Abstract
The de novo L-cysteine biosynthetic pathway is critical for the growth, antioxidative stress defenses, and pathogenesis of bacterial and protozoan pathogens, such as Salmonella typhimurium and Entamoeba histolytica. This pathway involves two key enzymes, serine acetyltransferase (SAT) and cysteine synthase (CS), which are absent in mammals and therefore represent rational drug targets. The human parasite E. histolytica possesses three SAT and CS isozymes; however, the specific roles of individual isoforms and significance of such apparent redundancy remains unclear. In the present study, we generated E. histolytica cell lines in which CS and SAT expression was knocked down by transcriptional gene silencing. The strain in which CS1, 2 and 3 were simultaneously silenced and the SAT3 gene-silenced strain showed impaired growth when cultured in a cysteine lacking BI-S-33 medium, whereas silencing of SAT1 and SAT2 had no effects on growth. Combined transcriptomic and metabolomic analyses revealed that, CS and SAT3 are involved in S-methylcysteine/cysteine synthesis. Furthermore, silencing of the CS1-3 or SAT3 caused upregulation of various iron-sulfur flavoprotein genes. Taken together, these results provide the first direct evidence of the biological importance of SAT3 and CS isoforms in E. histolytica and justify the exploitation of these enzymes as potential drug targets.
Introduction
Critical metabolic pathways that are unique to pathogens and are significantly divergent from their hosts are rational targets for the development of new chemotherapeutic agents. In particular, sulfur-containing amino acid metabolism, particularly the de novo L-cysteine biosynthetic pathway, is a promising target for drug development against bacterial and parasitic infections, such as those caused by Mycobacterium tuberculosis, Salmonella typhimurium, and Entamoeba histolytica 1–6.
Amebiasis is an intestinal infection caused by the protozoan pathogen E. histolytica and is widespread worldwide [CDC, https://www.cdc.gov/parasites/amebiasis/index.html], particularly in countries with inadequate sewage treatment and poor water quality7. According to the WHO, an estimated 50 million people are infected with E. histolytica worldwide, resulting in 40,000–100,000 deaths annually8. Metronidazole is the drug of choice for treating amebiasis despite its low efficacy against asymptomatic cyst carriers1. Moreover, metronidazole is also teratogenic and causes adverse side effects, such as nausea, vomiting, headache, insomnia, dizziness, drowsiness and hypersensitivity reactions (urticaria, pruritus, erythematous rash)9. In addition, E. histolytica is capable of tolerating sub-therapeutic levels of metronidazole in vitro 10,11. Therefore, new drugs that target parasite-specific metabolic pathways and enzymes distinct from those targeted by metronidazole are urgently needed.
Sulfur-containing amino acid metabolism in E. histolytica differs markedly from that in humans with respect to three main features: (i) the absence of forward and reverse transsulfuration pathways and thus does not convert L-methionine to L-cysteine12,13 or vice versa; (ii) the presence of a sulfur-assimilatory de novo L-cysteine biosynthetic pathway14–16; and (iii) the presence of a unique enzyme, methionine γ-lyase (MGL), which is involved in the degradation of sulfur-containing amino acids17–19. As MGL and two enzymes involved in the cysteine biosynthetic pathway, serine O-acetyltransferase (SAT) and cysteine synthase (CS, O-acetylserine sulfhydrylase), are absent in mammals, these enzymes are potential suitable targets for chemotherapeutic agents against amebiasis.
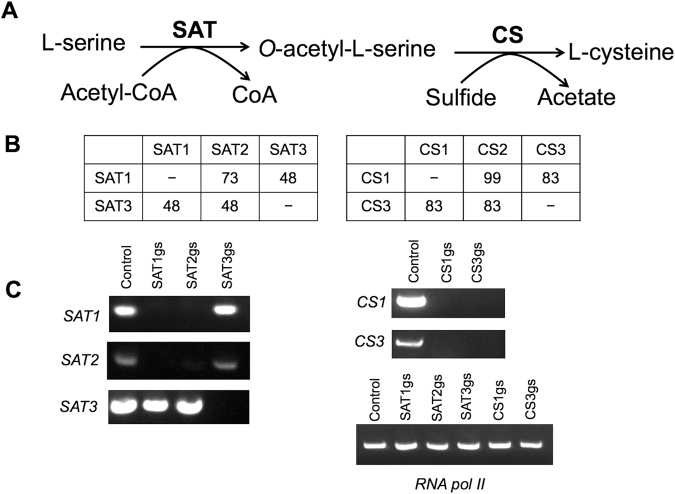
The cysteine biosynthetic pathway plays an important role in the incorporation of inorganic sulfur into organic compounds1 and has been extensively studied in bacteria, plants, and protozoa20–24. In this pathway, SAT (EC 2.3.1.30) catalyzes the formation of O-acetyl-L-serine (OAS) from L-serine and acetyl-CoA15,16 (Fig. 1A). CS [O-acetyl-L-serine (thiol)-lyase] (EC 4.2.99.8) then catalyzes the production of L-cysteine/S-methylcysteine (SMC) through the modification of sulfide/methanthiol with the alanyl moiety of O-acetylserine13,14. However, using a metabolomics approach, we previously showed that CS enzymes in E. histolytica trophozoites cultured in the absence of exogenous L-cysteine are predominantly involved in SMC formation, but not L-cysteine13. E. histolytica SAT and CS have several unique features with respect to localization, complex formation and homology. For example, isozymes of SAT (EhSAT1-3) and CS (EhCS1-3) are localized to the cytosol14–16, whereas plant isoforms of SAT and CS are found in the mitochondria, plastids, and cytosol25. In addition, EhCS1 and EhSAT1 do not form a heteromeric complex26, whereas bacterial and plant SAT and CS form complexes that are involved in cross-talk between sulfur assimilation, carbon and nitrogen metabolism via the generation of OAS27. Further, EhSAT1-3 are biochemically divergent, showing 48–73% mutual sequence identity (Fig. 1B) and markedly different sensitivities to allosteric feedback by L-cysteine16. EhCS1-3 also exhibit sequence divergence with CS1 and CS2 being identical with the exception of two amino acid changes and CS3 having 83% amino acid identity with CS1 and CS2 (Fig. 1B).
Figure 1.
Epigenetic repression of cysteine biosynthesis pathway genes in E. histolytica G3 strain. (A) Scheme of the sulfur assimilatory de novo cysteine biosynthetic pathway in E. histolytica. Abbreviations: SAT, serine O-acetyltransferase (EC 2.3.1.30); CS, cysteine synthase (O-acetyl-L-serine sulfhydrylase, EC 2.5.1.47); and CoA, coenzyme A. (B) Percent amino acid identity among E. histolytica SAT and CS isoforms by ClustalW multiple sequence alignment score. GenBank accession numbers: SAT1, BAA82868; SAT2, XP_650001; SAT3, XP_656373; CS1, XP_650965; CS2, XP_648291; and CS3, XP_653246. (C) Semi-quantitative RT-PCR analysis of SAT1, SAT2, SAT3, CS1 and CS3 gene transcript levels in trophozoites of strain G3 transfected with either empty vector (psAP2G) or the constructed gene silencing plasmids (psAP2G-SAT1, psAP2G-SAT2, psAP2G-SAT3, psAP2G-CS1 and psAP2G-CS3). cDNA from the generated cell lines (psAP2G, SAT1gs, SAT2gs, SAT3gs, CS1gs and CS3gs) was subjected to 30 cycles of PCR using specific primers for the SAT1, SAT2, SAT3, CS1 and CS3 genes. RNA polymerase II served as a control. PCR analysis of samples without reverse transcription was used to exclude the possibility of genomic DNA contamination.
Although the sulfur-assimilatory cysteine biosynthetic pathway in plants, bacteria, and protozoa has been extensively studied and exploited for drug development, the role of individual SAT and CS isozymes and significance of the apparent redundancy of this pathway in E. histolytica remain to be elucidated. In the present study, we investigated the role of the cysteine biosynthesis pathway in E. histolytica using parasites in which genes for the enzymes involved in cysteine biosynthesis were silenced by antisense RNA-mediated transcriptional attenuation. Using transcriptomic and metabolomic analyses, we demonstrated that EhCS and EhSAT3 are critical for SMC/cysteine production and cell growth. Furthermore, we examined the fate of SMC unique metabolite in E. histolytica and revealed that this unique metabolite is involved in the antioxidative stress mechanism.
Results
Establishment of CS and SAT gene-silenced strains
To investigate the role of the L-cysteine biosynthesis pathway in E. histolytica, we utilized antisense small RNA-mediated epigenetic gene silencing to repress the CS1/2 (CS1 and CS2 are 99% identical at the amino acid level), CS3, SAT1, SAT2, and SAT3 genes in E. histolytica strain G3 (Fig. 1C)28,29. In the CS1/2 and CS3 gene-silenced strains, CS1/2 and CS3 gene expression were simultaneously repressed, likely due to the high sequence similarity (83% amino acid identity) between these genes (Fig. 1B). Similarly, the SAT1 and SAT2 genes, whose products share 73% amino acid identity, were simultaneously silenced in the SAT1 and SAT2 gene-silenced transformants (Fig. 1B), whereas the SAT3 gene was not silenced in either of these transformants because of low (48% at the amino acid level) identity between SAT1 and SAT3 and between SAT2 and SAT3. In the SAT3 gene-silenced strain, only the SAT3 gene was silenced (Fig. 1C), and neither SAT1 nor SAT2 was affected. In subsequent analyses, the SAT1/2 and CS1/3 gene-silenced transformants, designated SAT1/2gs and CSgs, respectively, were used for further analyses.
Effects of CS and SAT gene silencing on E. histolytica growth
To examine if the L-cysteine biosynthesis plays a role in the proliferation of E. histolytica, the growth kinetics of trophozoites of the gene-silenced and control transformants (cell line transfected with psAP2G plasmid) were compared in normal BI-S-33 medium containing 8 mM L-cysteine (Fig. 2A) or BI-S-33 medium without L-cysteine which we called as L-cysteine lacking medium (Fig. 2B). However this medium may still contain trace amounts of cysteine from yeast extract and/or tryptone. When cultured in L-cysteine lacking medium, CS gene-disrupted transformants displayed a severe growth defect, whereas SAT3 gene-disrupted transformants showed a mild growth defect (Fig. 2B). In contrast, SAT1/2gs transformants appeared to grow normally in L-cysteine lacking medium (Fig. 2B). However, in normal BI-S-33 medium, none of the gene-silenced strains showed defective growth (Fig. 2A). These results indicate that CS and SAT3 are essential for growth in the absence of exogenous L-cysteine and therefore contribute to cell proliferation.
