Summary
Pluripotent stem cells hold great promise for regenerative medicine since they can differentiate into all somatic cells. MicroRNAs (miRNAs) could be important for the regulation of these cell-fate decisions. Profiling of miRNAs revealed 19 differentially expressed miRNAs in the endoderm and 29 in the mesoderm when analyzing FACS-purified cells derived from human embryonic stem cells. The mesodermal-enriched miR-483-3p was identified as an important regulator for the generation of mesodermal PDGFRA+ paraxial cells. Repression of its target PGAM1 significantly increased the number of PDGFRA+ cells. Furthermore, miR-483-3p, miR-199a-3p, and miR-214-3p might also have functions for the mesodermal progenitors. The endoderm-specific miR-489-3p and miR-1263 accelerated and increased endoderm differentiation upon overexpression. KLF4 was identified as a target of miR-1263. RNAi-mediated downregulation of KLF4 partially mimicked miR-1263 overexpression. Thus, the effects of this miRNA were mediated by facilitating differentiation through destabilization of pluripotency along with other not yet defined targets.
Keywords: microRNA, miRNome, human pluripotent stem cell, endoderm, mesoderm, cell-fate decision, purified population
Highlights
-
•
miRNome profiling of purified mesoderm and endoderm compared with human ESCs
-
•
19 and 29 differently expressed DE-specific and ME-specific miRNAs, respectively
-
•
miR-1263 overexpression increased DE differentiation via KLF4 repression
-
•
miR-483-3p regulates the development of PDGFRA+ ME cells via PGAM1 repression
In this article, Ishikawa and colleagues show that miRNAs can regulate early cell-fate decisions during differentiation of human ESCs. miRNome profiling in pure endoderm and mesoderm revealed differentially expressed miRNAs. The endoderm-specific miR-489-3p and miR-1263 increased endoderm specification. The miR-1263 effect was mediated by KLF4 repression. The mesoderm-specific miR-483-3p regulated the generation PDGFRA+ paraxial cells via PGAM1 repression.
Introduction
During vertebrate development one of the first cell-fate decisions is gastrulation, resulting in the formation of three embryonic germ layers (Murry and Keller, 2008). During gastrulation the definitive endoderm (DE) and mesoderm (ME) arise, with evidence that they originate from a mesendodermal progenitor (Murry and Keller, 2008, Yang et al., 2014). Human pluripotent stem cells (hPSCs) such as embryonic stem cells (hESCs) or induced pluripotent stem cells (hiPSCs) can differentiate under appropriate conditions into virtually any adult cell type. These cells can be used to study human development in vitro or provide a cell source for regenerative medicine. However, despite extensive studies of transcriptional networks and dynamics in model organisms and during hPSC differentiation, many aspects of gene regulation during germ layer formation are not well understood.
Endogenous non-coding RNAs, such as microRNAs (miRNAs), are regulatory elements that can control the expression of target genes on the post-transcriptional level (Bartel, 2009). They exert important functions in development, differentiation, cell-fate specification, and pathogenesis (Eliasson and Esguerra, 2014, Fiedler et al., 2014, Sayed and Abdellatif, 2011). Knockout of the miRNA-processing proteins Dicer1 or Dgcr8 results in lethality during embryogenesis and disturbed ESC differentiation, demonstrating that miRNAs possess essential functions for early development (Bernstein et al., 2003, Wang et al., 2007, Kanellopoulou et al., 2005). Additionally, miRNAs can facilitate reprogramming of somatic cells into iPSCs and help to maintain pluripotency (Leonardo et al., 2012). Several studies identified miRNA clusters that are highly enriched in PSCs with decreasing expression levels upon differentiation, such as the species-conserved miR-302/367 or the human miR-371–373 cluster (ortholog of the murine miR-290–295 cluster) (Chen et al., 2007, Diekmann et al., 2013, Lakshmipathy et al., 2010, Laurent et al., 2008, Stadler et al., 2010). However, miRNAs enriched in ESCs can exhibit additional functions during early differentiation, as shown across different species for the miR-430/427/302 family that is also important for proper endoderm and mesoderm development (Rosa et al., 2009).
Studies of the miRNA transcriptome (miRNome) during DE differentiation of hESCs revealed a unique miRNA expression profile (Fogel et al., 2015, Hinton et al., 2010, Hinton et al., 2014, Liao et al., 2013) but these studies analyzed heterogeneous cultures, which did not allow a reliable correlation between miRNA expression and the DE. Therefore, this study comparatively analyzed the miRNome of hESCs from fluorescence-activated cell sorting (FACS)-purified DE and ME to identify differentially expressed miRNAs. Identified miRNAs were functionally analyzed during differentiation, in silico predicted target mRNAs were analyzed by a luciferase reporter assay, and effects of these genes upon differentiation were investigated. Out of the DE candidate miRNAs miR-489-3p, miR-1263, and the miR-371–373 cluster were primarily expressed in DE cells. Transfection with miR-1263 and/or miR-489-3p mimics increased the number of CXCR4+ DE cells and accelerated DE differentiation. The pluripotency regulator KLF4 was regulated by miR-1263 on the mRNA and protein expression level. Additionally, repression of KLF4 by small interfering RNA (siRNA) partially mimicked this effect. The miRNAs miR-199a-3p, miR-214-3p, and miR-483-3p were highly enriched in ME cells. Functional analysis revealed that only miR-483-3p was able to alter the composition of the analyzed ME subpopulations. PGAM1 was identified as an mRNA target of miR-483-3p, which was also regulated on the protein level. The miR-483-3p effect was in part mimicked by PGAM1 repression. Thus, this study showed that miR-1263 facilitates DE differentiation likely by KLF4 repression, while miR-483-3p has an important function for subdividing the broad ME into progenitor subpopulations for further lineage specification.
Results
Characterization of Sorted Populations upon Differentiation
Initially, several protocols were tested to induce ME from hESCs, with highest expression values of mesodermal genes (KDR, PDGFRA, MEOX1, CD34) using protocol ME3 (Figures S1A and S1B). In line with the role of T (Bry) for early mesendo/mesoderm specification (Tan et al., 2013), ME3 induced its peak expression early if GSK3 inhibition by CHIR-99021 (CHIR), to activate Wnt/β-catenin signaling, was present, and a decreased expression thereafter (Figure S1C). GSK3 inhibition for more than 2 days (ME1, ME5) or together with fibroblast growth factor 2 supplementation (ME4) reduced the expression of KDR and PDGFRA (Figure S1B). A nearly identical expression profile was obtained with the second hESC line, HUES8 (Figure S1D). Thus, ME3 was used for the mesoderm differentiation in the following experiments.
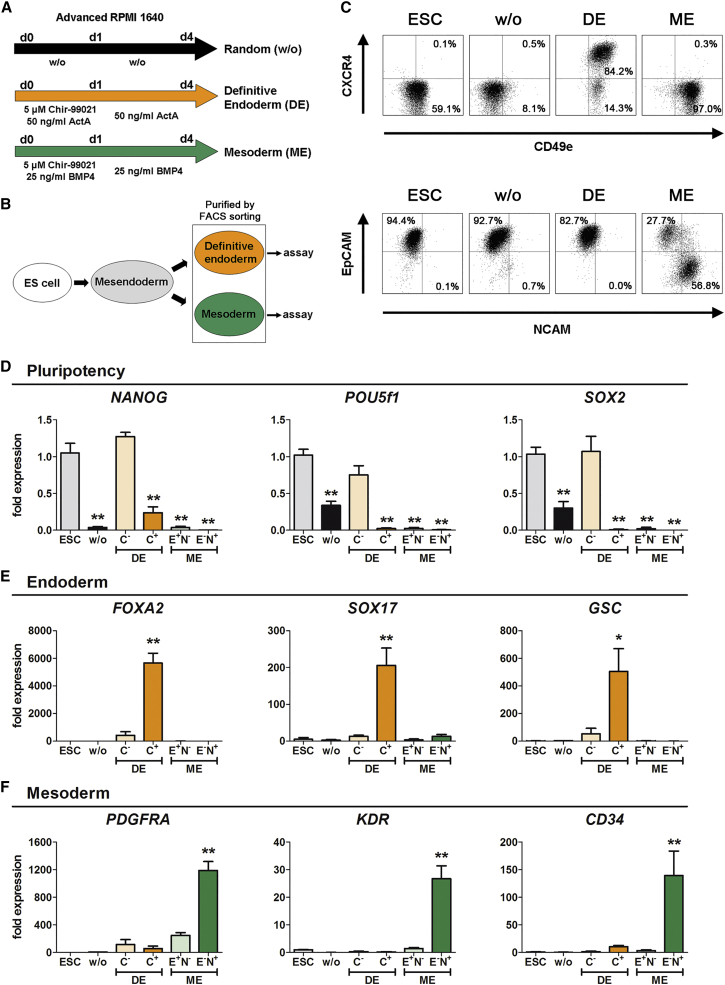
Figure 1A shows the applied differentiation protocols to purify endoderm and ME by FACS (Figure 1B). CXCR4 was solely induced upon differentiation toward the DE, while CD49e, a described marker for DE progeny (Wang et al., 2011), was additionally detected upon ME differentiation (Figure 1C). EpCAM was highly expressed on hESCs and maintained under DE or randomized conditions. Upon ME differentiation EpCAM decreased while NCAM+ cells appeared (Figure 1C). Time-course analysis during ME differentiation revealed early EpCAM/NCAM double-positive cells at day 2, which decreased their EpCAM positivity upon further differentiation (Figure S1E). Hence, EpCAM−/NCAM+ and EpCAM+/NCAM− cells after ME differentiation as well as CXCR4+ and CXCR4− cells upon DE differentiation were characterized in detail.
