Abstract
What does it take to convert a living organism into a truly productive biofactory? Apart from optimizing biosynthesis pathways as standalone units, a successful bioengineering approach must bend the endogenous metabolic network of the host, and especially its central metabolism, to support the bioproduction process. In practice, this usually involves three complementary strategies which include tuning-down or abolishing competing metabolic pathways, increasing the availability of precursors of the desired biosynthesis pathway, and ensuring high availability of energetic resources such as ATP and NADPH. In this review, we explore these strategies, focusing on key metabolic pathways and processes, such as glycolysis, anaplerosis, the TCA (tricarboxylic acid) cycle, and NADPH production. We show that only a holistic approach for bioengineering — considering the metabolic network of the host organism as a whole, rather than focusing on the production pathway alone — can truly mold microorganisms into efficient biofactories.
Keywords: anaplerosis, central metabolism, glycolysis, metabolic engineering, NADPH production, TCA cycle
Introduction
The bioproduction of value-added chemicals is gaining momentum, slowly but surely replacing environmentally unsustainable fossil-carbon-based chemical processes. Metabolic engineering of microbes now supports the production of food additives [1], plastic monomers [2] and polymers [3], solvents [4], aromatics [5], pharmaceuticals [6], pigments [7], hydrocarbons [8], fuels [9], and chemical building blocks [10]. Successful engineering of a microbe for the efficient bioproduction of a compound requires more than just the introduction of the biosynthesis enzymes. For instance, adaptation of foreign genes into the host organism, in terms of GC content or codon utilization, plays an important role in obtaining high protein expression efficiency and enzymatic activity [11]. Fine-tuning of enzyme levels using different plasmid backbones [12], promoters [13], and ribosome-binding sites [14] is also vital to ensure sufficient flux via the biosynthetic route, while minimizing protein burden and accumulation of toxic or wasteful intermediates [14].
Yet, apart from the properties of the bioproduction pathway itself, as a standalone unit, the endogenous metabolic network of the host organism plays a key role in determining biosynthesis efficiency. There are three primary means by which central metabolism can be modified to enhance a bioproduction process: decreasing and abolishing metabolic flux toward competing pathways that lead to waste byproducts, increasing the availability of the direct precursors of the desired biosynthesis routes, and ensuring high availability of cellular energetic resources, mainly ATP and NADPH. These metabolic strategies are the focus of this review. We argue that engineering of the cell as a whole is crucial for the optimization of any metabolic process. We find the term holistic bioengineering most appropriate to describe this approach.
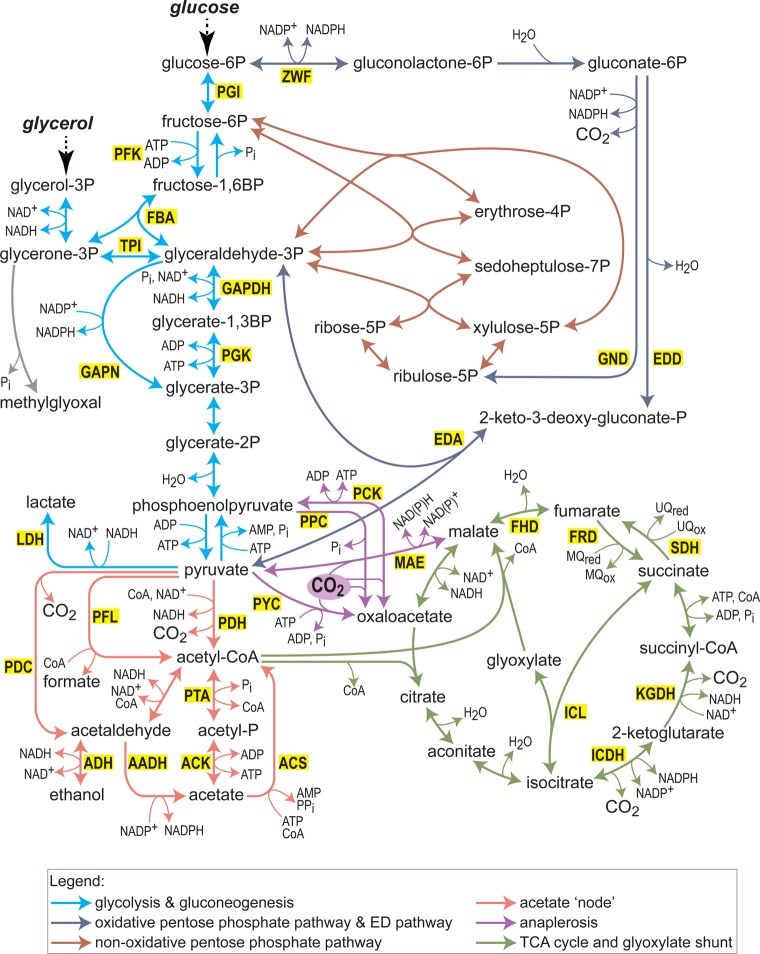
Figure 1 presents the canonical central metabolism, on which the networks of several model biotechnological organisms — mainly Escherichia coli, Corynebacterium glutamicum, and Saccharomyces cerevisiae — are overlaid (ignoring compartmental localization). Key pathways are marked by specific colors. Enzymes to which we directly refer below are marked with a yellow background. We divide this review into several sections, each discussing the modifications made to a different central pathway or process with the aim of enhancing a particular biosynthetic flux. As the topic is quite extensive, our review focuses on several primary examples which, we believe, demonstrate the key aspects.
Figure 1. An overview of the structure of central metabolism in model organisms, such as E. coli, C. glutamicum, and S. cerevisiae, as discussed in the present paper.
Each organism possesses only a subset of the enzymes shown in the figure. Compartmental separation (in case of eukaryotic organisms) is not shown. Glucose and glycerol are shown as representative carbon feedstocks. We have divided central metabolism into different generalized pathways, as indicated by the colors of the arrows. Enzymes mentioned in the text are shown with a yellow background. Some anaplerotic reactions use bicarbonate instead of CO2; for the sake of simplicity, we write CO2 as the substrate of all of them. Abbreviations: ACK, acetate kinase; ACS, acetyl-CoA synthetase; ADH, alcohol dehydrogenase; AADH, acetaldehyde dehydrogenase; EDA, 2-keto-3-deoxygluconate 6-phosphate aldolase; EDD, phosphogluconate dehydratase; FBA, fructose-bisphosphate aldolase; FHD, fumarate hydratase; FRT, fumarate reductase; GAPDH, glyceraldehyde 3-phosphate dehydrogenase (phosphorylating); GAPN, non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase; GND, 6-phosphogluconate dehydrogenase (decarboxylating); ICDH, isocitrate dehydrogenase; ICL, isocitrate lyase; KGDH, 2-ketoglutarate dehydrogenase; LDH, lactate dehydrogenase; MAE, malic enzyme; PKF, 6-phosphofructokinase; PGI, glucose-6-phosphate isomerase; PGK, phosphoglycerate kinase; PCK, phosphoenolpyruvate carboxykinase; PDC, pyruvate decarboxylase; PDH, pyruvate dehydrogenase; PFL, pyruvate formate-lyase; PPC, phosphoenolpyruvate carboxylase; PTA, phosphate acetyltransferase; PYC, pyruvate carboxylase; SDH, succinate dehydrogenase; TPI, triose-phosphate isomerase; ZWF, NADP+-dependent glucose-6-phosphate dehydrogenase.
Abolishing endogenous fermentation products channels flux toward desired pathways
Different organisms, grown under different conditions (e.g. carbon sources and presence or absence of molecular oxygen), produce different combinations of compounds. While it is clear that fermentation, in the absence of electron acceptors, results in the conversion of most carbon input into fermentation products, it is important to emphasize that many organisms also produce large amounts of such products under aerobic conditions, e.g. acetate production by E. coli, ethanol by yeasts, and lactate by cancerous cells. This ‘overflow metabolism’ phenomenon is the center of a wide range of research efforts (e.g. [15–18]). For biotechnological applications, these endogenous fermentation products mostly represent a net loss of carbon which could otherwise be converted into a desired compound. It is, therefore, a common metabolic engineering practice to eliminate competing fermentation pathways with the aim of increasing the conversion yield of feedstock to product [19,20].
Multiple fermentation-related enzymes are targets for disruption: (i) lactate dehydrogenase (LDH), to eliminate lactate production; (ii) alcohol dehydrogenase, to eliminate ethanol biosynthesis; (iii) acetate kinase and/or phosphate acetyltransferase (PTA), to eliminate acetate production; (iv) pyruvate formate-lyase (PFL), to eliminate formate production; and (v) fumarate reductase (FRD), to eliminate succinate production. Deletion of the genes encoding these enzymes, or a subset of them, was shown to dramatically increase the production of hydrogen [21], pyruvate [22–24], lactate [25], succinate [26–28], malate [22,29], 2,3-butanediol [30–32], acetoin [31], acetyl-CoA [33], ethanol [34,35], n-butanol [36], branched-chain alcohols [37], tryptophan [38], citramalate [39], and many other chemicals of interest. Disruption of other enzymes, such as phosphoenolpyruvate (PEP) synthetase, PEP carboxylase (PPC), pyruvate kinase, pyruvate dehydrogenase (PDH), pyruvate oxidase, threonine decarboxylase, and 2-ketobutyrate formate-lyase, is less common, but was also shown to increase the yield of some products (e.g. [23,25,28,29]).
Unsurprisingly, disruption of endogenous fermentation enzymes commonly resulted in lower growth rate and redistribution of central metabolism flux to compensate for the disturbance of cellular ATP and NAD(P)H balance. For example, the simultaneous disruption of multiple fermentation-related enzymes in E. coli has not only led to the accumulation of pyruvate but also resulted in the increased activity of the alternative anaplerotic enzyme PEP carboxykinase (PCK, see section below), as well as an enhanced oxidative phosphorylation, which was required to balance the increased cellular NADH/NAD+ ratio [23]. In another study, deletion of the genes encoding for LDH and PFL in E. coli led to a high NADH/NAD+ ratio which inhibited dihydrolipoamide dehydrogenase, an essential component of the PDH complex. The authors were able to isolate a mutant strain in which a point mutation in this enzyme (E354K) substantially reduced its sensitivity to NADH and therefore enabled a high rate of pyruvate oxidation to acetyl-CoA [40]. In some cases, the disruption of fermentation enzymes resulted in higher biomass yield, and, surprisingly, a higher consumption rate of the sugar feedstock [30]. The latter finding may be explained by the fact that more glucose must now be consumed via alternative pathways with lower ATP yields to supply the energy once provided by the deleted fermentation routes.
