissue images can be clearly appreciated from the integrated images in Fig 6. The bony and facial landmarks can be used for guiding invasive experiments on macaque brains. The coronal slices in Fig 6a and axial slices in Fig 6b are indicated by red lines across the 3D reconstructed whole head, which is semitransparent with the inside brain displayed in solid yellow. All atlas labels on the population-averaged aDWIs and orientation-encoded colormaps in evenly sampled coronal and axial planes are shown in Fig 7 and Fig 8, respectively. The indices and abbreviations of all 118 anatomical labels of brain neural structures including cortical gyri, subcortical nuclei, cerebral ventricles and WM tracts are listed in Table 1. The volumes of ventricle, caudate, putamen, thalamus, amygdala/hippocampus, cerebellum, cerebrum and brain stem are listed in Table 2.
Figure 6.
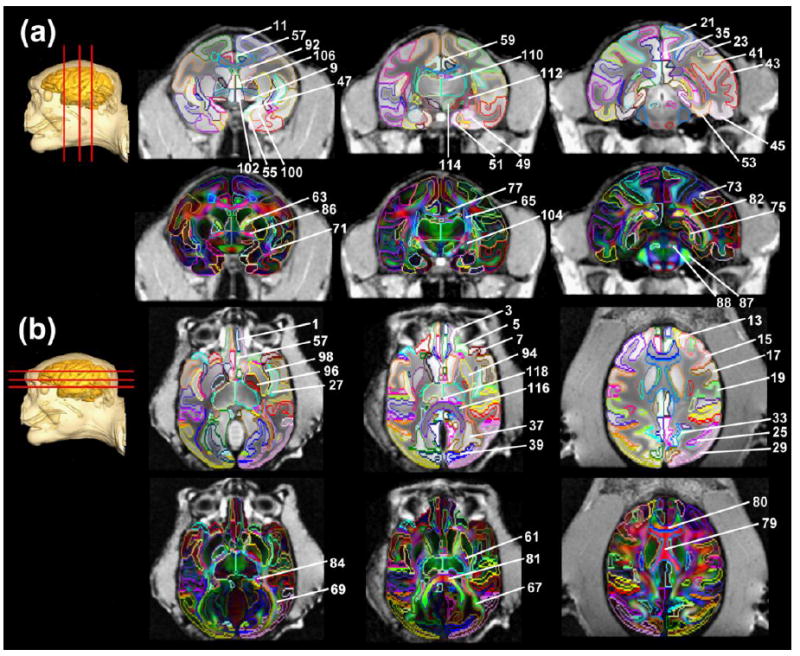
Annotation and labeling of all cortical gyri, subcortical nuclei and white matter tracts on coronal (a) and axial (b) T1 weighted images orientation-encoded aDWI (upper panels in a and b) and DTI colormaps (lower panels in a and b) of the populatin-averaged macaque brain. All images are presented in in vivo space after integration of ex vivo aDWI and DTI colormaps into the in vivo space. Numbers indicated in the figure are identical to the structure number listed in the Table 1. The geometric locations of these coronal (a) and axial (b) images are indicated by the red lines on top of a 3D reconstructed whole head with brain highlighted with solid yellow color.
Figure 7.
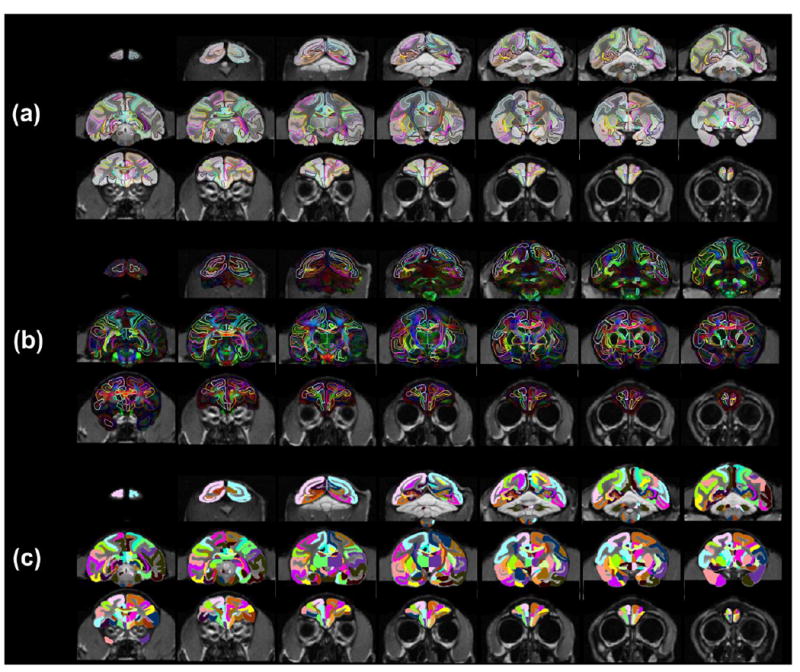
All atlas labels on the population-averaged aDWIs (a), population-averaged orientation-encoded colormaps (b) and single-subject T1 weighted image (c) in the evenly sampled coronal planes from anterior to posterior brain. Population-averaged aDWIs and orientation-encoded colormaps were integrated into in vivo space with bony and facial landmarks kept from in vivo T1 weighted images
Table 2.
