Abstract
Semi-invariant natural killer T (NKT) cells are innate-like lymphocytes with immunoregulatory properties. NKT cell survival during development requires signal processing by activated RelA/NF-κB. Nonetheless, the upstream signal(s) integrated by NF-κB in developing NKT cells remains incompletely defined. We show that the introgression of Bcl-xL-coding Bcl2l1 transgene into NF-κB signalling-deficient IκBΔN transgenic mouse rescues NKT cell development and differentiation in this mouse model. We reasoned that NF-κB activation was protecting developing NKT cells from death signals emanating either from high affinity agonist recognition by the T cell receptor (TCR) or from a death receptor, such as tumor necrosis factor receptor 1 (TNFR1) or Fas. Surprisingly, the single and combined deficiency in PKC-θ or CARMA-1—the two signal transducers at the NKT TCR proximal signalling node—only partially recapitulated the NKT cell deficiency observed in IκBΔN tg mouse. Accordingly, introgression of the Bcl2l1 transgene into PKC-θ null mouse failed to rescue NKT cell development. Instead, TNFR1-deficiency, but not the Fas-deficiency, rescued NKT cell development in IκBΔN tg mice. Consistent with this finding, treatment of thymocytes with an antagonist of the inhibitor of κB kinase —which blocks downstream NF-κB activation— sensitized NKT cells to TNF-α-induced cell death in vitro. Hence, we conclude that signal integration by NF-κB protects developing NKT cells from death signals emanating from TNFR1, but not from the NKT TCR or Fas.
Introduction
Natural killer T (NKT) cells are thymus-derived, innate-like lymphocytes that resemble both natural killer and T cells in phenotype and function. Based on the T cell receptor (TCR) α-chain expressed, NKT cells are broadly divided into two types: Type I NKT cells, the majority group, expresses an invariant TCR α-chain encoded by a conserved rearrangement between mouse Vα14 and Jα18 gene segments. This invariant α-chain predominantly pairs with mouse Vβ8.2 (and with Vβ7, 6, 2, and 14) TCR β-chain to express a semi-invariant NKT TCR. Type II NKT cells, the minority group, expresses more diverse TCR α- and β-chains. This report focuses on the semi-invariant NKT (hereinafter NKT) cells. In this regard, it is noteworthy that mouse, rat, human, and other mammalian species also develop functional NKT cells that express the orthologous semi-invariant TCR. Hence, findings in mice will have implications for understanding NKT cell development and function in other mammalian species including humans.
NKT cells control immune responses to microbial, allograft and self antigens. NKT cell functions are controlled by lipid ligand-presenting CD1d molecules. Consequently, NKT cells are capable of sensing cellular lipid homeostasis to maintain cardiovascular and metabolic health1. These seemingly divergent functions are mediated by the ability of NKT cells to rapidly produce pro-inflammatory and immunoregulatory cytokines, which permits these cells to interact with and transactivate cells of the innate and adaptive immune systems2. These properties of NKT cells are acquired during thymic development. Hence, insights into the mechanisms by which NKT cells develop, attain functional competence and are maintained can help understand the role these cells play in health and disease.
The unique phenotypic and functional features of NKT cells —especially their capacity to secrete cytokines promptly after thymic emigration3 —suggested that these cells might follow a distinct ontogenetic path. Commitment to the NKT cell lineage occurs at the CD4- and CD8-positive (DP) thymocyte stage4–7 whence self-lipid(s) bound CD1d/NKT TCR8–11 and homotypic/heterotypic SLAM-SLAM12,13 interactions lead to downstream PKCθ-NFκB14, NFAT-Egr215–17, and SAP-Fyn12,13,18 activation, which turns-on a unique transcriptional programming mediated by the Zbtb16-encoded master transcription factor PLZF (pro-myelocytic leukaemia zinc finger). PLZF controls multiple molecular events distinguishing NKT cells from conventional T cells19,20. NK1.1-negative NKT cell precursors undergo several rounds of cell division21,22, upon which some remain within the thymus whereas the majority emigrate and populate the peripheral lymphoid organs23. Both the thymic and peripheral NK1.1-negative NKT cells undergo further maturation and become NK1.1-positive NKT cells. During this maturation process, NKT cells differentiate into subsets and acquire distinct effector behaviours24–29,23.
Several research groups30–36, including ours14,37, have reported that signal integration by NF-κB was essential for NKT cell development, survival, and function. RelA and RelB were shown to confer cell autonomous and cell extrinsic activities of NF-κB, respectively30–32. The cell-extrinsic NF-κB activity was required for IL-15 induction32, a cytokine essential for survival as well as for terminal NK1.1-negative to NK1.1+ NKT cell differentiation and effector maturation38–47. Likewise, the cell-intrinsic NF-κB activity was essential for the survival of NK1.1-negative NKT cells, as introgression of Bcl2l1 transgene under the control of the Lck proximal promotor, which thereby encodes Bcl-xL in thymocytes, rescued NKT cell numbers but not function14. Despite this understanding of the role of NF-κB in NKT cell development, a few key questions have remained unanswered: (a) Is signal integration by NF-κB essential for the survival of both immature NK1.1-negative and mature NK1.1+ NKT cells?; (b) Does NF-κB activation protect against TCR-induced NKT cell death?; and (c) What other death receptor(s) trigger NKT cell death during ontogeny? In seeking answers to these questions, we report herein that signal integration by NF-κB is essential for the survival of both immature NK1.1-negative and mature NK1.1+ NKT cells and to protect these cells from death signals induced by tumor necrosis factor (TNF)-α and emanating from TNF receptor superfamily 1a (TNFR1), but not the NKT TCR or Fas.