In several studies, the ‘trimming’ of natural fermentation pathways was so intensive that the resulting strain became auxotrophic, as was shown in E. coli, where the disruption of PDH, PFL, and pyruvate oxidase not only enhanced pyruvate or lactate production but also rendered the cell dependent on external supply of acetate [24,25]. Such engineering is reasonable only if the externally supplied compound is considerably cheaper (and/or required in much smaller amounts) than the product. An advantage of this approach is that the concentration of the externally provided compound can control the growth of the organism, thereby enabling a tuning of cell growth versus product biosynthesis [25].
Diverting flux toward NADPH production enhances NADPH-consuming pathways
The biosynthesis of many economically interesting products requires high investment of reducing power in the form of NADPH, which, in turn, necessitates increasing the regeneration rate of this essential cofactor [41]. NADP+ is endogenously reduced to NADPH via several routes and enzymes (which not all microbes share): glucose-6-phosphate dehydrogenase (ZWF, NADP+-dependent glucose-6-phosphate dehydrogenase) and 6-phosphogluconate dehydrogenase (GND, 6-phosphogluconate dehydrogenase, decarboxylating) of the oxidative pentose phosphate pathway, NADP-dependent malic enzyme (MAE) working in the decarboxylation direction, isocitrate dehydrogenase (ICDH) of the TCA (tricarboxylic acid) cycle, and the membrane, proton-translocating transhydrogenase (mTH) [42]. Increasing the metabolic flux through these enzymes, via the overexpression of their corresponding genes and deletion of competing pathways, was shown in many studies to enhance the NADPH-dependent biosynthesis of various products.
For example, blockage of normal glycolytic flux, via the disruption of glucose-6-phosphate isomerase (PGI) or 6-phosphofructokinase (PFK), channeled glucose toward the oxidative pentose phosphate pathway and/or the Entner–Doudoroff (ED) pathway (enzymes EDA, 2-keto-3-deoxygluconate 6-phosphate aldolase, and EDD, phosphogluconate dehydratase, in Figure 1). This resulted in increased regeneration of NADPH that supported enhanced production of various commodities, including hydrogen [43,44], lysine [45], valine [46], arginine [47], ornithine [48], lycopene [49], 2-chloropropionic acid [49], and terpenoids [50]. In some of these cases, rather than completely removing PGI, which often results in serious growth retardation, it was possible to reduce the expression level of its gene via replacement of its start codon ATG with GTG [47,48]. In other studies, it was shown that overexpression of the genes of the oxidative pentose phosphate pathway (e.g. ZWF) is enough to channel significant flux toward NADPH regeneration [51–53]. Moreover, as ZWF and GND tend to be inhibited by NADPH, their replacement with NADPH-insensitive counterparts can support higher flux via the pathway [44].
Another study took the idea of diverting flux toward NADPH regeneration to the extreme: glycolytic flux in C. glutamicum was completely blocked by deleting the genes of both PFK and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), such that (almost) all glucose molecules were completely oxidized to CO2. This provided nearly stoichiometric amounts of NADPH via a cyclic activity of the oxidative pentose phosphate pathway [54]. (In reality, one-fourth of the glucose was converted into glycerol, thereby reducing NADPH yield from 12 — maximal stoichiometric yield — to 9.) This enabled the resting cells to serve as a highly efficient catalysts for the NADPH-dependent reduction of the methyl acetoacetate to (R)-methyl-3-hydroxybutyrate [54].
In E. coli growing on glucose under standard aerobic batch conditions, 35–45% of NADPH is produced via the mTH [42], which prompted many metabolic engineers to overexpress the corresponding gene in order to increase NADPH levels. For example, such overexpression resulted in enhanced NADPH-dependent production of poly(3-hydroxybutyrate) (PHB) in E. coli [55]. Similarly, the introduction of E. coli mTH to C. glutamicum improved lysine bioproduction [56]. Overexpression of NAD kinase, which increases the concentration of the overall NADP(H) pool, was further shown to enhance NADPH-dependent bioproduction routes [57–59].
Less ubiquitous enzymes can also channel flux toward NADPH regeneration. The most interesting of these is the NADP-dependent non-phosphorylating glyceraldehyde 3-phosphate dehydrogenase. Overexpression of the Clostridium acetobutylicum gene encoding this irreversible enzyme — directly oxidizing glyceraldehyde 3-phosphate to 3-phosphoglycerate [60] — was shown to enhance the NADPH-dependent production of compounds such as lysine [61] and PHB [62]. Finally, moving away from central metabolism, NADPH availability could be increased by providing reduced compounds that can donate their electrons to NADP, such as formate [63] or phosphite [64].
Alternative glycolytic architectures provide pathway precursors
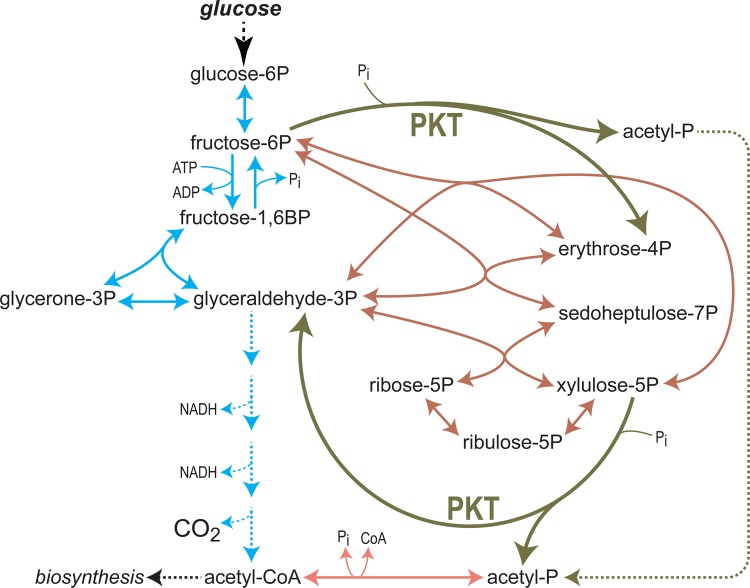
Apart from diverting flux toward increased NADPH production, glycolytic bypasses can also serve to channel carbon toward key precursors of bioproduction pathways, thus supporting their enhanced activity. Perhaps, the most famous example is the use of the enzyme phosphoketolase (PKT) to bypass most of central metabolism, generating activated acetyl moieties directly from phosphosugars [65]. PKT combines ketol cleavage, dehydration, and phosphorolysis to cleave xylulose 5-phosphate to glyceraldehyde 3-phosphate and acetyl phosphate, and fructose 6-phosphate to erythrose 4-phosphate and acetyl phosphate (Figure 2) [66]. Acetyl phosphate can, then, be converted into the central metabolite acetyl-CoA.
Figure 2. PKT enables direct conversion of sugar phosphates to acetyl phosphate and thus acetyl-CoA.
The two reactions catalyzed by the PKT are shown in dark green. Note that the conversion of sugars to acetyl-CoA via PKT is carbon- and redox-neutral, while glycolysis emits CO2 and generates two NADH molecules per each acetyl-CoA produced.
The use of PKT offers two main advantages for the biosynthesis of commodities for which acetyl-CoA serves as a precursor: (i) a direct route from upstream metabolism, i.e. phosphosugars, to acetyl-CoA, which forces high flux toward this central metabolite and (ii) conversion of phosphosugars into acetyl-CoA without the loss of carbon from pyruvate decarboxylation [67], which could potentially increase product yield. In theory, the PKT route could serve to convert glucose into three acetyl-CoA molecules, instead of two via natural glycolysis. However, unlike the endogenous glycolytic pathway, no reducing power is gained via the PKT route, which limits any downstream pathway that heavily relies on NAD(P)H consumption. For example, the biosynthesis of fatty acids requires two NAD(P)H molecules per one acetyl-CoA and therefore optimally operates with endogenous glycolysis that provides this exact ratio.
One of the first uses of PKT in metabolic engineering was to enhance ethanol production from xylose in S. cerevisiae [68]. The enzyme was further harnessed to support increased bioproduction of other acetyl-CoA-derived compounds, such as n-butanol [69], succinate [70], glutamate [71], isoprenoid [72], polyhydroxybutyrate [73], fatty acid ethyl esters [74], and 6-methylsalicylic acid [75]. Yet, the biosynthesis of these compounds also requires reducing power, hence limiting the contribution of the NAD(P)-free PKT route. On the other hand, as the conversion of acetyl-CoA into acetate [67,76] or acetone [77,78] is redox-free, the production of these chemicals can benefit the most from the PKT glycolytic bypass. One especially interesting idea is the use of PKT in cyanobacteria for increased acetone biosynthesis: the CO2 fixed into sugar phosphates was directly channeled to acetyl-CoA and acetone production without any loss of carbon [78].
Increasing the availability of acetyl-CoA can be done by other alternative glycolytic structures. For example, S. cerevisiae does not have a cytosolic pyruvate dehydrogenase, as is the case in E. coli. To increase acetyl-CoA production, the genes encoding acetaldehyde dehydrogenase and acetyl-CoA synthetase (ACS) were overexpressed. Together with the highly active pyruvate decarboxylase (endogenous), these enzymes constituted a ‘pyruvate dehydrogenase’ shunt that efficiently converts pyruvate into acetyl-CoA (Figure 3) [79]. Similarly, establishing high activity of ACS in the cytosol of the yeast Yarrowia lipolytica was shown to divert acetaldehyde and acetate (produced from pyruvate decarboxylase) toward acetyl-CoA, thereby supporting the biosynthesis of the downstream metabolite 2-ketoglutarate [80]. Interestingly, the authors also established high activity of ATP-citrate lyase in the yeast cytosol, regenerating acetyl-CoA from citrate, which was originally produced in the mitochondria and then transported to the cytosol [80].
Figure 3. A synthetic ‘pyruvate dehydrogenase shunt’ engineered in yeast for increased acetyl-CoA production.
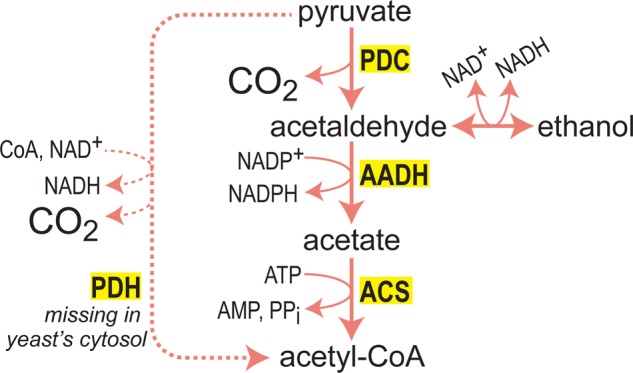
Overexpression of AADH and ACS, in a background of high pyruvate decarboxylase activity, results in a short pathway that converts pyruvate into acetyl-CoA, as is the case for the PDH complex. The PDH shunt, however, dissipates considerably more energy by hydrolyzing ATP.
