Abstract
Background
Photoperiod insensitive Ppd-1a alleles determine early flowering of wheat. Increased expression of homoeologous Ppd-D1a and Ppd-A1a result from deletions in the promoter region, and elevated expression of Ppd-B1a is determined by an increased copy number.
Results
In this study, using bread wheat cultivars Sonora and PSL2, which contrast in flowering time, and near-isogenic lines resulting from their cross, “Ppd-m” and “Ppd-w” with Ppd-B1a introgressed from Sonora, we investigated the putative factors that influence Ppd-B1a expression. By analyzing the Ppd-B1a three distinct copies, we identified an indel and the two SNPs, which distinguished the investigated allele from other alleles with a copy number variation.
We studied the expression of the Ppd-A1, Ppd-B1a, and Ppd-D1 genes along with genes that are involved in light perception (PhyA, PhyB, PhyC) and the flowering initiation (Vrn-1, TaFT1) and discussed their interactions. Expression of Ppd-B1a in the “Ppd-m” line, which flowered four days earlier than “Ppd-w”, was significantly higher. We found PhyC to be up-regulated in lines with Ppd-B1a alleles. Expression of PhyC was higher in “Ppd-m”. Microsatellite genotyping demonstrated that in the line “Ppd-m”, there is an introgression in the pericentromeric region of chromosome 5B from the early flowering parental Sonora, while the “Ppd-w” does not have this introgression. FHY3/FAR1 is known to be located in this region. Expression of the transcription factor FHY3/FAR1 was higher in the “Ppd-m” line than in “Ppd-w”, suggesting that FHY3/FAR1 is important for the wheat flowering time and may cause earlier flowering of “Ppd-m” as compared to “Ppd-w”.
Conclusions
We propose that there is a positive bidirectional regulation of Ppd-B1a and PhyC with an FHY3/FAR1 contribution.
The bidirectional regulation can be proposed for Ppd-A1a and Ppd-D1a. Using in silico analysis, we demonstrated that the specificity of the Ppd-B1 regulation compared to that of homoeologous genes involves not only a copy number variation but also distinct regulatory elements.
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1126-z) contains supplementary material, which is available to authorized users.
Keywords: Photoperiod sensitivity, Flowering time, Ppd-B1, Phytochrome, Common wheat
Background
Photoperiod sensitivity is an important agronomic trait that influences the wheat heading date, and the Ppd-1 (Photoperiod-1) genes are significant regulators of this process. The Ppd-1 genes are members of the Pseudo-Response Regulator (PRR) gene family [1]. There are three Ppd-1 genes in the hexaploid wheat Triticum aestivum, which are located on the short arms of chromosomes 2A, 2B and 2D [2–4] and are designated Ppd-A1, Ppd-B1 and Ppd-D1, respectively. Mutant alleles that are responsible for photoperiod insensitivity and, thus, early flowering in short day (SD) conditions are labeled with the suffix “a”.
To date, the photoperiod insensitive (PI) alleles of all the Ppd-1 genes have been identified. Ppd-D1a has a 2089 bp deletion in its promoter region [1]. This allele is widely used in wheat selection, and its influence on the phenotype has been well studied [5, 6]. Ppd-A1a dominant alleles were first described using T. durum (Triticum durum). There are two alleles, both of which are characterized by 1027 bp or 1117 bp deletions in the promoters [4]. The T. aestivum Ppd-A1a allele, which was described by Nishida et al. [7], has a 1085 bp deletion in the 5′ UTR region similarly to the other PI alleles.
Nishida et al. [7] revealed the PI Ppd-B1a allele in the cultivar Winter-Abukumawase with a 308 bp insertion in the promoter region. This insertion was suggested to be a MITE (miniature inverted-repeat transposable element). Another PI Ppd-B1 mutation is a copy number variation (CNV) [8]. The haploid copy number of the wild-type Ppd-B1 allele is one, while the PI alleles possess two to four copies. In addition to the copy number, these alleles may have different intercopy regions. Thus, there are three variants of the Ppd-B1a alleles based on the differences in the copy number and the intercopy junctions as follows: Recital type, Sonora64/Timstein/C591 type and Chinese Spring type [8]. Kiss et al. [9] ascertained that the phenotypical effect of the Ppd-B1 loci is associated not only with the copy number but also with the intercopy junction type. Under field conditions, the Ppd-B1 intercopy junction type was demonstrated to influence the heading date more than the number of copies.
The photoperiod sensitive (PS) Ppd-1 alleles are expressed during the light period, and the peak of the expression occurs 3 to 6 h after dawn; these alleles are not expressed during the dark period. In contrast, all PI alleles demonstrate a misexpression throughout the 24 period [1, 4, 8]. The Ppd-1a alleles are expressed both during the light and dark periods. During the night period, PI Ppd-1a alleles were shown to up-regulate TaFT1 (Triticum aestivum Flowering Locus T) expression under SD (Short Day) conditions [10, 11]. TaFT1 is expressed in the leaves and transfers to the floral meristems to induce flowering [12].
The aim of the current investigation is to study the Ppd-B1 PI allele with an increased number of copies and characterize the functional specifications of this allele and its interaction with other photoperiod genes. PCR analysis, molecular cloning and sequencing were used for the characterization of the Ppd-B1 sequence. To analyze the diurnal expression of genes that regulate the heading date, qPCR with SYBR Green I was performed. The PlantPAN 2.0 database [13] was used to determine the putative plant transcription factor binding sites.
Methods
Plant materials
Two pairs of the near-isogenic lines “Ppd-m” and its sister line “Ppd-0m”, “Ppd-w” and its sister line “Ppd-0w” and their parental forms Sonora (К-47942, Vrn-A1/Vrn-B1/Vrn-D1 and Ppd-A1b/Ppd-B1a/Ppd-D1b) and PSL2 (Photoperiod Sensitive Line 2, Vrn-A1/Vrn-B1/vrn-D1 and Ppd-A1b/Ppd-B1b/Ppd-D1b), which have different heading dates, were used in this investigation. Lines “Ppd-m” and “Ppd-w” differ from their sister lines by the introgressions on the 2B chromosome from the Sonora variety. This difference was shown previously using SSR analysis [14]. An analysis of specific molecular markers also demonstrated that the near-isogenic lines (NILs) have recessive alleles of Ppd-D1 and Ppd-A1 [14]. DNA was extracted from the seedlings using the GeneJET Plant Genomic DNA Purification Mini Kit (Thermo Fisher Scientific, Lithuania) according to the manufacturer’s protocol.
Ppd-B1 intercopy region analysis
Previously reported PCR primers [8] were used to define the Ppd-B1 intercopy region. The PCR was performed as previously described [8].
SSR genotyping
SSR genotyping of the full genome of NILs was performed in previous study [14]. Unfortunately, there was a mistake in illustration described “Ppd-w” and “Ppd-0w” lines. “Ppd-w” and “Ppd-0w” lines do not have an introgression on 5B chromosome. Therefore, to ascertain the 5B chromosome inheritance in the NILs, we used an additional set of markers (Xgpw358, Xbarc74, Xgwm213, Xgpw2124). The marker sequences and their annealing temperatures are available [15]. The amplified fragments were analyzed in 5% high-resolution agarose MS-8 (Dia-m, Russia) using ethidium bromide staining.
Primer design, PCR and sequencing of Ppd-B1
Previously, we have hypothesized that the Ppd-B1 allele in the investigated lines might be characterized by sequence mutations in addition to copy number variations [14]. To investigate all possible sequence variations, we developed primers with a coding region that is specific to all Ppd-1 genes. The primer sequences are as follows: F2-Ppd-exon2-ACCAGGCGTGGGCGTATCT; R2-Ppd-exon6-GCTCTAGCTGCCTGTTGGG; F3-Ppd-exon6-TGGAGATAGGTGCCCCTGG; and R3-Ppd-3UTR-GGACCGTCTCTGAATGATCCA. The primers for the promoter region were Ppd-B1 gene-specific due to the strong differences in the Ppd-1 promoter region and were designed according to the alignment of 184 sequences from the NCBI database. The primer sequences are as follows: F-Ppd-5UTR-CACTCTTATTCCCTCTATGCC and R-Ppd-5UTR-CTGTTATTATTGGAATCGTCAG. The reaction mixture was as follows: DNA in a concentration of 5 ng/μL, 1× buffer for Taq-polymerase (pH 8.6, 2.5 mM Mg2+), 200 μmoles dNTPs, 0.2 μmoles of each primer, 1 U Taq polymerase (Medigen, Russia), and sterile water up to a volume of 25 μL. The PCR conditions were as follows: 94°С for 3 min, and 35 cycles (94°С for 40 s; 55°С for 30 s; and 72°С for 60 s) and 72°С for 7 min.
The amplicons were recovered using a 1% agarose gel and purified with the kit for DNA elution from agarose gel (Biosilica, Russia). The purified amplicons were ligated into a pAL-TA vector (Evrogen, Russia) with 1 U of T4 DNA Ligase (Thermo Fisher Scientific, UK). The construct was used for the transformation of the E.coli Top-10 competent cells, which were prepared according to the CCMB80 protocol [16]. The colonies with the target insert were selected using a blue-white selection with X-Gal/IPTG and PCR with primers to target the sequences. Plasmid DNA was extracted with the Biosilica kit for pDNA extraction (Biosilica, Russia). The sequencing was performed using an ABI PRISM Dye Terminator Cycle Sequencing ready reaction kit (Perkin Elmer Cetus, USA) with M13 primers and extra primers for the following target sequences: F-5UTRad-TTCTTCACACTAGGGCTGGT; R-5UTRad-CGCATAATAGCACAACCAGC; F-ex4-GTGGCAGTGGTAGTGGAAGT; and F-7ex-ACGCCGCTCAGATGAAGCAA.