Results
Bcl-xL overexpression rescues immature and mature NKT cell development in NF-κB signalling deficient mice
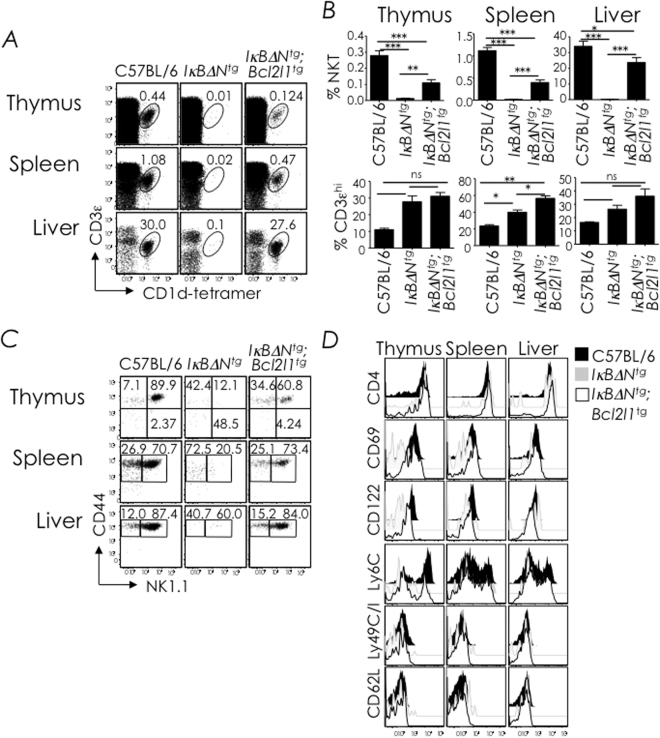
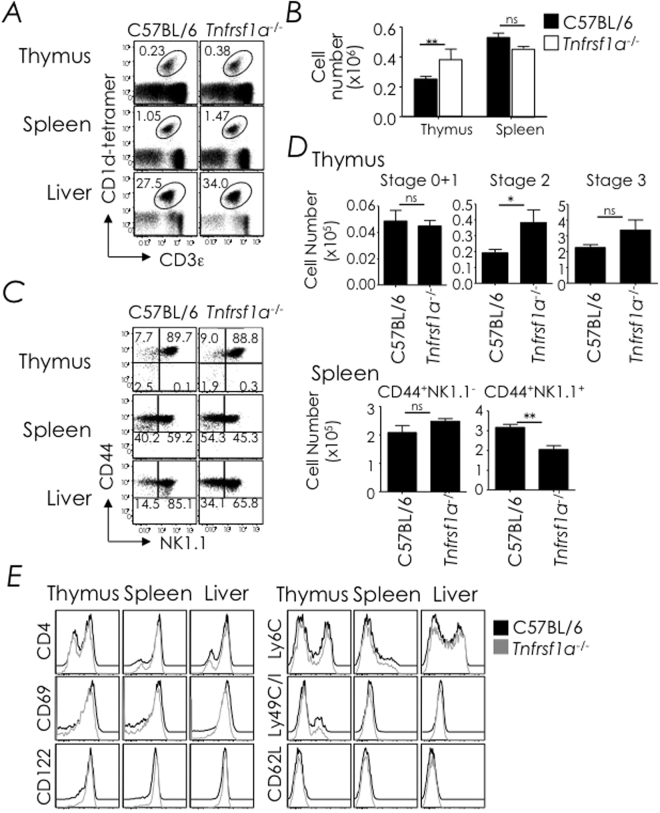
We previously showed that Bcl-xL overexpression rescued NKT cell development in mice expressing IκBΔN, a dominant-negative inhibitor of NF-κB activation37. Nonetheless, NKT cells that developed in these animals failed to respond to stimulation with αGalCer, a potent CD1d-restricted NKT cell agonist14. This lack of response to the agonist could be due to immaturity of the NKT cells that developed in Iκ BΔN tg;Bcl2l1 tg mice, their inability to process the TCR activation signals, or both. Because the genetic rescue experiment was performed in BALB/c mice, which do not express the Klrb1c allele that codes for NK1.1, maturation of NKT cells from NK1.1-negative to NK1.1+ NKT cells could not be determined37. Hence, we generated Iκ BΔN tg;Bcl2l1 tg mice in the C57BL/6 background (B6), which express the NK1.1-coding Klrb1c allele detectable with the PK136 mAb. Phenotyping of NKT cells from C57BL/6 and B6-Iκ BΔN tg;Bcl2l1 tg mice revealed that both NK1.1-negative and NK1.1+ NKT cells developed in B6-Iκ BΔN tg;Bcl2l1 tg mice (Fig. 1A–C). Further, the NKT cells of these mice showed phenotypic maturation as judged by expression of CD4, CD69, CD122 (IL-2/15 Rβ chain), Ly49 and Ly6C as well as the loss of CD62L expression similar to NKT cells that develop in C57BL/6 mice (Fig. 1D). Hence, both NK1.1-negative immature and NK1.1+ mature NKT cells escape death by thymocyte-specific Bcl-xL overexpression in NF-κB signalling-deficient mice.
Figure 1.
Bcl-xL overexpression rescues NKT cell development in NFκB-signalling deficient mice. (A) Thymic, splenic and hepatic NKT cells from C57BL/6 (n = 4), B6-IκBαΔN tg (n = 8) and B6-IκBαΔN tg;Bcl2l1 tg (n = 4) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes within each organ. (B) % of NKT cells (top) and CD3εhi cells in thymus spleen and liver, Data are mean ± standard error (sem) from 3 independent experiments. (C) NKT cell developmental stages were identified as CD44−NK1.1− stage 0 + 1, CD44+NK1.1− stage 2, or CD44+NK1.1+ stage 3 in the thymus or as NK1.1−tetramer+ or NK1.1+tetramer+ splenocytes and liver MNCs. Numbers are % cells among total NKT cells. n, as in A. (D) Expression of CD4, CD69, CD122, Ly6C, Ly49C/I, and CD62L by thymic, splenic, and hepatic NKT cells was determined by flow cytometry after surface staining with specific mAb. Data are representative of 2 independent experiments. n, as in A. ns, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.001, ***P ≤ 0.0001.
PKC-θ and CARMA1-deficient mice poorly develop NKT cells
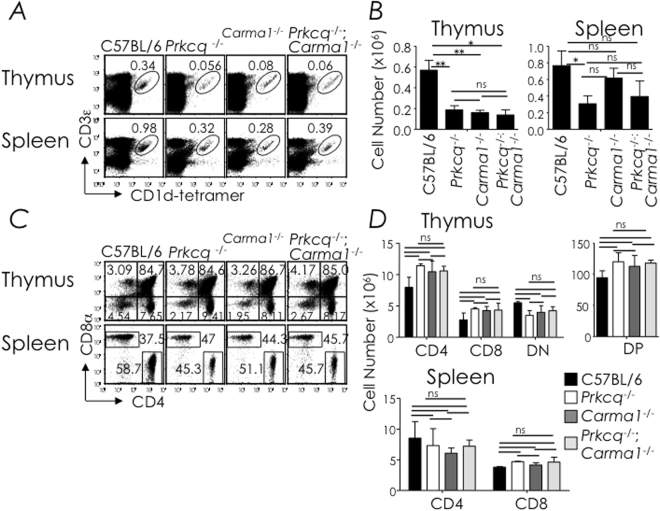
NF-κB activation integrates upstream signals from cell surface receptors, including both antigen receptors and death receptors. We initially focused on the antigen receptor because NKT TCRs are selected by agonistic self ligand(s). As well, we previously reported that a fraction of NK1.1-negative NKT cells that developed in Iκ BΔN tg mice expressed Nr4a1-encoded Nur7737. Nur77 is a TCR activation-induced pro-apoptotic transcription factor48, which was shown to be expressed in stage 0 precursor NKT cells49 and upon NKT TCR stimulation by αGalCer or bacterial glycosphingolipids50. Therefore, we determined whether the NKT TCR was the source of the death signal in developing NKT cells. Toward this goal, we determined the role of two NKT TCR proximal signal nodes, PKC-θ (encoded by Prkcq) and CARMA1. Additionally, the SLAM-SAP-FynT signalling axis partially integrates with PKC-θ51. As reported by us previously14, PKCθ-deficiency only partially impacts NKT cell development. This partial impact on NKT cell development was also reflected by CARMA1-deficiency and by combined PKC-θ and CARMA1 deficiencies (Fig. 2). Because NKT cell phenotype in PKCθ-deficient mice were reported previously (see Fig. 3 as well) and because NKT cell development in CARMA1-deficient mice reflected that of PKCθ-deficiency, NKT cell phenotype in neither type of mouse was further investigated. The results so far suggest that the membrane proximal NKT TCR-PKCθ-CARMA1 and/or SLAM-SAP-FynT-PKCθ-CARMA1 axes are unlikely required for transducing the death signal in developing NKT cells.
Figure 2.
CARMA1-deficiency only partially disrupts NKT cell development. (A) Thymic and splenic NKT cells from C57BL/6 (n = 9), Prkcq −/− (n = 9), Carma1 −/− (n = 5) and Prkcq −/−; Carma1 −/− (n = 3) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes within each organ. (B) Absolute cell numbers were calculated from % NKT cell and total cell count. n, as in A. (C) Thymic and splenic CD4+ and CD8+ T cells from C57BL/6 (n = 3), Prkcq −/− (n = 3), Carma1 −/− (n = 3) and Prkcq −/−;Carma1 −/− (n = 3) mice were identified as CD4+ or CD8+ cells within electronically gated thymocytes or B220lo CD3ε+ splenocytes. Numbers are % CD4+ or CD8+ T cells among total leukocytes. Data are mean ± sem (standard error of the mean) from 3 experiments; n, as in C. (D) Absolute cell numbers were calculated from % CD4, CD8, CD4+8+ DP or CD4−8− DN and total cell count. n, as in C. Data are mean ± sem from 3 (A,B) or 2 (C,D) independent experiments. ns, not significant (P > 0.05); *P ≤ 0.05, **P ≤ 0.001.
