Abstract
Background
Cardiorespiratory fitness (VO2max) is an excellent predictor of chronic disease morbidity and mortality risk. Guidelines recommend individuals undertake exercise training to improve VO2max for chronic disease reduction. However, there are large inter-individual differences between exercise training responses. This systematic review is aimed at identifying genetic variants that are associated with VO2max trainability.
Methods
Peer-reviewed research papers published up until October 2016 from four databases were examined. Articles were included if they examined genetic variants, incorporated a supervised aerobic exercise intervention; and measured VO2max/VO2peak pre and post-intervention.
Results
Thirty-five articles describing 15 cohorts met the criteria for inclusion. The majority of studies used a cross-sectional retrospective design. Thirty-two studies researched candidate genes, two used Genome-Wide Association Studies (GWAS), and one examined mRNA gene expression data, in addition to a GWAS. Across these studies, 97 genes to predict VO2max trainability were identified. Studies found phenotype to be dependent on several of these genotypes/variants, with higher responders to exercise training having more positive response alleles than lower responders (greater gene predictor score). Only 13 genetic variants were reproduced by more than two authors. Several other limitations were noted throughout these studies, including the robustness of significance for identified variants, small sample sizes, limited cohorts focused primarily on Caucasian populations, and minimal baseline data. These factors, along with differences in exercise training programs, diet and other environmental gene expression mediators, likely influence the ideal traits for VO2max trainability.
Conclusion
Ninety-seven genes have been identified as possible predictors of VO2max trainability. To verify the strength of these findings and to identify if there are more genetic variants and/or mediators, further tightly-controlled studies that measure a range of biomarkers across ethnicities are required.
Keywords: Cardiorespiratory fitness, VO2max, Predictor genes, Training
Background
The worldwide prevalence of chronic diseases, such as cardiovascular disease, cancers, stroke and diabetes is rising [1]. Low cardiorespiratory fitness is strongly associated with chronic diseases and premature mortality [2–7]. To alleviate the health and economic burden associated with low cardiorespiratory fitness, health guidelines across the world recommend individuals undertake regular exercise [1].
Exercise training can increase cardiorespiratory fitness and decrease chronic disease via a number of mechanisms [7]. Adaptations include improvements to cardiac size, stroke volume (increase in volume of blood pumped from the left ventricle), cardiac output (volume of blood pumped from the heart per minute), pulmonary blood flow and respiratory function, supply of oxygen-rich blood to working muscles (increased number of capillaries and blood volume), muscle mitochondrial function and content, oxidative enzyme capacity, vascular wall health and function, and biomechanical efficiency [2, 7]. It has been suggested that improvements in cardiorespiratory fitness in response to exercise training varies greatly between individuals, with some people responding well or very well (‘responders’ or ‘high-responders’) to exercise training, whereas others only have mild increases in their cardiorespiratory fitness following similar exercise training (‘low-responders’) [4, 5, 8–11]. Importantly, these responses need to be compared to within-subject random variation to ascertain true inter-individual differences [12]. The ability to change cardiorespiratory fitness is a multifactorial trait influenced by environmental factors (such as exercise training) and genetic factors [4, 5, 11]. Considering cardiorespiratory fitness is one of the best integrative predictors of morbidity and mortality risk, it may be important to understand how genetics predict the variability in response to exercise training. This knowledge could lead to targeted personalised exercise therapy to decrease the burden of chronic disease.
The gold standard measure for cardiorespiratory fitness is maximal oxygen uptake (VO2max), which is quantified as the maximal amount of oxygen the body can use in 1 min, during dynamic work with large muscle mass [13]. Research into human variation of VO2max was first undertaken over forty years ago, with several authors identifying a strong genetic influence on VO2max in twins [14, 15]. Subsequent studies have identified significant familial aggregation for VO2max trainability. For example, authors have found greater variance between pairs of monozygotic (MZ; identical) twins than within pairs of twins for VO2max training response after standardized aerobic training interventions [16, 17]. The strongest evidence to date on this topic was found in the HEalth, Risk factors, exercise training And GEnetics (HERITAGE) family study [18]. Four hundred seventy-three Caucasian adults from 99 nuclear families completed 20 weeks of Moderate Intensity Continuous Training (MICT). The average increase in VO2max was 400 mL O2/min, with a range from − 114 to + 1097 mL/min. This difference was two and half times greater between families than within families, with a 47% heritability estimate for VO2max training response [18]. A major limitation from these findings, however, is there was no comparator control group.
