Abstract
The 1,3-dipolar cycloaddition reaction between an alkene and a tetrazole represents one elegant and rare example of fluorophore-forming bioorthogonal chemistry. This is an attractive reaction for imaging applications in live cells that requires less intensive washing steps and/or needs spatiotemporal resolutions. In the present work, as an effort to improve the fluorogenic property of the alkene-tetrazole reaction, an aromatic alkene (styrene) was investigated as the dipolarophile. Over 30-fold improvement in quantum yield of the reaction product was achieved in aqueous solution. According to our mechanistic studies, the observed improvement is likely due to an insufficient protonation of the styrene-tetrazole reaction product. This finding provides a useful guidance to the future design of alkene-tetrazole reactions for biological studies. Fluorogenic protein labeling using the styrene-tetrazole reaction was demonstrated both in vitro and in vivo. This was realized by the genetic incorporation of an unnatural amino acid containing the styrene moiety. It is anticipated that the combination of styrene with different tetrazole derivatives can generally improve and broaden the application of alkene-tetrazole chemistry in real-time imaging in live cells.
Keywords: bioconjugation, photoclick chemistry, tetrazole, fluorogenic, protein labeling
TOC Image
INTRODUCTION
Bioorthogonal reaction-based fluorogenic protein labeling, where the removal of unreacted reagents is not necessary, could potentially enable truly real-time imaging experiments in live cells. The most widely used strategy for such fluorogenic labeling is based on the removal of a specific functional group that suppresses fluorescence through a designed chemical transformation. In this case, the fluorescence quencher is also the reactive group on the probe, e.g., azide,1,2 alkyne,3 or tetrazine.4 A few examples of fluorogenic protein labeling have been reported using this strategy.2,5 A second strategy for fluorogenic protein labeling is based on the simultaneous formation of a fluorophore in a bioorthogonal reaction. In comparison to the fluorescence quencher-removal strategy, which turns a weak fluorescence signal into a stronger one, the fluorophore-forming strategy gives lower background signal since the bioconjugation product is the only fluorescent species within the entire system. Due to the challenging aspects in the reaction design, this strategy is less explored.6–10.
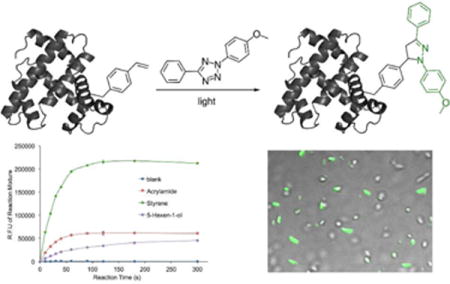
One elegant and rare example of the fluorophore-forming bioorthogonal reaction is the photoinducible 1,3-dipolar cycloaddition reaction between an alkene and a di-substituted tetrazole (Tz; Figure 1).11 Substituted pyrazoline, the product of this reaction, is a fluorophore. This reaction was initially reported in 1967 using benzene as solvent.12 In a series of seminal work, Lin and co-workers optimized this reaction and enabled its application in biological investigations, such as the labeling of proteins and the detection of alkene-containing metabolites.11, 13–17 Nitrile imine has been identified as the reactive intermediate from tetrazole under UV irradiation.18 Significant efforts have also been made to improve the reaction rate by modifying either the tetrazole19–21 or the alkene9,22,23 substrates. Here we report the first example to improve the quantum yield of the pyrazoline chromophore in aqueous solution by using an aromatic alkene, styrene, as the dipolarophile (Figure 1). Over 30-fold improvement was achieved in aqueous solution. According to our mechanistic studies, such improvement is likely due to an insufficient protonation of the pyrazoline ring from the styrene-based reaction. Our finding will likely enhance the biological utility of the alkene-tetrazole reaction. The fluorogenic and the photoinducible nature of this reaction should make it very attractive for imaging applications that requires less intensive washing steps and/or good spatiotemporal resolutions.
Figure 1.
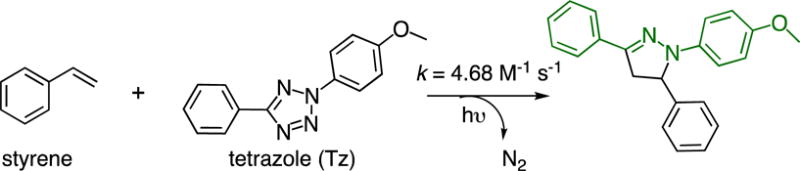
The fluorogenic styrene-tetrazole reaction.
