Abstract
Soil salinization and degradation is one of the consequences of climate change. Identification of major salt tolerance genes and marker assisted selection (MAS) can accelerate wheat breeding for this trait. We genotyped 154 wheat F2 lines derived from a cross between salt tolerant and susceptible cultivars using the Axiom Wheat Breeder’s Genotyping Array. A high-density linkage map of 988 single nucleotide polymorphisms (SNPs) was constructed and utilized for quantitative trait loci (QTL) mapping for salt tolerance traits and mineral concentrations under salinity. Of 49 mapped QTLs, six were for Na+ exclusion (NAX) and two QTLs (qSNAX.2 A.1, qSNAX.2 A.2) on chromosome 2 A coincided with a reported major NAX QTL (Nax1 or HKT1;4). Two other major NAX QTLs were mapped on 7 A, which contributed 11.23 and 18.79% of the salt tolerance respectively. In addition to Ca+2 and Mg+2 QTLs, twenty-seven QTLs for tissue Phosphorus, Zinc, Iron, Manganese, Copper, Sulphur and Boron concentrations under salinity were also mapped. The 1293 segregating SNPs were annotated/located within genes for various ion channels, signalling pathways, transcription factors (TFs), metabolic pathways and 258 of them showed differential expression in silico under salinity. These findings will create new opportunities for salt tolerance breeding programs.
Subject terms: Genetic linkage study, Plant genetics
Introduction
The effects of climate change are predicted to reduce the cultivated land area of the world by 2–9%1. This land loss or soil degradation is feared to be increased by soil salinization, which is caused both by natural processes and human activities such as saline irrigation and land clearing1,2. More than 800 million hectares of land including 20% of irrigated area worldwide is affected3,4. Soil salinity significantly reduces wheat growth and development at the seedling stage, resulting in lower grain yield as higher Na+ influx causes toxicity and disrupts leaf function5. Conversely, 100–110% extra food will be required by 2050 to feed the growing world population6. Therefore, utilization of saline soils through development of salt tolerant and/or climate resilient wheat is important for meeting increasing food demand.
However, despite a major focus on drought, comparatively little work has been performed on breeding wheat for salt tolerance7. Development of salt tolerant cultivars is hindered greatly by the complexity and severity of salt stress, which occurs in two phases, i.e. osmotic stress and ionic stress2. Osmotic stress, resulting from higher salt concentrations outside the root, inhibits water uptake, cell expansion and development7. Subsequently, high Na+ ion uptake into leaves promotes leaf chlorosis, necrosis and mortality due to reduced photosynthesis2,7. Wheat yield data collected from field experiments cannot easily be used as a salt tolerance index due to the range of interactions between variable Na+ in soil profile, differential salt responses depending on genotype, growth stage, and other factors such as high pH and drought7. However, hydroponics/pot screening performed in greenhouse conditions and physiological studies have indicated that wheat has significant genetic variation for salt tolerance4,8,9 which can be exploited for wheat breeding and genetics. There are few works has been directed to exploring the physiological and genetic complexity of multi-genic and multi-faceted salinity related traits.
Recent development in genomic knowledge and technology has provided new horizons and foundations for genetic improvement of complex traits such as drought and salt tolerance. The combination of genomic tools with MAS can be used to identify and select the preferred genes in breeding populations at a much faster rate than by classical breeding10–15. Until recently, progress in MAS for wheat was slowed by the limited availability of genomic data, but advances in genotyping techniques and DNA sequencing have now produced genome datasets that have been used to design sequence-based simple sequence repeats (SSRs) and SNP markers16–18. Among the various marker types, SNPs are increasingly used for germplasm characterization and gene mapping as they provide cost-effective, rapid and high-throughput genotyping18. As SNPs are co-dominant, sequence tagged and highly abundant, they are suitable for the dissection of complex traits using highly multiplexed marker microarrays such as the Affymetrix GeneChip19. For example, the Axiom Wheat Breeders’ Genotyping array is a recently developed, highly efficient system for screening large wheat populations. It contains 35,143 pre-validated SNPs which cover all 21 wheat chromosomes and is capable of genotyping 384 wheat samples simultaneously, making it a cost-effective high-throughput genotyping method. Recently, it has also been used for construction of a high-density linkage map and subsequent identification of genomic regions for drought-related traits in durum wheat20.
Genotipic data from multiplexed marker assays is used to construct high-density linkage maps, which is a prerequisite to mapping the QTLs for agronomically important traits21–24. High-density linkage maps also provide a genomic resource for positional cloning of important genes. Due to their construction from sequenced-tagged markers, they can also be used for comparative genomics to dissect chromosomal organization and evolution21. Markers in the linkage map provide tags for regions containing QTLs of targeted traits, and several QTLs for salt tolerance have previously been mapped in wheat23,25,26. Genc et al.25 mapped 40 QTLs for seven seedling traits including chlorophyll content, Na+ and K+ concentrations in shoot, and seedling biomass under salinity. A NAX QTL interval i.e. wPt-3114-wmc170, mapped on Chromosome 2A, was associated with 10% increase in seedling biomass. Two of the five QTLs for NAX were co-located with seedling biomass QTLs, however, all five QTLs contributed only 18% of the seedling biomass phenotypic variation. Therefore, a need remains for QTL mapping for salt tolerance in more populations to identify other major/novel QTLs.
It was suggested that in addition to NAX and K+, several other factors might be involved in wheat salt tolerance. For example, it has been reported that Mg2+ and Ca2+ accumulation also influence salt tolerance and several QTLs for Mg2+ and Ca2+ concentrations under salinity were identified in wheat27. Apart from K+, Mg2+ and Ca2+, genetic bases of other minerals such as P, Zn, Fe, Mn, Cu, S and Boron has been investigated under different water regimes, but not under salinity24. Therefore, we aimed to identify the genetic basis of accumulation of these minerals under salt stress. Additionally, we also report the functional annotation of linkage-mapped SNPs for an F2 population de for salt tolerance.
The objectives of the present study were: (a) construction of a high-density SNP linkage map for an F2 population with phenotypic variation for salt tolerance, (b) identification of QTLs and markers linked with various micronutrient concentrations and salt tolerance related seedling traits, and (c) functional annotation of segregating markers in the F2 population, and (d) validation of annotated SNPs/genes by differential expression analysis.
Materials and Methods
Plant material
The F2 population was developed from hybrids of bread wheat cultivars, with contrasting salt tolerance (WTSD91 and WN-64). WTSD91 and WN-64 were found to be moderately salt tolerant and highly susceptible respectively, under 300 mM Sodium Chloride (NaCl) treatment in hydroponics screening4. Crossed seeds were grown during 2012–13 to generate F1 hybrids. To ensure purity, heads were covered with paper bags during anthesis, and seeds for F2 lines were obtained.
Growth conditions and phenotyping
The experiment was conducted in Venlo-type greenhouse located at 40°53′25″N, 29°22′47″E in Istanbul, equipped with computerized climate control for supplemental lighting, evaporative cooling and heating. The day and night temperatures were kept at 25 ± 4 °C and 20 ± 4 °C during the experiment period. 250 F2 lines were germinated in perlite for 5 days and 180 seedlings were then transplanted into 2.7-L pots containing aerated nutrient solution28 after removing their residual endosperm. On the following day, 75 mM NaCl was added to the nutrient solution. The solution was replaced every four days and the salinity level increased successively to 150, 225 and 300 mM NaCl on the 4th, 8th and 12th day after transplantation. Plants remained under salinity for a total of 32 days including 20 days at 300 mM NaCl. Plants were then divided into four groups based on their phenotype: (i) tolerant (T) group had 5 fully extended green leaves without any signs of salt injury; (ii) moderately tolerant (MT) group had 4–5 fully extended green leaves with minor salt injury signs on leaf tips; (iii) susceptible (S) group had 2–3 leaves showing severe salt injury signs and 1–2 dead leaves; and (iv) highly susceptible (HS) group with 2–3 leaves having severe injury signs and death of 60–80% of leaves. Four representative plants from each group were selected for mineral analysis.