Diurnal quantitative expression of the photoperiod genes
Sonora, PSL2, “Ppd-m”, and “Ppd-w” plants were grown for 21 days after germination under controlled conditions in a climatic chamber Rubarth Apparate (RUMED GmbH) with short days (9 h of light, 20 °C). Three replicate samples from each genotype were harvested into liquid nitrogen at each three-hourly time point over 24 h since the beginning of the light period. RNA was extracted using the Plant RNA MiniPrep (Zymo Research, USA), followed by a DNase treatment with the RNase-Free DNase set (QIAGEN, Hilden, Germany). cDNA was synthesized using RevertAid First Strand cDNA Synthesis (Thermo Fisher Scientific, Lithuania) following the manufacturer’s protocol with 2 μg of total RNA as a template and Oligo(dT)18 as the primers. In total, 2 μl of the 20-fold dilution of the final cDNA were used for the following analysis.
For the expression analysis, previously published primers for the TaFT-1, Ppd-1 [11], Vrn-1 [17], PhyA, PhyB and PhyC [18] genes were used. The primers for the FHY3/FAR1 were designed using the URGI: Traes_5BS_BCC406654.2 sequence. The primer sequences are as follows: F-TaFHY3/FAR1-5B- GCAAACGTCATCAGGATACA and R-TaFHY3/FAR1-5B- CCTCTTCTCAGCTTTACTTGC. The primers for the 18S rRNA gene [1] were used for normalization. The fluorescence data were collected using ABI 7500fast Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) with SYBR Green I (Syntol, Russia) as the intercalating dye. The measurements were performed in three technical replicates. The reaction products were checked by a melting curve analysis and 2% agarose gel electrophoresis. The relative expression values were calculated according to the method proposed by Pfaffl [19]. The PCR efficiencies were determined using LinReg software [20]. The expression of target genes was normalized against 18S rRNA.
Statistical analysis
ANOVA with a post hoc Tukey test was used to compare the expression levels. Correlations between the patterns of gene expression were calculated using Pearson coefficient (significance level P = 0.001).
Bioinformatic analysis of the gene promoters
The PlantPAN 2.0 [13] database was used to determine the putative plant cis-acting regulatory elements. The 2000 bp upstream TSS (Transcription Start Site), first exon and first intron were analyzed for all sequences except for TaFT-5D due to the absence of the sequence of its upstream TSS in the databases.
Results
Sequence analysis of the photoperiod insensitive Ppd-B1 allele
We have hypothesized that the Ppd-B1 gene in the PI Sonora, “Ppd-m” and “Ppd-w” is characterized by not only an increased copy number but also by a nucleotide polymorphism in one of the copies because NILs “Ppd-m” and “Ppd-w” flowered slightly later than the line with the same background and introgressed Ppd-D1a gene [14]. Therefore, we analyzed the sequences of the Ppd-B1 distinct copies. To investigate the possible polymorphisms, the amplicons that overlapped gene and its promoter were obtained and inserted into a vector to transform E. coli and distinct colonies that were regarded to have different copies, which were then sequenced and analyzed.
The results are presented in Fig. 1. Every line in our investigation differed by a single nucleotide indel in the promoter region (−2373 bp) of the Ppd-B1 allele, which was detected in the cultivars Sonora64, Timstein (DQ885765.2), and Renan (DQ885764.2). Thus, the NILs and the parental cultivar Sonora had the same Ppd-B1 intercopy region as cultivars Sonora64, Timstein and C591. However, the detected polymorphism provided a distinction among these Ppd-B1 alleles.
Fig. 1.

Scheme of the Ppd-B1 gene features. Distances are presented in base pairs from the TSS (transcription start site). Black rectangles indicate exons. CS means Chinese Spring cultivar. Numbers on the right side of some sequences indicate number of gene copies in actual alleles
The other SNP (−630 bp) allows for the distinction between the PI and PS NILs. Thus, “Ppd-m” and “Ppd-w” have “C” and the sister lines have “G” upstream the coding region. In exon 3 (+546 bp), the detected SNP distinguished the investigated sequence from another allele by the increased copy number (Chinese Spring).
Diurnal expression analysis
To study the interactions between the Ppd-B1a PI allele and other genes that are important for wheat flowering, we used a qRT-PCR assay. We used 21-day-old plants from the late-flowering parental line PSL2, early flowering parental cultivar Sonora with the PI Ppd-B1a allele, and NILs “Ppd-m” and “Ppd-w”, representing PSL2 with different alleles at some loci, including Ppd-B1a, introgressed from Sonora.
The results of the diurnal expression analysis are presented in Fig. 2. Ppd-B1 was expressed in the PI lines during the dark period but was not expressed in the photoperiod sensitive PSL2 as expected. The TaFT1 gene was expressed only in the PI lines, supporting previous investigations [10, 11] in which the influence of the Ppd-1 dominant alleles on TaFT1 expression was shown.
Fig. 2.
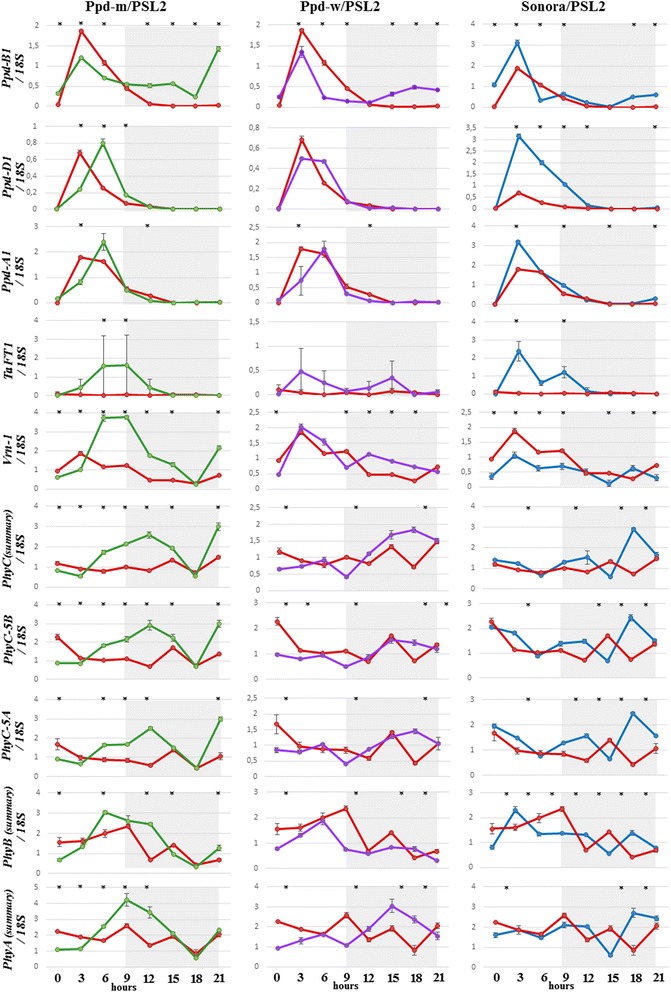
Patterns of the diurnal gene expression. Quantitative gene expression data from plants grown in short days (0–9 h light period) in а climatic chamber. Gray shadowing indicates the dark period (9–24 h). The graphs compare the expression between the photoperiod sensitive parental line PSL2 (red) and the photoperiod insensitive NILs (“Ppd-m” (green), “Ppd-w” (purple)) and parent Sonora (blue). Values are expressed as the relative levels normalized against 18S ribosomal RNA. Error bars indicate the SE of the means. Asterisks indicate significant (P < 0.05) differences in a one-way ANOVA with a post hoc Tukey test comparing the photoperiod insensitive NILs (“Ppd-m”, “Ppd-w”) and the parent Sonora with the photoperiod sensitive parental line PSL2 in each time point
The Ppd-D1 expression was increased in Sonora compare to that in PSL2 and the NILs. This finding may be due to the 5 bp deletion in exon 7. The specificity of Ppd-A1 is shown in the expression peak shift in “Ppd-m” and “Ppd-w” compared to that in Sonora and PSL2. The shift in the expression peaks in the “Ppd-m” line can be observed in Ppd-D1, TaFT1, Vrn-1 and the other genes. The expression patterns of Vrn-1 did not display any robust differences between Sonora and “Ppd-w” but was rather higher in Ppd-m. PhyC expression decreased since the beginning of the light period and displayed one or two peaks during the dark period. A tendency of an elevated expression during the dark period in the PI lines was observed. The expression patterns of PhyB and PhyA could be characterized by the two expression peaks as follows: one at the end of the light period and one during the dark period.
We analyzed the correlations (Additional file 1: Table S1) between the expression patterns of the genes involved in light perception (PhyA, PhyB, PhyC) and those involved in the flowering transaction (Ppd-A1, Ppd-B1, Ppd-D1, Vrn-1, TaFT1) separately during the dark and light periods in the photoperiod sensitive and insensitive lines.
