Abstract
Background
Dried blood spots (DBS) are a convenient tool to enable diagnostic testing for viral diseases due to transport, handling and logistical advantages over conventional venous blood sampling. A better understanding of the performance of serological testing for hepatitis C (HCV) and hepatitis B virus (HBV) from DBS is important to enable more widespread use of this sampling approach in resource limited settings, and to inform the 2017 World Health Organization (WHO) guidance on testing for HBV/HCV.
Methods
We conducted two systematic reviews and meta-analyses on the diagnostic accuracy of HCV antibody (HCV-Ab) and HBV surface antigen (HBsAg) from DBS samples compared to venous blood samples. MEDLINE, EMBASE, Global Health and Cochrane library were searched for studies that assessed diagnostic accuracy with DBS and agreement between DBS and venous sampling. Heterogeneity of results was assessed and where possible a pooled analysis of sensitivity and specificity was performed using a bivariate analysis with maximum likelihood estimate and 95% confidence intervals (95%CI). We conducted a narrative review on the impact of varying storage conditions or limits of detection in subsets of samples. The QUADAS-2 tool was used to assess risk of bias.
Results
For the diagnostic accuracy of HBsAg from DBS compared to venous blood, 19 studies were included in a quantitative meta-analysis, and 23 in a narrative review. Pooled sensitivity and specificity were 98% (95%CI:95%–99%) and 100% (95%CI:99–100%), respectively. For the diagnostic accuracy of HCV-Ab from DBS, 19 studies were included in a pooled quantitative meta-analysis, and 23 studies were included in a narrative review. Pooled estimates of sensitivity and specificity were 98% (CI95%:95–99) and 99% (CI95%:98–100), respectively. Overall quality of studies and heterogeneity were rated as moderate in both systematic reviews.
Conclusion
HCV-Ab and HBsAg testing using DBS compared to venous blood sampling was associated with excellent diagnostic accuracy. However, generalizability is limited as no uniform protocol was applied and most studies did not use fresh samples. Future studies on diagnostic accuracy should include an assessment of impact of environmental conditions common in low resource field settings. Manufacturers also need to formally validate their assays for DBS for use with their commercial assays.
Background
It is estimated that 71 million people are infected with hepatitis C (HCV) (defined as those with viraemic infection) and 257 million with hepatitis B (HBV) (defined as HBsAg positive) worldwide [1–4]. Both HBV and HCV infection predominantly affect persons in low and middle income countries [2, 5]. Late complications of HBV and HCV infection are cirrhosis and hepatocellular carcinoma. Overall, there were 1.34 million deaths attributable to these complications in 2015 with a trend towards increasing deaths since 1990 [2]. Most patients are not aware of their infection until they have advanced complications of the disease [6] due to lack of access to and affordability of testing. Just as an increase in access to treatment is important [7, 8] early identification is paramount and also cost-effective [9] to avoid disease complications.
HBsAg and HCV-Ab screening is traditionally done by serology using either laboratory based enzyme-immuno-assay (EIA) or a point-of-care rapid diagnostic test (RDT). While several rapid lateral flow tests exist for HBsAg and HCV-Ab detection, their quality is variable or unknown and in particular there is a paucity of good quality HBsAg RDTs available on the market [10, 11]. Four HCV-Ab RDTs are WHO prequalified as of August 2017 but none of the HBsAg RDTs have met the requirements for WHO prequalification [12]. The choice of which format of serological assays to use in a programmatic setting will depend on a variety of factors; most importantly, ease of use and the characteristics of the testing site, such as storage facilities, infrastructure, level of staff skills and cost. Confirmation of active viraemic infection is done using nucleic acid testing (NAT) for HCV RNA and HBV DNA or using serological assays for detection of HCV core antigen.
In order to facilitate more widespread uptake of testing for HBV and HCV, there needs to be greater access to diagnostic assays. The use of dried blood spots (DBS) for transportation using fingerstick (in adults and older children), or heel pricking (in neonates and infants) sampling of capillary blood and subsequent analysis with automated high-throughput laboratory-based EIA represents another affordable alternative to testing using RDTs, particularly in settings with limited infrastructure. Another advantage of DBS use in low resource settings [13] is that capillary blood collection does not require a trained health worker to perform venipuncture and that DBS utilizing capillary blood needs less volume than venepuncture. Furthermore, no centrifuge or even basic laboratory facilities with electrical power are needed to prepare plasma [14]; and since transport and handling do not require high skill or a cold chain, the risk of contamination is reduced [15]. The main disadvantage of DBS is that the existing commercial assays have not been validated or received regulatory approval with this method of sample collection and transport.
Despite this limitation, DBS have been increasingly used in recent years to screen for a number of viral diseases, including HIV and viral hepatitis [13, 16–19]. Some studies have suggested that the use of DBS may increase uptake of hepatitis testing among certain vulnerable risk groups [20–22]. While there have been several systematic reviews on diagnostic performance of RDTs for HBsAg and HCV-Ab [10, 11, 23] and on the use of POC tests in viral hepatitis testing [10, 24, 25], and various validation studies of diagnostic accuracy studies aiming to validate DBS have been performed [26, 27] including a systematic review of HCV RNA detection with DBS [28], to our knowledge there has been no summary of evidence on diagnostic accuracy for HBsAg and HCV-Ab testing using DBS. We have conducted two systematic reviews and meta-analyses: one on the diagnostic accuracy of HCV-Ab and the other on the diagnostic accuracy of HBsAg from DBS samples compared to venous samples in persons identified for hepatitis testing. This review informed the WHO guidelines on testing for chronic HBC and HCV in low and middle income countries [29] and was conducted in conjunction with systematic reviews on the diagnostic accuracy of the detection of HBV DNA and HCV RNA from DBS [30].
Methods
Search strategy and selection criteria
PRISMA reporting guidelines were followed [31] and the QUADAS-2 tool was used to estimate quality of studies as a risk of bias tool [32]. We conducted two systematic reviews and meta-analyses on the diagnostic accuracy of HBsAg and HCV antibody detection from DBS compared to venous blood. We searched English language studies using five databases (MEDLINE, Web of Science, EMBASE, Global Health and LILACS) with the following search terms: DBS, Dried blood spot, Dry blood spot, filter paper, Guthrie paper, hepatitis, hepatitis B, hepatitis C, HBsAg, HBV, HCV, HCV RNA, HBV DNA (adapted to databases) on 1.9.2015 and updated it on 22.8.2017.
Abstracts were included for fulltext review if inclusion criteria were fulfilled (namely DBS samples and plasma or serum samples used for detection of HCV-Ab and/or HBsAg) or if exclusion of the abstract could not be performed solely on the basis of the information of the abstract. Eligible studies included comparisons of an index test HCV-Ab and HBsAg using DBS with a reference test HCV-Ab and HBsAg using serum or plasma and following outcomes were analysed: correlation, regression coefficient, specificity, sensitivity and positive/negative predictive values. We included studies regardless of whether the assay used was commercially available or used an in-house technique, and testing on DBS and plasma/serum did not have to use the same assay. There were no date, geographic or population demographic restrictions, and individuals of all age groups were included.
Screening and data extraction
Two independent reviewers performed title, abstract and full-text review to identify eligible studies. Disagreements were resolved by consent of the reviewers. The references of articles selected for inclusion were also reviewed for additional articles to review. The same data extraction procedure was performed in duplicate for each study and included the following variables: author, publication and study dates, country, percentage of children and adults, age range, gender distribution, type of specimen used for DBS, specimen used as gold standard (plasma or serum), serological assay used, storage conditions and effect of storage conditions. Additional data and clarifications were sought by contacting study authors where necessary.
Risk of bias and quality assessment
The QUADAS-2 tool categories of study design, index and reference test conduct and reporting of patient flow were adapted for use to assess the risk of bias in included studies. In particular, studies where there was consecutive sampling of patients were rated as being at low risk of bias, and a case control design as at high risk of bias. Studies reporting use of a consistent protocol for index and reference testing for each sample and description of patient flow were rated at low risk of bias) while lack of reporting or inconsistent use of a protocol were considered at high risk of bias.
Statistical data analysis
Summary estimates of sensitivity and specificity were generated with a bivariate random effects meta-analysis using maximum likelihood estimate and 95% confidence intervals. We calculated positive (Sensitivity/(1-Specificity) and negative (1-Sensitivity/Specificity) likelihood ratios directly from the pooled sensitivity and specificity. Several studies did not have sufficient quantitative data to contribute to both sensitivity and specificity - for example no samples with a negative reference test. In such cases, we performed a univariate random effects meta-analysis of the sensitivity and/or specificity estimates separately to incorporate studies that did not report estimates for both. We then compared univariate analyses with the result of the bivariate analysis, in order to make complete use of all the available data.
Heterogeneity was assessed visually from forest plots and by reporting an estimate of τ2 corresponding to the variance of the logit-transformed specificity and sensitivity, which can be interpreted as a measure of between-study variability [33]. Stratified analyses were performed by type of assay used for the index test and by storage conditions. Some studies exposed individual samples or subsets of samples to varying storage conditions or used them to define limits of detection. Since these subsets were not necessarily part of the diagnostic accuracy evaluation we included them in the narrative analysis of the impact of storage conditions on the validity of results. Statistical analysis of the data was performed using STATA 14 (StataCorp. 2015. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP).