RESULTS AND DISCUSSION
General design strategy
Although the carbon skeleton of alkene is not part of the pyrazoline chromophore, the substituent at the vinyl position of an alkene may influence the overall chemical and/or fluorescence properties of the pyrazoline chromophore. Since the goal of this work is to use the alkene-tetrazole reaction for protein labeling through genetic incorporation of an unnatural amino acid (unAA) containing the alkene moiety, two criteria need to be considered: (1) the substituent at vinyl position of the alkene should not be too large. Otherwise, it would be challenging to incorporate such alkene-containing unAA into proteins; and (2) the reaction rate should still be fast enough for protein labeling in live cells. Styrene, which meets the aforementioned two criteria, was chosen for the present study.
Rate of the styrene-tetrazole reaction
We first conducted kinetics study of the styrene-tetrazole reaction in PBS/acetonitrile (v/v 1:1). The pseudo-first-order rate constant (kobs) was measured by monitoring the formation of the product in the presence of different concentrations of excess styrene and was used to determine the second-order rate constant (k = 4.68 M−1 s−1; Figure S1 of Supporting Information). It was approximately 8-fold faster than the reaction between an isolated alkene (5-hexen-1 -ol) and tetrazole (k = 0.60 M−1 s−1; Figure S1 of Supporting Information). This observation is consistent with the fact that the phenyl substituent in styrene lowers the LUMO energy of the alkene.20 The strained alkenes (cyclopropene and norbornene) and acrylamide were reported to have similar rate constants (k = 32 – 58 M−1 s−1),9 which are larger than that of styrene. To characterize reaction rates side-by-side, we conducted competitive studies using three unstrained alkenes, styrene, 5-hexen-1-ol, and acrylamide. They react with tetrazole (Tz) to form PZL-ST, PZL-HE, and PZL-AC, respectively (Figure 2A). In the first competitive study, tetrazole was mixed with a large excess of equal amount of styrene and acrylamide. A subsequent HPLC (Figure S2 of Supporting Information) analysis showed the formation of 16% PZL-ST (from styrene) and 84% PZL-AC (from acrylamide). In the second competitive study, tetrazole was mixed with a large excess of equal amount of styrene and 5-hexen-1-ol. PZL-ST was observed as the only product (Figure S3 of Supporting Information). From the above data, we believe that the styrene-tetrazole reaction has an appreciable reaction rate for protein labeling in live cells. In fact, its rate is faster than some common bioorthogonal reactions, such as the strain-promoted cycloaddition of fluorinated cyclooctynes with azides24 and the cyclopropene-tetrazine reaction,25,26 which have been successfully applied to the labeling of biomolecules in live cells.24,25,27–29
Figure 2. Fluorogenic reactions between tetrazole and various terminal alkenes.
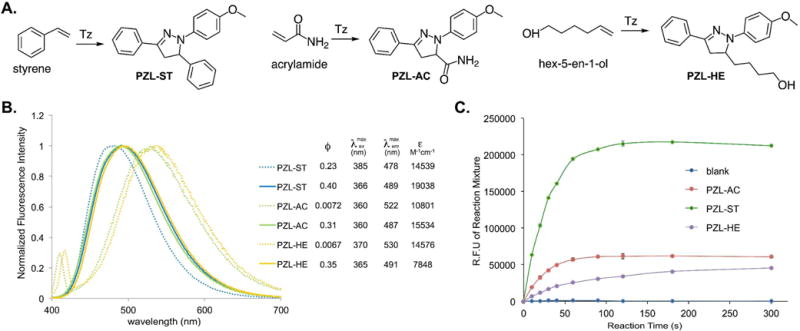
(A) Reactions between tetrazole and three terminal alkenes; (B) Fluorescence emission spectra of pyrazolines, PZL-ST, PZL-AC, and PZL-HE. Dashed lines: in PBS buffer (pH 7.4, 5% DMSO); Solid lines: in acetonitrile; (C) Fluorogenic progress of the three reactions.