Figure 2.
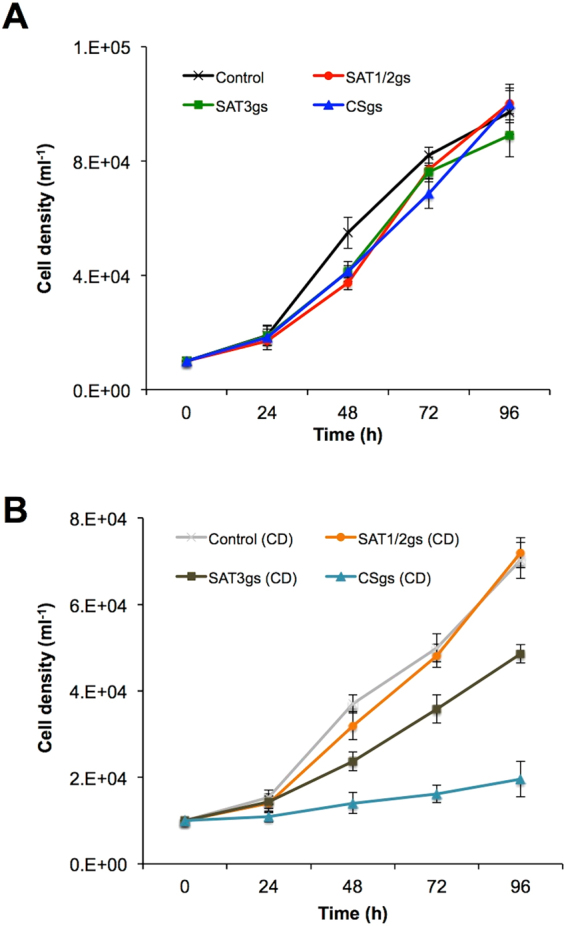
Effect of SAT1/2, SAT3 and CS gene silencing on the growth of trophozoites cultured under normal (A) and L-cysteine lacking (CD) BI-S-33 medium (B). Approximately 6000 amoebae in the logarithmic growth phase were inoculated into 6 mL fresh culture medium and amoebae were then counted every 24 h. Data shown are the means ± standard deviations of five biological replicates.
Metabolomic analysis of gene-silenced transformants cultured in normal and L-cysteine lacking BI-S-33 medium
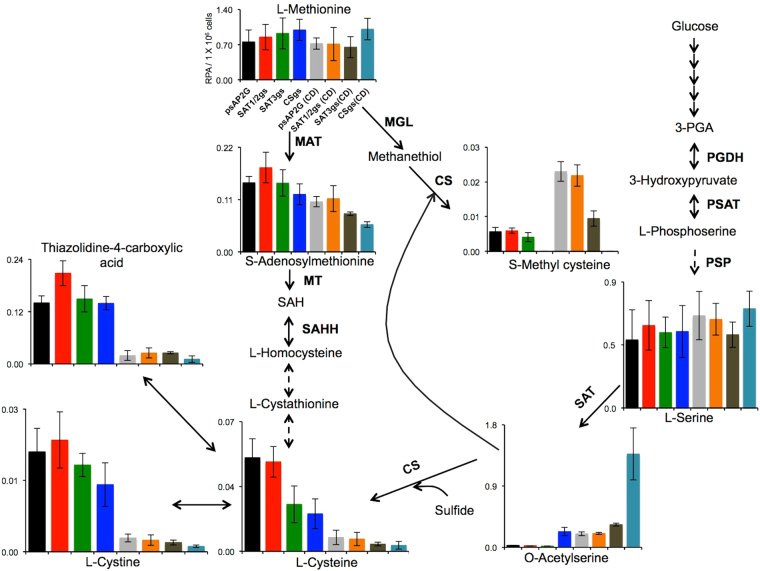
A total of 48 intermediary metabolites, including amino acids, nucleotides, and organic acids, were measured by CE-TOFMS-based metabolomic analysis in the SAT1/2gs, SAT3gs and CSgs transformants under different culture conditions (Supplementary Table S2). Silencing of the CS genes caused drastic changes in the metabolites involved in sulfur-containing amino acid metabolism (Fig. 3). Specifically, the L-cysteine concentration in CSgs trophozoites was approximately 60% lower than that in control trophozoites when cultured under normal BI-S-33 containing 8 mM L-cysteine and L-cysteine-lacking BI-S-33 medium, consistent with the speculation that CS is involved in L-cysteine production. CS gene silencing also resulted in a marked increase in OAS, an activated form of L-serine that is synthesized from L-serine and acetyl-CoA by SAT, in both normal and L-cysteine lacking conditions. In addition, SMC formation was completely abolished by CS gene silencing (Fig. 3), suggesting that CS enzymes are indispensable for SMC production.
Figure 3.
Effect of SAT1/2, SAT3 and CS3 gene silencing and L-cysteine depletion on sulfur-containing amino acid metabolism. Levels of metabolites extracted from SAT1/2, SAT3 and CS gene silenced (SAT1/2gs, SAT3gs, and CSgs) and control (harboring plasmid psAP2G) strains cultured under normal and L-cysteine lacking (CD) BI-S-33 medium conditions is shown. Data shown are the means ± standard deviations of three biological replicates. In the metabolic pathway schemes, solid arrows represent the steps catalyzed by the enzymes whose encoding genes are present in the E. histolytica genome, whereas an arrow with a dashed line indicates those likely absent in the genome or not yet identified so far. Abbreviations: RPA, relative peak area; 3PGA, 3-phosphoglycerate; SAH, S-adenosylhomocysteine; PGDH, phosphoglycerate dehydrogenase (EC 1.1.1.95); PSAT, phosphoserine aminotransferase (EC 2.6.1.52); PSP, phosphoserine phosphatase (EC 3.1.3.3); MGL, methionine γ-lyase (L-methioninase, EC 4.4.1.11); SAT, serine O-acetyltransferase (EC 2.3.1.30); CS, cysteine synthase (O-acetyl-L-serine sulfhydrylase, EC 2.5.1.47); MAT, methionine adenosyltransferase (S-adenosyl-L-methionine synthetase, EC 2.5.1.6); MT, various methyltransferases (EC 2.1.1.X); and SAHH, adenosylhomocysteinase (S-adenosyl-L-homocysteine hydrolase, EC 3.3.1.1).
In contrast to CS, silencing of SAT1/2 did not markedly alter the levels of sulfur-containing metabolites, particularly OAS, SMC, L-cysteine and L-methionine, in E. histolytica, suggesting that SAT3 can compensate for the loss of SAT1/2. However, upon silencing of the SAT3 gene, the levels of SMC and L-cysteine were decreased approximately 40–50% compared to the control strain despite the presence of high levels of the precursor metabolite OAS, which is formed by SAT1 and SAT2 in strain SAT3gs. The reduced level of SMC/cysteine with the concurrent higher OAS level (approximately 60% increase) in strain SAT3gs may be due to a decreased level of CS protein in strain SAT3gs. To investigate this possibility, we examined CS expression at the protein level in the SAT1/2gs, SAT3gs, and control transformant strains. Immunoblot analysis using anti-rEhCS1, anti-rEhCS3, and anti-rEhCPBF130 antibodies showed that the relative amounts of these proteins were comparable between these strains (Supplementary Fig. S1), suggesting that SAT3 may positively regulate CS activity, but not gene expression or protein stability, whereas SAT1/2 do not regulate CS activity.
Gene silencing of SAT1/2, SAT3, or CS1-3 caused global transcriptomic changes
To determine if the silencing of the CS and SAT genes affected the expression of other genes, global gene expression in the SAT1/2gs SAT3gs and CSgs transformants was analyzed using a whole-genome DNA microarray. However, the analysis revealed that after the removal of redundant or obsolete genes (those represented with probe sets with ‘_x_at’ and those for which corresponding NCBI entries were removed after genome reannotation)31, only a limited number of genes had three-fold or higher changes in expression (Supplementary Table S3).
In CSgs, 34 genes were up-regulated and 25 genes were down-regulated when compared to the control (Table 1). CS1-3 transcript levels were reduced by 104, 128 and 20.4 fold, respectively, in CSgs. Among the genes that were significantly down-regulated included those encoding for several hypothetical proteins (EHI_020830, EHI_196760, and EHI_066720), Rab family GTPase, RabH232 (EHI_128180), and a nonpathogenic pore-forming peptide precursor (EHI_169350), which may belong in the saposin-like protein31 (SAPLIP1) family. In contrast, Ras family GTPase (EHI_074750_at), methylene-fatty-acyl-phospholipid synthase (EHI_153710_at), and deoxyuridine 5′ triphosphate nucleotide hydrolase domain-containing protein (EHI_091670_at) were up-regulated in all three gene-silenced transformants (Supplementary Table S3), suggesting that the increased expression of these genes may compensate for the impairment of the cysteine biosynthetic pathway. The most highly upregulated gene related to sulfur metabolism in CSgs was a gene encoding a member of the NADPH-dependent FMN reductase domain-containing protein family (Table 1). Genes encoding NADPH-dependent oxidoreductase 2 (EHI_045340), which was previously shown to be involved in cystine reduction33, was also upregulated in CSgs strain (Table 1).
Table 1.