Activation of the ED pathway was found to support increased pyruvate and acetyl-CoA availability and PHB biosynthesis in E. coli [81]. This is likely because the ED pathway can sustain higher glycolytic flux, per amount of overall enzyme, when compared with the common (Embden–Meyerhof pathway, EMP) glycolysis [82]. Activation of serine degradation, providing yet another route for pyruvate production, further enhanced accumulation of PHB [81], demonstrating that multiple glycolytic bypasses can operate in parallel in a beneficial manner. The ED pathway was further shown to increase isoprenoid biosynthesis via the 2-C-methyl-d-erythritol 4-phosphate pathway (MEP), as it provides balanced amounts of glyceraldehyde 3-phosphate and pyruvate, both of which serve as the direct precursors of this metabolic route [83,84].
In many key biotechnological organisms, such as E. coli and C. glutamicum, the phosphotransferase system (PTS) serves as a key cellular entry point of sugars (e.g. [85,86]). The PTS transfers an activated phosphate group from PEP to the sugar feedstock — e.g. glucose and fructose — thus activating it for use in central metabolism. This forces half of the PEP molecules to be converted into pyruvate, instead of being used for other biosynthetic purposes, for example, anaplerosis and production of aromatic amino acids. (We further note that while C. glutamicum and S. cerevisiae possess an anaplerotic pyruvate carboxylase, PYC, E. coli does not have this enzyme and therefore solely relies on PEP carboxylation for anaplerosis.) Hence, a useful alteration of sugar fermentation results from the replacement of the PTS with a sugar-kinase enzyme that does not consume PEP [87,88], thus freeing this central intermediate to support other metabolic processes. For example, replacement of PTS by ATP-dependent phosphorylation increased the biosynthesis of shikimic acid — a PEP-derived intermediate of aromatic amino acid biosynthesis — in E. coli [89], and enhanced lysine yield in C. glutamicum [90].
Disruption of the endogenous TPI (triose-phosphate isomerase) is a powerful tool to manipulate sugar fermentation, such that only half of its flux proceeds via the normal glycolysis, while the other half (starting from dihydroxyacetone phosphate, DHAP) is channeled toward the biosynthesis of products of interest [91]. This approach was used, for example, to divert half of the glycolytic flux toward methylglyoxal and downstream production of 1,2-propanediol and 1-propanol [92]. Similarly, disruption of TPI resulted in high metabolic conversion of DHAP into glycerol and further into 1,3-propanediol [93].
In some cases, native glycolysis was not disrupted but rather enhanced by an elevated level of its enzymes, in an attempt to increase its flux and support the bioproduction of downstream compounds.1 For example, overexpression of the native components of the PDH complex (i.e. aceEF and lpd), as well as other glycolytic enzymes, e.g. GAPDH and phosphoglycerate kinase (PGK), was used to increase acetyl-CoA availability for the downstream production of butanol in E. coli [94,95]. In a similar manner, elevated levels of the native GAPDH and PGK increased the carbon flux toward malonyl-CoA, a direct product of acetyl-CoA carboxylation [96]. Overexpression of the endogenous genes encoding for TPI and fructose-bisphosphate aldolase led to increased production of acetyl-CoA and its downstream product PHB [97]. Finally, the biosynthesis of glycolytic products can also be enhanced by increasing the availability of essential cofactors, e.g. supplementation of pantothenic acid, a CoA-precursor, resulted in increased levels of CoA and acetyl-CoA and enhanced the production of downstream products, such as isoamyl acetate [98].
We conclude this section by noting that instead of replacing (EMP) glycolysis, it could be beneficial to introduce this pathway into organisms that do not possess it, as the ATP yield of the pathway is higher than the alternatives. For example, Pseudomonas putida metabolizes glucose through a cycle formed by enzymes of the ED, EMP, and pentose phosphate pathways, but cannot operate a linear glycolytic flux as it lacks PFK [99]. Introducing the enzymes of the EMP pathway to this organism enabled linear flux, increasing PHB production 3-fold, presumably due to increased ATP availability [100].
Alternative anaplerosis increases ATP availability and product yield
Anaplerotic reactions convert glycolytic intermediates into those of the TCA cycle, thus enabling the net biosynthesis of di- and tricarboxylic acids. Some of these compounds, such as succinate, malate, and citrate, are economically interesting by themselves, while others are precursors for the biosynthesis of chemicals of interest, e.g. aconitate which can be decarboxylated into itaconate, a top value-added chemical [101]. In most organisms, anaplerotic reactions are carried either by PPC (e.g. in E. coli) or by PYC (e.g. S. cerevisiae). These can be replaced by enzymes that usually support the reverse, decarboxylation direction, i.e. PCK or MAE. Such a replacement results in the net gain of an ATP molecule (Figure 4): while PPC releases the activated phosphate of PEP without generating ATP and PYC consumes the ATP produced by pyruvate kinase, PCK generates ATP from PEP during its carboxylation, and the malic enzyme carboxylates pyruvate without consuming ATP.
Figure 4. An overview of different anaplerotic reactions and their ATP economy.

PPC and PYK are very efficient carboxylating enzymes, but dissipate an ATP equivalent. PCK and the MAE can perform anaplerosis only at high CO2 and at low rate, but can support the conservation of an ATP equivalent, contributing to high cellular energetic state.
The potential to gain an additional ATP by the use of PCK or MAE has led several groups to replace PPC or PYC with these enzymes. It was expected that the increased ATP production would reduce the fraction of carbon channeled toward ATP supply (e.g. oxidation to provide reducing power for oxidative phosphorylation and acetate production) and increase the carbon flux toward the desired product. Indeed, replacement of PPC with PCK in E. coli approximately doubled the intracellular ATP concentration [102,103]. As expected by the increased ATP availability, production of succinate increased by up to 7-fold by such engineering, but only when bicarbonate was added [104,105]. Notably, replacing PPC with PCK resulted in a marked decrease in the growth rate of the engineered E. coli strains [102,103]. Likewise, overexpression of PCK in a PYC-disrupted S. cerevisiae strain sustained growth only at elevated CO2 concentration and only at half the normal growth rate [106]. It is interesting to note that a long-term cultivation of a succinate-producer E. coli strain resulted in increased activity of PCK, which has become the major anaplerotic enzyme, vividly demonstrating the advantage of increasing ATP yield during the production of a TCA cycle intermediate [107].
Replacement of PYC or PPC with MAE has led to similar results. For example, increasing the expression of NADP-dependent MAE in E. coli enhanced the production of succinate and other C4 metabolites but only at increased inorganic carbon concentration [108,109]. An NAD-dependent MAE from E. coli was able to rescue the growth of a PYC-disrupted S. cerevisiae under high CO2 concentration, but only after the emergence of a point mutation that switched the cofactor preference of MAE to NADPH [110].
The low growth rate of the PCK- and the MAE-dependent strains, as well as their dependence on high concentrations of inorganic carbon and NADPH (in the case of the MAE), points to a general phenomenon: a trade-off between efficiency and rate. Specifically, unlike PPC and PYC, which prefer the carboxylation direction (ΔrG′m approximately −40 kJ/mol and approximately −14 kJ/mol, respectively, at pH 7 and ionic strength of 0.25 mM [111]), where ΔrG′m corresponds to the reaction change in Gibbs energy under metabolite concentration of 1 mM [112]), PCK and MAE prefer the decarboxylation direction (ΔrG′m approximately −6 kJ/mol and approximately −4 kJ/mol, respectively). Therefore, operating at high concentration of inorganic carbon and with the reduced NADP pool (rather than the oxidized NAD pool [113]) is essential to provide sufficient driving force to push PCK and MAE toward carboxylation. Even then, the relatively low driving force [114,115] and poor kinetics of these enzymes in the carboxylation direction — low affinity toward inorganic carbon and low kcat — limit the anaplerotic flux and constrain the growth of the engineered strains. Hence, metabolic engineers trying to maximize C4 production are faced with a dilemma: should they increase product yield at the expense of biosynthesis rate (PCK/MAE) or should they favor high flux even if a considerable fraction of the carbon feedstock is wasted to energize the cell (PPC/PYC)?
Disruptions of the TCA cycle direct flux toward metabolites of interest
The enzymes of the TCA cycle are also a common target of disruption for many purposes. A common gene deletion within the TCA cycle, as discussed in the first section, is that of FRD, typically used to abolish the unwanted production of succinate. Yet, it is important to note that this deletion is only partially useful, as succinate dehydrogenase (SDH, preferentially using ubiquinone instead of menaquinone, as is the case for FRD) can also operate in the reductive direction [116]. Another common disruption is that of 2-ketoglutarate dehydrogenase (KGDH), in order to avoid carbon loss via the oxidative cyclic flux and further limit the consumption of acetyl-CoA, thus freeing this key precursor to be used in bioproduction pathways (e.g. [117,118]). Notably, the disruption of KGDH does not interfere with the production of essential cellular building blocks, e.g. oxaloacetate, 2-ketoglutarate, and succinyl-CoA, as they can be produced via the independent operation of the two branches of the TCA cycle, that is, reductive flux from pyruvate/PEP to oxaloacetate and succinyl-CoA and oxidative flux from citrate to 2-ketoglutarate.
As is the case in glycolysis, it was shown that enhancing the flux via the native TCA cycle could serve to support increased bioproduction of its intermediates. For example, succinate biosynthesis was shown to increase in E. coli by overexpression of FRD — supporting higher flux via the reductive arm of the cycle [119–121]. Production of succinate was also shown to benefit from the disruption of KGDH and redirection of TCA flux toward the glyoxylate shunt. For example, disruption of KGDH in S. cerevisiae resulted in isocitrate cleavage to succinate, which, together with the deletion of the SDH gene to prevent recycling of succinate, led to a 4.8-fold higher titer of this dicarboxylic acid [122]. Disruption of SDH and activation of the glyoxylate shunt, via the deletion of its repressor (IclR) and/or deletion of the KGDH gene, were also shown to substantially increase succinate production in E. coli [123–126]. Similarly, increased fumarate production was observed upon deletion of the iclR gene and disruption of all fumarate hydratase isozymes [127].