The expression patterns of all the phytochromes significantly correlated with each other, indicating that common factors influence their expression. The Ppd-1 genes also significantly correlated with each other (ρ = 0.55–0.85 in the photoperiod insensitive and ρ = 0.89–0.97 in the photoperiod sensitive lines). TaFT1 expression correlated with the Ppd-1 genes in the PI lines, and in PSL2, TaFT1 was not expressed at all. We observed significant correlations between the expression levels of TaFT1 and PhyС-5A (ρ = 0.75) and between TaFT1 and PhyC-5B (ρ = 0.8) during the light period. The expression patterns of Vrn-1 significantly correlated with the Ppd-1 genes (ρ = 0.89–0.95) in PSL2. This finding may suggest that there are common factors influencing their expression. In the PI lines, Vrn-1 correlated with all the phytochromes during the light (ρ = 0.65–0.79) and dark (ρ = 0.56–0.72) periods. These data are consistent with [21], in which the influence of PhyC and PhyB on Vrn-1 expression was shown using RNA-Seq analysis. The correlation between Ppd-B1 and PhyC (PhyC in combination and PhyC-5B and PhyC-5A individually) was significant during the dark period in the PI lines (ρ = 0.67).
Features of the expression patterns in the lines with the photoperiod insensitive Ppd-B1 allele
The expression peaks of all genes except for Ppd-B1 were shifted in the “Ppd-m” line. The parental forms and “Ppd-w” did not display these expression patterns. Most likely, the genes located in the additional introgressions from Sonora, which are specific to “Ppd-m” but not “Ppd-w”, interact with the PSL2 background genes, which may result in such an effect. Both the “Ppd-m” and “Ppd-w” lines have introgression of Ppd-B1a from Sonora, but they differ by four days in their heading date under SD conditions (“Ppd-m” flowered earlier) [14]. “Ppd-m” has the following two supplemental loci from Sonora that were not found in “Ppd-w”: on chromosome 4D nearby the Xbarc165, on chromosome 5A nearby the Xgwm154 and in the pericentromeric region of chromosome 5B (markers Xbarc74, Xgwm67, Xgwm371, and Xgwm213). QTLs associated with the heading date were previously identified in the 4D and 5B chromosomes. In chromosome 4D, the QTL was located in the region between markers wPt8836 and Xgwm165 [22]. No genes that are located in these areas are known to correspond to the heading time regulation.
In chromosome 5B, the heading date QTL was associated with the Xgwm371 marker [23]. Later, a QTL associated with candidate genes FHY3/FAR1, AP2/ERF and WRKY was identified in the 5B pericentromeric region [24]. Therefore, chromosome 5B was genotyped using an additional set of SSR markers to ascertain the length of the introgression region. As a result, chromosome 5B in the “Ppd-w” and “Ppd-0w” lines was completely inherited from PSL2. In the line “Ppd-m”, there is an introgression from the early flowering parental Sonora precisely in the region that carries the FHY3/FAR1 locus, while in “Ppd-w”, which flowers four days later than Ppd-m, does not have this introgression (Fig. 3). FHY3/FAR1 is proposed as a good candidate to explain the difference in the heading date.
Fig. 3.
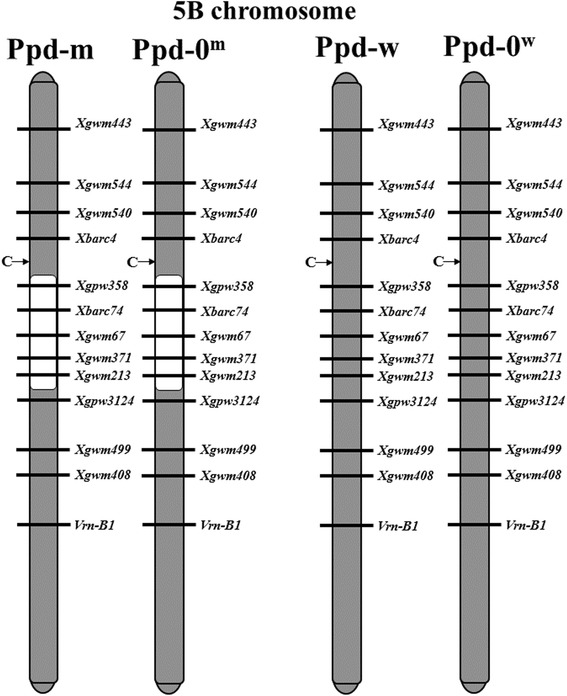
5B chromosome genotyping of NILs. Genotypes of the 5B chromosome of the near-isogenic “Ppd-m” and “Ppd-0m”, “Ppd-w” and “Ppd-0w” lines with introgressed chromosome regions of the Sonora variety (white color) into the genetic background of the recipient PSL2 parent (gray color). C indicates centromere region
To examine whether there are any differences in FHY3/FAR1 expression between lines “Ppd-m” and “Ppd-w”, we applied the diurnal expression analysis and demonstrated significant differences of FHY3/FAR1 expression. Expression of this gene in “Ppd-m” was higher in 6–9 h after dawn indicating putative reason for the heading time variation (Fig. 4). FHY3/FAR1 expression pattern correlated with phytochromes and Vrn-1 at significant levels. In night FHY3/FAR1 correlated with Ppd-B1 in PI lines (“Ppd-m”, “Ppd-w” and Sonora).
Fig. 4.
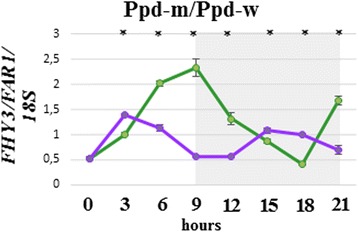
Patterns of the FHY3/FAR1 diurnal expression. Quantitative gene expression data from plants grown in short days (0–9 h light period) in а climatic chamber. Gray shadowing indicates the dark period (9–24 h). The graphs compare the expression between the photoperiod insensitive NILs “Ppd-m” (green), “Ppd-w” (purple). Values are expressed as the relative levels normalized against 18S ribosomal RNA. Error bars indicate the SE of the means. Asterisks indicate significant (P < 0.05) differences in a one-way ANOVA with a post hoc Tukey test comparing the “Ppd-m” and “Ppd-w” in each time point
Sequence analysis of Ppd-D1 and Ppd-A1
Ppd-D1 of Sonora has a 5 bp deletion that is specific to haplotype IV based on the classification suggested by Guo [25]. Phenotypically, this haplotype is much weaker than haplotype I, which is represented by the dominant PI allele with a 2 kb deletion in the promoter region, but slightly reduces the heading time [25]. However, in contrast to [25], who showed that the expression of the Ppd-D1 haplotype IV was only 5% higher, Sonora’s Ppd-D1 expression was statistically significantly higher during the light period than the expression of the PSL’s allele (corresponding to haplotype II). During the dark period, no expression was observed. Most likely, the high expression of Sonora Ppd-D1 can partially explain the fact that the PI lines flower 7–12 days later than Sonora, because their Ppd-D1 alleles are inherited from PSL2.
The Ppd-A1 sequence of PSL2 and the NILs has a SNP (A/C) that has not been previously described in any published Ppd-A1 sequence. The comparison included 286 sequences of tetra- and hexaploid wheat varieties and was accomplished using the Blastn algorithm.
Discussion
Ppd-B1 is the only Ppd-1 gene with a dominant PI allele that is associated with the copy number variation. However, little is known about the mechanisms underlying its misexpression. Previously, using near-isogenic lines resulting from a cross of the cultivars Sonora and PSL2, which contrast in flowering time, we have shown that the difference in flowering time was associated with an introgression on chromosome 2B between the markers Xgwm148 and Xgwm388 from the PI Sonora variety [14]. Ppd-B1 is known to be located in this position [26]. Early flowering NILs do not have the PI Ppd-B1a allele with a 318 bp insertion in the promoter region that was identified by Nishida et al. [7]. Using Real-Time PCR, we revealed that the NILs and Sonora are characterized by increased copy numbers of Ppd-B1 [14].
In this study, we have shown that Ppd-B1a in the PI lines is characterized by an increased number of copies and possesses an intercopy junction similarly to cultivars Sonora64, Timstein and C591. However, the investigated Ppd-B1a allele differs from Sonora64, Timstein and C591 allele by an indel in the promoter region.
We have also found two SNPs. The SNP in exon 3 (+546 bp) distinguished the investigated sequence from the Chinese Spring allele with an increased copy number.
The SNP (−630 bp) distinguished between the PI and PS NILs. This SNP (“G”) was previously detected in cultivars Recital, Paragon and Winter-Abukumawase [1, 8]. It is interesting to note that the Ppd-B1 gene of the parental cultivar Sonora and all the other tetra- and hexaploid wheats is characterized by “C” in this position. Thus, “G” in this position is rare (4 cultivars, including PSL2). However, no association between photoperiod sensitivity and this SNP was shown [1, 7, 8].
To study the possible influence of the detected SNPs on the expression of Ppd-B1a, the interactions between the investigated Ppd-B1a allele and other wheat flowering genes and the activity of these genes in the presence of Ppd-B1a, we performed a diurnal expression analysis.
We analyzed the correlations between the expression patterns of genes involved in light perception (PhyA, PhyB, PhyC) and those involved in the flowering transaction (Ppd-A1, Ppd-B1, Ppd-D1, Vrn-1, TaFT1) separately during the dark and light periods in the photoperiod sensitive and insensitive lines.
TaFT1 expression correlated with the Ppd-1 genes in the PI lines, but in PSL2, TaFT1 was not expressed at all. We identified the following cis-elements that corresponded to the RR (Response Regulator) or PRR (Pseudo Response Regulator) regulation: ARR14, RR14, PRR4, APRR2, ARR1, RR1 (Myb/SANT, MYB, ARR-B families) and binding site ARR1AT. These findings propose the putative sites for the TaFT1 regulation through the Ppd-1 genes.
