Abstract
Isolated Complex I (CI) deficiency is the most commonly observed mitochondrial respiratory chain biochemical defect, affecting the largest OXPHOS component. CI is genetically heterogeneous; pathogenic variants affect one of 38 nuclear-encoded subunits, 7 mitochondrial DNA (mtDNA)-encoded subunits or 14 known CI assembly factors. The laboratory diagnosis relies on the spectrophotometric assay of enzyme activity in mitochondrially-enriched tissue homogenates, requiring at least 50 mg skeletal muscle, as there is no reliable histochemical method for assessing CI activity directly in tissue cryosections. We have assessed a validated quadruple immunofluorescent OXPHOS (IHC) assay to detect CI deficiency in the diagnostic setting, using 10 µm transverse muscle sections from 25 patients with genetically-proven pathogenic CI variants. We observed loss of NDUFB8 immunoreactivity in all patients with mutations affecting nuclear-encoding structural subunits and assembly factors, whilst only 3 of the 10 patients with mutations affecting mtDNA-encoded structural subunits showed loss of NDUFB8, confirmed by BN-PAGE analysis of CI assembly and IHC using an alternative, commercially-available CI (NDUFS3) antibody. The IHC assay has clear diagnostic potential to identify patients with a CI defect of Mendelian origins, whilst highlighting the necessity of complete mitochondrial genome sequencing in the diagnostic work-up of patients with suspected mitochondrial disease.
Introduction
Mitochondrial diseases are a group of clinically heterogeneous disorders caused by dysfunction of the oxidative phosphorylation (OXPHOS) system. Accounting for approximately 30% of all cases, isolated Complex I (CI) deficiency is the most frequently observed biochemical manifestation of an OXPHOS defect in patients, particularly within a paediatric setting1. With a relative molecular mass of 1MDa, mitochondrial CI (NADH: ubiquinone oxidoreductase) is the largest complex of the respiratory chain, oxidising NADH to NAD+ and playing a central role in both electron transfer and cellular energy (ATP) production. The complex is L-shaped in structure, consisting of 2 fragments; a hydrophilic peripheral arm, which extends into the matrix of the mitochondria and a hydrophobic arm which is embedded into the inner mitochondrial membrane. The complex is comprised of 45 subunits, 7 of which are encoded by the mitochondrial DNA (mtDNA) and the remaining 38 subunits encoded by the nuclear DNA (nDNA)2. This structure can be further divided into 3 functional modules: within the peripheral arm the N module (which binds and oxidizes NADH, providing electrons to the Fe-S clusters) and the Q module (where ubiquinone is reduced to ubiquinol) whilst the membrane arm contains the third module known as the P module which catalyses proton transfer3–8.
The assembly of CI is a multistep process; recent studies propose a modular assembly model, in which five assembly intermediates - termed Q/Pp-a, Pp-b, PD-a, PD-b and N - form separately before assembling into higher molecular mass intermediates to subsequently form the completed holoenzyme5. Despite differences in the nomenclature of assembly intermediates, all proposed models described in the recent literature follow the same sequence of steps9–12. Briefly, the first step involves formation of the Q module, followed by the addition of the mitochondrial encoded ND1 core subunit (early stage of assembly). Next, the membrane arm is assembled, starting with the insertion of ND2, ND3, ND6, ND4L (PP-b module), ND4 (PD-a) and ND5 (PD-b) (early-mid stage assembly and mid-late stage assembly). The membrane arm comes together with the Q module (Q-P module) prior to the addition of the pre-assembled N module and remaining subunits (last stage of assembly)5,9,13.
To date, pathogenic variants have been reported in all 7 mtDNA core structural subunits (ND1–6 and ND4L), 21 of the nuclear-encoded subunits and 10 of the known CI assembly factors (reviewed in refs14,15). Further to this genetic heterogeneity, CI deficiency also exhibits considerable clinical heterogeneity with the spectrum ranging from severe presentations including Leigh Syndrome (LS) and fatal infantile mitochondrial disease in early childhood through to Leber Hereditary Optic Neuropathy (LHON) and exercise-induced muscle weakness which develop during young adult life.
The diagnosis of mitochondrial disease often uses a multidisciplinary approach including the histopathological and biochemical assessment of tissue biopsy samples, usually skeletal muscle, as well as molecular genetic testing. Current histochemical methods to investigate mitochondrial disease are available for some OXPHOS components (e.g. the sequential assessment of cytochrome c oxidase (Complex IV, COX) and succinate dehydrogenase (Complex II, SDH)) but not all; no reliable histochemical assay is available to assess CI activity. Whilst validated biochemical assays can determine the activity of individual respiratory chain enzyme complexes, for example, CI by the spectrometric assay of rotenone-sensitive NADH oxidation, consensus diagnostic protocols are not widely adopted. Furthermore, most diagnostic laboratories request at least 50 mg of muscle tissue to reliably measure enzyme activities, which represents a considerable proportion of the total amount of patient material available, particularly in children. Additionally, the biochemical assay is known to only measure the redox activity of the peripheral arm of CI (containing the N-module and Q- module). Therefore, patients with mutations residing in the membrane arm (P-module) - where only the proton pumping ability is affected rather than electron transport - may show a ‘normal’ enzyme profile following biochemical assessment despite an underlying CI defect, although this may reflect the level of mtDNA heteroplasmy in muscle tissue8. A confirmed diagnosis of isolated CI deficiency can facilitate appropriate molecular genetic testing to elucidate the underlying genetic cause, with screening of the mitochondrial genome preceding high throughput analysis of Mendelian candidates either by targeted gene panels, whole exome sequencing (WES) or whole genome sequencing (WGS)16.
Given the vast clinical and genetic heterogeneity associated with CI deficiency, accurate diagnosis is essential. A recently developed quadruple immunofluorescent assay to assess mitochondrial respiratory chain defects through the immunodetection of NDUFB8 (a CI subunit), COX-1 (a Complex IV subunit), porin (a mitochondrial mass marker) and laminin (a myofibre membrane marker) has been shown to provide an objective and reliable quantitative method for the assessment of CI and CIV protein abundance relative to the mitochondrial mass in individual muscle fibres within a single transversely-orientated, 10 µm muscle section17. We have extended this original study to assess CI status in skeletal muscle biopsies from 25 patients with proven pathogenic variants in proteins leading to a biochemical defect of isolated CI activity, including nuclear-encoded CI structural subunits, CI assembly factors, or one of the 7 mtDNA-encoded CI structural subunits. We demonstrate that the IHC assay has clear diagnostic potential, particularly for patients with Mendelian-inherited defects and propose that the IHC assay should form part of the multidisciplinary approach for the diagnostic investigation of patients with suspected CI deficiency.
Results
Mitochondrial Respiratory Chain (MRC) profiles
We assessed 25 skeletal muscle biopsies taken from patients with genetically-confirmed pathogenic variants either shown or predicted to cause isolated CI deficiency to validate a recently-developed immunofluorescence assay (IHC) within a diagnostic setting. The clinical, biochemical and molecular genetic characteristics of this patient cohort are shown in Table 1. The IHC assay, detecting CI subunit NDUFB8, CIV subunit COX-1 and porin (a marker of mitochondrial mass), was performed in all muscle sections (Supplementary Figs S1, S2 and S3), and fibres were classified according to Z scores – where any fibres with a Z score under -3SD were classified as deficient. The IHC results (NDUFB8 immunoreactivity), presented as a percentage of fibres deficient in CI – calculated by totalling the percentage of fibres classified as negative, intermediate negative and intermediate positive fibres (Table 2) - were subsequently correlated with the diagnostic biochemical findings, presented as residual CI activity (Table 1).
Table 1.