Fluorescence and other optical properties
We examined optical properties of PZL-ST, PZL-HE, and PZL-AC. The fluorescence emission maximum of PZL-ST is 478 nm in PBS (with 5% DMSO). In comparison to PZL-HE (530 nm) and PZL-AC (522 mm), PZL-ST displayed significant blue shift (Figure 2B). On the other hand, PZL-ST, PZL-HE, and PZL-AC showed very similar emission maxima in acetonitrile (Figure 2B). While PZL-ST (ϕ = 0.40) displayed only slightly better quantum yield than that of PZL-HE (ϕ = 0.35) and PZL-AC (ϕ = 0.31) in acetonitrile, we were glad to observe that PZL-ST (ϕ = 0.23) had over 30-fold higher quantum yield than that of PZL-HE (ϕ = 0.0067) and PZL-AC (ϕ = 0.0072) in PBS buffer (Figure 2B). To further demonstrate the benefit of a higher quantum yield, we conducted reactions in PBS buffer side-by-side with different alkenes as dipolarophiles and measured fluorescence intensities during reaction progress. As shown in Figure 2C, the styrene-tetrazole reaction displayed much stronger fluorescence intensity than those reactions in which 5-hexen-1-ol or acrylamide were used. As our goal is to use styrene-tetrazole reaction for protein labeling, a brighter bioconjugation product in aqueous solution is desirable.
Mechanistic study
We observed that the absorption spectra of PZL-ST, PZL-HE, and PZL-AC were significantly different in different solvents (acetonitrile and PBS buffer; Figure S4 of Supporting Information). To better understand this observation, computational studies were conducted. The optimized geometries of PZL-ST, PZL-HE, and PZL-AC are shown in Figure S5 of Supporting Information. The calculated vertical excitation energies (S0→Sn) are shown in Table S1 of Supporting Information. These calculated values are in good agreement with experimentally measured absorption spectra. By analyzing experimental and computational data (details can be found in the Computational Results section of Supporting Information), we discovered that PZL-HE and PZL-AC were likely protonated in PBS buffer. On the other hand, PZL-ST was not protonated or protonated to an insignificant extent. To understand if such protonation could affect fluorescence properties of the three molecules, we calculated S1→S0 de-excitation energies. Near-zero oscillator strengths were obtained for all three molecules after protonation (Table S2 of Supporting Information), indicating that no fluorescence should be observed when they were protonated. The above data strongly suggest that the observed higher quantum yield of PZL-ST is due to a significant portion of the neutral form (not protonated) of PZL-ST in the PBS buffer at the physiological pH (7.4). This finding provides a useful guidance to the future design of the fluorogenic alkene-tetrazole reaction for biological applications under physiological conditions.
Protein labelling in vitro
In order to apply the fluorogenic styrene-tetrazole reaction to protein labeling, a lysine-derived unAA containing styrene moiety (KStyr;10 Figure 3A) was used in this work. In our previously report, an inverse electron-demand Diels-Alder (iEDDA) reaction between KStyr and tetrazine was examined for fluorogenic protein labeling.10 It was intriguing that styrene could serve as a versatile reagent for two different fluorophore-forming bioorthogonal reactions. By employing our previously identified pyrrolysyl-tRNA synthetase (PylRS) mutant-tRNAPyl pair,10 we were able to obtained 25 mg/L of sfGFP mutant (sfGFP-Asn149KStyr) that contains KStyr at position Asn149. In a typical labeling experiment, sfGFP-Asn149KStyr was incubated with 500 μM tetrazole in PBS buffer under UV irradiation at 302 nm. A robust fluorogenic protein labeling was observed. As shown in Figure 3B, fluorescence was detected 20s after the reaction was initiated with the addition of tetrazole. The fluorescence intensity increased gradually in a time-dependent manner. As a control experiment, no fluorescence was observed when sfGFP-Asn149KStyr was irradiated in the absence of tetrazole (Figure 3B). As a second control experiment, wild-type sfGFP did not show any fluorescence labeling (Figure 3B). Above results confirmed the notion that the fluorogenic styrene-tetrazole reaction could be used as an efficient tool to selectively label purified proteins.
Figure 3. Protein labelling with the fluorogenic styrene-tetrazole reaction.
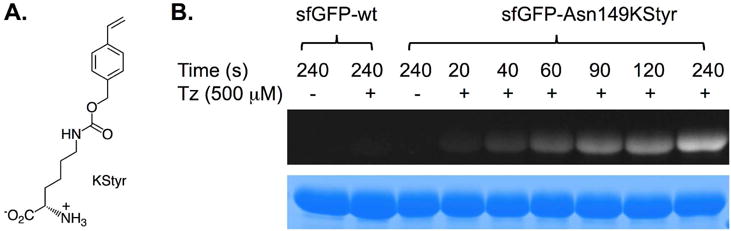
(A) The structure of KStyr; (B) Protein labeling experiments. These experiments were conducted with the indicated irradiation time. Wild-type sfGFP was included as the negative control. Following labeling reactions, protein samples were denatured by heating and analyzed by SDS-PAGE. The bottom panel shows Coomassie blue stained gel and the top panel shows the fluorescent image of the same gel before Coomassie blue treatment.