Results
Included studies
For HBsAg detection, our search identified 521 abstracts, of which 65 full text articles were assessed for eligibility, and 23 [34–56] met criteria for inclusion in the review (Fig. 1 & Table 1). Six studies came from Africa [34, 36, 39, 41, 47, 51], nine from Europe [37, 38, 42–44, 50, 53, 55, 56], four from the Americas [35, 45, 46, 54], three from Asia [40, 48, 49]and one from Australia [52]. Four studies did not have sufficient data for sensitivity and specificity analysis [40, 43, 44, 53]. Most studies provided limited information on the characteristics of participants. One study only included pregnant women [35], one only children [51] and one only HIV-infected persons [39]. All studies used the same commercial assays for their reference and their index test, respectively. Five studies from the 1980s used Ausria II (Abbott), whereas newer studies used diverse commercial assays, including Enzygnost (Behring) and ARCHITECT (Abbott) (see also Table 1).
Fig. 1.
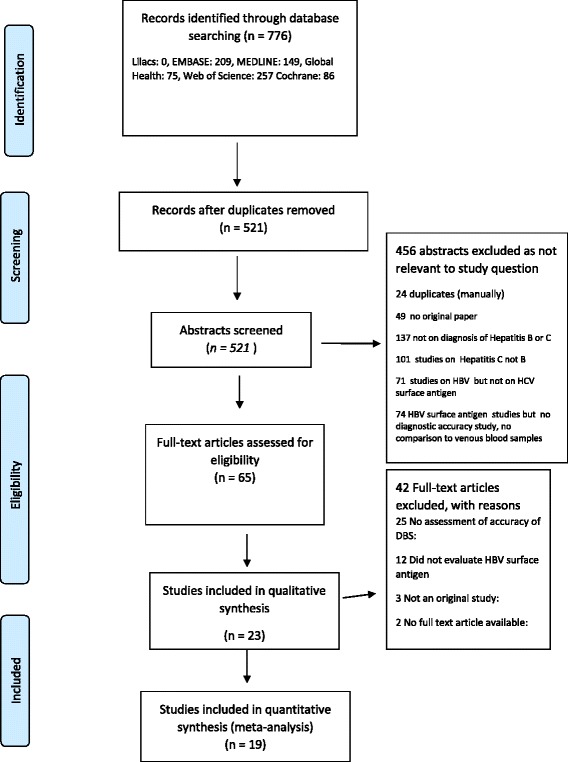
PRISMA flow chart of studies included in the systematic review of detection of hepatitis B Surface antigen from DBS samples compared to venous blood sampling (plasma/serum)
Table 1.
Characteristics of studies included in the systematic review on HBV surface antigen detection from DBS compared to venous blood sampling
| Author, Year, Country | Study design | Study pop, Sample size | Storage conditions | DBS collection method | Serum and plasma antigen test | DBS antigen test | Suggested Cut-off | Specificity | Sensitivity | Correlation/Agreement | Effect of storage conditions |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Alidjinou 2013 Congo-Brazzaville | Cross-sectional | Attending hepatology clinic, 32 | Temperature: Room temperature (25C) Time: 2 months | Whole blood by venipuncture, 30 μl of plasma onto filter paper | Enzygnost ELISA (Siemens) | Enzygnost ELISA (Siemens) | NR | 100 | 100 | Spearman correlation coefficient r of 0.89 | No significant change |
| Boa-Sorte 2014 Brazil |
Cross-sectional | Pregnant women, 692 | Temperature: Refrigerated Time: Less than 5 days | Unclear amount, venipuncture, Schleicher & Schuell 903© filter paper | IMUNOSCREENHBSAG–SS (Mbiolog Diag.), and Murax HB Sag (Murax BioTech Unlmtd.) | IMUNOSCREENHBSAG–SS (Mbiolog Diag.), and Murax HB Sag (Murax BioTech Unlmtd.) | NR | 100 | 100 | NR | NR |
| Brown, 2012, UK | Cross sectional | Unclear | NR | NR | Abbott Architect I2000 | Abbott Architect I2000 | NR | 100 (unclear N) | 98% (unclear N) | NR | NR |
| Farghaly AM 1990 Egypt | Cross sectional | 29 HBsAg pos sera 10 sera positive for anticore only 25 sera negative for any marker of HBV | Temperature: Refrigerated (−20 °C) | 5 ml by venepuncture, three drops of blood (50-100 μl) on Whatman No.3 | Enzygnost HBsAg | Enzygnost HBsAg | NR | 100 (25/25) | 100 (29/29) | NR | NR |
| Farzadegan 1978 Iran |
Cross-sectional | 10 carriers of HBsAg, 10 | NR | Capillary (>0.025 ml) Whatman 4 | RIA (Austria II, Abbot), Abbot’s Auscell | RIA (Austria II, Abbot), Abbot’s Auscell | NR | NR | 100 (10/10) | NR | Samples stored for 1, 3, 7, 14, and 30 days at 4 °C and 37 °C. Best storage temp °4. |
| Forbi 2010 Nigeria | Cross-sectional | Broad, 300 | Temperature: 4C Time: Overnight | Venipuncture, 25 μl of whole blood, Whatman filter paper | Shantest TM- HBsAg ELISA | Shantest TM- HBsAg ELISA | NR | 88.6 | 78.6 | NR | NR |
| Grune 2015 Germany | Cross-sectional | Inpatients 299 | Temperature: −20C, 4C or ambient temperature Time: Up to 14 days | Venipuncture, 100 μl whole blood on paper card | Not specified | Not specified | 0.15 IU/ml Derived from different sample | 99.8 | 91.7 | NR | NR |
| Halfon 2012 | Cohort | Broad, 200 | Temperature: -20C (within 48 h of collection) Time: Not specified | Finger prick, 3 drops whole blood on paper card | BV Cobas Taqman (Roche), Abbott Architect | BV Cobas Taqman (Roche), Abbott Architect | 2.8 IU/mL | 100 | 98 | NR | NR |
| Kania 2013 Burkina-Faso | Cross-sectional | Attendees of HIV testing center, 218 | Temperature: Ambient temperature Time: Not specified | 5 spots whole blood (50 μl) on Whatman 903 | Determine, SD Bioline, ETI-MAK-4 HBsAg EIA (DiaSorin S.p.A.), Architect HBsAg QT (Abbott) | Determine, SD Bioline, ETI-MAK-4 HBsAg EIA (DiaSorin S.p.A.), Architect HBsAg QT (Abbott) | Optical density: MAPC (mean absorbance of positive control)/2 + 0.3 standard deviations = 0.825. | 100 | 96 | Kappa: 0.98 | NR |
| Khan 1996 Pakistan |
Cross sectional | Broad, 90 | Temperature: 2–8 °C Time: overnight |
Venepuncture, NR | Enzygnost, HBsAg monoclonal II by Behring | Enzygnost, HBsAg monoclonal II by Behring | Cut off value (according to figures: 0.116 mean absorbance value) | 100 (86/86) | 100 (4/4) | NR | NR |
| Lukacs Germany 2005 | Unclear | 70 random samples from newborns (as negative controls), 8 hepatitis b patients | Temperature: NR Time: NR |
Venipuncture, NR | NR | Luminex 100 reader, SREPE | Cut-off median fluorescence intensity 233 (mean + 2 Standard deviations for healthy controls) | NR | 100% (8/8) | NR | NR |
| Lee 2011 Malaysia | Cross-sectional | Patients at a tertiary hospital, 150 | Temperature: -20C Time: NR |
Venipuncture, 50 μl whole blood on filter paper | Abbott, Pearson Test | Abbott, Pearson Test | Cut-off point of 1.72 Relative light units |
98 | 97 | Pearson value, r = 0.43 | NR |
| Mayer 2012 US |
Cross-sectional | Temprature: ambient temperatures Time: Not specified |
50 μl on Whatman (8 mm) | ADVIA Centaur 5634 W | ADVIA Centaur 5634 W | 100 19/19 |
100 44/44 |
r = 0.99 | Samples stored at ambient temperature and assayed for month four months every week with HBsAg concentration stable | ||
| Mendy 2005 The Gambia |
Cohort | Broad, 166 | Temperature: 30-33C (humid conditions) Time: Up to 4 weeks |
Three to five drops of blood (20 μl) on filter paper | Determine HBsAg (Abbott Laboratories) | Determine HBsAg (Abbott Laboratories) | 100 | 96 | NR | NR | |
| Mohamed 2013 France |
Cohort | Broad, 200 | Temperature: Room temperature Time: 1, 3, 7 and 14 days |
Venipuncture 50 μl on 12 mm discs (Whatman) |
Chemiluminescent microparticle immunoassay (Abbott) | Chemiluminescent microparticle immunoassay (Abbott) | 0.30 +/−0.81 IU/mL. | 100 | 98 | r = 0.85 | No significant change |
| Mossner Denmark 2016 |
Cohort | 404 Prospective patients from hepatitis clinic and blood donors | Temperature: Room temperature Time 1–5 days |
Finger prick, 75 μ on Whatman filter paper | Architect HBs Ab and Architect HBc Ab assays using the Architect system (Abbott Diagnostics, Delkenheim, Germany) |
Architect HBs Ab and Architect HBc Ab assays using the Architect system (Abbott Diagnostics, Delkenheim, Germany) |
The DBS negative patient had serum quantitative level of HbsAg 175 IU/mL. | 100% 318/318 | 98% 84/85 |
NR | Variation of 24 h to up to 7 d found no difference in stability of samples |
| Nielsen, 1980, Germany | Cross sectional | 110 patients of both sexes older than 6 years attending outpatient department of the General Hospital Barmenda, Cameroon | Temperature | Venipuncture, 50 μl on 16 mm diameter filter paper, Schleicher-Schüll | RIA (Ausria II-test) | RIA (Ausria II-test) | NR | NR | 100 11/11 |
NR | NR |