Clinical, biochemical and molecular genetic characteristics of our patient cohort with isolated Complex I deficiency.
| Patient | Gender | Adult/Paediatric | Clinical Presentation | Gene | Genetic Defect | Residual Complex I activity | Mutation Load |
|---|---|---|---|---|---|---|---|
| Nuclear-encoded Complex I structural subunits | |||||||
| P1a | F | Paediatric | IUGR and oligohydramnios, FTT, mild hypertrophic cardiomyopathy | NDUFB3 | Homozygous c.64 T > C, p.(Trp22Arg) | 33% | n.a. |
| P2b | F | Paediatric | IUGR. Acute life-threatening event, age 20 days, required intubation. Hypertrophic cardiomyopathy | NDUFB3 | Homozygous c.64 T > C, p.(Trp22Arg) | 32% | n.a. |
| P3c | F | Paediatric | Oligohydramnios. IUGR. Poor feeding at birth. MRI brain and echocardiogram normal. Age-appropriate skills. Family history of previous neonatal death | NDUFB3 | Homozygous c.64 T > C, p.(Trp22Arg) | 35% | n.a. |
| P4 | F | Paediatric | Leigh syndrome | NDUFS4 | Compound heterozygous c.99-1 G > A + c.416_417delinsC, p.(Glu139Alafs*50) | 39% | n.a. |
| P5 | F | Paediatric | Consanguineous, first cousin parents; Leigh-like syndrome; elevated lactates | NDUFS4 | Homozygous exon 3 and 4 deletion | 37% | n.a. |
| P6 | M | Paediatric | Infantile-onset mitochondrial disease; marked lactic acidosis | NDUFS6 | Homozygous c.316_319delGAAA, p.(Glu106Glnfs*41) | 5% | n.a. |
| P7 | F | Paediatric | Leigh syndrome | NDUFS2 | Homozygous c.998 G > A, p.(Arg333Gln) | 42% | n.a. |
| P8 | F | Paediatric | Leigh-like syndrome; elevated serum lactates | NDUFS3 | Homozygous c.642_644delTGA, p.(Asp214del) | 26% | n.a. |
| Nuclear-encoded Complex I assembly factors | |||||||
| P9 | F | Paediatric | Leigh-like syndrome; elevated lactates | NDUFAF6 | Compound heterozygous c.805 C > T, p.(His269Tyr) and c.581-7 A > G, p.(Leu193_Gly194insValIle) | 26% | n.a. |
| P10 | F | Paediatric | Lethal infantile mitochondrial disease presentation; presented day 1 with persistent lactic acidosis; died at 9 weeks | NDUFAF6 | Homozygous c.659 C > A, p.(Thr220Lys) | 45% | n.a. |
| P11 | F | Paediatric | Presented at 8 months; developmental regression, rotatory nystagmus bilaterally; elevated blood and CSF lactate; extensive basal ganglia and brainstem changes on MRI | NDUFAF5 | Compound heterozygous c.826 C > T, p.(Arg276*) and c.848 C > T, p.(Ala283Val) | 44% | n.a. |
| P12 | M | Paediatric | Myoclonic seizures, developmental delay | FOXRED1 | Compound heterozygous c.612_615dup, p.(Ala206Serfs*15) and c.1261 G > A, p.(Val421Met) | 31% | n.a. |
| P13 | M | Paediatric | Hypertrophic cardiomyopathy at birth; severe metabolic acidosis (18–30 mmol/L); died at 2 days of age | ACAD9 | Compound heterozygous c.868 G > A, p.(Gly290Arg) and c.976 G > C, p.(Ala326Pro) | 13% | n.a. |
| P14 | M | Adult | Exercise intolerance, muscle cramps, elevated serum lactate | ACAD9 | Compound heterozygous c.1150 G > A, p.(Val384Met) and c.1168 G > A, p.(Ala390Thr) | 13% | n.a. |
| P15d | M | Adult | Exercise intolerance, unable to perform sustained aerobic exercise; normal strength; normal ECG and echocardiogram; normal resting lactate, normal CK | TMEM126B | Homozygous c.635 G > T, p.(Gly212Val) | 36% | n.a. |
| Mitochondrial DNA-encoded Complex I structural subunits | |||||||
| P16e | F | Adult | Exercise intolerance, persistent lactic acidaemia | MTND1 | m.3356 T > C, p.(Met17Thr) | 3% | 92% |
| P17 | M | Paediatric | Leigh syndrome | MTND3 | m.10158 T > C, p.(Ser34Pro) | 44% | 90% |
| P18 | M | Paediatric | Leigh syndrome | MTND3 | m.10197 G > A, p.(Ala47Thr) | n.d. | 93% |
| P19 | M | Paediatric | Leigh syndrome | MTND5 | m.13514 A > G, p.(Asp393Gly) | 27% | 66% |
| P20f | F | Paediatric | Chronic renal failure, myopathy and persistent lactic acidosis | MTND5 | m.12425delA, p.(Asn30Thrfs*7) | 16% | 85% |
| P21 | F | Paediatric | Bilateral ptosis, ophthalmoplegia, pyramidal tract signs, elevated blood and CSF lactates | MTND5 | m.13094 T > C, p.(Val253Ala) | 59% | 58% |
| P22 | M | Adult | Mitochondrial myopathy, elevated lactates | MTND5 | m.13513 G > A, p.(Asp393Asn) | 39% | 60% |
| P23 | M | Paediatric | Leigh syndrome | MTND5 | m.13513 G > A, p.(Asp393Asn) | 38% | 77% |
| P24 | F | Adult | Elevated CK, muscle pain and fatigue, myopathy | MTND5 | m.13513 G > A, p.(Asp393Asn) | 100% | 45% |
| P25 | M | Paediatric | Failure to strive, myopathy, increased brainstem signal on MRI, lactic acidosis | MTND5 | m.13513 G > A, p.(Asp393Asn) | 100% | 63% |
Residual Complex I activities, normalised to the activity of the matrix marker enzyme citrate synthase, are expresses as a percentage of mean control values.
Residual Complex I activity and mtDNA mutation load measured in muscle. Key: IUGR, intrauterine growth restriction; FTT, failure to thrive; ECG, electrocardiogram; CK, creatinine kinase; a,b,c,d,epublished patients: aP1 = Patient 3 in Alston et al.16, bP2 = Patient 2 in Alston et al.16, cP3 = Patient 6 in Alston et al.16, dP15 = Patient S1 in Alston et al.20, eP16 = Patient 1 in Gorman et al.27, eP20 = Patient published in Alston et al.26, F; Female, M; Male, n.a.; not applicable, n.d.; not determined.
Table 2.
Percentage of Complex I (CI) deficient fibres detected with the IHC assay.
