Abstract
Letermovir is a human cytomegalovirus (CMV) terminase inhibitor that was clinically effective in a Phase III prevention trial. In vitro studies have shown that viral mutations conferring letermovir resistance map primarily to the UL56 component of the terminase complex and uncommonly to UL89. After serial culture of a baseline CMV laboratory strain under letermovir, mutation was observed in a third terminase component in 2 experiments, both resulting in amino acid substitution P91S in gene UL51 and adding to a pre-existing UL56 mutation. Recombinant phenotyping indicated that P91S alone conferred 2.1-fold increased letermovir resistance (EC50) over baseline, and when combined with UL56 mutation S229F or R369M, multiplied the level of resistance conferred by those mutations by 3.5 to 7.7-fold. Similarly a combination of UL56 mutations S229F, L254F and L257I selected in the same experiment conferred 54-fold increased letermovir EC50 over baseline, but 290-fold when combined with UL51 P91S. The P91S mutant was not perceptibly growth impaired. Although pUL51 is essential for normal function of the terminase complex, its biological significance is not well understood. Letermovir resistance mutations mapping to 3 separate genes, and their multiplier effect on the level of resistance, suggest that the terminase components interactively contribute to the structure of a letermovir antiviral target. The diagnostic importance of the UL51 P91S mutation arises from its potential to augment the letermovir resistance of some UL56 mutations at low fitness cost.
Keywords: Cytomegalovirus, antiviral drug resistance, letermovir, terminase
1. Introduction
The preventive antiviral management of human cytomegalovirus (CMV) infection and disease has been a major factor in improving patient outcomes of solid organ and hematopoietic cell transplantation (HCT) over recent decades. During this time, anti-CMV therapy has strongly relied on the nucleoside analog ganciclovir and its oral prodrug valganciclovir, with foscarnet and cidofovir in secondary roles. All of these drugs have dose limiting toxicities and ultimately target the same UL54 CMV DNA polymerase, leading to problems of cross-resistance. Compounds with different CMV antiviral targets are being developed, including the UL97 kinase inhibitor maribavir and the terminase inhibitor letermovir. The latter drug recently showed significant prophylactic efficacy in a Phase III CMV prevention trial in HCT recipients (clinicaltrials.gov NCT02137772) and is a strong candidate to be the first licensed systemic CMV drug with an alternative antiviral target.
As letermovir comes into clinical use, the incidence and genetic pathways of CMV drug resistance to this and other terminase inhibitors require detailed characterization for diagnosis, treatment selection and design of inhibitors with improved genetic barriers to resistance. In vitro mapping of letermovir resistance is largely focused on the UL56 component of the terminase complex, which has homology to the small subunit of the two-component bacteriophage terminases and is important in recognizing the genomic termini for processing and translocation of unit length viral genomes (Bogner, 2010; Neuber et al., 2017). Diverse UL56 mutations conferring letermovir resistance are mainly located in the codon range 231–369 (Chou, 2015; Goldner et al., 2014). Notable examples are amino acid substitution V236M, which is the first one observed in a treated human subject (Lischka et al., 2016), and various substitutions at the C325 residue that confer absolute letermovir resistance with minor impact on viral growth fitness. Additional in vitro selection experiments with letermovir showed the occasional emergence of UL89 mutations that confer cross-resistance to older terminase inhibitors (Chou, 2017). UL89 is homologous to the bacteriophase terminase large subunit and encodes nuclease activity essential to the cleavage of replicated viral DNA into unit length genomes, in addition to other incompletely understood functions (Bogner, 2010; Neuber et al., 2017).
Here, several new in vitro drug selection experiments revealed mutation in a third component of the terminase complex (UL51), which combined with certain UL56 mutations to increase the overall level of letermovir resistance significantly.