After three washings in dH2O, shoots and roots were dried at 65 °C for 3 days, dry root weight (DRW) and dry shoot weights (DSW) were recorded and tissues analyzed for mineral concentrations as previously described28. Dry roots and shoots were ground to powder in an agate vibrating cup mill (Pulverisette 9; Fritsch GmbH; Germany) and ~0.15–0.2 g powder from each sample was digested in 5 ml of 65% HNO3 and 2 ml of 30% H2O2 using closed-vessel microwave system (Mars Express; CEM Corp; NC, USA). Digested solutions were diluted with milli-Q water to 20 ml final volume for measurement of Ca, Cu, Fe, K, Mg, Mn, P, S, Zn and Boron concentrations by inductively coupled plasma optical emission spectrometry (ICP-OES; Vista-Pro Axial; Varian Pty Ltd; Mulgrave, Australia)29. The mineral concentration data was checked against standard values for standard reference material (SRM 8436 Durum Wheat Flour, NIST, Gaithersburg, MD). For measuring Na+ concentration, solutions were further diluted 1:50. The data for 22 traits was obtained by multiplying ICP-OES values by the dilution factor and dividing the result by the DRW or DSW used for digestion. As reduction in the concentration of Na+ means higher NAX so Na+ values were multiplied by −1 to get root Na exclusion (RNAX) and shoot Na exclusion (SNAX) values. Correlation coefficients among the phenotypic data were calculated using Statistix 8.1 program.
DNA extraction and genotyping
DNA extraction of 164 F2 lines and the two parents was carried out using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA). DNA concentrations were measured with the Quant-iT PicoGreen dsDNA Assay Kit (ThermoFisher Scientific, Waltham, MA, USA) and 1.5 µg of gDNA from each line was dissolved in 10 mM Tris-HCl pH 8.0 to a final volume of 30 µl. Genotyping was performed using the Axiom Wheat Breeder’s Genotyping Array (Affymetrix, Santa Clara, CA, USA) with 35,143 SNPs for each sample (hereafter referred to as the wheat 35 K array). Genotyping was done at the Bristol Genomics Facility (Bristol University, UK) utilizing the Affymetrix GeneTitan MT system following the Affymetrix procedure (Axiom 2.0 Assay Manual). We used Axiom Analysis Suite 1.1.0.616 software for SNP calling by following the Axiom Best Practices Genotyping Workflow with default wheat SNP call rate cut-off = 97%, QC call rate cut-off = 92% and DQC cut-off = 0.82. However, 10 lines failed the QC and DQC cut-offs; therefore, SNP call codes for the remaining 154 lines were used for downstream analysis. This software uses cluster separation, call rate and deviation from expected cluster positions and classifies the SNPs into six performance categories19.
Construction of SNP linkage map
The markers with significant segregation distortion (P < 0.05) were removed through chi-square test combined with sequential Bonferroni correction30. A genetic linkage map was constructed using MapDisto 2.0 b9322. In MapDisto, the markers were grouped with a recombination fraction of 0.3 and logarithm of the odds ratio (LOD) score of six using the Kosambi mapping function. The Seriation algorithm was used for ordering the linkage groups. Linkage groups were assigned to chromosomes by comparing shared markers with a recent wheat consensus linkage map19. Chromosomes were found to be divided into multiple linkage groups, so these linkage groups were combined and re-ordered. Rippling of marker order with a window size of five markers and checking for inversions were performed to improve the marker order and produce the shortest map of each chromosome.
QTL mapping
Additive QTLs for all 22 traits were mapped by the composite interval mapping (CIM) method using single-treatment phenotypic data and QTL IciMapping V4.1.0 with 1-cM walking speed and LOD thresholds of 2.531. The linkage maps and QTLs were drawn using MapChart 2.30 software32. The contribution of single QTL in phenotypic variation of mineral concentrations and salt tolerance was calculated by following Zhang et al.33. The DRW and DSW are considered to be the direct measure of salt tolerance25,34 so the contribution of QTLs to salt tolerance was calculated by using DRW and DSW data.
Functional annotation of segregating SNPs
The flanking sequences of 3381 ‘Poly High Resolution’ (PHR) or polymorphic SNPs were mapped to gene coding sequences (CDS) from the International Wheat Genome Sequencing Consortium (IWGSC) database35 using BLAST+ 2.2.30. From 1448 initial hits, basic local alignment search tool (BLAST) alignments with less than 60 bp length and/or 95% identity were removed. In this way, 1323 SNPs were found to be located in 1257 IWGSC CDS which were functionally annotated by using Blast2GO V4.036. Functional annotation was done by using NCBI blast, mapping and annotation commands of Blast2Go using default parameters, and annotations with E-value ≥1 × 10−30 were discarded.
In-silico expression analysis of annotated genes
For the purpose, above mentioned 1257 IWGS CDS were aligned with transcriptome reads expressed under normal and saline conditions3 by using BLASTN 2.6.1+. Significant alignments with >200 alignment scores were counted and salt/normal alignment counts ratio was divided by 3.25 (347,200/106,600 spots for salt/control) to get the expression value. Genes with at least a 2-fold change in expression up or down were considered to be differentially regulated.
Results
SNP calling yielded six categories of SNPs
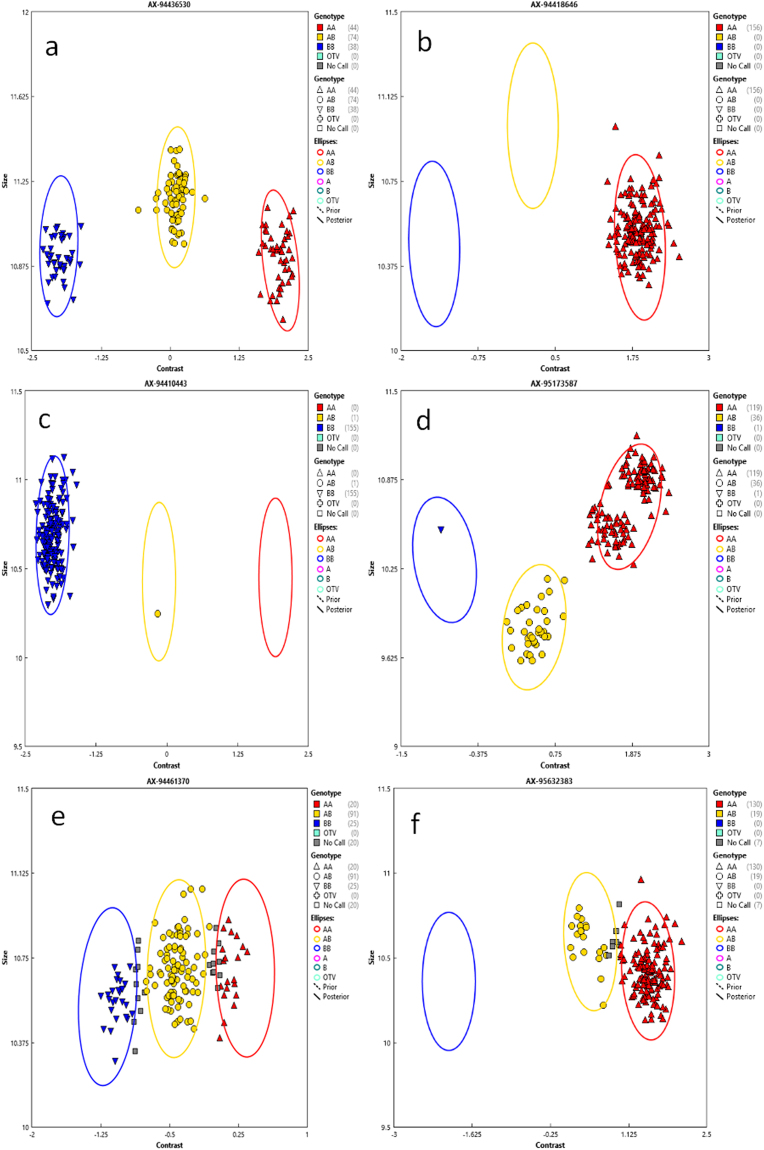
From 164 F2 lines analysed on the wheat 35 K array, 154 gave good quality data and were used for SNP clustering. The SNPs were categorized into six groups: (i) PHR SNPs, which were polymorphic and co-dominant with a minimum of two samples containing the minor allele; (ii) Mono High Resolution (MHR) or monomorphic SNPs had only one cluster/allele; (iii) No Minor Homozygote (NMH), these polymorphic and dominant SNPs had only two clusters, one being the heterozygote; (iv) Off-Target Variants (OTV) had four clusters including one for a null allele; (v) Call Rate Below Threshold (CRBT) had all cluster properties above the threshold except for the call rate cut-off; and (vi) Other type SNPs, which had one or more cluster properties below quality thresholds (Fig. 1). Out of 35,143 array SNPs, 46.1% or 16,210 were MHR/monomorphic and 51 (0.15%) SNPs were OTVs. Meanwhile 3,381 (9.6%) SNPs were PHR/polymorphic (Table 1) whose call codes were used for linkage map construction.
Figure 1.
Representative of six SNPs calling categories: (a) Poly High Resolution; (b) Mono High Resolution; (c) No Minor Homozygote; (d) Off-Target Variants (e) Call Rate Below Threshold; and (f) Other.
Table 1.