Since this familial longitudinal research, the Human Genome Project completed sequencing of the human genome resulting in significant advancements in genetic analysis capabilities. This led to a better understanding of genetic variations of large populations. Analyzing genetic variants on a population level using techniques such as candidate gene analysis, GWAS, whole genome and exome sequencing and RNA expression analysis (RNA-seq, or microarrays) has resulted in the possibility of developing ‘personalized genomics’. This aims for biological profiling to provide more effective health management and treatment [5]. However, research in the field of exercise genomics it still in its infancy and much work is needed before genomic tools could be utilized to personalize exercise training programs [19].
The aim of this study was to systematically review the literature and identify genetic variants that have been associated with VO2max trainability following an aerobic exercise training intervention. Given the infancy of this research field, results should only be used to provide the basis for future research. This research should aim to confirm previous findings and investigate mediators that can influence gene expression. Importantly, future genetic studies in this area should attempt to investigate the physiological functions that contribute to improving VO2max training response and overall health outcomes. Findings from ongoing research may assist clinical professionals to provide personalized evidenced-based medicine centered on phenotype, contributing to the fight against chronic disease.
Methods
A comprehensive search of four databases (PubMed, Embase, Cinahl, Cochrane) was completed from their inception until October 2016. Studies focusing on genes and their VO2max/VO2peak response to supervised aerobic training were sought with the following search terms: genetic profiling, polymorphism, single nucleotide polymorphisms, SNPs, genetic variants, predictor genes, trainability, endurance training, cardiovascular fitness, cardiorespiratory fitness, VO2max, VO2peak, aerobic power, aerobic fitness, aerobic capacity. A full list of search terms can be found at the end of this review.
Two authors (CW and JC) agreed on the criteria for inclusion. Articles were incorporated if they were: original, peer-reviewed research; included an aerobic intervention, with minimum 75% supervision; included genetic variant testing; included a maximal VO2max/peak using direct gas analysis from an incremental test (pre and post intervention); conducted on humans; and written in English.
Using an extraction grid, one author (CW) conducted the initial screening analysis. After removing duplicates and scanning the titles and abstract of articles, those meeting the inclusion criteria were reviewed. Data recorded from the review consisted of the author’s name and place of study, study design, study sample, tissue source, genotyping method used, gene and variant examined, genotype, gene expression (if examined), intervention used, possible mediators (such as medications and health concerns), and the influence of the genetic variant investigated on VO2max change. Further articles were retrieved from snowballing included articles from their reference lists. Articles included in the review are in Table 1.
Table 1.