| Parkinson, 1996, Alaska | Cross sectional | Alaska Native carriers of HBsAg, volunteer and pregnant females, 10 | Temperature and Time NR | Venipuncture, 100 μl applied to 1,5 cm diameter filter paper (No 903, Schleicher &Schuell) | RIA (Ausria II-test) | RIA (Ausria II-test) | Limit of detection approximately 1.25 ng/ml in blood spots | NR | 100 (10/10) | NR | Detectable after 8 weeks of storage at ambient temperatures |
| Ross 2013 Germany |
Cross-sectional | Broad, 299 | Time and temperature not specified | Venipuncture, 100 μl applied to filter paper (Whatman) | Abbott ARCHITECT HBsAg | Abbott ARCHITECT HBsAg | 15 IU/ml (HBsAg) | 100 | 99 | r = 0.86, good agreement in Bland-Altmann plot | NR |
| Villa Italy 1981 |
Cross-sectional | Patients attending liver clinic, 24 | Temperature: -20C 4C and room temperature. Time: 1,7, 15, 30, 60, and 180 days |
Capillary blood on Whatman filter paper | Ausria II, Ausab, Corab, e-anti e RIA kit (Abbott) | Ausria II, Ausab, Corab, e-anti e RIA kit (Abbott) | Unknown | 100 | 100 | NR | Temperature: Storage at room temperature resulted in no significant change compared to samples stored at 4C or -20C. Time: Storage greater than 15 days negatively affected sensitivity. |
| Villar Brazil 2011 |
Cross sectional | Patients attending HCVlinic, 133 | Temperature:-20C, 4–8C, 22–25C, and 22–25C. Time:1, 7, 14, 21, 42, 63, 112, and 183 days. |
either a finger prick or from 70 μl of a whole blood sample onto the 903 Specimen Collection Paper, Whatman Protein Saver Card | ETI-MAK-4 (Diasorin) | ETI-MAK-4 (Diasorin) | Absorbance value 0.115 | 97 | 98 | NR | Accuracy of DBS samples was stable over 63 days at all temperatures evaluated but after 63 days, accuracy diminished when stored at 22–25C |
| Zhuang Australia 1982 |
Cross-sectional | 90 sera selected from serum collection at Fairfield Hospital, Australia | Temperature: incubated at room temperature Time: 1 night |
200 μl on filter-paper (glass-fibre paper, cut into 3x3cm squares) | Ausria II-125 | RIA | NR | NR | 100 90/90 |
NR | Storage at 4 °C, ambient temperatures and 37 °C up to 1 month did not lower sensitivity |
| Zoulek Sao Tome and Principe 1983 |
Cross sectional | 86 children (48 boys, 38 girls, mean age: 8,3 + − 8,5) | Temperature: stored at 4 °C for 2 weeks, −20 °C for weeks Time: 2 weeks, then unspecified |
Venepuncture 2–4 drops (50-100 μl) on 1 cm filter paper discs | RIA Abbot (unspecified) | RIA Abbot (unspecified) | Lowest concentration analysed 0,1 mg/l | NR | 100 24/24 | NR | NR |
For HCV-Ab detection, our search identified 521 abstracts, of which 101 full text articles were assessed for eligibility, 23 studies met the criteria for inclusion in the review [26, 27, 37, 39, 40, 43, 55–71]. Nineteen had sufficient data for inclusion in the quantitative meta-analysis (Fig. 2 & Table 2). Four studies did not have sufficient data for sensitivity and specificity calculations [60, 61, 67] or described a Receiver Operating Characteristic (ROC) curve only [40], and contacting the authors did not yield further information. Two of the 19 studies provided sufficient data to calculate sensitivity but did not have any negative samples to assess specificity [37, 56] (Fig. 2 & Table 2). Of the 23 studies included in the review, most originated from Europe, North America and Australia (16 in total). Four studies were from South America [26, 59, 69, 70] and three from South-East and Central Asia (India [62], Mongolia [60] and Malaysia [40]). All included studies had been published between 1997 to 2017. Most studies used 50 μl to 100 μl of whole blood on filter paper to test for HCV-Ab. Six studies reported using capillary blood samples [26, 55, 57, 58, 63, 66], while others either did not report on this or used venous blood. One study included children [68], however, age ranges or gender for patients were not always reported. Various assays for detection of HCV-Ab in serum and DBS were used (see also Table 2).
Fig. 2.
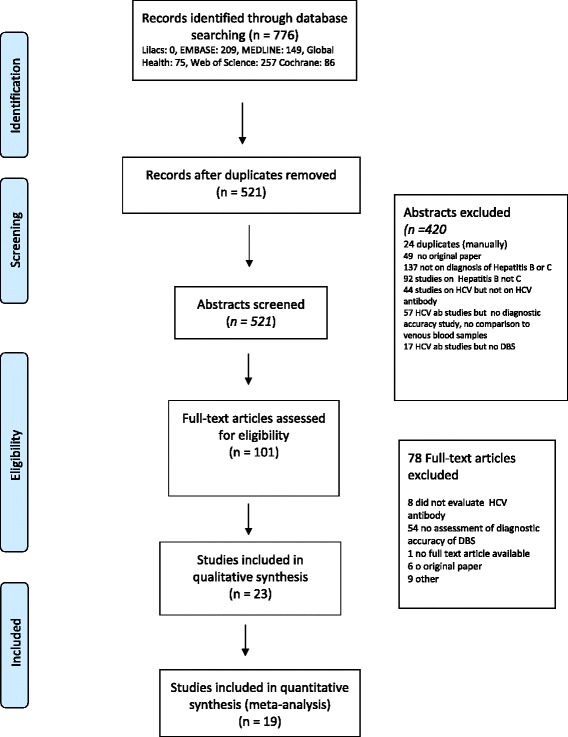
PRISMA flow chart of studies included in the systematic review on detection of hepatitis C antibody detection from DBS samples compared to venous blood sampling (plasma/serum)
Table 2.
Characteristics of studies included in the systematic review on HCV antibody detection from DBS compared to venous blood sampling
| Author, Country | Study design | Study pop, Sample size | Storage conditions | DBS collection method | Serum and plasma antibody test | DBS antibody test | Suggested Cut-off | Specificity | Sensitivity | Correlation/Agreement | Effect of storage conditions |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Brandao Brazil 2013 |
Cross sectional | 386 persons, 40 anti-HCV pos, 346 blood donoers HCV non-reactive, | DBS samples air dried at room temperature for 4 h, stored at −20 °C. Temperature: −20 °C Time: Not specified |
capillary blood by finger prick 75 μl onto Whatman filter paper | MonolisaTM HCV AgAb ULTRA, Bio-Rad (Marnes-la-Coquette, France), and Murex HCV AgAb, Abbott (Kyalami, Republic of South Africa). | MonolisaTM HCV AgAb ULTRA, Bio-Rad (Marnes-la-Coquette, France), and Murex HCV AgAb, Abbott (Kyalami, Republic of South Africa). |
ROC cutoff: 0.287 nm for Monolisa assay ROC cut-off for Murex assay 0.238 nm Derived from the same sample |
99.7 (98.4–99.9) 95.9 (93.3–97.8) |
97.5 (86.8–99.9) 97.5 (86.8–99.9) |
PPV and NPV calculated Kappa = 0.99 (with ROC cut-off), |
stability up to 60 days of storage at room temperature, but less variation at −20 °C |
| Croom Australia 2006 | Cross sectional | 103 samples from high risk groups, negative samples from 94 indivdiuals tested at Haematology Lab | Air dried at room temperature, storage at −20 °C, plasma at −20 °C, time of storage 1 week −11 months Temperature: −20 °C Time: 1 week −11 months |
Venipuncture, 80 μl of each whole blood sample spotted onto Schleicher and Schuell cards (Grade 903) | Monolisa EIA, confirmation test: Murex anti HCV (version 4.0), EIA | Monolisa EIA, confirmation test: Murex anti HCV (version 4.0), EIA | NR | 100% (96–100) 108/108 |
100% (94–100) 75/75 |
NR | NR |
| Chevaliez France 2014 |
Unclear | 529 patients, 183 HCV seronegative, 346 seropositive | NR | NR | EIA HCV assay | EIA HCV assay | 0.2 | 98.9 (96.1–99.7) | 99.1 (97.4–99.7) | R = 0.56 | NR |
| Dokubo US 2014 |
Cross sectional within a prospective cohort study of those HCV positive being followed | 148 participants in a prospective study of HCV | DBS air-dried for 2 h, then sent to another insitute, then stored at −70 °C | Fingerstick on Whatman 903 cards 0.5 ml blood | Standard diagnostics HCV TMA (Norvatis®) ELISA v3.0(Ortho®). |
Standard diagnostics HCV TMA (Norvatis®) ELISA v3.0(Ortho®). |
100% (71/71) |
70% (54/77) |
Kappa 0.69 | NR | |
| Flores 2016 Brazil |
Cross sectional | Participants recruited from ambulatory and general hospital, known HCV/HIV serological status | DBS airdried 4 h, then frozen at -20C | Venipuncture, Whatman filter paper, 3–5 drops (~75 μl) | HCV Murex AB, Diasorin | HCV Murex AB, Diasorin | NR | 100% (99/99) |
94% (213/230) |
Spearman correlation r = 0.520 | NR |
| Gruner 2015 Germany |
Cross-sectional | Inpatients, 299 | Temperature: -20C, 4C or ambient temperature Time: Up to 14 days |