Protein labelling in live cells
Finally, we demonstrated that the fluorogenic styrene-tetrazole reaction could be used to label an intracellular stress response protein, HdeA, in live cells. Plasmid pHdeA was constructed and used to express an HdeA mutant containing KStyr at position 28 (HdeA-F28KStyr). E. coli cells expressing HdeA-F28KStyr was washed and incubated with 100 μM tetrazole for one hour at 37 °C. Cells were collected, resuspended in PBS buffer, and placed between two coverslips. Visualization using fluorescence microscope was followed immediately after the slips-cells-slips sandwich was irradiated at 302 nm for 60 s. As shown in Figure 4A, strong fluorescent signals were detected and co-localized nicely with cells. As a control experiment, no fluorescence labeling was observed when cells expressing the wild-type HdeA (HdeA-wt) protein were treated with tetrazole under the same conditions (Figure 4B).
Figure 4. Selective labeling of E. coli cells expressing HdeA-F28KStyr.
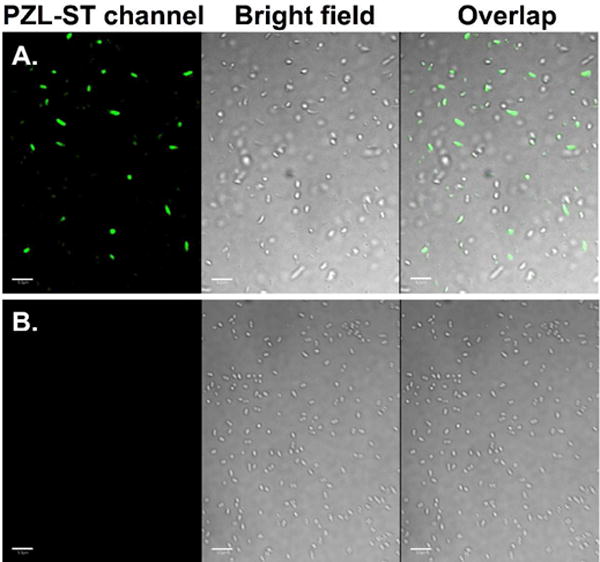
(A) HdeA-F28KStyr mutant that was expressed in the presence of KStyr; (B) Wild-type HdeA that was expressed in the presence of KStyr. For all images, the left panel shows fluorescent images of E. coli cells in the PZL-ST channel (405 nm excitation and 510 nm emission), the middle panel shows bright-field images of the same E. coli cells, and the right panel shows composite images of bright-field and fluorescent images. Scale bars, 10 μm.
In conclusion, we have shown that the styrene-tetrazole reaction afforded a fluorophore (PZL-ST) with a significantly higher quantum yield than those using aliphatic terminal alkenes as the dipolarophile. Our mechanistic studies suggest that the higher quantum yield is likely due to the less protonation of PZL-ST in PBS buffer. This finding provides insights and a useful guidance for the future design of alkene-tetrazole reaction for biological applications. We also demonstrated that the styrene-tetrazole reaction could be used to accomplish efficient protein labeling both in vitro and in vivo through the genetic incorporation of a styrene-containing unAA. We believe that styrene represents as an excellent choice of dipolarophile, which could facilitate further developments of the fluorogenic and photoinduced alkene-tetrazole reaction and broaden its application in live cell imaging.
EXPERIMENTAL PROCEDURES
Materials and general methods
Unless otherwise noted, starting materials, solvents, and reagents for chemical synthesis were obtained from commercial suppliers (Acros, Alfa Aesar, Sigma-Aldrich, or Chem-impex) and used without further purification. Dry solvents were either freshly distilled by following standard methods or directly purchased from Acros. Deuterated solvents were obtained from Sigma-Aldrich. Flash chromatography (FC) was carried out using SiliaFlash P60 (0.04–0.063 mm, 230–400 mesh) from Silicycle or Amberlite XAD4 from Sigma-Aldrich. Thin layer chromatography (TLC) was performed on glass-backed, precoated silica gel plates (Analtech). NMR spectra were recorded at 25 °C using a Bruker Advance III-HD 400 MHz NMR. Chemical shifts were reported in ppm with deuterated solvents as internal standards (CDCl3, H 7.26, C 77.0; DMSO-d6, H 2.50, C 39.5; D2O, 4.79). Multiplicity was reported as follows: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, b = broad. Kinetic studies by HPLC were performed on Agilent 1260 Infinity system. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on Bio-Rad miniPROTEAN electrophoresis system using 15% or 18% homemade SDS-PAGE gels. Bio-Rad Prestained Protein Ladder was applied to at least one lane of each gel for the estimation of apparent molecular weights. Protein gels were stained by Coomassie Brilliant Blue staining solution and visualized under Bio-rad Molecular Imager ChemiDoc XRS+ System. For in-gel fluorescence imaging of labeled protein bands, Bio-rad Molecular Imager ChemiDoc XRS+ System was used. Fluorescence characterization was performed on BioTek Synergy H1 Hybrid Multi-mode Monochromater Fluorescence Microplate reader or Horiba FluoroMax 4 spectrometer. Live cells were imaged on Olympus FV500 inverted (Olympus IX-81) confocal microscope.