List of genes down and up regulated ≥3 fold upon CS gene silencing.
| ProbeSetID | Accession Numbers | Common Name | Basal Expression (log2) | Fold change | Regulation | p-value |
|---|---|---|---|---|---|---|
| EHI_160930_s_at | XM_643199 | cysteine synthase 2 | 11.4 | 127.7 | down | 0.000 |
| EHI_024230_s_at | XM_645873 | cysteine synthase 1 | 10.4 | 104.3 | down | 0.019 |
| EHI_060340_at | XM_648154 | cysteine synthase 3 | 6.7 | 20.4 | down | 0.000 |
| EHI_020830_s_at | XM_001913952 | hypothetical protein | 7.8 | 19.9 | down | 0.016 |
| EHI_128180_s_at | XM_649666 | Rab family GTPase | 8.1 | 14.9 | down | 0.004 |
| EHI_196760_s_at | XM_643708 | hypothetical protein | 7.7 | 10.2 | down | 0.005 |
| EHI_133210_s_at | XM_001914244 | peptidase S54 (rhomboid) family protein | 7.9 | 6.1 | down | 0.009 |
| EHI_169350_at | XM_650744 | nonpathogenic pore-forming peptide precursor, putative | 7.6 | 6.0 | down | 0.005 |
| EHI_056700_at | XM_643998 | hypothetical protein | 7.1 | 5.1 | down | 0.006 |
| EHI_066720_at | XM_646043 | hypothetical protein, conserved | 7.2 | 5.0 | down | 0.000 |
| EHI_161970_at | XM_644065 | leucyl-tRNA synthetase, putative | 8.7 | 4.6 | down | 0.009 |
| EHI_153670_at | XM_651265 | U1 small nuclear ribonucleoprotein subunit, putative | 7.9 | 4.0 | down | 0.004 |
| EHI_059870_s_at | XM_647804 | WH2 domain containing protein | 9.8 | 4.0 | down | 0.000 |
| EHI_187280_at | XM_651366 | transcription initiation factor SPT5, putative | 8.7 | 3.8 | down | 0.015 |
| EHI_185620_at | XM_644513 | protein kinase, putative | 5.7 | 3.8 | down | 0.033 |
| EHI_029600_at | XM_644990 | leucine rich repeat-containing protein | 7.9 | 3.7 | down | 0.002 |
| EHI_197440_at | XM_646593 | hypothetical protein | 10.6 | 3.6 | down | 0.000 |
| EHI_180940_at | XM_646942 | lipase, putative | 4.9 | 3.5 | down | 0.010 |
| EHI_060350_at | XM_648153 | splicing factor Prp8, putative | 6.9 | 3.5 | down | 0.016 |
| EHI_155220_at | XM_643278 | T-complex protein 1, alpha subunit, putative | 8.3 | 3.4 | down | 0.010 |
| EHI_065670_at | XM_648551 | cation-transporting P-typeATPase, putative | 10.8 | 3.3 | down | 0.019 |
| EHI_178610_at | XM_651172 | tyrosine kinase, putative | 7.6 | 3.3 | down | 0.047 |
| EHI_177660_at | XM_650844 | isoleucyl-tRNA synthetase, putative | 8.6 | 3.2 | down | 0.006 |
| EHI_005050_at | XM_647746 | sucrose transporter, putative | 4.5 | 3.2 | down | 0.039 |
| EHI_167130_at | XM_649685 | filopodin, putative | 9.3 | 3.2 | down | 0.000 |
| EHI_074750_at | XM_644490 | Ras family GTPase | 5.5 | 10.1 | up | 0.003 |
| EHI_091670_at | XM_644055 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 2.3 | 4.9 | up | 0.009 |
| EHI_126550_at | XM_643463 | AIG1 family protein, putative | 6.5 | 4.6 | up | 0.003 |
| EHI_022270_s_at | XM_644774 | NADPH-dependent FMN reductase domain containing protein | 7.2 | 4.5 | up | 0.001 |
| EHI_159660_at | XM_645152 | hypothetical protein | 5.9 | 4.4 | up | 0.005 |
| EHI_151780_at | XM_652309 | hypothetical protein | 2.4 | 4.1 | up | 0.017 |
| EHI_067720_s_at | XM_643101 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 4.0 | up | 0.015 |
| EHI_045340_s_at | XM_648481 | NADPH-dependent oxidoreductases 2 | 9.0 | 4.0 | up | 0.003 |
| EHI_022600_s_at | XM_643169 | NADPH-dependent FMN reductase domain containing protein | 7.0 | 4.0 | up | 0.003 |
| EHI_134710_at | XM_647029 | hypothetical protein | 4.4 | 4.0 | up | 0.013 |
| EHI_059320_s_at | XM_001914076 | hypothetical protein | 2.6 | 3.9 | up | 0.024 |
| EHI_072960_s_at | XM_001914509 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 4.3 | 3.9 | up | 0.010 |
| EHI_153710_at | XM_001913338 | methylene-fatty-acyl-phospholipid synthase, putative | 6.5 | 3.9 | up | 0.007 |
| EHI_182540_at | XM_651612 | Protein tyrosine phosphatases domain containing protein | 3.8 | 3.9 | up | 0.000 |
| EHI_150660_s_at | XM_642980 | hypothetical protein | 3.7 | 3.9 | up | 0.028 |
| EHI_022990_at | XM_648401 | hypothetical protein | 4.8 | 3.8 | up | 0.008 |
| EHI_174570_at | XM_648228 | hypothetical protein | 2.3 | 3.7 | up | 0.001 |
| EHI_025710_at | XM_644279 | iron-sulfur flavoprotein, putative | 5.5 | 3.7 | up | 0.001 |
| EHI_146130_at | XM_644793 | hypothetical protein | 3.3 | 3.7 | up | 0.035 |
| EHI_046630_at | XM_645444 | Rho family GTPase | 4.1 | 3.7 | up | 0.011 |
| EHI_172510_at | XM_643770 | acid sphingomyelinase-like phosphodiesterase 3a precursor, putative | 3.5 | 3.6 | up | 0.000 |
| EHI_174970_at | XM_648244 | hypothetical protein | 5.4 | 3.5 | up | 0.010 |
| EHI_103260_s_at | XM_001913434 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 3.5 | up | 0.007 |
| EHI_181710_s_at | XM_001914510 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 3.5 | up | 0.013 |
| EHI_121870_at | XM_646700 | ADP-ribosylation factor 1, putative | 5.4 | 3.5 | up | 0.000 |
| EHI_069590_at | XM_001913469 | hypothetical protein | 5.1 | 3.4 | up | 0.005 |
| EHI_125910_at | XM_651393 | double-strand break repair protein MRE11, putative | 3.1 | 3.4 | up | 0.006 |
| EHI_052130_at | XM_650257 | PQ loop repeat protein | 4.6 | 3.3 | up | 0.018 |
| EHI_155430_s_at | XM_650657 | hypothetical protein | 4.7 | 3.3 | up | 0.012 |
| EHI_001800_at | XM_644530 | hypothetical protein | 4.7 | 3.3 | up | 0.019 |
| EHI_192550_at | XM_001913649 | hypothetical protein | 2.3 | 3.2 | up | 0.009 |
| EHI_101260_at | XM_651922 | Ras family GTPase | 5.6 | 3.1 | up | 0.008 |
| EHI_105080_at | XM_648821 | zinc finger protein, putative | 8.2 | 3.1 | up | 0.031 |
| EHI_159470_at | XM_648174 | hypothetical protein | 2.7 | 3.0 | up | 0.018 |
In strain SAT1/2gs, 39 genes were up-regulated and 13 genes were down-regulated compared to the control strain (Table 2). The SAT1 and SAT2 transcript levels were reduced by 529 and 4.2 fold, respectively, whereas the expression of the SAT3 gene remained unchanged. The genes encoding phosphoserine aminotransferase (EHI_026360), which catalyzes the formation of L-phosphoserine from 3-phosphohydroxypyruvate in the phosphorylated pathway of L-serine biosynthesis34, were down-regulated more than five fold (Table 2). Among the most highly upregulated genes was sulfotransferase (EHI_031640), which was up-regulated more than 8 fold, and Fe hydrogenase, which was induced more than 4 fold in strain SAT1/2gs (Table 2). In strain SAT3gs, 16 genes were up-regulated and 19 were down-regulated compared to the control (Table 3). The most highly repressed gene in SAT3gs was SAT3, which had 187-fold lower transcript levels compared the control, whereas SAT1 and SAT2 gene expression remained unchanged. Among the genes that were up-regulated by SAT3 gene silencing were several genes encoding NADPH-dependent FMN reductase domain-containing protein and iron-sulfur flavoprotein (ISF) genes, which were among the most highly up-regulated genes by CS3 gene silencing (Table 1).
Table 2.
List of genes down and up regulated ≥3 fold upon SAT1/2 gene silencing.
