Abstract
Simultaneous monitoring of biomarkers as well as single-cell analyses based on flow cytometry and mass cytometry are important for investigations of disease mechanisms, drug discovery, and signaling-network studies. Flow cytometry and mass cytometry are complementary to each other; however, probes that can satisfy all the requirements for these two advanced technologies are limited. In this study, we report a probe of lanthanide-coordinated semiconducting polymer dots (Pdots), which possess fluorescence and mass signals. We demonstrated the usage of this dual-functionality probe for both flow cytometry and mass cytometry in a mimetic cell mixture and human peripheral blood mononuclear cells as model systems. The probes not only offer high fluorescence signal for use in flow cytometry, but also show better performance in mass cytometry than the commercially available counterparts.
Keywords: flow cytometry, mass cytometry, lanthanide, Pdots
Graphical Abstract
Lanthanide-coordinated semiconducting polymer dots with bright fluorescence was demonstrated the outstanding performance in both flow cytometry and mass cytometry for cell analyses.
Highly multiplexed detection of biomarkers in individual cells will impact cell research,[1] drug discovery,[2] disease diagnosis, and treatment.[3] Clinical samples are scarce so high-throughput assays are critical for efficiently extracting as much information as possible from limited samples.[4] Immunologists, hematologists, and oncologists are looking for the highly efficient multiplexed assays that will satisfy their requirements for cell analysis with small samples.[5] Biologists, for example, would like to classify heterogeneous cell populations using multiplexed assays that permit simultaneous detection of multiple biomarkers in a single sample.[6] Moreover, multiple biomarkers detection on a single cancer cell are critical for diagnosis and treatment since only one kind of marker can’t be relied on to identify the cancer type and stage.[7] Immunologists’ studies of cellular features, diverse cellular networks, and immune-system responses heavily rely on high-throughput, single-cell analyses.[3c,8]
Flow cytometry is a widely used single-cell assay, and has well-established commercially available reagents and instruments.[9] However, the broad-emission spectra of the fluorescent tags limit flow cytometry when it comes to highly multiplexed assays.[10] Mass cytometry was developed to further push the multiplexing capability of flow cytometry. Mass cytometry can accomplish the simultaneous detection of dozens of biomarkers, which is useful for multi-parameter, single-cell analyses in heterogeneous small samples, such as immune and cancer cells.[11] Mass cytometry uses antibodies labeled with metal isotopes to stain the cells in a way similar to what is done for flow cytometry. After staining, the cells are injected individually into the plasma torch of an inductively coupled plasma mass spectrometer. By quantitatively analyzing the abundance of metal ions for each cell, the relative levels of biomarkers on a single cell can be quantified.[3a, 12] Currently, the commercially available mass-cytometry tags are mainly based on the lanthanide isotopes, which have almost zero background signal in biological samples. Considering the availability of additional metal isotopes with masses in the range of 75 to 209 atomic mass unit, over 100 parameters potentially can be simultaneously measured in a single sample without requiring compensation between mass signals.[5a,13] The reported probes for mass cytometry are antibodies modified with metal-chelating polymers (MCPs), which carry 150–250 metal atoms per antibody.[11a,14]
Mass cytometry and flow cytometry are highly complementary methods. While mass cytometry offers a high degree of multiplexing, flow cytometry has better sensitivity and higher throughput, and cells are not destroyed during the process of detection and analysis. With optical detection, it is also possible to sort cells, a feature inherently not possible with mass cytometry. Finally, flow cytometry is a better-established method, and thus can be used to troubleshoot issues encountered in mass cytometry. A probe that can be used in both flow and mass cytometry, therefore, will find broad utility. Indeed, Majonis et al. reported the first dual-purpose probe with fluorescent and mass tags contained in a single polymer.[15] While this work represents an important first demonstration, as the authors pointed out, the developed probes showed borderline performance because of the relatively low mass and optical signals. Here, we describe a new strategy to design and implement dual fluorescent-mass probes that offer excellent optical and mass signals, and which overcome limitations of existing probes.