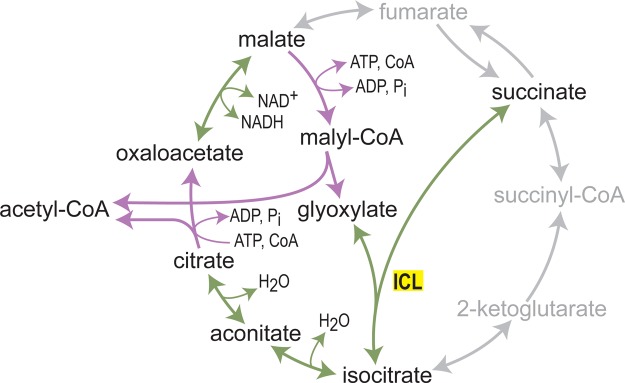
A disruption of citrate synthase resulted in a 2-ketoglutarate auxotrophic strain (required for the biosynthesis of glutamate, glutamine, proline, and arginine), whose prototrophic growth could be reinstituted by the reverse activity of isocitrate lyase in the presence of succinate and glyoxylate [128]. The authors of this study have further demonstrated that glyoxylate can be produced in situ via the combined activity of heterologously expressed malyl-CoA synthetase and lyase, and that a foreign ATP-citrate lyase can serve as the sole source of oxaloacetate (for the biosynthesis of aspartate, asparagine, lysine, methionine, threonine, and isoleucine). Together, this work serves as a preliminary step for the establishment of a reverse glyoxylate shunt, that is, cleavage of succinate into two acetyl-CoA moieties (Figure 5) [128,129].
Figure 5. A reverse glyoxylate shunt, metabolizing succinate into two acetyl-CoA molecules.
Reactions specific to this reverse activity are shown in purple: malyl-CoA synthetase and lyase, and ATP-citrate synthase.
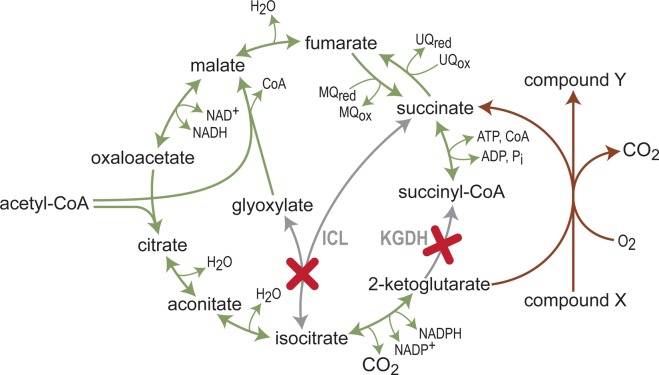
An interesting rewiring of the TCA cycle was established by disrupting both KGDH and ICDH. This strain was not able to grow on a minimal medium with glucose or glycerol as carbon sources, as the downstream flux from citrate was completely blocked [130]. Growth of this strain was restored only when an alternative sink for 2-ketoglutarate was established via a 2-ketoglutarate-dependent dioxygenase (Figure 6) [131], therefore enabling the coupling of cellular growth to the biosynthesis of 4-hydroxyisoleucine (isoleucine + 2-ketoglutarate + O2 → 4-hydroxyisoleucine + succinate + CO2) [130] or 4-hydroxyproline (proline + 2-ketoglutarate + O2 → 4-hydroxyproline + succinate + CO2) [132]. In both these studies, high conversion yields of substrate to product were achieved, vividly demonstrating a central lesson in holistic bioengineering: if it is possible to couple the biosynthesis of a target product to cellular growth, the cell will naturally adapt its metabolism toward increased biosynthetic rate.
Figure 6. Coupling cellular growth to a desired bioconversion.
A strain disrupted in isocitrate lyase (ICL) and KGDH cannot grow on a minimal medium as it is blocked in all routes for citrate/isocitrate/2-ketoglutarate recycling. Overexpression of a 2-ketoglutarate-dependent dioxygenase (brown line) provides a sink for 2-ketoglutarate and thus enables growth, while catalyzing the conversion of a feedstock compound X to a desired compound Y.
Concluding remarks
This review explores and exemplifies the importance of considering the entire metabolic network, and especially central metabolism, when optimizing the biosynthesis of a product of interest. Through the examples presented in this review, several general principles of holistic bioengineering emerge. First and foremost, regardless of the exact pathway or process in question, the manipulation of cellular metabolism is mostly aimed at one of three key goals: (i) deleting endogenous pathways that compete with the biosynthesis of a desired product. These include fermentation pathways in which carbons leak out of the cell, oxidative pathways — such as the TCA cycle — that release carbon as CO2, or simply metabolic highways that channel flux away from the required biosynthetic route. (ii) Increasing flux toward a key precursor of the biosynthesis route. This can be performed by the overexpression of endogenous enzymes or the introduction of foreign enzymes and pathways that directly convert feedstock to precursor. The most commonly pursued biosynthetic precursor is acetyl-CoA; numerous studies have developed innovative strategies to increase the level of this central metabolite toward enhancing the production of a myriad of downstream products. Other metabolic precursors, such as pyruvate or 2-ketoglutarate, have received less attention; however, considering their importance for the biosynthesis of multiple chemicals, we can expect more efforts to be directed toward their enhanced synthesis. (iii) Manipulating the energy and redox state of the cell to rewire cellular fluxes toward bioproduction. Here, we differentiate between two complementary approaches. A general strategy is to increase the ATP levels in the cell — e.g. via ATP-generating anaplerosis or more efficient glycolysis — such that less of the carbon feedstock is directed toward energy production and more toward biosynthesis. A more focused approach is to increase the availability of an energy carrier that is required in high amounts for a specific biosynthetic process. The most common example is NADPH, a high supply of which is essential for the production of numerous chemicals.
Another important principle is the inherent trade-off between rate and yield, especially from a thermodynamic point of view. That is, ATP-efficient pathways tend to operate under a low thermodynamic driving force. This leads to one of two outcomes: low pathway rate or high investment in pathway enzymes, leading to protein burden [114]. One of the most illuminating examples of this phenomenon is the prevalence of the ED pathway. While this pathway generates only half the ATP molecules the EMP glycolysis does, its higher thermodynamic driving force enables high flux at low protein investment and hence, it is the preferred pathway in many microorganisms that can produce ATP via means other than glycolysis, e.g. respiration or photosynthesis [82].
An additional fundamental lesson relates to harnessing natural selection for increased bioproduction. In most cases, sustaining high flux via a biosynthetic route does not benefit the microbe, but rather consumes resources that could otherwise be used to sustain growth, i.e. the activity of the pathway has a negative impact on fitness. As such, strain instability, where short-term cultivation results in the rise of mutations that abolish the activity of a biosynthetic route, is a common hurdle that limits many bioproduction projects (e.g. [133,134]). Yet, in some cases, it is possible to engineer the bioproduction strain such that the activity of the biosynthetic route will be beneficial, or even essential for microbial growth. As growth is coupled to pathway activity, strain evolution is highly unlikely to disrupt it. As discussed above, a common example for this is the deletion of competing fermentation pathways, such that ATP-producing, redox-balanced fermentation can proceed only via a specific pathway leading to a desired product. Another interesting example, discussed in the TCA cycle section, is the use of 2-ketoglutarate-dependent dioxygenase as sole way to recycle 2-ketoglutarate, thus coupling growth to a required biocatalysis. Harnessing natural selection in such a way can serve more than to just ensure the stability of a biosynthetic route: it can also be used to enhance its activity. While most metabolic engineering efforts require multiple design-test cycles — tweaking gene expression levels and monitoring the effect on production — these can be avoided if direct selection for high pathway activity is possible, thus saving time and labor.
In most cases, the rewiring of cellular metabolism was rationally designed based on the knowledge and experience of the authors. In other cases, a computational strategy was taken to systematically explore all possible alternations of central metabolism that could lead to beneficial results (e.g. [135–141]). While both approaches are valid, it is fair to say that in the long run, precise fine-tuning of the endogenous cellular metabolism will vastly benefit from specialized software and computational tools. Yet, for such tools to truly reinvent the field of metabolic engineering, they should involve much more than a stoichiometric analysis, as is commonly the case now. Instead, thermodynamics and kinetics of reactions should be considered [142–145], alongside known regulatory effects, such as allosteric inhibition or activation, as well as measured ranges of enzyme and metabolite concentrations (e.g. [113]). While obtaining sufficient data on these parameters is a not an easy task, the availability of -omics tools and databases can considerably assist in addressing this challenge.
We emphasize that while we focused on central metabolism in this review, a truly holistic bioengineering encompasses other aspects of cellular physiology, including amino acid, nucleotide and fatty acid metabolism, assimilation of inorganic elements such as nitrogen and sulfur, passive and active cellular transport, as well as transcription and translation. Each of these cellular processes can be — and should be — modified or rewired as to support the most efficient conversion of feedstock into product. We would like to finish by noting that even if we keep focusing on central metabolism, the challenge of wholly redrawing it — as opposed to mildly rewiring it, as is commonly done — is still open. To what extent can we reinvent key metabolic pathways? Can such dramatic changes be useful for biotechnological applications? Some pioneering studies, e.g. engineering an active Calvin Cycle in E. coli [146] — may start providing answers to these questions soon.
Acknowledgements
The authors thank Charles A.R. Cotton for helpful discussions, critical reading of the manuscript, and formulation of the holistic bioengineering terminology.
Abbreviations
- ACS
acetyl-CoA synthetase
- C. glutamicum
Corynebacterium glutamicum
- DHAP
dihydroxyacetone phosphate
- E. coli
Escherichia coli
- ED
Entner–Doudoroff
- EMP
Embden–Meyerhof–Parnas
- GAPDH
glyceraldehyde 3-phosphate dehydrogenase (phosphorylating)
- GND
6-phosphogluconate dehydrogenase (decarboxylating)
- ICDH
isocitrate dehydrogenase
- KGDH
2-ketoglutarate dehydrogenase
- LDH
lactate dehydrogenase
- MAE
malic enzyme
- MEP
2-C-methyl-d-erythritol 4-phosphate pathway
- mTH
membrane, proton-translocating transhydrogenase
- PFK
6-phosphofructokinase
- PGI
glucose-6-phosphate isomerase
- PGK
phosphoglycerate kinase
- PCK
phosphoenolpyruvate carboxykinase
- PDH
pyruvate dehydrogenase
- PEP
phosphoenolpyruvate
- PFL
pyruvate formate-lyase
- PHB
poly(3-hydroxybutyrate)
- PTS
phosphotransferase system
- PKT
phosphoketolase
- PPC
phosphoenolpyruvate carboxylase
- PYC
pyruvate carboxylase
- S. cerevisiae
Saccharomyces cerevisiae
- SDH
succinate dehydrogenase
- TCA
tricarboxylic acid
- TPI
triose-phosphate isomerase
- ZWF
NADP+-dependent glucose-6-phosphate dehydrogenase
Footnotes
We note that overexpression of an enzyme does not necessarily lead to an increased pathway flux. The effect of such overexpression will be dependent on the control the enzyme exerts on pathway activity, which is beyond the scope of this review.
Author Contribution
S.A., E.N., and A.B.-E. performed the literature search. S.A. and A.B.-E. wrote the paper. E.N. and A.B.-E. prepared the figures.
Funding
The work was funded by the Max Planck Society. This work was also funded by the Swiss Initiative in Systems Biology (SystemsX.ch) TPdF fellowship [2014-230] (to E.N.)