SNP calling distribution for 154 bread wheat F2 lines identified using thewheat 35 K Array.
| SNPs calling categories | No. of Markers | Percent SNPs calling (%) |
|---|---|---|
| Mono High Resolution | 16210 | 46.1 |
| Poly High Resolution | 3381 | 9.6 |
| Other | 8141 | 23.2 |
| No Minor Homozygote | 3017 | 8.6 |
| Call Rate Below Threshold | 4343 | 12.35 |
| OTV | 51 | 0.15 |
| Total | 35143 | 100 |
Whole genome wheat genetic linkage map
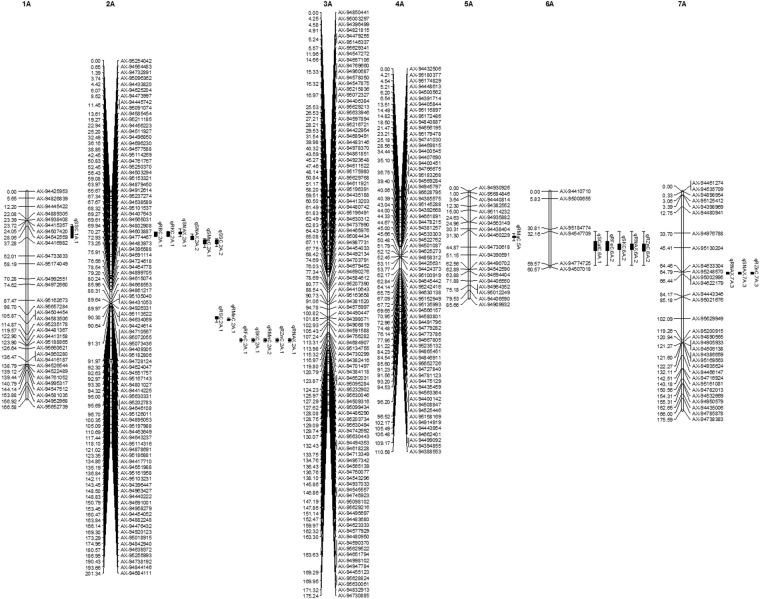
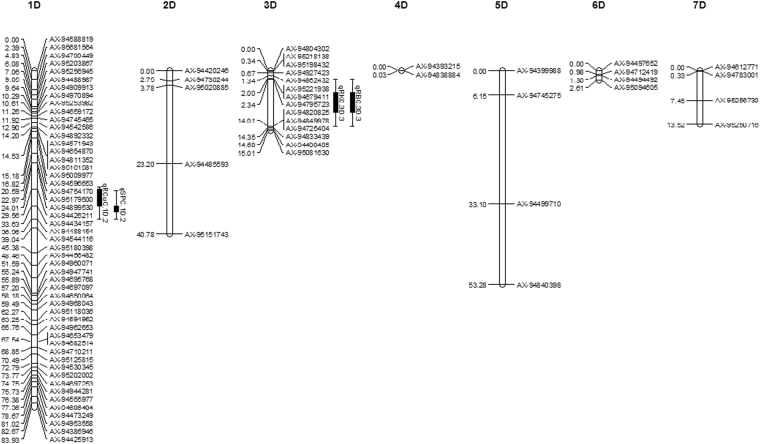
Out of 3,381 PHR markers, 1072 passed the chi-square test for segregation distortion and were used to construct the linkage map. We assigned 988 markers to 21 chromosomes and 84 markers were unlinked. The B genome had the highest number of markers assigned (562) followed by the A (342) and D (84) genomes. 183 markers were assigned to chromosome 1B while the fewest in the B genome (31) were assigned to 4B. For the A genome, maximum (100) and minimum (6) markers were assigned to chromosomes 3A and 6A respectively. In the D genome, chromosome 1D had 51 linked markers while 4D had only 2. The total length of the whole genome map was 2317.88 cM consisting of 975.56, 1133.16 and 209.16 cM for A, B and D genomes respectively. The maximum length was 221.26 cM for chromosome 2B and minimum was 0.03 cM for chromosome 4D. Average length per marker was 3.71, 4.43, 2.68 and 4.03 cM for whole genome, A, B and D genome (Table 2; Figs 2, 3 and 4).
Table 2.
Distribution of 988 assigned markers to 21 wheat linkage groups and chromosomes lengths for an F2 population of 154 lines.
| Chromosome | No. of Markers | Chromosome length (cM) | Length/ marker (cM) | Consensus map lengths (cM)19 |
|---|---|---|---|---|
| 1A | 32 | 166.58 | 5.21 | 182.07 |
| 2A | 88 | 201.34 | 2.29 | 203.99 |
| 3A | 100 | 175.24 | 1.75 | 136.11 |
| 4A | 66 | 110.58 | 1.48 | 75.68 |
| 5A | 19 | 85.66 | 4.51 | 221.39 |
| 6A | 6 | 60.57 | 10.1 | 189.4 |
| 7A | 31 | 175.59 | 5.66 | 231.64 |
| A Genome | 342 | 975.56 | 4.43 | 1240.28 |
| 1B | 183 | 173.30 | 0.95 | 182.35 |
| 2B | 151 | 221.26 | 1.47 | 216.96 |
| 3B | 59 | 187.64 | 3.18 | 234.56 |
| 4B | 31 | 100.04 | 3.23 | 76.67 |
| 5B | 66 | 201.77 | 3.06 | 208.75 |
| 6B | 33 | 101.58 | 3.08 | 165.99 |
| 7B | 39 | 147.57 | 3.78 | 279.28 |
| B Genome | 562 | 1133.16 | 2.68 | 1364.56 |
| 1D | 51 | 83.93 | 1.65 | 151.29 |
| 2D | 5 | 40.78 | 8.16 | 177.47 |
| 3D | 14 | 15.01 | 1.07 | 234.87 |
| 4D | 2 | 0.03 | 0.014 | 162.07 |
| 5D | 4 | 53.28 | 13.32 | 167.57 |
| 6D | 4 | 2.61 | 0.65 | 167.78 |
| 7D | 4 | 13.52 | 3.38 | 73.34 |
| D Genome | 84 | 209.16 | 4.03 | 1134.39 |
| Total | 988 | 2317.88 | 3.71 | 3739.23 |
Figure 2.
Additive QTLs mapped on A genome for salt tolerance traits and mineral concentrations under salt stress in wheat F2 population.
Figure 3.
Additive QTLs mapped on B genome for salt tolerance traits and mineral concentrations under salt stress in wheat F2 population.
Figure 4.
Additive QTLs mapped on D genome for salt tolerance traits and mineral concentrations under salt stress in wheat F2 population.
Comparison of linkage map with consensus map
By comparing our map with a recently published consensus map using these markers19, it was found that a total of 511 of 988 (51.7%) markers were assigned to the same chromosome as consensus map and 398 of 988 or 40.28% of the SNPs were mapped for the first time. The number of markers newly assigned to the A, B and D genomes was 132, 247 and 19 respectively. The maximum of these markers in the A genome i.e. 45 were located on chromosome 2A while 1B harboured 153 of these markers (Table S1). Remaining 79 markers were assigned to different chromosomes from the consensus map. The biggest such group consisted of 32 markers assigned to chromosome 2B in our map. While 17 such markers were assigned to chromosome 2A and 29 of total 79 SNPs were assigned to their respective homoeologous chromosomes (Table S2).
QTL mapping for salt tolerance and mineral nutrient concentrations
The phenotypic data for 22 traits and correlation coefficients among different traits are presented in Table S3 and S4 respectively. We identified 49 additive QTLs for 20 single-treatment phenotypic traits on 12 wheat chromosomes. These QTLs were mapped on two chromosomes of the D genome and 5 chromosomes each from the A and B genomes (Table 3; Figs 2, 3 and 4). Among the six QTLs detected for RNAX and SNAX, the QTL on 7A (qRNAX.7 A.3 and qSNAX.7A.3) contributed 13.69 and 15.35% of phenotypic variation of NAX from root and shoot. They contributed 11.23 and 19.79% of DRW and DSW (salt tolerance) respectively. Other RNAX and SNAX QTLs were located on 2A and 6A. Shoot K+ concentration (SKC) and root K+ concentration (RKC) QTLs were mapped on 2A, 6A, 4B and 3D, and the most important SKC QTL on 6A contributed 7.46 and 9.76% in K concentration and salt tolerance. Among the four root Zinc Conc. (RZnC) and shoot Zinc Conc. (RZnC) QTLs, RZnC QTL on 7A contributed 12.08 and 11.23% of Zn phenotype and salt tolerance respectively. For root Ca2+ concentration (RCalC) and shoot Ca2+ concentration (SCalC), QTLs qRCalC.6B.3 and qSCalC.6B.2 presented 10.91 and 6.52% of Ca conc. in wheat root and shoot. These also contributed 5.92 and 11.87% of salt tolerance. Among Mg2+ concentration (RMgC and SMgC) QTLs, 2A QTL contributed maximum 6.37% of phenotypic variation of SMgC and qSMgC.6B.2 contributed maximum 8.36% to salt tolerance (DSW).