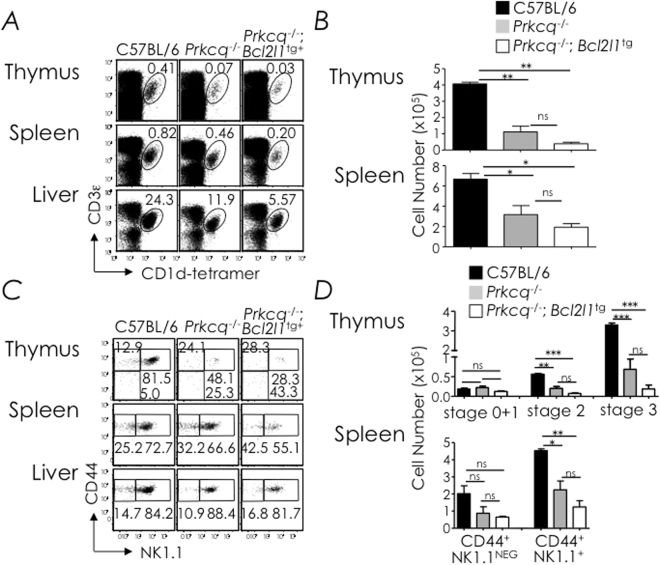
Figure 3.
Bcl-xL overexpression in PKCθ-deficient mice fails to rescue NKT cell development. (A) Thymic, splenic and hepatic NKT cells from C57BL/6 (n = 5), Prkcq −/− (n = 5) and Prkcq −/−;Bcl2l1 tg (n = 7) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes within each organ. (B) Absolute cell numbers were calculated from % NKT and total cell count. n, as in A. (C) NKT cell developmental stages in the thymus, spleen and liver were identified as in Fig. 1A. Numbers are % cells among total NKT cells. n, as in A. (D) Absolute numbers of thymic stages 0 + 1, 2 and 3 NKT cells; they were calculated from the absolute NKT cell numbers and % stage 0 + 1, stage 2, or stage 3 cells in C. n, as in A. Data are mean ± sem from 3 independent experiments. ns, not significant (P > 0.05), *P ≤ 0.05, **P ≤ 0.001, **P ≤ 0.0001.
Bcl-xL overexpression does not rescue NKT cell development in PKCθ-deficient mice
To provide further evidence that PKCθ-CARMA1 activation does not induce a key death signal in NKT cell development, we tested whether Bcl-x L transgene introgression into PKCθ mutant mice rescues NKT cell development. Hence, we generated Prkcq −/−;Bcl2l1 tg mice and analysed NKT cell development in them. Surprisingly, Prkcq −/−;Bcl2l1 tg mice poorly developed NKT cells akin to Prkcq −/− and Carma1 −/− mice (Figs 2 and 3). Together, the data indicated that the NKT TCR and its downstream signalling modules PKC-θ and CARMA1 unlikely transduce death signals within developing NKT cells. Or alternatively, it is quite possible that Bcl-xL overexpression was unable to rescue the loss of NKT cells from the extrinsic apoptotic pathway that may be induced by the NKT TCR.
NKT cells constitutively express several TNF receptor superfamily members
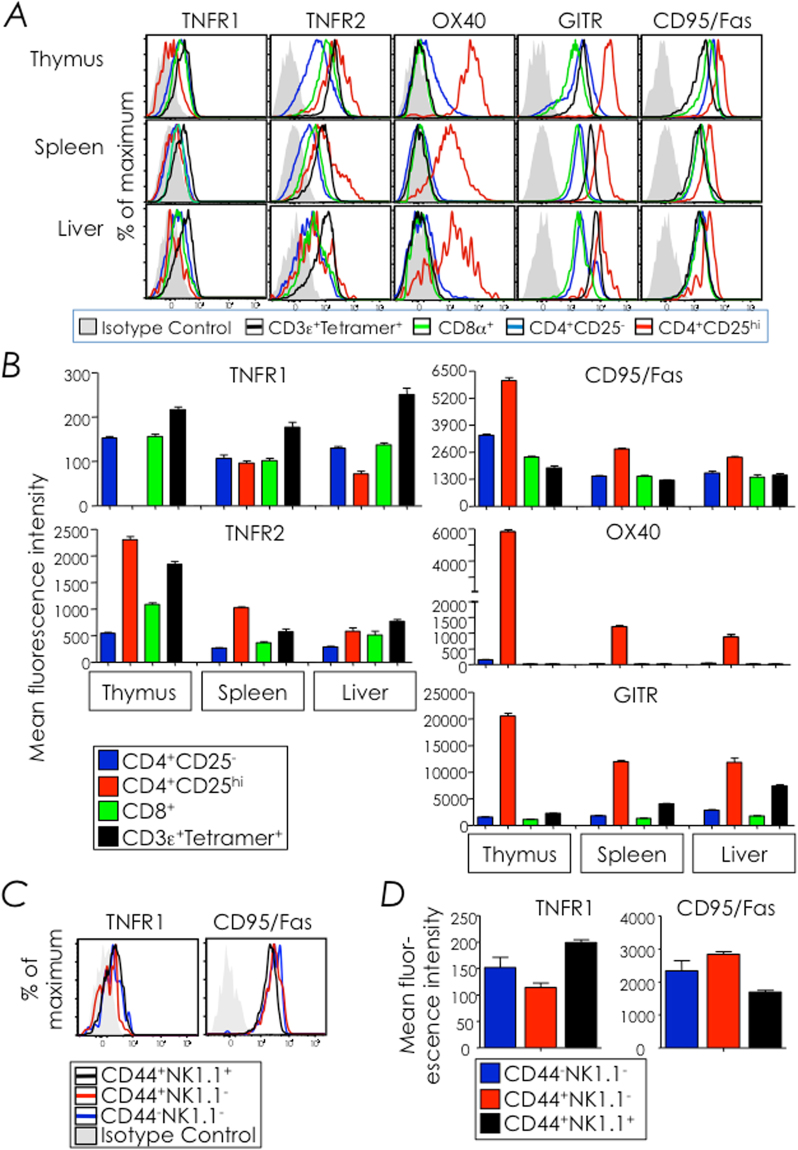
We next turned to death domain-containing cell surface receptors as the source of death signalling in developing NKT cells. To lay focus on a particular TNFR superfamily (TNFRSF) member, we first defined TNFR1 (TNFRSF1A/CD120a), TNFR2 (TNFRSF1B/CD120b), Fas (TNFRSF6/CD95), OX40 (TNFRSF4/CD134), and GITR (TNFRSF18/CD357) expression by thymic, splenic and hepatic NKT cells. Thymic and splenic NKT cells constitutively expressed TNFR1, TNFR2, Fas, and GITR but not OX40. Of these, TNFR1 expression by NKT cells was highest amongst thymocyte subsets tested (Fig. 4A). We then focused on TNFR1 and Fas expression on the three developmental stages (stages 0 + 1, 2 and 3) of thymic NKT cells because these two TNFRSF members contain a cytoplasmic death domain. Both TNFR1 and Fas were equally expressed by all three thymic NKT cell developmental stages (Fig. 4B). Thus, NKT cells indiscriminately express the two tested TNFRSF members that contain death domains.
Figure 4.
NKT cells constitutively express TNFR1 and Fas (CD95). (A,B) The expression of TNFR1, TNFR2, OX-40, GITR and Fas (CD95) by thymic, splenic, and hepatic NKT cells, CD4+CD25+, CD4+CD25hi and CD8+ cells was determined by flow cytometry after surface staining with specific mAb and corresponding isotype control mAb. NKT cells from C57BL/6 (n = 9) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. CD4+ and CD8+ T cells from mice were identified as CD4+ or CD8+ cells within electronically gated thymocytes or B220lo CD3ε+tetramerneg splenocytes and hepatic leukocytes. CD4+ T cells were further gated to identify CD4+CD25− and CD4+CD25hi cells. (A) Overlays show expression of the tested TNFRSF member on different T cell subsets. Filled grey histogram are representative isotype controls; note that all T cell subsets had nearly similar isotype control staining and, therefore, the staining of isotype control in CD4+CD25− is overlaid here. (B) Difference in mean fluorescence intensity (ΔMFI) between specific mAb and corresponding isotype control staining on each T cell subset is plotted as mean ΔMFI ± sem; n and replicates as in A. (C,D) TNFR1 and Fas expression on thymic stage 0 + 1, stage 2 and stage 3 NKT cells was determined by flow cytometry after surface staining as in A. Thymic NKT cell developmental stages were identified as in Fig. 1. (C) Filled grey histograms are isotype controls; note that isotype control for stage 3 NKT cells are shown here as stage 1 and stage 2 NKT cells showed similar isotype control staining intensity. Representative of 3 independent experiments; n = 3 mice/experiment. (D) A plot of mean ΔMFI ± sem, computed as in B; n and replicates as in C.