Competing Interests
The Authors declare that there are no competing interests associated with the manuscript.
References
- 1.Gu P., Su T. and Qi Q. (2016) Novel technologies provide more engineering strategies for amino acid-producing microorganisms. Appl. Microbiol. Biotechnol. 100, 2097–2105 10.1007/s00253-015-7276-8 [DOI] [PubMed] [Google Scholar]
- 2.Chu H.S., Ahn J.-H., Yun J., Choi I.S., Nam T.-W. and Cho K.M. (2015) Direct fermentation route for the production of acrylic acid. Metab. Eng. 32, 23–29 10.1016/j.ymben.2015.08.005 [DOI] [PubMed] [Google Scholar]
- 3.Lin Z., Zhang Y., Yuan Q., Liu Q., Li Y., Wang Z. et al. (2015) Metabolic engineering of Escherichia coli for poly(3-hydroxybutyrate) production via threonine bypass. Microb. Cell Fact. 14, 185 10.1186/s12934-015-0369-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li L., Li K., Wang Y., Chen C., Xu Y., Zhang L. et al. (2015) Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R,3R)-2,3-butanediol from lignocellulose-derived sugars. Metab. Eng. 28, 19–27 10.1016/j.ymben.2014.11.010 [DOI] [PubMed] [Google Scholar]
- 5.Zhao S., Jones J.A., Lachance D.M., Bhan N., Khalidi O., Venkataraman S. et al. (2015) Improvement of catechin production in Escherichia coli through combinatorial metabolic engineering. Metab. Eng. 28, 43–53 10.1016/j.ymben.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 6.Galanie S., Thodey K., Trenchard I.J., Filsinger Interrante M. and Smolke C.D. (2015) Complete biosynthesis of opioids in yeast. Science 349, 1095–1100 10.1126/science.aac9373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li X.-R., Tian G.-Q., Shen H.-J. and Liu J.-Z. (2015) Metabolic engineering of Escherichia coli to produce zeaxanthin. J. Ind. Microbiol. Biotechnol. 42, 627–636 10.1007/s10295-014-1565-6 [DOI] [PubMed] [Google Scholar]
- 8.Menon N., Pásztor A., Menon B.R.K., Kallio P., Fisher K., Akhtar M.K. et al. (2015) A microbial platform for renewable propane synthesis based on a fermentative butanol pathway. Biotechnol. Biofuels 8, 61 10.1186/s13068-015-0231-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Generoso W.C., Schadeweg V., Oreb M. and Boles E. (2015) Metabolic engineering of Saccharomyces cerevisiae for production of butanol isomers. Curr. Opin. Biotechnol. 33, 1–7 10.1016/j.copbio.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 10.Kruyer N.S. and Peralta-Yahya P. (2017) Metabolic engineering strategies to bio-adipic acid production. Curr. Opin. Biotechnol. 45, 136–143 10.1016/j.copbio.2017.03.006 [DOI] [PubMed] [Google Scholar]
- 11.Quax T.E.F., Claassens N.J., Söll D. and van der Oost J. (2015) Codon bias as a means to fine-tune gene expression. Mol. Cell 59, 149–161 10.1016/j.molcel.2015.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lutz R. and Bujard H. (1997) Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 10.1093/nar/25.6.1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braatsch S., Helmark S., Kranz H., Koebmann B. and Jensen P.R. (2008) Escherichia coli strains with promoter libraries constructed by Red/ET recombination pave the way for transcriptional fine-tuning. BioTechniques 45, 335–337 10.2144/000112907 [DOI] [PubMed] [Google Scholar]
- 14.Zelcbuch L., Antonovsky N., Bar-Even A., Levin-Karp A., Barenholz U., Dayagi M. et al. (2013) Spanning high-dimensional expression space using ribosome-binding site combinatorics. Nucleic Acids Res. 41, e98 10.1093/nar/gkt151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basan M., Hui S., Okano H., Zhang Z., Shen Y., Williamson J.R. et al. (2015) Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 10.1038/nature15765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vemuri G.N., Eiteman M.A., McEwen J.E., Olsson L. and Nielsen J. (2007) Increasing NADH oxidation reduces overflow metabolism in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. U.S.A. 104, 2402–2407 10.1073/pnas.0607469104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paczia N., Nilgen A., Lehmann T., Gätgens J., Wiechert W. and Noack S. (2012) Extensive exometabolome analysis reveals extended overflow metabolism in various microorganisms. Microb. Cell Fact. 11, 122 10.1186/1475-2859-11-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liberti M.V. and Locasale J.W. (2016) The Warburg effect: how does it benefit cancer cells? Trends Biochem. Sci. 41, 211–218 10.1016/j.tibs.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsumoto T., Tanaka T. and Kondo A. (2017) Engineering metabolic pathways in Escherichia coli for constructing a ‘microbial chassis’ for biochemical production. Bioresour. Technol. 245(Pt B), 1362–1368 10.1016/j.biortech.2017.05.008 [DOI] [PubMed] [Google Scholar]
- 20.Wendisch V.F., Bott M. and Eikmanns B.J. (2006) Metabolic engineering of Escherichia coli and Corynebacterium glutamicum for biotechnological production of organic acids and amino acids. Curr. Opin. Microbiol. 9, 268–274 10.1016/j.mib.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 21.Maeda T., Sanchez-Torres V. and Wood T.K. (2007) Enhanced hydrogen production from glucose by metabolically engineered Escherichia coli. Appl. Microbiol. Biotechnol. 77, 879–890 10.1007/s00253-007-1217-0 [DOI] [PubMed] [Google Scholar]
- 22.Dong X., Chen X., Qian Y., Wang Y., Wang L., Qiao W. et al. (2017) Metabolic engineering of Escherichia coli W3110 to produce L-malate. Biotechnol. Bioeng. 114, 656–664 10.1002/bit.26190 [DOI] [PubMed] [Google Scholar]
- 23.Yang M. and Zhang X. (2017) Construction of pyruvate producing strain with intact pyruvate dehydrogenase and genome-wide transcription analysis. World J. Microbiol. Biotechnol. 33, 59 10.1007/s11274-016-2202-5 [DOI] [PubMed] [Google Scholar]
- 24.Zelic B., Gostovic S., Vuorilehto K., Vasic-Racki D. and Takors R. (2004) Process strategies to enhance pyruvate production with recombinant Escherichia coli: from repetitive fed-batch to in situ product recovery with fully integrated electrodialysis. Biotechnol. Bioeng. 85, 638–646 10.1002/bit.10820 [DOI] [PubMed] [Google Scholar]
- 25.Zhu Y., Eiteman M.A., DeWitt K. and Altman E. (2007) Homolactate fermentation by metabolically engineered Escherichia coli strains. Appl. Environ. Microbiol. 73, 456–464 10.1128/AEM.02022-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sánchez A.M., Bennett G.N. and San K.-Y. (2005) Novel pathway engineering design of the anaerobic central metabolic pathway in Escherichia coli to increase succinate yield and productivity. Metab. Eng. 7, 229–239 10.1016/j.ymben.2005.03.001 [DOI] [PubMed] [Google Scholar]
- 27.Lee S.J., Song H. and Lee S.Y. (2006) Genome-based metabolic engineering of Mannheimia succiniciproducens for succinic acid production. Appl. Environ. Microbiol. 72, 1939–1948 10.1128/AEM.72.3.1939-1948.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jantama K., Zhang X., Moore J.C., Shanmugam K.T., Svoronos S.A. and Ingram L.O. (2008) Eliminating side products and increasing succinate yields in engineered strains of Escherichia coli C. Biotechnol. Bioeng. 101, 881–893 10.1002/bit.22005 [DOI] [PubMed] [Google Scholar]
- 29.Zhang X., Wang X., Shanmugam K.T. and Ingram L.O. (2011) l-malate production by metabolically engineered Escherichia coli. Appl. Environ. Microbiol. 77, 427–434 10.1128/AEM.01971-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Z.-J., Jian J., Wei X.-X., Shen X.-W. and Chen G.-Q. (2010) Microbial production of meso-2,3-butanediol by metabolically engineered Escherichia coli under low oxygen condition. Appl. Microbiol. Biotechnol. 87, 2001–2009 10.1007/s00253-010-2676-2 [DOI] [PubMed] [Google Scholar]
- 31.Nielsen D.R., Yoon S.-H., Yuan C.J. and Prather K.L.J. (2010) Metabolic engineering of acetoin and meso-2, 3-butanediol biosynthesis in E. coli. Biotechnol. J. 5, 274–284 10.1002/biot.200900279 [DOI] [PubMed] [Google Scholar]
- 32.Jung M.-Y., Mazumdar S., Shin S.H., Yang K.-S., Lee J. and Oh M.-K. (2014) Improvement of 2,3-butanediol yield in Klebsiella pneumoniae by deletion of the pyruvate formate-lyase gene. Appl. Environ. Microbiol. 80, 6195–6203 10.1128/AEM.02069-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krivoruchko A., Zhang Y., Siewers V., Chen Y. and Nielsen J. (2015) Microbial acetyl-CoA metabolism and metabolic engineering. Metab. Eng. 28, 28–42 10.1016/j.ymben.2014.11.009 [DOI] [PubMed] [Google Scholar]
- 34.Papanek B., Biswas R., Rydzak T. and Guss A.M. (2015) Elimination of metabolic pathways to all traditional fermentation products increases ethanol yields in Clostridium thermocellum. Metab. Eng. 32, 49–54 10.1016/j.ymben.2015.09.002 [DOI] [PubMed] [Google Scholar]
- 35.Desai S.G., Guerinot M.L. and Lynd L.R. (2004) Cloning of l-lactate dehydrogenase and elimination of lactic acid production via gene knockout in Thermoanaerobacterium saccharolyticum JW/SL-YS485. Appl. Microbiol. Biotechnol. 65, 600–605 10.1007/s00253-004-1575-9 [DOI] [PubMed] [Google Scholar]
- 36.Atsumi S., Cann A.F., Connor M.R., Shen C.R., Smith K.M., Brynildsen M.P. et al. (2008) Metabolic engineering of Escherichia coli for 1-butanol production. Metab. Eng. 10, 305–311 10.1016/j.ymben.2007.08.003 [DOI] [PubMed] [Google Scholar]
- 37.Atsumi S., Hanai T. and Liao J.C. (2008) Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451, 86–89 10.1038/nature06450 [DOI] [PubMed] [Google Scholar]