Table 3.
Additive QTLs mapped on 12 chromosomes for various mineral concentrations under 300 mM NaCl salt condition.
| Trait | QTL | Marker Interval | Position (cM) | LOD | PQCMC | PQCST |
|---|---|---|---|---|---|---|
| RBC | qRBC.2A.1 | AX-94496850–AX-94696230 | 32.49–36.16 | 3.59 | 1.29 | 0.14 |
| qRBC.2B.2 | AX-94909085–AX-95120904 | 95.80–100.2 | 2.50 | 0.16 | 0.10 | |
| qRBC.3D.3 | AX-94795723–AX-94820825 | 2.34–14.01 | 5.68 | 2.29 | 0.21 | |
| RCalC | qRCalC.3B.1 | AX-95232967–AX-94969522 | 37.39–45.65 | 2.86 | 5.27 | 0.68 |
| qRCalC.3B.2 | AX-94457592–AX-94518159 | 169.44–180.24 | 2.84 | 5.38 | 0.98 | |
| qRCalC.6B.3 | AX-94668676–AX-94883829 | 51.44–57.07 | 7.09 | 10.91 | 5.92 | |
| RCuC | qRCuC.7B.1 | AX-94409804–AX-94566622 | 20.46–24.15 | 2.75 | 2.83 | 0.08 |
| qRCuC.1D.2 | AX-94426211–AX-94434157 | 29.56–33.63 | 2.53 | 6.06 | 0.39 | |
| RFeC | qRFeC.2A.1 | AX-95114316–AX-94878691 | 118.10–121.02 | 2.51 | 3.98 | 3.81 |
| qRFeC.6A.2 | AX-94547709–AX-94774725 | 32.16–59.57 | 26.7 | 12.96 | 3.15 | |
| qRFeC.6B.3 | AX-94668676–AX-94883829 | 51.44–57.07 | 18.10 | 8.73 | 5.92 | |
| RKC | qRKC.2A.1 | AX-94496850–AX-94696230 | 32.49–36.16 | 3.70 | 4.79 | 0.14 |
| qRKC.4B.2 | AX-95103748–AX-94957045 | 70.35–71.34 | 21.4 | 11.31 | 1.40 | |
| qRKC.3D.3 | AX-94795723–AX-94820825 | 2.34–14.01 | 4.01 | 7.96 | 0.21 | |
| RMgC | qRMgC.5 A | AX-94460229–AX-94730618 | 31.30–44.87 | 6.20 | 4.96 | 5.58 |
| RMnC | qRMnC.2A.1 | AX-94895053–AX-95197988 | 100.35–105.09 | 5.96 | 8.13 | 0.37 |
| qRMnC.2 A.2 | AX-95114316–AX-94878691 | 118.10–121.02 | 3.01 | 5.17 | 3.81 | |
| qRMnC.6B.3 | AX-94668676–AX-94883829 | 51.44–57.07 | 19.20 | 14.16 | 5.92 | |
| RNAX | qRNAX.2A.1 | AX-95114316–AX-94878691 | 118.10–121.02 | 2.53 | 4.85 | 3.81 |
| qRNAX.6A.2 | AX-94547709–AX-94774725 | 32.16–59.57 | 9.35 | 6.46 | 3.15 | |
| qRNAX.7 A.3 | AX-95248570–AX-95002995 | 64.79–66.44 | 2.51 | 13.69 | 11.23 | |
| RPC | qRPC.7B | AX-94409804–AX-94566622 | 20.46–24.15 | 2.59 | 2.73 | 0.08 |
| RSC | qRSC.2A.1 | AX-94895053–AX-95197988 | 100.35–105.09 | 3.50 | 5.65 | 0.37 |
| qRSC.3B.2 | AX-95232967–AX-94969522 | 37.39–45.65 | 2.96 | 4.61 | 0.68 | |
| qRSC.3B.3 | AX-94457592–AX-94518159 | 169.44–180.24 | 2.92 | 6.42 | 0.98 | |
| qRSC.6B.4 | AX-94668676–AX-94883829 | 51.44–57.07 | 16.18 | 10.04 | 5.92 | |
| qRSC.7B.5 | AX-94538131–AX-94660701 | 144.46–147.7 | 2.52 | 2.89 | 1.97 | |
| RZnC | qRZnC.2A.1 | AX-95114316–AX-94878691 | 118.10–121.02 | 2.83 | 5.25 | 3.81 |
| qRZnC.6A.2 | AX-94547709–AX-94774725 | 32.16–59.57 | 11.22 | 7.45 | 3.15 | |
| qRZnC.7 A.3 | AX-95248570–AX-95002995 | 64.79–66.44 | 2.52 | 12.08 | 11.23 | |
| SBC | qSBC.3B.1 | AX-94402393–AX-95232967 | 34.72–37.39 | 2.70 | 5.86 | 0.59 |
| qSBC.3B.2 | AX-94811682–AX-94445993 | 163.83–164.82 | 2.63 | 5.53 | 0.46 | |
| qSBC.3B.3 | AX-94457592–AX-94518159 | 169.44–180.24 | 2.82 | 4.91 | 1.01 | |
| SCalC | qSCalC.6A.1 | AX-94547709–AX-94774725 | 32.16–59.57 | 29.1 | 8.98 | 3.08 |
| qSCalC.6B.2 | AX-94668676–AX-94883829 | 51.44–57.07 | 11.41 | 6.52 | 11.87 | |
| SKC | qSKC.2A.1 | AX-95114316–AX-94878691 | 118.10–121.02 | 2.51 | 4.34 | 5.18 |
| qSKC.6A.2 | AX-94547709–AX-94774725 | 32.16–59.57 | 9.15 | 7.46 | 9.76 | |
| SMgC | qSMgC.2A.1 | AX-94577588–AX-95114269 | 38.86–42.45 | 2.79 | 6.37 | 1.23 |
| qSMgC.6B.2 | AX-94668676–AX-94883829 | 51.44–57.07 | 2.58 | 5.90 | 8.36 | |
| SMnC | qSMnC.4B | AX-94842084–AX-94446850 | 29.71–30.05 | 4.75 | 3.12 | 1.03 |
| SNAX | qSNAX.2A.1 | AX-94496850–AX-94696230 | 32.49–36.16 | 2.89 | 5.14 | 0.95 |
| qSNAX.2A.2 | AX-94696230–AX-94577588 | 36.16–38.86 | 3.10 | 7.10 | 1.45 | |
| qSNAX.7 A.3 | AX-95248570–AX-95002995 | 64.79–66.44 | 2.92 | 15.35 | 18.79 | |
| SPC | qSPC.4B.1 | AX-94699353–AX-94987788 | 12.80–16.52 | 3.38 | 2.10 | 1.62 |
| qSPC.1D.2 | AX-94434157–AX-94488154 | 33.63–3696 | 2.58 | 4.73 | 0.20 | |
| SSC | qSSC.1 A.1 | AX-94542559–AX-94416982 | 28.06–37.28 | 2.55 | 1.38 | 0.96 |
| qSSC.2A.2 | AX-94577588–AX-95114269 | 38.86–42.45 | 4.13 | 2.13 | 0.91 | |
| qSSC.4B.3 | AX-94957045–AX-95257129 | 71.34–72.34 | 4.55 | 2.46 | 0.01 | |
| SZnC | qSZnC.7B | AX-94409804–AX-94566622 | 20.46–24.15 | 2.78 | 3.28 | 0.13 |
QTL: quantitative trait loci, LOD: logarithm of the odds ratio; PQCMC: percent QTL contribution for mineral concentration; PQCST: percent QTL contribution for salt tolerance.
Although no QTL was detected for shoot Cu and Fe conc. (SCuC and SFeC), a root Cu conc. (RCuC) QTL on 1D and root Fe Conc. (RFeC) QTL on 6A contributed 6.06% and 12.96% of Cu and Fe phenotypic variation. Another RFeC QTL on 6B contributed 5.92% to salt tolerance. In the same region, a QTL for Mn contributed 14.16% of Mn phenotypic variation and 5.92% of salt tolerance. Although the QTLs for Boron, P and S contributed as much as 10.04% to mineral concentration variation, they made little contribution to salt tolerance (Table 3). We found five QTL clusters, on chromosomes 2A, 6A, 7A, 3B and 6B (Figs 2 and 3).