Venipuncture, 100 μl whole blood on paper card | Not specified | Not specified | Derived from different sample | NR | 99% 339/343 |
NR | NR |
| Kania Burkina-Faso 2013 | Cross sectional | 218 HIV screening participants, 5 anti-HCV pos, 213 anti-HCV neg | NR | Venipuncture, 5 spots whole blood (50 μl) on Whatman 903 | Monolisa HCV-Ab-Ag ULTRA assay (Bio-Rad), further assessment with Inno-Lia HCV Score assay (Innogenetics) | Monolisa HCV-Ab-Ag ULTRA assay (Bio-Rad), further assessment with Inno-Lia HCV Score assay (Innogenetics) | 0.439 | 100% (97,8–100%) | 100% (46,3–100%) | kappa: 1,00 (0,93–1,00) | NR |
| Larrat France 2012 |
Cross sectional | One hundred thirteen HCV-positive cases consecutively recruited 17 HIV/HCV co-infected patients (15%) |
DSB dried 24 h at room temperature | Finger prick blood on Whatman card | Monolisa® HCV-Ag-Ab-ULTRA, Bio-Rad) |
Oraquick HCV CEIA Biorad |
0.1 0.2 cEIA |
100 (95.8–100) 88/88 100 (95.8–100) 88/88 |
97.4(92.5–99.1) 110/113 98.2 (93.8–99.5) 111/113 |
ROC AUC OMT cEIA Biorad: 0.99 ROC AUC FSB cEIA Biorad: 0.918 |
At 3 days room temperature 3/3 HCV negative samples NR, ODs lower in HIV co-infected patients |
| Lee Malaysia 2011 | Cross sectional | 150 paired samples | Left to dry overnight at room temperature, then stored −20 °C | Venipuncture, 50 μl whole blood on filter paper | Abbott | Abbott | ROC cut off 0.10 RLU | 100% | 97.3% | ROC curve AUC: 0.99 R = 0.631 |
NR |
| Lukacs Germany 2005 | Unclear | 7 samples from known HCV patients | NR | NR | NR | Luminex | NR | NR | 100% 7/7 | NR | NR |
| McCarron UK 1999 |
Case control | NR | NR | NR | NR | NR | 0.99 1.99 |
87.5% 100% |
100% 97.2% |
NR | NR |
| Marques Brazil 2012 |
Cross sectional | 21 and 24 HCV reactive patients, 234 individual and 132 HCV negative |
serum stored at −20 °C | Venipuncture, 75 μl whole blood on Whatman paper | Two methods: HCV-Ab Radim, Pomezzia, Italy and ETI-AB-HCVK-4 DiaSorin, Vercelli, Italy | Two methods: HCV-Ab Radim, Pomezzia, Italy and ETI-AB-HCVK-4 DiaSorin, Vercelli, Italy | Radim: manu-facturer’s cut off ROC curve for DiaSorin EIA using the same sample population |
99.5% (98–99.9) 98.9% (96.80–99.55) |
97.5% (86.84–99.94) 88.9% (75.95–96.29) |
NR | 2–8 degrees C, 20–25 degrees C, and −20 degrees were evaluated, −20 resulted in lowest variation Methods of cut off determination: the receiver operating characteristic curve(AUROC) |
| Marques Brazil 2016 |
Recruited at Viral Hepatitis Lab | 99 (59 anti HCV/HCV RNA pos, 40 neg samples) | NR | Venipuncture, 3–5 drops on Whatman filter paper | HCV Ab Radim, Pomezia, Italy | HCV Ab Radim, Pomezia, Italy | 100% 40/40 |
94.9% 56/59 |
|||
| Mossner Denmark 2016 | Cohort | 404 Prospective patients from hepatitis clinic and blood donors | Temperature: Room temperature Time 1–5 days | Finger prick, 75 μ on Whatman filter paper | Architect HCV Ab, using the Architect system (Abbott Diagnostics, Delkenheim, Germany) |
Architect HCV Ab, using the Architect system (Abbott Diagnostics, Delkenheim, Germany) |
NR | 100% 288/288 |
97% 112/116 |
NR | Variation of 24 h to up to 7 d found no difference in stability of samples |
| Nandagopal India 2014 | Unclear | Murex | 60 samples | Venipuncture,50 μl of whole blood 903 Whatman card | NR | NR | NR | 100 (29/29) | 100 (31/31) | Pearson correlation coefficient 0.98 | NR |
| O Brien US 2001 |
Multicenter prospective trial (one-arm only) | 1286 subjects enrolled in multi-centre study, | Air dry for 30 min, sent in FedEx envelope | Self collected capillary blood with at home kit | NR | HCV Check, Home Access Corp. self use DBS home kit | NR Several inconclusive and indeterminate results not included in diagnostic accuracy calculations |
100% 686/686 |
99.5% 402/404 | NR | NR |
| Parker UK 1997 |
Case control design | 80 anti HCV positive samples, 52 negative 569 dB sample fields from South African neonates |
Air dry at room temperature before storage at 4 °C | NR, Dried blood field samples | In house IgG ELISA, immunoblot RIBA 3.0 | In house IgG ELISA, immunoblot RIBA 3.0 | T/N 5.0 T/N10.0 |
541/569 95.1% | 78/80 98% 69/80 86.2% |
NR | |
| Ross Germany 2013 |
Unclear | 339 samples | Dried overnight at room temperature | Venipuncture 100 μl of whole blood applied to Whatman 903 filter paper |
ARCHITECT system (Abbott Diagnostics, Delkenheim, Germany). |
ARCHITECT system (Abbott Diagnostics, Delkenheim, Germany). |
NR | 100% (97.7–100) 160/160 |
97.8% (96–100) 175/179 |
NR | NR |
| Soulier France 2017 |
Cross-sectional | 511 patients recruited, with known serostatus for HCV | Temperature:-80 Time: NR |
Venipuncture, 50 μl on Whatman filter paper | EIA; aHCV Vitros ECi; Diagnostics, Raritan, New Jersey). | EIA; aHCV Vitros ECi; Ortho-Clinical Diagnostics, Raritan, New Jersey). | NR | 312/315 98% |
183/186 99% |
NR | 25 dB samples stored at ambient temperatures (24 °C) for a mean duration (±SD) of 19 ± 1 months Sensitivity for genotype detection in DBS 84.5% |
| Sheperd UK 2013 |
Cross sectional | 19 recently infected 300 chronic carrier 82 resolved infection |
DBS stored at 4 °C until use | NR, 50 μl on 903 Whatman Protein Saver cards | ORTHO HCV 3.0 ELISA Test System with Enhanced SAVekit (Ortho Clinical Diagnostics) was used to detect anti-HCV in DBS | NR | Avidity cut-off AI < 30 | NR | NR | NR | NR |
| Tejada-Strop US 2015 |
Case control | 103 patients with known HCV result, 33 adult patients with chronic HepC with stored samples | -20 °C until 5 years later | NR, 75 μl of whole blood on 12 mm DBS | Two immunoassays, the VITROS anti-HCV IgG chemi-luminescence assay (CIA) and the HCV 3.0 enzyme immunoassay(EIA), both from Ortho Clinical Diagnostics (Rochester, NY), | Two immunoassays, the VITROS anti-HCV IgG chemi-luminescence assay (CIA) and the HCV 3.0 enzyme immunoassay(EIA), both from Ortho Clinical Diagnostics (Rochester, NY), | 3.26 CIA 1.5 EIA Derived in the same sample |
Not calculated | CIA 48/52 92% EIA 90% 47/52 For stored samples CIA: 100% (33/33) EIA: 32/33 97% |
100% (CIA and EIA) | NR |
| Tuaillon France 2010 |
Case control | 100 anti HCV pos serum samples and 100 anti HCV neg samples | 18 h dried at room temperature, stored at −20°c for 1–8 weeks | NR, 50 μl of whole blood on Whatman 12 mm paper discs | Ortho HCV 3.0 ELISA, immunoblot assay INNO-LIA HCV Score as confirmatory test | Ortho HCV 3.0 ELISA, immunoblot assay INNO-LIA HCV Score as confirmatory test | Threshold value 0.380 | 98% (97–100) | 99% 97–99) | NR | Stability of anti HCV and HCV RNA investigated by varying room temperature exposure 2–12 days until freezing, after 6 days at room temperature ODs > than cut off values |
| Waterboer Mongolia 2011 |
Cross sectional | 1022 sexually active women from cross sectional study (response rate 69%) | Room temperature up to 8 h, then −20 °C up to 1 month (serum + DBS) | Venipuncture, Whole blood applied to 5 spots on DBS filter paper cards (Whatman 903) | In house, the HCV (strain H77, subtype 1a) Core and NS3 proteins |
In house, the HCV (strain H77, subtype 1a) Core and NS3 proteins | Sera 1492 (Core) 371 (NS3) DBS 967 (Core) 310 (NS3) |
Not calculable from the data | Not calculable from the data | 98% Agreement (kappa 0.94) for Core 96.1% Agreement (kappa 0.90) for NS3 |
NR |
Assessment of study quality and risk of bias (Tables 3 & 4)
Table 3.