Fas-deficiency fails to recue NKT cell development in NF-κB signalling-deficient mice
We next determined whether the expression of TNFRSF members containing a death domain altered NKT cell development. Because Fas expression was high, the development of NKT cells in B6-lpr/lpr mice, which express a nonfunctional mutant form of Fas molecule, was determined first. Thymic, splenic and hepatic NKT cells developed in the lpr/lpr mutant mice in frequencies similar to wt/lpr mice (Fig. 5). Further, the introgression of the lpr/lpr mutation into NF-κB signalling deficient IκBΔN tg mice (B6-lpr/lpr;Iκ BΔN tg) mice did not rescue NKT cell development in these mice because very few if any NKT cells were observed in the thymus, spleen or liver of B6-lpr/lpr;Iκ BΔN tg mice (Fig. 5). From these data, we concluded that Fas unlikely transmits the death signal counteracted by NF-κB activation.
Figure 5.
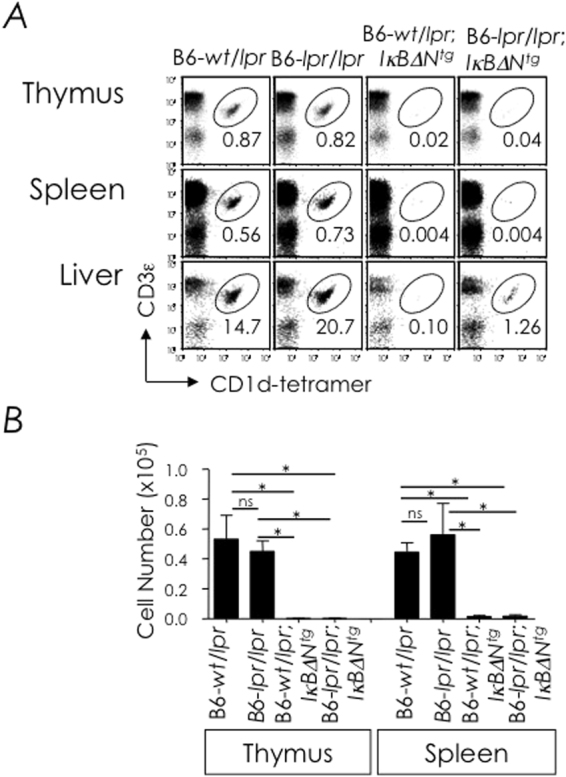
Fas-deficiency fails to rescue NKT cell development in IκBαΔNtg. (A) Thymic, splenic and hepatic NKT cells from B6-wt/lpr (n = 2), B6-lpr/lpr (n = 2), B6-wt/lpr;IκBαΔN tg (n = 2) and B6-lpr/lpr;IκBαΔN tg (n = 2) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes in each organ. (B) Absolute cell numbers were calculated from % NKT cell and total cell count. n, as in A. Data are mean ± sem from two independent experiments. ns, not significant (P > 0.05), *P ≤ 0.05.
Thymic NKT cells develop unhindered in TNFR1-deficient mice
In a similar experiment, we next determined whether TNFR1-deficiency impacted NKT cell development. The data revealed that NKT cell development proceeded unhindered in the thymus of TNFR1-deficient mice and these cells emigrated to the spleen and liver as in wild type mice (Fig. 6A,B). We noticed that the absolute NKT cell number was increased by 25–30% in the thymus, which was statistically significant, but only modestly, albeit not significantly, reduced in the spleen (Fig. 6B). Further, NKT cells proceeded to phenotypic maturity in TNFR1-deficient mice with regards to CD44 and NK1.1 expression as well as CD4, CD69, CD122 (IL-2/15 Rβ chain), Ly6C, Ly49 and CD62L (L-selectin) expression (Fig. 6C–E). Notably, the number of splenic NK1.1+ NKT cells was reduced in a statistically significant manner (Fig. 6D). Thus, TNFR1-deficiency marginally impacted thymic NKT cell development, whilst TNFR1 signalling was partially required for splenic NK1.1+ NKT cell maturation.
Figure 6.
TNFR1-deficiency does not impact NKT cell development. (A) Thymic, splenic and hepatic NKT cells from C57BL/6 (n = 5) and B6.129-Tnfrsf1a −/− (n = 5) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes within each organ. (B) Absolute cell numbers were calculated from % NKT and total cell count. Data are mean ± sem from 3 independent experiments; n, as in A. (C) Thymic NKT cell developmental stages were identified as in Fig. 1 or NKT cells were identified as splenic and hepatic NK1.1-tetramer+ or NK1.1+tetramer+ cells. Numbers are % cells among total NKT cells. n, as in A. (D) Absolute numbers of thymic (upper panel) stage 0 + 1, stage 2 and stage 3 NKT cells and absolute numbers of splenic (lower panel) CD44+NK1.1NEG and CD44+NK1.1+ NKT cells; they were calculated from the absolute NKT cell numbers in B and % stage 0 + 1, stage 2 and stage 3 cells or CD44+NK1.1NEG and CD44+NK1.1+ cells in C. n, as in A. Data are mean ± sem from 3 independent experiments. (E) Expression of CD4, CD69, CD122, Ly6C, Ly49C/I, and CD62L by thymic, splenic, and hepatic NKT cells was determined by flow cytometry after surface staining with specific mAb. Data are representative of 2 independent experiments. n, as in A. ns, not significant (P > 0.05), *P ≤ 0.05, **P ≤ 0.001.
NF-κB activation protects developing NKT cells from TNFR1-induced death
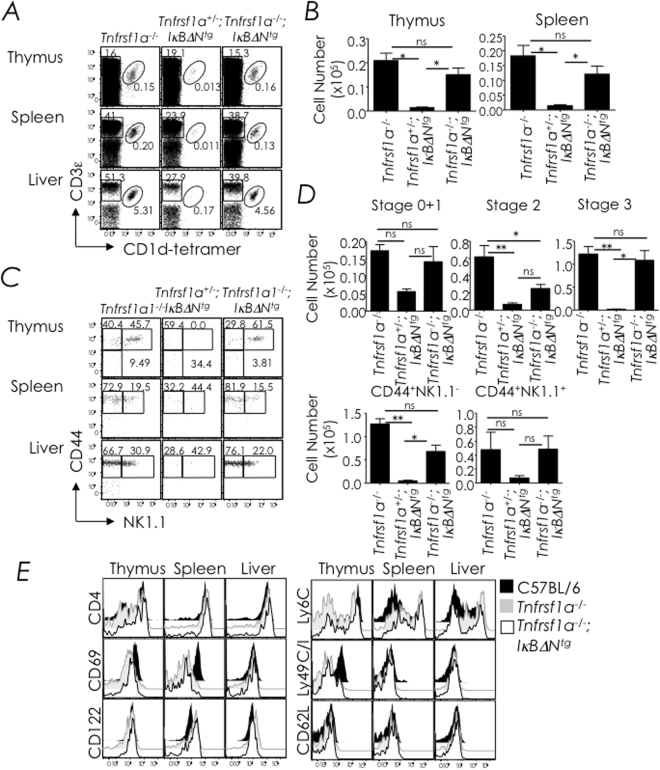
Deficiency in the inhibitor of κB kinase (IKK)-β, which is essential for NF-κB activation, results in poor lymphopoiesis due to enhanced sensitivity to TNF-α52. Consequently, normal thymopoiesis and conventional CD4 and CD8 T cell development ensues when mice are doubly deficient in TNFR1 and IKK-β52 suggesting that TNF-α might be a thymic inducer of NKT cell death. Therefore, we determined whether TNFR1-null mutation introgressed into IκBαΔN tg mice rescues NKT cells from death. For this purpose, we bred Tnfrsf1a −/− mice with IκBαΔNtg mice and the resulting progenies were analysed for NKT cell development.