Candidate gene identification for segregating SNPs
Out of 3381 polymorphic SNPs, 1306 were functionally annotated based on 20 BLAST alignments for each SNP; the best/top BLAST alignments for these SNPs were associated with sequences from 22 species. With 480 homologous sequences, Aegilops tauschii had the maximum number of BLAST hits, whereas only 1 BLAST hit was retrieved for nine species. While 290, 280, 136 and 77 coding sequences containing SNPs were homologous to sequences from Triticum urartu, Hordeum vulgare, T. aestivum and Brachypodium distachyon, respectively (Table 4).
Table 4.
Top BLAST hit distribution of 1306 wheat sequences/SNPs in 22 different species by Blast2GO alignment.
| Species | BLAST top hits | Species | BLAST top hits |
|---|---|---|---|
| Aegilops tauschii | 480 | O. brachyantha | 3 |
| Triticum urartu | 290 | Zea mays | 2 |
| Hordeum vulgare | 280 | B. sylvaticum | 1 |
| T. aestivum | 136 | Gossypium hirsutum | 1 |
| Brachypodium distachyon | 77 | Phyllostachys edulis | 1 |
| T. durum | 9 | P. praecox | 1 |
| Sorghum bicolor | 6 | Secale cereale | 1 |
| Oryza sativa Japonica | 5 | Zootermopsis nevadensis | 1 |
| Setaria italic | 3 | Agropyron mongolicum | 1 |
| T. monococcum | 3 | Avena longiglumis | 1 |
| Dichanthelium oligosanthes | 3 | A. sativa | 1 |
| Total Species | 22 | Total Annotations | 1306 |
Annotated sequences carrying polymorphic SNPs varied widely in function. For instance, 44 SNPs were annotated to ion transporters/channels linked to wheat salt tolerance, including seven potassium, two Chloride, four calcium, five zinc, four magnesium, two sulfate, three nitrate transporters/channels along with one proton pump, four proton transporters, cationic, anionic and metal channels (Table 5, Table S5). Another 50 SNPs were annotated for protein, glucose, malate, drug, fatty acid and proline transport along with endocytosis/exocytosis (Table S5). Importantly, 92 SNPs were located on genes involved in Auxin, ABA and ethylene-activated and Jasmonic acid, gibberellic acid and sugar mediated signaling pathways; G-protein, apoptotic, cell surface receptor/Wnt signaling, signal recognition and transduction (Table S5). A further 63 SNPs were associated with genes from 35 different classes of TFs (Table S6). In total, 166 SNPs were located on genes with enzymatic activity (ligases, transferases, hydrolases), particularly kinases, which carried 77 SNPs. Some of these SNPs were closely related to post-translational modifications, such as phosphorylation and glycosylation, and to protein localization (Table S6). Genes involved in core cellular machineries, such as transcription, translation and replication, carried 198 SNPs (Table S7). Fifty-one SNPs were found within genes with Zn, Ca, Mg, Fe and metal ion binding activities, and 111 SNPs were associated with structural molecules, and thus, were part of different cellular organelles (Table S8). The highest number of SNPs (275) were confined to genes of various metabolic processes including photosynthesis, carbohydrate metabolism, oxidation-reduction, the Krebs cycle/respiration, protein catabolism, antioxidant activity, sugar, protein and lipid biosynthesis (Table S9).
Table 5.
List of 44 ion transporters/channels annotated to polymorphic SNPs in a segregating wheat population developed for salt tolerance.
| SNP ID | IWGSC Sequence Hit | Blast Top Hit Spp. | Annotated Channel/Transporter | Transports |
|---|---|---|---|---|
| AX-94778362 | lcl|Traes_4BL_5A58CACB2.1 | T. durum | HKT transporter | K |
| AX-95224228 | lcl|Traes_6BS_E420DDD6D.1 | H. vulgare | K+-H+ exchange | K |
| AX-94995317 | lcl|Traes_1DS_07F02E427.1 | A. tauschii | K transporter 12 | K |
| AX-94388980 | lcl|Traes_5DS_49CF8A4C4.1 | H. vulgare | K (+) efflux antiporter | K |
| AX-95215612 | lcl|Traes_XX_D8915FD17.1 | A. tauschii | Voltage-gated K channel | K |
| AX-95654644 | lcl|Traes_1BL_611DF0433.1 | H. vulgare | Jacalin-related lectin 3 | K |
| AX-94699167 | lcl|Traes_5AL_51E31BF07.1 | T. urartu | Out-rectifying K channel | K |
| AX-94484138 | lcl|Traes_3B_81DB429AC.1 | T. urartu | Chloride channel CLC-e | Cl |
| AX-94546397 | lcl|Traes_2DL_A591AC867.1 | A. tauschii | Chloride channel CLC-g | Cl |
| AX-95126745 | lcl|Traes_4BL_E0ABA8471.1 | A. tauschii | Cation Ca exchanger 4 | Ca |
| AX-95069958 | lcl|Traes_1AS_429D67C42.1 | A. tauschii | Ca stress-gated channel 1 | Ca |
| AX-94849975 | lcl|Traes_7BL_13D715DB0.1 | T. urartu | Ca homeostasis ER | Ca |
| AX-94662401 | lcl|Traes_4BS_06DC8C269.1 | A. tauschii | Ca-transporting ATPase | Ca |
| AX-94635693 | lcl|Traes_XX_0593C741B.1 | H. vulgare | Zn transporter 6 | Zn |
| AX-94414919 | lcl|Traes_1DS_C2EFEFBB9.1 | T. aestivum | Zn transporter 7 | Zn |
| AX-95172326 | lcl|Traes_1DS_D28FA6FF2.1 | A. tauschii | Zn transporter At3g08650 | Zn |
| AX-94495517 | lcl|Traes_XX_79F99051D.1 | H. vulgare | Metal tolerance C2 | Zn |
| AX-95634832 | lcl|Traes_1AS_0B179C27B.1 | T. urartu | IQM1 | Zn |
| AX-94755145 | lcl|Traes_2DL_5C445EE47.1 | H. vulgare | Mg transporter NIPA3 | Mg |
| AX-95159756 | lcl|Traes_2AL_065DBAB56.1 | H. vulgare | Mg transporter NIPA4 | Mg |
| AX-94692118 | lcl|Traes_2BL_D5156A4A5.1 | H. vulgare | Mg transporter NIPA4 | Mg |
| AX-94624155 | lcl|Traes_4AL_7541D0C33.1 | T. urartu | ER membrane body 2-X2 | Mg, Fe |
| AX-95142803 | lcl|Traes_2DL_CCAE7B431.1 | A. tauschii | Sulfate transporter | Sulfate |
| AX-94518655 | lcl|Traes_3AL_224FB10D3.1 | T. aestivum | Sulfate transporter | Sulfate |
| AX-94852973 | lcl|Traes_XX_7D456E213.1 | T. urartu | Nitrate transporter | Nitrate |
| AX-94991110 | lcl|Traes_2BL_0E87D8729.1 | A. tauschii | NRT1 PTR FAMILY | Nitrate |
| AX-95216700 | lcl|Traes_2BS_88803DFE6.1 | A. tauschii | NRT1 PTR FAMILY | Nitrate |
| AX-94863332 | lcl|Traes_3B_91715BB56.1 | A. tauschii | Anion transporter 7 | Anions |
| AX-94775993 | lcl|Traes_2BL_007AADDF1.1 | A. tauschii | G-3-Phosphate transporter1 | Anions |
| AX-94550729 | lcl|Traes_4BL_32F50466D.1 | T. urartu | Mo-anion transporter | Anions |
| AX-94583481 | lcl|Traes_3DS_50B54D1FC.1 | A. tauschii | WPP domain-associated | Cations |
| AX-94752371 | lcl|Traes_2DL_51FF05F66.1 | A. tauschii | Cu-transporting HMA5 | Copper |
| AX-94936984 | lcl|Traes_7DS_D439AB891.1 | H. vulgare | Pyrophosphate H+ pump | Proton Pump |
| AX-94384299 | lcl|Traes_XX_796D903AA.1 | H. vulgare | ATP synthase Mitochon. | Proton |
| AX-95118708 | lcl|Traes_7DL_41A6D7A34.1 | A. tauschii | Cytochrome-c oxidase | Proton |
| AX-94486290 | lcl|Traes_3AL_06CDB999D.1 | G. hirsutum | ATP synthase Mitochon. | Proton |
| AX-94982994 | lcl|Traes_2DL_B4C9A5695.1 | B. distachyon | H+-exporting ATPase | Proton |
| AX-94713620 | lcl|Traes_XX_DFFB37624.1 | B. distachyon | Anthranilate BTase 1 | Non-selective |
| AX-94909932 | lcl|Traes_5AS_D7A8B1D1B.1 | H. vulgare | Mechanosensitive channel | Ions |
| AX-95257567 | lcl|Traes_2DL_C065A5C4A.1 | A. tauschii | Solute carrier 22–15 | Metal ions |
| AX-94985111 | lcl|Traes_7AS_705BE4B61.1 | T. urartu | Solute carrier fam 35-F1 | Metal ions |
| AX-94757270 | lcl|Traes_1AL_CEA78C84D.1 | T. urartu | S deficiency-induced 1 | S |
| AX-94560970 | lcl|Traes_1BL_6741F0C8B.1 | T. urartu | S deficiency-induced 1 | S |
| AX-94869513 | lcl|Traes_1DL_36CEA53FD.1 | A. tauschii | S deficiency-induced 1 | S |
Furthermore, 92 SNPs were associated with growth-related processes, including cell division/growth, meristem growth, autophagy, apoptosis, cell wall biogenesis/organization, lateral root development, leaf and shoot morphogenesis, flowering time; and flower, pollen, ovule and embryo sac development, while 48 more were located on genes related to stress responses, including disease resistance, drought, wounding, toxicity and freezing tolerance (Table S10). Finally, 109 SNPs were linked to hypothetical or uncharacterized proteins (Table S10).