Summary of included articles
| Author, Year, Country | Gene/s tested for VO2max trainability | Study Design | Study Sample | Tissue source | Method for Genotyping | Intervention |
|---|---|---|---|---|---|---|
| Xu, 2015, China | ALAS2 | Single group, longitudinal. VO2max and venous blood samples taken pre & post intervention. | N = 244 healthy Chinese males; 18-22 years (20 ± 1.76); wt 65.06 ± 9.59 kg; ht. 174.37 ± 6.16 cm. N = 72 randomly selected for HiHiLo training (69.8 ± 7.8 kg and 177.93 ± 5.26 cm). | Peripheral blood leucocytes | PCR protocol + separation on polyacrylamide gel | 4 weeks; supervised HiLo training in hypoxia-training centre. Hi = bicycle ergometer for 30 mins at 75% VO2max, in 15.4% O2 concentrated environment, 3×/week for 4 weeks. Lo = same training but at lower elevation. |
| Yu, 2014, China | APOE | Single group, longitudinal. VO2max, anthropometric and serum levels tested pre & post intervention. | N = 360; 180 Chinese males and females; age 32.8 ± 11.9 yrs.; BMI 25.4 ± 5.6 kg/m2 M; BMI 26 ± 6.2 kg/m2 F; no health concerns; inactive. | Peripheral blood leucocytes | PCR-(polymerase chain reaction)-RFLP (restriction fragment length polymorphism) assay | 6 mths; progressive; supervised aerobic training; 60–85% VO2max. |
| Zarebska, 2014, Poland | GSTP1 | Single group, longitudinal. VO2max, HRmax, VEmax, AT and body composition tested pre & post intervention; balanced diet prior to intervention (2000 kcal) | N = 66 Polish females; 19–24 yrs.; BMI 21.8 ± 2.1 kg/m2; no health concerns; inactive; no supplements or medications; non-smokers. | Buccal cells | TaqMan allelic discrimination assay using qPCR | 3 mths; supervised; progressive MICT; 3×/wk.; 50–75% HRmax; 30–60 min. |
| Ghosh, 2013, Singapore | GWAS | Retrospective, single-group longitudinal. V02max tested pre & post intervention. | HERITAGE WHITES: n = 473 Caucasians; 230 male & 243 females; no major health concerns; inactive. | Lymphoblastoid cell lines | Illumina Human CNV370-Quad Bead Chips | HERITAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min. |
| Bouchard, 2011, USA | GWAS | Retrospective HERITAGE: Single group, longitudinal; VO2max tested pre & post intervention. DREW: RCT; VO2max tested pre & post intervention. STRRIDE 1 & 2: RCT; VO2max tested pre & post intervention. | HERITAGE WHITES: n = 473 Caucasians (252 women); 17–65 yrs.; inactive; no major health concerns HERITAGE BLACKS: n = 259 (177 women); 17–65 years; inactive; no major health concerns HERITAGE average age = 35.7 ± 14.5 yrs., BMI 25.8 ± 4.9 kg/m2. DREW study: n = 464 overweight or obese postmenopausal women; inactive; no major health concerns. STRRIDE 1 study: M&F; 40–65 yrs.; inactive; overweight, dyslipidemic and postmenopausal (F). STRRIDE 2 study: 18–70 yrs.; inactive; overweight, dyslipidemic. N = 183 for STRRIDE 1&2 studies. |
Lymphoblastoid cell lines | Illumina Human CNV370-Quad Bead Chips | HERITAGE 20 wks; supervised; progressive, MICT; 3×/wk.; 55–75% VO2max; 30–50 min. DREW: 6 mths; supervised; exercise groups: 4, 8 or 12 kcal/kg/week (MICT); 3-4×/week; progressive training intensity started at 50% VO2max. Each group expended 4 kcal/kg/week for first week. Group 1: maintained 4 kcal/kg/week for 6 months. Group 2: increased by 1 kcal/kg/week until 8ckal/week reached – maintain for remaining time. Group 3: increased by 1 kcal/kg/week until 8ckal/week reached – maintain for remaining time. STRRIDE 1: 8–9 mths; supervised exercise sessions. Three groups: 1. High-amount/vigorous intensity exercise (170 min/week/2000 kcal/week) or the calorie equivalent of jogging for ~20 miles per week at 55–85% VO2max. 2. Low amount/vigorous-intensity exercise/1200 kcal/week (~120 min/week) or the equivalent of 12 miles/week for jogging at 65–80%. 