Mean and standard deviation (SD) of volumetric measurements of each subcortical nuclei, ventricle, cerebrum, cerebellum and brain stem from T1 weighted images of ten macaque brains.
| Structures | Mean volumes (mm3) | Standard deviation (mm3) | |
|---|---|---|---|
| 1 | ventricle | 510.95 | 74.99 |
| 2 | caudate | 1290.50 | 109.70 |
| 3 | putamen | 1870.12 | 99.42 |
| 4 | thalamus | 1641.23 | 135.14 |
| 5 | amygdala/hippocampus | 1057.43 | 74.01 |
| 6 | cerebellum | 8788.49 | 510.90 |
| 7 | cerebrum | 84023.86 | 3943.30 |
| 8 | brain stem | 3592.64 | 289.64 |
Microstructural profiles of the white matter tracts in different tract groups
Fig 9 shows the profiles of FA, MD, AxD and RD of all major tracts and relationships among these different DTI-derived metrics for live macaque brains. The measurements of FA, MD, AxD and RD of all the tracts are displayed in Table 3. The WM tract groups with smallest to largest averaged FA in order are limbic tracts, association tracts, thalamic tracts, commissural tracts and projection tracts. The plots with almost linear relationship between FA and AxD (Fig 9b) and between FA and RD (Fig 9c) indicate that the variations of both AxD and RD measurements contribute to FA differences among these WM tracts. For example, the smallest FA values of the limbic tracts are contributed by smallest AxD and highest RD values. On the other hand, the left and right corticospinal tracts (cst) have the largest FA values contributed by the highest AxD and smallest RD values.
Figure 9.
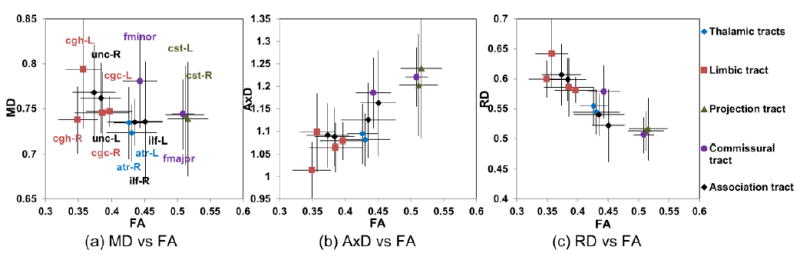
Scatter plots of FA versus MD (a), FA versus AxD (b) and FA versus RD (c) for major macaque white matter tracts in the left (L) and right (R) hemispheres. The white matter tract groups with smallest to largest FA in order are limbic tracts, association tracts, thalamic tracts, commissural tracts and projection tracts. The relationship plot of FA and AxD (b) and that of FA and RD (c) indicate uniformity of both AxD and RD measurements that contribute to FA differences among these white matter tracts. Different colors indicate different tract groups. Please see legend of Fig 4 for abbreviations.
Table 3.