| Patient ID | IHC findings - % of CI deficient fibres | Total number of fibres analysed | |
|---|---|---|---|
| Nuclear-encoded Complex I structural subunits | P1 | 79% | 4372 |
| P2 | 93% | 559 | |
| P3 | 39% | 13422 | |
| P4 | 98% | 5964 | |
| P5 | 89% | 4683 | |
| P6 | 79% | 880 | |
| P7 | 99% | 5337 | |
| P8 | 96% | 7154 | |
| Nuclear-encoded Complex I assembly factors | P9 | 99% | 9504 |
| P10 | 96% | 5355 | |
| P11 | 26% | 7352 | |
| P12 | 96% | 1708 | |
| P13 | 100% | 2684 | |
| P14 | 100% | 239 | |
| P15 | 100% | 131 | |
| Mitochondrial DNA-encoded Complex I structural core subunits | P16 | 92% | 696 |
| P17 | 90% | 5842 | |
| P18 | 0% | 7302 | |
| P19 | 0% | 2427 | |
| P20 | 100% | 3795 | |
| P21 | 0% | 1311 | |
| P22 | 0% | 3341 | |
| P23 | 0% | 2730 | |
| P24 | 0% | 675 | |
| P25 | 0% | 785 |
Group 1: Nuclear-encoded CI structural subunits
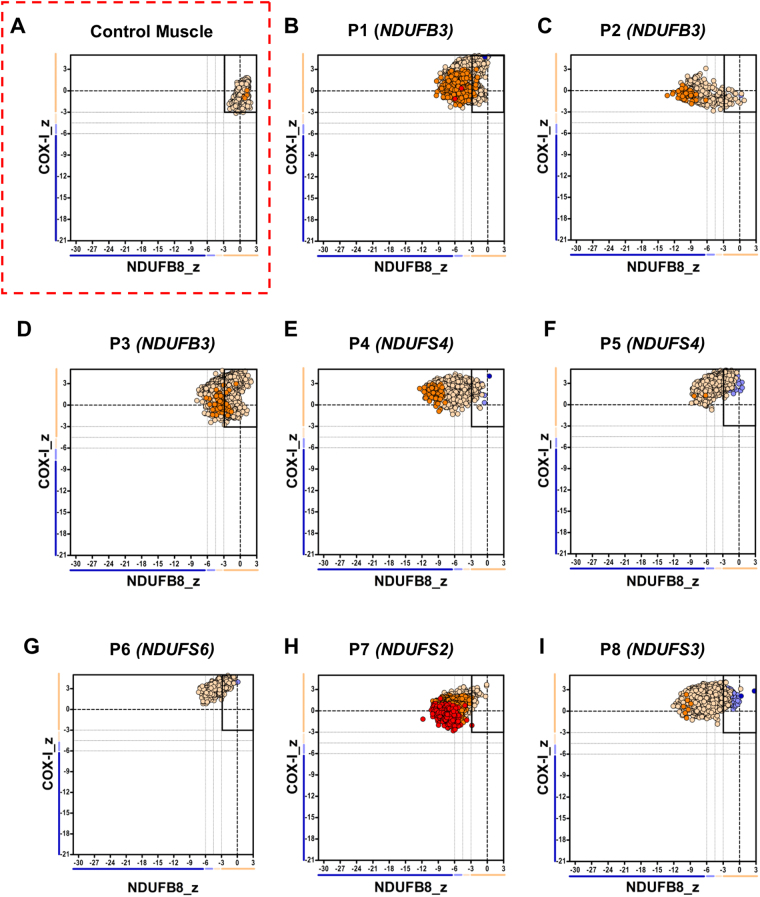
All eight patients who have pathogenic variants in nuclear-encoded CI structural core or accessory subunits (Group 1) showed varying levels of decreased NDUFB8 immunoreactivity (representing a decrease in NDUFB8 protein abundance) when compared to control muscle (Supplementary Fig. S1). Further analysis revealed that the proportion of CI-immunodeficient fibres ranged between 39% and 99% across this group of patients. The mitochondrial respiratory chain profiles (Fig. 1), showing the NDUFB8 and COX-1 protein abundance in conjunction with mitochondrial mass in individual fibres, also illustrates this as most fibres analysed were outside of the normal range for NDUFB8 (Z-score from -3SD to 3 SD) – causing a shift in their distribution on the plot to the left - despite the normal levels of COX-1 (subunit of complex IV) in virtually all fibres assessed (n = 131–13422 fibres analysed; dependent on the size of muscle section). These findings were in agreement with the respiratory chain biochemical findings, where a decrease in residual CI activity was observed (Table 1).
Figure 1.
Mitochondrial respiratory chain expression profile linking complex I, complex IV and porin levels in patients with isolated Complex I deficiency caused by defects in nuclear-encoded Complex I subunits. Graphs show complex I and complex IV expression profile from (A) Normal adult control and patients with (B–D) homozygous c.64T>C, p.(Trp22Arg) NDUFB3 variant, P1–n = 4372 fibres analysed, P2–n = 559, P3–n = 13422 (E) compound heterozygous NDUFS4 variant, P4, n = 5964 (F) Homozygous exon 3 and 4 deletion in NDUFS4, P5, n = 4683 (G) homozygous NDUFS6 variant, P6, n = 880 (H) Homozygous NDUFS2 variant, P7, n = 5337 (I) Homozygous NDUFS3 variant, P8, n = 7154. Each dot represents a single muscle fibre, colour co-ordinated according to its mitochondrial mass: very low – blue, low – light blue, normal – beige, high – orange, very high - red. Black dashed lines represent the SD limits for the classification of the fibres. Lines adjacent to X and Y axis represent the levels of NDUFB8 and COX-1: beige: normal (<−3), light beige: intermediate (+) (−3 to −4.5), light blue: intermediate (−) (−4.5 to −6) and blue: deficient (>−6). Bold dashed lines indicate the mean expression level of normal fibres.
Group 2: Nuclear-encoded CI assembly factors
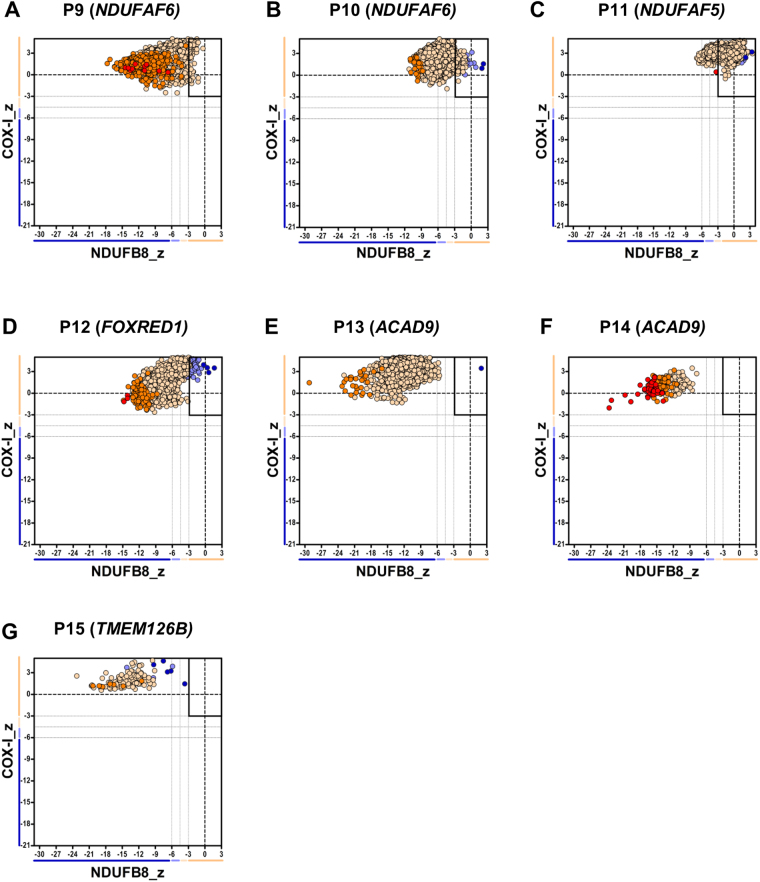
Similar to patients in Group 1, all seven patients with pathogenic variants in nuclear-encoded assembly factors (Group 2), displayed a severe loss of NDUFB8 subunit immunoreactivity (Supplementary Fig. S2). With the exception of P11, further quantification showed that the percentage of CI-deficient fibres was > 96%, highlighting the severe loss of NDUFB8. Specifically, the assay detected a complete loss of NDUFB8 immunoreactivity (100% CI-deficient fibres) in patients P13, P14 and P15 all of whom had pathogenic variants in either the ACAD9 or TMEM126B genes; these encode CI assembly factors involved in the biogenesis of the proximal part of the P module (PP). Only P11, who has compound heterozygous pathogenic variants in the NDUFAF5 gene, encoding a CI assembly factor involved in an early step of the biogenesis of the holoenzyme, maintained a relatively normal level of NDUFB8 (26% CI-deficient fibres). As previously observed in patients from Group 1, the mitochondrial respiratory chain profiles from patients in Group 2 (Fig. 2) similarly showed a “shifting to the left”, but to a greater extent. Again, the IHC findings correlated with available biochemical results which show a more severe decrease in muscle CI activity (Table 1).
Figure 2.