Kinetic measurement of Alkene-tetrazole cycloaddition
Reactions were carried out in PBS buffer/acetonitrile (1:1, v/v) solution under pseudo first order conditions with 100 μM tetrazole and a large excess of alkene. Handheld UV lamp (302 nm, UVP, 0.16 Amps, 8 watt) was used to irradiate five separate reaction mixtures (1 mL) with stirring in quartz test tubes at a distance of 4 cm. At the indicated time point, 50 μL of mixture was withdrawn and added into 50 μL of 500 μM 2-nitrobenzyl alcohol (ONB) solution in PBS buffer/acetonitrile (1:1, v/v). ONB was used as an internal reference for quantification using HPLC. Calibration curves were generated by linear plotting the ratio between integrated areas of pyrazoline and ONB at 350 nm against known concentrations of purified cycloadducts. Reactions mixtures (10 μL each) from different time point containing ONB as internal reference were analyzed by HPLC. Product concentrations ([P]t) at different time point were calculated based on their integrated area using calibration curves. The pseudo first-order rate constant kobs (s−1) was determined from the slope of a linear plot of ln(100 μM - [P]t) at different time point (t). Three independent measurements were performed and the second-order rate constant k2 (M−1s−1) was derived from the reaction using 10 μM of alkene.
Fluorogenic measurement of alkene-tetrazole reactions
Reactions were carried out as aforementioned. Reaction mixtures (150 μL) from each reaction were withdrawn at the specified time. Fluorescence intensity of each sample was measured at 510 nm with 405 nm excitation using BioTek Synergy H1 Hybrid Multi-mode Monochromater Fluorescence Microplate reader.
Emission spectra and quantum yield determination
Emission spectra of each pyrazoline derivatives in specified solvents were recorded from 400 nm to 700 nm using Horiba FluoroMax 4 spectrometer with 365 nm excitation. Quantum yields were determined by following a reported procedure.30 Quinine sulfate in 0.5 M H2SO4 was used as the reference in the calculation.31
Protein expression and purification
Plasmid pBK-KStyrRS harboring KStyrRS gene was cotransformed into E. coli BL21(DE3) cells with plasmid psfGFP-Asn149TAG.10 Cells were grown on a LB agar plate containing 100 mg/L ampicillin and 34 mg/L chloramphenicol at 37 °C overnight. A single colony from the plate was inoculated into 5 mL LB media supplemented with 100 mg/L ampicillin and 34 mg/L chloramphenicol. After incubation with shaking at 37 °C overnight, 1 mL cells were transferred into 50 mL LB medium containing 100 mg/L ampicillin and 34 mg/L chloramphenicol, and grown with shaking at 37 °C. Protein expression was induced at OD600 of 0.8 by the additions of IPTG (0.5 mM) and KStyr (1 mM). After being cultivated at 37 °C for an additional 16 h, cells were collected by centrifugation at 5000 g and 4 °C for 15 min. Harvested cells were re-suspended in the loading buffer for affinity chromatography and were lysed by sonication. After centrifugation (21000 g, 30 min, 4 °C) to remove cellular debris, the cell-free supernatant was applied to Ni Sepharose 6 Fast Flow resin (GE Healthcare) and the protein was purified by following manufacture’s instruction. The purified protein was buffer exchanged into 10 mM PBS buffer prior to assays and mass spectrometry analysis. Protein concentrations were determined by Bradford assay (Bio-Rad).
Protein mass spectrometry
The sample was analyzed by a Q-Exactive HF mass spectrometer under the positive mode.