Mean and standard deviation (SD) of FA, MD, AxD and RD of major tracts measured from in vivo DTI of ten macaque brains.
| White matter tracts | Mean FA | SD of FA | Mean MD | SD of MD | Mean AxD | SD of AxD | Mean RD | SD of RD | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| (×10-3mm2/s) | (×10-3mm2/s) | (×10-3mm2/s) | |||||||
| Thalamic tracts | atr_L | 0.430 | 0.038 | 0.723 | 0.031 | 1.081 | 0.059 | 0.544 | 0.038 |
| atr_R | 0.426 | 0.023 | 0.734 | 0.040 | 1.094 | 0.066 | 0.554 | 0.032 | |
|
| |||||||||
| Limbic tracts | cgh_L | 0.357 | 0.027 | 0.794 | 0.066 | 1.098 | 0.087 | 0.642 | 0.058 |
| cgh_R | 0.349 | 0.030 | 0.738 | 0.037 | 1.014 | 0.062 | 0.600 | 0.031 | |
| cgc_L | 0.397 | 0.035 | 0.747 | 0.017 | 1.078 | 0.041 | 0.581 | 0.022 | |
| cgc_R | 0.385 | 0.041 | 0.745 | 0.046 | 1.064 | 0.055 | 0.586 | 0.049 | |
|
| |||||||||
| Projection tracts | cst_L | 0.512 | 0.030 | 0.743 | 0.055 | 1.203 | 0.114 | 0.513 | 0.032 |
| cst_R | 0.516 | 0.031 | 0.739 | 0.063 | 1.240 | 0.156 | 0.516 | 0.052 | |
|
| |||||||||
| Commissural tracts | Fmajor | 0.509 | 0.017 | 0.744 | 0.039 | 1.220 | 0.066 | 0.506 | 0.030 |
| Fminor | 0.443 | 0.027 | 0.781 | 0.052 | 1.185 | 0.079 | 0.579 | 0.045 | |
|
| |||||||||
| Association tracts | ilf_L | 0.451 | 0.027 | 0.736 | 0.067 | 1.163 | 0.118 | 0.522 | 0.060 |
| ilf_R | 0.435 | 0.042 | 0.735 | 0.026 | 1.125 | 0.083 | 0.540 | 0.036 | |
| unc_L | 0.384 | 0.030 | 0.762 | 0.039 | 1.088 | 0.065 | 0.599 | 0.036 | |
| unc_R | 0.373 | 0.032 | 0.768 | 0.053 | 1.091 | 0.071 | 0.607 | 0.052 | |
Discussion
The population-averaged digital macaque atlas includes a total 118 white and gray matter labels, broken down into 60 labels of cortical gyri, 31 labels of WM tracts and 27 labels of subcortical nuclei or ventricle. The atlas is characterized with integration of high resolution ex vivo DTI data into in vivo space. Furthermore, the included facial and bony landmarks provide critical neuroanatomical guidance for invasive neurophysiological experiments. The presented macaque brain atlas is population-averaged, comprehensive, three-dimensional, high-resolution and digital with high contrast, established to be a next-generation atlas compared to the previous atlases based on histology. 3D pathways of the WM tracts were revealed by DTI tractography. The WM tracts in the cross-sectional 2D planes were delineated with high contrasts from DTI-derived maps. Unique insights into in vivo microstructural profiles of major WM tracts and the microstructural organization among the major WM tract groups were revealed.
Population-averaged digital macaque brain DTI atlas with comprehensive white and gray matter labels
Besides integration of in vivo and ex vivo information elaborated in the next discussion section, the most prominent features of the presented atlases are 1) population-averaged, 2) comprehensive (offering both white and gray matter labels with high contrasts of DTI-derived maps), 3) three-dimensional rooted from data generation (three-dimensional instead of two-dimensional MRI acquisition), 4) high-resolution and 5) digital. Compared to the atlases based on 2D histological images, the presented atlas is adapted to the current needs of mapping the parcellated information in a template space to the users’ macaque MRI data in the native space. High contrasts from DTI in the atlas can be used to delineate WM tracts, which cannot be distinguished with previous macaque atlases based on conventional relaxation-based MRI. The higher resolution of this atlas, in contrast to those from in vivo DTI (e.g. Adluru et al., 2012; Zakszewski et al., 2014; Liu et al., 2015; Shi et al., 2017), is likely to offer anatomical information in finer detail. For example, the delineation of multiple tracts in a small region is clearly demonstrated in the most right panel of Fig 2a. Comprehensive white and gray matter neural structures were characterized with 118 labels, based on contrasts of DTI-derived maps, cross-sections of the traced fibers (similar to those used in Wakana et al., 2004) and existing macaque brain atlases (Martin and Bowden, 1996; Martin and Bowden, 2000). Despite that the major delineation was performed on the coronal slides, extensive efforts by neuroanatomists (LF, QY) have been made to refine all 118 labels to achieve naturally smooth boundaries in all three (axial, coronal and sagittal) planes (Figs 5-8) as most registration software requires image transformation in 3D. With its population-averaged nature, the anatomical information from this atlas may be used not only for anatomical guidance for neurobiological research, but also for evolution studies.