2. Materials and Methods
2.1 Letermovir
Letermovir was obtained commercially (MedChemExpress HY-15233) and was dissolved in dimethyl sulfoxide (DMSO) to make 100 mM drug stocks, from which further 10-fold dilutions were made in DMSO, except that solutions of 10 μM and 1 μM were made in water for immediate use. Letermovir stock solutions were diluted into culture media to final letermovir concentrations of 1 nM to 4 μM as needed, while keeping final DMSO concentrations below 0.1%.
2.2 Viral clones and strains
The BD1 bacterial artificial chromosome (BAC) clone of CMV strain AD169 containing a secreted alkaline phosphatase (SEAP) reporter gene (Chou, 2015) was used to derive the corresponding live virus strain T4175, representing baseline virus with wild type terminase gene sequences, which was used for the letermovir selection experiments. Unlike some previous work, this strain has an intact UL54 DNA polymerase gene without an exonuclease defect designed to accelerate the evolution of drug resistance (Chou, 2015, 2017). Other BD1-derived baseline strains T4190 and T4272 contain a silent Frt motif upstream of UL56 and UL89 exon 2 respectively (Chou, 2015, 2017), for insertion and removal of a Kan selection marker during BAC mutagenesis and were used as the parental strain for comparisons of growth and drug susceptibility with mutant strains.
2.3 Cell and viral cultures
In vitro selection experiments with letermovir were performed using human foreskin fibroblasts (HFF) cultured in Eagle minimal essential medium and 3% fetal bovine serum. Beginning at a concentration of 5 nM, letermovir was added to culture medium at the time of viral inoculation. At weekly intervals, the cells were trypsinized and ~30% of the infected cell suspension was transferred to a subconfluent uninfected HFF monolayer seeded the prior day. The letermovir concentration was increased as tolerated by the amount of visible cytopathology at the time of passage, with the goal of cytopathic effect involving about 30% of the monolayer a week later. Weekly passages were continued for at least 20 weeks. At intervals of about 5 passages, or with a notable increase in cytopathic effect, DNA extracts of infected cells were prepared for PCR amplification and standard dideoxy sequencing of UL51, UL56 and UL89 coding sequences involved in the viral terminase complex (Neuber et al., 2017). Mutant recombinant viruses were constructed as described below for phenotypic assays of specific mutations and combinations of interest. Drug susceptibility and growth assays were performed in retinal epithelial cells transduced to over-express platelet-derived growth factor receptor α (ARPEp), which facilitate assay standardization as described (Chou, 2017; Chou et al., 2017).
2.4 Recombinant phenotyping
Mutagenesis of BAC clones to introduce specific amino acid substitutions in UL51 and UL56 was performed as previously described (Chou, 2015, 2017). New UL56 mutations were introduced by recombination of a transfer vector containing a selectable Kan resistance marker and subsequent removal by Flp recombinase, while the one UL51 mutation studied was introduced by the markerless en passant procedure (Tischer et al., 2010) into BAC clones containing either wild type UL56 and UL89 sequences, or pre-existing UL56 or UL89 mutations. BAC cloned viral DNA was transfected into HFF or ARPEp to yield cell-free CMV stocks, which were sequenced throughout the mutagenized gene for the presence of the intended mutation(s). Phenotypic assays for letermovir susceptibility were performed as recently detailed (Chou, 2017), using SEAP activity in culture supernatants as a measure of viral growth. The drug concentration required to reduce supernatant SEAP activity by 50% (EC50) at 6 to 7 days was determined by assaying growth under no drug and at 5 two-fold increasing concentrations centered on the estimated EC50 value. The mean and standard deviation of EC50 values and the number of replicates (at least 10 replicates set up on at least 4 separate dates) were used to estimate a 95% confidence interval for the EC50 under the prevailing assay conditions (Chou, 2017). Statistical significance of the difference in EC50s between mutant and baseline viral strains was evaluated by the Student t test, using values obtained for the two strains on the same setup dates. Growth fitness of mutant viruses was compared using growth curves resulting from assay of culture supernatant SEAP activity at each of days 4 to 8 after inoculation of ARPEp cells at equivalent low multiplicity of 0.02, as previously described for other terminase mutants (Chou, 2015, 2017).