Mitochondrial respiratory chain expression profile linking complex I, complex IV and porin levels in patients with isolated Complex I deficiency caused by defects in nuclear-encoded Complex I assembly factors. Graphs show complex I and complex IV expression profile from patients with (A) Compound heterozygous NDUFAF6 variant, P9, n = 9504 fibres analysed (B) Homozygous NDUFAF6 variant, P10, n = 5355 (C) Compound heterozygous NDUFAF5 variant, P11, n = 7352 (D) Compound heterozygous FOXRED1 variant, P12, n = 1708 (E–F) Compound heterozygous ACAD9 variant, (E = P13, n = 2684, F = P14, n = 239) (G) Homozygous TMEM126B variant, P15, n = 131. Each dot represents a single muscle fibre, colour co-ordinated according to its mitochondrial mass: very low – blue, low - light blue, normal – beige, high – orange, very high - red. Black dashed lines represent the SD limits for the classification of the fibres. Lines adjacent to X and Y axis represent the levels of NDUFB8 and COX-1: beige: normal (<−3), light beige: intermediate (+) (−3 to -4.5), light blue: intermediate (−) (−4.5 to −6) and blue: deficient (>−6). Bold dashed lines indicate the mean expression level of normal fibres.
IHC shows variable results in patients with mutations in mtDNA-encoded CI subunits (Group 3)
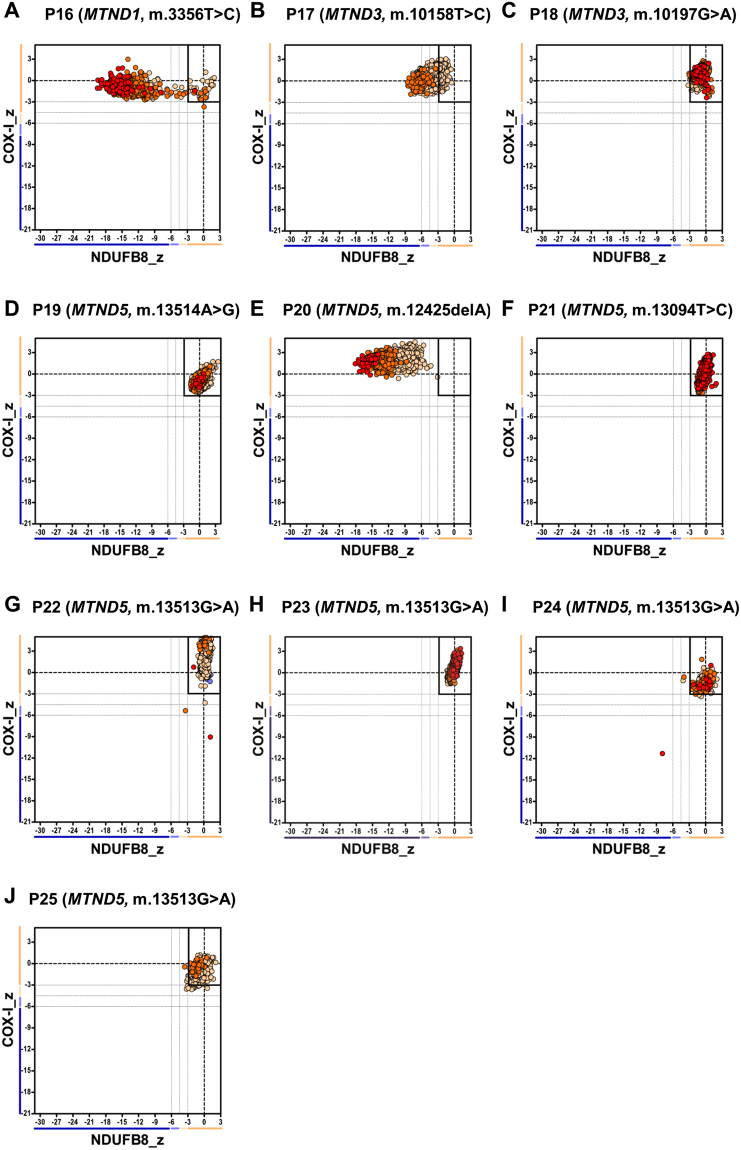
Patients harbouring mutations in mtDNA genes encoding core structural subunits of Complex I (Group 3, n = 10) showed more heterogeneous results. The IHC assay detected 100% CI-deficient fibres, representing a complete loss of NDUFB8 immunoreactivity, in three previously-reported patients with high levels of pathogenic mtDNA variants in muscle; P16 who has a m.3356 T > C, p.(Met17Thr) MTND1 variant, P17 who harbours a m.10158 T > C, p.(Ser34Pro) MTND3 variant and P20 who harbours a m.12425delA MTND5 frameshift mutation (see Supplementary Fig. S3 and Table 2). As expected, the mitochondrial respiratory chain profiles for these three patients were all shifted to the left, consistent with the loss of NDUFB8 immunoreactivity associated with preserved COX-I immunoreactivity (Fig. 3).
Figure 3.
Mitochondrial respiratory chain expression profile linking complex I (NDUFB8), complex IV and porin levels in patients with isolated Complex I deficiency caused by defects in mtDNA-encoded Complex I subunits. Graphs show complex I and complex IV expression profile from patients with (A) m.3356T>C MTND1 variant, P16, n = 696 fibres analysed (B) m.10158T>C MTND3 variant, P17, n = 5842 (C) m.10197G>A MTND3 variant, P18, n = 7302 (D) m.13514A>G MTND5 variant, P19, n = 2427 (E) m.12425delA MTND5 variant, P20, n = 3795 (F) m.13094T>C MTND5 variant, P21, n = 1311 (G) m.13513G>A MTND5 variant, P22, n = 3341 (H) m.13513G>A MTND5 variant, P23, n = 2730 (I) m.13513G>A MTND5 variant, P24, n = 675 (J) m.13513G>A MTND5 variant, P25, n = 785. Each dot represents a single muscle fibre, colour co-ordinated according to its mitochondrial mass: very low – blue, low - light blue, normal – beige, high – orange, very high - red. Black dashed lines represent the SD limits for the classification of the fibres. Lines adjacent to X and Y axis represent the levels of NDUFB8 and COX-1: beige: normal (<−3), light beige: intermediate (+) (−3 to −4.5), light blue: intermediate (−) (−4.5 to −6) and blue: deficient (>−6). Bold dashed lines indicate the mean expression level of normal fibres.
The remaining 7 patients (P18, P19, P21, P22, P23, P24 and P25) showed NDUFB8 immunoreactivity similar to control muscle (Supplementary Fig. S4 and Table 2). The mitochondrial respiratory chain profiles demonstrated normal respiratory chain function as all fibres fell within the normal range (with Z scores between -3SD and 3 SD) (Fig. 3). Of these 7 patients, P22- P25 all harbour a common pathogenic m.13513 G > A, p.(Asp393Asn) MTND5 variant, P19 harbours a m.13514 A > G, p.(Asp393Gly) variant, P21 harbours a m.13094 T > C, p.(Val253Ala) variant whilst P18 has high levels of a m.10197 G > A, p.(Ala393Thr) MTND3 variant. Detection of normal levels of immunoreactive NDUFB8 subunit in these patients could be due to these variants only affecting the catalytic function of CI rather than the assembly of the enzyme complex.