Figure 1.
Characterization of Sorted Cell Populations
(A) Protocols used for randomized, endoderm (DE), and mesoderm (ME) differentiation.
(B) Schematic overview of the experimental setup to purify the different populations.
(C) Representative flow-cytometric dot plot diagrams of HES3 cells in the undifferentiated state (ESC) or after 4 days of differentiation with the indicated protocols. Numbers represent the respective percentages of cells in the indicated quadrant. Depicted are CXCR4 and CD49e (upper panel) or EpCAM and NCAM (lower panel).
(D–F) Normalized expression of marker genes for pluripotency (NANOG, POU5f1, SOX2) (D), DE (FOXA2, SOX17, GSC) (E), and ME (PDGFRA, KDR, CD34) (F) scaled to hESC. Values are mean ± SEM, n = 4–7. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05, ∗∗p < 0.01 compared with hESC. C−, CXCR4− cells; C+, CXCR4+ cells; E+N−, EpCAM+/NCAM− cells; E−N+, EpCAM−/NCAM+ cells.
See also Figures S1 and S2.
The CXCR4− population exhibited similar expression levels of NANOG, POU5f1, and SOX2 compared with hESCs, suggesting that these cells resisted endodermal differentiation and maintained pluripotency (Figure 1D). In contrast, CXCR4+ cells and both populations upon ME differentiation showed very low expression levels of these pluripotency marker genes (Figure 1D). The DE marker genes SOX17, FOXA2, GSC, and MIXL1 were highly expressed only in CXCR4+ cells (Figures 1E and S2A), while the ME marker genes PDGFRA, KDR, CD34, MEOX1, and OSR1 were significantly increased solely in EpCAM−/NCAM+ cells without induction of neuroectodermal marker genes (SOX1, NES) (Figures 1F, S2B, and S2C). These findings were reproduced with the second hESC line, HUES8 (Figures S2D–S2F). Thus, CXCR4+ cells represented highly purified DE cells, whereas sorted EpCAM−/NCAM+ cells represented purified ME cells.
miRNA Profiling in Purified Endoderm, Purified Mesoderm, and hESCs
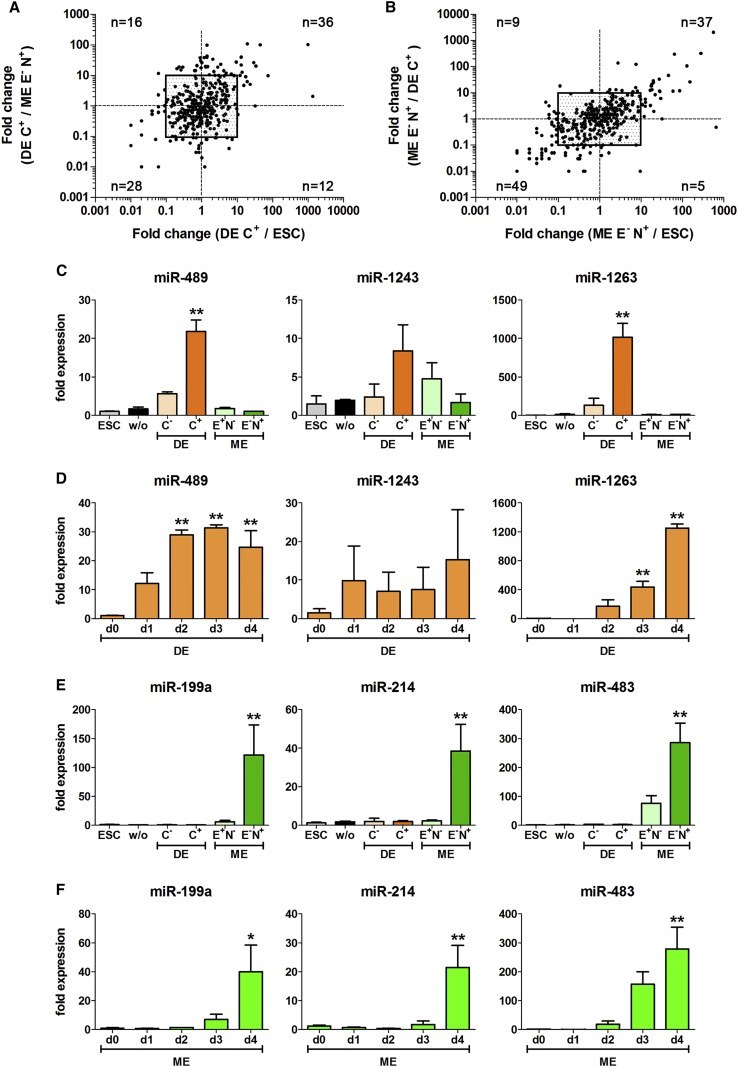
Sorted CXCR4+ DE, EpCAM−/NCAM+ ME, and undifferentiated cells of the hESC line HUES8 were analyzed by qRT-PCR array cards (Figures 1B, 2A, and 2B). Out of the 754 tested miRNAs, 188 were not expressed in the analyzed populations, 369 were expressed in all populations, and 200 were expressed in two or only one population. Stringent criteria were used to identify differentially expressed miRNAs with a Ct value of <28 and an exclusive or a 10-fold altered expression compared with the other populations (Table 1 and Figures 2A and 2B).
Figure 2.
Comparative Analysis of the miRNomes (HUES8) and Validation of Selected miRNAs in a Second hESC Cell Line (HES3)
(A) Comparison of the miRNA expression ratios of purified DE cells versus ME cells (y axis) or versus hESCs (x axis) using the HUES8 line.
(B) miRNA expression ratios of ME cells versus DE cells (y axis) or versus hESCs (x axis) (HUES8 cell line). (A) and (B) present results from pooled samples each comprising 4 independent experiments. Plotted are the 369 miRNAs expressed in all analyzed populations. The central quadrant represents the chosen 10-fold altered expression difference and the indicated numbers represent the miRNAs in the respective quadrant.
(C–F) Normalized expressions of the indicated miRNAs in the different populations (C and E) or during DE (D) or ME (F) differentiation, respectively. Values are mean ± SEM, n = 4–8. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05, ∗∗p < 0.01 compared with all other populations in (C) and (E), and compared with day 0 in (D) and (F). C+, CXCR4+ cells; E−N+, EpCAM−/NCAM+ cells; C−, CXCR4− cells; E+N−, EpCAM+/NCAM− cells.
See also Figure S3.
Table 1.