Computational Methods
Density functional theory (DFT) and time-dependent DFT (TDDFT) methods32,33 were used in the calculation. The solvent was described by using the FixSol34 solvation model (with dielectric constant as 78.39) implemented in the Quantum Chemistry Polarizable force field program (QuanPol)35 integrated in the General Atomic and Molecular Electronic Structure System (GAMESS)36,37 package. The B3LYP (Becke, three-parameter, Lee-Yang-Parr) exchange-correlation functional38 and the 6–31++G(d,p)39 basis set were used in both DFT and TDDFT calculations. The GAMESS TDDFT program implemented by Chiba et al40 was used together with the QuanPol FixSol model. All S0 ground state structures (Figure S5 of Supporting Information) were optimized using FixSol/B3LYP, then single point energy calculations using FixSol/TD-B3LYP were performed to obtain the vertical excitation energies (S0→Sn). The S1 excited state geometry optimizations were performed using FixSol/TD-B3LYP41 to obtain the S1→S0 de-excitation energies.
Labeling of purified sfGFP-Asn149KStyr by tetrazole
One microliter tetrazole stock solution in DMSO (7.5 mM, 5 mM, 2.5 mM and 1 mM) was added to sfGFP-wt and sfGFP-Asn149KStyr (9 μL at a concentration of 1 mg/mL) in PBS buffer, respectively. The reaction mixtures were irradiated at 302 nm with a handheld UV lamp for indicated time (20, 60, 90, 120, 240 s). As a control, sfGFP-Asn149KStyr was treated with tetrazole (0.75 mM final concentration) without UV irradiation at room temperature for 60 min. Reactions were stopped by the removal of UV lamp and the addition of SDS-PAGE sample loading buffer. After being heated at 95 °C in water bath for 15 minutes, the protein samples were analyzed by SDS-PAGE. Fluorescence was detected before coomassie blue staining. Bio-rad Molecular Imager ChemiDoc XRS+ was used for imaging coomassie blue stained and Tz labeled protein gels.
Fluorescence microscope of live E. coli cells labeled with tetrazole
E. coli cells expressing either HdeA-wt or HdeA-F28KStyr were collected by centrifugation (21000 g) for 5 min at 4 °C. The cell pellets were washed three times with PBS buffer and re-suspended in PBS buffer containing 5% glycerol. Tz stock solution was added to give a final concentration of 100 μM. After 1 hour of incubation, 10 μL of the cell suspension was placed on top of a glass slip and covered by a glass cover slip. The glass slip-cell-slip sandwich was irradiated at 302 nm using a handheld UV lamp for 60 s. Cells were immediately imaged using Olympus FV500 inverted (Olympus IX-81) confocal microscope. Cells were excited with DAPI excitation wavelength (405 nm) and imaged using GFP emission filter (510nm).
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health (grant 1R01A1111862 to J.G. and W.N.) and National Science Foundation (grant 1553041 to J.G.). The authors thank Dr. Y. Zhou and Microscopy facility in the Center for Biotechnology at the University of Nebraska - Lincoln for help in fluorescence microscope analysis. The authors thank Dr. Sophie Alvarez (Proteomics and Metabolomics Facility) for mass spectrometry analysis. All calculations were performed with resources at the University of Nebraska Holland Computing Center.
Footnotes
ASSOCIATED CONTENT
Supporting Information. Synthetic procedures, computational calculation details, additional figures and tables. This material is available free of charge via the Internet at http://pubs.acs.org.
Notes
The authors declare no competing financial interest.
References
- 1.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 2.Friscourt F, Fahrni CJ, Boons GJ. A fluorogenic probe for the catalyst-free detection of azide-tagged molecules. J Am Chem Soc. 2012;134:18809–18815. doi: 10.1021/ja309000s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Z, Fahrni CJ. A fluorogenic probe for the Copper(I)-catalyzed azide-alkyne ligation reaction: Modulation of the fluorescence emission via 3(n,π)-1(π,π) inversion. J Am Chem Soc. 2004;126:8862–8863. doi: 10.1021/ja049684r. [DOI] [PubMed] [Google Scholar]