Risk of bias in studies included in the systematic review on detection of HB surface antigen
| Author | Patient selection | Bias | Index test | Bias | Reference standard | Bias | Flow and timing | Bias |
|---|---|---|---|---|---|---|---|---|
| Was a case control design avoided? Consecutive or random sample of patients? Inappropriate exclusions? |
Blinded to reference standard Could the conduct or interpretation of the index test have introduced bias? |
Blinded to index? Could the reference standard have introduced bias? |
There is an appropriate interval between the index test and reference standard? All patients receive the same reference standard and are included in the analysis? |
|||||
| Alidjinou | NR, but no case control design | UR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Boa-Sorte | No case control design, consecutive recruitment | LR | blinded | LR | blinded | LR | Same reference standard, all patients included in analysis | LR |
| Brown | Not reported | UR | Not reported | UR | Not reported | UR | Not reported | UR |
| Farzadegan | Only cases, no consecutive sampling | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Farghaly | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Forbi | No case control design | LR | Not blinded, interpretation unbiased | UR | Not blinded, interpretation unbiased | UR | Sampling not reported, same reference standard | UR |
| Gruner | NR | UR | Not blinded, NR | UR | NR | UR | NR | UR |
| Halfon | NR, probably case control design | UR | Not blinded, NR | UR | Not blinded, NR | UR | NR | UR |
| Kania | Consecutive recruitment | LR | Not blinded, interpretation unbiased | UR | Not blinded, interpretation unbiased | UR | Sampling reported | LR |
| Khan | No case control design | LR | Not reported, unclear whether blinded | UR | Not reported, unclear whether blinded | UR | Sampling not reported | UR |
| Lee | Consecutive recruitment | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard | LR |
| Lukacs | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling not reported, same reference standard | UR |
| Mayer | NR, probably case control | UR | Not reported, interpretation unbiased | LR | Not reported, interpretation unbiased | LR | Sampling not reported | UR |
| Mendy | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported | LR |
| Mohamed | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Mossner | Sampling from high-risk and low risk groups | HR | Not blinded, interpretation unbiased | UR | Not blinded, interpretation unbiased | UR | Sampling reported, same reference standard, all patients included in analysis | LR |
| Nielsen | Only cases | HR | Not reported | UR | Not reported | UR | Sampling not reported | UR |
| Parkinson | Only cases | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling not reported, same reference standard | UR |
| Ross | Sampling not reported, probable case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Flow reported | LR |
| Villa | Case control design | HR | NR | UR | NR | UR | NR | UR |
| Villar | Case control design | HR | Bias possible, as selective samples by OD values | HR | Not blinded, interpretation unbiased | LR | Sampling reported | LR |
| Zhuang | No case control design | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling not reported | UR |
| Zoulek | Unclear, but no case control design, probably random or successive | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling not reported, same reference standard | LR |
Abbreviations: LR: low risk, HR: high risk, UR: unknown risk, NR: not reported; shaded: low risk of bias
Table 4.
Risk of bias table for HCV antibody
| Patient selection | Bias | Index test | Bias | Reference standard | Bias | Flow | Bias | |
|---|---|---|---|---|---|---|---|---|
| Was a case control design avoided? Consecutive or random sample of patients? Inappropriate exclusions? |
Blinded to reference standard Could the conduct or interpretation of the index test have introduced bias? |
Blinded to index? Could the reference standard have introduced bias? |
There is an appropriate interval between the index test and reference standard? All patients receive the same reference standard? All patients recruited into the study are included in the analysis? |
|||||
| Brandao | No case control design, consecutive sample, no exclusions | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard | LR |
| Croom | Sampling from high-risk and low risk groups | UR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | All patients included, same reference standard | LR |
| Chevaliez | NR | UR | NR | UR | NR | UR | NR | UR |
| Dokubo | No case control, concurrent sampling from a prospective cohort | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard, all patients recruited included in analysis | LR |
| Flores | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard, all patients recruited included in analysis | LR |
| Gruner | NR | UR | Not blinded, NR | UR | NR | UR | NR | UR |
| Kania | Consecutive recruitment | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported | LR |
| Larrat | Consecutive recruitment, but of known cases and known negative controls | HR | blinded | LR | Blinded | LR | Sampling reported, same reference standard | LR |
| Lee | Consecutive recruitment | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard | LR |
| Lukacs | NR | UR | NR | UR | NR | UR | Sampling reported, same reference standard | LR |
| McCarron | Case control, known positive and negative cases from prevalence survey | HR | NR | UR | NR | UR | NR | UR |
| Marques 2012 | No case control design | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling reported, same reference standard | LR |
| Marques 2016 | NR | UR | Not blinded, interpretation unbiased | UR | Not blinded, interpretation unbiased | UR | NR, same reference standard, NR | UR |
| Mossner | Sampling from high-risk and low risk groups | UR | Not blinded, interpretation unbiased | UR | Not blinded, interpretation unbiased | UR | Sampling reported, same reference standard, all patients included in analysis | LR |
| Nandagopal | NR | UR | NR | UR | NR | UR | NR | UR |
| O Brien | No case control design, | LR | Blinded | LR | Blinded | LR | Sampling partly reported, same reference standard | LR |
| Parker | Case control design | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Sampling partly reported, same reference standard | LR |
| Ross | Possible case control design, sampling NR | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | Flow reported | LR |
| Sheperd | No case control design, but partly sampling from patients with known disease | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Soulier | Sampling from high-risk and low risk-groups | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR, same reference standard, NR | LR |
| Tejada-Strop | Case control | HR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
| Tuaillon, E | Case control | HR | Blinded | LR | Blinded | LR | Sampling reported, same reference standard | LR |
| Waterboer, T | No case control | LR | Not blinded, interpretation unbiased | LR | Not blinded, interpretation unbiased | LR | NR | UR |
Abbreviations: LR: low risk, HR: high risk, UR: unknown risk, NR: not reported; shaded: low risk of bias
Ten studies investigating HBsAg did not use a random or consecutive sampling method [41–48, 50, 55, 56]. Only one study reported on blinding of laboratory personnel to results of the reference test while performing diagnostic tests [35]. Overall, the study quality was rated as moderate. For HCV-Ab detection, eight of the included studies used case-control designs [27, 43, 64, 66–69, 71] and only two reported consecutive sampling [39, 40]. Only three studies reported blinding of laboratory personnel to results of the reference test [27, 57, 66]. However, most of the other studies (as with HBsAg detection) used and reported a clear and consistent protocol for both reference and index test, so this was not judged as a major cause of bias.
Diagnostic performance
In general, based on the 19 studies evaluated, testing for HBsAg using DBS maintained good accuracy compared to testing using plasma or serum from venipuncture. Reported sensitivity estimates ranged from 79%–100% with a pooled estimate from a meta-analysis of 98% (95%CI 95%–99%), and specificity ranged from 89%–100%, with a pooled estimate of 100% (95%CI 99–100%) (Fig. 3). The positive likelihood ratio was 703 (95%CI 107–4615) and the negative likelihood ratio was 0.02 (95% CI 0.008–0.04). Bivariate and univariate estimates of pooled sensitivity and specificity were similar.
Fig. 3.
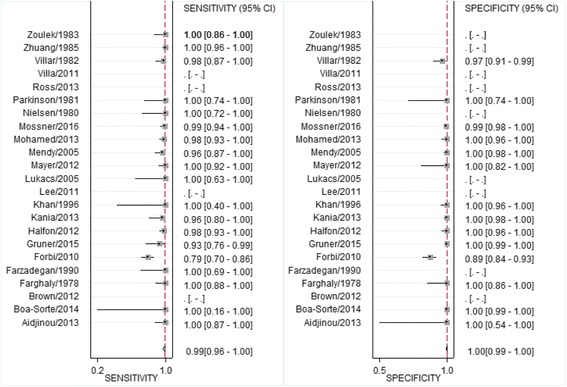
Forest plot of sensitivity and specificity of hepatitis B Surface antigen serological diagnosis in DBS compared to venous blood samples
Based on the 19 studies included in the quantitative meta-analysis for HCV antibody, the reported sensitivity for HCV-Ab using DBS ranged from 70% to 100% and specificity ranged from 95 to 100%. The pooled bivariate estimate of sensitivity and specificity was 98% (95%CI 95–99) and 99% (95%CI 98–100), respectively (Fig. 4). The positive likelihood ratio was 361 (95%CI 61–2163) and the negative likelihood ratio 0.02 (95%CI 0.01–0.05). Four included studies reported also measures of agreement with kappa values ranging from 0.87–1 between DBS and venous blood samples [26, 39, 59, 60]. Bivariate and univariate estimates of pooled sensitivity and specificity were similar.
Fig. 4.
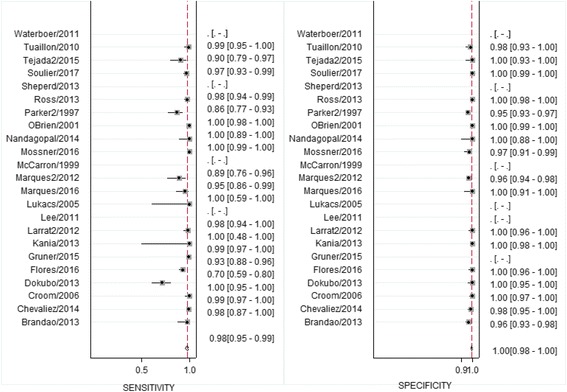
Forest plot of sensitivity and specificity of hepatitis C antibody detection in DBS samples compared to venous blood samples
Visual assessment as well τ2 and its p-value showed moderate heterogeneity in the bivariate analysis of studies. To further assess the heterogeneity, we evaluated different parameters with potential to affect accuracy, including assay type and cut-off used, and storage conditions.
Effect of test and cut-off used
There are no standardised cut-offs for HBsAg detection using DBS. Eight of the studies reported a cut-off based on ROC analysis from the same set of samples as the validation set. Several of the included studies noted that the ideal cut-off (as suggested by ROC curves) for determining test positivity should be higher for DBS samples than for plasma or serum samples. Other studies have indicated that this was due to the small sample volume used in DBS (commonly 50 μl). Indeed the one study with low sensitivity (79%) and specificity (89%) [36] only used 25 μl of blood on filter paper (37).