| ProbeSetID | Accession Number | Common Name | Basal Expression (log2) | Fold change | Regulation | p-value |
|---|---|---|---|---|---|---|
| EHI_202040_at | AB023954 | serine acetyltransferase 1 | 12.2 | 528.6 | down | 0.002 |
| EHI_020830_s_at | XM_001913952 | hypothetical protein | 7.8 | 11.7 | down | 0.001 |
| EHI_196760_s_at | XM_643708 | hypothetical protein | 7.7 | 10.6 | down | 0.014 |
| EHI_026360_s_at | XM_650291 | phosphoserine aminotransferase, putative | 8.5 | 5.1 | down | 0.004 |
| EHI_187090_at | XM_651385 | Rab family GTPase | 10.8 | 4.4 | down | 0.007 |
| EHI_021570_at | XM_644909 | serine acetyltransferase 2 | 4.2 | 3.6 | down | 0.005 |
| EHI_169350_at | XM_650744 | nonpathogenic pore-forming peptide precursor, putative | 7.6 | 3.5 | down | 0.024 |
| EHI_066720_at | XM_646043 | hypothetical protein, conserved | 7.2 | 3.3 | down | 0.000 |
| EHI_003950_at | XM_643818 | hypothetical protein | 8.3 | 3.2 | down | 0.027 |
| EHI_094060_s_at | XM_001913553 | actin binding protein, putative | 10.1 | 3.2 | down | 0.015 |
| EHI_199170_s_at | XM_648207 | hypothetical protein, conserved | 6.4 | 3.1 | down | 0.049 |
| EHI_183120_s_at | XM_649872 | centromeric protein E, putative | 8.0 | 3.1 | down | 0.014 |
| EHI_073980_s_at | XM_648468 | surface antigen ariel1, putative | 4.0 | 3.0 | down | 0.001 |
| EHI_031640_at | XM_648447 | sulfotransferase, putative | 7.5 | 8.5 | up | 0.000 |
| EHI_193640_s_at | XM_643661 | hypothetical protein | 2.3 | 8.2 | up | 0.002 |
| EHI_074750_at | XM_644490 | Ras family GTPase | 5.5 | 7.8 | up | 0.004 |
| EHI_018140_s_at | XM_001914260 | deoxyuridine 5′-triphosphate nucleotidohydrolase domain containing protein | 5.5 | 6.7 | up | 0.002 |
| EHI_072960_s_at | XM_001914509 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 4.3 | 6.0 | up | 0.007 |
| EHI_070690_at | XM_001913839 | Ras GTPase domain conting protein | 2.3 | 5.7 | up | 0.020 |
| EHI_046630_at | XM_645444 | Rho family GTPase | 4.1 | 5.3 | up | 0.005 |
| EHI_174970_at | XM_648244 | hypothetical protein | 5.4 | 5.2 | up | 0.000 |
| EHI_126550_at | XM_643463 | AIG1 family protein, putative | 6.5 | 5.1 | up | 0.002 |
| EHI_068270_s_at | XM_646627 | Rho guanine nucleotide exchange factor, putative | 4.1 | 4.9 | up | 0.001 |
| EHI_146680_s_at | XM_001914548 | hypothetical protein | 2.4 | 4.8 | up | 0.009 |
| EHI_004520_at | XM_651631 | hypothetical protein | 5.1 | 4.6 | up | 0.013 |
| EHI_046040_s_at | XM_645992 | hypothetical protein | 5.0 | 4.5 | up | 0.001 |
| EHI_095910_at | XM_001913730 | lipase, putative | 4.7 | 4.4 | up | 0.001 |
| EHI_134850_at | XM_647045 | Fe-hydrogenase, putative | 7.5 | 4.4 | up | 0.011 |
| EHI_120580_at | XM_646886 | hypothetical protein | 3.3 | 4.3 | up | 0.026 |
| EHI_067910_at | XM_651687 | competence protein ComEC, putative | 6.3 | 4.3 | up | 0.011 |
| EHI_146130_at | XM_644793 | hypothetical protein | 3.3 | 4.1 | up | 0.001 |
| EHI_151440_at | XM_652272 | cysteine proteinase, putative | 7.5 | 4.1 | up | 0.000 |
| EHI_182540_at | XM_651612 | Protein tyrosine phosphatases domain containing protein | 3.8 | 4.1 | up | 0.001 |
| EHI_074580_at | XM_645859 | hypothetical protein | 3.2 | 3.9 | up | 0.025 |
| EHI_019630_at | XM_643344 | hypothetical protein | 6.9 | 3.9 | up | 0.002 |
| EHI_123700_at | XM_648695 | hypothetical protein | 3.7 | 3.9 | up | 0.019 |
| EHI_028940_at | XM_645826 | hypothetical protein | 8.8 | 3.8 | up | 0.000 |
| EHI_191730_at | XM_643923 | cysteine protease binding protein family 10 | 5.3 | 3.7 | up | 0.017 |
| EHI_022990_at | XM_648401 | hypothetical protein | 4.8 | 3.7 | up | 0.002 |
| EHI_153710_at | XM_001913338 | methylene-fatty-acyl-phospholipid synthase, putative | 6.5 | 3.6 | up | 0.007 |
| EHI_059320_s_at | XM_001914076 | hypothetical protein | 2.6 | 3.4 | up | 0.037 |
| EHI_105080_at | XM_648821 | zinc finger protein, putative | 8.2 | 3.4 | up | 0.026 |
| EHI_126560_at | XM_001914189 | AIG1 family protein, putative | 7.5 | 3.4 | up | 0.004 |
| EHI_091670_at | XM_644055 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 2.3 | 3.4 | up | 0.030 |
| EHI_180390_at | XM_648725 | AIG1 family protein, putative | 9.1 | 3.4 | up | 0.016 |
| EHI_139400_at | XM_646219 | TATA-binding protein-associated phosphoprotein, putative | 3.7 | 3.3 | up | 0.002 |
| EHI_154270_at | XM_645351 | cell division control protein 42, putative | 6.1 | 3.3 | up | 0.008 |
| EHI_172510_at | XM_643770 | acid sphingomyelinase-like phosphodiesterase 3a precursor, putative | 3.5 | 3.2 | up | 0.000 |
| EHI_050570_at | XM_651510 | cysteine proteinase, putative | 11.3 | 3.2 | up | 0.001 |
| EHI_052130_at | XM_650257 | PQ loop repeat protein | 4.6 | 3.1 | up | 0.016 |
| EHI_009910_at | XM_652020 | TBC domain containing protein | 5.6 | 3.0 | up | 0.011 |
| EHI_121750_at | XM_646688 | hypothetical protein | 5.5 | 3.0 | up | 0.006 |
Table 3.
List of genes down and up regulated ≥3 fold upon SAT3 gene silencing.
| ProbeSetID | Accession Number | Common Name | Basal Expres-sion (log2) | Fold change | Regula-tion | p-value |
|---|---|---|---|---|---|---|
| EHI_153430_at | XM_651281 | serine acetyltransferase 3 | 10.5 | 186.7 | down | 0.007 |
| EHI_153420_at | XM_651282 | hypothetical protein | 7.9 | 14.0 | down | 0.011 |
| EHI_196760_s_at | XM_643708 | hypothetical protein | 7.7 | 12.5 | down | 0.004 |
| EHI_020830_s_at | XM_001913952 | hypothetical protein | 7.8 | 7.6 | down | 0.002 |
| EHI_128180_s_at | XM_649666 | Rab family GTPase | 8.1 | 6.5 | down | 0.004 |
| EHI_147860_at | XM_646798 | hypothetical protein | 5.6 | 5.2 | down | 0.011 |
| EHI_133210_s_at | XM_001914244 | peptidase S54 (rhomboid) family protein | 7.9 | 5.1 | down | 0.011 |
| EHI_051430_at | XM_652271 | Ras guanine nucleotide exchange factor, putative | 5.0 | 4.6 | down | 0.032 |
| EHI_128190_s_at | XM_649665 | peptidase S54 (rhomboid) family protein | 5.8 | 4.5 | down | 0.005 |
| EHI_079870_at | XM_647774 | NTP pyrophosphatase domain containing protein | 7.3 | 4.1 | down | 0.024 |
| EHI_164410_at | XM_649301 | DNA double-strand break repair Rad50 ATPase, putative | 4.4 | 3.9 | down | 0.008 |
| EHI_111210_at | XM_652499 | DNA double-strand break repair Rad50 ATPase, putative | 4.2 | 3.6 | down | 0.000 |
| EHI_178130_at | XM_646412 | hypothetical protein | 8.8 | 3.5 | down | 0.002 |
| EHI_079970_at | XM_001913704 | leucine rich repeat protein, BspA family | 5.2 | 3.2 | down | 0.027 |
| EHI_005930_at | XM_648325 | hypothetical protein | 4.1 | 3.1 | down | 0.011 |
| EHI_161970_at | XM_644065 | leucyl-tRNA synthetase, putative | 8.7 | 3.1 | down | 0.010 |
| EHI_170940_at | XM_001913890 | lipase, putative | 8.9 | 3.1 | down | 0.002 |
| EHI_074480_s_at | XM_001914032 | hypothetical protein | 3.9 | 3.0 | down | 0.002 |
| EHI_054660_at | XM_646970 | apyrase, putative | 3.9 | 3.0 | down | 0.047 |
| EHI_022600_s_at | XM_643169 | NADPH-dependent FMN reductase domain containing protein | 7.0 | 8.9 | up | 0.002 |
| EHI_022270_s_at | XM_644774 | NADPH-dependent FMN reductase domain containing protein | 7.2 | 8.5 | up | 0.002 |
| EHI_067720_s_at | XM_643101 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 8.3 | up | 0.001 |
| EHI_181710_s_at | XM_001914510 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 7.9 | up | 0.004 |
| EHI_103260_s_at | XM_001913434 | NADPH-dependent FMN reductase domain containing protein | 7.3 | 7.2 | up | 0.001 |
| EHI_153710_at | XM_001913338 | methylene-fatty-acyl-phospholipid synthase, putative | 6.5 | 5.8 | up | 0.002 |
| EHI_029930_at | XM_001914435 | hypothetical protein | 3.6 | 4.7 | up | 0.003 |
| EHI_155430_s_at | XM_650657 | hypothetical protein | 4.7 | 4.0 | up | 0.028 |
| EHI_091670_at | XM_644055 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 2.3 | 3.9 | up | 0.007 |
| EHI_146130_at | XM_644793 | hypothetical protein | 3.3 | 3.8 | up | 0.000 |
| EHI_072960_s_at | XM_001914509 | deoxyuridine 5′triphosphate nucleotidohydrolase domain containing protein | 4.3 | 3.7 | up | 0.011 |
| EHI_158010_at | XM_645706 | hypothetical protein | 4.0 | 3.5 | up | 0.024 |
| EHI_034530_s_at | XM_643791 | hypothetical protein | 2.5 | 3.4 | up | 0.036 |
| EHI_074750_at | XM_644490 | Ras family GTPase | 5.5 | 3.4 | up | 0.011 |
| EHI_198440_s_at | XM_001914343 | hypothetical protein | 2.5 | 3.3 | up | 0.013 |
| EHI_018140_s_at | XM_001914260 | deoxyuridine 5′-triphosphate nucleotidohydrolase domain containing protein | 5.5 | 3.3 | up | 0.016 |
Confirmation of differential gene expression by qRT–PCR
The microarray results were validated by qRT–PCR. Table 4 shows a comparison of the qRT-PCR and microarray data of six representative differentially expressed genes identified by the transcriptome analysis, with the RNA polymerase II gene used as reference33. The results of the qRT-PCR analysis agreed well with the microarray data for all examined gene transcripts (Table 4).
Table 4.