3. Results
3.1 Mutations detected after serial culture passage under letermovir
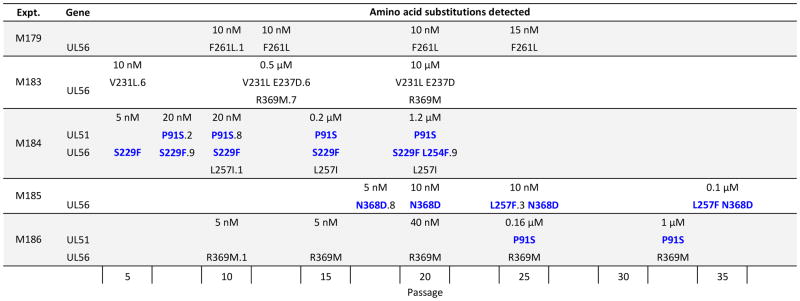
The mutations that evolved in 5 selection experiments are shown in a time-line format (Fig. 1), and included UL56 amino acid substitutions that have been observed previously: V231L, E237D, L257I, F261L and R369M (Chou, 2015; Goldner et al., 2014). Several novel UL56 substitutions were also observed, including S229F, L254F, L257F and N368D, which are at or near the loci of other established letermovir resistance mutations. No UL89 mutations were detected. The same UL51 mutation resulting in substitution P91S was observed in two experiments, in one instance by passage 7 and another by passage 25, in both cases adding to a pre-existing UL56 mutation (S229F or R369M). Eventually viral cytopathic effect was readily observed at 1 μM letermovir (>200-fold baseline EC50) in both experiments; in one case (M184) after the emergence of additional UL56 substitutions L257I and L254F. Letermovir concentrations could not be increased to these levels in the 3 other experiments because of suppression of viral growth. As expected, the tempo of evolution of letermovir resistance was much slower with baseline CMV strain T4175 than with an error prone exonuclease mutant (Chou, 2015). Two of the 5 experiments had detectable UL56 mutations by passage 5, but progression to absolute letermovir resistance (typically by mutation at codon 325) did not occur within 20 passages as happened routinely with the exonuclease mutant.
Figure 1. Evolution of detected mutations in letermovir selection experiments.
Baseline strain T4175 was propagated serially under increasing letermovir concentrations beginning with 5 nM (5 experiments). Letermovir concentrations are shown in the top row and amino acid substitutions are listed from left to right as detected during serial cell culture passage. Novel substitutions are shown in color. Others have been previously published (Goldner et al., 2014, Chou, 2015). Numeric suffix denotes estimated subpopulation in tenths. No suffix denotes a complete sequence population.
3.2 Phenotypic characterization of newly detected mutations
The genotypes and phenotypes of recombinant viral strains representing the newly detected mutations are shown in Table 1, along with those of calibrating control strains. Mutant strains were generated by mutagenesis of BAC clones as in previous studies (Chou, 2015, 2017). A sufficient number of letermovir EC50 assay replicates were performed such that there was no overlap of 95% confidence intervals of EC50s between parental virus and any mutant strain. All mutant virus letermovir EC50s were significantly different from those of parental controls from the same setup dates, with p values of <2×10−5. The letermovir EC50 values and ratios for baseline and mutant control strains are consistent with published data (Chou, 2015, 2017; Goldner et al., 2014). Among the novel UL56 mutations studied, substitutions S229F and N368D conferred slight increases in letermovir EC50, while L254F and L257F conferred modestly higher EC50 increases of 3.2 to 8.6-fold, similar to the 5-fold increase conferred by L257I (Chou, 2015). Also consistent with previous observations (Chou, 2015), a combination of UL56 substitutions at codons 229, 254 and 257 conferred 54-fold increased letermovir EC50, considerably higher than any of the individual substitutions. The UL51 substitution P91S alone conferred a low-grade but statistically significant 2.1-fold increased EC50 over baseline (p=2×10−21), but notably (Table 1, right column) increased by 3.5- to 7.7-fold the level of resistance conferred by the UL56 mutations present in the same selection experiments (S229F, L254F, L257I and R369M). To study the effect of the UL51 P91S substitution in combination with mutations at other well-known sites in the terminase complex, the double mutants were constructed with UL51 P91S and UL56 V236M or UL89 D344E. Results (Table 1) indicated that the addition of P91S had no significant effect on the degree of letermovir resistance conferred by V236M, but did augment the degree of resistance conferred by D344E by 2.6-fold.