Upregulated and Downregulated miRNAs in Differentiated Cells Compared with Undifferentiated Cells
| Endoderm |
Mesoderm |
||||
|---|---|---|---|---|---|
| miRNA | Fold Change of Expression |
miRNA | Fold Change of Expression |
||
| Compared with ESCs | Compared with ME | Compared with ESCs | Compared with DE | ||
| Upregulated | Upregulated | ||||
| miR-1263 | n.d. in ESC | n.d. in ME | miR-196b-5p | n.d. in ESC | n.d. in DE |
| miR-1243 | 1354.3 | 2.1 | miR-483-3p | n.d. in ESC | n.d. in DE |
| miR-375 | 988.6 | 102.8 | miR-675-5p | n.d. in ESC | n.d. in DE |
| miR-888-5p | 74.6 | 9.7 | miR-214-3p | 1387.6 | n.d. in DE |
| miR-489-3p | 44.4 | 101.2 | miR-10a-5p | 563.9 | 2043.6 |
| miR-371-3p | 33.9 | 20.2 | miR-199a-3p | 281.5 | 320.2 |
| miR-7-2-3p | 29.3 | 26.3 | miR-145-5p | 129.3 | 105.6 |
| miR-373-3p | 19.2 | 6.3 | miR-193-5p | 64.4 | 300.0 |
| miR-372-3p | 17.5 | 9.1 | miR-143-3p | 57.2 | n.d. in DE |
| miR-661 | 13.8 | 12.7 | miR-483-5p | 46.1 | 45.7 |
| miR-200a-5p | 9.4 | 27.3 | miR-532-3p | 28.5 | 11.0 |
| miR-1260a | 2.4 | 10.1 | miR-455-3p | 28.0 | 11.6 |
| miR-365-3p | 20.0 | 24.4 | |||
| miR-15a-5p | 18.4 | 13.2 | |||
| miR-362-5p | 16.6 | 12.4 | |||
| Downregulated | Downregulated | ||||
| miR-154-3p | n.d. in DE | n.d. in DE | miR-98-5p | n.d. in ME | n.d. in ME |
| miR-518e-3p | 5.8 × 10−3 | 5.5 × 10−2 | miR-367-3p | 1.2 × 10−2 | 6.0 × 10−2 |
| miR-187-3p | 3.9 × 10−2 | 6.7 × 10−1 | miR-302c-5p | 1.2 × 10−2 | 1.0 × 10−2 |
| miR-520g-3p | 4.2 × 10−2 | 5.3 × 10−2 | miR-302b-3p | 2.4 × 10−2 | 2.7 × 10−2 |
| miR-517a-3p | 5.7 × 10−2 | 6.0 × 10−2 | miR-302d-3p | 3.0 × 10−2 | 3.3 × 10−2 |
| miR-512-3p | 5.8 × 10−2 | 9.3 × 10−2 | miR-302d-5p | 3.1 × 10−2 | 2.6 × 10−2 |
| miR-100-5p | 6.2 × 10−2 | 8.1 × 10−3 | miR-302a-5p | 3.3 × 10−2 | 2.3 × 10−2 |
| miR-200b-3p | 3.9 × 10−2 | 3.8 × 10−2 | |||
| miR-302a-3p | 5.4 × 10−2 | 5.6 × 10−2 | |||
| miR-31-5p | 6.5 × 10−2 | 5.1 × 10−2 | |||
| miR-520c-3p | 1.7 × 10−1 | 7.5 × 10−2 | |||
| miR-559 | 3.0 × 10−1 | 2.9 × 10−2 | |||
| miR-200a-5p | 3.4 × 10−1 | 3.7 × 10−2 | |||
| miR-489-3p | 4.4 × 10−1 | 1.0 × 10−2 | |||
n.d., not detected; DE, definitive endoderm; ME, mesoderm.
According to these criteria 12 upregulated and 7 downregulated miRNAs were detected in the CXCR4+ DE population (Table 1). miR489-3p was expressed in all populations but exhibited a strong induction (>44-fold) in DE cells, while miR-1263 was exclusively expressed in this population. In line with earlier studies, miR-375 (>100-fold) and the hESC-enriched miR-371-373 cluster were also upregulated in DE cells (Fogel et al., 2015, Hinton et al., 2010, Joglekar et al., 2009, Lakshmipathy et al., 2007, Laurent et al., 2008, Stadler et al., 2010). Similar to earlier results (Fogel et al., 2015, Hinton et al., 2014), three of the seven downregulated miRNAs are part of an miRNA cluster on chromosome 19 (miR-517a-3p, miR518e-3p, and miR-520g-3p), suggesting an inhibitory role of this cluster for DE development.
Sorted EpCAM−/NCAM+ ME cells exhibited 15 upregulated and 14 downregulated miRNAs compared with DE and ESCs according to our criteria (Table 1). Seven out of the 15 upregulated miRNAs showed either an exclusive or a more than 100-fold induced expression in ME. Interestingly, the 3p and 5p form of miR-483 were included in the list of upregulated miRNAs but only miR-483-3p showed an exclusive expression in sorted ME. Six out of the 14 downregulated miRNAs are part of the miR-302 cluster that is linked to hESCs and DE development (Leonardo et al., 2012, Rosa and Brivanlou, 2011, Rosa et al., 2009). Interestingly, the endodermal-enriched miR-489-3p is also in the list of repressed miRNAs, indicating its inverse regulation during DE and ME formation.
Validation of Endoderm- and Mesoderm-Specific miRNAs
For further validation we focused on top-ranked upregulated miRNAs (Table 1), and analyzed their expression in sorted populations and during differentiation using a second hESC line (HES3) to exclude cell-line-dependent effects.
The miRNAs miR-489-3p and miR-1263 were highly expressed only in CXCR4+ DE cells, while miR-1243 could not be validated as endoderm specific (Figure 2C). Time-course analysis revealed that miR-489-3p was induced at day 1 and plateaued from day 2 on, whereas miR-1263 started to be increased from day 2 on and peaked at day 4 with a ∼40-fold stronger induction compared with miR-489-3p (Figure 2D). Thus, miR-489-3p might be important during early events of DE formation, whereas miR-1263 seems to be important for later stages. The miR-371-373 cluster was already described as hESC enriched (Lakshmipathy et al., 2007, Laurent et al., 2008, Stadler et al., 2010), but CXCR4+ DE cells exhibited even a ∼5-fold increased expression of this cluster compared with hESCs and all other populations (Figure S3A). Their expression was induced from day 2 on and plateaued (miR-372-3p) or peaked at day 4 (Figure S3B), indicating additional roles of these miRNAs during DE formation.
Mesodermal-enriched miRNAs with an exclusive or more than 100-fold induced expression in ME cells were chosen for further validation. Out of these seven miRNAs miR-199a-3p, miR-214-3p, and miR-483-3p showed a strong induction nearly exclusively in EpCAM−/NCAM+ ME cells (Figure 3E). Their expressions were detectable from day 2/3 on and significantly increased until day 4, with the strongest induction rate of more than 200-fold detected for miR-483-3p (Figure 3F). Expression of miR-143-3p and miR-145-5p was significantly induced in ME cells, whereas miR-10a-5p and miR-196b-5p were induced in ME cells and in the EpCAM+/NCAM− population (Figures S3C and S3D). However, these four miRNAs were also induced by a treatment with CHIR alone (Figure S3E), suggesting that they are potentially regulated by Wnt/β-catenin signaling independent of the developmental stage.
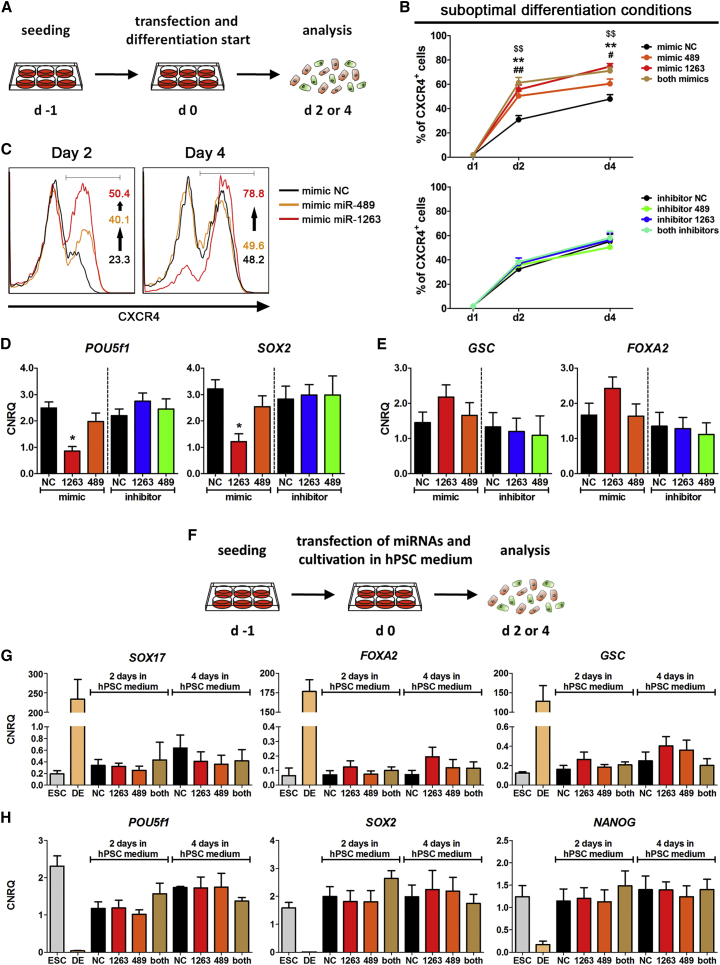
Figure 3.
Functional Analysis of Endoderm-Specific miRNAs (HES3)
(A) Schematic overview of the transfection of miRNA mimics/inhibitors.
(B) Time courses of the derivation of CXCR4+ cells during suboptimal DE differentiation after transfection with mimics or inhibitors. Values are mean ± SEM, n = 5–6. ANOVA plus Bonferroni's post hoc test, #p < 0.05 or ##p < 0.05 (miR-489-3p), ∗∗p < 0.01 (miR-1263), $$p < 0.05 (both) compared with the negative control (NC) on day 2 or day 4, respectively.
(C) Representative flow-cytometric histograms of CXCR4 staining at days 2 and 4.
(D and E) Normalized expression of marker genes for pluripotency (POU5f1, SOX2) (D) and primitive streak/DE (GSC, FOXA2) (E) after 4 days of differentiation post transfection. Values are mean ± SEM, n = 5–6. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05 compared with the respective NC.
(F) Schematic overview of the transfection of miRNA mimics/inhibitors under hESC cultivation condition.