Validation of microarray data by qRT-PCR and microarray analysis.
| Common Name | Accession Number | Fold Change by qRT-PCR (by microarray) | ||
|---|---|---|---|---|
| SAT1/2gs | SAT3gs | CSgs | ||
| Fe-hydrogenase | XM_647045 | 5.0 (4.4) | ND | ND |
| Sulfotransferase | XM_648447 | 9.1 (8.5) | ND | ND |
| Phosphoserine aminotransferase | XM_650291 | −4.6 (−5.1) | ND | ND |
| NADPH-dependent oxidoreductases 2 | XM_648481 | 1.4 (1.9) | 1.3 (1.7) | 4.7 (4.0) |
| NADPH-dependent FMN reductase domain-containing protein | XM_643169 | 2.4 (3.0) | 9.4 (8.9) | 5.1 (4.0) |
| Methylene-fatty-acyl-phospholipid synthase | XM_001913338 | 4.1 (3.6) | 4.7 (5.8) | 4.5 (3.9) |
| RNA polymerase II | XM_643999 | 1.2 (1.4) | 1.2 (1.1) | 1.1 (1.0) |
The common names, accession numbers, and fold changes of the selected genes are shown. The values are the fold changes in the expression obtained from qRT-PCR and the corresponding fold changes in the expression values obtained from Affymetrix analysis are shown in parentheses. ND, not detected.
S-Methylcysteine production leads to increased oxidative stress tolerance
To investigate whether the SMC accumulation observed in trophozoites cultured in L-cysteine lacking BI-S-33 medium protects E. histolytica against oxidative stress, the CSgs, which does not produce SMC and control (harboring plasmid psAP2G) transformants were compared for oxidative stress sensitivity by culturing the two strains in L-cysteine lacking medium. After 48-h cultivation in L-cysteine lacking medium, SMC had accumulated in the control transformant, but remained absent in the CSgs transformant (Fig. 3). The CSgs and control transformants were next exposed to different concentrations of H2O2 (0–6.4 mM) for 1 h and viability was then determined. The CSgs transformant showed slightly, but significantly (Student’s t-test), lower sensitivity to 0.8–4.0 mM H2O2 compared to the control transformant, suggesting that CS1–3 or SMC synthesis may be involved in protecting the cells against oxidative stress (Fig. 4).
Figure 4.

Effect of CS gene silencing on oxidative stress tolerance. Trophozoites of CSgs and control (harboring plasmid psAP2G) strains were exposed to different concentrations of H2O2 for 1 h and viability was then determined. Survival rates are shown as percent of untreated control cells (mean ±S.D. of three independent experiments conducted in triplicate). Statistical comparisons were made by the Student’s t test (**P <0.01, ***P <0.001).
Discussion
The identification and functional characterization of the molecular components involved in essential metabolic pathways contribute to the overall understanding of parasite biology, but also aid in the rational design of novel therapeutics. L-Cysteine is indispensable for the survival of virtually all living organisms and plays a major role in maintaining the redox balance of thiol compounds in microaerophiles18. The cysteine biosynthetic pathway exists in bacteria, plants, and several parasitic protozoa, including Leishmania major, Trypanosoma cruzi, and Trichomonas vaginalis 1, and enzymes involved in this pathway are suitable targets for the development of novel drugs to prevent disease caused by these parasites2–6.
In the present study, we investigated the specific role of individual isotypes of SAT and CS using a gene silencing approach. Although SAT26 and CS35 have been biochemically13–16 and structurally characterized, the specific role of individual SAT and CS isotypes in proliferation, pathogenesis, and parasitism remains to be elucidated. Although we attempted to silence the expression of single genes, all the three CS isotypes were simultaneously silenced in E. histolytica due to their high similarity at the nucleotide and amino acid levels (CS1-3, 83–99%). The global repression of CS expression resulted in impaired trophozoites proliferation in L-cysteine lacking BI-S-33 medium, but not in normal BI-S-33 medium containing 8 mM L-cysteine (Fig. 2B). Metabolomic analysis of the CS-silenced transformant under the two culture conditions further revealed that SMC was not detectable and that the level of OAS was markedly reduced, demonstrating that CS is involved in SMC synthesis from OAS (Fig. 3). We previously showed that E. histolytica trophozoites produce SMC, rather than L-cysteine, when cultured in L-cysteine lacking BI-S-33 medium13. The present metabolomic analysis further revealed that L-cysteine levels were also decreased when the CSgs (and SATgs) transformants were cultured in normal BI-S-33 medium containing 8 mM L-cysteine, suggesting that E. histolytica synthesizes both L-cysteine and SMC, and that the flux towards cysteine synthesis likely depends upon the intracellular availability of sulfides (i.e., methanethiol and sulfide).
In contrast to CS, silencing of SAT1/2 and SAT3 was specific to the targeted SAT isotypes. Unlike other organisms, E. histolytica possesses three apparently redundant SAT isozymes16. These three SAT isotypes differ from one other in their regulatory properties. The isoenzymes SAT1 and SAT2 are regulated through allosteric feedback by L-cysteine15,16, whereas SAT3 is relatively insensitive to L-cysteine inhibition16. Consistent with these findings, EhSAT1-3 showed different levels of sensitivity to allosteric feedback by L-cysteine16 [inhibition constant (Ki) values of EhSAT1-3 are 4.7, 28, and 460 μM, respectively]. We previously showed that under cysteine lacking conditions, OAS and SMC expression levels in E. histolytica increase, whereas the expression of SAT and CS isotypes are not affected by L-cysteine depletion13,36. As OAS and SMC were undetectable under normal conditions, OAS, which is produced by SAT1-3, appears to be readily converted to cysteine, but not SMC. Alternatively, the in-vivo activities of SAT3 and cysteine-sensitive SAT1/2 may be repressed by unknown mechanisms. Under cysteine lacking conditions, L-cysteine-sensitive SAT1 and SAT2, together with cysteine-insensitive SAT3, were derepressed, leading to increased production of OAS. The mechanism by which SMC, but not cysteine, accumulates in response to cysteine deprivation in strains SAT1/2gs and SAT3gs remains unknown. However, it is conceivable that in strain SAT1/2gs, SAT3 compensates for the loss of SAT1/2 by producing sufficient cellular OAS and thereby contributes to the maintenance of high SMC levels under cysteine lacking conditions.
Another unique aspect of E. histolytica SAT1 is the lack of protein-protein interaction with CS26. It is well known that in bacteria and plants, CS and SAT form a heteromeric complex with a molecular mass of several hundred kilodaltons37. However, EhCS1 and EhSAT1 form a homodimer and homotrimer, respectively26, but these proteins do not interact under physiological conditions26. The lack of interaction between EhCS1 and EhSAT1 was structurally elucidated26,35. The apparent reduction of cysteine/SMC production in SAT3gs strain despite high level of OAS may explain the possible formation of a SAT3-CS complex that regulates cellular CS activity. Here, silencing of SAT3 resulted in the loss of complex formation, reduction of CS activity, and decreased production of SMC and L-cysteine (Fig. 3). Metabolomic analysis confirmed that the similar response occurred in the CS gene-silenced transformants. SAT3 possesses a unique 25–30 amino acids extension at the carboxyl terminus and has a low isoelectric point compared to SAT1 and SAT216. These features may favor the interaction with E. histolytica CS, particularly EhCS3, which possesses the highest pI (8.17) among the three CS isotypes. However, this hypothesis needs to be experimentally proven.
The present metabolomic analyses combined with the results of the growth kinetic assay demonstrated that neither the concentrations of OAS, L-cysteine or SMC in the two culture conditions, nor trophozoites growth under L-cysteine lacking conditions were affected by SAT1/2 gene silencing (Fig. 3). These data suggest that SAT3 is a robust enzyme that likely compensated for the loss of SAT1/2 under in-vitro conditions. In contrast to SAT1/2, repression of SAT3 had more marked effects on growth than the repression of SAT1/2, suggesting that SAT3 is critical for survival under stressful conditions, whereas SAT1/2 are involved in more general house-keeping roles. This speculation is also supported by the fact that the levels of both L-cysteine and SMC were decreased in strain SAT3gs.
To determine whether other genes, particularly those involved in sulfur metabolism, compensate for the loss of CS and SAT gene expression, we compared the transcriptomes of the CSgs, SAT1/2, SAT3gs, and control strains grown in normal medium. Notably, several genes from a family of the NADPH-dependent FMN reductase domain-containing proteins, also known as iron-sulfur flavoproteins (ISFs), which are commonly found in anaerobic prokaryotes, were highly upregulated in CSgs and SAT3gs (Tables 1 and 3). To date, the only eukaryotic species that have been found to possess ISF homologs are E. histolytica and Trichomonas vaginalis 38,39. A search of the genome database of E. histolytica revealed the presence of seven independent ISF genes40, which were previously shown to be upregulated in E. histolytica cells cultured in L-cysteine lacking BI-S-33 media, suggesting that these genes are regulated in response to L-cysteine deprivation36.
In contrast to CSgs strain, we found that in SAT1/2 gene-silenced strain one of the sulfotransferase, SULT9 (XP_653539, EHI_031640) (Table 2), was up-regulated more than eight fold, suggesting its involvement in L-cysteine biosynthesis and/or redox-related metabolism. The E. histolytica genome contains 10 genes that encode putative sulfotransferases (SULTs), which are localized in the cytosol and are involved in the production of sulfated molecules41. For example, SULT6 (XP_649714, EHI_146990) is responsible for synthesizing cholesteryl sulfate, an important compound for the encystation process in the Entamoeba life cycle41. However, the function of other SULTs in E. histolytica remains largely unknown. In Arabidopsis roots, a plasma membrane sulfate ion transporter (SULTR) physically interacts with CS to coordinate internalization of sulfate ions based on the energetic/metabolic state of root cells42. Here, we also determined that Fe hydrogenase, which belongs to a distinct class of hydrogen-producing metalloenzymes and is found in a wide variety of prokaryotes and eukaryotes43, was up-regulated more than four fold in strain SAT1/2gs. Fe hydrogenase contributes to the utilization of hydrogen as a growth substrate and for the disposal of excess electrons through combination with protons to form hydrogen43. Although the role of Fe hydrogenase in Entamoeba is unclear, it is possible that this enzyme is regulated in response to oxygen levels, as was shown in Chlamydomonas reinhardtii, which contained increased transcript levels of Fe hydrogenase upon shifting from an aerobic to anaerobic atmosphere44.