Table 1.
Genotypes and phenotypes of recombinant viruses
| Strain | Genotype | Letermovir |
||||
|---|---|---|---|---|---|---|
| EC501 | SD2 | N3 | Fold4 | Δ5 | ||
| Control Strains | ||||||
| 4190 | WT UL56 Frt | 3.7 | 0.77 | 79 | ||
| 4272 | WT UL89 Frt | 3.5 | 0.65 | 26 | ||
| 4194 | UL56 V231L | 21 | 3.4 | 81 | 5.6 | |
| 4296 | UL56 V236M | 115 | 13 | 11 | 32 | |
| 4192 | UL56 L241P | 633 | 136 | 25 | 173 | |
| 4315 | UL56 R369M | 60 | 8.6 | 20 | 16 | |
| 4274 | UL89 D344E | 5.5 | 1.1 | 24 | 1.6 | |
| Newly Phenotyped Mutants | ||||||
| 4244 | UL56 S229F | 6.6 | 1.1 | 11 | 1.8 | |
| 4324 | UL56 S229F L254F L257I | 198 | 25 | 12 | 54 | |
| 4292 | UL56 L254F | 12 | 2.1 | 10 | 3.2 | |
| 4346 | UL56 L257F | 31 | 6.1 | 10 | 8.6 | |
| 4312 | UL56 N368D | 7.4 | 1.5 | 10 | 2.0 | |
| 4317 | UL51 P91S | 7.6 | 1.7 | 43 | 2.1 | |
| 4318 | UL51 P91S UL56 S229F | 23 | 3.3 | 10 | 6.3 | 3.5 |
| 4348 | UL51 P91S UL56 S229F L254F L257I | 1060 | 137 | 12 | 290 | 5.4 |
| 4319 | UL51 P91S UL56 V236M | 101 | 10 | 10 | 28 | 0.9 |
| 4325 | UL51 P91S UL56 R369M | 460 | 53 | 10 | 126 | 7.7 |
| 4400 | UL51 P91S UL89 D344E | 14 | 1.2 | 17 | 4.0 | 2.6 |
Mean letermovir EC50 in nM
Standard deviation of EC50 values
Number of replicates (performed on at least 4 separate dates)
Ratio of EC50 value to that of wild type control strain (values >1.5 in bold)
Fold change in EC50 value resulting from addition of UL51 P91S to UL56 or UL89 mutation
3.3 Effects of mutations on growth fitness
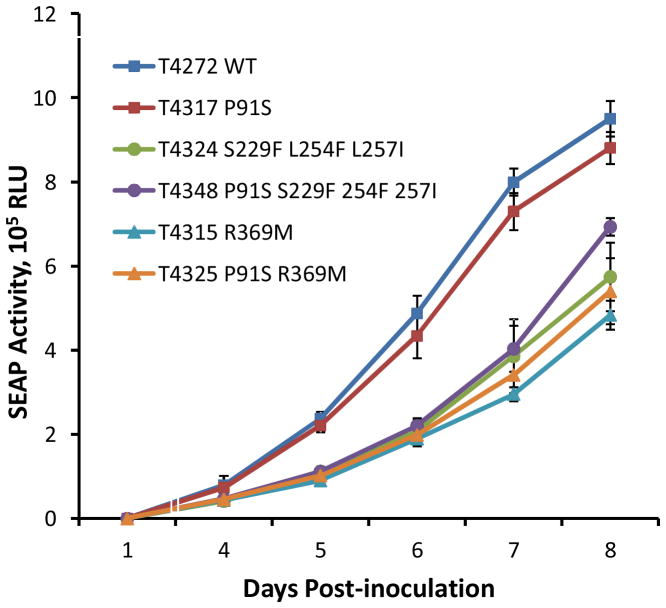
Samples of supernatant assayed daily on days 4 to 8 after inoculation at equal multiplicities of infection confirmed that the growth curves of baseline virus and the UL51 mutant P91S were similar (Fig. 2). The addition of P91S to pre-existing UL56 substitutions such as R369M or a combination of S229F, L254F and L257I did not attenuate viral growth more than the UL56 mutations alone.