(G and H) Normalized expression of marker genes for primitive streak/DE (SOX17, FOXA2, GSC) (G) and pluripotency (POU5f1, SOX2, NANOG) (H) 2 or 4 days after transfection and cultivation in hESC medium. Values are mean ± SEM, n = 4.
See also Figures S4A–S4D.
miR-1263 and miR-489 Facilitate Endoderm Differentiation
Next, effects of the endoderm-specific miR-489-3p and miR-1263 on DE formation were analyzed by transfecting hESCs prior to differentiation (Figure 3A). Under optimal DE conditions (∼70% CXCR4+ cells), transfection of miR-489-3p or miR-1263 mimic showed slightly increased numbers of CXCR4+ cells at day 2, whereas at day 4 only miR-1263 mimic maintained this tendency with a ∼10% increased number of DE cells (Figure S4A). We also tested a suboptimal DE condition (2.5 μM CHIR instead of 5 μM for the first 24 hr) to assess the miRNA effect in a setting of less extrinsic signal strength (Figure 3B).
Under the suboptimal DE condition, transfection of miR-489-3p or miR-1263 mimic significantly increased the number of CXCR4+ cells compared with the negative control (NC). This effect was especially more pronounced at day 4 for miR-1263 than for miR-489-3p (Figures 3B and 3C). Simultaneous transfection of both mimics did not further enhance the number of CXCR4+ cells but showed a profile similar to that of miR-1263 alone (Figure 3B). Upon transfection with the miR-1263 mimic, pluripotency marker genes (POU5f1, SOX2, NANOG) were significantly downregulated at day 4 without affecting the extraembryonic endoderm marker SOX7 (Figures 3D and S4B). Contrarily, the endodermal marker genes GSC, MIXL1, and FOXA2 were slightly increased, while SOX17 showed no differences (Figures 3E and S4C). Two days after transfection a slight but not significantly reduced expression of the pluripotency marker genes was detected, while all other genes showed no differences (Figure S4D). Interestingly, transfection of the respective miRNA inhibitors alone or together showed no effects on DE formation (Figures 3B–3E and S4A–S4D). Taken together, miR-1263 facilitates and accelerates DE differentiation while miR-489-3p promotes initial cell-fate decisions with a less pronounced effect on the overall efficiency compared with miR-1263.
Next, the effects of miR-489-3p and miR-1263 on hESCs were analyzed by transfecting their mimics alone or together followed by cultivation in hPSC medium (Figure 3F). Under this condition, transfection of the mimics alone or together did not induce the expression of primitive streak/DE marker genes (SOX17, FOXA2, GSC) or alter the expression of pluripotency marker genes (POU5f1, SOX2, NANOG) (Figures 3G and 3H). Thus, miR-489-3p and/or miR-1263 were not able to induce differentiation of hESCs under hPSC cultivation conditions.
Functional Characterization of Mesodermal miRNAs
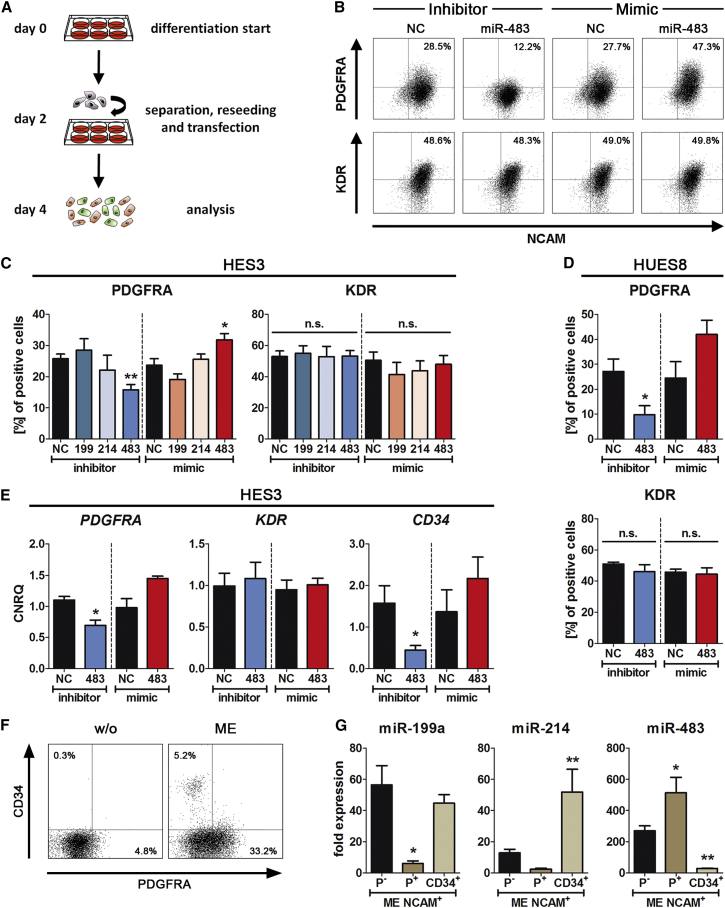
Earlier publications demonstrated that KDR+ cells represent lateral plate ME, whereas PDGFRA+ cells exhibit a paraxial ME character (Sakurai et al., 2006, Tan et al., 2013). Thus, the distribution of these cells was analyzed in EpCAM−/NCAM+ cells upon transfection with mimics or inhibitors of identified ME-enriched miRNAs (Figure 4A). Inhibition of miR-483-3p significantly reduced the number of PDGFRA+ cells, while inversely transfection of miR-483-3p mimic significantly increased their number compared with the respective NC (Figures 4B and 4C). Inhibiting or mimicking miR-199a-3p or miR-214-3p did not yield any significant changes (Figure 4C). Also, the number of KDR+ cells was not altered under all tested conditions (Figure 4C). Transfection of a second hESC line (HUES8) with the miR-483-3p inhibitor or mimic resulted in nearly identical results (Figure 4D) excluding a cell-line-dependent effect. In line with these flow-cytometric results, inhibition of miR-483-3p significantly reduced the expression of PDGFRA while mimicking slightly increased PDGFRA expression without affecting KDR in both hESC lines (Figures 4E and S4E). Interestingly, the expression of CD34 decreased significantly upon inhibition of miR-483-3p (Figures 4E and S4E). Thus, the observed changes of the PDGFRA+ population upon miR-483-3p modulation were confirmed on the protein and mRNA levels (Figures 4C–4E and S4E), demonstrating an important role of miR-483-3p for the formation of PDGFRA+ cells during ME development.
Figure 4.
Functional Analysis of Mesoderm-Specific miRNAs
(A) Schematic overview of the experimental setup.
(B) Representative flow-cytometric dot plot diagrams after 4 days of ME differentiation (HES3). Transfected miRNA inhibitors or mimics are marked. Indicated numbers represent the percentages of double-positive cells (upper panel: NCAM+/PDGFRA+; lower panel: NCAM+/KDR+).
(C and D) Quantification of NCAM+/PDGFRA+ and NCAM+/KDR+ cells by flow cytometry at day 4 using the HES3 (C) or HUES8 (D) cell line. Transfection with the indicated mimics/inhibitors was performed at day 2. Values are mean ± SEM, n = 4–8. ANOVA plus Dunnett’s post hoc test compared with the respective NC (C) or Student's t test (D), ∗p < 0.05 and ∗∗p < 0.01; n.s., not significant.
(E) Normalized gene expressions of ME markers (PDGFRA, KDR, CD34) at day 4 (HES3). Values are means ± SEM, n = 4. Student's t test, ∗p < 0.05.
(F) Depicted is a PDGFRA and CD34 staining of the mesodermal NCAM+ population at day 4 after randomized or ME differentiation (HES3). Indicated percentages represent the numbers of cells within the quadrant of this particular experiment.
(G) Normalized miRNA expression in the three purified populations of mesodermal NCAM+ cells scaled to undifferentiated hESCs (HES3). Values are mean ± SEM, n = 3–6. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05 and ∗∗p < 0.01 compared with P− cells. P−, NCAM+/PDGFRA−/CD34− cells; P+, NCAM+/PDGFRA+/CD34− cells; CD34+, NCAM+/CD34+/PDGFRA− cells.
See also Figure S4E.
Next, the expression of miR-199a-3p, miR-214-3p, and miR-483-3p was analyzed in the different NCAM+ subpopulations (Figures 4F and 4G). The miR-483-3p was already expressed in the PDGFRA− (P−) population and increased in PDGFRA+ (P+) cells, whereas its expression was nearly abolished in CD34+ cells (Figure 4G). Also miR-199a-3p and miR-214-3p were expressed in P− cells but their expression was passed to CD34+ cells with a nearly absent expression in the P+ population (Figure 4G). Taken together, all of these three miRNAs were already expressed in the P− population, which might contain a potential progenitor cell population for P+ and CD34+ cells. Furthermore, miR-483-3p was passed from the P− population to P+ cells, whereas miR-199a-3p and miR-214-3p were nearly abolished in these cells but passed to the CD34+ population.