The present metabolome data of strain CSgs suggest that in addition to L-cysteine, CS enzymes are involved in SMC production (Fig. 3). SMC is a sulfur-containing amino acid that is found in relatively large amounts in several legumes, where it is considered to be a sulfur storage compound45. However, the fate and physiological significance of SMC in protozoa, particularly E. histolytica, is not yet fully understood. Previously, we investigated metabolic responses to hydrogen peroxide - and paraquat-mediated oxidative stress in E. histolytica trophozoites and reported that SMC levels are increased more than two fold under both stress conditions46, suggesting the involvement of this metabolite in the oxidative stress response. To confirm this speculation, we compared the oxidative stress tolerance between the control and CSgs transformant because SMC was undetected in CSgs strain (Fig. 3), and demonstrated that the CSgs transformant was more sensitive to oxidative stress. In Brassica exposed to H2O2 or O2 stress, SMC is non-enzymatically converted to SMC sulfoxide47, which is further enzymatically catabolized into pyruvate, ammonia, and alkylthiosulfinates48. The enzyme that catalyzes the last reaction is cystine lyase (EC 4.4.1.8) and behaves similarly to allinase (EC 4.4.1.4) in garlic, with the exception that cystine lyase also has the ability to cleave L-cystine49. Based on this observation, we propose that under oxidative stress conditions, SMC is converted to SMC sulfoxide and is further degraded by a lyase enzyme, such as methionine γ-lyase17 (MGL), to pyruvate and sulfenic acid.
In summary, the present metabolomic analysis revealed that CS and SAT3 are key enzymes for cysteine/SMC production in E. histolytica and are also essential for parasite survival under oxidative stress conditions. Transcriptomic analysis of the constructed CSgs and SAT3gs strains revealed that compensatory mechanisms in which ISFs play key roles operate under conditions where the CS and SAT3 pathway(s) are inactivated. These findings corroborate the metabolic and physiological importance of the L-cysteine pathway in E. histolytica and suggest that CS and SAT3 represent good targets for drug development. Further work is needed to demonstrate the specific role of these ISFs in E. histolytica.
Methods
Microorganisms and cultivation
In-vitro cultures of E. histolytica strains HM-1:IMSS cl6 and G3 were routinely maintained in Diamond’s BI-S-33 medium at 35.5 °C, as described previously50,51.
Gene silencing
Strain G3 and plasmid psAP2 were kindly provided by Dr. David Mirelman (Weisman Institute, Israel)28,29. Gene silencing was performed as previously described28,29,36,52. Briefly, 420-bp fragments containing the entire open reading frames of the E. histolytica CS1, CS3, SAT1 SAT2 and SAT3 genes starting at the initiation codon were amplified by PCR from cDNA using the oligonucleotide primers listed in Supplementary Table S1. The obtained PCR products were digested with StuI and SacI and inserted into StuI/SacI-digested pSAP2G to produce psAP2G-CS1, psAP2G-CS3, psAP2G-SAT1, psAP2G-SAT2 and psAP2G-SAT3. The constructed plasmids were introduced into E. histolytica strain G3 by liposome-mediated transfection15, and the resulting transformants (designated psAP2G [control], CS1gs, CS3gs, SAT1gs, SAT2gs and SAT3gs) were selected and maintained in normal BI-S-33 medium supplemented with 7 μg/ml geneticin (Invitrogen). The expression of the respective genes was confirmed by semi-quantitative RT-PCR as described previously using RNA polymerase II mRNA (GenBankTM accession number XM_643999) as a reference33, as the expression of this gene was invariant in all of the transformants. The transformants were designated psAP2G (control), CS1gs, CS3gs, SAT1gs, SAT2gs and SAT3gs.
Extraction of metabolites from E. histolytica
E. histolytica trophozoites were cultured for approximately 24 h in standard BI-S-33 medium containing 8 mM L-cysteine. The medium was replaced with either normal BI-S-33 medium or medium lacking L-cysteine13, and trophozoites were cultured for a further 48 h. To extract metabolites, approximately 1.5 × 106 cells were then harvested and immediately suspended in 1.6 mL of −75 °C methanol to quench metabolic activity. To minimize the effects of experimental artifacts, such as ion suppression, on metabolite levels, 2-(N-morpholino) ethanesulfonic acid, methionine sulfone, and D-camphor-10-sulfonic acid were added to each sample as internal standards13,53,54. The samples were sonicated for 30 s and then mixed with 1.6 mL chloroform and 640 µl deionized water. After vortexing, the mixed samples were centrifuged at 4600 g for 5 min at 4 °C. The aqueous layer (1.6 mL) was filtrated using an Amicon Ultrafree-MC ultrafilter (Millipore Co., Massachusetts, USA) and the collected sample was centrifuged at 9100 g at 4 °C for approximately 2 h. The filtrate was vacuum dried and stored at −80 °C until needed for mass spectrometric analysis55. Prior to the analysis, the sample was dissolved in 20 μl de-ionized water containing 200 μmol/L of two reference compounds (3-aminopyrrolidine and trimesic acid).
Instrumentation and capillary electrophoresis-time-of-flight mass spectrometry (CE-TOFMS)
CE-TOFMS was performed using an Agilent CE Capillary Electrophoresis System equipped with an Agilent 6210 Time-of-Flight mass spectrometer, Agilent 1100 isocratic HPLC pump, Agilent G1603A CE-MS adapter kit, and Agilent G1607A CE-ESI-MS sprayer kit (Agilent Technologies, Waldbronn, Germany). The system was controlled by Agilent G2201AA ChemStation software for CE. Data acquisition was performed using Analyst QS software for Agilent TOF (Applied Biosystems, CA, USA; MDS Sciex, Ontario, Canada).
CE-TOFMS conditions for cationic metabolite analysis
Cationic metabolites were separated in a fused-silica capillary (50 μm i.d. × 100 cm total length) filled with 1 mol/L formic acid as the reference electrolyte56. Sample solution (~3 nL) was injected at 50 mbar for 3 s, and a positive voltage of 30 kV was applied. The capillary and sample trays were maintained at 20 °C and below 5 °C, respectively. Sheath liquid composed of methanol/water (50% v/v) and 0.1 μmol/L hexakis (2,2- difluorothoxy) phosphazene was delivered at 10 μL/min. ESI-TOFMS was operated in positive ion mode. The capillary voltage was set at 4 kV and the flow rate of nitrogen gas (heater temperature 300 °C) was set at 10 psig. For TOFMS, the fragmenter voltage, skimmer voltage, and octapole radio frequency voltage (Oct RFV) were set at 75, 50, and 125 V, respectively. An automatic recalibration function was performed using the masses of two reference compounds, protonated 13C methanol dimer (m/z 66.063061) and protonated hexakis (2,2-difluorothoxy) phosphazene (m/z 622.028963), which provided the lock mass for exact mass measurements. Exact mass data were acquired at the rate of 1.5 cycles/s over 50 to 1,000 m/z.
CE-TOFMS conditions for anionic metabolite analysis
Anionic metabolites were separated in a cationic-polymer–coated COSMO(+) capillary (50 μm i.d. × 110 cm) (Nacalai Tesque) filled with a 50 mmol/L ammonium acetate solution (pH 8.5) as the reference electrolyte57,58. Sample solution (~30 nL) was injected into the system at 50 mbar for 30 s and a negative voltage of −30 kV was applied. Ammonium acetate (5 mmol/L) in methanol/water (50% v/v) containing 0.1 μmol/L hexakis (2,2-difluorothoxy) phosphazene was delivered as sheath liquid at 10 μL/min. ESI-TOFMS was performed in negative ion mode at a capillary voltage of 3.5 kV. For TOFMS, the fragmenter voltage, skimmer voltage, and Oct RFV were set at 100, 50, and 200 V, respectively58. An automatic recalibration function was performed using the masses of two reference compounds: deprotonated 13C acetate dimer (m/z 120.038339) and an acetate adduct of hexakis (2,2-difluorothoxy) phosphazene (m/z 680.035541). The other conditions were identical to those used for the cationic metabolome analysis.
CE-TOFMS data processing
Raw data were processed using in-house Masterhands software59. The overall data processing flow consisted of the following steps: noise-filtering, baseline-removal, migration time correction, peak detection, and peak area integration from a 0.02 m/z-wide slice of the electropherograms. The data processing resembled the common strategies used for LC-MS and GC-MS data analysis software, such as MassHunter (Agilent Technologies) and XCMS60. Accurate m/z values for each peak were calculated by Gaussian curve fitting in the m/z domain, and migration times were normalized using alignment algorithms based on dynamic programming61,62. All target metabolites were identified by matching their m/z values and normalized migration times with those of standard compounds in the in-house library.
RNA isolation and Affymetrix microarray hybridization
Trophozoites were grown in BI-S-33 medium containing 8 mM L-cysteine for approximately 48 h. The collected cell pellets were resuspended in Trizol reagent (Invitrogen, Carlsbad, CA, USA) and RNA was isolated according to the manufacturer’s protocol. The RNA concentration for each sample was measured using a Nanodrop Spectrophotometer 1000 (Thermo Scientific, Wilmington, DE, USA). RNA integrity was checked using an Experion Automated Electrophoresis System (RNA StdSens analysis kit, Bio-Rad). All reagents and protocols followed those described in the Affymetrix user manuals. Using the One-Cycle cDNA synthesis kit, 5 μg total RNA was reverse transcribed using a T7-Oligo (dT) primer for first strand cDNA synthesis. After second strand synthesis, the double-stranded cDNA template was used for in-vitro transcription (IVT) in the presence of biotinylated nucleotides (GeneChip IVT labeling kit) to produce Biotin-labeled cRNA. Unincorporated NTPs were removed from the biotinylated cRNA (GeneChip sample cleanup module), which was then purified, quantified and fragmented. A hybridization cocktail consisting of eukaryotic hybridization controls and fragmented, labeled cRNA (GeneChip Hybridization, Wash and Stain Kit) were hybridized for 16 h at 45 °C in a Hybridization Oven 640 (Affymetrix) onto a custom-generated Affymetrix platform microarray (49–7875) with probe sets consisting of 11 probe pairs, each representing 12,384 E. invadens 63 (Eh_Eia520620F_Ei) and 9,327 E. histolytica 36 (Eh_Eia520620F_Eh) open reading frames. The array chips were washed and stained (GeneChip Hybridization, Wash and Stain Kit) with Streptavidin–phycoerythrin Biotinylated anti-streptavidin antibody using a GeneChip Fluidics Station 450 (Affymetrix) for 1.5 h. After washing and staining, the GeneChip arrays were scanned using a Hewlett-Packard Affymetrix Scanner 3000.