- 4.Devaraj NK, Hilderbrand S, Upadhyay R, Mazitschek R, Weissleder R. Bioorthogonal turn-on probes for imaging small molecules inside living cells. Angew Chem, Int Ed. 2010;49:2869–2872. doi: 10.1002/anie.200906120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lang K, Davis L, Wallace S, Mahesh M, Cox DJ, Blackman ML, Fox JM, Chin JW. Genetic encoding of bicyclononynes and trans-cyclooctenes for site-specific protein labeling in vitro and in live mammalian cells via rapid fluorogenic diels-alder reactions. J Am Chem Soc. 2012;134:10317–10320. doi: 10.1021/ja302832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jewett JC, Bertozzi CR. Synthesis of a fluorogenic cyclooctyne activated by Cu-free click chemistry. Org Lett. 2011;13:5937–5939. doi: 10.1021/ol2025026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu Z, Ho LY, Lin Q. Rapid, photoactivatable turn-on fluorescent probes based on an intramolecular photoclick reaction. J Am Chem Soc. 2011;133:11912–11915. doi: 10.1021/ja204758c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song W, Wang Y, Qu J, Madden MM, Lin Q. A photoinducible 1,3-dipolar cycloaddition reaction for rapid, selective modification of tetrazole-containing proteins. Angew Chem Int Ed. 2008;47:2832–2835. doi: 10.1002/anie.200705805. [DOI] [PubMed] [Google Scholar]
- 9.Yu Z, Pan Y, Wang Z, Wang J, Lin Q. Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. Angew Chem, Int Ed. 2012;51:10600–10604. doi: 10.1002/anie.201205352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shang X, Song X, Faller C, Lai R, Li H, Cerny R, Niu W, Guo J. Fluorogenic protein labeling using a genetically encoded unstrained alkene. Chem Sci. 2017;8:1141–1145. doi: 10.1039/c6sc03635j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lim RKV, Lin Q. Photoinducible bioorthogonal chemistry: a spatiotemporally controllable tool to visualize and perturb proteins in live cells. Acc Chem Res. 2011;44:828–839. doi: 10.1021/ar200021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clovis JS, Eckell A, Rolf H, Sustmann R. 1,3-Dipolar cycloadditions. XXV Confirmation of free diphenylnitrilimine as an intermediate in cycloadditions. Chem Ber. 1967;100:60–70. [Google Scholar]
- 13.Wang YZ, Vera CIR, Lin Q. Convenient synthesis of highly functionalized pyrazolines via mild, photoactivated 1,3-dipolar cycloaddition. Org Lett. 2007;9:4155–4158. doi: 10.1021/ol7017328. [DOI] [PubMed] [Google Scholar]
- 14.Wang YZ, Hu WJ, Song WJ, Lint RKV, Lin Q. Discovery of long-wavelength photoactivatable diaryltetrazoles for bioorthogonal 1,3-dipolar cycloaddition reactions. Org Lett. 2008;10:3725–3728. doi: 10.1021/ol801350r. [DOI] [PubMed] [Google Scholar]
- 15.Song W, Wang Y, Qu J, Lin Q. Selective functionalization of a genetically encoded alkene-containing protein via “photoclick chemistry” in bacterial cells. J Am Chem Soc. 2008;130:9654–9655. doi: 10.1021/ja803598e. [DOI] [PubMed] [Google Scholar]
- 16.Song WJ, Wang YZ, Yu ZP, Vera CIR, Qu J, Lin Q. A metabolic alkene reporter for spatiotemporally controlled imaging of newly synthesized proteins in mammalian cells. ACS Chem Biol. 2010;5:875–885. doi: 10.1021/cb100193h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang JY, Zhang W, Song WJ, Wang YZ, Yu ZP, Li JS, Wu MH, Wang L, Zang JY, Lin Q. A biosynthetic route to photoclick chemistry on proteins. J Am Chem Soc. 2010;132:14812–14818. doi: 10.1021/ja104350y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zheng SL, Wang Y, Yu Z, Lin Q, Coppens P. Direct observation of a photoinduced nonstabilized nitrile imine structure in the solid state. J Am Chem Soc. 2009;131:18036–18037. doi: 10.1021/ja9094523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houk KN, Sims J, Watts CR, Luskus LJ. Origin of reactivity, regioselectivity, and periselectivity in 1,3-dipolar cycloadditions. J Am Chem Soc. 1973;95:7301–7315. [Google Scholar]
- 20.Wang YZ, Song WJ, Hu WJ, Lin Q. Fast alkene functionalization in vivo by photoclick chemistry: HOMO lifting of nitrile imine dipoles. Angew Chem Int Ed. 2009;48:5330–5333. doi: 10.1002/anie.200901220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu ZP, Lim RKV, Lin Q. Synthesis of macrocyclic tetrazoles for rapid photoinduced bioorthogonal 1,3-dipolar cycloaddition reactions. Chem Eur J. 2010;16:13325–13329. doi: 10.1002/chem.201002360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li FH, Zhang H, Sun Y, Pan YC, Zhou JZ, Wang JY. Expanding the Genetic Code for Photoclick Chemistry in E. coli, Mammalian Cells, and A. thaliana. Angew Chem Int Ed. 2013;52:9700–9704. doi: 10.1002/anie.201303477. [DOI] [PubMed] [Google Scholar]