The data revealed that NKT cell development was rescued in Tnfrsf1a −/−;IκBαΔN tg mice when compared with IκBαΔN tg mice (Fig. 7A,B). Even though Tnfrsf1a −/−;IκBαΔN tg NKT cells matured to the CD44hiNK1.1+ stage 3 (Fig. 7C,D), they expressed lower levels of Ly49C/I (Fig. 7E) while expressing normal levels of other developmentally regulated markers (e.g., CD4, CD62L, CD122 and Ly6C) when compared to thymic Tnfrsf1a −/−; IκBαΔN tg NKT cells (Fig. 7E). Because introgression of the lpr/lpr mutation into IκBαΔN tg mice did not rescue NKT cells from death (Fig. 5), we conclude that the effect of TNFR1-deficiency on NKT cell development in Tnfrsf1a −/−;IκBαΔN tg mice is specific. The data also demonstrate that NF-κB activation controls NKT cell homeostasis by overcoming TNFR1-induced death.
Figure 7.
TNFR1-deficiency rescues NKT cell development in IκBαΔN tg mice. (A) Thymic and splenic NKT cells from Tnfrsf1a −/− (n = 3), Tnfrsf1a +/−;IκBαΔN tg (n = 4) and Tnfrsf1a −/−;IκBαΔN tg (n = 15) mice were identified as CD3ε+tetramer+ cells within electronically gated CD8lo thymocytes or B220lo splenocytes. Numbers are % NKT cells among total leukocytes within each organ. (B) Absolute cell numbers were calculated from % NKT cells and total cell count. n, as in A. (C) Thymic NKT cell developmental stages were identified as in figure, whilst splenic and hepatic NKT cells were identified as NK1.1−tetramer+ or NK1.1+tetramer+ cells. Numbers are % cells among total NKT cells. n, as in A. (D) Absolute numbers of thymic stage 0 + 1, stage 2 and stage 3 NKT cells; they were calculated from the absolute NKT cell numbers and % stage 0 + 1, stage 2 and stage 3 cells in C. n, as in A. (E) Expression of CD4, CD69, CD122, Ly6C, Ly49C/I, and CD62L by thymic, splenic, and hepatic NKT cells was determined as in Fig. 1D. Data are representative of 3 independent experiments. n, as in A. ns, not significant (P > 0.05), *P ≤ 0.05, **P ≤ 0.001.
NF-κB signalling deficiency renders NKT cells sensitive to TNF-α-induced apoptosis
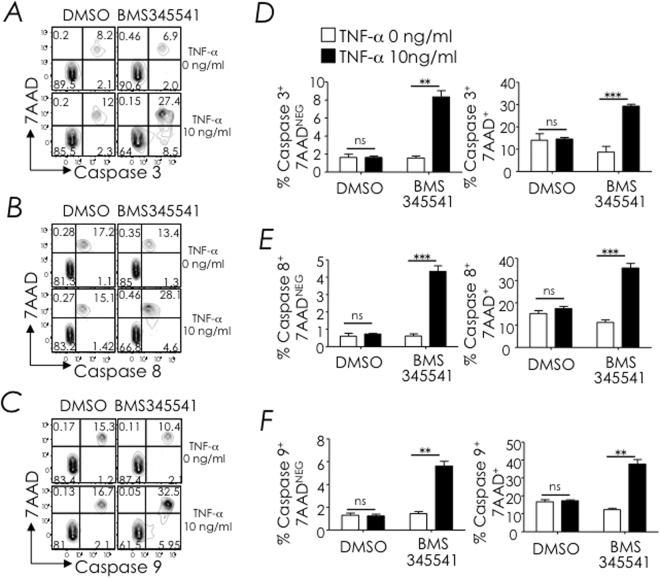
The data so far indicate that NF-κB signalling protects NKT cells from TNFR1-induced cell death. Further, in a previous study, we reported that NKT cells in Iκ BΔN tg mice, in which thymocytes are unable to activate NF-κB, expressed higher frequency of caspase 8 and caspase 9 positive cells. Forced expression of Bcl-xL in Iκ BΔN tg NKT cells lowered the frequency of caspase 8 and caspase 9 positive cells to levels observed in wild type non-transgenic mice37. Guided by that finding, we designed the following experiment: C57BL/6 mouse thymocytes were cultivated in the presence of 3–24 μM BMS345541 —an inhibitor of IKK, a key node upstream of NF-κB activation53— or DMSO —the vehicle used to dissolve BMS345541— in the presence or absence of 10 or 100 ng/ml mouse TNF-α. After 14 hours, cells were labelled with fluorescently-tagged inhibitors for active caspase 3, caspase 8, and caspase 9. The binding of fluorescently-tagged inhibitors to active caspases allowed their detection within NKT cells by flow-cytometry. Caspase substrate tagged cells were co-stained with 7-aminoactinomycin D (7-AAD) to reveal dying/dead cells. The data presented herein are those performed with 6 μM BMS345541 treatment. This is because 12 μM BMS345541 was partially and 24 μM BMS345541 was fully toxic to thymocytes in vitro even in the absence of TNF-α stimulation (data not shown). Further, 3 μM BMS345541-treated thymocytes showed suboptimal caspase activation upon stimulation with 10 ng/ml and even 100 ng/ml TNF-α (data not shown).
We found that 10 ng/ml TNF-α activated caspase 3, caspase 8, and caspase 9 within thymic NKT cells in which IKK activation was inhibited with 6 μM BMS345541. In contrast, the same amount of TNF-α poorly induced cell death in NKT cells in which IKK signalling was intact (Fig. 8). This finding is consistent with the notion that NF-κB signalling is critical for protecting developing NKT cells from TNFR1-induced cell death. Furthermore, the finding that TNF-α induces both caspase 8 and caspase 9 suggested that, under the conditions tested, thymic NKT cell death is mediated by both extrinsic and intrinsic apoptotic pathways.
Figure 8.
Blocking NFκB-signalling sensitises NKT cells to TNF-α-induced apoptosis. Thymocytes were treated with the IKK inhibitor BMS345541 or DMSO. After an hour, cells were washed and stimulated with indicated amount of TNF-α. After 14 hours, cells were washed and treated with fluorescently-tagged, active caspase 3 (A,D), caspase 8 (B,E) or caspase 9 (C,F) substrates separately. After four hours, NKT cells were identified by staining with anti-CD3ε and CD1d-tetramer and dying/dead cells were identified by staining with 7-AAD. Representative dot plots and quantification of caspase+7-AAD+ cells within electronically gated CD8lo thymocytes are shown. Data are mean + sem from over three independent experiments. ns, not significant (P > 0.05), **P ≤ 0.001, ***P ≤ 0.0001.
Discussion
Unlike conventional T cells that are selected by antagonistic ligands54, several thymic T cell lineages —such as NKT cells and T regulatory cells— are selected by agonistic ligands. Agonistic ligands that positively select NKT cells in the thymus are similar or identical to the ligands that activate these cells in the periphery55–57. In addition to NKT TCR agonistic interactions, NKT cells persistently interact with the selecting DP cells through SLAM-SLAM interactions, which activates PKC-θ via the SAP-FynT signalling module12,13,18,51,58–60. Hence, we reasoned that developing NKT cells need a survival signal to protect them from death induced by high affinity NKT TCR interaction with cognate agonistic ligand and potentially SLAM-SLAM molecules as well. Consistent with this line of thinking, several laboratories30–35,61, including ours37, have found a critical requirement for NF-κB in providing a survival signal during NKT cell development. Similarly, NKT cell development partially depended on signals relayed through the TCR proximal signalling nodes, PKC-θ14 (this report) and CARMA1 (this report). Nonetheless, even though forced overexpression of Bcl-xL partially rescued NKT development in NF-κB signalling deficient mice, the introgression of the same transgene into PKCθ-deficient mice failed to phenocopy the rescue seen in B6-IκBαΔN tg;Bcl2l1 tg mice. Hence, NF-κB provides a survival signal to NKT cells, in part through the TCR-PKCθ-CARMA1-axis, although other receptor(s) may play a role as well. This conclusion is consistent with other reports61–64 including the role of IL-15 in controlling NKT cell homeostasis41–43,46,47. Together these findings suggest that NKT cells narrowly escape death during thymic development.