Lanthanide-doped microparticles and lanthanide inorganic nanoparticles are two types of reported probes that contain higher loading of metal atoms to allow for more sensitive detection by mass cytometry.[16] However, the non-specific binding of these probes and their large sizes (>100 nm) caused low detection efficiency in mass cytometry and even lower staining capacity compared to MCP probes.[16a, 16b] To improve the performance of these inorganic nanoparticles, Winnik et al. developed small uniform lanthanide nanoparticles with a diameter of about 80nm.[16c, 16d] Therefore, the design of small nanoparticles containing a large amount of metal atoms with low non-specific binding remains a challenge for mass cytometry.
Based on our development of a series of semiconducting polymer dot (Pdots) as fluorescent probes for cell analysis by flow cytometry,[17] we have four reasons to believe that Pdots can be a promising candidate for mass cytometry if enough metal atoms were loaded into them. First, Pdots show low non-specific binding, which has been demonstrated in our previous work with flow cytometry.[18] Second, the size (typically 10 – 30 nm in diameter) of Pdots ensures that they will carry enough metal atoms for the purposes of mass cytometry. Third, the small size of metal-loaded Pdots will be completely converted into mass signals in the ICP torch, which will increase sensitivity. Finally, the extraordinary fluorescence properties of Pdots[17] will make them as ideal fluorescence and mass reporters for single-cell analyses.
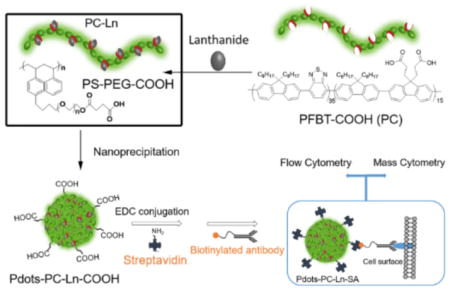
We designed and synthesized lanthanide-coordinated Pdots with high fluorescence and mass signals (Scheme 1). The lanthanide ions were first chelated by the carboxyl-functionalized poly[(9,9-dioctylfluorenyl-2,7-diyl)-co-(1,4-benzo-{2,10,3}-thiadazole)] polymer (abbreviated as PC) in THF for an hour. Immediately after the mixing with the block copolymer, polystyrene-grafted ethylene oxide functionalized with carboxyl groups (PS-PEG-COOH), the THF solution was injected into water to form lanthanide-coordinated Pdots (Pdot-PC-Ln-COOH). The carboxyl groups on Pdots were used for the bioconjugation with streptavidin (SA) to form Pdot-PC-Ln-SA for labeling the cell-surface receptors. The excellent fluorescence properties of the Pdots ensured that we had a bright fluorescence tag for flow cytometry and imaging. The heavily loaded lanthanides in the Pdots provided an excellent metal probe for mass cytometry. Furthermore, we can adjust the fluorescence color by using different Pdots synthesized with carboxyl-functional groups.[18b,19] Also, by changing the lanthanide salt, different Ln atoms can be coordinated into Pdots. Combining these two features, we potentially can create a series of dual-functionality probes for both flow and mass cytometry.
Scheme 1.

Schematic depiction of dual-functionality Pdots’ preparation, bioconjugation, and cell labeling for flow cytometry and mass cytometry.
In this design, the polymer PC not only acted as a fluorescent unit, but also was used to chelate with lanthanides to form a stable lanthanide-polymer complex. The feeding ratio of lanthanides to polymer played an important role because it affected the diameters and loading density of lanthanides into the Pdots. From our previous studies, we know that smaller-diameter Pdots improved the labeling efficiency in cells,[20] which is important for both flow and mass cytometry. However, the smaller Pdots would decrease the amounts of lanthanides loaded per Pdot, which might sacrifice the sensitivity of the mass-cytometric studies. Therefore, using europium (Eu) as a model lanthanide, we found out that the optimal Eu density and Pdots’ diameter was achieved when the molar feeding ratio between the number of carboxyl groups on the polymer to the number of Eu3+ was 2 (Figure S1a). The best ratio of PS-PEG-COOH over PC to form ideal Pdots with adequately small size and optimal Eu loading density was 40% wt (Figure S1b).