Different assays were used in HCV-Ab detection. Cut-offs varied widely and as no standardized cut-offs exist, investigators for many studies determined their own cut-off via ROC curves using the same set of samples. Nine of the included studies reported on cut-offs used for DBS [26, 27, 40, 59, 60, 64–66]. Stratification by type of test or cut-off used was not possible as the number of strata would have been large and the results difficult to interpret. Stratification in a pooled analysis based on amount of blood (50 μl versus >50 μl) or sampling method (venous blood vs capillary blood) showed similar sensitivity and specificity estimates (data not shown).
Effect of storage conditions
For HBsAg detection, the effect of a range of storage conditions was evaluated in six studies, including storage temperature ranging from −20 to 33 °C and storage duration ranging from overnight to 180 days. In general, storage at room temperature or higher (30–33 °C) did not clearly affect accuracy of testing and no decrease in sensitivity was found with prolonged storage at ambient temperatures [34, 44–46, 48, 51, 54].
Four studies of diagnostic accuracy for HCV-Ab using DBS samples evaluated different storage conditions in a subset of samples that did not contribute to the diagnostic accuracy evaluations. In one study, three out of three previously negative samples exceeded cut-off values (i.e. would have been interpreted as positive) after storage for 3 days at room temperature [66]. Similarly, one of the included studies showed that after 6 days of storage at room temperature, the cut-off values were exceeded and previously negative samples became positive [63]. Two studies showed relative stability at room temperature for up to 60 days, but found that storage at −20 °C was associated with less variation in quantitative values [26, 59].
One of the 19 studies contributing to the diagnostic accuracy calculations kept study samples at room temperature for more than 24 h [55]. A pooled analysis stratifying studies according to whether samples had been left at room temperature for longer than 4 h or not did not find any difference in performance and so did not explain the heterogeneity observed in the meta-analysis (data not shown).
Sensitivity analysis
In a sensitivity analysis, we found no difference in estimates of diagnostic performance between those studies that reported consecutive sampling and those with case-control study design (data not shown).
Discussion
To our knowledge, this is the first comprehensive and systematic review to summarize the utility of DBS for HCV-Ab and HBsAg testing. Overall, we found very good diagnostic accuracy and precision for detection of HCV-Ab and HBsAg using DBS samples. These findings were based on a larger number of studies compared to the companion systematic reviews undertaken for HBV DNA and HCV RNA using DBS [30] and were rated as moderate quality. This provides support for the use of DBS from capillary blood for diagnostic testing of HBV or HCV, especially in community settings and hard to reach populations where there is limited access to venipuncture or inadequate laboratory infrastructure to prepare or transport plasma samples or limited access to rapid diagnostic tests.
The WHO 2017 testing guidelines recommended the use of a single quality-assured serological assay (i.e either a laboratory-based EIA or RDT to detect HBsAg and anti-HCV that meet minimum performance standards, and where possible delivered at the point of care to improve access and linkage to care and treatment. The guidelines also made a conditional recommendation to consider the option of DBS specimens for HBsAg and HCV-Ab serology testings in settings where there are no facilities or expertise to take venous whole blood specimens; or RDTs are not available or their use is not feasible, or there are persons with poor venous access (e.g. drug treatment programmes, prisons) [29]. This recommendation was rated as conditional mainly because of the slightly lower accuracy using DBS compared to venous blood sampling and uncertainty regarding the generalizability of studies included in this review (in particular regarding impact of different storage and transport conditions).
There were several limitations to this review. First, we did not include studies in languages other than English and no unpublished data from laboratories were included. Second, while only a few studies were rated as having a low risk of bias, overall the quality of evidence from studies was rated as moderate. Third, most studies did not use fresh samples and no uniform protocol was applied. Fourth, there is also a risk of overestimation of pooled sensitivity and specificity by including studies that applied cut-off levels derived from the same study population. No stratified analysis was possible for the type of test within this review. Therefore, we are unable to recommend the use of certain commercial tests over others using DBS testing of HCV-Ab or HBsAg or to suggest a cut-off specific to a test that should be used for DBS samples.
Several studies found that when assessing stability in separate sample sets samples became false-positive after longer exposure at ambient temperatures for both HCV-Ab [27, 66] and HBsAg [44, 45]. Further data is needed to understand the stability of DBS with different environmental conditions (i.e. temperature and humidity). The review highlights the need for standardized validation of specific tests with DBS.
While some studies published detailed protocols on how to collect and analyze DBS [37], no manufacturers to date have provided instructions on how to use their assays with DBS (including processing methods and possibly different cut-offs). There is a need for manufacturers to validate their assays and provide instructions for the use of DBS even if no claim for regulatory approval is made.
Implementation of DBS and future work:
Consideration of the use DBS sampling for HBV and C serological or nucleic acid testing or both will depend on the health-care setting and infrastructure, and epidemiological context. If good-quality RDTs are available that can be performed using capillary blood then the focus may be more on prioritizing DBS for NAT testing of HBV DNA and HCV RNA. However, if RDTs are not available and there are no facilities or expertise to take venous blood samples, then DBS testing may be equally important to increase access to serological testing as well as NAT. Conveniently, both could be performed from the same specimen. A further situation where DBS may be applicable is where large numbers of individuals are being tested simultaneously. DBS may also be useful where polyvalent screening for multiple diseases is done, but where multiplex RDTs for this purpose are not available or are more costly. The adoption of DBS sampling in a hepatitis testing programme requires the availability of a centralized laboratory competent at handling and processing this sample type. The current lack of validation of assays on this sample type for HBsAg or HCV-Ab and manufacturer’s guidance on the optimal pre-analytical treatment of specimens makes quality control challenging.
Further data is also needed on demonstrating the stability of DBS with individual tests under different field transport and storage conditions likely to be encountered in low resource settings. as well as on the type of test used and how long DBS samples can be left at different temperature or humidity levels. Since DBS requires only a small sample of blood to maintain sensitivity, a higher analytical cut-off may be required to maintain sensitivity and overcome variability at the lower end of the dynamic range of the test as compared to higher volume plasma samples – even if in our limited stratified analysis on this we did not find an effect of blood volume on diagnostic accuracy of HBV or HCV. Further insight can also be gained from individual patient analyses of the existing data, which was beyond the scope of this review.
Conclusion
While diagnostic accuracy of DBS for HCV-Ab and HBsAg testing is adequate in studies included in this review, lack of standardization of testing protocols and uncertainty about their use in field conditions and the appropriate assay cut-offs limits the wider application of DBS.
Acknowledgments
Funding
Funding for this study was provided by the World Health Organization, Global Hepatitis Program. Publication of this article was funded by the World Health Organization.
Availability of data and materials
Not applicable.
About this supplement
This article has been published as part of BMC Infectious Diseases Volume 17 Supplement 1, 2017: Testing for chronic hepatitis B and C – a global perspective. The full contents of the supplement are available online at https://bmcinfectdis.biomedcentral.com/articles/supplements/volume-17-supplement-1.
Authors’ contributions
BL, JCo and CMD designed the study according to PICO questions stated during the WHO guideline process. BL, JCa, CP designed the search strategy and performed the search, and data extraction for HCV-Ab DBS studies. JCo, JCh, NG, AN and JG performed the search, screening and data extraction for the HBV surface antigen DBS studies. TR, AI, ET PvdP and PE provided important design and technical insight. BL and JCo performed the meta-analyses. BL, CMD and PE wrote the manuscript. All authors helped with revisions and the final version of the manuscript.
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol. 2014;61(1 Suppl):S45–S57. doi: 10.1016/j.jhep.2014.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Stanaway JD, Flaxman AD, Naghavi M, Fitzmaurice C, Vos T, Abubakar I, et al. The global burden of viral hepatitis from 1990 to 2013: findings from the global burden of disease study 2013. Lancet. 2016;388(10049):1081–1088. doi: 10.1016/S0140-6736(16)30579-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basnayake SK, Easterbrook PJ. Wide variation in estimates of global prevalence and burden of chronic hepatitis B and C infection cited in published literature. J Viral Hepat. 2016;23(7):545–559. doi: 10.1111/jvh.12519. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization. Global hepatitis report. Geneva: World Health Organization; 2017.