Significant correlations were found between the expression levels of TaFT1 and PhyС-5A and TaFT1 and PhyC-5B during the light period. During the night, the TaFT1 expression level was very low, and phytochromes are inactive during the dark. It was suggested that phytochromes influence TaFT1 expression. Altogether, this finding demonstrated that Ppd-1 is important but not essential for flowering, and based on the triple Ppd-1 loss-of-function mutant analysis [27], it was proposed that TaFT1 is directly regulated by the phytochromes. Most likely, this process is performed with the assistance of Ppd-1. In the TaFT1 promoter, the following binding sites for the transcription factor that is associated with the phytochrome regulation were found: PIL5, POC1, PIF3, PAP3 (bHLH/bZIP), FHY3/FAR1 (FAR1), VOZ2 (VOZ), SORLIP1AT, SORLIP2AT, SORLREP5AT, SORLIP5AT, and RE1ASPHYA3. The expression correlation and the identification of TFBSs for Phytochrome-regulation in the TaFT1 promoter suggest that phytochromes are other regulators of TaFT1.
We detected a significant correlation between Ppd-B1 and PhyC (PhyC in combination and PhyC-5B and PhyC-5A individually) during the dark period in the PI lines.
Phytochromes express in the dark and produce inactive Pr (Red light-absorbing phytochrome) molecules [28, 29] that cannot influence the expression of other genes; thus, the correlation should imply that the expression of Ppd-B1 during the night period positively influences PhyC expression, but PhyC does not influence Ppd-1 during the night. Regarding PhyA, the same tendency can be observed, but no significant correlation was found. Because wild type Ppd-1 reveals no expression during the dark period, it is unable to influence PhyC in general. We may hypothesize that Ppd-1 influences PhyC even during the day period, but PhyC mRNA degradation in light conceals this putative effect. Previously, it was shown that the NILs with the PI Ppd-1a alleles (Ppd-D1a or Ppd-B1a) have an increased level of phytochrome protein compared to the sister lines with recessive Ppd-1 alleles [30]. Thus, we suggest that the Ppd-1a genes may influence the PhyC expression directly or indirectly. To verify this hypothesis, we analyzed the promoter sequences of the PhyC genes to detect the possible binding sites for Ppd-1.
The binding sites for the response regulators (TFmatrixID_0348 for ARR2, RR2 and TF_motif_seq_0268 for ARR1AT) and the pseudo-response regulators (TF_motif_seq_0252 for APRR4) may be regulated by these pseudo-response regulators, such as Ppd-1 genes. Some of these TFs correspond to phytochrome-regulation (EPR1/RVE7, POC1/PIF3, FHY3/FAR1, SORLIP2AT, and SORLIP1AT), suggesting their self-regulation. There were some other TFBSs that were associated with flowering or photoperiod regulation as follows: GATA12, AtHB33, AtDOF1, SPL3, TEM1, STM, GBOX10NT, BS1EGCCR, and IBOXCORE.
Taken together, the expression correlation data, the PhyC promoter analysis and the fact that there are phytochrome protein increments in the lines with the PI Ppd-1a alleles, as demonstrated by [30], suggest that Ppd-B1a is expressed during night period and positively regulates PhyC expression.
“Ppd-m” flowered four days earlier than “Ppd-w”. Using SSR genotyping, we demonstrated that the loci in the 5B pericentromeric region were different in the “Ppd-m” and “Ppd-w” lines. FHY3/FAR1 is known to be located in this locus [24]. Binding sites for FHY3/FAR1 were identified in the Ppd-1, PhyC and TaFT1 promoters.
Using the diurnal expression analysis, we found that the FHY3/FAR1 expression was higher in “Ppd-m” than in “Ppd-w”. FHY3/FAR1 expression pattern correlated with phytochromes during the 24-h period in all the lines and with Ppd-B1 in night in PI lines.
FHY3/FAR1, known to be involved in phytochrome signaling in Arabidopsis and rice [31, 32], control phytochrome accumulation through the FHY1 regulation in Arabidopsis [33]. However, there is no data about FHY3/FAR1 functions in wheat.
In wheat, the phytochromes (PhyB and PhyC) influence the Ppd-1 expression [18, 21] and flowering time. FHY3/FAR1, which is involved in flowering time control, may contribute to this process. Ppd-B1a was suggested to increase PhyC expression. Thus, the data propose a bidirectional regulation between the PhyC and Ppd-B1a genes with a putative FHY3/FAR1 contribution. This hypothesis requires further investigation and verification. The near-isogenic lines used in this study differ in the 5B pericentromeric region and are a good source for this work.
The lines with Ppd-D1a also demonstrated an increase in the phytochrome protein [30]. Therefore, this mechanism is proposed to be common for all Ppd-1a alleles. However, the regulation of Ppd-B1 expression should be different.
Previously, it was suggested that a deletion in the promoter region of Ppd-A1 and Ppd-D1 causes the misexpression of the corresponding genes due to the disappearance of the CHE-binding site and impossibility of the repressor CHE (CCA1 HIKING EXPEDITION) to associate with its regulatory element [7]. Then, it could be hypothesized that an increase in the Ppd-B1 copy number, which was shown to occur in its dominant alleles, results in a misexpression because of the increase in the Ppd-1 genes with the same quantity of the repressor. However, in this case, an altered expression should be observed in all three Ppd-1 genes. Previously, published data [11] demonstrated that the Ppd-B1a allele shows a misexpression by itself but does not influence Ppd-A1 and Ppd-D1 expression. Thus, about the hypothesis regarding the role of the Ppd-B1 copy number against the repressor’s quantity should be rejected. It is interesting to note that there is no Ppd-B1 allele with a deletion in the promoter region, while all PI alleles of Ppd-A1 and Ppd-D1 are characterized by deletions.
To identify the probable factors involved in Ppd-B1 but not Ppd-A1 and Ppd-D1 regulation, we investigated the sequences of the promoter regions of these genes and determined (1) TFBSs (Transcription Factor Binding Sites) that are common to the promoters of all Ppd-1 homoeologous genes and (2) TFBSs that are specific to Ppd-B1.
To detect the TFBSs that are common to the Ppd-1 genes in the A, B and D genomes, a PlantPan2 multiple promoter analysis was utilized. An in silico promoter sequence analysis revealed that that the Ppd-1 promoters have many cis-acting elements that are associated with flowering that could be divided into the following three groups according to their input signal nature: phytochrome-regulated, circadian clock-regulated and other binding sites that are involved in light-regulated development. A detailed description of these transcription factors is summarized in Table 1. The positions of these cis-elements on the Ppd-1 sequences are presented in Fig. 5.
Table 1.
Characterization of the selected transcription factors and the TFBSs with unknown transcription factors that were identified in the promoter regions of the Ppd-1 genes
| Family | Transcription factors/TFBSs with unknown TF | Description |
|---|---|---|
| Phytochrome regulated transcription factors | ||
| VOZ | VOZ2 | VOZ2 was identified as one of the highly conserved transcriptional factors in land plant genes that are PhyB-interacting factors [58]. |
| bHLH | Phytochrome Interacting Factor3 (PIF3) | The G-box, CACGTG, is a target sequence for PIF3 [59]. PIF3 plays important roles in the Phy-mediated light responses. This factor can regulate the downstream genes either positively or negatively [60]. In Arabidopsis, PIF3 was suggested to play an important role in the control of flowering by the regulation of CO and FT gene expression [61]. |
| bHLH | Phytochrome Interacting Factor3-Like5 (PIL1) | PIL5 interacts with the Pfr forms of Phytochrome A (PhyA) and Phytochrome B (PhyB) [62]. |
| FAR1 | Far-Red Elongated Hypokotyls3/Far-Red-Impaired Response1 (FHY3/FAR1) | Transcription factor FHY3/FAR1 modulates PhyA-signaling in higher plants [63]. Other investigations demonstrated that FHY3 plays a principal role in the circadian clock, heading date control and regulation of heading time through ELF4 (EARLY FLOWERING4) [64]. |
| Myb/SANT; MYB-related | RVE7/EPR1 | RVE7/EPR1 is regulated by both PhyA and PhyB and negatively regulates flowering [65]. |
| – | SORLIP2AT and SORLIP1AT | Sequences over-represented in the light-induced promoters SORLIP2AT and SORLIP1AT were identified in the PhyA-induced promoters [66]. |
| – | RE1ASPHYA3 | RE1ASPHYA3 (RE1, putative repressor element) is a highly conserved motif in the most monocot PhyA promoters and is responsible for the Pfr-directed repression [67]; this motif was detected in certain other genes [68]. |
| Circadian-clock regulated transcription factors | ||
| AP2/RAV/B3 | RAV1 | RAV1 is a negative component in the regulation of plant development [69]. |
| AP2/RAV/B3 | TEMPRANILLO1 (TEM1) TEMPRANILLO2 (TEM2) | TEM1 and TEM2 genes act as direct repressors of FT [70]. |
| MYB-related | REVEILLE1 (RVE1) | RVE1 is a morning-phased transcription factor that integrates the circadian clock and auxin pathways [71]. |
| Myb/SANT; MYB-related | REVEILLE 8 (RVE8) | RVE8 promotes the expression of some evening element that contains clock genes and forms a negative feedback loop with PRR5 [72, 73]. |
| MYB | Circadian Clock Associated 1 (CCA1); Late Elongated Hypocotyl (LHY) | The MYB transcription factors CCA1 and LHY are some of the key genes in the central oscillator of the plant circadian clock [74]. LHY and CCA1 negatively regulate TOC1 expression. |
| TCP | CCA1 HIKING EXPEDITION (CHE) | The TCP transcription factor CHE is a clock component that is partially redundant with LHY in the repression of CCA1 [75]. |
| Other light regulated transcription factors | ||
| GATA/tify | GATA2 | GATA2 directly regulates genes that respond to light [76]. |
| GATA/tify | GATA12 | GATA12 is involved in the regulation of many light-responsive genes [77, 78]. |
| MADF; Trihelix | GT-1 | GT-1 may act as a light-responsive transcription factor [79]. |
| bHLH | AtMYC2 | AtMYC2 acts as a negative regulator of blue light–mediated photomorphogenic growth and blue and far-red-light–regulated gene expression [80]. |
| – | BOXIIPCCHS | BOXIIPCCHS was suggested to be essential for light regulation [29, 81]. |
| – | TBOXATGAPB | Mutations in TBOXATGAPB cause a reduction in light-activated transcription [82, 83]. |
| – | IBOXCORENT | I-box core motif IBOXCORENT in the conserved DNA modular arrays is associated with the light-responsive promoter regions [84]. |
| – | LREBOXIIPCCHS1 | Light responsive element LREBOXIIPCCHS1, which was detected in parsley, is required for light responsiveness [85]. |
| – | IBOXCORE | Conserved sequence in the 5′ UTR of light-regulated genes IBOXCORE may be involved in the regulation of transcription by light and the circadian clock. This sequence was detected in both monocots and dicots [86]. |
Fig. 5.