To test the generality of the preparation protocol for making other lanthanide-coordinated Pdots, five different lanthanide ions, Nd3+, Eu3+, Tb3+, Ho3+ and Er3+, were used to prepare Pdots-PC-Ln-COOH. The diameters of the Pdots-PC-Ln-COOH were measured to be in the range of 20 – 30 nm (Figure 1a–b, and Figure S2 a–c) as determined by high-resolution transmission electron microscopy (TEM) imaging (Figure 1c–d, and Figure S2 d–f). The z-average hydrodynamic diameters measured by DLS were slightly larger while the number-average diameters from DLS were slightly smaller than the size determined by TEM. The absorption spectra and fluorescence spectra showed the same profile among the different lanthanides (Figure 1e and f). The quantum yields (QY) of these Pdots-PC-Ln-COOH varied depending on the coordinated lanthanides (Table 1 and Table S1), but were higher than that of the pure Pdots-PC-COOH. The low quantum yield of pure Pdots-PC-COOH is because of the high percentage of carboxyl groups on the polymer based on our previous studies.[21]
Figure 1.
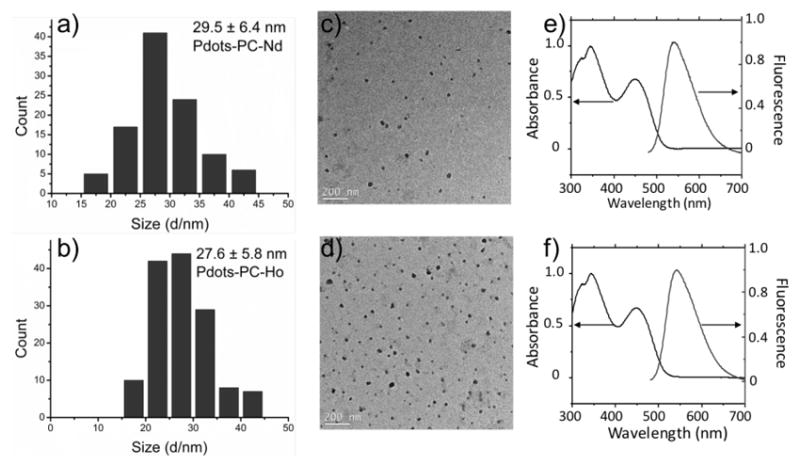
(a–b) Histograms of the distribution of the sizes of Pdot-PC-Nd-COOH and Pdot-PC-Ho-COOH, respectively, measurement from more than 100 particles in TEM images (the mean size is (a) 29.5 ± 6.4 nm, and (b) 27.6 ± 5.8 nm). (c–d) TEM images of Pdot-PC-Nd-COOH and Pdot-PC-Ho-COOH, respectively. (e–f) Absorption and fluorescence spectra of Pdot-PC-Nd-COOH and Pdot-PC-Ho-COOH, respectively.
Table 1.
Size, single particle absorption cross section, quantum yield, and average loading number of lanthanides in Pdots-PC-Ln.
| Size[a] (nm) | σ[b] (×10−13 cm2) | QY[c] (%) | Average Ln/Pdot[d] | Ln/nm3[e] | |
|---|---|---|---|---|---|
| Pdot-PC-Nd | 29.5±6.4 | 4.77 | 11.9±0.1 | 1990 | 0.148 |
| Pdot-PC-Ho | 27.6±5.8 | 3.96 | 11.5±0.2 | 1500 | 0.138 |
Size measured by DLS.