- 5.Ott JJ, Stevens GA, Groeger J, Wiersma ST. Global epidemiology of hepatitis B virus infection: new estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine. 2012;30(12):2212–2219. doi: 10.1016/j.vaccine.2011.12.116. [DOI] [PubMed] [Google Scholar]
- 6.Mathurin P. HCV burden in Europe and the possible impact of current treatment. Digestive and liver disease : official journal of the Italian Society of Gastroenterology and the Italian Association for the Study of the Liver. 2013;45(Suppl 5):S314–S317. doi: 10.1016/j.dld.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Guidelines for the prevention, care, and treatment of persons with chronic hepatitis B infection. Geneva; 2015. http://www.who.int/hiv/pub/hepatitis/hepatitis-b-guidelines/en/. Accessed 10 Sept 2017. [PubMed]
- 8.World Health Organization. Guidelines for the screening, care and treatment of persons with chronic hepatitis C infection. Geneva; 2016. http://www.who.int/hepatitis/publications/hepatitis-c-guidelines-2016/en/. Accessed 10 Sept 2017. [PubMed]
- 9.Eckman MH, Kaiser TE, Sherman KE. The cost-effectiveness of screening for chronic hepatitis B infection in the United States. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2011;52(11):1294–1306. doi: 10.1093/cid/cir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khuroo MS, Khuroo NS, Khuroo MS. Diagnostic accuracy of point-of-care tests for hepatitis C virus infection: a systematic review and meta-analysis. PLoS ONE [Electronic Resource] 2015;10(3):e0121450. doi: 10.1371/journal.pone.0121450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shivkumar S, Peeling R, Jafari Y, Joseph L, Pant PN. Accuracy of rapid and point-of-care screening tests for hepatitis C: a systematic review and meta-analysis. Ann Intern Med. 2012;157(8):558–566. doi: 10.7326/0003-4819-157-8-201210160-00006. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization. WHO list of prequalified in vitro diagnostic products. Geneva; 2017. [cited 2017 27.08.2017]. Available from: http://www.who.int/diagnostics_laboratory/evaluations/161003_prequalified_product_list.pdf?ua=1
- 13.Snijdewind IJM, van Kampen JJA, Fraaij PLA, van der Ende ME, Osterhaus ADME, Gruters RA. Current and future applications of dried blood spots in viral disease management. Antivir Res. 2012;93(3):309–321. doi: 10.1016/j.antiviral.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 14.McDade TW, Williams S, Snodgrass JJ. What a drop can do: dried blood spots as a minimally invasive method for integrating biomarkers into population-based research. Demography. 2007;44(4):899–925. doi: 10.1353/dem.2007.0038. [DOI] [PubMed] [Google Scholar]
- 15.Hirtz C, Lehmann S. Blood sampling using "dried blood spot": a clinical biology revolution underway? Ann Biol Clin. 2015;73(1):25–37. doi: 10.1684/abc.2014.1007. [DOI] [PubMed] [Google Scholar]
- 16.Sherman GG, Stevens G, Jones SA, Horsfield P, Stevens WS. Dried blood spots improve access to HIV diagnosis and care for infants in low-resource settings. J Acquir Immune Defic Syndr. 2005;38(5):615–617. doi: 10.1097/01.qai.0000143604.71857.5d. [DOI] [PubMed] [Google Scholar]
- 17.Bravo MJ, Vallejo F, Barrio G, Brugal MT, Molist G, Pulido J, et al. HCV seroconversion among never-injecting heroin users at baseline: no predictors identified other than starting injection. Int J Drug Policy. 2012;23(5):415–419. doi: 10.1016/j.drugpo.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Allen EJ, Palmateer NE, Hutchinson SJ, Cameron S, Goldberg DJ, Taylor A. Association between harm reduction intervention uptake and recent hepatitis C infection among people who inject drugs attending sites that provide sterile injecting equipment in Scotland. Int J Drug Policy. 2012;23(5):346–352. doi: 10.1016/j.drugpo.2012.07.006. [DOI] [PubMed] [Google Scholar]
- 19.McLeod A, Weir A, Aitken C, Gunson R, Templeton K, Molyneaux P, et al. Rise in testing and diagnosis associated with Scotland's action plan on hepatitis C and introduction of dried blood spot testing. J Epidemiol Community Health. 2014;68(12):1182–1188. doi: 10.1136/jech-2014-204451. [DOI] [PubMed] [Google Scholar]
- 20.Hickman M, McDonald T, Judd A, Nichols T, Hope V, Skidmore S, et al. Increasing the uptake of hepatitis C virus testing among injecting drug users in specialist drug treatment and prison settings by using dried blood spots for diagnostic testing: a cluster randomized controlled trial. J Viral Hepat. 2008;15(4):250–254. doi: 10.1111/j.1365-2893.2007.00937.x. [DOI] [PubMed] [Google Scholar]
- 21.McAllister G, Innes H, McLeod A, Dillon JF, Hayes PC, Fox R, et al. Uptake of hepatitis C specialist services and treatment following diagnosis by dried blood spot in Scotland. J Clin Virol. 2014;61(3):359–364. doi: 10.1016/j.jcv.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Hutchinson S. Translating research into public health policy: The scottish national hepatitis C action plan. Suchtmedizin in Forschung und Praxis. 2013; Conference: 3rd International Symposium on Hepatitis Care in Substance Users Munich Germany. Conference Start: 20130905 Conference End: 20130906. Conference Publication: (var.pagings). 15(4):217.
- 23.Jones L, Bates G, McCoy E, Beynon C, McVeigh J, Bellis MA. Effectiveness of interventions to increase hepatitis C testing uptake among high-risk groups: a systematic review. Eur J Pub Health. 2014;24(5):781–788. doi: 10.1093/eurpub/ckt156. [DOI] [PubMed] [Google Scholar]
- 24.Tang W, Chen W, Amini A, Boeras D, Falconer J, Kelly H, et al. Diagnostic accuracy of tests to detect hepatitis C antibody: a meta-analysis and review of the literature. BMC Infect Dis. 2017;17(Suppl 1). doi:10.1186/s12879-017-2773-2. [DOI] [PMC free article] [PubMed]
- 25.Amini A, Varsaneux O, Kelly H, Tang W, Chen W, Boeras D, et al. Diagnostic accuracy of tests to detect hepatitis B surface antigen: a systematic review of the literature and meta-analysis. BMC Infect Dis. 2017;17(Suppl 1). doi:10.1186/s12879-017-2772-3. [DOI] [PMC free article] [PubMed]
- 26.Brandao CP, Marques BL, Marques VA, Villela-Nogueira CA, Do OK, de Paula MT, et al. Simultaneous detection of hepatitis C virus antigen and antibodies in dried blood spots. J Clin Virol. 2013;57(2):98–102. doi: 10.1016/j.jcv.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 27.Tuaillon E, Mondain AM, Meroueh F, Ottomani L, Picot MC, Nagot N, et al. Dried blood spot for hepatitis C virus serology and molecular testing. Hepatology. 2010;51(3):752–758. doi: 10.1002/hep.23407. [DOI] [PubMed] [Google Scholar]
- 28.Greenman J, Roberts T, Cohn J, Messac L. Dried blood spot in the genotyping, quantification and storage of HCV RNA: a systematic literature review. J Viral Hepat. 2015;22(4):353–361. doi: 10.1111/jvh.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization, Guidelines on Hepatitis B and C Testing. Geneva; 2017. http://www.who.int/hepatitis/publications/guidelines-hepatitis-c-b-testing/en/. Accessed 10 Sept 2017.
- 30.Lange B, Roberts T, Cohn J, Greenman J, Camp J, Ishizaki A, et al. Diagnostic accuracy of detection and quantification of HBV-DNA and HCV-RNA using dried blood spot (DBS) samples – a systematic review and meta-analysis. BMC Infect Dis. 2016;17(Suppl 1). doi:10.1186/s12879-017-2776-z. [DOI] [PMC free article] [PubMed]
- 31.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 33.Naaktgeboren CA, Ochodo EA, Van Enst WA, de Groot JAH, Hooft L, Leeflang MMG, et al. Assessing variability in results in systematic reviews of diagnostic studies. BMC Med Res Methodol. 2016;16(1):6. doi: 10.1186/s12874-016-0108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alidjinou EK, Moukassa D, Sane F, Nyenyeli ST, Akoko EC, Mountou MV, et al. Detection of hepatitis B virus infection markers in dried plasma spots among patients in Congo-Brazzaville. Diagn Microbiol Infect Dis. 2014;78(3):229–231. doi: 10.1016/j.diagmicrobio.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Boa-Sorte N, Purificacao A, Amorim T, Assuncao L, Reis A, Galvao-Castro B. Dried blood spot testing for the antenatal screening of HTLV, HIV, syphilis, toxoplasmosis and hepatitis B and C: prevalence, accuracy and operational aspects. Braz J Infect Dis. 2014;18(6):618–624. doi: 10.1016/j.bjid.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forbi JC, Obagu JO, Gyar SD, Pam CR, Pennap GR, Agwale SM. Application of dried blood spot in the sero-diagnosis of hepatitis B infection (HBV) in an HBV hyper-endemic nation. Ann Afr Med. 2010;9(1):44–45. doi: 10.4103/1596-3519.62625. [DOI] [PubMed] [Google Scholar]
- 37.Gruner N, Stambouli O, Ross RS. Dried blood spots--preparing and processing for use in immunoassays and in molecular techniques. Journal of visualized experiments : JoVE. 2015;97 [DOI] [PMC free article] [PubMed]
- 38.Halfon P, Raimondo A, Ouzan D, Bourliere M, Khiri H, Cohen-Bacrie S, et al. Dried blood spot for Hepatitis B virus serology and molecular testing. Journal of hepatology. 2012;Conference: 47th Annual Meeting of the European Association for the Study of the Liver, International Liver Congress 2012 Barcelona Spain. Conference Start: 20120418 Conference End: 20120422. Conference Publication: (var.pagings). 56:S62.