In-silico expression analysis of candidate genes
Gene expression was categorised into six groups: up-regulated, expression not conclusive, down-regulated, expressed under salinity only, data not conclusive and no expression with 122, 425, 136, 156, 241 and 209 genes falling into these groups respectively (Table S11).
Discussion
The Wheat 35 K array is suitable for construction of genetic linkage map
We have constructed a high-density SNP linkage map of bread wheat using the wheat 35 K array, which consisted of probes for 35,143 exome-captured SNPs. Of these SNPs, 46.1% or 16,210 were monomorphic in our population and only ‘PHR’ SNPs (3,381 or 9.6%) were used for linkage map construction contrary to the recent paper19 which used ‘NMH’ and ‘OTV’ SNPs as well. This was because we genotyped an F2 population, so only PHR SNPs could be tested for typical F2 segregation. Removal of markers with segregation distortion/bias is essential for getting a good quality linkage map, which was performed by Chi-square test with sequential Bonferroni correction30. From the 1072 SNPs passing the test, 988 were assigned to 21 chromosomes. Only 84 polymorphic markers were assigned to the D genome as compared to 342 and 562 markers assigned to A and B genome. This has been linked to lower nucleotide diversity of the D genome due to its relatively recent evolutionary origin37,38. Our genetic map has a total length of 2317.88 cM in comparison to 3739.23 cM for the published wheat consensus map19, largely due to the lack of diversity/segregation in the D genome under salinity. Of 988 SNPs, 398 SNPs were assigned to chromosomes for the first time, and 79 markers were assigned to different chromosomes from the consensus map. These novel and conflicting markers show genetic diversity originating from genetic differences between European and Pakistani wheat lines. Secondly, we used an F2 population instead of inbred lines, which may have contributed in different segregation patterns. Even so, the large majority (511 of 590) of markers common to both maps were assigned to the same chromosomes19.
Several novel and major QTLs were mapped for minerals and salt tolerance
We mapped 49 QTLs for 22 traits under salt stress on 12 chromosomes, including four QTLs on two chromosomes of the D genome. Salt tolerance in wheat is mainly conferred by NAX or reduced Na+ uptake, as Na+ influx leads to reduce photosynthesis, growth, development and yield5,7,9. The genetic basis of NAX was unknown until the identification of a major NAX QTL on chromosome 2A39; and QTL mapping for salt tolerance has focused on NAX in recent years23,25,26. Among the six mapped RNAX and SNAX QTLs, two closely located QTLs (qSNAX.2A.1 and qSNAX.2A.2) and qRNAX.2A.1 coincided with the major NAX locus Nax1 or HKT1;4 in durum wheat39 and three NAX QTLs found on 2A in bread wheat25. Similarly, the qRNAX.6A.2 QTL has also been reported previously25,26. We mapped two novel and major QTLs (qSNAX.7A.3 and qRNAX.7A.3) on chromosome 7A, which contributed 13.69 and 15.35% of the phenotypic variation of RNAX and SNAX and accounted for 11.23 and 19.79% of the observed salt tolerance (DRW and DSW respectively). The HKT genes are well characterized regulators of K+ and/or Na+ transport in plants and encode proteins that reduce Na+ transport to the shoot, thus conferring salt tolerance5. Accordingly, QTL mapping for K+ conc. under salinity has also been performed by some groups23,25–27. We identified a major SKC QTL on 6A contributing 7.46 and 9.76% of the phenotypic variation in K+ conc. and salt tolerance. Another novel and major QTL (qRKC.4B.2) presented 11.31% of phenotypic variation of RKC. Another RKC QTL mapped on 3D was consistent with a reported QTL25 and the remaining SKC and RKC QTLs were co-located with the SNAX and RNAX QTLs. This co-localization of QTLs is explained by the functional correlation between these traits.
Due to the focus on NAX and K+ QTLs, to our knowledge the genetics of Ca2+ and Mg2+ accumulation under salinity has only been investigatedonce27. We mapped two major QTLs for RCalC (qRCalC.6B.3 and qSCalC.6B.2), which contributed 5.92 and 11.87% of salt tolerance and 10.91 and 6.52% of Ca phenotypic variation. The above mentioned and two RCalC QTLs on 3B coincided with previously reported QTLs27, but a novel QTL on 6A also presented 8.98% of the observed SCalC phenotypic variance. Among Mg2+ (RMgC and SMgC) QTLs, a 2A QTL presented 6.37% of SMgC phenotypic variation and qSMgC.6B.2 contributed 8.36% to salt tolerance. Another novel QTL on 5A accounted for 5.58% of salt tolerance. We also mapped 27 novel QTLs for P, Zn, Fe, Mn, Cu, S and Boron concentrations in wheat root and shoot under salinity for the first time as QTLs for these mineral were previously reported under different water regimes only24. Among them, the most important major QTL for RZnC on 7A contributed 11.23 and 12.08% to salt tolerance and RZnC phenotypic variation. One each of the FeC and MnC QTLs, found on 6B, presented 5.92% of salt tolerance and the Mn QTL contributed 14.16% to Mn phenotypic variation. Similarly, a major qRFeC.6A.2 contributed 12.96% of Fe phenotypic variation. Despite the low contribution of Boron, P and S QTLs to salt tolerance, their contribution to mineral phenotypic variation under salinity was high, which could be useful information for future wheat breeding.
We found five QTL clusters on the A and B genome containing QTLs for several minerals. Such clusters are expected as changes in cellular Na+ could affect concentrations of many other ions; indeed, previous studies have shown clustering of QTLs for closely correlated salt traits23,24,26. Each cluster may represent either a single gene or several closely linked genes; e.g. the occurrence of two closely linked NAX QTLs on 2A and the co-localization of Zn and NAX QTLs. This co-localization or QTL clustering is due to high the correlation coefficient among the traits of the respective QTLs as highlighted (Table S4).
Functional annotation of segregating SNPs highlighted complexity of salt tolerance
Segregating F2 populations are valuable material for the dissection of the genetic architecture of complex traits, as they depict maximum segregation in phenotype and polymorphism at molecular level14,39. The Wheat 35 K Array contains exome-captured SNPs; thus, flanking sequences of segregating SNPs could be used for functional annotation, which revealed that genes of various biological processes and molecular functions are likely to be involved in salt tolerance mechanisms. SNP-carrying sequences were highly similar to those from A. tauschii and T. urartu, reflecting their close ancestral relationships with bread wheat37. Another top species from BLAST comparisons was H. vulgare, the most salt tolerant cereal15; indicting shared tolerance genes between wheat and barley. We annotated 44 ion transporters responsible for ion homeostasis. Among the seven annotated K transporters/channels, the role of HKTs i.e. TmHKT1;5-A Nax2, HKT1;4 Nax1 as sodium excluders5,25, K+/H+ exchanger, TaNHX2, as K+/H+ antiporter40 and Jacalin-related lectin 3 (TaJRL3) in salt tolerance response41 was reported previously. The K+ outward-rectifying channel (KORC) negatively regulates salt tolerance by K+ efflux from plant roots under salinity42, but the functions of the remaining K+ channels are not known. The annotated chloride channels (CLC-g and CLC-e) could be linked to previously mapped Cl− QTLs including a major 5A QTL27. The annotated Pyrophosphate-energized proton pump (H+-PPase or TVP1) and four proton transporters were previously found to induce the sequestration of Na+ ions into the vacuole and act as Na+/H+ antiporters to confer salt tolerance in wheat43. However, none of the four Ca, five Zn transporters, three nitrate transporters and two each of Mg, Sulfate and Cu-transporters have been characterized for their salt tolerance response in wheat. Another 50 SNPs annotated for transport of proteins, mRNA, glucose, malate, fatty acids and proline need further investigation under salinity.