Analysis of microarray data
A minimum of two arrays was used for each test condition. Raw probe intensities were generated using Gene Chip Operating Software (GCOS) and the Gene Titan Instrument from Affymetrix. Normalized expression values for each probe set were obtained from R 2.7.0 downloaded from the Bioconductor project (http://www.bioconductor.org) using robust multiarray averaging with correction for oligosequence (gcRMA). Standard correlation coefficients were calculated using GeneSpring GX 10.0.2. One-way ANOVA analysis with Tukey’s Post Hoc test was performed to extract differentially expressed genes. P values were calculated using Welch’s t-test after multiple test correction by the Benjamini–Hochberg method. A post-hoc test using Tukey’s Honestly Significant Difference test was conducted to determine significant differences between samples.
Quantitative real-time PCR (qRT–PCR)
Total RNA extracted above were used for qRT–PCR. cDNA synthesis was performed using the SuperScript III First-Strand Synthesis System (Invitrogen) following the manufacturer’s instructions. cDNA was synthesized from 5 μg total RNA and oligo (dT) 20 primers using the Superscript III First-Strand Synthesis System (Invitrogen). PCR was performed with cDNA as the template and gene-specific primers using the ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). The genes whose expression was verified by qRT–PCR are listed in Supplementary Table S4. The RNA polymerase II gene was used as a control. The parameters for PCR were: an initial denaturation step at 95 °C for 9 min followed by 40 cycles of denaturation at 94 °C for 30 s, annealing at 50 °C for 30 s and extension at 65 °C for 1 min. A final step at 95 °C for 9 s, 60 °C for 9 s and 95 °C for 9 s was used to remove primer dimers. All test samples were run in triplicate. An RT-negative control was also used for each sample set along with a blank control consisting of nuclease-free water in place of cDNA.
Growth assay of E. histolytica trophozoites
A cell-growth assay was performed as described previously36. Briefly, approximately 6 × 104 exponentially growing SAT1/2, SAT3, or CS gene-silenced trophozoites and control transformants were inoculated into 6 mL normal BI-S-33 medium with and without L-cysteine supplemented with 7 μg/mL geneticin, and the number of parasites was counted every 24 h using a haemocytometer.
Hydrogen peroxide (H2O2) sensitivity assay
To examine sensitivity to H2O2, E. histolytica CS gene-silenced and control (harboring plasmid psAP2G) transformants were cultured in L-cysteine lacking BI-S-33 media containing 7 μg/mL geneticin for 48 h at 35.5 °C. After 48 h, approximately 104 trophozoites per well were seeded into the wells of a 96-well plate containing BI-S-33 medium supplemented with 7 μg/mL geneticin and further incubated for 1 h at 35.5 °C. The trophozoites were then exposed to H2O2 (0, 0.8, 1.6, 2.4, 3.2, 4, 4.8 and 6.4 mM) for 1 h. After incubation, the medium was removed and 90 μl of pre-warmed Opti-MEM I (Life Technologies) and 10 μl WST-1 solution64 (Roche Diagnostics, Mannheim, Germany) were added to each well. Viability of trophozoites was detected by measuring absorbance at 450 nm using a microplate reader (Model 550, Bio-Rad, Tokyo, Japan). The sensitivity assays were performed in triplicate and repeated at least three times.
Electronic supplementary material
Acknowledgements
This work was supported in part by a grant for Research on Emerging and Re-emerging Infectious Diseases from the Japan Agency for Medical Research and Development (AMED), Grants-in-Aid for Scientific Research on Innovative Areas (23117001, 23117005), a Grant-in-Aid for Scientific Research (23390099) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), and a grant for Science and Technology Research Partnership for Sustainable Development from AMED and the Japan International Cooperation Agency to TN.
Author Contributions
G.J. and T.N. conceived and designed the experiments. G.J. and D.S. performed the experiments. G.J. analyzed the data. T.N. and T.S. contributed reagents/materials/analysis tools. G.J. and T.N. wrote the paper. All authors reviewed the manuscript prior to submission.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15923-3.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ali V, Nozaki T. Current therapeutics, their problems, and sulfur-containing amino acid metabolism as a novel target against infections by “amitochondriate” protozoan parasites. Clin Microbiol Rev. 2007;20:164–187. doi: 10.1128/CMR.00019-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunner K, et al. Inhibitors of the Cysteine Synthase CysM with Antibacterial Potency against Dormant Mycobacterium tuberculosis. J Med Chem. 2016;59:6848–59. doi: 10.1021/acs.jmedchem.6b00674. [DOI] [PubMed] [Google Scholar]
- 3.Spyrakis F, et al. Isozyme-specific ligands for O-acetylserine sulfhydrylase, a novel antibiotic target. PLoS ONE. 2013;8:e77558. doi: 10.1371/journal.pone.0077558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mori M, et al. Identification of natural inhibitors of Entamoeba histolytica cysteine synthase from microbial secondary metabolites. Front Microbiol. 2015;6:962. doi: 10.3389/fmicb.2015.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nagpal I, Raj I, Subbarao N, Gourinath S. Virtual Screening, Identification and In Vitro Testing of Novel Inhibitors of O-Acetyl-L-Serine Sulfhydrylase of Entamoeba histolytica. PLoS ONE. 2012;7:e30305. doi: 10.1371/journal.pone.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal SM, Jain R, Bhattacharya A, Azam A. Inhibitors of Escherichia coli serine acetyltransferase block proliferation of Entamoeba histolytica trophozoites. Int J Parasitol. 2008;38:137–41. doi: 10.1016/j.ijpara.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Salles JM, Salles MJ, Moraes LA, Silva MC. Amebiasis: an update on diagnosis and management. Expert Rev Anti Infect Ther. 2007;5:893–901. doi: 10.1586/14787210.5.5.893. [DOI] [PubMed] [Google Scholar]
- 8.Ximenez C, Moran P, Rojas L, Valadez A, Gomez A. Reassessment of the epidemiology of amebiasis: state of the art. Infect Gen Evol. 2009;9:1023–1032. doi: 10.1016/j.meegid.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 9.Cudmore SL, Delgaty KL, Hayward-McClelland SF, Petrin DP, Garber GE. Treatment of infections caused by metronidazole-resistant Trichomonas vaginalis. Clin Microbiol Rev. 2004;17:783–793. doi: 10.1128/CMR.17.4.783-793.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wassmann C, Hellberg A, Tannich E, Bruchhaus I. Metronidazole resistance in the protozoan parasite Entamoeba histolytica is associated with increased expression of iron-containing superoxide dismutase and peroxiredoxin and decreased expression of ferredoxin 1 and flavin reductase. J Biol Chem. 1999;274:26051–26056. doi: 10.1074/jbc.274.37.26051. [DOI] [PubMed] [Google Scholar]
- 11.Penuliar GM, Nakada-Tsukui K, Nozaki T. Phenotypic and transcriptional profiling in Entamoeba histolytica reveal costs to fitness and adaptive responses associated with metronidazole resistance. Front Microbiol. 2015;6:354. doi: 10.3389/fmicb.2015.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeelani G, Nozaki T. Metabolomic analysis of Entamoeba: applications and implications. Curr Opin Microbiol. 2014;20:118–124. doi: 10.1016/j.mib.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 13.Husain A, et al. Metabolome analysis revealed increase in S-methylcysteine and phosphatidylisopropanolamine synthesis upon L-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. J Biol Chem. 2010;285:39160–39170. doi: 10.1074/jbc.M110.167304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nozaki T, et al. Molecular cloning and characterization of the genes encoding two isoforms of cysteine synthase in the enteric protozoan parasite Entamoeba histolytica. Mol Biochem Parasitol. 1998;97:33–44. doi: 10.1016/S0166-6851(98)00129-7. [DOI] [PubMed] [Google Scholar]
- 15.Nozaki T, et al. Characterization of the gene encoding serine acetyltransferase, a regulated enzyme of cysteine biosynthesis from the protist parasites Entamoeba histolytica and Entamoeba dispar. Regulation and possible function of the cysteine biosynthetic pathway in Entamoeba. J Biol Chem. 1999;274:32445–32452. doi: 10.1074/jbc.274.45.32445. [DOI] [PubMed] [Google Scholar]
- 16.Hussain S, Ali V, Jeelani G, Nozaki T. Isoform-dependent feedback regulation of serine O-acetyltransferase isoenzymes involved in L- cysteine biosynthesis of Entamoeba histolytica. Mol Biochem Parasitol. 2009;163:39–47. doi: 10.1016/j.molbiopara.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 17.Tokoro M, Asai T, Kobayashi S, Takeuchi T, Nozaki T. Identification and characterization of two isoenzymes of methionine γ-lyase from Entamoeba histolytica: a key enzyme of sulfur-amino acid degradation in an anaerobic parasitic protest that lacks forward and reverse transsulfuration pathways. J Biol Chem. 2003;278:42717–42727. doi: 10.1074/jbc.M212414200. [DOI] [PubMed] [Google Scholar]
- 18.Nozaki T, Ali V, Tokoro M. Sulfur-containing amino acid metabolism in parasitic protozoa. Adv Parasitol. 2005;60:1–99. doi: 10.1016/S0065-308X(05)60001-2. [DOI] [PubMed] [Google Scholar]
- 19.Sato D, Yamagata W, Harada S, Nozaki T. Kinetic characterization of methionine gamma-lyases from the enteric protozoan parasite Entamoeba histolytica against physiological substrates and trifluoromethionine, a promising lead compound against amoebiasis. FEBS J. 2008;275:548–560. doi: 10.1111/j.1742-4658.2007.06221.x. [DOI] [PubMed] [Google Scholar]
- 20.Campanini B, et al. Moonlighting O-acetylserine sulfhydrylase: New functions for an old protein. Biochim Biophys Acta. 2015;1854:1184–93. doi: 10.1016/j.bbapap.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero LC, et al. Cysteine and cysteine-related signaling pathways in Arabidopsis thaliana. Mol Plant. 2014;7:264–76. doi: 10.1093/mp/sst168. [DOI] [PubMed] [Google Scholar]
- 22.Williams RA, Westrop GD, Coombs GH. Two pathways for cysteine biosynthesis in Leishmania major. Biochem J. 2009;420:451–62. doi: 10.1042/BJ20082441. [DOI] [PubMed] [Google Scholar]
- 23.Marciano D, Santana M, Nowicki C. Functional characterization of enzymes involved in cysteine biosynthesis and H2S production in Trypanosoma cruzi. Mol Biochem Parasitol. 2012;185:114–20. doi: 10.1016/j.molbiopara.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 24.Jeelani, G., Sato, D. & Nozaki, T. Metabolomic analysis of Enatmoeba biology, In: Nozaki, T. & Bhattacharya, A. (Eds), Amebiasis: Biology and Pathogenesis of Entamoeba, Springer Verlag, Japan, 331–349 (2015).