Patients with mutations in mtDNA-encoded CI subunits and normal NDUFB8 profile also display normal NDUFS3 levels
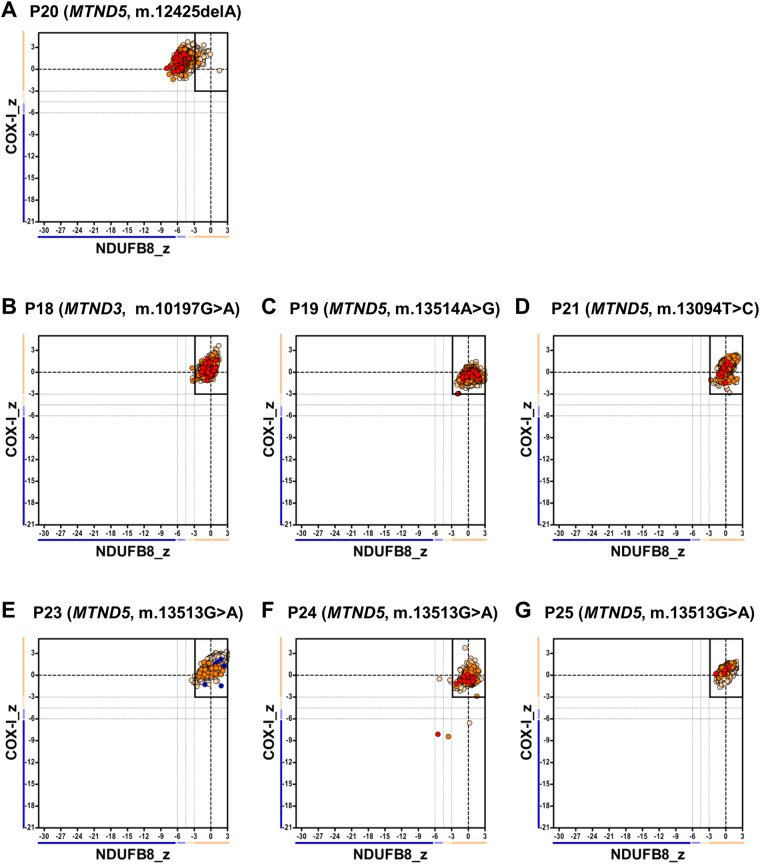
Since interrogation of NDUFB8 immunoreactivity failed to detect CI deficiency in 7 of the 10 patients with mtDNA-encoded structural CI mutations, we further investigated whether any deficiency could be detected using an alternative, but commercially-available CI antibody targeting NDUFS3 – a subunit which is integrated during the early stage of complex I assembly. The IHC assay was performed on 10μm muscle sections from P18, P19, P20, P21, P23, P4 and P25 using antibodies against NDUFS3, porin, COX-I and laminin, and showed normal NDUFS3 immunoreactivity (Supplementary Fig. S5). As shown in Fig. 4, the mitochondrial respiratory chain profile for P20 shows a shift of fibres to the left whilst for all the remaining patients also tested with NDUFS3 fall into the normal range – these findings are the same as the assessment with the NDUFB8 antibody
Figure 4.
Mitochondrial respiratory chain expression profile linking complex I (NDUFS3), complex IV and porin levels in patients with Isolated complex I deficiency caused by defects in mtDNA-encoded Complex I subunits. Graphs show complex I and complex IV expression profile from patients with (A) m.12425delA MTND5 variant, P20, n = 5536 fibres analysed (B) m.10197G>A MTND3 variant, P18, n = 6645 (C) m.13514A>G MTND5 variant, P19, n = 2730 (D) m.13094T>C MTND5 variant, P21, n = 3979 (E) m.13513G>A MTND5 variant, P23, n = 10009 (F) m.13513G>A MTND5 variant, P24, n = 575 (G) m.13513G>A MTND5 variant, P25, n = 1168. Each dot represents a single muscle fibre, colour co-ordinated according to its mitochondrial mass: very low – blue, low - light blue, normal – beige, high – orange, very high - red. Black dashed lines represent the SD limits for the classification of the fibres. Lines adjacent to X and Y axis represent the levels of NDUFB8 and COX-1: beige: normal (<−3), light beige: intermediate (+) (−3 to −4.5), light blue: intermediate (−) (−4.5 to −6) and blue: deficient (>−6). Bold dashed lines indicate the mean expression level of normal fibres.
BN-PAGE assessing the steady-state levels of fully-assembled Complex I
To further interrogate the IHC results observed in the subset of patients displaying normal NDUFB8 and NDUFS3 immunoreactivity, we performed BN-PAGE analysis to assess the steady-state levels of fully assembled CI (980 kDa) where muscle was available (P18, P19, P21, P22, P23 and P25). P17 was also included as a positive control in this analysis given the IHC assay had shown down-regulated levels of NDUFB8 immunoreactivity. Samples prepared for BN-PAGE analysis retain OXPHOS complexes in their structural and active form, permitting the investigation of any effects on the assembly of the holoenzyme or catalytic activity. Since we had previously measured residual CI activity spectrophotometrically, we only assessed the levels of fully assembled complex I, using complex II assembly as a control.
Analysis of muscle mitochondrial fractions by BN-PAGE revealed a decrease in steady-state levels of fully-assembled CI in P17, P18, P23 and, P24 (Fig. 5) when compared to controls, indicating that the mutations harboured by these patients affect levels of fully-assembled CI. As complex II activity is normal in these patients, we targeted SDHA subunit and used the steady-state levels of assembled complex II (detected by immunoreactivity against the SDHA subunit) as a loading control. By contrast, patients P19, P21 and P22 showed normal levels of fully assembled CI, in line with the IHC assay results, therefore likely indicating that these mutations affect the activity of the complex rather than CI assembly.
Figure 5.
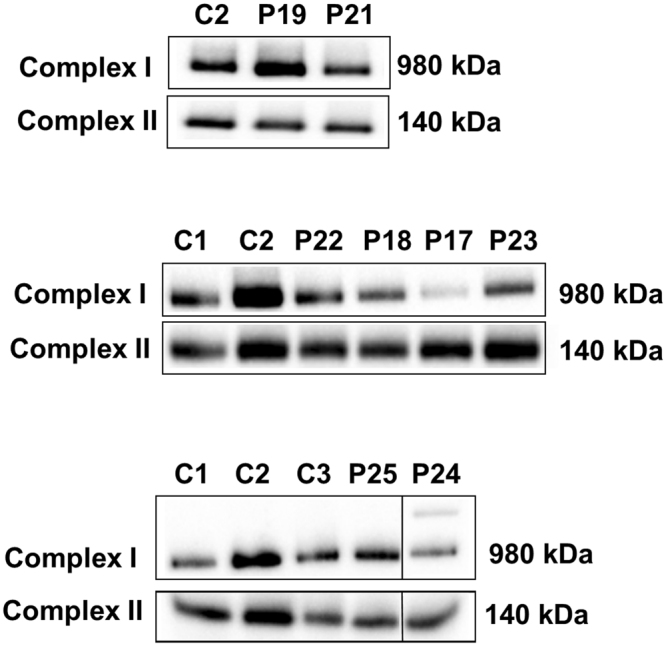
Analysis of Complex I assembly by BN-PAGE. Complex I assembly profiles were analysed using one dimensional blue native polyacrylamide gel electrophoresis (BN-PAGE) (4-16% gradient). Analysis showed a decrease in fully-assembled CI in patients P17, P18, P23 and P24, whilst normal assembly is seen in patients P19, P21, P22 and P25. Complex II was used a loading control. Both OXPHOS complexes were detected by immunoblotting using subunit specific antibodies – NDUFB8 (Complex I) and SDHA (complex II). The original, full length blots are included in the Supplementary Information File (Supplementary Fig. S6).
The level of CI was also normal in P25, correlating with the findings from the IHC (Fig. 5). Furthermore, the residual biochemical CI activity was also 100%, indicative that the m.13513 G > A, p.(Asp393Asn) variant in this case has no major effect on the activity or assembly of CI despite the relatively high level of mtDNA heteroplasmy (Table 1).
Discussion
This study aimed to assess and validate the application of a recently developed quadruple immunofluorescent OXPHOS assay to the diagnosis of patients with isolated mitochondrial CI deficiency. The IHC assay, which detects NDUFB8 as a marker of CI integrity, was successful in detecting CI deficiency in 18 of the 25 patients tested, including all patients with pathogenic variants in nuclear genes encoding CI structural subunits and assembly factors. However, only 3 of the 10 patients harbouring pathogenic mtDNA variants showed a consistent decrease in NDUFB8 immunoreactivity and thus protein abundance, later confirmed using a further commercially-available antibody against NDUFS3, another key component of CI. We chose to optimise the IHC assay using a commercially-available and widely-used antibody against NDUFB8. The subunit is assembled at the mid/late stage of assembly, alongside the mtDNA-encoded core structural subunit, ND5. Although, it would have been preferable to use an antibody targeting a mtDNA-encoded subunit, such as ND1 (interrogating the early stage of CI assembly), there are currently no robust, commercially-available antibodies raised against this or other mtDNA subunits, important to consider if the assay is to be adopted across diagnostic centres.