Parcellation of WM tracts with various intensities (index 61-91 in Table 1) is uniquely presented in this atlas. The atlas-making procedures for WM tracts are similar to those used in human brain WM parcellation (Mori et al., 2008). It is noteworthy that population-averaged DTI color-encoded maps that preserve sharp contrasts were used for manual delineation of white and gray matter labels. Delineation on the population-averaged DTI-derived maps instead of individual maps could represent the common neural structures with less variability among a population of subjects. The reproducibility of the labeled major WM tracts was further confirmed by probabilistic tract maps in Fig 4 showing the reproducible core of major tract bundles among different macaque brains. Cerebral cortical gyri, subcortical nuclei, brain stem structures and ventricle were manually labeled with population-averaged aDWI to yield an atlas with comprehensive anatomical information.
Significance of integration of high resolution ex vivo DTI data into in vivo space
The high DTI resolution benefits from the lengthy postmortem scan that has several apparent advantages. Postmortem MRI data (Lerch et al., 2012; Calabrese et al., 2015) usually has a high signal-to-noise ratio and is not complicated by physiologic factors, subject motion, EPI-16 induced eddy current distortions or susceptibility artifacts (e.g. Huang et al., 2008) which are especially severe with the big sinuses in the macaque head. However, there are several tradeoffs with the high resolution postmortem DTI, compared to the macaque brain atlases based on in vivo DTI (Adluru et al., 2012; Zakszewski et al., 2014; Liu et al., 2015, Shi et al., 2017). The postmortem samples were shrunk due to the fixation process. Furthermore, to minimize the potential motion of the samples during the lengthy MRI scan, the samples were usually tightly fitted into the container and the confinement from the container would cause further distortion of the macaque brain in the MR data. The above-mentioned processes are unavoidable and influence anatomical accuracy. With invasive experiments on live macaques irreplaceable in neurobiological studies (e.g. Hendry and Yoshioka 1994; Hendry and Calkins 1998; Steinmetz et al, 2000), the bony and facial landmarks need to be preserved in macaque atlases for guiding these procedures, but are usually removed in postmortem scans. To overcome these tradeoffs and still take advantage of the high resolution of postmortem brain scans, we integrated high resolution ex vivo DTI data and in vivo T1-weighted images (Fig 1) to keep accurate live brain anatomy and bony/facial landmarks of the head (Figs 6-8). Such an integration approach is made possible by the high quality of the image registration. This procedure can be potentially used for brain atlas making of other species.
Morphological and microstructural properties of macaque brain white matter tracts
The topology of the corpus callosum (Hofer et al., 2008) of macaque brain (Fig 3c) is similar to that of the human brain (3D presentations not shown). Of the note, arcuate fasciculus, related to language and other functions unique to humans, could not be reliably traced in the macaque brain and is not shown. Other association tracts were successfully traced, including the uncinate fasciculus, inferior fronto-occipital fasciculus and inferior longitudinal fasciculus, and appear thinner and narrower in the macaque brain (Fig 3d) compared to those in the human brain (Wakana et al., 2004). This suggests that the connected cortical regions of these association tracts are not as widely distributed as those in the human brain (Huang et al., 2011).
This study has provided unique insights into the microstructural profiles of major WM tracts and into the microstructural organization among the major WM tract groups. The distinctive microstructural properties of the five tract groups can be seen as the clusters of data points in the same colors (Fig 9), with different colors indicating different tract groups. FA can be considered approximately as the ratio of AxD over RD. The almost linear relationship of AxD-FA and RD-FA plots in Fig 9b and 9c suggest FA changes are contributed by uniform variations of AxD and RD among the tracts. Specifically, a high FA is characterized by a simultaneously high AxD and a low RD, and vice versa. Similar to human brain microstructural properties in the literature (Wakana et al., 2007), the limbic and association tracts have relatively low FA (low AxD and high RD) while the projection and commissural tracts have relatively high FA (high AxD and low RD). The FA of macaque projection tract is the highest, differing from FA profiles in the human brain where FA values of the callosal tracts are the highest (Wakana et al., 2007). These measurements (FA, MD, AxD and RD) obtained from in vivo DTI represent live brain WM tract microstructural properties which can be significantly altered by fixation procedures (Dyrby et al., 2011).