Figure 2. Comparative growth curves of viral strains.
Baseline control virus with wild type (WT) UL56, UL89 and UL51 sequences, and mutants containing UL51 P91S and/or UL56 substitutions as shown were inoculated at equal multiplicity of infection of 0.02 as calibrated by culture supernatant SEAP activity at 24 hours. At each of 4 to 8 days post-inoculation, samples were collected for assay of SEAP activity (as relative light units, RLU) as a reporter for viral growth. Data points are the mean and standard deviation of 8 replicates in two independent setups. Amino acid substitutions refer to UL56, except for P91S which refers to UL51. Strain numbers are as listed in Table 1.
4. Discussion
Additional in vitro selection experiments with letermovir added to the extensive list of known mutations in the UL56 gene, the principal genetic locus of letermovir resistance. Unexpectedly, UL51 mutation emerged in two cases, making it the third gene to be implicated in letermovir resistance after some UL89 mutations were recently described (Chou, 2017). The level of resistance conferred by the UL51 P91S substitution by itself is minor (2.1-fold), but it multiplied the level of resistance conferred by some other mutations by 3- to 8-fold and for this reason may have diagnostic significance considering that the mutation is not substantially growth attenuating.
The CMV UL51 gene product (pUL51) has been given little attention until recently (Borst et al., 2013) as it does not have a homologous bacteriophage protein, from which much of the early knowledge on viral packaging proteins was derived. Interaction of pUL51 with pUL56 and pUL89 was demonstrated in co-immunoprecipitation experiments (Borst et al., 2013), and suggests that the three together constitute the CMV terminase complex. The pUL51 component has been shown to be essential for viral viability and the normal localization, stability and packaging function of the CMV terminase complex (Borst et al., 2013; Neuber et al., 2017; Wang et al., 2012). It is a small protein of 157 residues, with an amino terminal end notable for runs of acidic amino acids. The P91S substitution affects one of 10 to 11 proline residues (strain dependent) in pUL51. Along with other proline residues P68, P80 and P83, it is located in a block of residues (66 to 122) that is completely conserved among over 100 CMV UL51 sequences available in Genbank, suggesting the structural importance of this region. However, the P91S change did not cause significant growth impairment (Fig. 2).
In previous in vitro selection experiments with letermovir, the UL51 gene was screened for emergent mutations but none were found (Chou, 2015; Goldner et al., 2014). The present finding of a UL51 letermovir resistance mutation supports the evidence cited above that the three terminase components physically and functionally interact. The P91S substitution may transmit conformational changes that differentially affect the interaction of letermovir with specific pUL56 or pUL89 residues and mutations (e.g. more so with UL56 R369M than with V236M), but there is very limited information currently available on the actual structure of the CMV terminase complex (Chou, 2017). It seems less likely that incremental resistance arising from UL51 mutation results from a mechanism independent of the drug target, such as the ganciclovir resistance conferred by UL97 mutations that affect the initial phosphorylation of the drug, or the maribavir resistance conferred by UL27 mutations that alter host cell cycle conditions.