- 38.Xu Q., Bai F., Chen N. and Bai G. (2017) Gene modification of the acetate biosynthesis pathway in Escherichia coli and implementation of the cell recycling technology to increase L-tryptophan production. PLoS ONE 12, e0179240 10.1371/journal.pone.0179240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parimi N.S., Durie I.A., Wu X., Niyas A.M.M. and Eiteman M.A. (2017) Eliminating acetate formation improves citramalate production by metabolically engineered Escherichia coli. Microb. Cell Fact. 16, 114 10.1186/s12934-017-0729-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y., Ingram L.O. and Shanmugam K.T. (2008) Dihydrolipoamide dehydrogenase mutation alters the NADH sensitivity of pyruvate dehydrogenase complex of Escherichia coli K-12. J. Bacteriol. 190, 3851–3858 10.1128/JB.00104-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spaans S.K., Weusthuis R.A., van der Oost J. and Kengen S.W.M. (2015) NADPH-generating systems in bacteria and archaea. Front. Microbiol. 6, 742 10.3389/fmicb.2015.00742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sauer U., Canonaco F., Heri S., Perrenoud A. and Fischer E. (2004) The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J. Biol. Chem. 279, 6613–6619 10.1074/jbc.M311657200 [DOI] [PubMed] [Google Scholar]
- 43.Seol E., Ainala S.K., Sekar B.S. and Park S. (2014) Metabolic engineering of Escherichia coli strains for co-production of hydrogen and ethanol from glucose. Int. J. Hydrogen Energy 39, 19323–19330 10.1016/j.ijhydene.2014.06.054 [DOI] [Google Scholar]
- 44.Sundara Sekar B., Seol E. and Park S. (2017) Co-production of hydrogen and ethanol from glucose in Escherichia coli by activation of pentose-phosphate pathway through deletion of phosphoglucose isomerase (pgi) and overexpression of glucose-6-phosphate dehydrogenase (zwf) and 6-phosphogluconate dehydrogenase (gnd). Biotechnol. Biofuels 10, 85 10.1186/s13068-017-0768-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marx A., Hans S., Möckel B., Bathe B., de Graaf A.A., McCormack A.C. et al. (2003) Metabolic phenotype of phosphoglucose isomerase mutants of Corynebacterium glutamicum. J. Biotechnol. 104, 185–197 PMID: [DOI] [PubMed] [Google Scholar]
- 46.Blombach B., Schreiner M.E., Bartek T., Oldiges M. and Eikmanns B.J. (2008) Corynebacterium glutamicum tailored for high-yield l-valine production. Appl. Microbiol. Biotechnol. 79, 471–479 10.1007/s00253-008-1444-z [DOI] [PubMed] [Google Scholar]
- 47.Park S.H., Kim H.U., Kim T.Y., Park J.S., Kim S.-S. and Lee S.Y. (2014) Metabolic engineering of Corynebacterium glutamicum for l-arginine production. Nat. Commun. 5, 4618 10.1038/ncomms5618 [DOI] [PubMed] [Google Scholar]
- 48.Kim S.Y., Lee J. and Lee S.Y. (2015) Metabolic engineering of Corynebacterium glutamicum for the production of l-ornithine. Biotechnol. Bioeng. 112, 416–421 10.1002/bit.25440 [DOI] [PubMed] [Google Scholar]
- 49.Wang Y., San K.-Y. and Bennett G.N. (2013) Improvement of NADPH bioavailability in Escherichia coli through the use of phosphofructokinase deficient strains. Appl. Microbiol. Biotechnol. 97, 6883–6893 10.1007/s00253-013-4859-0 [DOI] [PubMed] [Google Scholar]
- 50.Ng C.Y., Farasat I., Maranas C.D. and Salis H.M. (2015) Rational design of a synthetic Entner–Doudoroff pathway for improved and controllable NADPH regeneration. Metab. Eng. 29, 86–96 10.1016/j.ymben.2015.03.001 [DOI] [PubMed] [Google Scholar]
- 51.Perez-Zabaleta M., Sjöberg G., Guevara-Martínez M., Jarmander J., Gustavsson M., Quillaguamán J. et al. (2016) Increasing the production of (R)-3-hydroxybutyrate in recombinant Escherichia coli by improved cofactor supply. Microb. Cell Fact. 15, 91 10.1186/s12934-016-0490-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang H., Xie X., Xu Q., Zhang C. and Chen N. (2013) Enhancement of cytidine production by coexpression of gnd, zwf, and prs genes in recombinant Escherichia coli CYT15. Biotechnol. Lett. 35, 245–251 10.1007/s10529-012-1068-3 [DOI] [PubMed] [Google Scholar]
- 53.Lim S.-J., Jung Y.-M., Shin H.-D. and Lee Y.-H. (2002) Amplification of the NADPH-related genes zwf and gnd for the oddball biosynthesis of PHB in an E. coli transformant harboring a cloned phbCAB operon. J. Biosci. Bioeng. 93, 543–549 10.1016/S1389-1723(02)80235-3 [DOI] [PubMed] [Google Scholar]
- 54.Siedler S., Lindner S.N., Bringer S., Wendisch V.F. and Bott M. (2013) Reductive whole-cell biotransformation with Corynebacterium glutamicum: improvement of NADPH generation from glucose by a cyclized pentose phosphate pathway using pfkA and gapA deletion mutants. Appl. Microbiol. Biotechnol. 97, 143–152 10.1007/s00253-012-4314-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez A.M., Andrews J., Hussein I., Bennett G.N. and San K.-Y. (2006) Effect of overexpression of a soluble pyridine nucleotide transhydrogenase (UdhA) on the production of poly(3-hydroxybutyrate) in Escherichia coli. Biotechnol. Prog. 22, 420–425 10.1021/bp050375u [DOI] [PubMed] [Google Scholar]
- 56.Kabus A., Georgi T., Wendisch V.F. and Bott M. (2007) Expression of the Escherichia coli pntAB genes encoding a membrane-bound transhydrogenase in Corynebacterium glutamicum improves l-lysine formation. Appl. Microbiol. Biotechnol. 75, 47–53 10.1007/s00253-006-0804-9 [DOI] [PubMed] [Google Scholar]
- 57.Lindner S.N., Niederholtmeyer H., Schmitz K., Schoberth S.M. and Wendisch V.F. (2010) Polyphosphate/ATP-dependent NAD kinase of Corynebacterium glutamicum: biochemical properties and impact of ppnK overexpression on lysine production. Appl. Microbiol. Biotechnol. 87, 583–593 10.1007/s00253-010-2481-y [DOI] [PubMed] [Google Scholar]
- 58.Cui Y.-Y., Ling C., Zhang Y.-Y., Huang J. and Liu J.-Z. (2014) Production of shikimic acid from Escherichia coli through chemically inducible chromosomal evolution and cofactor metabolic engineering. Microb. Cell Fact. 13, 21 10.1186/1475-2859-13-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shi F., Li K., Huan X. and Wang X. (2013) Expression of NAD(H) kinase and glucose-6-phosphate dehydrogenase improve NADPH supply and l-isoleucine biosynthesis in Corynebacterium glutamicum ssp. lactofermentum. Appl. Biochem. Biotechnol. 171, 504–521 10.1007/s12010-013-0389-6 [DOI] [PubMed] [Google Scholar]
- 60.Komati Reddy G., Lindner S.N. and Wendisch V.F. (2015) Metabolic engineering of an ATP-neutral Embden-Meyerhof-Parnas pathway in Corynebacterium glutamicum: growth restoration by an adaptive point mutation in NADH dehydrogenase. Appl. Environ. Microbiol. 81, 1996–2005 10.1128/AEM.03116-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takeno S., Murata R., Kobayashi R., Mitsuhashi S. and Ikeda M. (2010) Engineering of Corynebacterium glutamicum with an NADPH-generating glycolytic pathway for l-lysine production. Appl. Environ. Microbiol. 76, 7154–7160 10.1128/AEM.01464-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Centeno-Leija S., Huerta-Beristain G., Giles-Gómez M., Bolivar F., Gosset G. and Martinez A. (2014) Improving poly-3-hydroxybutyrate production in Escherichia coli by combining the increase in the NADPH pool and acetyl-CoA availability. Antonie Van Leeuwenhoek 105, 687–696 10.1007/s10482-014-0124-5 [DOI] [PubMed] [Google Scholar]
- 63.Ihara M., Kawano Y., Urano M. and Okabe A. (2013) Light driven CO2 fixation by using cyanobacterial photosystem I and NADPH-dependent formate dehydrogenase. PLoS ONE 8, e71581 10.1371/journal.pone.0071581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johannes T.W., Woodyer R.D. and Zhao H. (2005) Directed evolution of a thermostable phosphite dehydrogenase for NAD(P)H regeneration. Appl. Environ. Microbiol. 71, 5728–5734 10.1128/AEM.71.10.5728-5734.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henard C.A., Freed E.F. and Guarnieri M.T. (2015) Phosphoketolase pathway engineering for carbon-efficient biocatalysis. Curr. Opin. Biotechnol. 36, 183–188 10.1016/j.copbio.2015.08.018 [DOI] [PubMed] [Google Scholar]
- 66.Tittmann K. (2014) Sweet siblings with different faces: the mechanisms of FBP and F6P aldolase, transaldolase, transketolase and phosphoketolase revisited in light of recent structural data. Bioorg. Chem. 57, 263–280 10.1016/j.bioorg.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 67.Bogorad I.W., Lin T.-S. and Liao J.C. (2013) Synthetic non-oxidative glycolysis enables complete carbon conservation. Nature 502, 693–697 10.1038/nature12575 [DOI] [PubMed] [Google Scholar]
- 68.Sonderegger M., Schumperli M. and Sauer U. (2004) Metabolic engineering of a phosphoketolase pathway for pentose catabolism in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 70, 2892–2897 10.1128/AEM.70.5.2892-2897.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anfelt J., Kaczmarzyk D., Shabestary K., Renberg B., Rockberg J., Nielsen J. et al. (2015) Genetic and nutrient modulation of acetyl-CoA levels in Synechocystis for n-butanol production. Microb. Cell Fact. 14, 167 10.1186/s12934-015-0355-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jo S., Yoon J., Lee S.-M., Um Y., Han S.O. and Woo H.M. (2017) Modular pathway engineering of Corynebacterium glutamicum to improve xylose utilization and succinate production. J. Biotechnol. 258, 69–78 10.1016/j.jbiotec.2017.01.015 [DOI] [PubMed] [Google Scholar]
- 71.Chinen A., Kozlov Y.I., Hara Y., Izui H. and Yasueda H. (2007) Innovative metabolic pathway design for efficient l-glutamate production by suppressing CO2 emission. J. Biosci. Bioeng. 103, 262–269 10.1263/jbb.103.262 [DOI] [PubMed] [Google Scholar]
- 72.Meadows A.L., Hawkins K.M., Tsegaye Y., Antipov E., Kim Y., Raetz L. et al. (2016) Rewriting yeast central carbon metabolism for industrial isoprenoid production. Nature 537, 694–697 10.1038/nature19769 [DOI] [PubMed] [Google Scholar]