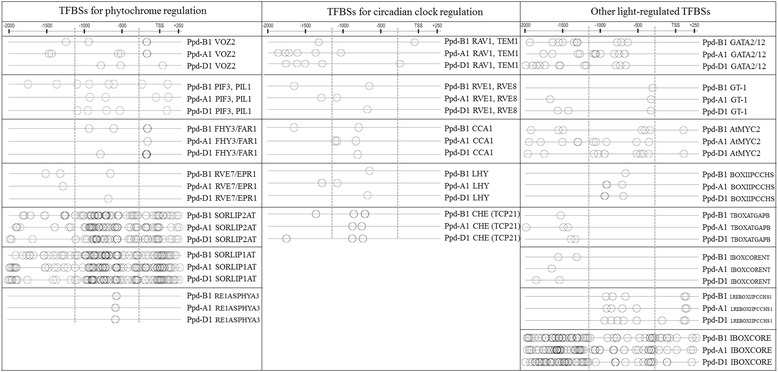
Groups of transcription factor binding sites that are common in the Ppd-1 genes. Vertical dotted lines indicate the borders of a 900-bp region that is commonly deleted in the Ppd-A1 and Ppd-D1 photoperiod insensitive alleles [4, 7]. TSS indicates Transcription Start Site; the distances are presented in base pairs
PhyC and PhyB were recently shown to regulate Ppd-1 expression [18, 21]. These genes were suggested to be important regulators of the photoperiod sensitivity and flowering transaction in wheat [18, 21] and barley [34–36]. Although the chromosome position of PhyA in wheat was identified [37], no association between these loci and flowering was described. However, PhyA is an important regulator of flowering time in certain other species, such as Arabidopsis [38, 39]. Therefore, an investigation of the PhyA sequence and functions in cereals may complement the mechanisms of the photoperiod sensitivity and flowering. Here, we may hypothesize that Ppd-1 expression is regulated by the phytochromes with the assistance of transcription regulators, such as PIF3, PIL5, FHY3/FAR1, RVE7/EPR1, and VOZ2, and certain TFBSs with unknown transcription factors, such as SORLIP2AT, SORLIP1AT, and RE1ASPHYA3.
The Ppd-1 genes were suggested to be regulated by the circadian oscillator in wheat [18] and barley [40]. In barley, some genes that are involved in this process are known, while in wheat, no putative circadian affecters are proposed. We identified the TFBSs that might be involved in the circadian clock regulation of the Ppd-1 genes as follows: RAV1, TEM1, TEM2, RVE1, CCA1, LHY, and CHE.
The Ppd-1 activation by PhyC was proposed to be light sensitive throughout the day [41]. Most likely, this process can be mediated by some of the following detected elements: GATA2, GATA12, GT-1, and AtMYC2 and some binding sites with unknown transcription factors, such as BOXIIPCCHS, TBOXATGAPB, IBOXCORENT, LREBOXIIPCCHS1, and IBOXCORE.
Binding sites that are specific to Ppd-B1 and not Ppd-A1 and Ppd-D1 are likely associated with the circadian rhythms and flowering time and regulatory elements from the following families: AP2 (ANT), C2H2 (Zat12), MADS box/MIKC (AGL19, FLC, MAF2, AGL69, AGL68, FLM, AGL6, AGL18, AGL14, and AGL15), and Lyase Aromatic (PAL1) and TFBSs with unknown TF RBCSGBOXPS, REBETALGLHCB21, LEAFYATAG, SORLIP5AT, and MNF1ZMPPC1. A detailed characterization of these transcription factors is presented in Table 2. The positions of these TFBSs on the Ppd-B1 sequence are presented in Fig. 6.
Table 2.
Characterization of the selected transcription factors and the TFBSs with unknown transcription factors that were identified in the promoter regions of Ppd-B1
| Family | Transcription factors/TFBSs with unknown TF | Description |
|---|---|---|
| AP2 | AINTEGUMENTA (ANT) | ANT initiates floral organ development [87]. It was shown to play a critical role in regulating the ovule and female gametophyte development [88]. |
| C2H2 | RESPONSIVE TO HIGH LIGHT 41 (Zat2) | Zat12 was originally isolated as a light stress-response cDNA [89]; then, it was suggested to be able to regulate transcripts involved in the response to high-light, cold and oxidative stress [90]. |
| MADS box/MIKC | AGL19 | AGL19 controls (promotes) flowering downstream of a cold-perception pathway and acts independently of FT and SOC1 [42]. |
| MADS box/MIKC | FLOWERING LOCUS C (FLC) | FLC acts as an inhibitor of flowering [45]. |
| MADS box/MIKC | MAF2 (AGL31) | MAF2 (AGL31), a paralog of FLC, is another flowering repressor that acts in non-inductive photoperiods [46, 47]. |
| MADS box/MIKC | AGL69 (MAF5) | MAF5 is normally repressed. Overexpression of MAF5 under a non-inductive day length causes late-flowering [48]. |
| MADS box/MIKC | AGL68 (MAF4) | MAF4 represses the transition to flowering [49, 50]. |
| MADS box/MIKC | FLM (AGL27, MAF1) | FLM acts as a flowering inhibitor [51]. |
| MADS box/MIKC | AGL6 | AGL6 was suggested to be able to act as a flowering repressor or activator, depending on the context [43]. |
| MADS box/MIKC | AGL14 (XAANTAL2, XAL2) | XAL2 is essential for flowering induction. XAL2 promotes flowering in response to different signals and is important for the maintenance and differentiating of flowering meristems [44]. |
| MADS box/MIKC | AGAMOUS-like 15 (AGL15) | AGL15 and AGL18 are floral transition repressors. The agl15 agl18 mutants were characterized by a partial suppression of the photoperiod pathway [52]. |
| MADS box/MIKC | AGAMOUS-like 18 (AGL18) | |
| Lyase Aromatic | Phenylalanine Ammonia-Lyase (PAL1) | PAL1 is a light response element. These motifs are conserved at similar positions in several elicitor or light-responsive genes from different species [91]. |
| – | RBCSGBOXPS | RBCSGBOXPS binding site, identified in Parsley, is involved in light responsiveness [92]. |
| – | REBETALGLHCB21 | REBETALGLHCB21, first found in the Lemna dibba Lhcb genes, is necessary for phytochrome regulation. These elements are likely to function by repressing the promoter activity in the dark [93]. |
| – | SORLIP5AT | SORLIP5AT are PhyA-induced motifs that are overrepresented in light-induced genes. These elements, which predominate in the early responsive promoters, are more likely to have the fewest steps in the signal transduction cascade to gene expression [66]. |
| – | MNF1ZMPPC1 | MNF1ZMPPC1 is involved in the light-dependent transcriptional control of gene expression [94]. |
Fig. 6.

Groups of transcription factor binding sites that are specific to Ppd-B1. TSS indicates Transcription Start Site; the distances are presented in base pairs. Different colors designate different TFBSs or TFBSs with unknown TF
The transcription factors that were associated with the flowering transaction included MADS box and the MIKC family genes. Some of these transcription factors contribute to flowering induction, and others contribute to flowering repression. For example, AGL19, AGL6 and AGL14 are positive regulators of the flowering transition [42–44]. In contrast, the majority of the identified MADS transcription factors, i.e., FLC, MAF2, AGL69, AGL68, FLM, AGL15 and AGL18, negatively regulate the transition from vegetative to reproductive development [45–52]. Gu et al. [50] demonstrated that FLC, MAF3 (AGL70), FLM (MAF1,AGL27), MAF2 (AGL31) and MAF4 (AGL69) interact with each other and form nuclear complexes that are responsible for flowering repression.