Using this assay, we could detect CI deficiency in patients that harboured mutations in nuclear genes encoding either the accessory, (P1, P2, P3 = NDUFB3 subunit of the P-D region of the P-module, P4 and P5 = NDUFS4 subunit of the N-module, P6 = NDUFS6 subunit of the N-Module, P7 = NDUFS2 subunit of the Q-module) or core subunits (P8 = NDUFS3 subunit of the Q-module) of CI, in line with the biochemical findings. Of interest, we note that despite patients 1, 2 and 3 harbouring the same genetic variant (p.Trp22Arg in NDUFB3) and similar residual CI activities, the IHC findings showed variable levels of NDUBF8 immunoreactivity (P1 = 79%, P2 = 93% P3 = 39%). However, these findings are consistent with the steady-state levels of the NDUFB8 subunit of CI on western and BN-PAGE originally reported by Alston and colleagues, where it was shown that P1 and P2 (P3 and P2 respectively in ref.16) had decreased protein levels when compared to P3 (P6 in ref.16). Taken together, the collective data highlight the potential of the IHC assay to be a diagnostic tool for such cases and shows that all mutations in Group 1 exerted a similar effect on steady-state levels of fully assembled CI as previously documented16,18,19.
Similarly, the IHC assay was able to diagnose all patients with genetic variants in CI assembly factors. As expected, mutation of these genes led to a decrease in CI assembly and accordingly our IHC assay recorded a marked loss of CI immunoreactivity in many of the fibres analysed; a complete loss of NDUFB8 immunoreactivity (100% CI-deficient fibres) was noted in P13, P14 (ACAD9 variants) and P15 (TMEM126B variant), consistent with previous reports of severe CI deficiency in ACAD9 and TMEM126B mutations20–23. These two assembly factors, alongside with NDUFAF1, ECSIT and TIMMDC1 form the ‘Mitochondrial CI assembly’ (MCIA) complex24 which is associated with the assembly of the P-P submodule (that contains the ND2, ND3, ND4-L and ND6 mitochondrial- encoded subunits). Since depletion of TMEM126B leads to the accumulation of the Q/PP-a (Q module-ND1) intermediate, it seems that NDUFB8 is unstable and as such degraded before the complete assembly of the holoenzyme, explaining why a complete loss of NDUFB8 protein is observed in all fibres24,25.
For patients with pathogenic variants in mtDNA-encoded CI structural subunits, the IHC assay was able to detect CI deficiency in 3 patients (P16, P17 and P20). The m.12425delA MTND5 mutation in P20 leads to a truncated form of the ND5 subunit which has previously been shown to have a severe impact on CI assembly26. This was supported by our IHC results where we observed a complete loss of NDUFB8 immunoreactivity (100% CI-deficient fibres). Similarly, P16 harbours a m.3356 T > C, p.(Met17Thr) MTND1 variant at high levels in muscle associated with a severe defect in CI assembly27, corroborating our IHC results ( > 90% CI-deficient fibres). Finally, P17 harbours a m.10158 T > C, p.(Ser34Pro) MTND3 variant, leading to a marked decrease in NDUFB8 immunoreactivity and steady-state levels of fully-assembled CI; together these results show that the IHC assay confidently detects the deficiency associated with mtDNA variants which severely impact upon CI assembly.
The remaining 7 patients, where normal NDUFB8 (and, where tested, NDUFS3) immunoreactivity was detected, harbour well-characterised pathogenic variants in either the MTND5 (P19, P21, P22, P23, P24 and P25) or MTND3 genes (P18). It has been shown that missense variants in these proteins predominantly affect CI redox activity whereas mutations in the MTND1, MTND2 and MTND6 genes have a more severe impact on complex assembly28–33. We speculate that the patient mutations in which we found normal levels of CI based on both IHC and BN-PAGE analysis (P19: MTND5, m.13514 A > G, p.(Asp393Gly); P21: MTND5, m.13094 T > C, p.(Val253Thr); P22: MTND5, m.13513 G > A, p.(Asp393Asn)) are likely to only affect the catalytic site of the holoenzyme without disturbing CI stability and assembly. In line with this, the m.13514 A > G MTND5 mutation (P19) has been shown to affect the redox activity of CI34–36. The change of amino acid at position 393 (which is part of a putative quinone-reactive site of the enzyme), causes a loss of the putative quinone-reactive site, thereby leading to a decline in CI activity37,38 and validating the IHC findings. Patients harbouring mtDNA variants which only affect CI catalytic activity are likely to show normal IHC profiles.
Furthermore, many studies have also shown that defects in the MTND3 and MTND5 genes display variable biochemical phenotypes; either decreased redox activities as determined by biochemical spectrophotometric assays, decreased levels of fully assembled CI or both18,31,34–36,39–45. This supports the variability found in our results as the m.13513 G > A, p.(Asp393Asn) variant, the most frequently occurring mutation in the MTND5 gene43, can manifest differing effects: either a decrease in fully assembled CI (P23 and P24) or normal levels of assembled CI (P25)18,34,35,38–43. Somewhat surprisingly, P25 (mean m.13513 G > A heteroplasmy level of 63% in muscle) also presented with normal redox activity and CI assembly profiles, likely due to the confounding factor of mutant load. While most pathogenic mtDNA variants cause a biochemical defect only when the levels of mutations mtDNA exceeds 80–90%46, lower thresholds are reported in patients with CI deficiency due to structural subunit mutation35,41. In the case of P25, it could be possible that the pathogenic threshold in this patient is higher, thereby explaining the absence of a biochemical defect despite the presence of relatively high mutation levels and an associated clinical phenotype.
Finally, we detected normal NDUFB8 immunoreactivity but decreased levels of fully-assembled CI in patient P18 harbouring a pathogenic m.10197 G > A, p.(Ala47Thr) MTND3 variant. This has previously been associated with decreased redox activity and diminished levels of fully-assembled CI31,44,45; unfortunately, due to inadequate amounts of skeletal muscle tissue from this patient, we were unable to verify if an effect on activity was also present. Our IHC data indicate, however, that the subunit must be present as the NDUFB8 level was normal. Interrogating NDUFS3 levels also showed normal results, suggesting that our IHC assay is less able to identify CI defects associated with pathogenic variants causing milder defects in CI assembly.
We believe our assay contributes to available diagnostic tools for studying mitochondrial diseases which are typified by extensive biochemical, genetic and clinical heterogeneity. Whilst all patients in our cohort with Mendelian CI defects showed a decrease or complete loss of NDUFB8 immunoreactivity and redox activity, patients with mtDNA variants affecting CI subunits displayed variable biochemical phenotypes. Consequently, we emphasise the importance of complete mitochondrial genome sequencing in the diagnostic work up of patients with suspected mitochondrial disease, to identify possible pathogenic variants associated with clinical symptoms that may result from mutation of mtDNA-encoded structural CI subunits.
In conclusion, our study has accessed and validated the use of a quadruple IHC assay in a diagnostic setting for identifying patients with suspected CI deficiency using a single 10 µm transverse skeletal muscle section. We have been able to directly detect varying levels of NDUFB8 protein abundance at a single cell level, a key advantage of the IHC assay in comparison to current histochemical and histological methodologies. Given these data, we have been able to provide evidence that this assay has clear diagnostic potential for patients with CI deficiency, particularly for those with mutations affecting nuclear-encoded genes, which account for ~75–80% of genetic causes of CI deficiency. Whilst the assay is not sufficiently sensitive to identify a biochemical defect associated with some very well-characterised mtDNA – or rare catalytic defects affecting either mtDNA-encoded or nuclear-encoded CI subunits – we believe that a combination of the quadruple IHC assay, in tandem with full mitochondrial genome sequencing and standard biochemical assays, can be used to investigate likely genetic causes of CI deficiency in patients with mitochondrial disease, especially when muscle biopsy sample sizes are small necessitating the analysis of cryosectioned material.