- 39.Kania D, Bekale AM, Nagot N, Mondain AM, Ottomani L, Meda N, et al. Combining rapid diagnostic tests and dried blood spot assays for point-of-care testing of human immunodeficiency virus, hepatitis B and hepatitis C infections in Burkina Faso, West Africa. Clin Microbiol Infect. 2013;19(12):E533–E541. doi: 10.1111/1469-0691.12292. [DOI] [PubMed] [Google Scholar]
- 40.Lee CE, Sri Ponnampalavanar S, Syed Omar SF, Mahadeva S, Ong LY, Kamarulzaman A. Evaluation of the dried blood spot (DBS) collection method as a tool for detection of HIV Ag/Ab, HBsAg, anti-HBs and anti-HCV in a Malaysian tertiary referral hospital. Ann Acad Med Singap. 2011;40(10):448–453. [PubMed] [Google Scholar]
- 41.Mendy M, Kirk GD, van der Sande M, Jeng-Barry A, Lesi OA, Hainaut P, et al. Hepatitis B surface antigenaemia and alpha-foetoprotein detection from dried blood spots: applications to field-based studies and to clinical care in hepatitis B virus endemic areas. J Viral Hepat. 2005;12(6):642–647. doi: 10.1111/j.1365-2893.2005.00641.x. [DOI] [PubMed] [Google Scholar]
- 42.Mohamed S, Raimondo A, Penaranda G, Camus C, Ouzan D, Ravet S, et al. Dried blood spot sampling for hepatitis B virus serology and molecular testing. PLoS ONE [Electronic Resource] 2013;8(4):e61077. doi: 10.1371/journal.pone.0061077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross RS, Stambouli O, Gruner N, Marcus U, Cai W, Zhang W, et al. Detection of infections with hepatitis B virus, hepatitis C virus, and human immunodeficiency virus by analyses of dried blood spots--performance characteristics of the ARCHITECT system and two commercial assays for nucleic acid amplification. Virol J. 2013;10:72. doi: 10.1186/1743-422X-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villa E, Cartolari R, Bellentani S, Rivasi P, Casolo G, Manenti F. Hepatitis-B virus markers on dried blood spots - a new tool for epidemiological research. J Clin Pathol. 1981;34(7):809–812. doi: 10.1136/jcp.34.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villar LM, de Oliveira JC, Cruz HM, Yoshida CFT, Lampe E, Lewis-Ximenez LL. Assessment of dried blood spot samples as a simple method for detection of hepatitis B virus markers. J Med Virol. 2011;83(9):1522–1529. doi: 10.1002/jmv.22138. [DOI] [PubMed] [Google Scholar]
- 46.Parkinson AJ, McMahon BJ, Zanis L, Lanier AP, Wainwright RB. Detection of alpha-fetoprotein and hepatitis-B surface antigen in blood spotted on filter paper: use as a screen for hepatocellular carcinoma in Alaska natives. Arctic Med Res. 1996;55(3):123–128. [PubMed] [Google Scholar]
- 47.Farghaly AM, Kotkat AM. Study of the sensitivity of blood spotted on filter paper in the detection of HBsAg and anticore using ELISA technique. J Egypt Public Health Assoc. 1990;65(3–4):391–400. [PubMed] [Google Scholar]
- 48.Farzadegan H, Noori KH, Ala F. Detection of hepatitis-B surface antigen in blood and blood products dried on filter paper. Lancet. 1978;1(8060):362–363. doi: 10.1016/S0140-6736(78)91085-1. [DOI] [PubMed] [Google Scholar]
- 49.Khan NR, Sadiq F. HBsAg ELISA: comparison of serum and uncentrifuged plasma samples dried on filter paper. East Afr Med J. 1996;73(9):592–593. [PubMed] [Google Scholar]
- 50.Nielsen G, Bienzle U. Evaluation of the hepatitis BS-antigen determination in serum, red blood cells and whole blood dried onto filter paper. Tropenmed Parasitol. 1980;31(1):128–130. [PubMed] [Google Scholar]
- 51.Zoulek G, Burger P, Deinhardt F. Markers of hepatitis viruses a and B: direct comparison between whole serum and blood spotted on filter-paper. Bull World Health Organ. 1985;63(5):935–939. [PMC free article] [PubMed] [Google Scholar]
- 52.Zhuang H, Coulepis AG, Locarnini SA, Gust ID. Detection of markers of hepatitis B infection in serum dried on to filter-paper: an application to field studies. Bull World Health Organ. 1982;60(5):783–787. [PMC free article] [PubMed] [Google Scholar]
- 53.Brown BS, Klapper PE, Guiver M. Development of diagnostic serological and molecular screening from dried blood spots for HCV, HIV, HBV and syphilis. Programme and abstracts of European Society for Clinical Virology (ESCV) winter meeting. J Clin Virol. 2012;44:S27–S28.
- 54.Mayer TK, Vargas RL, Knebel AE, Williams SA, Culver SP, Clark DM, et al. Hepatitis B assays in serum, plasma and whole blood on filter paper. BMC Clin Pathol. 2012;12(8) [DOI] [PMC free article] [PubMed]
- 55.Mossner BK, Staugaard B, Jensen J, Lillevang ST, Christensen PB, Holm DK. Dried blood spots, valid screening for viral hepatitis and human immunodeficiency virus in real-life. World J Gastroenterol. 2016;22(33):7604–7612. doi: 10.3748/wjg.v22.i33.7604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lukacs Z, Dietrich A, Ganschow R, Kohlschutter A, Kruithof R. Simultaneous determination of HIV antibodies, hepatitis C antibodies, and hepatitis B antigens in dried blood spots - a feasibility study using a multi-analyte immunoassay. Clin Chem Lab Med. 2005;43(2):141–145. doi: 10.1515/CCLM.2005.023. [DOI] [PubMed] [Google Scholar]
- 57.JM O’B, Kruzel KE, Wandell MG, Vinogradov IV, Sheagren JN, Frank AP. Detection of hepatitis C antibody with at-home collection kits using an innovative laboratory algorithm. Infect Dis Clin Pract. 2001;10(9):474–480. doi: 10.1097/00019048-200112000-00004. [DOI] [Google Scholar]
- 58.Croom HA, Richards KM, Best SJ, Francis BH, Johnson EI, Dax EM, et al. Commercial enzyme immunoassay adapted for the detection of antibodies to hepatitis C virus in dried blood spots. J Clin Virol. 2006;36(1):68–71. doi: 10.1016/j.jcv.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Marques BL, Brandao CU, Silva EF, Marques VA, Villela-Nogueira CA, Do OK, et al. Dried blood spot samples: optimization of commercial EIAs for hepatitis C antibody detection and stability under different storage conditions. J Med Virol. 2012;84(10):1600–1607. doi: 10.1002/jmv.23379. [DOI] [PubMed] [Google Scholar]
- 60.Waterboer T, Dondog B, Michael KM, Michel A, Schmitt M, Vaccarella S, et al. Dried blood spot samples for seroepidemiology of infections with human papillomaviruses, helicobacter pylori, hepatitis C virus, and JC virus. Cancer Epidemiol Biomark Prev. 2012;21(2):287–293. doi: 10.1158/1055-9965.EPI-11-1001. [DOI] [PubMed] [Google Scholar]
- 61.Shepherd SJ, Kean J, Hutchinson SJ, Cameron SO, Goldberg DJ, Carman WF, et al. A hepatitis C avidity test for determining recent and past infections in both plasma and dried blood spots. J Clin Virol. 2013;57(1):29–35. doi: 10.1016/j.jcv.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 62.Nandagopal P, Iqbal HS, Saravanan S, Solomon SS, Mehta S, Selvakumar M, et al. Evaluation of dried blood spot as an alternative specimen for the diagnosis of anti-HCV in resource-limited settings. Indian J Med Microbiol. 2014;32(2):208–210. doi: 10.4103/0255-0857.129867. [DOI] [PubMed] [Google Scholar]
- 63.Dokubo EK, Evans J, Winkelman V, Cyrus S, Tobler LH, Asher A, et al. Comparison of hepatitis C virus RNA and antibody detection in dried blood spots and plasma specimens. J Clin Virol. 2014;59(4):223–227. doi: 10.1016/j.jcv.2014.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tejada-Strop A, Drobeniuc J, Mixson-Hayden T, Forbi JC, Le NT, Li L, et al. Disparate detection outcomes for anti-HCV IgG and HCV RNA in dried blood spots. J Virol Methods. 2015;212:66–70. doi: 10.1016/j.jviromet.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 65.Chevaliez S, Soulier A, Poiteau L, Pawlotsky JM. Dried blood spots (DBS), a promising tool for large-scale hepatitis c screening, diagnosis and treatment monitoring. Journal of hepatology. 2014;Conference: 49th Annual Meeting of the European Association for the Study of the Liver, International Liver Congress 2014 London United Kingdom. Conference Start: 20140409 Conference End: 20140413. Conference Publication: (var.pagings). 60 (1 SUPPL. 1):S325-S6.
- 66.Larrat S, Bourdon C, Baccard M, Garnaud C, Mathieu S, Quesada JL, et al. Performance of an antigen-antibody combined assay for hepatitis C virus testing without venipuncture. J Clin Virol. 2012;55(3):220–225. doi: 10.1016/j.jcv.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 67.McCarron B, Fox R, Wilson K, Cameron S, McMenamin J, McGregor G, et al. Hepatitis C antibody detection in dried blood spots. J Viral Hepat. 1999;6(6):453–456. doi: 10.1046/j.1365-2893.1999.00197.x. [DOI] [PubMed] [Google Scholar]
- 68.Parker SP, Cubitt WD, Ades AE. A method for the detection and confirmation of antibodies to hepatitis C virus in dried blood spots. J Virol Methods. 1997;68(2):199–205. doi: 10.1016/S0166-0934(97)00127-4. [DOI] [PubMed] [Google Scholar]
- 69.Flores GL, Cruz HM, Potsch DV, May SB, Brandao-Mello CE, Pires MMA, et al. Evaluation of HBsAg and anti-HBc assays in saliva and dried blood spot samples according HIV status. J Virol Methods. 2017;247:32–37. doi: 10.1016/j.jviromet.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 70.Marques BL, do Espirito-Santo MP, Marques VA, Miguel JC, da Silva EF, Villela-Nogueira CA, et al. Evaluation of dried blood spot samples for hepatitis C virus detection and quantification. J Clin Virol. 2016;82:139–144. doi: 10.1016/j.jcv.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 71.Soulier A, Poiteau L, Rosa I, Hezode C, Roudot-Thoraval F, Pawlotsky JM, et al. Dried blood spots: a tool to ensure broad access to hepatitis C screening, diagnosis, and treatment monitoring. J Infect Dis. 2016;213(7):1087–1095. doi: 10.1093/infdis/jiv423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.