Most of these TFs contribute to the same binding site, TFmatrixID_0503, except for AGL15, which is associated with the TF_motif_seq_0105 sequence. Motif TFmatrixID_0503 corresponded to the MADS transcription factors not only in Arabidopsis but also in Brachpodium distachyon, Oryza sativa and Sorgum bicolor. These transcription factors were shown to be involved in the flowering transaction and flowering time formation in rice [53, 54]. For example, OsMADS50 is a positive regulator of flowering, and OsMADS56 negatively influences the flower transition [54]. OsMADS7 and OsMADS8 are involved in flowering time modulation [55]. MADS genes were identified in wheat [56]. Some of them are homologs of rice OsMADS8 (OsMADS24) and OsMADS7 (OsMADS45), which can influence the flowering time [57]. However, no data are available regarding the binding sites for the wheat MADS genes; thus, we can only propose that their association is at the same sites as those for the Arabidopsis and rice MADS genes.
Based on the data regarding the regulatory elements in the Ppd-B1 promoter and the common TFs in the homoeologous Ppd-1 genes, we may propose that the MADS genes play a major role in the misexpression of Ppd-B1a with an increased number of copies. Many of these genes are known to be flowering repressors. Most likely, Ppd-B1 with an increased copy number continues to express during the night period; thus, the quantity of the repressors remains the same.
Thus, we have identified a set of putative transcription factors that regulate all homoeologous Ppd-1 genes. We divided these transcription factors into three groups according to the input signal as follows: phytochrome-regulated, circadian clock-regulated and other light-regulated. However, there are several Ppd-B1 specific factors and MADS genes that are known to be flowering repressors and are most likely Ppd-B1 regulators. Our future prospects include the verification of the involvement of the detected transcription factors in Ppd-B1 regulation, and the discussed NILs are a relevant model for such studies.
Conclusions
The results of this study suggest that there is a positive bidirectional regulation of Ppd-B1a and PhyC with an FHY3/FAR1 contribution. The bidirectional regulation can be proposed for Ppd-A1a and Ppd-D1a. Using in silico analysis, we demonstrated that the specificity of the Ppd-B1 regulation compared to that of homoeologous genes involves not only a copy number variation but also distinct regulatory elements.
Additional files
Correlation coefficients of the expression patterns of the flowering genes. Bold font indicates significant values (P = 0.001). PI means Photoperiod Insensitive and PSL means Photoperiod Sensitive samples. (XLSX 26 kb)
Acknowledgments
We are grateful to Prof. V. A. Koshkin for providing seeds of Sonora, PSL2 and NILs, used in this study. We thank SB RAS Genomics Core Facility and Joint Access Center for Artificial Plant Cultivation ICG SB RAS.
Funding
This study is supported by Russian Scientific Foundation (http://rscf.ru/) grant № 14–14-00161. The work at the Joint Access Center for Artificial Plant Cultivation ICG SB RAS was supported by the IC&G Budgetary Project No. 0324–2016-0001. Publication of this article has been funded by the Russian Scientific Foundation (Project No. 14–14-00161).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
About this supplement
This article has been published as part of BMC Plant Biology Volume 17 Supplement 1, 2017: Selected articles from PlantGen 2017. The full contents of the supplement are available online at https://bmcplantbiol.biomedcentral.com/articles/supplements/volume-17-supplement-1.
Abbreviations
- AGL
Agamous-Like
- ANT
Aintegumenta
- ARR
Arabidopsis Response Regulator
- CCA1
Circadian Clock Associated 1
- cDNA
DNA complementary to RNA
- CHE
CCA1 Hiking Expedition
- CNV
Copy number variation
- FHY3/FAR1
Far-Red Elongated Hypokotyls3/Far-Red-Impaired Response1
- FLC
Flowering Locus C
- FLM
Flowering Locus M
- LHY
Late Elongated Hypocotyl
- MAF
Mads Affecting Flowering
- MITE
Miniature inverted-repeat transposable element
- NILs
Near-Isogenic Lines
- PAL1
Phenylalanine Ammonia-Lyase
- Phy
PhytochromeA
- PI
Photoperiod insensitive
- PIF3
Phytochrome nteracting Factor3
- PIL1
Phytochrome Interacting Factor3-Like5
- Ppd-1
Photoperiod-1
- PRR
Pseudo-Response Regulator
- PS
Photoperiod sensitive
- PSL2
Photoperiod Sensitive Line 2
- qPCR
Quantitative PCR
- QTLs
Quantitative Trait Loci
- RAV1
Related to ABI3/VP1
- RE1ASPHYA3
RE1 putative repressor element
- RR
Response Regulator
- rRNA
DNA coding for rRNA
- RVE1
Reveille1
- RVE7/EPR1
Reveille 7/Early Phytochrome Responsive 1
- RVE8
Reveille8
- SD
Short day
- SORLIP2AT: SORLIP1AT: SORLIP5AT
Sequences over-represented in the light-induced promoters
- SSR
Simple Sequence Repeat
- TaFT1
Triticum aestivum Flowering Locus T
- TEM1
Tempranillo1
- TEM2
Tempranillo2
- TFBS
Transcription Factor Binding Sites
- TSS
Transcription Start Site
- VOZ2
Vascular Plant One–Zinc Finger1
- Vrn-1
Vernalization-1
- Zat2
Responsive To High Light 41
Authors’ contributions
Conceptualization: AK, EP, ES. Formal analysis and investigation: AK. Project administration, supervision: ES. Writing – original draft preparation: AK. Writing – review and editing: AK, EP, ES. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12870-017-1126-z) contains supplementary material, which is available to authorized users.
References
- 1.Beales J, Turner A, Griffiths S, Snape JW, Laurie DAA. Pseudo-response regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.) Theor Appl Genet. 2007;115:721–733. doi: 10.1007/s00122-007-0603-4. [DOI] [PubMed] [Google Scholar]
- 2.Law CN, Sutka J, Worland AJA. Genetic study of day-length response in wheat. Heredity (Edinb). 1978;41:185–191. doi: 10.1038/hdy.1978.87. [DOI] [Google Scholar]
- 3.Scarth R, Law CN. The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat. Heredity (Edinb) 1983;51:607–619. doi: 10.1038/hdy.1983.73. [DOI] [Google Scholar]
- 4.Wilhelm EP, Turner AS, Laurie DA. Photoperiod insensitive Ppd-A1a mutations in tetraploid wheat (Triticum durum Desf.) Theor Appl Genet. 2009;118:285–294. doi: 10.1007/s00122-008-0898-9. [DOI] [PubMed] [Google Scholar]
- 5.Worland T, Snape JW. Genetic basis of worldwide wheat varietal improvement. In: William A, Alain B, Maarten G, editors. World wheat book, a history of wheat breeding. Paris: Lavoisier Publishing; 2001. pp. 61–67. [Google Scholar]
- 6.Seki M, Chono M, Matsunaka H, Fujita M, Oda S, Kubo K, et al. Distribution of photoperiod-insensitive alleles Ppd-B1a and Ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed Sci. 2011;61:405–412. doi: 10.1270/jsbbs.61.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishida H, Yoshida T, Kawakami K, Fujita M, Long B, Akashi Y, et al. Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time. Mol Breed. 2013;31:27–37. doi: 10.1007/s11032-012-9765-0. [DOI] [Google Scholar]
- 8.Díaz A, Zikhali M, Turner A, Isaac P, Laurie D. Copy number variation affecting the Photoperiod-B1 and Vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum) PLoS One. 2012;7:e33234. doi: 10.1371/journal.pone.0033234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiss T, Balla K, Bányai J, Veisz O, Karsai I. Effect of different sowing times on the plant developmental parameters of wheat (Triticum aestivum L.) Cereal Res Commun. 2014;42:239–251. doi: 10.1556/CRC.2013.0064. [DOI] [Google Scholar]
- 10.Kitagawa S, Shimada S, Murai K. Effect of Ppd-1 on the expression of flowering-time genes in vegetative and reproductive growth stages of wheat. Genes Genet Syst. 2012;87:161–168. doi: 10.1266/ggs.87.161. [DOI] [PubMed] [Google Scholar]
- 11.Shaw LM, Turner AS, Laurie DA. The impact of photoperiod insensitive Ppd-1a mutations on the photoperiod pathway across the three genomes of hexaploid wheat (Triticum aestivum) Plant J. 2012;71:71–84. doi: 10.1111/j.1365-313X.2012.04971.x. [DOI] [PubMed] [Google Scholar]
- 12.Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, et al. The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci U S A. 2006;103:19581–19586. doi: 10.1073/pnas.0607142103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Plant Promoter Analysis Navigator. http://plantpan2.itps.ncku.edu.tw/index.html. Accessed 5 May 2017.
- 14.Kiseleva AA, Eggi EE, Koshkin VA, Sitnikov MN, Roder M, Salina EA, et al. Detection of genetic determinants that define the difference in photoperiod sensitivity of Triticum aestivum L. near-isogenic lines. Russ J Genet. 2014;50:701–711. doi: 10.1134/S102279541405007X. [DOI] [PubMed] [Google Scholar]
- 15.GrainGenes 2.0. http://wheat.pw.usda.gov/GG3/. Accessed 5 May 2017.