Potential Candidate Genes and Their Functions during Differentiation
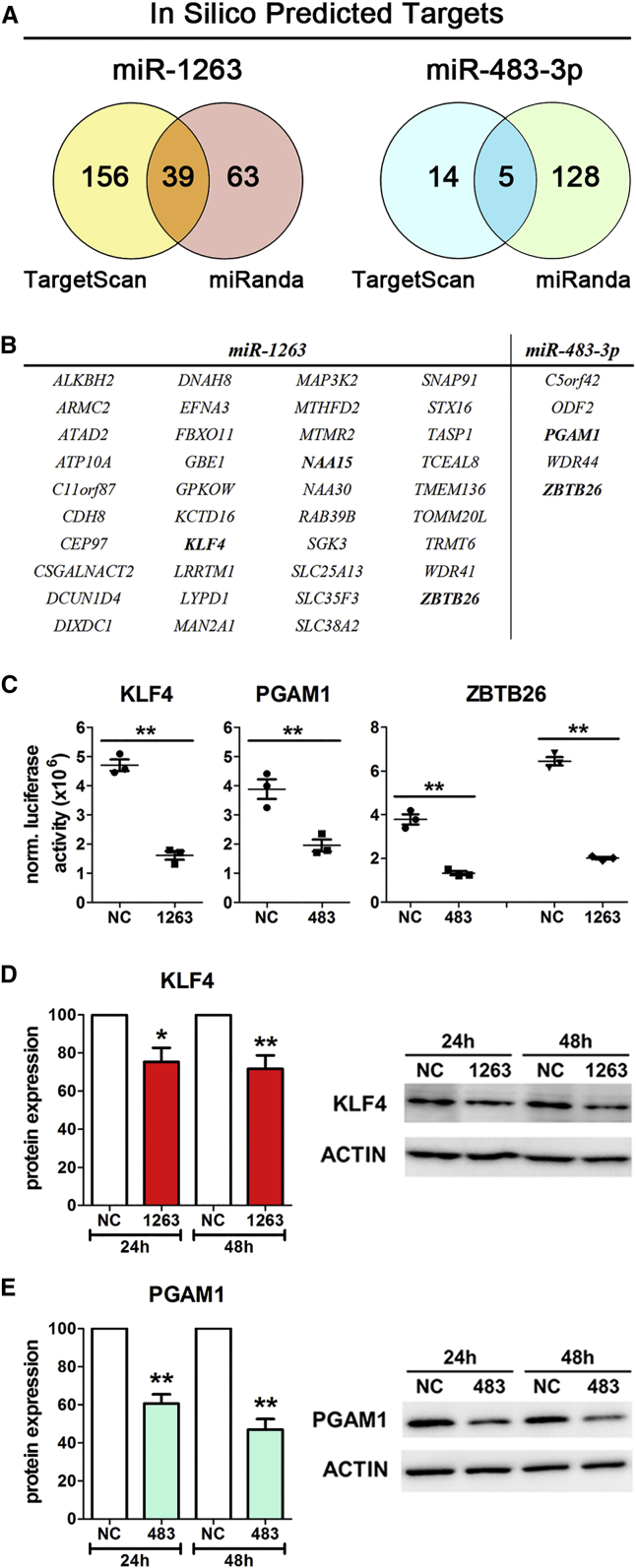
In silico prediction of potential miRNA binding sites in the 3′ UTR region of mRNAs was performed with the TargetScan and miRanda algorithms by choosing a total context++ score < −0.4 and a mirSVR score < −1.0 as cutoffs. For the endodermal miR-1263, 195 (TargetScan) and 102 (miRanda) potential targets were predicted with 39 overlapping targets (Figures 5A and 5B). In silico prediction for miR-483-3p revealed 19 (TargetScan) and 133 (miRanda) targets, with five candidates included in both predictions (Figures 5A and 5B). Target prediction for miR-199a-3p and miR-214-3p resulted in an overlap of 29 and 25 potential targets, respectively (Figures S5A–S5D).
Figure 5.
Target Prediction and Verification of Their Regulation via Endoderm- and Mesoderm-Specific miRNAs
(A) Predicted targets of miR-1263 and miR-483-3p calculated by the TargetScan or miRanda algorithm.
(B) Tabular presentation of the candidate genes predicted by both algorithms for miR-1263 and miR-483-3p. Names written in bold were experimentally analyzed by luciferase reporter assays.
(C) Luciferase activity normalized to the β-galactosidase activity for the subcloned 3′ UTRs of KLF4, PGAM1, and ZBTB26 in HEK293 cells upon transfection of the indicated mimics and the reporter plasmids. Values are mean ± SEM, n = 4–6. Two-tailed Student's t test, ∗∗p < 0.01.
(D and E) Normalized protein expression of KLF4 (D) and PGAM1 (E) 24 hr and 48 hr post transfection with the indicated miRNA mimics in HEK293. The NC of each experiment was set as 100%. For each protein one representative western blots is shown on the right. Values are mean ± SEM, n = 4–5. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05 and ∗∗p < 0.01 compared with the respective NC.
See also Figure S5.
Of the predicted targets for miR-1263, KLF4, a regulator for pluripotency, exhibited one potential binding site in its 3′ UTR. Combined transfection of a luciferase reporter vector and miR-1263 mimic strongly repressed luciferase activity (Figure 5C), indicating binding to this site. Of the predicted targets for miR-483-3p, phosphoglycerate mutase 1 (PGAM1) contained two potential binding sites and binding of miR-483-3p was demonstrated with the luciferase reporter assay (Figure 5C). Interestingly, the so far less characterized gene zinc finger and BTB domain-containing 26 (ZBTB26) was proved as a target of miR-1263 and miR-483-3p (Figure 5C), indicating a negative influence of this gene on DE and ME differentiation. The activin A receptor type 2A (ACVR2A) was determined as a possible target of miR-199a-3p, while N-α-acetyltransferase 15 (NAA15) is a potential target of miR-214-3p and miR-1263 according to the luciferase assay results (Figure S5E).
Next, changes of the KLF4 and PGAM1 protein expression after transfection with the respective miRNA mimics were examined. For this purpose HEK293 cells were used as a model cell line because these cells had a similar expression of KLF4 and PGAM1 on the mRNA and protein levels compared with hESCs (HES3) and a nearly undetectable expression of miR-1263 and miR-483-3p (Figures S5F–S5I). The transfection of miR-1263 mimic significantly decreased KLF4 protein levels 24 hr and 48 hr post transfection compared with the respective NC (Figure 5D). Also, transfection of the miR-483-3p mimic was able to significantly decrease the protein amount of PGAM1 at both time points compared with the NC (Figure 5E).
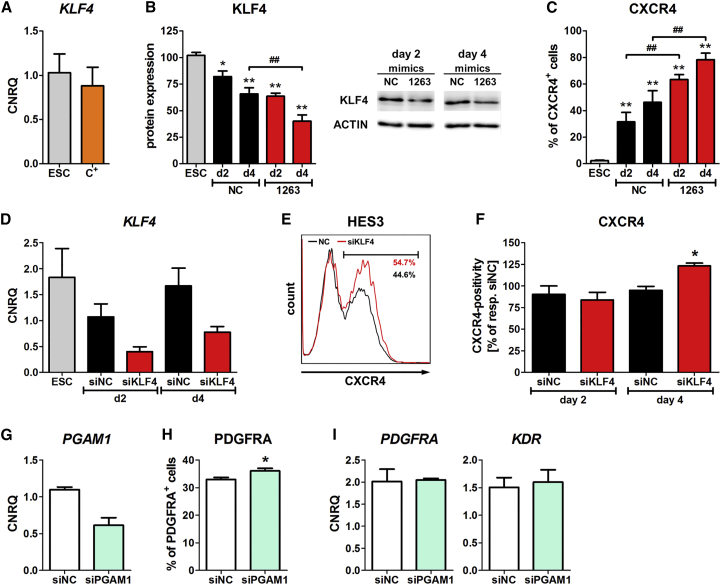
Surprisingly, the mRNA expression of KLF4 in sorted CXCR4+ DE cells was comparable with that of hESCs (Figure 6A). Certainly the KLF4 protein expression significantly decreased upon differentiation, and transfection of the miR-1263 mimic further decreased the KLF4 protein amount compared with the NC (Figure 6B). Inversely to the amount of KLF4 protein, the number of CXCR4+ DE cells increased (Figures 6B and 6C), suggesting that lower KLF4 protein levels are beneficial for DE formation. Inhibition of miR-1263 had no effects on the differentiation efficiency into the DE and the amount of KLF4 protein was maintained on the same level as the NC (Figures 3B, S4A, and S6). We speculate that the amount of inhibitor may not be able to quench the strong miR-1263 expression.
Figure 6.
Influence of KLF4 and PGAM1 on Endoderm or Mesoderm Differentiation (HES3)
(A) Normalized gene expression of KLF4 in undifferentiated hESCs (HES3) and sorted CXCR4+ DE cells. Values are mean ± SEM, n = 4.