Methods
Tissue Samples and patient cohort
25 skeletal muscle biopsies (quadriceps muscle) of patients -both paediatrics (<16 years of age) and adults (>16 years of age), who have been investigated for mitochondrial disease and shown to have genetically-confirmed pathogenic variants attributed to isolated CI deficiency were included in this study (see Table 1 for detailed information). Measurements of the enzymatic activities of respiratory chain complexes were undertaken at one of two Mitochondrial Diagnostic laboratories, the NHS Highly Specialised Services located within the Wellcome Centre for Mitochondrial Research at Newcastle University or the Neurometabolic Laboratory at University College London Hospitals (UCLH). Muscle biopsy referral, enzyme measurements and genetic studies were all undertaken as part of the diagnostic work-up of these patients for suspected mitochondrial disease. All samples were obtained and used with informed consent. This study was approved and performed under the ethical guidelines issued by the Newcastle and North Tyneside Local Research Ethics Committees (reference 09/H0906/75) and complied with the Declaration of Helsinki. Control muscle was obtained from patients who were undergoing anterior cruciate ligament (ACL) operations and shown to have normal respiratory enzyme activities.
Quadruple Immunohistochemistry
Quadruple immunohistochemistry was undertaken on 10 µm sections as described by Rocha et al.17. Briefly, sections were incubated with the primary antibodies (Supplementary Table S1) overnight at 4 °C followed by a three wash steps for 10 minutes. This was followed by incubation with secondary antibodies for 2 hours in 4 °C and a further incubation step with streptavidin conjugated with Alexa 647 for 2 hours at 4 °C (Supplementary Table S1). Sections were then mounted using ProLong gold antifade reagent. No-primary antibody control (NPC) sections (incubated only with the anti-laminin primary antibody), were processed alongside each muscle sample.
Image Acquisition
Tiled fluorescent images were taken at x20 magnification using a Zen 2011 (blue edition) software and Zeiss Axio imager MI microscope, equipped with a motorised stage, an AxioCam digital camera and filter cubes detecting wavelengths at 488 nm, 546 nm, 647 nm and 750 nm. Exposure times were set for each channel to avoid over saturation – the same exposure times were then maintained across all cases.
Statistical Analysis
Densitometry measurements
Fluorescent images were analysed using an in-house analysis software coded for by MatLab 2015a. The laminin immunofluorescence (750 channel) was used to detect fibres automatically. Any unwanted surfaces including those over background, fibres with poor morphology or folded were removed. The surfaces allowed for the measurement of mean intensity/optical density (OD) of 488 (COX-1), 546 (Porin) and 647 (NDUFB8) in each individual fibre. The same procedure was repeated for each NPC in order to determine the levels of non-specific binding
Data Analysis
Following image analysis, excel files containing the mean ODs for each case were analysed using an in-house web-based tool as previously described14, along with the statistical calculations within the manuscript Rocha and colleagues17. This analysis determined the Z scores for porin, COX-1 and NDUFB8 based on their expected levels, which were derived using data obtained from control muscle sections. Fibres were classified based on the SD limits into groups of NDUFB8 and COX-1 levels; >−3 = normal, −3 to −4.5 = intermediate positive, −4.5 to −6 = intermediate negative and <−6 = deficient/negative. Fibres were also classified into levels of porin according to Z scores (Z-score: “very low” (porin_Z < −3 SD), “low” (porin_Z between −3 SD and −2 SD), “normal” (porin_Z between −2 SD and +2 SD), “high” (porin_Z between +2 SD and +3 SD) and “very high” (porin_Z above +3 SD)).
Blue Native–Polyacrylamide Gel Electrophoresis (BN-PAGE)
Mitochondrial fractions from both controls and patient muscle were prepared for Blue Native–Polyacrylamide Gel Electrophoresis (BN-PAGE) as previously described in detail by Olahova et al., 2015. The protein concentration of samples was determined using the Pierce BCA protein assay kit and absorption spectrophotometry measured at 562 nm. A minimum of 150ug of muscle mitochondria extracts were loaded on a native 4–16% BisTris gel (Life technologies) and electrophoretically separated in first dimension according to the NOVEX NativePAGETM Bis-Tris Gel system instructions (2 hours, 250 volts). Proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane (Immobilon-P, Millipore Corporation) through wet transfer. Thereafter, the membrane was fixed in 8% acetic acid, washed and blocked with 5% milk for 1 hour at room temperature. Membrane was then subjected to standard immunoblotting analysis of OXPHOS complexes using primary and horseradish peroxidise conjugated secondary antibodies against NDUFB8 (980 kDa CI holoenzyme) and SDHA (140 kDa) (Supplementary Table S2).
Electronic supplementary material
Acknowledgements
This work is supported by the Wellcome Centre for Mitochondrial Research (203105/Z/16/Z), the Medical Research Council (MRC) Centre for Translational Research in Neuromuscular Disease, Mitochondrial Disease Patient Cohort (UK) (G0800674), the Lily Foundation, the UK NIHR Biomedical Research Centre for Ageing and Age-related disease award to the Newcastle upon Tyne Foundation Hospitals NHS Trust, the MRC/EPSRC Molecular Pathology Node and the UK NHS Highly Specialised Service for Rare Mitochondrial Disorders of Adults and Children. CLA is supported by a National Institute for Health Research (NIHR) doctoral fellowship (NIHR-HCS-D12–03–04).
Author Contributions
S.T.A.: acquisition of data, analysis and interpretation of data, statistical analysis, wrote the first draft of the manuscript. C.L.A., L.H., I.P.H., G.F., M.O.: acquisition of data, analysis and interpretation of data. S.H.: acquisition of data, analysis and interpretation of data, statistical analysis. R.M.: acquisition of clinical data. M.C.R.: critical revision of the manuscript for important intellectual content, statistical analysis, study supervision. D.M.T., R.W.T.: study concept and design, critical revision of the manuscript for important intellectual content, study supervision and coordination, obtaining funding.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-14623-2.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Swalwell H, et al. Respiratory chain complex I deficiency caused by mitochondrial DNA mutations. European Journal of Human Genetics. 2011;19:769–775. doi: 10.1038/ejhg.2011.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu J, Vinothkumar KR, Hirst J. Structure of mammalian respiratory complex. I.Nature. 2016;536:354–358. doi: 10.1038/nature19095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mimaki M, Wang X, McKenzie M, Thorburn DR, Ryan MT. Understanding mitochondrial complex I assembly in health and disease. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2012;1817:851–862. doi: 10.1016/j.bbabio.2011.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Sánchez-Caballero L, Guerrero-Castillo S, Nijtmans L. Unraveling the complexity of mitochondrial complex I assembly: A dynamic process. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2016;1857:980–990. doi: 10.1016/j.bbabio.2016.03.031. [DOI] [PubMed] [Google Scholar]
- 5.Guerrero-Castillo S, et al. The Assembly Pathway of Mitochondrial Respiratory Chain Complex I. Cell metabolism. 2017;25:128–139. doi: 10.1016/j.cmet.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Stroud DA, et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123–126. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- 7.Sazanov LA. A giant molecular proton pump: structure and mechanism of respiratory complex I. Nat Rev Mol Cell Biol. 2015;16:375–388. doi: 10.1038/nrm3997. [DOI] [PubMed] [Google Scholar]
- 8.Fassone E, Rahman S. Complex I deficiency: clinical features, biochemistry and molecular genetics. J Med Genet. 2012;49:578–590. doi: 10.1136/jmedgenet-2012-101159. [DOI] [PubMed] [Google Scholar]
- 9.McKenzie M, Ryan MT. Assembly factors of human mitochondrial complex I and their defects in disease. IUBMB life. 2010;62:497–502. doi: 10.1002/iub.335. [DOI] [PubMed] [Google Scholar]
- 10.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Molecular and cellular biology. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonicka H, et al. Identification and characterization of a common set of complex I assembly intermediates in mitochondria from patients with complex I deficiency. The Journal of biological chemistry. 2003;278:43081–43088. doi: 10.1074/jbc.M304998200. [DOI] [PubMed] [Google Scholar]