- 16.Hanahan D, Jessee J, Bloom FR. Plasmid transformation of Escherichia Coli and other bacteria. Methods Enzymol. 1991;204:63–113. doi: 10.1016/0076-6879(91)04006-A. [DOI] [PubMed] [Google Scholar]
- 17.Shcherban AB, Khlestkina EK, Efremova TT, Salina EA. The effect of two differentially expressed wheat VRN-B1 alleles on the heading time is associated with structural variation in the first intron. Genetica 2013;141:133–41. [DOI] [PubMed]
- 18.Chen A, Li C, Hu W, Lau MY, Lin H, Rockwell NC, et al. Phytochrome C plays a major role in the acceleration of wheat flowering under long-day photoperiod. Proc Natl Acad Sci U S A. 2014;111:10037–10044. doi: 10.1073/pnas.1409795111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ramakers C, Ruijter JM, Lekanne Deprez RH, Moorman AFM. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. doi: 10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 21.Pearce S, Kippes N, Chen A, Debernardi JM, Dubcovsky J. RNA-seq studies using wheat PHYTOCHROME B and PHYTOCHROME C mutants reveal shared and specific functions in the regulation of flowering and shade-avoidance pathways. BMC Plant Biol. 2016;16:141. doi: 10.1186/s12870-016-0831-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffiths S, Simmonds J, Leverington M, Wang Y, Fish L, Sayers L, et al. Meta-QTL analysis of the genetic control of ear emergence in elite European winter wheat germplasm. Theor Appl Genet. 2009;119:383–395. doi: 10.1007/s00122-009-1046-x. [DOI] [PubMed] [Google Scholar]
- 23.Hanocq E, Niarquin M, Heumez E, Rousset M, Le Gouis J. Detection and mapping of QTL for earliness components in a bread wheat recombinant inbred lines population. Theor Appl Genet. 2004;110:106–115. doi: 10.1007/s00122-004-1799-1. [DOI] [PubMed] [Google Scholar]
- 24.Kiseleva AA, Shcherban AB, Leonova IN, Frenkel Z, Salina EA. Identification of new heading date determinants in wheat 5B chromosome. BMC Plant Biol. 2016;16(s1):35–46. doi: 10.1186/s12870-015-0688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Z, Song Y, Zhou R, Ren Z, Jia J. Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 2010;185:841–851. doi: 10.1111/j.1469-8137.2009.03099.x. [DOI] [PubMed] [Google Scholar]
- 26.Mohler V, Lukman R, Ortiz-Islas S, William M, Worland AJ, van Beem J, et al. Genetic and physical mapping of photoperiod insensitive gene Ppd-B1 in common wheat. Euphytica. 2004;138:33–40. doi: 10.1023/B:EUPH.0000047056.58938.76. [DOI] [Google Scholar]
- 27.Shaw LM, Turner AS, Herry L, Griffiths S, Laurie DA. Mutant alleles of Photoperiod-1 in wheat (Triticum aestivum L.) that confer a late flowering phenotype in long days. PLoS One. 2013;8:e79459. doi: 10.1371/journal.pone.0079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kendrick RE, Kronenberg GHM. Photomorphogenesis in plants. Dordrecht: Springer Netherlands; 1994. [Google Scholar]
- 29.Terzaghi WB, Cashmore AR. Light-regulated transcription. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:445–474. doi: 10.1146/annurev.pp.46.060195.002305. [DOI] [Google Scholar]
- 30.Koshkin VA, Lisker IS, Merezhko AF, Kosareva IA, Dragavtsev VA. Influence of genes Ppd on phytochrome, photoperiodic sensitivity, growth and development of isogenic lines in wheat. Dokl. Russ. Acad. Agric Sci. 2004;1:3–4. [Google Scholar]
- 31.Li J, Li G, Wang H, Wang Deng X. Phytochrome signaling mechanisms. The. Arabidopsis Book. 2011;29:e0148. doi: 10.1199/tab.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mongkolsiriwatana C, Pongtongkam P, Peyachoknagul S. silico promoter analysis of photoperiod-responsive genes identified by DNA microarray in rice (Oryza sativa L.) J Nat Sci. 2009;43:164–177. [Google Scholar]
- 33.Genoud T, Schweizer F, Tscheuschler A, Debrieux D, Casal JJ, Schäfer E, et al. FHY1 mediates nuclear import of the light-activated phytochrome a photoreceptor. PLoS Genet. 2008;4:e1000143. doi: 10.1371/journal.pgen.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanumappa M, Pratt L, Cordonnier-Pratt M, Deitzer GA. Photoperiod-insensitive barley line contains a light-labile phytochrome B. Plant Physiol. 1999;119:1033–1040. doi: 10.1104/pp.119.3.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szucs P, Karsai I, von Zitzewitz J, Mészáros K, Cooper LLD, YQ G, et al. Positional relationships between photoperiod response QTL and photoreceptor and vernalization genes in barley. Theor Appl Genet. 2006;112:1277–1285. doi: 10.1007/s00122-006-0229-y. [DOI] [PubMed] [Google Scholar]
- 36.Nishida H, Ishihara D, Ishii M, Kaneko T, Kawahigashi H, Akashi Y, et al. Phytochrome C is a key factor controlling long-day flowering in barley. Plant Physiol. 2013;163:804–814. doi: 10.1104/pp.113.222570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ogihara Y, Shimizu H, Hasegawa K, Tsujimoto H, Sasakuma T. Chromosome assignment of four photosynthesis-related genes and their variability in wheat species. Theor Appl Genet. 1994;88:383–394. doi: 10.1007/BF00223649. [DOI] [PubMed] [Google Scholar]
- 38.Johnson E, Bradley M, Harberd NP, Whitelam CC. Photoresponses of light-crown phyA mutants of Arabidopsis. Plant Physiol. 1994;105:141–149. doi: 10.1104/pp.105.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neff MM, Chory J. Genetic interactions between phytochrome a, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faure S, Turner AS, Gruszka D, Christodoulou V, Davis SJ, von Korff M, et al. Mutation at the circadian clock gene EARLY MATURITY 8 adapts domesticated barley (Hordeum vulgare) to short growing seasons. Proc Natl Acad Sci U S A. 2012;109:4–9. doi: 10.1073/pnas.1120496109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Song YH, Shim JS, Kinmonth-Schultz HA, Imaizumi T. Photoperiodic flowering: time measurement mechanisms in leaves. Annu Rev Plant Biol. 2015;66:441–464. doi: 10.1146/annurev-arplant-043014-115555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schönrock N, Bouveret R, Leroy O, Borghi L, Köhler C, Gruissem W, et al. Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway. Genes Dev. 2006;20:1667–1678. doi: 10.1101/gad.377206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koo SC, Bracko O, Park MS, Schwab R, Chun HJ, Park KM, et al. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6. Plant J. 2010;62:807–816. doi: 10.1111/j.1365-313X.2010.04192.x. [DOI] [PubMed] [Google Scholar]
- 44.Pérez-Ruiz RV, García-Ponce B, Marsch-Martínez N, Ugartechea-Chirino Y, Villajuana-Bonequi M, de Folter S, et al. XAANTAL2 (AGL14) is an important component of the complex gene regulatory network that underlies Arabidopsis shoot apical meristem transitions. Mol Plant. 2015;8:796–813. doi: 10.1016/j.molp.2015.01.017. [DOI] [PubMed] [Google Scholar]
- 45.Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scortecci KC, Michaels SD, Amasino RM. Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 2001;26:229–236. doi: 10.1046/j.1365-313x.2001.01024.x. [DOI] [PubMed] [Google Scholar]
- 47.Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell. 2003;15:1159–1169. doi: 10.1105/tpc.009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim DH, Sung S. The plant homeo domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci U S A. 2010;107:17029–17034. doi: 10.1073/pnas.1010834107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu X, Jiang D, Wang Y, Bachmair A, He Y. Repression of the floral transition via histone H2B monoubiquitination. Plant J. 2009;57:522–533. doi: 10.1111/j.1365-313X.2008.03709.x. [DOI] [PubMed] [Google Scholar]
- 50.Gu X, Le C, Wang Y, Li Z, Jiang D, Wang Y, et al. Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun. 2013, 1947;4 [DOI] [PMC free article] [PubMed]
- 51.Scortecci K, Michaels SD, Amasino RM. Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol Biol. 2003;52:915–922. doi: 10.1023/A:1025426920923. [DOI] [PubMed] [Google Scholar]
- 52.Adamczyk BJ, Lehti-Shiu MD, Fernandez DE. The MADS domain factors AGL15 and AGL18 act redundantly as repressors of the floral transition in Arabidopsis. Plant J. 2007;50:1007–1019. doi: 10.1111/j.1365-313X.2007.03105.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee S, Kim J, Han J-J, Han M, An G. Functional analyses of the flowering time gene OsMADS50, the putative SUPPRESSOR OF OVEREXPRESSION OF CO 1/AGAMOUS-LIKE 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004;38:754–764. doi: 10.1111/j.1365-313X.2004.02082.x. [DOI] [PubMed] [Google Scholar]
- 54.Ryu C-H, Lee S, Cho L-H, Kim SL, Lee Y-S, Choi SC, et al. OsMADS50 and OsMADS56 function antagonistically in regulating long day (LD)-dependent flowering in rice. Plant. Cell Environ. 2009;32:1412–1427. doi: 10.1111/j.1365-3040.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- 55.Kang HG, Jang S, Chung JE, Cho YG, An G. Characterization of two rice MADS box genes that control flowering time. Mol Cells. 1997;7:559–566. [PubMed] [Google Scholar]
- 56.Zhao T, Ni Z, Dai Y, Yao Y, Nie X, Sun Q. Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.) Mol Gen Genomics. 2006;276:334–350. doi: 10.1007/s00438-006-0147-3. [DOI] [PubMed] [Google Scholar]