- 23.Yu ZP, Lin Q. Design of spiro[2.3]hex-1-ene, a genetically encodable double-strained alkene for superfast photoclick chemistry. J Am Chem Soc. 2014;136:4153–4156. doi: 10.1021/ja5012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baskin JM, Prescher JA, Laughlin ST, Agard NJ, Chang PV, Miller IA, Lo A, Codelli JA, Bertozzi CR. Copper-free click chemistry for dynamic in vivo imaging. Proc Natl Acad Sci U S A. 2007;104:16793–16797. doi: 10.1073/pnas.0707090104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patterson DM, Nazarova LA, Xie B, Kamber DN, Prescher JA. Functionalized cyclopropenes as bioorthogonal chemical reporters. J Am Chem Soc. 2012;134:18638–18643. doi: 10.1021/ja3060436. [DOI] [PubMed] [Google Scholar]
- 26.Kamber DN, Nazarova LA, Liang Y, Lopez SA, Patterson DM, Shih HW, Houk KN, Prescher JA. Isomeric cyclopropenes exhibit unique bioorthogonal reactivities. J Am Chem Soc. 2013;135:13680–13683. doi: 10.1021/ja407737d. [DOI] [PubMed] [Google Scholar]
- 27.Lee YJ, Kurra Y, Yang Y, Torres-Kolbus J, Deiters A, Liu WR. Genetically encoded unstrained olefins for live cell labeling with tetrazine dyes. Chem Commun. 2014;50:13085–13088. doi: 10.1039/c4cc06435f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rieder U, Luedtke NW. Alkene-tetrazine ligation for imaging cellular DNA. Angew Chem, Int Ed. 2014;53:9168–9172. doi: 10.1002/anie.201403580. [DOI] [PubMed] [Google Scholar]
- 29.Niederwieser A, Spaete AK, Nguyen LD, Juengst C, Reutter W, Wittmann V. Two-color glycan labeling of live cells by a combination of Diels-Alder and click chemistry. Angew Chem, Int Ed. 2013;52:4265–4268. doi: 10.1002/anie.201208991. [DOI] [PubMed] [Google Scholar]
- 30.Fery-Forgues S, Lavabre D. Are fluorescence quantum yields so tricky to measure? a demonstration using familiar stationery products. J Chem Educ. 1999;76:1260–1264. [Google Scholar]
- 31.Melhuish WH. Quantum efficiencies of fluorescence of organic substances: effect of solvent and concentration of the fluorescent solute. J Phys Chem. 1961;65:229–235. [Google Scholar]
- 32.Casida ME. Time-dependent density functional response theory for molecules. Recent advances in density functional methods. 1995:155. [Google Scholar]
- 33.Casida ME, Jamorski C, Casida KC, Salahub DR. Molecular excitation energies to high-lying bound states from time-dependent density-functional response theory: Characterization and correction of the time-dependent local density approximation ionization threshold. J Chem Phys. 1998;108:4439–4449. [Google Scholar]
- 34.Thellamurege NM, Li H. Note: FixSol solvation model and FIXPVA2 tessellation scheme. J Chem Phys. 2012;137:246101. doi: 10.1063/1.4773280. [DOI] [PubMed] [Google Scholar]
- 35.Thellamurege NM, Si D, Cui F, Zhu H, Lai R, Li H. QuanPol: A full spectrum and seamless QM/MM program. J Comput Chem. 2013;34:2816–2833. doi: 10.1002/jcc.23435. [DOI] [PubMed] [Google Scholar]
- 36.Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S. General atomic and molecular electronic structure system. J Comput Chem. 1993;14:1347–1363. [Google Scholar]
- 37.Gordon MS, Schmidt MW. Advances in electronic structure theory: GAMESS a decade later. Theory and Applications of Computational Chemistry: the first forty years. 2005:1167–1189. [Google Scholar]
- 38.Becke AD. Density - functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- 39.Francl MM, Pietro WJ, Hehre WJ, Binkley JS, Gordon MS, DeFrees DJ, Pople JA. Self - consistent molecular orbital methods. XXIII. A polarization - type basis set for second - row elements. J Chem Phys. 1982;77:3654–3665. [Google Scholar]
- 40.Chiba M, Tsuneda T, Hirao K. Excited state geometry optimizations by analytical energy gradient of long-range corrected time-dependent density functional theory. J Chem Phys. 2006;124:144106. doi: 10.1063/1.2186995. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Li H. Excited state geometry of photoactive yellow protein chromophore: a combined conductorlike polarizable continuum model and time-dependent density functional study. J Chem Phys. 2010;133:034108. doi: 10.1063/1.3462248. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