Understanding of complex abiotic stress signalling pathways is essential for a successful breeding program34,44. In total 92 SNPs were located within genes involved in several signalling pathways, e.g. signal recognition, transduction, cell surface receptor and Wnt signalling to identify stress stimuli. Annotation of apoptotic signalling genes could be linked to the hypersensitive response to avoid stress injury45. Similarly, a total of 14 genes were identified for ABA and ethylene activated signalling and for JA and SA-mediated signalling pathways, which are thought to be involved in conferring salt tolerance3. Moreover, some genes for ABA, ethylene and JA signalling also confer salt tolerance in wheat46,47. We identified 22 genes for auxin biosynthesis, transport and auxin-activated signalling pathways, but the role of auxin signalling under salinity is not known. TFs are proteins that regulate the expression of several stress-related genes34 and 63 annotated SNPs were linked to 35 classes of 52 TFs e.g. MYB44, WRKY16, WRKY70, bZIP17, NAC17, NAC78 etc. However, bHLH140, GATA26, ZNFX1-NFXL1, EIN3, ABI3, ARF3, ARF5 and ARF21 appear to be major salinity responsive TFs based on their GO annotations. Only the TFs ABI3 and ARF3 have previously been reported to be involved in salt tolerance mechanisms and signalling46,48 in wheat.
Interestingly, 198 SNPs were located on genes involved in various nucleic acid processes including chromatin modifications, helicases, DNA repair mechanisms, DNA replication, transcription and translation regulation. Some of these genes were over-expressed under salinity3. Other such gene groups coded for epigenetic (DNA, tRNA, rRNA and histone-lysine methylation) or epi-transcriptomic (splicosomal complexes, splicing site recognition, mRNA splicing, mRNA based gene silencing) processes. As these reactions are core cellular functions they may regulate the expression of salinity related genes49. Additionally, 166 SNPs were located on genes with enzymatic (ligases, transferases, hydrolases, isomerase and kinase) or protein modification activity. These included 77 kinases, protein kinases and protein serine/threonine kinases coding for protein phosphorylation or post-translational protein modification. Protein serine/threonine kinases and protein kinases play a role in wheat salt tolerance and ABA signaling respectively48,50. Genes for other post-translational processes (protein de-phosphorylation, dimerization and glycosylation) were also identified. Another 51 SNPs were linked to genes involved in Zn, Ca, Mg, Fe and metal ion binding, as metal ions are part of several enzymes that may have direct roles in conferring salt tolerance.
The largest number of annotated SNPs, 275, were linked to genes of metabolic processes. The biggest subgroup consisted of 65 SNPs associated with photosynthesis regulation, chloroplast fission/organization, chlorophyll biosynthesis/catabolism, Photosystem (PS) I and II complex, PS II assembly, light reaction, Carbon/energy pathway, chloroplast DNA synthesis/translation; and synthesis of photosynthetic sugars (glucose, galactose, fructose, mannose). The role of these genes in photosynthesis under salinity in wheat has not been thoroughly investigated, but 22 photosynthetic proteins showed differential expression under salinity in wheat51. Additionally, 45 genes were identified for oxidation-reduction (Redox) processes, which are the backbone of cell mechanisms. A redox gene, 12-oxophytodienoate reductase 1 (OPR1), confers salt tolerance to wheat by enhanced ABA signalling and reactive oxygen species (ROS) scavenging52. The redox genes coding for peroxidase 1, 2 and 12, and 19 genes for synthesis of antioxidants (glutathione, flavonoid, carboxylic acid, lactate, cytokinin, vitamin B and E) could be involved in ROS scavenging under salinity induced osmotic stress. Genes for ubiquitin, proteasome and proteolysis dependent protein catabolism could destroy the unwanted proteins under salinity. However, roles of 21 respiratory genes (photorespiration, respiratory chain complex I and II, glycolysis, and Kreb cycle) and 64 genes for biosynthesis of carbohydrates, lipids and proteins needs to be investigated. Among the 92 SNPs associated with genes of several growth stages such as root/shoot development to reproductive growth (flowering time, pollen germination, ovule development etc.), inheritance of flowering time under salinity has been reported15.
Several annotated genes were transcriptionally expressed
In order to validate the role of annotated SNPS/genes in conferring salt tolerance, they were aligned with published transcriptome data describing genes that are differentially expressed under salinity3. A total of 122 of the annotated genes were up-regulated in silico, which included protein, mRNA, nitrate and cations transporters; several Wnt/ABA/auxin activated signaling molecules; signal transduction, kinases, proteolysis, REDOX process, flavonoid metabolism, defense response, cell wall/xylum development genes etc. On the other hand, 136 genes were down-regulated under salinity. The only available salt-expressed transcriptome in wheat root is based on a single genotype; therefore, several of the annotated genes, such as photosynthetic genes, were not expected to show differential expression.
Conclusions and prospects
We have identified two novel major QTLs on wheat chromosome 7A which contributed 11.23 and 15.79% to salt tolerance, and 13.69–15.35% to NAX in our population. Another major Zn QTL contributed 12.08 and 11.23% to Zn phenotypic variation and salt tolerance respectively. A major SCaIC QTL also accounted for 11.87% of the observed salt tolerance trait. We have also identified other novel QTLs which contributed 10.91, 12.96,11.31 and 14.16% of Ca, Fe, K and Mn phenotypic variation. Other mapped QTLs represented 2.1–8.98% of phenotypic variation of different minerals. These novel QTLs could be used for could be used for MAS breeding for salt tolerance and breeding for biofortification of wheat for Ca, Zn, Mg, Fe and Mn. We have also annotated 1293 segregating SNPs, which were located within genes for various ion channels, signalling pathways, TFs, metabolic pathways etc. and 258 of them were differentially expressed under salinity, indicating that they may have a role in salt tolerance. As the published transcriptome used for this analysis is based on Roche 454-GS FLX sequencing, future transcriptome data fromsalt stressed wheat using Illumina technology could help to further understand gene expression under salinity. The characterization of the annotated genes described here will help to dissect salt tolerance mechanisms, guiding future breeding for this important trait.
Electronic supplementary material
Acknowledgements
B.H. is thankful to Scientific and Technological Research Council of Turkey (TUBITAK) BIDEB-2215 program for his graduate scholarship. This research was partially funded by Sabanci University and Montana State University Endowment.
Author Contributions
H.B. conceived and designed the experiment and drafted manuscript. B.H. conducted the experiment, did the data analysis and drafted the manuscript. S.J.L. helped with DNA extraction and data analysis. L.O. helped in mineral analysis.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
10/22/2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-15726-6.