(B and C) Normalized protein expression of KLF4 (B) and quantification of CXCR4+ DE cells by flow cytometry (C) after suboptimal DE differentiation and transfection with the indicated miRNA mimics (HES3). Values are mean ± SEM, n = 4. ANOVA plus Bonferroni's post hoc test, ∗p < 0.05, ∗∗p < 0.01 compared with hESCs and ##p < 0.01 between miR-1263 and the NC on the particular day. A representative western blot of KLF4 is depicted in (B).
(D) Normalized gene expression of KLF4 in hESCs and after transfection with the indicated siRNAs plus subsequent suboptimal DE differentiation. Values are mean ± SEM, n = 4–6.
(E and F) Representative flow-cytometric histogram of CXCR4+ cells at day 4 (E) and quantification of CXCR4+ cells at days 2 and 4 (F) upon siRNA transfection and subsequent suboptimal DE differentiation. Values in (F) are mean ± SEM and one respective NC of each experiment is scaled to 100%, n = 4–6. Student's t test, ∗p < 0.05.
(G–I) Normalized gene expression of PGAM1 (G), flow-cytometric quantification of PDGFRA+ cells (H), and normalized gene expression of PDGFRA and KDR (I) at day 5 of ME differentiation. The cells were reseeded on day 2 and transfected with the indicated siRNAs. Values are mean ± SEM, n = 4–6. Student's t test, ∗p < 0.05.
See also Figure S6.
To further examine the influence of KLF4 on DE formation, we transfected siRNA prior to suboptimal DE differentiation. This resulted in a ∼50% decrease of the KLF4 gene expression (Figure 6D) and, confirmatory to the mimic transfections, in a significant increase of CXCR4+ cells compared with the NC (Figures 6E and 6F). Thus, downregulation of KLF4 alone increased the overall differentiation efficiency but did not accelerate DE formation.
To elucidate potential roles of the miR-483-3p target PGAM1 during ME differentiation, we performed transfection of siRNA. This reduced the gene expression of PGAM1 by ∼50% at day 5 (Figure 6G) and resulted in a small but significant induction of PDGFRA+ cells compared with the NC, without effects on the gene expression of KDR or PDGFRA (Figures 6H and 6I).
Discussion
One of the earliest cell-fate decisions during development is the formation of the three embryonic germ layers. The study of human development using hPSCs represents a promising in vitro model. Although many signaling pathways are known that contribute to in vitro germ layer formation, the differentiation yields are often inefficient and variable. Next to gene regulatory networks, miRNAs have the potential to influence cell-fate decisions (Kanellopoulou et al., 2005, Rosa et al., 2009, Wang et al., 2007) by fine-tuning the expression of their target genes on the post-transcriptional level (Bartel, 2009). In this report three different embryonic populations were analyzed to study the differential expression of miRNAs and their functions during gastrulation. In contrast to previous reports (Fogel et al., 2015, Hinton et al., 2010, Hinton et al., 2014, Liao et al., 2013), purified DE and ME populations were compared with hESCs to exclude unwanted cell lineages. This permitted a distinct correlation of miRNA expression to a specific cell fate.
DE differentiation was performed using an established protocol (Diekmann et al., 2015). For ME induction different conditions were tested, and subsequently a protocol was used that is similar to that of Tan et al. (2013). Specific expression of surface proteins was used to purify the desired cell populations from heterogeneously composed differentiation cultures. Pure DE cells were obtained using the specific DE marker CXCR4 (Davenport et al., 2016, Diekmann and Naujok, 2016, Kroon et al., 2008, Naujok et al., 2014). In contrast to a recent publication (Wang et al., 2011), CD49e could not be validated as truly DE specific because it was also detectable upon ME differentiation. In line with Evseenko et al. (2010), the EpCAM−/NCAM+ population represented early ME cells. Additionally, we showed that EpCAM was passed from hPSCs to the DE but not to ME cells. Thus, EpCAM could be an additional surface marker to distinguish DE from ME.
Analysis of the specific miRNomes from purified populations and hESCs identified differentially expressed miRNAs, from which some were already described. The DE-associated miR-375 (Fogel et al., 2015, Hinton et al., 2010, Hinton et al., 2014, Joglekar et al., 2009, Liao et al., 2013) was highly enriched in purified DE cells and served as positive control for our purification and characterization strategy. Earlier studies associated the miR-371-373 cluster with the pluripotent state (Lakshmipathy et al., 2007, Laurent et al., 2008, Stadler et al., 2010), while recent reports showed its upregulation also in heterogeneously composed DE populations (Fogel et al., 2015, Hinton et al., 2010, Hinton et al., 2014, Liao et al., 2013). Here we verified that this cluster is upregulated specifically in DE cells. Thus, the miR-371-373 cluster is important for the maintenance of pluripotency and also for proper DE development. This might be mediated by modulation of the Nodal/transforming growth factor β (TGF-β) signaling, similar to the miR-302/367 cluster (Rosa et al., 2009, Rosa et al., 2014, Barroso-delJesus et al., 2011).
Recently, miR-489-3p and miR-1263 were described to be upregulated in mixed DE populations, but these reports did not characterize their specific functions (Liao et al., 2013, Fogel et al., 2015, Hinton et al., 2014). In this study we confirmed their specific expression in DE cells. Functional characterization demonstrated that both miRNAs accelerated and increased DE formation. However, miR-1263 was more effective in increasing the overall DE efficiency. Simultaneous transfection of both miRNA mimics showed a profile similar to that of the miR-1263 mimic alone, suggesting that the effect on the overall DE efficiency is predominantly mediated by miR-1263. However, both miRNAs under hPSC cultivation conditions were not potent enough alone or together to induce differentiation by themselves. In silico target prediction and functional evaluation confirmed KLF4, among others, as a target mRNA of miR-1263, which is regulated also on the protein level. Interestingly, KLF4 was expressed in hESCs and purified DE cells on a similar level, but downregulation of the KLF4 protein correlated with a more efficient DE formation. This effect could be partially mimicked by siRNA-mediated downregulation of KLF4, albeit without accelerating DE formation. Thus, miR-1263 mediates its positive effects on DE formation partially by destabilization of the pluripotency transcriptional network via KLF4 downregulation. The comparison of the siRNA- and miR mimic-mediated effects suggests that other miR-1263 target genes are also involved in this process. Future studies should address the effects of further miR-1263 targets.
Our characterization of the identified ME-enriched miRNAs revealed different roles during ME formation. miR-199a-3p, miR-214-3p, and miR-483-3p were validated as miRNAs that may influence ME development or specification due to their strong induction and nearly exclusive expression in EpCAM−/NCAM+ ME cells. Even though miR-143-3p and miR-145-5p were exclusively expressed in purified ME cells, these two miRNAs, like miR-10a-5p and miR-196b-5p, were also induced upon CHIR treatment alone. A recent publication strengthens a potential role of miR-145 for ME formation because its overexpression inhibits hESC self-renewal and induces differentiation with an increased expression of ME and ectodermal genes (Xu et al., 2009). However, our data also suggest that these four miRNAs are potentially regulated by Wnt/β-catenin signaling independent of the developmental stage or that they might be expressed in a different ME sublineage induced by treatment with CHIR alone (Mendjan et al., 2014, Lian et al., 2013).
The early hPSC-derived ME can be subdivided into paraxial (PDGFRA+/CD34−) and lateral plate (KDR+/CD34+) ME (Tan et al., 2013). Here we provide evidence that miR-483-3p modulates the number of PDGFRA+ paraxial ME cells without affecting the number of KDR+ cells. In contrast, miR-199a-3p and miR-214-3p showed no effects on these two ME subpopulations although they may have functions in the PGFRA− ME sublineages. This is indicated by their maintained/increased expression in the CD34+ population, while being nearly abolished in the PGFRA+ paraxial ME lineage. We speculate that the two analyzed ME populations arise from an exclusively NCAM+ ancestor that serves as progenitor pool. In the NCAM+ cells all three miRNAs were expressed, suggesting additional functions for all three miRNAs already in the early ME progenitor pool. Consequently, this report identified miR-483-3p as an important regulator of the PDGFRA+ paraxial ME that is capable of further differentiation into chondrocytes, osteocytes, and myocytes (Tan et al., 2013, Evseenko et al., 2010). All three miRNAs seem to be important for the NCAM+ population.
Target predication and experimental validation identified PGAM1 as a target gene of miR-483-3p that is regulated on the mRNA and protein levels. Inhibition of PGAM1 by siRNA could partially mimic the effect of miR-483-3p. Nonetheless it is likely that additional genes are regulated by this miRNA, which results in the more potent effect of miR-483-3p compared with the siRNA-mediated PGAM1 reduction. PGAM1 is part of the glycolysis pathway, which indicates a role of metabolic flux regulation to influence cell-fate decisions. This is in line with a recent publication showing that PSCs exhibited a higher glycolytic flux than early differentiated cells and that inhibition of glycolysis in PSCs leads to differentiation and histone deacetylation (Moussaieff et al., 2015).