The finding that TNF-α induces both caspase 8 and caspase 9 suggested that, under the conditions tested, thymic NKT cell death is mediated by both extrinsic and intrinsic apoptotic pathways. This result is consistent with our previous observation in IκBΔN tg mice wherein active caspase 8 and caspase 9 expression was increased within residual NKT cells37. How TNF-α induces both the intrinsic and extrinsic apoptotic pathways in NKT cells is unknown. Based on studies in hepatocytes and cancer cells, we speculate that TNFR1 ligation in NKT cells deficient in NF-κB signalling activates caspase 8 within complex II death-inducing signalling complex. Activated caspase 8 in addition to activating the executioner caspases 3 and 7 then activates caspase 9 perhaps by cleaving BH3-interacting domain death agonist (BID) to truncated BID. Truncated BID formation induces mitochondrial outer membrane permeabilization (MOMP) via B cell lymphoma 2 antagonist or killer activation. Proteins released by MOMP triggers death receptor-induced apoptosis mediated by caspase 9 apoptosome and the activation of the executioner caspases 3 and 765–67. Our finding that forced BCL-xL overexpression partially rescues NKT cell development in NF-κB signalling deficient IκBΔN tg mice (this report and ref.37) lends credence to this hypothesis.
NF-κB activation is tightly controlled in NKT cells and too little or too much signal is detrimental. Hence, the absence of RelA31 or inability to activate NF-κB as in the IκBΔN tg mice prevents NKT cell development14,30. In contrast, when the NF-κB pathway is overactive, as in mice expressing the constitutively activated IκB kinase in hematopoietic cells or mice deficient in the negative regulator of NF-κB signalling CYLD, NKT cells develop but these cells fail to mature and to populate the periphery61. These findings suggest that NF-κB functions as a rheostat to control the development and survival of NKT cells. In doing so, NF-κB activation perhaps sets the threshold for peripheral NKT cell activation. Setting such a threshold may be critical for agonistic ligand(s)-dependent NKT cell selection in the thymus and function in the periphery so as not to set off an autoreactive response should the agonistic ligand(s) be displayed by CD1d-positive cells in peripheral tissues such as hepatocytes and intestinal epithelial cells68–70. The exact mechanism by with NF-κB functions as a rheostat and sets the activation threshold remains to be elucidated.
Materials and Methods
Mice
Age-matched, 8–12-week old mice were used in the experiments described herein. C57BL/6, B6-lpr/lpr and B6.129-Tnfrsf1 −/− mice were purchased from the Jackson Laboratory. B6.129-Prkcq −/− 71 and B6.129-Carma1 −/− 72 were generous gifts from Drs. D.R. Littman (NYU) and V.M. Dixit (Genentech). B6-Iκ BΔN tg;Bcl2l1 tg mice were generated by crossing B6-IκBΔN tg with C-Bcl2l1 tg, both previously described mice14,37, and backcrossed to the C57BL/6 background for eight generations. B6.129-Tnfrsf1 −/−;IκBΔN tg mice were generated by crossing B6.129-Tnfrsf1 −/− mice with B6-IκBΔN tg mice and inter-crossing the F1 B6.129-Tnfrsf1 +/−;IκBΔN tg heterozygotes to generate B6.129-Tnfrsf1 −/−;IκBΔN tg homozygotes with the Tnfrsf1 null mutation. A similar strategy was used to generate B6.129-Prkcq −/−;Bcl2l1 tg and B6-lpr/lpr;IκBΔN tg mice. All mouse crosses and experiments complied with the approved protocol M/13/193 in accordance with relevant guidelines and regulations of Vanderbilt University Institutional Animal Care and Use Committee.
Antibodies and Reagents
All antibodies (Abs) used for the identification of NKT cells and developmental stages are listed in Table 1 as described73. α-Galactosylceramide (αGalCer; KRN7000) was generously provided by Kirin Brewery Company (Gunma, Japan) or purchased (Funakoshi, Japan). Mouse CD1d monomer was obtained from the NIH Tetramer Facility (Emory University). Preparation of CD1d tetramers from monomers and their use are described elsewhere73. IKK inhibitor BMS 345541 was purchased from Sigma. Recombinant mouse TNF-α and 7AAD were purchased from BD Biosciences. CaspGLOW fluorescein Active Caspase 3 (FITC-DEVD-FMK), Caspase 8 (FITC-LETD-FMK) and Caspase 9 (FITC-LEHD-FMK) staining kit were purchased from Invitrogen.
Table 1.
List of antibodies used for multiparametric NKT cells analyses in this study.
| Antigen | Fluorochrome* | Clone | Source** |
|---|---|---|---|
| CD1d-tetramer | APC, BV421 | na | in-house73; NIH Tetrmer Core (Emory University) |
| CD3ε | PE, PerCP-Cy5.5, APC, PE-Cy7 | 145-2C11 | BD Biosciences, BioLegend, Tonbo Biosciences |
| CD8α | FITC, PerCP-Cy5.5, APC-Cy7 | 53–6.7 | BD Biosciences, BioLegend, eBiosciences |
| CD44 | FITC | IM7 | BD Biosciences |
| NK1.1 | PE, PerCP-Cy5.5, APC, APC-Cy7 | PK136 | BD Biosciences |
| B220 | FITC, PerCP-Cy5.5, APC-Cy7 | RA3-6B2 | BD Biosciences |
| CD4 | FITC, PE, PerCP-Cy5.5 | RM4-4 | BD Biosciences, BioLegend |
| CD69 | FITC, PE, PE-Cy7 | H1.2F3 | BD Biosciences |
| CD122 | PE | TM-b1 | BioLegend |
| Ly6C | FITC, APC-Cy7 | AL-21 | BD Biosciences |
| Ly49C/I | FITC | 5E6 | BD Biosciences |
| CD62L | APC-Cy7 | MEL-14 | BD Biosciences |
| CD25 | PerCP-Cy5.5, PE-Cy7 | PC61 | BD Biosciences |
| TNFR1 | PE | 55R-286 | BioLegend |
| TNFR2 | PE | TR75-89 | BioLegend |
| Isotype control | PE | HTK888 | BioLegend |
| OX40 | PE | OX-86 | BioLegend |
| Isotype control | PE | RTK2071 | BioLegend |
| GITR | PE | DTA-1 | BioLegend |
| Isotype control | PE | RTK4530 | BioLegend |
| CD95/Fas | PE | Ja2 | BD Biosciences |
| Isotype control | PE | Ha4/8 | BD Biosciences |
*APC, allophycocyannin; Cy, cyanine; PE, phycoerythrin; PerCP, peridinin chlorophyll protein; APC-Cy7, tandem allophycocyannin-cyanine 7 dye; **where multiple sources are indicated, the reagent worked equally well from the different sources.
Caspase Assay
Approximately 3 × 106 C57BL/6 mouse thymocytes were treated with 3–24 μM BMS345541 or DMSO control for 60 min. As 12 μM BMS345541 was partially and 24 μM BMS345541 fully toxic to thymocytes in vitro, and because 3 μM BMS345541 showed suboptimal caspase activation, all experiments were performed with 6 μM BMS345541 treatment. BMS345541-Treated cells were cultivated in the presence or absence of 10 ng/ml TNF-α for 14 hours. Cells were then incubated with fluorescently labelled caspase 3, caspase 8 or caspase 9 inhibitors for an additional 60 min. Cells were washed twice with wash buffer provided with CaspGLOW caspase assay kits according to the Manufacturer’s instructions. Washed cells were surface stained with CD1d tetramer and with antibodies against mouse CD3ε. Finally, cells were incubated with 7-AAD for 15 min and analyzed by FACS.
Statistics
Comparisons between three or more groups were performed by one-way ANOVA with Tukey’s post test to determine significance. Comparison between two groups was performed by two-tailed, unpaired t test. All statistical analyses were performed using Prism (GraphPad software).