- 73.Kocharin K., Siewers V. and Nielsen J. (2013) Improved polyhydroxybutyrate production by Saccharomyces cerevisiae through the use of the phosphoketolase pathway. Biotechnol. Bioeng. 110, 2216–2224 10.1002/bit.24888 [DOI] [PubMed] [Google Scholar]
- 74.de Jong B.W., Shi S., Siewers V. and Nielsen J. (2014) Improved production of fatty acid ethyl esters in Saccharomyces cerevisiae through up-regulation of the ethanol degradation pathway and expression of the heterologous phosphoketolase pathway. Microb. Cell Fact. 13, 39 10.1186/1475-2859-13-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Panagiotou G., Andersen M.R., Grotkjaer T., Regueira T.B., Nielsen J. and Olsson L. (2009) Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl. Environ. Microbiol. 75, 2212–2220 10.1128/AEM.01461-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Papini M., Nookaew I., Siewers V. and Nielsen J. (2012) Physiological characterization of recombinant Saccharomyces cerevisiae expressing the Aspergillus nidulans phosphoketolase pathway: validation of activity through 13C-based metabolic flux analysis. Appl. Microbiol. Biotechnol. 95, 1001–1010 10.1007/s00253-012-3936-0 [DOI] [PubMed] [Google Scholar]
- 77.Yang X., Yuan Q., Zheng Y., Ma H., Chen T. and Zhao X. (2016) An engineered non-oxidative glycolysis pathway for acetone production in Escherichia coli. Biotechnol. Lett. 38, 1359–1365 10.1007/s10529-016-2115-2 [DOI] [PubMed] [Google Scholar]
- 78.Chwa J.-W., Kim W.J., Sim S.J., Um Y. and Woo H.M. (2016) Engineering of a modular and synthetic phosphoketolase pathway for photosynthetic production of acetone from CO2 in Synechococcus elongatus PCC 7942 under light and aerobic condition. Plant Biotechnol. J. 14, 1768–1776 10.1111/pbi.12536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shiba Y., Paradise E.M., Kirby J., Ro D.-K. and Keasling J.D. (2007) Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab. Eng. 9, 160–168 10.1016/j.ymben.2006.10.005 [DOI] [PubMed] [Google Scholar]
- 80.Zhou J., Yin X., Madzak C., Du G. and Chen J. (2012) Enhanced α-ketoglutarate production in Yarrowia lipolytica WSH-Z06 by alteration of the acetyl-CoA metabolism. J. Biotechnol. 161, 257–264 10.1016/j.jbiotec.2012.05.025 [DOI] [PubMed] [Google Scholar]
- 81.Zhang Y., Lin Z., Liu Q., Li Y., Wang Z., Ma H. et al. (2014) Engineering of Serine-Deamination pathway, Entner-Doudoroff pathway and pyruvate dehydrogenase complex to improve poly(3-hydroxybutyrate) production in Escherichia coli. Microb. Cell Fact. 13, 172 10.1186/s12934-014-0172-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flamholz A., Noor E., Bar-Even A., Liebermeister W. and Milo R. (2013) Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl Acad. Sci. U.S.A. 110, 10039–10044 10.1073/pnas.1215283110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu H., Sun Y., Ramos K.R.M., Nisola G.M., Valdehuesa K.N.G., Lee W.-K. et al. (2013) Combination of Entner-Doudoroff pathway with MEP increases isoprene production in engineered Escherichia coli. PLoS ONE 8, e83290 10.1371/journal.pone.0083290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li C., Ying L.-Q., Zhang S.-S., Chen N., Liu W.-F. and Tao Y. (2015) Modification of targets related to the Entner–Doudoroff/pentose phosphate pathway route for methyl-D-erythritol 4-phosphate-dependent carotenoid biosynthesis in Escherichia coli. Microb. Cell Fact. 14, 117 10.1186/s12934-015-0301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Saier M.H. Jr and Reizer J. (1994) The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 13, 755–764 10.1111/j.1365-2958.1994.tb00468.x [DOI] [PubMed] [Google Scholar]
- 86.Parche S., Burkovski A., Sprenger G.A., Weil B., Krämer R. and Titgemeyer F. (2001) Corynebacterium glutamicum: a dissection of the PTS. J. Mol. Microbiol. Biotechnol. 3, 423–428 PMID: [PubMed] [Google Scholar]
- 87.Hernández-Montalvo V., Martínez A., Hernández-Chavez G., Bolivar F., Valle F. and Gosset G. (2003) Expression of galP and glk in a Escherichia coli PTS mutant restores glucose transport and increases glycolytic flux to fermentation products. Biotechnol. Bioeng. 83, 687–694 10.1002/bit.10702 [DOI] [PubMed] [Google Scholar]
- 88.Moon M.-W., Kim H.-J., Oh T.-K., Shin C.-S., Lee J.-S., Kim S.-J. et al. (2005) Analyses of enzyme II gene mutants for sugar transport and heterologous expression of fructokinase gene in Corynebacterium glutamicum ATCC 13032. FEMS Microbiol. Lett. 244, 259–266 10.1016/j.femsle.2005.01.053 [DOI] [PubMed] [Google Scholar]
- 89.Chandran S.S., Yi J., Draths K.M., von Daeniken R., Weber W. and Frost J.W. (2003) Phosphoenolpyruvate availability and the biosynthesis of Shikimic acid. Biotechnol. Prog. 19, 808–814 10.1021/bp025769p [DOI] [PubMed] [Google Scholar]
- 90.Lindner S.N., Seibold G.M., Henrich A., Kramer R. and Wendisch V.F. (2011) Phosphotransferase system-independent glucose utilization in Corynebacterium glutamicum by inositol permeases and glucokinases. Appl. Environ. Microbiol. 77, 3571–3581 10.1128/AEM.02713-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bujara M., Schümperli M., Pellaux R., Heinemann M. and Panke S. (2011) Optimization of a blueprint for in vitro glycolysis by metabolic real-time analysis. Nat. Chem. Biol. 7, 271–277 10.1038/nchembio.541 [DOI] [PubMed] [Google Scholar]
- 92.Jain R., Sun X., Yuan Q. and Yan Y. (2015) Systematically engineering Escherichia coli for enhanced production of 1,2-propanediol and 1-propanol. ACS Synth. Biol. 4, 746–756 10.1021/sb500345t [DOI] [PubMed] [Google Scholar]
- 93.Nakamura C.E. and Whited G.M. (2003) Metabolic engineering for the microbial production of 1,3-propanediol. Curr. Opin. Biotechnol. 14, 454–459 10.1016/j.copbio.2003.08.005 [DOI] [PubMed] [Google Scholar]
- 94.Bond-Watts B.B., Bellerose R.J. and Chang M.C.Y. (2011) Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 7, 222–227 10.1038/nchembio.537 [DOI] [PubMed] [Google Scholar]
- 95.Chen X., Zhu P. and Liu L. (2016) Modular optimization of multi-gene pathways for fumarate production. Metab. Eng. 33, 76–85 10.1016/j.ymben.2015.07.007 [DOI] [PubMed] [Google Scholar]
- 96.Xu P., Ranganathan S., Fowler Z.L., Maranas C.D. and Koffas M.A.G. (2011) Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab. Eng. 13, 578–587 10.1016/j.ymben.2011.06.008 [DOI] [PubMed] [Google Scholar]
- 97.Lee S.H., Kang K.-H., Kim E.Y., Chae T.U., Oh Y.H., Hong S.H. et al. (2013) Metabolic engineering of Escherichia coli for enhanced biosynthesis of poly(3-hydroxybutyrate) based on proteome analysis. Biotechnol. Lett. 35, 1631–1637 10.1007/s10529-013-1246-y [DOI] [PubMed] [Google Scholar]
- 98.Vadali R.V., Bennett G.N. and San K.-Y. (2004) Applicability of CoA/acetyl-CoA manipulation system to enhance isoamyl acetate production in Escherichia coli. Metab. Eng. 6, 294–299 10.1016/j.ymben.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 99.Nikel P.I., Chavarría M., Fuhrer T., Sauer U. and de Lorenzo V. (2015) Pseudomonas putida KT2440 strain metabolizes glucose through a cycle formed by enzymes of the Entner-Doudoroff, Embden-Meyerhof-Parnas, and pentose phosphate pathways. J. Biol. Chem. 290, 25920–25932 10.1074/jbc.M115.687749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sánchez-Pascuala A., de Lorenzo V. and Nikel P.I. (2017) Refactoring the Embden–Meyerhof–Parnas pathway as a whole of portable GlucoBricks for implantation of glycolytic modules in gram-negative bacteria. ACS Synth. Biol. 6, 793–805 10.1021/acssynbio.6b00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Werpy T., Petersen G., Aden A., Bozell J., Holladay J., White J. et al. (2004) Top Value Added Chemicals From Biomass. Volume 1—Results of Screening for Potential Candidates From Sugars and Synthesis Gas. Department of Energy, Washington, DC [Google Scholar]
- 102.Kwon Y.-D., Lee S.Y. and Kim P. (2008) A physiology study of Escherichia coli overexpressing phosphoenolpyruvate carboxykinase. Biosci. Biotechnol. Biochem. 72, 1138–1141 10.1271/bbb.70831 [DOI] [PubMed] [Google Scholar]
- 103.Na Y.-A., Lee J.-Y., Bang W.-J., Lee H.J., Choi S.-I., Kwon S.-K. et al. (2015) Growth retardation of Escherichia coli by artificial increase of intracellular ATP. J. Ind. Microbiol. Biotechnol. 42, 915–924 10.1007/s10295-015-1609-6 [DOI] [PubMed] [Google Scholar]
- 104.Kim P., Laivenieks M., Vieille C. and Zeikus J.G. (2004) Effect of overexpression of Actinobacillus succinogenes phosphoenolpyruvate carboxykinase on succinate production in Escherichia coli. Appl. Environ. Microbiol. 70, 1238–1241 10.1128/AEM.70.2.1238-1241.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kwon Y.D., Lee S.Y. and Kim P. (2006) Influence of gluconeogenic phosphoenolpyruvate carboxykinase (PCK) expression on succinic acid fermentation in Escherichia coli under high bicarbonate condition. J. Microbiol. Biotechnol. 16, 1448 [Google Scholar]
- 106.Zelle R.M., Trueheart J., Harrison J.C., Pronk J.T. and van Maris A.J. (2010) Phosphoenolpyruvate carboxykinase as the sole anaplerotic enzyme in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 76, 5383–5389 10.1128/AEM.01077-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang X., Jantama K., Moore J.C., Jarboe L.R., Shanmugam K.T. and Ingram L.O. (2009) Metabolic evolution of energy-conserving pathways for succinate production in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 106, 20180–20185 10.1073/pnas.0905396106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kwon Y.-D., Kwon O.-H., Lee H.-S. and Kim P. (2007) The effect of NADP-dependent malic enzyme expression and anaerobic C4 metabolism in Escherichia coli compared with other anaplerotic enzymes. J. Appl. Microbiol. 103, 2340–2345 10.1111/j.1365-2672.2007.03485.x [DOI] [PubMed] [Google Scholar]