Faial et al. (2015) showed that induction of ME genes required BMP4 signaling or Nodal/TGF-β antagonism, while upregulation of DE genes was strictly dependent on Nodal/TGF-β signaling. In this study we also functionally validated the type-2 activin receptor ACVR2A as target of the mesodermal miR-199a-3p, which is important for Nodal/TGF-β signaling. Together with the verified downregulation of the Nodal/TGF-β-regulating miR-302 cluster (Rosa et al., 2009, Barroso-delJesus et al., 2011), this suggests that miRNAs lower Nodal/TGF-β signaling to permit ME differentiation. Finally, we identified that miRNAs enriched either in DE or ME share identical target mRNAs as shown here for ZBTB26 and NAA15. Both genes were not yet described to be functionally relevant for DE or ME specification, but both germ layers repressed these two genes. This raises the question of whether these unknown genes might be inhibitory for DE and ME differentiation or even for gastrulation. Further experiments will be needed to uncover these mechanisms.
In summary, this comprehensive analysis of purified DE and ME permitted the correlation of miRNA expression to particular cell fates. We found 12 upregulated and 7 downregulated DE-specific miRNAs as well as 15 upregulated and 14 downregulated ME-specific miRNAs to match stringent criteria. The ME-enriched miR-199a-3p, miR-214-3p, and miR-483-3p seem to have functions already within the early ME progenitor pool, while miR-483-3p is additionally an important regulator for the development of PDGFRA+ paraxial ME cells. The miR-483-3p mediates its effects on the formation of PDGFRA+ cells in part by regulating PGAM1. In the DE population miR-489-3p and miR-1263 were validated as highly enriched. Both miRNAs could accelerate and increase the efficiency of DE differentiation upon overexpression, while the effect on the overall DE efficiency was predominantly mediated by miR-1263. KLF4 was validated as a functional target of miR-1263, and siRNA-mediated downregulation of KLF4 mimicked the miR-1263 effect to some extent. This demonstrates that miR-1263 mediates its effects by interference with the pluripotent transcriptional network, although other targets will likely play a role.
Experimental Procedures
For more detailed information of the experimental procedures and the used antibodies, please refer to Supplemental Experimental Procedures.
Human ESC Culture and Differentiation
HUES8 and HES3 cells were cultivated under feeder-free conditions and the differentiation was induced from a defined number of seeded singe cells as described earlier (Diekmann et al., 2015, Diekmann and Naujok, 2016, Naujok et al., 2014). Advanced RPMI-1640 supplemented with 1% penicillin-streptomycin, 1% Glutamax, and 0.2% fetal bovine serum was used as base medium and for randomized differentiation. DE differentiation was performed with 50 ng/mL activin A (Peprotech) and 5 μM (optimal condition) or 2.5 μM (suboptimal DE condition) CHIR (Cayman Chemicals) for 24 hr followed by 72 hr in the same medium without CHIR. Induction of ME differentiation was performed in base medium supplemented with 25 ng/mL BMP4 (Peprotech) plus 5 μM CHIR for 24 hr and subsequently for 72 hr in the same medium without CHIR. Media were changed daily during differentiation.
Flow Cytometry and Cell Sorting
Cells were washed, dissociated with trypsin/EDTA, resuspended in PBS plus 2% fetal calf serum, and counted. They were then stained with primary conjugated antibodies following standard protocols and measured on a CyFlow ML flow cytometer (Partec) or sorted at the central cell sorting facility of Hannover Medical School. Data analysis was performed with FlowJo.
Gene and miRNA Expression Analysis
Total RNA was isolated using the peqGOLD RNAPure kit (Peqlab) and total RNA plus small RNAs using the miRNeasy kit (Qiagen) following the manufacturer's instructions. From 1 to 2 μg total RNA cDNA was synthesized and subsequently 5–10 ng was used for each qRT-PCR reaction. Samples were measured in triplicates using specific primer pairs or TaqMan assays (Table S1) on a ViiA7 system (Thermo Fisher Scientific). Data normalization was performed with qBasePlus (Biogazelle) against the geometric mean of three stably expressed housekeeping genes (G6PD, TBP, TUBA1A; M < 0.5).
Reverse transcription for miRNA expression analysis was performed with either 25 ng (single miRNA assays) or 500 ng (array cards) of total RNA plus small RNAs. Sequence-specific primers (Table S2) or the respective Megaplex RT primer pools (array cards, Thermo Fisher Scientific) were used. For miRNA array cards, each analyzed population was a pooled sample from four independent experiments. Data normalization of single miRNA assays was performed with qBasePlus against three stably expressed controls (RNU48, U6 snRNA, mir-425-5p; M < 0.5) and for the array cards using the modified global mean normalization strategy (D'Haene et al., 2012).
miRNA and siRNA Transfection Experiments
For transfection of miRNA inhibitors/mimics (Thermo Fisher Scientific), Lipofectamine 2000 was used. Transfection during DE differentiation was performed on day 0 (30 pmol/24-well) directly after changing to the first DE medium. During ME differentiation, transfection with 30 pmol miRNA inhibitors/mimics per 12-well was performed on day 2 upon dissociation and reseeding (7.5 × 104 cells/cm2) of the cells in ME differentiation medium plus 10 μM Y-27632 (Selleck Chemicals) to ensure acceptable transfection efficiencies. Transfection of siRNAs (Thermo Fisher Scientific) was performed accordingly using 50 pmol or 30 pmol per 12-well during DE or ME differentiation, respectively. Transfection efficiencies were examined by flow cytometry 24 hr later with an FAM-conjugated NC, and a 60% cutoff was set for further analysis.
Luciferase Reporter Assay
Partial 3′ UTRs harboring at least one potential binding site for the respective miRNA were individually cloned into the pMIR-REPORT vector (Thermo Fisher Scientific). Lipofectamine 2000 was used to co-transfect 20 ng of pMIR-REPORT, 20 ng of β-galactosidase control plasmid (Promega), and 30 nM miRNA mimic or control into HEK293 cells. After 24 hr, luciferase and β-galactosidase activity were measured (Promega) and normalization was performed against the β-galactosidase activity of the respective sample.
Western Blot
Cells were detached, collected, centrifuged, and resuspended in RIPA buffer (Thermo Fisher Scientific). These whole-cell extracts were sonicated and a protease inhibitor mixture (Roche Diagnostic) was added. The protein content was determined by BCA assay (Thermo Fisher Scientific). Ten to twenty micrograms of total protein were separated by SDS-PAGE and transferred via electroblotting to a PVDF membrane. Blocking was performed with 2%–5% nonfat dry milk in PBS plus 0.1% Tween 20. Primary antibodies were incubated overnight at 4°C and subsequently washed and incubated with the respective peroxidase-labeled secondary antibodies. The bands were visualized by chemiluminescence using the ECL select or ECL detection kit (GE Healthcare Europe) on a chemiluminescence imager (INTAS Science Imaging). Densitometric analysis was performed with Image Studio Lite software (LI-COR).
Statistics
Unless stated otherwise, all data values represent mean ± SEM. The number of independent experiments (n) is stated in each figure legend. Statistical analyses were performed using GraphPad Prism software.
Author Contributions
D.I. and U.D. designed, performed, and analyzed experiments, interpreted data, and prepared the figures. O.N. and U.D. carried out conceptual design of the study and interpreted the data. D.I., U.D., S.L., and O.N. wrote and edited the manuscript. J.F., A.J., and T.T. cloned the luciferase constructs, performed the reporter assays, and analyzed the data. All authors proofread and edited the manuscript.
Acknowledgment
D.I. was on leave from the Department of Surgery, Tokushima University, Japan and received a Global Grant from Rotary International. The skillful technical assistance of J. Kresse and M. Funck is gratefully acknowledged. We acknowledge the assistance of the Cell Sorting Facility of Hannover Medical School supported by the Braukmann-Wittenberg-Herz-Stiftung and the Deutsche Forschungsgemeinschaft. T.T. acknowledges the support funding from the German Ministry for Education and Research, IFB-Tx, and the REBIRTH Excellence Cluster.
Published: November 14, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and two tables and can be found with this article online at https://doi.org/10.1016/j.stemcr.2017.10.011.