The level of letermovir resistance conferred by UL51 P91S alone (~2-fold) is minor, but its potential clinical significance lies in the disproportionate degree (>2-fold and up to ~8-fold) to which it can increase the level of resistance conferred by certain other UL56 or UL89 mutations, thus significantly impairing antiviral action. Pending further information on the relative frequency of UL51 mutation as a letermovir resistance pathway in vivo, it is appropriate to screen for UL51 mutation during clinical trials and in cases of suspected treatment failure. This should be fairly straightforward considering the small size of this gene.
Highlights.
Additional CMV mutations selected in vitro under letermovir exposure were studied
Four new UL56 mutations were identified to add to many others already published
Mutation in a terminase gene UL51 is newly shown to cause letermovir resistance
By itself, the UL51 P91S amino acid substitution confers low level resistance
Adding P91S increases the letermovir resistance of some UL56 and UL89 mutants by 2- to 8-fold at low fitness cost
Acknowledgments
Gail Marousek, Ronald J. Ercolani, Michelle A. Hendrick and L. Elizabeth Satterwhite provided technical assistance. Gregory A. Smith provided E. coli strain GS1783 for en passant BAC mutagenesis. This work was supported by the National Institutes of Health (grant R01-AI116635) and Department of Veterans Affairs research funds. Presented in part at the 16th International CMV/betaherpesvirus Workshop, Noordwijkerhout, Netherlands, May 2017.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bogner E. Human cytomegalovirus packaging: an update on structure-function relationships. Future Virol. 2010;5:397–404. [Google Scholar]
- Borst EM, Kleine-Albers J, Gabaev I, Babic M, Wagner K, Binz A, Degenhardt I, Kalesse M, Jonjic S, Bauerfeind R, Messerle M. The human cytomegalovirus UL51 protein is essential for viral genome cleavage-packaging and interacts with the terminase subunits pUL56 and pUL89. J Virol. 2013;87:1720–1732. doi: 10.1128/JVI.01955-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Rapid in vitro evolution of human cytomegalovirus UL56 mutations that confer letermovir resistance. Antimicrob Agents Chemother. 2015;59:6588–6593. doi: 10.1128/AAC.01623-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S. Comparison of Cytomegalovirus Terminase Gene Mutations Selected after Exposure to Three Distinct Inhibitor Compounds. Antimicrob Agents Chemother. 2017 doi: 10.1128/AAC.01325-17. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou S, Ercolani RJ, Vanarsdall AL. Differentiated Levels of Ganciclovir Resistance Conferred by Mutations at Codons 591 to 603 of the Cytomegalovirus UL97 Kinase Gene. J Clin Microbiol. 2017;55:2098–2104. doi: 10.1128/JCM.00391-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldner T, Hempel C, Ruebsamen-Schaeff H, Zimmermann H, Lischka P. Geno- and phenotypic characterization of human cytomegalovirus mutants selected in vitro after letermovir (AIC246) exposure. Antimicrob Agents Chemother. 2014;58:610–613. doi: 10.1128/AAC.01794-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lischka P, Michel D, Zimmermann H. Characterization of cytomegalovirus breakthrough events in a Phase 2 prophylaxis trial of letermovir (AIC246, MK 8228) J Infect Dis. 2016;213:23–30. doi: 10.1093/infdis/jiv352. [DOI] [PubMed] [Google Scholar]
- Neuber S, Wagner K, Goldner T, Lischka P, Steinbrueck L, Messerle M, Borst EM. Mutual Interplay between the Human Cytomegalovirus Terminase Subunits pUL51, pUL56, and pUL89 Promotes Terminase Complex Formation. J Virol. 2017:91. doi: 10.1128/JVI.02384-16. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer BK, Smith GA, Osterrieder N. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol. 2010;634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- Wang JB, Zhu Y, McVoy MA, Parris DS. Changes in subcellular localization reveal interactions between human cytomegalovirus terminase subunits. Virol J. 2012;9:315. doi: 10.1186/1743-422X-9-315. [DOI] [PMC free article] [PubMed] [Google Scholar]