- 57.Shitsukawa N, Tahira C, Kassai KI, Hirabayashi C, Shimizu T, Takumi S, et al. Genetic and epigenetic alteration among three homoeologous genes of a class. E MADS Box gene in hexaploid wheat Plant Cell. 2007;19:1723–1737. doi: 10.1105/tpc.107.051813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yasui Y, Mukougawa K, Uemoto M, Yokofuji A, Suzuri R, Nishitani A, et al. The Phytochrome-interacting VASCULAR PLANT ONE-ZINC FINGER1 and VOZ2 redundantly regulate flowering in Arabidopsis. Plant Cell. 2012;24:3248–3263. doi: 10.1105/tpc.112.101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin J, Park E, Choi G. PIF3 regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J. 2007;49:981–994. doi: 10.1111/j.1365-313X.2006.03021.x. [DOI] [PubMed] [Google Scholar]
- 60.Kim J, Yi H, Choi G, Shin B, Song P, Choi G. Functional characterization of PIF3 in phytochrome-mediated lightsignal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Oda A, Fujiwara S, Kamada H, Coupland G, Mizoguchi T. Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett. 2004;557:259–264. doi: 10.1016/S0014-5793(03)01470-4. [DOI] [PubMed] [Google Scholar]
- 62.Oh E, Kim J, Park E, Kim J-I, Kang C, Choi G. PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell. 2004;16:3045–3058. doi: 10.1105/tpc.104.025163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hudson ME, Lisch DR, Quail PH. The FHY3 and FAR1 genes encode transposase-related proteins involved in regulation of gene expression by the phytochrome A-signaling pathway. Plant J. 2003;34:453–471. doi: 10.1046/j.1365-313X.2003.01741.x. [DOI] [PubMed] [Google Scholar]
- 64.Li G, Siddiqui H, Teng Y, Lin R, Wan X, Li J, et al. Coordinated transcriptional regulation underlying the circadian clock in Arabidopsis. Nat Cell Biol. 2011;13:616–622. doi: 10.1038/ncb2219. [DOI] [PubMed] [Google Scholar]
- 65.Kuno N, Møller SG, Shinomura T, Xu X, Chua N-H, Furuya M. The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell. 2003;15:2476–2488. doi: 10.1105/tpc.014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hudson ME, Quail PH. Identification of promoter motifs involved in the network of Phytochrome A-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol. 2003;133:1605–1616. doi: 10.1104/pp.103.030437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bruce WB, Deng XW, Quail PHA. Negatively acting DNA sequence element mediates phytochrome-directed repression of phyA gene transcription. EMBO J. 1991;10:3015–3024. doi: 10.1002/j.1460-2075.1991.tb07852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ngai N, Tsai FY, Coruzzi G. Light-induced transcriptional repression of the pea AS1 gene: identification of cis-elements and transfactors. Plant J. 1997;12:1021–1034. doi: 10.1046/j.1365-313X.1997.12051021.x. [DOI] [PubMed] [Google Scholar]
- 69.YX H, Wang YH, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- 70.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18:1338–1343. doi: 10.1016/j.cub.2008.07.075. [DOI] [PubMed] [Google Scholar]
- 71.Rawat R, Schwartz J, Jones MA, Sairanen I, Cheng Y, Andersson CR, et al. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc Natl Acad Sci U S A. 2009;106:16883–16888. doi: 10.1073/pnas.0813035106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rawat R, Takahashi N, Hsu PY, Jones MA, Schwartz J, Salemi MR, et al. REVEILLE8 and PSEUDO-REPONSE REGULATOR5 form a negative feedback loop within the Arabidopsis circadian clock. PLoS Genet. 2011;7:e1001350. doi: 10.1371/journal.pgen.1001350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Farinas B, Mas P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011;66:318–329. doi: 10.1111/j.1365-313X.2011.04484.x. [DOI] [PubMed] [Google Scholar]
- 74.Mizoguchi T, Wheatley K, Hanzawa Y, Wright L, Mizoguchi M, Song HR, et al. LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell. 2002;2:629–641. doi: 10.1016/s1534-5807(02)00170-3. [DOI] [PubMed] [Google Scholar]
- 75.Pruneda-Paz JL, Breton G, Para A, Kay SAA. Functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science. 2009;323:1481–1485. doi: 10.1126/science.1167206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Luo XM, Lin WH, Zhu S, Zhu JY, Sun Y, Fan XY, et al. Integration of light- and brassinosteroid-signaling pathways by a GATA transcription factor in Arabidopsis. Dev. Cell. 2010;19:872–883. doi: 10.1016/j.devcel.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Franco-Zorrilla JM, López-Vidriero I, Carrasco JL, Godoy M, Vera P, Solano R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc Natl Acad Sci U S A. 2014;111:2367–2372. doi: 10.1073/pnas.1316278111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Manfield IW, Devlin PF, Jen C, Westhead DR, Gilmartin PM. Conservation, convergence, and divergence of light-responsive, circadian-regulated, and tissue-specific expression patterns during evolution of the Arabidopsis GATA gene family. Plant Physiol. 2007;143:941–958. doi: 10.1104/pp.106.090761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Green PJ, Yong MH, Cuozzo M, Kano-Murakami Y, Silverstein P, Chua NH. Binding site requirements for pea nuclear protein factor GT-1 correlate with sequences required for light-dependent transcriptional activation of the rbcS-3A gene. EMBO J. 1988;7:4035–4044. doi: 10.1002/j.1460-2075.1988.tb03297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yadav V, Mallappa C, Gangappa SN, Bhatia S, Chattopadhyay SA. Basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell. 2005;17:1953–1966. doi: 10.1105/tpc.105.032060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Block A, Dangl JL, Hahlbrock K, Schulze-Lefert P. Functional borders, genetic fine structure, and distance requirements of cis elements mediating light responsiveness of the parsley chalcone synthase promoter. Proc Natl Acad Sci U S A. 1990;87:5387–5391. doi: 10.1073/pnas.87.14.5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chan CS, Guo L, Shih MC. Promoter analysis of the nuclear gene encoding the chloroplast glyceraldehyde-3-phosphate dehydrogenase B subunit of Arabidopsis thaliana. Plant Mol Biol. 2001;46:131–141. doi: 10.1023/A:1010602031070. [DOI] [PubMed] [Google Scholar]
- 83.Yamamoto YY, Ichida H, Matsui M, Obokata J, Sakurai T, Satou M, et al. Identification of plant promoter constituents by analysis of local distribution of short sequences. BMC Genomics. 2007;8:67. doi: 10.1186/1471-2164-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martinez-Hernandez A, Lopez-Ochoa L, Arguello-Astorga G, Herrera-Estrella L. Functional properties and regulatory complexity of a minimal rbcs light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 2002;128:1223–1233. doi: 10.1104/pp.010678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schulze-Lefert P, Dangl JL, Becker-André M, Hahlbrock K, Schulz W. Inducible in vivo DNA footprints define sequences necessary for UV light activation of the parsley chalcone synthase gene. EMBO J. 1989;8:651–656. doi: 10.1002/j.1460-2075.1989.tb03422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Borello U, Ceccarelli E, Giuliano G. Constitutive, light-responsive and circadian clock-responsive factors compete for the different l box elements in plant light-regulated promoters. Plant J. 1993;4:611–619. doi: 10.1046/j.1365-313X.1993.04040611.x. [DOI] [PubMed] [Google Scholar]
- 87.Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, et al. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Klucher KM, Chow H, Reiser L, Fischer RL. The AINTEGUMENTA gene of Arabidopsis required for ovule and female gametophyte development is related to the floral homeotic gene APETALA2. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Iida A, Kazuoka T, Torikai S, Kikuchi H, Oeda KA. Zinc finger protein RHL41 mediates the light acclimatization response in Arabidopsis. Plant J. 2000;24:191–203. doi: 10.1046/j.1365-313x.2000.00864.x. [DOI] [PubMed] [Google Scholar]
- 90.Davletova S, Davletova S, Schlauch K, Schlauch K, Coutu J, Coutu J, et al. The zinc-finger protein zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol. 2005;139:847–856. doi: 10.1104/pp.105.068254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lois R, Dietrich A, Hahlbrock K, Schulz WA. Phenylalanine ammonia-lyase gene from parsley: structure, regulation and identification of elicitor and light responsive cis-acting elements. EMBO J. 1989;8:1641–1648. doi: 10.1002/j.1460-2075.1989.tb03554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilmartin PM, Sarokin L, Memelink J, Chua NH. Molecular light switches for plant genes. Plant Cell. 1990;2:369–378. doi: 10.1105/tpc.2.5.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Degenhardt J, Tobin EMADNA. Binding activity for one of two closely defined phytochrome regulatory elements in an Lhcb promoter is more abundant in etiolated than in green plants. Plant Cell. 1996;8:31–41. doi: 10.1105/tpc.8.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Morishima A. Identification of preferred binding sites of a light-inducible DNA-binding factor (MNF1) within 5′-upstream sequence of C4-type phosphoenolpyruvate carboxylase gene in maize. Plant Mol Biol. 1998;38:633–646. doi: 10.1023/A:1006085812507. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Correlation coefficients of the expression patterns of the flowering genes. Bold font indicates significant values (P = 0.001). PI means Photoperiod Insensitive and PSL means Photoperiod Sensitive samples. (XLSX 26 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