Single-particle absorption cross-section,
Absolute photoluminescence quantum yield.
Average lanthanide amount per Pdots measured by ICP-MS.
Lanthanide loading density in Pdots.
The lanthanide content in Pdots were measured by ICP-MS. Table 1 and Table S1 summarize the photophysical properties and lanthanide content of the five different Ln-coordinated Pdots. The lanthanides’ amount per Pdot on average changed from 1100 to 2000, which was at least 7 times higher than the commercial probe for each antibody (150 Ln/antibody).[22] We therefore anticipated that our Pdots would provide a higher signal in mass cytometry compared to commercial probes. Although the lanthanide amount per Pdot changed over a large range, the metal’s density (number of Ln per unit volume of Pdots) showed similar values, indicating that amount of Ln per Pdot could be adjusted by varying the size of the Pdots. Moreover, the Pdots showed no noticeable leakage of the metal ions in both HEPES and PBS buffer, indicating good stability (Figure S4).
To test the performance of the lanthanide-coordinated Pdots for both flow and mass cytometry, a cell mixture of MCF-7 breast-cancer cells and Jurkat T cells was chosen as a model system. Cell-surface biomarker, EpCAM, highly-expressed on the MCF-7 cell was the target biomarker. The T cell, which has no EpCAM, was used as a negative internal control. The biotinylated primary anti-CD326 (EpCAM) antibody was first incubated with the cell mixture, followed by the incubation of Pdot-PC-Nd-SA to examine the specific labeling of MCF-7 cells. The labeled MCF-7 cells showed strong fluorescence compared to the control T cells (Figure S5a), indicating effective and specific discrimination of the MCF-7 over T cells. Moreover, light-scattering results seen from the two distinctive peaks clearly showed the presence of the two different cells (Figure S5b and S5c). Fluorescence imaging was further conducted to confirm the specific cellular labeling by the Pdot-PC-Nd-SA (Figure S5d). High levels of fluorescence were observed on MCF-7 cells. No fluorescence was detected from the internal negative-control T cells, which indicated that there was little or none non-specific binding of the Pdots.
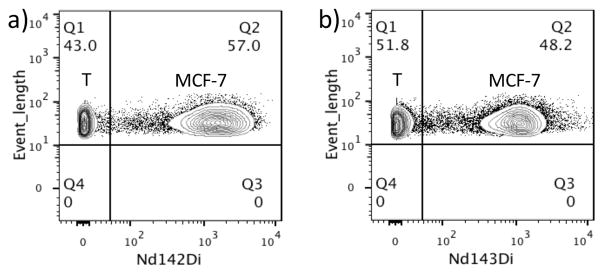
After the flow-cytometric analysis, the fixed cells were incubated with a Rh103-containing DNA intercalator to identify intact cells during mass-cytometric analysis. Cells were selected from the Rh103-positive clusters (Figure S6), and then plotted for the Nd signals. Taking costs into consideration, all lanthanides used in this work were natural species containing naturally existing isotopes. Therefore, for Nd signal, we chose the most abundant isotope, Nd142 (27.2%), to plot the results of mass cytometry. For comparison, a commercial probe, anti-biotin-Nd143, was used to label the same cell mixture in a side-by-side experiment. Figure 2 shows the two separated populations were clearly formed along the Nd142 and Nd143 axis, representing negative-control T cells and positive MCF-7 cells. The mean intensity of Nd142 from our Pdot-stained system was 1598, which was higher than that of the commercial probe (1185 for Anti-Biotin-Nd143 from Fluidigm) (Table S2). Meanwhile, the mean mass intensity of the negative control T cells in our Pdots stained system was 2.04, with standard deviation of 5.32, both of which were smaller than that of the commercial probe (4.40 for mean intensity and 6.73 for standard deviation). The results indicated that the Pdots-PC-Nd-SA possessed a better discrimination of the cells and higher sensitivity.