Flow Cytometry
NKT cells were identified as CD3ε+tetramer+ cells among B220lo splenocytes and hepatic mononuclear cells or CD8lo thymocytes by using four-colour flow cytometry, which was performed with a FACSCalibur instrument (Becton-Dickinson). NKT cell developmental stages were phenotyped by using six-colour flow cytometry, which was performed with a LSR-II instrument (Becton-Dickinson). Data were analyzed with FlowJo software (Treestar Inc.). Absolute NKT cell numbers were calculated from % tetramer+ cells and total number of cells recovered from each organ.
Acknowledgements
We thank the NIH Tetramer Facility in Atlanta, GA, for α-galactosylceramide-loaded mouse CD1d tetramers and Kirin Brewery Company for KRN7000. This work was supported by VA Merit Award (BX001444) to SJ and by NIH training (HL069765 & AI007611), research (AI042284, AI061721, AI070305, HL089667, AI068149, AI074754 & AI064639) and core (CA068485 & DK058404) grants.
Author Contributions
J.S.B., A.S.K. and S.J. conceived the project; A.K., L.G., J.S.B., A.S.K., T.M.H. and S.J. planned experiments; A.K., L.G., J.S.B. and A.S.K. performed experiments; A.K., L.G., J.S.B. and S.J. analysed data; M.R.B. and L.V.K. provided critical reagents; A.K., J.S.B. and S.J. wrote the manuscript; A.K., J.S.B., A.S.K., T.M.H., M.R.B., L.V.K. and S.J. edited and revised the manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lynch L. Adipose invariant natural killer T cells. Immunology. 2014;142:337–346. doi: 10.1111/imm.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nature reviews. Immunology. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 3.Bendelac A, Matzinger P, Seder RA, Paul WE, Schwartz RH. Activation events during thymic selection. J Exp Med. 1992;175:731–742. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Egawa T, et al. Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity. 2005;22:705–716. doi: 10.1016/j.immuni.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 5.Bezbradica JS, Hill T, Stanic AK, Van Kaer L, Joyce S. Commitment toward the natural T (iNKT) cell lineage occurs at the CD4 + 8 + stage of thymic ontogeny. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:5114–5119. doi: 10.1073/pnas.0408449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu T, Simmons A, Yuan J, Bender TP, Alberola-Ila J. The transcription factor c-Myb primes CD4+ CD8+ immature thymocytes for selection into the iNKT lineage. Nature immunology. 2010;11:435–441. doi: 10.1038/ni.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D’Cruz LM, Knell J, Fujimoto JK, Goldrath AW. An essential role for the transcription factor HEB in thymocyte survival, Tcra rearrangement and the development of natural killer T cells. Nature immunology. 2010;11:240–249. doi: 10.1038/ni.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–467. doi: 10.1016/S1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 9.Kawano T, et al. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science (New York, N.Y.) 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 10.Mendiratta SK, et al. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–477. doi: 10.1016/S1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 11.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E Production in the Absence of Interleukin-4-Secreting CD1-Dependent Cells. Science (New York, N.Y.) 1997;275:977–979. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 12.Griewank K, et al. Homotypic interactions mediated by Slamf1 and Slamf6 receptors control NKT cell lineage development. Immunity. 2007;27:751–762. doi: 10.1016/j.immuni.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang N, et al. The cell surface receptor SLAM controls T cell and macrophage functions. J Exp Med. 2004;199:1255–1264. doi: 10.1084/jem.20031835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanic AK, et al. Cutting edge: the ontogeny and function of Va14Ja18 natural T lymphocytes require signal processing by protein kinase C theta and NF-kappa B. Journal of immunology (Baltimore, Md.: 1950) 2004;172:4667–4671. doi: 10.4049/jimmunol.172.8.4667. [DOI] [PubMed] [Google Scholar]
- 15.Lazarevic V, et al. The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nature immunology. 2009;10:306–313. doi: 10.1038/ni.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seiler MP, et al. Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nature immunology. 2012;13:264–271. doi: 10.1038/ni.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qi Q, Kannan AK, August A. Tec family kinases: Itk signaling and the development of NKT alphabeta and gammadelta T cells. FEBS J. 2011;278:1970–1979. doi: 10.1111/j.1742-4658.2011.08074.x. [DOI] [PubMed] [Google Scholar]
- 18.Nichols KE, et al. Regulation of NKT cell development by SAP, the protein defective in XLP. Nature medicine. 2005;11:340–345. doi: 10.1038/nm1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Savage AK, et al. The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity. 2008;29:391–403. doi: 10.1016/j.immuni.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovalovsky D, et al. The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nature immunology. 2008;9:1055–1064. doi: 10.1038/ni.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dose M, et al. Intrathymic proliferation wave essential for Valpha14 + natural killer T cell development depends on c-Myc. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:8641–8646. doi: 10.1073/pnas.0812255106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mycko MP, et al. Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. Journal of immunology (Baltimore, Md.: 1950) 2009;182:4641–4648. doi: 10.4049/jimmunol.0803394. [DOI] [PubMed] [Google Scholar]
- 23.Godfrey DI, Stankovic S, Baxter AG. Raising the NKT cell family. Nature immunology. 2010;11:197–206. doi: 10.1038/ni.1841. [DOI] [PubMed] [Google Scholar]
- 24.Lee YJ, et al. Tissue-Specific Distribution of iNKT Cells Impacts Their Cytokine Response. Immunity. 2015;43:566–578. doi: 10.1016/j.immuni.2015.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watarai H, et al. Development and function of invariant natural killer T cells producing T(h)2- and T(h)17-cytokines. PLoS Biol. 2012;10:e1001255. doi: 10.1371/journal.pbio.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee YJ, Holzapfel KL, Zhu J, Jameson SC, Hogquist KA. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nature immunology. 2013;14:1146–1154. doi: 10.1038/ni.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammond KJ, et al. CD1d-restricted NKT cells: an interstrain comparison. Journal of immunology (Baltimore, Md.: 1950) 2001;167:1164–1173. doi: 10.4049/jimmunol.167.3.1164. [DOI] [PubMed] [Google Scholar]
- 28.Lynch L, et al. Regulatory iNKT cells lack expression of the transcription factor PLZF and control the homeostasis of Treg cells and macrophages in adipose tissue. Nature immunology. 2015;16:85–95. doi: 10.1038/ni.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sag D, Krause P, Hedrick CC, Kronenberg M, Wingender G. IL-10-producing NKT10 cells are a distinct regulatory invariant NKT cell subset. J Clin Invest. 2014;124:3725–3740. doi: 10.1172/JCI72308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vallabhapurapu S, Hammond KJL, Howells N, Pfeffer K, Weih F. Differential Requirement for Rel/Nuclear Factor κB Family Members in Natural Killer T Cell Development. J Exp Med. 2003;197:1613–1621. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vallabhapurapu S, et al. Rel/NF-kappaB family member RelA regulates NK1.1- to NK1.1+ transition as well as IL-15-induced expansion of NKT cells. European journal of immunology. 2008;38:3508–3519. doi: 10.1002/eji.200737830. [DOI] [PubMed] [Google Scholar]
- 32.Elewaut D, et al. NIK-dependent RelB activation defines a unique signaling pathway for the development of V alpha 14i NKT cells. J Exp Med. 2003;197:1623–1633. doi: 10.1084/jem.20030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmidt-Supprian M, et al. Differential dependence of CD4+ CD25+ regulatory and natural killer-like T cells on signals leading to NF-kappaB activation. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:4566–4571. doi: 10.1073/pnas.0400885101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stankovic S, et al. Distinct roles in NKT cell maturation and function for the different transcription factors in the classical NF-kappaB pathway. Immunology and cell biology. 2011;89:294–303. doi: 10.1038/icb.2010.93. [DOI] [PubMed] [Google Scholar]