Supplemental Information
References
- Barroso-delJesus A., Lucena-Aguilar G., Sanchez L., Ligero G., Gutierrez-Aranda I., Menendez P. The Nodal inhibitor Lefty is negatively modulated by the microRNA miR-302 in human embryonic stem cells. FASEB J. 2011;25:1497–1508. doi: 10.1096/fj.10-172221. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Kim S.Y., Carmell M.A., Murchison E.P., Alcorn H., Li M.Z., Mills A.A., Elledge S.J., Anderson K.V., Hannon G.J. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Chen C., Ridzon D., Lee C.T., Blake J., Sun Y., Strauss W.M. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm. Genome. 2007;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- D'Haene B., Mestdagh P., Hellemans J., Vandesompele J. miRNA expression profiling: from reference genes to global mean normalization. Methods Mol. Biol. 2012;822:261–272. doi: 10.1007/978-1-61779-427-8_18. [DOI] [PubMed] [Google Scholar]
- Davenport C., Diekmann U., Budde I., Detering N., Naujok O. Anterior-posterior patterning of definitive endoderm generated from human embryonic stem cells depends on the differential signaling of retinoic acid, Wnt-, and BMP-signaling. Stem Cells. 2016;34:2635–2647. doi: 10.1002/stem.2428. [DOI] [PubMed] [Google Scholar]
- Diekmann U., Elsner M., Fiedler J., Thum T., Lenzen S., Naujok O. MicroRNA target sites as genetic tools to enhance promoter-reporter specificity for the purification of pancreatic progenitor cells from differentiated embryonic stem cells. Stem Cell Rev. 2013;9:555–568. doi: 10.1007/s12015-012-9416-1. [DOI] [PubMed] [Google Scholar]
- Diekmann U., Lenzen S., Naujok O. A reliable and efficient protocol for human pluripotent stem cell differentiation into the definitive endoderm based on dispersed single cells. Stem Cells Dev. 2015;24:190–204. doi: 10.1089/scd.2014.0143. [DOI] [PubMed] [Google Scholar]
- Diekmann U., Naujok O. Generation and purification of definitive endoderm cells generated from pluripotent stem cells. Methods Mol. Biol. 2016;1341:157–172. doi: 10.1007/7651_2015_220. [DOI] [PubMed] [Google Scholar]
- Eliasson L., Esguerra J.L. Role of non-coding RNAs in pancreatic beta-cell development and physiology. Acta Physiol. 2014;211:273–284. doi: 10.1111/apha.12285. [DOI] [PubMed] [Google Scholar]
- Evseenko D., Zhu Y., Schenke-Layland K., Kuo J., Latour B., Ge S., Scholes J., Dravid G., Li X., MacLellan W.R. Mapping the first stages of mesoderm commitment during differentiation of human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2010;107:13742–13747. doi: 10.1073/pnas.1002077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faial T., Bernardo A.S., Mendjan S., Diamanti E., Ortmann D., Gentsch G.E., Mascetti V.L., Trotter M.W., Smith J.C., Pedersen R.A. Brachyury and SMAD signalling collaboratively orchestrate distinct mesoderm and endoderm gene regulatory networks in differentiating human embryonic stem cells. Development. 2015;142:2121–2135. doi: 10.1242/dev.117838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler J., Batkai S., Thum T. MicroRNA-based therapy in cardiology. Herz. 2014;39:194–200. doi: 10.1007/s00059-013-4047-0. [DOI] [PubMed] [Google Scholar]
- Fogel G.B., Kai Z.S., Zargar S., Hinton A., Jones G.A., Wong A.S., Ficici S.G., Lopez A.D., King C.C. MicroRNA dynamics during human embryonic stem cell differentiation to pancreatic endoderm. Gene. 2015;574:359–370. doi: 10.1016/j.gene.2015.08.027. [DOI] [PubMed] [Google Scholar]
- Hinton A., Afrikanova I., Wilson M., King C.C., Maurer B., Yeo G.W., Hayek A., Pasquinelli A.E. A distinct microRNA signature for definitive endoderm derived from human embryonic stem cells. Stem Cells Dev. 2010;19:797–807. doi: 10.1089/scd.2009.0224. [DOI] [PubMed] [Google Scholar]
- Hinton A., Hunter S.E., Afrikanova I., Jones G.A., Lopez A.D., Fogel G.B., Hayek A., King C.C. sRNA-seq analysis of human embryonic stem cells and definitive endoderm reveals differentially expressed microRNAs and novel IsomiRs with distinct targets. Stem Cells. 2014;32:2360–2372. doi: 10.1002/stem.1739. [DOI] [PubMed] [Google Scholar]
- Joglekar M.V., Joglekar V.M., Hardikar A.A. Expression of islet-specific microRNAs during human pancreatic development. Gene Expr. Patterns. 2009;9:109–113. doi: 10.1016/j.gep.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Kanellopoulou C., Muljo S.A., Kung A.L., Ganesan S., Drapkin R., Jenuwein T., Livingston D.M., Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E., Martinson L.A., Kadoya K., Bang A.G., Kelly O.G., Eliazer S., Young H., Richardson M., Smart N.G., Cunningham J. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 2008;26:443–452. doi: 10.1038/nbt1393. [DOI] [PubMed] [Google Scholar]
- Lakshmipathy U., Davila J., Hart R.P. miRNA in pluripotent stem cells. Regen. Med. 2010;5:545–555. doi: 10.2217/rme.10.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakshmipathy U., Love B., Goff L.A., Jornsten R., Graichen R., Hart R.P., Chesnut J.D. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2007;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent L.C., Chen J., Ulitsky I., Mueller F.J., Lu C., Shamir R., Fan J.B., Loring J.F. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- Leonardo T.R., Schultheisz H.L., Loring J.F., Laurent L.C. The functions of microRNAs in pluripotency and reprogramming. Nat. Cell Biol. 2012;14:1114–1121. doi: 10.1038/ncb2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Zhang J., Azarin S.M., Zhu K., Hazeltine L.B., Bao X., Hsiao C., Kamp T.J., Palecek S.P. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/beta-catenin signaling under fully defined conditions. Nat. Protoc. 2013;8:162–175. doi: 10.1038/nprot.2012.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X., Xue H., Wang Y.C., Nazor K.L., Guo S., Trivedi N., Peterson S.E., Liu Y., Loring J.F., Laurent L.C. Matched miRNA and mRNA signatures from an hESC-based in vitro model of pancreatic differentiation reveal novel regulatory interactions. J. Cell Sci. 2013;126:3848–3861. doi: 10.1242/jcs.123570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S., Mascetti V.L., Ortmann D., Ortiz M., Karjosukarso D.W., Ng Y., Moreau T., Pedersen R.A. NANOG and CDX2 pattern distinct subtypes of human mesoderm during exit from pluripotency. Cell Stem Cell. 2014;15:310–325. doi: 10.1016/j.stem.2014.06.006. [DOI] [PubMed] [Google Scholar]
- Moussaieff A., Rouleau M., Kitsberg D., Cohen M., Levy G., Barasch D., Nemirovski A., Shen-Orr S., Laevsky I., Amit M. Glycolysis-mediated changes in acetyl-CoA and histone acetylation control the early differentiation of embryonic stem cells. Cell Metab. 2015;21:392–402. doi: 10.1016/j.cmet.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Murry C.E., Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Naujok O., Diekmann U., Lenzen S. The generation of definitive endoderm from human embryonic stem cells is initially independent from activin A but requires canonical Wnt-signaling. Stem Cell Rev. 2014;10:480–493. doi: 10.1007/s12015-014-9509-0. [DOI] [PubMed] [Google Scholar]
- Rosa A., Brivanlou A.H. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. EMBO J. 2011;30:237–248. doi: 10.1038/emboj.2010.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Papaioannou M.D., Krzyspiak J.E., Brivanlou A.H. miR-373 is regulated by TGFbeta signaling and promotes mesendoderm differentiation in human Embryonic Stem Cells. Dev. Biol. 2014;391:81–88. doi: 10.1016/j.ydbio.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A., Spagnoli F.M., Brivanlou A.H. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell. 2009;16:517–527. doi: 10.1016/j.devcel.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Sakurai H., Era T., Jakt L.M., Okada M., Nakai S., Nishikawa S., Nishikawa S. In vitro modeling of paraxial and lateral mesoderm differentiation reveals early reversibility. Stem Cells. 2006;24:575–586. doi: 10.1634/stemcells.2005-0256. [DOI] [PubMed] [Google Scholar]
- Sayed D., Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- Stadler B., Ivanovska I., Mehta K., Song S., Nelson A., Tan Y., Mathieu J., Darby C., Blau C.A., Ware C. Characterization of microRNAs involved in embryonic stem cell states. Stem Cells Dev. 2010;19:935–950. doi: 10.1089/scd.2009.0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J.Y., Sriram G., Rufaihah A.J., Neoh K.G., Cao T. Efficient derivation of lateral plate and paraxial mesoderm subtypes from human embryonic stem cells through GSKi-mediated differentiation. Stem Cells Dev. 2013;22:1893–1906. doi: 10.1089/scd.2012.0590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Rodriguez R.T., Wang J., Ghodasara A., Kim S.K. Targeting SOX17 in human embryonic stem cells creates unique strategies for isolating and analyzing developing endoderm. Cell Stem Cell. 2011;8:335–346. doi: 10.1016/j.stem.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Medvid R., Melton C., Jaenisch R., Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat. Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- Yang D., Lutter D., Burtscher I., Uetzmann L., Theis F.J., Lickert H. miR-335 promotes mesendodermal lineage segregation and shapes a transcription factor gradient in the endoderm. Development. 2014;141:514–525. doi: 10.1242/dev.104232. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.