Figure 2.
Mass cytometry of the MCF-7/T cell mixture using anti-human CD326 EpCAM antibody as the primary antibody, followed by labeling with (a) 10 ppm Pdots-PC-Nd-SA, and (b) commercially available anti-biotin-Nd143. Q1 represents T cells and Q2 represents MCF-7 cells.
Next, we titrated the MCF-7/T cell mixture using Pdot-PC-Nd-SA at concentrations of 1 ppm, 2 ppm, 5 ppm, and 10 ppm. As shown in Figure 3, the results of flow and mass cytometry were compared to each other. Stain index (SI) for flow cytometry and mean mass intensity for mass cytometry were calculated and shown in Table S3. With the increase in Pdot-PC-Nd-SA concentration, the SI and mean mass intensity increased in these two modes, with the highest SI and mean mass intensity at 10 ppm of Pdot-PC-Nd-SA. We also found that different neodymium isotope channels showed different mean mass intensity, which is proportional to the abundance of the different isotopes (Figure S7). Taking the isotopic composition into consideration (27.2 % Nd142), the mean intensity in Figure 2a would be ~5875, which is about 4 times higher than that shown in Figure 2b if pure Nd142 was used. Thus, we hypothesized that the Pdots will generate a better performance in mass cytometry if a pure isotope was used for the preparation of the Pdots-PC-Ln-SA. We also investigated the quantitative detection ability of our Pdot-PC-Ln by mass cytometry. In the MCF-7/T cells system, we fixed the number of MCF-7 cells to 1 million and changed the T- cell population to 0.2, 0.5, 0.75 and 1 million. Figure S8 shows both flow cytometry and mass cytometry measurements using Pdots-PC-Nd-SA had a relative increase of the negative peak of T cells; this indicated that our Pdots-PC-Ln-SA had the ability to quantitatively measure a population of a certain cell type. Furthermore, we demonstrated our Pdot system could be used for the multiplexed detection of different targets (Figure S9). In the MCF-7/T cell system, EpCAM on MCF-7 cells and CD45 on T cells were successfully stained with Pdot-PC-Tb and Pdot-PC-Ho, respectively.
Figure 3.
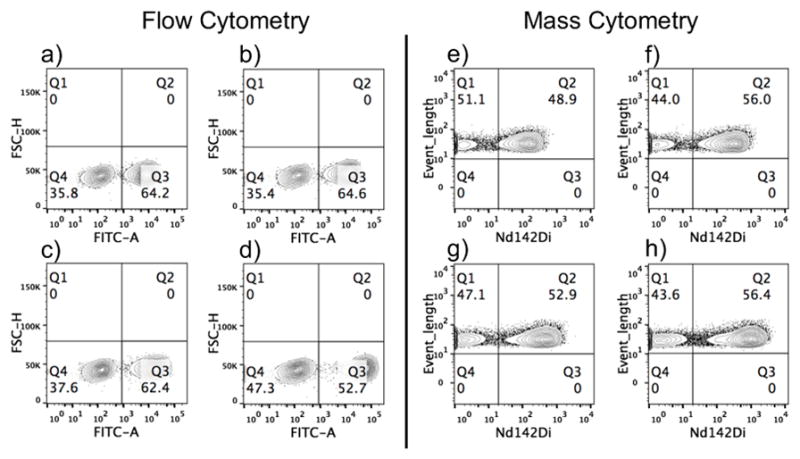
Titration of MCF-7/T cell mixtures using Pdots-PC-Nd-SA with flow cytometry (a–d) and mass cytometry (e–g). a) and e): 1 ppm; b) and f): 2 ppm; c) and g): 5 ppm; d) and h): 10 ppm. In (a–d), Q4 represents T cells and Q3 represents MCF-7 cells; in (e–h), Q1 represents T cells and Q2 represents MCF-7 cells.