Data sharing, technical considerations and future directions
All data will be shared with the public through the website www.brainmrimap.org. These shared datasets include population-averaged and single-subject high resolution DTI data in in vivo space with or without bony and facial landmarks as well as in vivo T1 weighted image. We expect these datasets will be used for parcellating neuroanatomical structures of users’ macaque subject MRI by transforming the delineated labels from these digital atlases. We also expect the bony and facial landmarks will be able to help design the most convenient and effective invasive protocol for in vivo macaque studies such as neurophysiological experiments. The microstructural measures of the macaque WM tract were probably affected by the partial volume effects. However, the partial volume effects may shift the AxD, RD and MD values of all tracts uniformly and therefore less likely affect the relationship of the microstructures among various tracts, namely the microstructural profile or organization observed in Fig 9. An analysis pipeline tailored for macaque brain and similar to tract-based spatial statistics (Smith et al, 2006) in measuring human brain WM microstructure at its center (”core”) may need to be established in the future to improve the accuracy of macaque brain WM measurements. To fully utilize the atlas, the future direction of this work would include developing a user-friendly software platform with the established atlas at the center to guide invasive procedures and analysis of experimental data. The platform will include both a surgical guidance toolkit and a data organization/registration toolkit. The software platform will be highly interactive and incorporate visualization of invasive needles to aid the surgical plan and help achieve minimum invasiveness. MR images and atlas labels bundled in the platform will also be able to be used as templates for neurobiological data reports in cases that no experimental macaque MRI data is available.
Acknowledgments
This study was supported by NIH grants R21 EB009545, R01MH092535 and U54 HD086984.
References
- Adluru N, Zhang H, Fox AS, Shelton SE, Ennis CM, Bartosic AM, Oler JA, Tromp do PM, Zakszewski E, Gee JC, Kalin NH, Alexander AL. A diffusion tensor brain template for rhesus macaques. Neuroimage. 2012;59(1):306–318. doi: 10.1016/j.neuroimage.2011.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Estimation of the effective self-diffusion tensor from the NMR spin echo. J Magn Reson. 1994;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Beaulieu C. The basis of anisotropic water diffusion in the nervous system - a technical review. NMR Biomed. 2002;15:435–455. doi: 10.1002/nbm.782. [DOI] [PubMed] [Google Scholar]
- Behrens TE, Berg HJ, Jbabdi S, Rushworth MF, Woolrich MW. Probabilistic diffusion tractography with multiple fibre orientations: What can we gain? Neuroimage. 2007;34(1):144–155. doi: 10.1016/j.neuroimage.2006.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KJ, Koller JM, Snyder AZ, Perlmutter JS. Template images for nonhuman primate neuroimaging: 2. Macaque. Neuroimage. 2001;4(3):744–748. doi: 10.1006/nimg.2001.0871. [DOI] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. Neuroimage. 2015;117:408–416. doi: 10.1016/j.neuroimage.2015.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catani M, Howard RJ, Pajevic S, Jones DK. Virtual in vivo interactive dissection of white matter fasciculi in the human brain. Neuroimage. 2002;17:77–94. doi: 10.1006/nimg.2002.1136. [DOI] [PubMed] [Google Scholar]
- Dyrby TB, Baare WFC, Alexander DC, Jelsing J, Garde E, Sogaard LV. An ex vivo imaging pipeline for producing high-quality and high-resolution diffusion weighted imaging datasets. Hum Brain Mapp. 2011;32:544–563. doi: 10.1002/hbm.21043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey S, Pandya DN, Chakravarty MM, Bailey L, Petrides M, Collins DL. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space) Neuroimage. 2011;55(4):1435–1442. doi: 10.1016/j.neuroimage.2011.01.040. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Calkins DJ. Neuronal chemistry and functional organization in the primate visual system. Trends Neurosci. 1998;21:344–349. doi: 10.1016/s0166-2236(98)01245-4. [DOI] [PubMed] [Google Scholar]