- 35.Qi Q, et al. A unique role for ITK in survival of invariant NKT cells associated with the p53-dependent pathway in mice. Journal of immunology (Baltimore, Md.: 1950) 2012;188:3611–3619. doi: 10.4049/jimmunol.1102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.August A. IL-2-inducible T-cell kinase (ITK) finds another (dance) partner…TFII-I. European journal of immunology. 2009;39:2354–2357. doi: 10.1002/eji.200939813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stanic AK, et al. Journal of immunology (Baltimore, Md.: 1950) 2004. NF-kappa B controls cell fate specification, survival, and molecular differentiation of immunoregulatory natural T lymphocytes; pp. 2265–2273. [DOI] [PubMed] [Google Scholar]
- 38.Boesteanu A, et al. Distinct roles for signals relayed through the common cytokine receptor gamma chain and interleukin 7 receptor alpha chain in natural T cell development. J Exp Med. 1997;186:331–336. doi: 10.1084/jem.186.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Journal of immunology (Baltimore, Md.: 1950) 1997. Role for IL-15/IL-15 receptor beta-chain in natural killer 1.1 + T cell receptor-alpha beta + cell development; pp. 5931–5935. [PubMed] [Google Scholar]
- 40.Ohteki T, et al. The Transcription Factor Interferon Regulatory Factor 1 (IRF-1) Is Important during the Maturation of Natural Killer 1.1 + T Cell Receptor–α/β + (NK1 + T) Cells, Natural Killer Cells, and Intestinal Intraepithelial T Cells. J Exp Med. 1998;187:967–972. doi: 10.1084/jem.187.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lodolce JP, et al. IL-15 Receptor Maintains Lymphoid Homeostasis by Supporting Lymphocyte Homing and Proliferation. Immunity. 1998;9:669–676. doi: 10.1016/S1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 42.Kennedy MK, et al. Reversible Defects in Natural Killer and Memory Cd8 T Cell Lineages in Interleukin 15–Deficient Mice. J Exp Med. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsuda JL, et al. Homeostasis of V[alpha]14i NKT cells. Nature immunology. 2002;3:966–974. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda JL, et al. T-bet concomitantly controls migration, survival, and effector functions during the development of Vα14i NKT cells. Blood. 2006;107:2797–2805. doi: 10.1182/blood-2005-08-3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Townsend MJ, et al. T-bet Regulates the Terminal Maturation and Homeostasis of NK and Vα14i NKT Cells. Immunity. 2004;20:477–494. doi: 10.1016/S1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 46.Castillo EF, Acero LF, Stonier SW, Zhou D, Schluns KS. Thymic and peripheral microenvironments differentially mediate development and maturation of iNKT cells by IL-15 transpresentation. Blood. 2010;116:2494–2503. doi: 10.1182/blood-2010-03-277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordy LE, et al. IL-15 Regulates Homeostasis and Terminal Maturation of NKT Cells. Journal of immunology (Baltimore, Md.: 1950) 2011;187:6335–6345. doi: 10.4049/jimmunol.1003965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng LEC, Chan FKM, Cado D, Winoto A. Functional redundancy of the Nur77 and Nor‐1 orphan steroid receptors in T‐cell apoptosis. EMBO J. 1997;16:1865–1875. doi: 10.1093/emboj/16.8.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moran AE, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med. 2011;208:1279–1289. doi: 10.1084/jem.20110308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Holzapfel KL, Tyznik AJ, Kronenberg M, Hogquist KA. Antigen-Dependent versus -Independent Activation of Invariant NKT Cells during Infection. Journal of immunology (Baltimore, Md.: 1950) 2014;192:5490–5498. doi: 10.4049/jimmunol.1400722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cannons JL, et al. SAP Regulates TH2 Differentiation and PKC-θ-Mediated Activation of NF-κB1. Immunity. 2004;21:693–706. doi: 10.1016/j.immuni.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 52.Senftleben U, Li ZW, Baud V, Karin M. IKKbeta is essential for protecting T cells from TNFalpha-induced apoptosis. Immunity. 2001;14:217–230. doi: 10.1016/S1074-7613(01)00104-2. [DOI] [PubMed] [Google Scholar]
- 53.Gerondakis S, Fulford TS, Messina NL, Grumont RJ. NF-[kappa]B control of T cell development. Nature immunology. 2014;15:15–25. doi: 10.1038/ni.2785. [DOI] [PubMed] [Google Scholar]
- 54.Stritesky G, Jameson S, Hogquist KA. Selection of self-reactive T cells in the thymus. Ann Rev Immunol. 2012;30:95–114. doi: 10.1146/annurev-immunol-020711-075035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei DG, et al. Expansion and long-range differentiation of the NKT cell lineage in mice expressing CD1d exclusively on cortical thymocytes. J Exp Med. 2005;202:239–248. doi: 10.1084/jem.20050413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kain L, et al. The Identification of the Endogenous Ligands of Natural Killer T Cells Reveals the Presence of Mammalian α-Linked Glycosylceramides. Immunity. 2014;41:543–554. doi: 10.1016/j.immuni.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wieland Brown LC, et al. Production of α-Galactosylceramide by a Prominent Member of the Human Gut Microbiota. PLOS Biology. 2013;11:e1001610. doi: 10.1371/journal.pbio.1001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang B, et al. CRISPR-Mediated Triple Knockout of SLAMF1, SLAMF5 and SLAMF6 Supports Positive Signaling Roles in NKT Cell Development. PLOS ONE. 2016;11:e0156072. doi: 10.1371/journal.pone.0156072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen, S. et al. Dissection of SAP-dependent and SAP-independent SLAM family signaling in NKT cell development and humoral immunity. J Exp Med (2017). [DOI] [PMC free article] [PubMed]
- 60.Michel M-L, et al. SLAM-associated protein favors the development of iNKT2 over iNKT17 cells. European journal of immunology. 2016;46:2162–2174. doi: 10.1002/eji.201646313. [DOI] [PubMed] [Google Scholar]
- 61.Lee AJ, et al. Regulation of natural killer T‐cell development by deubiquitinase CYLD. EMBO J. 2010;29:1600–1612. doi: 10.1038/emboj.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Crawford G, et al. DOCK8 is critical for the survival and function of NKT cells. Blood. 2013;122:2052–2061. doi: 10.1182/blood-2013-02-482331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamura K, et al. Survival of mature T cells depends on signaling through HOIP. Sci Rep. 2016;6:36135. doi: 10.1038/srep36135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lan, P. et al. TNF superfamily receptor OX40 triggers invariant NKT cell pyroptosis and liver injury. J Clin Invest127 (2017). [DOI] [PMC free article] [PubMed]
- 65.Tait SWG, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol. 2010;11:621–632. doi: 10.1038/nrm2952. [DOI] [PubMed] [Google Scholar]
- 66.Green DR. Apoptotic Pathways: The Roads to Ruin. Cell. 1998;94:695–698. doi: 10.1016/S0092-8674(00)81728-6. [DOI] [PubMed] [Google Scholar]
- 67.Yin X-M, et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 68.Fuss IJ, et al. Nonclassical CD1d-restricted NK T cells that produce IL-13 characterize an atypical Th2 response in ulcerative colitis. J Clin Invest. 2004;113:1490–1497. doi: 10.1172/JCI19836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Blumberg RS, et al. Expression of a nonpolymorphic MHC class I-like molecule, CD1D, by human intestinal epithelial cells. Journal of immunology (Baltimore, Md.: 1950) 1991;147:2518–2524. [PubMed] [Google Scholar]
- 70.Mandal M, et al. Tissue distribution, regulation and intracellular localization of murine CD1 molecules. Mol Immunol. 1998;35:525–536. doi: 10.1016/S0161-5890(98)00055-8. [DOI] [PubMed] [Google Scholar]
- 71.Sun Z, et al. PKC-theta is required for TCR-induced NF-kappaB activation in mature but not immature T lymphocytes. Nature. 2000;404:402–407. doi: 10.1038/35006090. [DOI] [PubMed] [Google Scholar]
- 72.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003;13:1247–1251. doi: 10.1016/S0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 73.Kumar, A., Bezbradica, J. S., Stanic, A. K. & Joyce, S. Characterization and Functional Analysis of Mouse Semi-invariant Natural T Cells. Curr Protoc Immunol117, 14 13 11–14 13 55 (2017). [DOI] [PubMed]