- 25.Sirko A, Błaszczyk A, Liszewska F. Overproduction of SAT and/or OASTL in transgenic plants: a survey of effects. J Exp Bot. 2004;55:1881–8. doi: 10.1093/jxb/erh151. [DOI] [PubMed] [Google Scholar]
- 26.Kumar S, Raj I, Nagpal I, Subbarao N, Gourinath S. Structural and biochemical studies of serine acetyltransferase reveal why the parasite Entamoeba histolytica cannot form a cysteine synthase complex. J Biol Chem. 2011;286:12533–41. doi: 10.1074/jbc.M110.197376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hesse H, Nikiforova V, Gakière B, Hoefgen R. Molecular analysis and control of cysteine biosynthesis: integration of nitrogen and sulphur metabolism. J Exp Bot. 2004;55:1283–92. doi: 10.1093/jxb/erh136. [DOI] [PubMed] [Google Scholar]
- 28.Bracha R, Nuchamowitz Y, Mirelman D. Transcriptional silencing of an amoebapore gene in Entamoeba histolytica: molecular analysis and effect on pathogenicity. Eukaryot Cell. 2003;2:295–305. doi: 10.1128/EC.2.2.295-305.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bracha R, Nuchamowitz Y, Anbar M, Mirelman D. Transcriptional silencing of multiple genes in trophozoites of Entamoeba histolytica. PLoS Pathog. 2006;2:e48. doi: 10.1371/journal.ppat.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakada-Tsukui K, Tsuboi K, Furukawa A, Yamada Y, Nozaki T. A novel class of cysteine protease receptors that mediate lysosomal transport. Cell Microbiol. 2012;14:1299–1317. doi: 10.1111/j.1462-5822.2012.01800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorenzi HA, et al. New assembly, reannotation and analysis of the Entamoeba histolytica genome reveal new genomic features and protein content information. PLoS Negl Trop Dis. 2010;4:e716. doi: 10.1371/journal.pntd.0000716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakada-Tsukui K, Saito-Nakano Y, Husain A, Nozaki T. Conservation and function of Rab small GTPases in Entamoeba: annotation of E. invadens Rab and its use for the understanding of Entamoeba biology. Exp Parasitol. 2010;126:337–47. doi: 10.1016/j.exppara.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 33.Jeelani G, et al. Two atypical L-cysteine-regulated NADPH-dependent oxidoreductases involved in redox maintenance, L-cystine and iron reduction, and metronidazole activation in the enteric protozoan Entamoeba histolytica. J Biol Chem. 2010;285:26889–26899. doi: 10.1074/jbc.M110.106310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiba Y, Makiuchi T, Jeelani G, Nozaki T. Heterogeneity of the serine synthetic pathway in Entamoeba species. Mol Biochem Parasitol. 2016;207:56–60. doi: 10.1016/j.molbiopara.2016.06.002. [DOI] [PubMed] [Google Scholar]
- 35.Chinthalapudi K, et al. Crystal structure of native O-acetyl-serine sulfhydrylase from Entamoeba histolytica and its complex with cysteine: structural evidence for cysteine binding and lack of interactions with serine acetyl transferase. Proteins. 2008;72:1222–32. doi: 10.1002/prot.22013. [DOI] [PubMed] [Google Scholar]
- 36.Husain A, Jeelani G, Sato D, Nozaki T. Global analysis of gene expression in response to L-cysteine deprivation in the anaerobic protozoan parasite Entamoeba histolytica. BMC Genomics. 2011;12:275. doi: 10.1186/1471-2164-12-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feldman-Salit A, Wirtz M, Hell R, Wade RC. A mechanistic model of the cysteine synthase complex. J Mol Biol. 2009;386:37–59. doi: 10.1016/j.jmb.2008.08.075. [DOI] [PubMed] [Google Scholar]
- 38.Krauth-Siegel RL, Leroux AE. Low molecular mass antioxidants in parasites. Antioxid Redox Signal. 2012;17:583–607. doi: 10.1089/ars.2011.4392. [DOI] [PubMed] [Google Scholar]
- 39.Smutná T, Pilarová K, Tarábek J, Tachezy J, Hrdý I. Novel functions of an iron-sulfur flavoprotein from Trichomonas vaginalis hydrogenosomes. Antimicrob Agents Chemother. 2014;58:3224–32. doi: 10.1128/AAC.02320-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeelani G, Nozaki T. Entamoeba thiol-based redox metabolism: A potential target for drug development. Mol Biochem Parasitol. 2016;206:39–45. doi: 10.1016/j.molbiopara.2016.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Mi-ichi F, et al. Entamoeba mitosomes play an important role in encystation by association with cholesteryl sulfate synthesis. Proc Natl Acad Sci USA. 2015;112:E2884–90. doi: 10.1073/pnas.1423718112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibagaki N, Grossman AR. Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J Biol Chem. 2010;285:25094–102. doi: 10.1074/jbc.M110.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das D, et al. Role of Fe-hydrogenase in biological hydrogen production. Current Science. 2006;90:1627–1637. [Google Scholar]
- 44.Forestier M, et al. Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur J Biochem. 2003;270:2750–8. doi: 10.1046/j.1432-1033.2003.03656. [DOI] [PubMed] [Google Scholar]
- 45.Rebeille F, et al. Methionine catabolism in Arabidopsis cells is initiated by a gamma-cleavage process and leads to S-methylcysteine and isoleucine syntheses. Proc Natl Acad Sci USA. 2006;103:15687–15692. doi: 10.1073/pnas.0606195103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Husain A, Sato D, Jeelani G, Soga T, Nozaki T. Dramatic increase in glycerol biosynthesis upon oxidative stress in the anaerobic protozoan parasite Entamoeba histolytica. PLoS Negl Trop Dis. 2012;6:e1831. doi: 10.1371/journal.pntd.0001831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mae T, Ohira K, Fujiwara A. Metabolism of S-methylcysteine and its sulfoxide in chinese cabbage, Brassica pekinensis Rupr. Plant cell Physiol. 1971;12:881–887. doi: 10.1093/oxfordjournals.pcp.a074692. [DOI] [Google Scholar]
- 48.Marks HS, Hilaon JA, Leichtweis HC, Stoewsand GS. S-Methylcysteine Sulfoxide in Brassica Vegetables and Formation of Methyl Methanethiosulfinate from Brussels Sprouts. J Agric Food Chem. 1992;40:2096–2101. doi: 10.1021/jf00023a012. [DOI] [Google Scholar]
- 49.Ukai K, Sekiya J. Rapid purification and characterization of cystine lyase b from broccoli inflorescence. Phytochemistry. 1999;51:853–859. doi: 10.1016/S0031-9422(99)00067-9. [DOI] [PubMed] [Google Scholar]
- 50.Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-X. [DOI] [PubMed] [Google Scholar]
- 51.Clark CG, Diamond LS. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev. 2002;15:329–341. doi: 10.1128/CMR.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Foda BM, Singh U. Dimethylated H3K27 Is a Repressive Epigenetic Histone Mark in the Protist Entamoeba histolytica and Is Significantly Enriched in Genes Silenced via the RNAi Pathway. J Biol Chem. 2015;290:21114–30. doi: 10.1074/jbc.M115.647263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jeelani G, et al. Metabolic profiling of the protozoan parasite Entamoeba invadens revealed activation of unpredicted pathway during encystation. PLoS One. 2012;7:e37740. doi: 10.1371/journal.pone.0037740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jeelani G, Sato D, Soga T, Watanabe H, Nozaki T. Mass spectrometric analysis of L-cysteine metabolism: physiological role and fate of L-cysteine in the enteric protozoan parasite Entamoeba histolytica. MBio. 2014;5:e01995. doi: 10.1128/mBio.01995-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi Y, et al. Depiction of metabolome changes in histidine starved Escherichia coli by CE-TOFMS. Mol Biosyst. 2008;4:135–147. doi: 10.1039/B714176A. [DOI] [PubMed] [Google Scholar]
- 56.Soga T, Heiger DN. Amino acid analysis by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2000;72:1236–1241. doi: 10.1021/ac990976y. [DOI] [PubMed] [Google Scholar]
- 57.Soga T, et al. Simultaneous determination of anionic intermediates for Bacillus subtilis metabolic pathways by capillary electrophoresis electrospray ionization mass spectrometry. Anal Chem. 2002;74:2233–2239. doi: 10.1021/ac020064n. [DOI] [PubMed] [Google Scholar]
- 58.Soga T, et al. Metabolomic profiling of anionic metabolites by CE-MS. Anal Chem. 2009;81:6165–6174. doi: 10.1021/ac900675k. [DOI] [PubMed] [Google Scholar]
- 59.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. 2006;78:779–787. doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 61.Baran R, et al. MathDAMP: a package for differential analysis of metabolite profiles. BMC Bioinformatics. 2006;7:530. doi: 10.1186/1471-2105-7-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soga T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J Biol Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 63.De Cádiz AE, Jeelani G, Nakada-Tsukui K, Caler E, Nozaki T. Transcriptome analysis of encystation in Entamoeba invadens. PLoS One. 2013;8:e74840. doi: 10.1371/journal.pone.0074840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeelani G, et al. Biochemical and functional characterization of novel NADH kinase in the enteric protozoan parasite Entamoeba histolytica. Biochimie. 2013;95:309–319. doi: 10.1016/j.biochi.2012.09.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.