- Hendry SH, Yoshioka T. A neurochemically distinct third channel in the macaque dorsal lateral geniculate nucleus. Science. 1994;264:575–577. doi: 10.1126/science.8160015. [DOI] [PubMed] [Google Scholar]
- Hofer S, Merboldt KD, Tammer R, Frahm J. Rehsus monkey and human share a similar topography of the corpus callosum as revealed by diffusion tensor MRI in vivo. Cereb Cortex. 2008;18:1079–1084. doi: 10.1093/cercor/bhm141. [DOI] [PubMed] [Google Scholar]
- Huang H, Ceritoglu C, Li X, Qiu A, Miller MI, van Zijl PCM, Mori S. Correction of B0 susceptibility induced distortion in diffusion-weighted images using large-deformation diffeomorphic metric mapping. Magn Reson Imaging. 2008;26:1294–1302. doi: 10.1016/j.mri.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Fan X, Weiner M, Martin-Cook K, Xiao G, Davis J, Devous M, Rosenberg R, Diaz-Arrastia R. Distinctive disruption patterns of white matter tracts in Alzheimer’s disease with full diffusion tensor characterization. Neurobiol Aging. 2012a;33:2029–2045. doi: 10.1016/j.neurobiolaging.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gundapuneedi T, Rao U. White matter disruptions in adolescents exposed to childhood maltreatment and vulnerability to psychopathology. Neuropsychopharmacology. 2012b;37:2693–2701. doi: 10.1038/npp.2012.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Prince JL, Mishra V, Carass A, Landman B, Park DC, Tamminga C, King R, Miller MI, van Zijl PC, Mori S. A framework on surface-based connectivity quantification for the human brain. J Neurosci Methods. 2011;197(2):324–32. doi: 10.1016/j.jneumeth.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Yamamoto A, Hossain MA, Younes L, Mori S. Quantitative cortical mapping of fractional anisotropy in developing rat brains. J Neurosci. 2008;28:1427–1433. doi: 10.1523/JNEUROSCI.3194-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Van Zijl PC, Kim J, Pearlson GD, Mori S. DtiStudio: resource program for diffusion tensor computation and fiber bundle tracking. Comput Methods Programs Biomed. 2006;81:106–116. doi: 10.1016/j.cmpb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Jones DK, Horsfield MA, Simmons A. Optimal strategies for measuring diffusion in anisotropic systems by magnetic resonance imaging. Magn Reson Med. 1999;42:515–525. [PubMed] [Google Scholar]
- Lerch JP, Gazdzinski L, Germann J, Sled JG, Henkelman RM, Nieman BJ. Wanted dead or alive? The tradeoff between in-vivo versus ex-vivo MR brain imaging in the mouse. Front Neuroinform. 2012;66 doi: 10.3389/fninf.2012.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Tian X, Liu H, Mo Y, Bai F, Zhao X, Ma Y, Wang J. Rehsus monkey brain development during late infancy and the effect of phencyclidine: A longitudinal MRI and DTI study. Neuroimage. 2015;107:65–75. doi: 10.1016/j.neuroimage.2014.11.056. [DOI] [PubMed] [Google Scholar]
- Martin R, Bowden DM. A stereotaxic template atlas of the macaque brain for digital imaging and quantitative neuroanatomy. NeuroImage. 1996;4:119–150. doi: 10.1006/nimg.1996.0036. [DOI] [PubMed] [Google Scholar]
- Martin R, Bowden DM. Primate brain maps: structure of the macaque brain. Amsterdam: Elsevier; 2000. [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC. A population-average MRI-based atlas collection of the rhesus macaque. Neuroimage. 2009;45(1):52–59. doi: 10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MI, Trouve A, Younes L. On the metrics and Euler-Lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC. Three-dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol. 1999;45:265–269. doi: 10.1002/1531-8249(199902)45:2<265::aid-ana21>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Mori S, Oishi K, Jiang H, Jiang L, Li X, Akhter K, Hua K, Faria AV, Mahmood A, Woods R, Toga AW. Stereotaxic white matter atlas based on diffusion tensor imaging in an ICBM template. Neuroimage. 2008;40(2):570–582. doi: 10.1016/j.neuroimage.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Huang XF, Toga AW. The Rhesus Monkey Brain I Stereotaxic Coordinates. 2. Academic Press; San Diego: 2009. [Google Scholar]