Because mass cytometry is often employed for studies in immunology, we investigated the performance of lanthanide-coordinated Pdots in human peripheral blood mononuclear cells (PBMCs). Figure S10 shows the live and dead cells could be distinguished by cis-platin and Ir intercalator, and followed by the staining of CD45-specific Pdot-PC-Ho. CD3+ expressed on T cells was used as the target biomarker to be labeled with Pdots. To overcome the effect of isotope mixtures (Figure S7), we prepared the Pdot-PC-Ho-SA because natural holmium has 100% abundance of Ho165. PBMCs were incubated with different concentrations of Pdot-PC-Ho-SA after an incubation with the biotinylated primary anti-CD3 antibody. Figure 4 and Table S4 show data from both flow (Figure S11 and Table S5) and mass cytometry (Figure S12 and Table S6) had clear separation of CD3+ T cells from the rest of the negative-control cells in PBMCs, which indicated that Pdots-PC-Ho-SA could effectively and specifically label immune cells and analyze the target biomarkers in both flow cytometry and mass cytometry.
Figure 4.
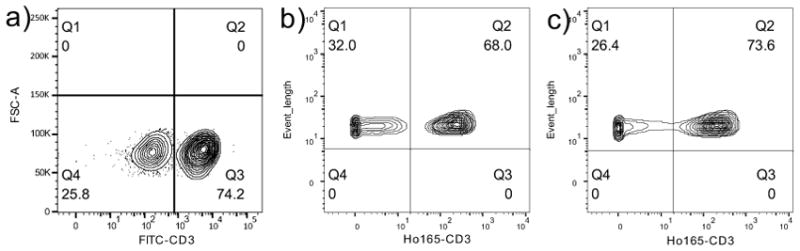
Flow cytometry (a) and mass cytometry (b–c) detection of CD3+ T cells in PBMCs using biotinlyated anti-human CD3 antibody as the primary antibody, followed by labeling with (a–b) 2 ppm Pdots-PC-Ho-SA, and (c) commercial available anti-biotin-Ho165 probe.
By staining with the commercially available anti-biotin-Ho165 probe for CD3+ T cells (Figures 4c, S13, and Tables S7), the highest mean mass intensity for the commercial anti-biotin-Ho165 probe was lower than most of the mean mass intensities of our Pdot-PC-Ho-SA in the mass-cytometric analyses with different concentrations. The enhancement of the higher positive signal intensity of Pdot-PC-Ho-SA was because there were more Ho atoms in each Pdot-PC-Ho-SA than that in the commercial anti-biotin-Ho165 probe. We believe that the Pdots coordinated with lanthanides will be very useful probes for cell analyses and cell separation by two advanced analytical methods: flow cytometry and mass cytometry.
In conclusion, we designed and synthesized dual- functionality Pdots with high loading of lanthanide atoms and demonstrated their applications in flow and mass cytometry to detect specific biomarkers. The high density of lanthanides and small size of the Pdots enabled efficient and specific cell labeling with low non-specific binding. However, the relatively broad size distribution of the Pdots may limit their performance; we are currently working to address this issue by developing new methods to generate Pdots with monodisperse size distributions. Nevertheless, the ability of the Pdots to load more metal atoms enabled them to offer comparable or higher sensitivity and mean mass intensity when compared with commercially available mass tags in both cancer cell lines and PBMCs. The Pdots showed similar background signal as the commercial probe, which indicated low non-specific binding with cells. Additionally, the high brightness offered by Pdots also make them valuable probes in flow cytometry. This Pdots-based system could potentially be expanded to form a library of probes that combine different fluorescence colors and metal isotopes, which will facilitate the development of multiplexed detection for single-cell analyses.
Supplementary Material
Acknowledgments
We are grateful to the NIH (CA186798 and DK097653) for the support of this work.
Footnotes
Supporting information is given via a link at the end of the paper.