- 109.Stols L. and Donnelly M.I. (1997) Production of succinic acid through overexpression of NAD+-dependent malic enzyme in an Escherichia coli mutant. Appl. Environ. Microbiol. 63, 2695–2701 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zelle R.M., Harrison J.C., Pronk J.T. and van Maris A.J.A. (2011) Anaplerotic role for cytosolic malic enzyme in engineered Saccharomyces cerevisiae strains. Appl. Environ. Microbiol. 77, 732–738 10.1128/AEM.02132-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Alberty R.A., Cornish-Bowden A., Goldberg R.N., Hammes G.G., Tipton K. and Westerhoff H.V. (2011) Recommendations for terminology and databases for biochemical thermodynamics. Biophys. Chem. 155, 89–103 10.1016/j.bpc.2011.03.007 [DOI] [PubMed] [Google Scholar]
- 112.Flamholz A., Noor E., Bar-Even A. and Milo R. (2012) eQuilibrator—the biochemical thermodynamics calculator. Nucleic Acids Res. 40, D770–D775 10.1093/nar/gkr874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bennett B.D., Kimball E.H., Gao M., Osterhout R., Van Dien S.J. and Rabinowitz J.D. (2009) Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 10.1038/nchembio.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Noor E., Bar-Even A., Flamholz A., Reznik E., Liebermeister W. and Milo R. (2014) Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 10, e1003483 10.1371/journal.pcbi.1003483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Noor E., Flamholz A., Liebermeister W., Bar-Even A. and Milo R. (2013) A note on the kinetics of enzyme action: a decomposition that highlights thermodynamic effects. FEBS Lett. 587, 2772–2777 10.1016/j.febslet.2013.07.028 [DOI] [PubMed] [Google Scholar]
- 116.Maklashina E., Cecchini G. and Dikanov S.A. (2013) Defining a direction: electron transfer and catalysis in Escherichia coli complex II enzymes. Biochim. Biophys. Acta, Bioenerg. 1827, 668–678 10.1016/j.bbabio.2013.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Causey T.B., Zhou S., Shanmugam K.T. and Ingram L.O. (2003) Engineering the metabolism of Escherichia coli W3110 for the conversion of sugar to redox-neutral and oxidized products: homoacetate production. Proc. Natl Acad. Sci. U.S.A. 100, 825–832 10.1073/pnas.0337684100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang J., Niyompanich S., Tai Y.-S., Wang J., Bai W., Mahida P. et al. (2016) Engineering of a highly efficient Escherichia coli strain for mevalonate fermentation through chromosomal integration. Appl. Environ. Microbiol. 82, 7176–7184 10.1128/AEM.02178-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Goldberg I., Lonberg-Holm K., Bagley E.A. and Stieglitz B. (1983) Improved conversion of fumarate to succinate by Escherichia coli strains amplified for fumarate reductase. Appl. Environ. Microbiol. 45, 1838–1847 PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang X., Gong C.S. and Tsao G.T. (1998) Bioconversion of fumaric acid to succinic acid by recombinant E. coli. Appl. Biochem. Biotechnol. 70–72, 919–928 10.1007/BF02920202 [DOI] [PubMed] [Google Scholar]
- 121.Yu C., Cao Y., Zou H. and Xian M. (2011) Metabolic engineering of Escherichia coli for biotechnological production of high-value organic acids and alcohols. Appl. Microbiol. Biotechnol. 89, 573–583 10.1007/s00253-010-2970-z [DOI] [PubMed] [Google Scholar]
- 122.Raab A.M., Gebhardt G., Bolotina N., Weuster-Botz D. and Lang C. (2010) Metabolic engineering of Saccharomyces cerevisiae for the biotechnological production of succinic acid. Metab. Eng. 12, 518–525 10.1016/j.ymben.2010.08.005 [DOI] [PubMed] [Google Scholar]
- 123.Lin H., Bennett G.N. and San K.-Y. (2005) Fed-batch culture of a metabolically engineered Escherichia coli strain designed for high-level succinate production and yield under aerobic conditions. Biotechnol. Bioeng. 90, 775–779 10.1002/bit.20458 [DOI] [PubMed] [Google Scholar]
- 124.Lin H., Bennett G.N. and San K.-Y. (2005) Genetic reconstruction of the aerobic central metabolism in Escherichia coli for the absolute aerobic production of succinate. Biotechnol. Bioeng. 89, 148–156 10.1002/bit.20298 [DOI] [PubMed] [Google Scholar]
- 125.Lin H., Bennett G.N. and San K.-Y. (2005) Metabolic engineering of aerobic succinate production systems in Escherichia coli to improve process productivity and achieve the maximum theoretical succinate yield. Metab. Eng. 7, 116–127 10.1016/j.ymben.2004.10.003 [DOI] [PubMed] [Google Scholar]
- 126.Cox S.J., Shalel Levanon S., Sanchez A., Lin H., Peercy B., Bennett G.N. et al. (2006) Development of a metabolic network design and optimization framework incorporating implementation constraints: a succinate production case study. Metab. Eng. 8, 46–57 10.1016/j.ymben.2005.09.006 [DOI] [PubMed] [Google Scholar]
- 127.Song C.W., Kim D.I., Choi S., Jang J.W. and Lee S.Y. (2013) Metabolic engineering of Escherichia coli for the production of fumaric acid. Biotechnol. Bioeng. 110, 2025–2034 10.1002/bit.24868 [DOI] [PubMed] [Google Scholar]
- 128.Mainguet S.E., Gronenberg L.S., Wong S.S. and Liao J.C. (2013) A reverse glyoxylate shunt to build a non-native route from C4 to C2 in Escherichia coli. Metab. Eng. 19, 116–127 10.1016/j.ymben.2013.06.004 [DOI] [PubMed] [Google Scholar]
- 129.Bar-Even A., Noor E. and Milo R. (2012) A survey of carbon fixation pathways through a quantitative lens. J. Exp. Bot. 63, 2325–2342 10.1093/jxb/err417 [DOI] [PubMed] [Google Scholar]
- 130.Smirnov S.V., Kodera T., Samsonova N.N., Kotlyarova V.A., Rushkevich N.Y., Kivero A.D. et al. (2010) Metabolic engineering of Escherichia coli to produce (2S, 3R, 4S)-4-hydroxyisoleucine. Appl. Microbiol. Biotechnol. 88, 719–726 10.1007/s00253-010-2772-3 [DOI] [PubMed] [Google Scholar]
- 131.Wu L.-F., Meng S. and Tang G.-L. (2016) Ferrous iron and α-ketoglutarate-dependent dioxygenases in the biosynthesis of microbial natural products. Biochim. Biophys. Acta, Proteins Proteomics 1864, 453–470 10.1016/j.bbapap.2016.01.012 [DOI] [PubMed] [Google Scholar]
- 132.Theodosiou E., Breisch M., Julsing M.K., Falcioni F., Bühler B. and Schmid A. (2017) An artificial TCA cycle selects for efficient α-ketoglutarate dependent hydroxylase catalysis in engineered Escherichia coli. Biotechnol. Bioeng. 114, 1511–1520 10.1002/bit.26281 [DOI] [PubMed] [Google Scholar]
- 133.Dien B.S., Cotta M.A. and Jeffries T.W. (2003) Bacteria engineered for fuel ethanol production: current status. Appl. Microbiol. Biotechnol. 63, 258–266 10.1007/s00253-003-1444-y [DOI] [PubMed] [Google Scholar]
- 134.Terpe K. (2006) Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72, 211–222 10.1007/s00253-006-0465-8 [DOI] [PubMed] [Google Scholar]
- 135.Ranganathan S., Suthers P.F. and Maranas C.D. (2010) OptForce: an optimization procedure for identifying all genetic manipulations leading to targeted overproductions. PLoS Comput. Biol. 6, e1000744 10.1371/journal.pcbi.1000744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tee T.W., Chowdhury A., Maranas C.D. and Shanks J.V. (2014) Systems metabolic engineering design: fatty acid production as an emerging case study. Biotechnol. Bioeng. 111, 849–857 10.1002/bit.25205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pharkya P., Burgard A.P. and Maranas C.D. (2004) Optstrain: a computational framework for redesign of microbial production systems. Genome Res. 14, 2367–2376 10.1101/gr.2872004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Burgard A.P., Pharkya P. and Maranas C.D. (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol. Bioeng. 84, 647–657 10.1002/bit.10803 [DOI] [PubMed] [Google Scholar]
- 139.Chowdhury A. and Maranas C.D. (2015) Designing overall stoichiometric conversions and intervening metabolic reactions. Sci. Rep. 5, 16009 10.1038/srep16009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Venayak N., Anesiadis N., Cluett W.R. and Mahadevan R. (2015) Engineering metabolism through dynamic control. Curr. Opin. Biotechnol. 34, 142–152 10.1016/j.copbio.2014.12.022 [DOI] [PubMed] [Google Scholar]
- 141.Suástegui M., Yu Ng C., Chowdhury A., Sun W., Cao M., House E. et al. (2017) Multilevel engineering of the upstream module of aromatic amino acid biosynthesis in Saccharomyces cerevisiae for high production of polymer and drug precursors. Metab. Eng. 42, 134–144 10.1016/j.ymben.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 142.Bar-Even A., Noor E., Lewis N.E. and Milo R. (2010) Design and analysis of synthetic carbon fixation pathways. Proc. Natl Acad. Sci. U.S.A. 107, 8889–8894 10.1073/pnas.0907176107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Bar-Even A., Flamholz A., Noor E. and Milo R. (2012) Rethinking glycolysis: on the biochemical logic of metabolic pathways. Nat. Chem. Biol. 8, 509–517 10.1038/nchembio.971 [DOI] [PubMed] [Google Scholar]
- 144.Bar-Even A., Noor E., Flamholz A. and Milo R. (2013) Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim. Biophys. Acta, Bioenerg. 1827, 1039–1047 10.1016/j.bbabio.2012.10.013 [DOI] [PubMed] [Google Scholar]
- 145.Lan E.I. and Liao J.C. (2012) ATP drives direct photosynthetic production of 1-butanol in cyanobacteria. Proc. Natl Acad. Sci. U.S.A. 109, 6018–6023 10.1073/pnas.1200074109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Antonovsky N., Gleizer S., Noor E., Zohar Y., Herz E., Barenholz U. et al. (2016) Sugar synthesis from CO2 in Escherichia coli. Cell 166, 115–125 10.1016/j.cell.2016.05.064 [DOI] [PMC free article] [PubMed] [Google Scholar]