References
- 1.Zhang X, Cai X. Climate change impacts on global agricultural land availability. Environ. Res. Lett. 2011;6:1–8. [Google Scholar]
- 2.Horie T, Karahara I, Katsuhara M. Salinity tolerance mechanisms in glycophytes: An overview with the central focus on rice plants. Rice. 2012;5:1–18. doi: 10.1186/1939-8433-5-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goyal E, Amit SK, Singh RS, Mahato AK, Chand S. Transcriptome profiling of the salt-stress response in Triticum aestivum cv. Kharchia Local. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep27752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hussain B, Khan AS, Ali Z. Genetic variation in wheat germplasm for salinity tolerance at seedling stage: improved statistical inference. Turkish J. Agric. For. 2015;39:182–192. [Google Scholar]
- 5.Munns R, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat. Biotechnol. 2012;30:360–364. doi: 10.1038/nbt.2120. [DOI] [PubMed] [Google Scholar]
- 6.Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Munns R, James RA, Läuchli A. Approaches to increasing the salt tolerance of wheat and other cereals. J. Exp. Bot. 2006;57:1025–1043. doi: 10.1093/jxb/erj100. [DOI] [PubMed] [Google Scholar]
- 8.Ali Z, Khan AS, Azhar FM, Khan IA, Khan AA. The response of genetically distinct bread wheat genotypes to salinity stress. Plant Breed. 2012;131:707–715. [Google Scholar]
- 9.Oyiga B, et al. Identification and characterization of salt tolerance of wheat germplasm using a multivariable screening approach. J. Agron. Crop Sci. 2016;2016:1–14. [Google Scholar]
- 10.Budak H, et al. Molecular characterization of Buffalograss germplasm using sequence-related amplified polymorphism markers. Theor. Appl. Genet. 2004;108:328–334. doi: 10.1007/s00122-003-1428-4. [DOI] [PubMed] [Google Scholar]
- 11.Budak H, Shearman RC, Gulsen O, Dweikat I. Understanding ploidy complex and geographic origin of the Buchloe dactyloides genome using cytoplasmic and nuclear marker systems. Theor. Appl. Genet. 2005;111:1545–1552. doi: 10.1007/s00122-005-0083-3. [DOI] [PubMed] [Google Scholar]
- 12.Castillo A, et al. Transferability and polymorphism of barley EST-SSR markers used for phylogenetic analysis in Hordeum chilense. BMC Plant Biol. 2008;9:1–9. doi: 10.1186/1471-2229-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogel JP, et al. Development of SSR markers and analysis of diversity in Turkish populations of Brachypodium distachyon. BMC Plant Biol. 2009;11:1–11. doi: 10.1186/1471-2229-9-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hussain B. Modernization in plant breeding approaches for improving biotic stress resistance in crop plants. Turkish J. Agric. For. 2015;39:515–530. [Google Scholar]
- 15.Saade S, et al. Yield-related salinity tolerance traits identified in a nested association mapping (NAM) population of wild barley. Sci. Reports. 2016;6:1–9. doi: 10.1038/srep32586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filiz E, et al. Molecular, morphological, and cytological analysis of diverse Brachypodium distachyon inbred lines. Genome. 2009;52:876–890. doi: 10.1139/g09-062. [DOI] [PubMed] [Google Scholar]
- 17.Lucas SJ, et al. Functional features of a single chromosome arm in wheat (1AL) determined from its structure. Funct. Integr. Genomics. 2012;12:173–182. doi: 10.1007/s10142-011-0250-3. [DOI] [PubMed] [Google Scholar]
- 18.Akpinar BA, Lucas S, Budak H. A large-scale chromosome-specific SNP discovery guideline. Funct. Integr. Genomics. 2017;7:97–105. doi: 10.1007/s10142-016-0536-6. [DOI] [PubMed] [Google Scholar]
- 19.Winfield MO, et al. High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol. J. 2015;14:1–12. doi: 10.1111/pbi.12485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucas, S. J., Salantur, A., Yazar, S. & Budak, H. High-throughput SNP genotyping of modern and wild emmer wheat for yield and root morphology using a combined association and linkage analysis. Funct. Integr. Genomics 1–19, 10.1007/s10142-017-0563-y (2017). [DOI] [PubMed]
- 21.Leonforte A, et al. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.) BMC Plant Biol. 2013;13:161. doi: 10.1186/1471-2229-13-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lorieux M. MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 2012;30:1231–1235. [Google Scholar]
- 23.Masoudi B, Mardi M, Hervan EM. QTL mapping of salt tolerance traits with different effects at the seedling stage of bread wheat. Plant Mol. Biol. Report. 2015;33:1790–1803. [Google Scholar]
- 24.Peleg Z, et al. Quantitative trait loci conferring grain mineral nutrient concentrations in durum wheat 3 wild emmer wheat RIL population. Theor. Appl. Genet. 2009;119:353–369. doi: 10.1007/s00122-009-1044-z. [DOI] [PubMed] [Google Scholar]
- 25.Genc Y, et al. Sodium exclusion QTL associated with improved seedling growth in bread wheat under salinity stress. Theor. Appl. Genet. 2010;121:877–894. doi: 10.1007/s00122-010-1357-y. [DOI] [PubMed] [Google Scholar]
- 26.Xu Y, et al. Mapping QTLs for salt tolerance with additive, epistatic and QTL x treatment interaction effects at seedling stage in wheat. Plant Breed. 2013;132:276–283. [Google Scholar]
- 27.Genc Y, Taylor J, Rongala J, Oldach K. A major locus for chloride accumulation on chromosome 5A in bread wheat. PLoS One. 2014;9:e98845. doi: 10.1371/journal.pone.0098845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ova EA, Kutman UB, Ozturk L. High phosphorus supply reduced zinc concentration of wheat in native soil but not in autoclaved soil or nutrient solution. Plant Soil. 2015;393:147–162. [Google Scholar]
- 29.Zarcinas BA, Cartwright B, Spouncer LR. Original Articles Nitric acid digestion and multi‐element analysis of plant material by inductively coupled plasma spectrometry. Commun. Soil Sci. Plant Anal. 1987;18:131–146. [Google Scholar]
- 30.Rice WR. Analysing tables of statistical tests. Evolution (N. Y). 1989;43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 31.Meng L, Li H, Zhang L, Wang J. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015;3:269–283. [Google Scholar]
- 32.Voorrips RE. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, Z. et al. QTL analysis of kernel-related traits in maize using an immortalized F 2 population. PLoS One9 (2014). [DOI] [PMC free article] [PubMed]
- 34.Budak H, Hussain B, Khan Z, Ozturk NZ, Ullah N. From genetics to functional genomics: improvement in drought signaling and tolerance in wheat. Front. Plant Sci. 2015;6:1–13. doi: 10.3389/fpls.2015.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.IWGSC. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science (80-.). 345, 286–287 (2014). [DOI] [PubMed]
- 36.Götz S, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akpinar BA, et al. Molecular organization and comparative analysis of chromosome 5B of the wild wheat ancestor Triticum dicoccoides. Sci. Rep. 2015;5:1–13. doi: 10.1038/srep10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Akpinar BA, Lucas SJ, Vrana J, Dole J, Budak H. Sequencing chromosome 5D of Aegilops tauschii and comparison with its allopolyploid descendant bread wheat (Triticum aestivum) Plant Biotechnol. J. 2015;13:740–752. doi: 10.1111/pbi.12302. [DOI] [PubMed] [Google Scholar]
- 39.Lindsay MP, Lagudah ES, Hare RA, Munns R. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct. Plant Biol. 2004;31:1105–1114. doi: 10.1071/FP04111. [DOI] [PubMed] [Google Scholar]
- 40.Xu Y, et al. Functional characterization of a wheat NHX antiporter gene TaNHX2 that encodes a K+/H+ exchanger. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0078098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song M, Xu W, Xiang Y. Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol. Biol. 2014;84:95–110. doi: 10.1007/s11103-013-0121-5. [DOI] [PubMed] [Google Scholar]
- 42.Cuin TA, Betts SA, Chalmandrier R, Shabala S. A root’s ability to retain K+ correlates with salt tolerance in wheat. J. Exp. Bot. 2008;59:2697–2706. doi: 10.1093/jxb/ern128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brini F, Gaxiola RA, Berkowitz GA, Masmoudi K. Cloning and characterization of a wheat vacuolar cation/proton antiporter and pyrophosphatase proton pump. Plant Physiol. Biochem. 2005;43:347–354. doi: 10.1016/j.plaphy.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 44.Akpinar BA, Avsar B, Lucas SJ, Budak H. Plant abiotic stress signaling. Plant Signal. Behav. 2012;7:1450–1455. doi: 10.4161/psb.21894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuzuoglu-Ozturk D, et al. Autophagy-related gene, TdAtg8, in wild emmer wheat plays a role in drought and osmotic stress response. Planta. 2012;236:1081–1092. doi: 10.1007/s00425-012-1657-3. [DOI] [PubMed] [Google Scholar]
- 46.Li S, Xu C, Yang Y, Xia G. Functional analysis of TaDi19A, a salt-responsive gene in wheat. Plant Cell Environ. 2010;33:117–129. doi: 10.1111/j.1365-3040.2009.02063.x. [DOI] [PubMed] [Google Scholar]
- 47.Zhao Y, et al. A wheat allene oxide cyclase gene enhances salinity tolerance via jasmonate signaling. Plant Physiol. 2014;164:1068–1076. doi: 10.1104/pp.113.227595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang H, Mao X, Wang C, Jing R. Overexpression of a common wheat gene TaSnRK2.8 enhances tolerance to drought, salt and low temperature in Arabidopsis. PLoS One. 2010;5:2–13. doi: 10.1371/journal.pone.0016041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karijolich J, Yu Y. The new era of RNA modification. RNA. 2015;21:659–660. doi: 10.1261/rna.049650.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen Ping G, et al. Isolation and characterization of TaGSK1 involved in wheat salt tolerance. Plant Sci. 2003;165:1369–1375. [Google Scholar]
- 51.Xu W, et al. Proteomic comparison reveals the contribution of chloroplast to salt tolerance of a wheat introgression line. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep32384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong W, et al. Wheat oxophytodienoate reductase gene TaOPR1 confers salinity tolerance via enhancement of Abscisic Acid signaling and reactive oxygen species scavenging. Plant Physiol. 2013;161:1217–1228. doi: 10.1104/pp.112.211854. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.