Contributor Information
Xu Wu, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Dr. Prof. Quinn DeGottardi, Benaroya Research Institute at Virginia Mason, Seattle, WA 98101 (USA). Department of Medicine, University of Washington, Seattle, WA 98195 (USA)
I-Che Wu, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Jiangbo Yu, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Li Wu, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Fangmao Ye, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Prof. Chun-Ting Kuo, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA)
William W. Kwok, Benaroya Research Institute at Virginia Mason, Seattle, WA 98101 (USA). Department of Medicine, University of Washington, Seattle, WA 98195 (USA)
Daniel T. Chiu, Department of Chemistry, University of Washington Seattle, WA 98195 (USA)
References
- 1.Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK. Trends Immunol. 2012;33:323–332. doi: 10.1016/j.it.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkuri KR, Stevens JC, Neubert H. Drug Metab Dispos. 2015;43:227–233. doi: 10.1124/dmd.114.060798. [DOI] [PubMed] [Google Scholar]
- 3.a) Spitzer MH, Nolan GP. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Kingsmore SF. Nat Rev Drug Discov. 2006;5:310–320. doi: 10.1038/nrd2006. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Maecker HT, Lindstrom TM, Robinson WH, Utz PJ, Hale M, Boyd SD, Shen-Orr SS, Fathman CG. Nat Rev Rheumatol. 2012;8:317–328. doi: 10.1038/nrrheum.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.a) Abdelrahman AI, Dai S, Thickett SC, Ornatsky O, Bandura D, Baranov V, Winnik MA. J Am Chem Soc. 2009;131:15276–15283. doi: 10.1021/ja9052009. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Chattopadhyay PK, Price DA, Harper TF, Betts MR, Yu J, Gostick E, Perfetto SP, Goepfert P, Koup RA, De Rosa SC, Bruchez MP, Roederer M. Nat Med. 2006;12:972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 5.a) Newell EW, Davis MM. Nat Biotechnol. 2014;32:149–157. doi: 10.1038/nbt.2783. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Bodenmiller B, Zunder ER, Finck R, chen TJ, Savig ES, Bruggner RV, Simonds EF, Bendall SC, Sachs K, Krutzik PO, Nolan GP. Nat Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Angelo M, Bendall SC, Finck R, Hale MB, Hitzman C, Borowsky AD, Levenson RM, Lowe JB, Liu SD, Zhao S, Natkunam Y, Nolan GP. Nat Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B. Nat Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 7.Majonis D, Herrera I, Ornatsky O, Schulze M, Lou X, Soleimani M, Nitz M, Winnik MA. Anal Chem. 2010;82:8961–8969. doi: 10.1021/ac101901x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Harvey CJ, Wucherpfennig KW. Nat Biotechnol. 2013;31:609–610. doi: 10.1038/nbt.2626. [DOI] [PubMed] [Google Scholar]; b) Bendall SC, Simonds EF, Qiu P, Amir el AD, Krutzik PO, Finck R, Bruggner RV, Melamed R, Trejo A, Ornatsky OI, Balderas RS, Plevritis SK, Sachs K, Pe’er D, Tanner SD, Nolan GP. Science. 2011;332:687–696. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shapiro H. Practical Flow Cytometry. 4. John Wiley & Sons, Inc; 2003. [Google Scholar]