- 12.Vogel RO, Smeitink JAM, Nijtmans LG. J. Human mitochondrial complex I assembly: A dynamic and versatile process. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2007;1767:1215–1227. doi: 10.1016/j.bbabio.2007.07.008. [DOI] [PubMed] [Google Scholar]
- 13.Leman G, et al. Assembly defects induce oxidative stress in inherited mitochondrial complex I deficiency. The International Journal of Biochemistry & Cell Biology. 2015;65:91–103. doi: 10.1016/j.biocel.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 14.Alston CL, Rocha MC, Lax NZ, Turnbull DM, Taylor RW. The genetics and pathology of mitochondrial disease. The Journal of pathology. 2017;241:236–250. doi: 10.1002/path.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Craven, L., Alston, C. L., Taylor, R. W. & Turnbull, D. M. Recent Advances in Mitochondrial Disease. Annual Review of Genomics and Human Genetics. 18, 257–275 (2017). [DOI] [PubMed]
- 16.Alston CL, et al. A recurrent mitochondrial p.Trp22Arg <em>NDUFB3</em> variant causes a distinctive facial appearance, short stature and a mild biochemical and clinical phenotype. Journal of Medical Genetics. 2016;53:634. doi: 10.1136/jmedgenet-2015-103576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rocha MC, et al. A novel immunofluorescent assay to investigate oxidative phosphorylation deficiency in mitochondrial myopathy: understanding mechanisms and improving diagnosis. Scientific reports. 2015;5:15037. doi: 10.1038/srep15037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petruzzella V, et al. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Human molecular genetics. 2001;10:529–535. doi: 10.1093/hmg/10.5.529. [DOI] [PubMed] [Google Scholar]
- 19.Kirby DM, et al. NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. The Journal of clinical investigation. 2004;114:837–845. doi: 10.1172/JCI20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alston CL, et al. Biallelic Mutations in TMEM126B Cause Severe Complex I Deficiency with a Variable Clinical Phenotype. American journal of human genetics. 2016;99:217–227. doi: 10.1016/j.ajhg.2016.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nouws J, et al. Acyl-CoA dehydrogenase 9 is required for the biogenesis of oxidative phosphorylation complex I. Cell metabolism. 2010;12:283–294. doi: 10.1016/j.cmet.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 22.Nouws J, te Brinke H, Nijtmans LG, Houten SM. ACAD9, a complex I assembly factor with a moonlighting function in fatty acid oxidation deficiencies. Human molecular genetics. 2014;23:1311–1319. doi: 10.1093/hmg/ddt521. [DOI] [PubMed] [Google Scholar]
- 23.Haack TB, et al. Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet. 2010;42:1131–1134. doi: 10.1038/ng.706. [DOI] [PubMed] [Google Scholar]
- 24.Heide H, et al. Complexome profiling identifies TMEM126B as a component of the mitochondrial complex I assembly complex. Cell metabolism. 2012;16:538–549. doi: 10.1016/j.cmet.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Guarani V, et al. TIMMDC1/C3orf1 functions as a membrane-embedded mitochondrial complex I assembly factor through association with the MCIA complex. Molecular and cellular biology. 2014;34:847–861. doi: 10.1128/MCB.01551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alston CL, et al. A novel mitochondrial MTND5 frameshift mutation causing isolated complex I deficiency, renal failure and myopathy. Neuromuscular disorders: NMD. 2010;20:131–135. doi: 10.1016/j.nmd.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 27.Gorman GS, et al. Novel MTND1 mutations cause isolated exercise intolerance, complex I deficiency and increased assembly factor expression. Clinical Science (London, England: 1979) 2015;128:895–904. doi: 10.1042/CS20140705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lazarou M, Thorburn DR, Ryan MT, McKenzie M. Assembly of mitochondrial complex I and defects in disease. Biochimica et biophysica acta. 2009;1793:78–88. doi: 10.1016/j.bbamcr.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 29.Perales-Clemente E, et al. Five entry points of the mitochondrially encoded subunits in mammalian complex I assembly. Molecular and cellular biology. 2010;30:3038–3047. doi: 10.1128/MCB.00025-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews B, Carroll J, Ding S, Fearnley IM, Walker JE. Assembly factors for the membrane arm of human complex I. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:18934–18939. doi: 10.1073/pnas.1319247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirby DM, et al. Mutations of the mitochondrial ND1 gene as a cause of MELAS. J Med Genet. 2004;41:784–789. doi: 10.1136/jmg.2004.020537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugalde C, et al. Impaired complex I assembly in a Leigh syndrome patient with a novel missense mutation in the ND6 gene. Annals of neurology. 2003;54:665–669. doi: 10.1002/ana.10734. [DOI] [PubMed] [Google Scholar]
- 33.McFarland R, et al. De novo mutations in the mitochondrial ND3 gene as a cause of infantile mitochondrial encephalopathy and complex I deficiency. Annals of neurology. 2004;55:58–64. doi: 10.1002/ana.10787. [DOI] [PubMed] [Google Scholar]
- 34.Bugiani M, et al. Clinical and molecular findings in children with complex I deficiency. Biochimica et biophysica acta. 2004;1659:136–147. doi: 10.1016/j.bbabio.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 35.Corona P, et al. A novel mtDNA mutation in the ND5 subunit of complex I in two MELAS patients. Annals of neurology. 2001;49:106–110. doi: 10.1002/1531-8249(200101)49:1<106::AID-ANA16>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 36.Lebon S, et al. Recurrent de novo mitochondrial DNA mutations in respiratory chain deficiency. J Med Genet. 2003;40:896–899. doi: 10.1136/jmg.40.12.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fisher N, Rich PR. A motif for quinone binding sites in respiratory and photosynthetic systems. Journal of molecular biology. 2000;296:1153–1162. doi: 10.1006/jmbi.2000.3509. [DOI] [PubMed] [Google Scholar]
- 38.Blok MJ, et al. Mutations in the ND5 subunit of complex I of the mitochondrial DNA are a frequent cause of oxidative phosphorylation disease. Journal of Medical Genetics. 2007;44:e74–e74. doi: 10.1136/jmg.2006.045716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chol M, et al. The mitochondrial DNA G13513A MELAS mutation in the NADH dehydrogenase 5 gene is a frequent cause of Leigh-like syndrome with isolated complex I deficiency. J Med Genet. 2003;40:188–191. doi: 10.1136/jmg.40.3.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanna MG, Nelson IP, Morgan-Hughes JA, Wood NW. MELAS: a new disease associated mitochondrial DNA mutation and evidence for further genetic heterogeneity. Journal of neurology, neurosurgery, and psychiatry. 1998;65:512–517. doi: 10.1136/jnnp.65.4.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kirby DM, et al. Low mutant load of mitochondrial DNA G13513A mutation can cause Leigh’s disease. Annals of neurology. 2003;54:473–478. doi: 10.1002/ana.10687. [DOI] [PubMed] [Google Scholar]
- 42.Santorelli FM, et al. Identification of a novel mutation in the mtDNA ND5 gene associated with MELAS. Biochemical and biophysical research communications. 1997;238:326–328. doi: 10.1006/bbrc.1997.7167. [DOI] [PubMed] [Google Scholar]
- 43.Shanske S, et al. The G13513A mutation in the ND5 gene of mitochondrial DNA as a common cause of MELAS or Leigh syndrome: evidence from 12 cases. Archives of neurology. 2008;65:368–372. doi: 10.1001/archneurol.2007.67. [DOI] [PubMed] [Google Scholar]
- 44.Chae JH, et al. A novel ND3 mitochondrial DNA mutation in three Korean children with basal ganglia lesions and complex I deficiency. Pediatric research. 2007;61:622–624. doi: 10.1203/pdr.0b013e3180459f2d. [DOI] [PubMed] [Google Scholar]
- 45.Sarzi E, et al. A novel recurrent mitochondrial DNA mutation in ND3 gene is associated with isolated complex I deficiency causing Leigh syndrome and dystonia.American journal of medical genetics. Part A. 2007;143A:33–41. doi: 10.1002/ajmg.a.31565. [DOI] [PubMed] [Google Scholar]
- 46.Mariotti C, et al. Defective respiratory capacity and mitochondrial protein synthesis in transformant cybrids harboring the tRNA(Leu(UUR)) mutation associated with maternally inherited myopathy and cardiomyopathy. The Journal of clinical investigation. 1994;93:1102–1107. doi: 10.1172/JCI117061. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.