- Pierpaoli C, Basser PJ. Toward a quantitative assessment of diffusion anisotropy. Magn Reson Med. 1996;36:893–906. doi: 10.1002/mrm.1910360612. [DOI] [PubMed] [Google Scholar]
- Rohlfing T, Kroenke CD, Sullivan EV, Dubach MF, Bowden DM, Grant KA, Pfefferbaum A. The INIA19 template and neuromaps atlas for primate brain image parcellation and spatial normalization. Front Neuroinform. 2012;6:27. doi: 10.3389/fninf.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem KS, Logothetis NK. A combined MRI and histology atlas of the rhesus monkey brain in stereotaxic coordinates. San Diego: Elsevier; 2007. [Google Scholar]
- Schmahmann D, Pandya DN. Fiber pathways of the brain. Oxford Univ Press; USA: 2006. [Google Scholar]
- Shi Y, Budin F, Yapuncich E, Rumple A, Young JT, Payne C, Zhang X, Hu X, Godfrey J, Howell B, Sanchez MM, Styner MA. UNC-Emory infant atlases for macaque brain image analysis: Postnatal brain development through 12 months. Front Neurosci. 2017;10:617. doi: 10.3389/fnins.2016.00617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, Nichols TE, Mackay CE, Watkins KE, Ciccarelli O, Cader MZ, Matthews PM, Behrens TE. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage. 2006;31:1487–1505. doi: 10.1016/j.neuroimage.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Song SK, Yoshino J, Le TQ, Lin SJ, Sun SW, Cross AH, Armstrong RC. Demyelination increases radial diffusivity in corpus callosum of mouse brain. NeuroImage. 2005;26:132–140. doi: 10.1016/j.neuroimage.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Steinmetz PN, Roy A, Fitzgerald PJ, Hsiao SS, Johnson KO, Niebur E. Attention modulates synchronized neuronal firing in primate somatosensory cortex. Nature. 2000;404:187–190. doi: 10.1038/35004588. [DOI] [PubMed] [Google Scholar]
- Stieltjes B, Kaufmann WE, van Zijl PC, Fredericksen K, Pearlson GD, Solaiyappan M, Mori S. Diffusion tensor imaging and axonal tracking in the human brainstem. Neuroimage. 2001;14:723–735. doi: 10.1006/nimg.2001.0861. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Dierker DL. Surface-based and probabilistic atlases of primate cerebral cortex. Neuron. 2007;56(2):209–225. doi: 10.1016/j.neuron.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Wakana S, Jiang H, Nagae-Poetscher LM, Van Zijl PC, Mori S. Fiber tract-based atlas of human white matter anatomy. Radiology. 2004;230:77–87. doi: 10.1148/radiol.2301021640. [DOI] [PubMed] [Google Scholar]
- Wakana S, Caprihan A, Panzenboeck MM, Fallon JH, Perry M, Gollub RL, Hua K, Zhang J, Jiang H, Dubey P, Blitz A, Van Zijl PC, Mori S. Reproducibility of quantitative tractography methods applied to cerebral white matter. NeuroImage. 2007;36:630–644. doi: 10.1016/j.neuroimage.2007.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods RP, Grafton ST, Holmes CJ, Cherry SR, Mazziotta JC. Automated image registration: I. General methods and intrasubject, intramodality validation. J Comput Assist Tomogr. 1998;22:139–152. doi: 10.1097/00004728-199801000-00027. [DOI] [PubMed] [Google Scholar]
- Xu D, Mori S, Shen D, van Ziji PC, Davatzikos C. Spatial normalization of diffusion tensor fields. Magn Reson Med. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- Zakszewski E, Adluru N, Tromp do PM, Kalin N, Alexander AL. A diffusion-tensor-based white matter atlas for rhesus macaques. PLoS One. 2014;9(9):e107398. doi: 10.1371/journal.pone.0107398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Richards LJ, Yarowsky P, Huang H, van Zijl PC, Mori S. Three-dimensional anatomical characterization of the developing mouse brain by diffusion tensor microimaging. Neuroimage. 2003;20:1639–1648. doi: 10.1016/s1053-8119(03)00410-5. [DOI] [PubMed] [Google Scholar]