- 10.Perfetto SP, Chattopadhyay PK, Roederer M. Nat Rev Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 11.a) Lou X, Zhang G, Herrera I, Kinach R, Ornatsky O, Baranov V, Nitz M, Winnik MA. Angew Chem Int Ed. 2007;46:6111–6114. doi: 10.1002/anie.200700796. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2007;119:6223–6226. [Google Scholar]; b) Frei AP, Bava FA, Zunder ER, Hsieh EW, Chen SY, Nolan GP, Gherardini PF. Nat Methods. 2016;13:269–275. doi: 10.1038/nmeth.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanner SD, Baranov VI, Ornatsky OI, Bandura DR, George TC. Cancer Immunol Immunother. 2013;62:955–965. doi: 10.1007/s00262-013-1416-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newell EW, Sigal N, Nair N, Kidd BA, Greenberg HB, Davis MM. Nat Biotechnol. 2013;31:623–629. doi: 10.1038/nbt.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Illy N, Majonis D, Herrera I, Ornatsky O, Winnik MA. Biomacromolecules. 2012;13:2359–2369. doi: 10.1021/bm300613x. [DOI] [PubMed] [Google Scholar]
- 15.Majonis D, Ornatsky O, Weinrich D, Winnik MA. Biomacromolecules. 2013;14:1503–1513. doi: 10.1021/bm4001662. [DOI] [PubMed] [Google Scholar]
- 16.a) Lin W, Hou Y, Lu Y, Abdelrahman AI, Cao P, Zhao G, Tong L, Qian J, Baranov V, Nitz M, Winnik MA. Langmuir. 2014;30:3142–3153. doi: 10.1021/la403627p. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Vancaeyzeele C, Ornatsky O, Baranov V, Shen L, Abdelrahman A, Winnik MA. J Am Chem Soc. 2007;129:13653–13660. doi: 10.1021/ja073970w. [DOI] [PubMed] [Google Scholar]; c) Tong L, Lu E, Pichaandi J, Zhao G, Winnik MA. J Phys Chem C. 2016;120:6269–6280. [Google Scholar]; d) Pichaandi J, Tong L, Bouzekri A, Yu Q, Ornatsky O, Baranov V, Winnik MA. Chem Mater. 2017;29:4980–4990. [Google Scholar]
- 17.a) Wu C, Chiu DT. Angew Chem Int Ed. 2013;52:3086–3109. doi: 10.1002/anie.201205133. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:3164–3190. [Google Scholar]; b) Wu C, Schneider T, Zeigler M, Yu J, Schiro PG, Burnham DR, McNeill JD, Chiu DT. J Am Chem Soc. 2010;132:15410–15417. doi: 10.1021/ja107196s. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wu I, Yu J, Ye F, Rong Y, Gallina ME, Fujimoto BS, Zhang Y, Chan Y, Sun W, Zhou X, Wu C, Chiu DT. J Am Chem Soc. 2014;137:173–178. doi: 10.1021/ja5123045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.a) Peng HS, Chiu DT. Chem Soc Rev. 2014;44:4699–722. doi: 10.1039/c4cs00294f. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Rong Y, Wu C, Yu J, Zhang X, Ye F, Zeigler M, Gallina ME, Wu IC, Zhang Y, Chan YH, Sun W, Uvdal K, Chiu DT. ACS Nano. 2013;7:376–384. doi: 10.1021/nn304376z. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Sun W, Yu J, Deng R, Rong Y, Fujimoto B, Wu C, Zhang H, Chiu DT. Angew Chem Int Ed. 2013;52:11294–11297. doi: 10.1002/anie.201304822. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem. 2013;125:11504–11507. [Google Scholar]
- 19.Rong Y, Yu J, Zhang X, Sun W, Ye F, Wu IC, Zhang Y, Hayden S, Zhang Y, Wu C, Chiu DT. ACS Macro Lett. 2014;3:1051–1054. doi: 10.1021/mz500383c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun K, Chen H, Wang L, Yin S, Wang H, Xu G, Chen D, Zhang X, Wu C, Qin W. ACS Appl Mater Interfaces. 2014;6:10802–10812. doi: 10.1021/am502733n. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Yu J, Wu C, Jin Y, Rong Y, Ye F, Chiu DT. ACS Nano. 2012;6:5429–5439. doi: 10.1021/nn301308w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mei HE, Leipold MD, Maecker HT. Cytometry Part A. 2016;89:292–300. doi: 10.1002/cyto.a.22778. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.