3. Low amount, moderate intensity exercise (1200 kcal/week (170 min/week) or the equivalent of 12 miles/week at 40–55% VO2max. STRRIDE 2: 8–9 mths; supervised; four groups: 1: Aerobic training – 1300 cal – 65-80%; 2: Resistance training only with 3 sets of 12–15 reps 3 x /week. 3: Combination of the first 2 protocols; 4: High anaerobic training – 2200 cal – 3 x week – 65-80%. First 2–3 months ‘ramp up period’. Following 6 mths using appropriate protocol. |
| McKenzie, 2011, USA | AKT | Single group, longitudinal. VO2max tested pre & post intervention; dietary stabilisation. | N = 51 M and 58 F Caucasians; 50–75 yrs.; no major health concerns; non-smoking; BMI <37; haematocrit >35; BP between 120/80 but less than 160/100 mmHg; at least one lipid abnormality; not any medication for blood pressure, cholesterol or glucose; F post-menopausal for at least 2 years (stable HRT or non HRT); inactive. | Peripheral blood leucocytes | TaqMan allelic discrimination assay using qPCR | 24 wks; supervised; progressive MICT; 3×/wk.; 50–70% HRR; 20–40 min. |
| Thomaes, 2011, Belgium | AMPD1; GR; CNTF | Retrospective, single group, longitudinal. VO2peak tested pre & post intervention. | N = 935 coronary artery disease patients (CAD); 76 females; Caucasian; age 56 ± 0.3 yrs.; BMI 25.8 ± 0.1 kg/m2; 5% smokers; 85% cardiac medications; 5% diabetes; 27% hypertension. | Peripheral blood leucocytes | Invader TM assay (third wave technologies) | 3 mths; supervised; 2-3×/wk.; 80% HRmax; 90 mins/session. |
| Onkelinx, 2011, Belgium | NOS3; Catalase; VEGF; Eco-SOD; GPX; P22Phox; PPARGC1; PPARα | Retrospective, single group, longitudinal. VO2peak tested pre & post intervention. | N = 935 coronary artery disease patients (CAD); 76 females; Caucasian; age 56 ± 0.3 yrs.; BMI 25.8 ± 0.1 kg/m2; 5% smokers; 85% cardiac medications; 5% diabetes; 27% hypertension. | Peripheral blood leucocytes | Invader TM Assay (third wave technologies) | CARAGENE: 3 mths; supervised; 3×/week; 90 mins; ~ intensity = 80% (HR/peakHRx100) |
| Silva, 2011, Brazil | NOS3 | Single group, longitudinal. VO2peak tested pre & post intervention. | N = 80 Portuguese police recruits; 20–35 years; BMI 23.3 ± 3.6 kg/m2; no health concerns; inactive. | Peripheral blood leucocytes | PCR-RFLP | 18 weeks; supervised; 3×/week/ 80 mins; intensity graded to VT HR. |
| Timmons, 2010, UK | GWAS | 1: Single group, longitudinal. VO2max & muscle biopsies tested pre & post intervention; 2: Blind test. VO2max & muscle biopsies tested pre & post intervention; 3: Retrospective: HERITAGE WHITES data | 1: N = 24 sedentary healthy Caucasian men (23 ± 1 yrs., 1.82 ± 0.02 m, 78.6 ± 2.7 kg); 2: 17 active & healthy Caucasian men (29 ± 6 yrs., 81.8 ± 9 kg, 1.8 ± 0.5 m); 3: HERITAGE Caucasians (as described in Bouchard 2011). | Lymphoblastoid cell lines from venous blood | Illumina Human CNV370-Quad Bead Chips | 1: 6 weeks; supervised MICT; 4 × 45 min cycling sessions/week @ 70% VO2max. 2:12 weeks; cycle ergometer 5×/week. Peak power test performed every Mon to determine intensity for week: Tues: 3 min intervals at 85%. Pmax separated by 3 min intervals at 40% Pmax; Thurs: 8 min intervals at 85% Pmax separated by 3 min intervals at 40% Pmax; Fri: 120 min at 55% Pmax continuously; duration increased by 5%/wk.; last 6 wks duration maintained but intensity increased by 1%/week; 3: HERITAGE WHITES Study (as described in Bouchard 2011). |
| Jenkins, 2010, USA | PLIN haplotypes | Retrospective, single group, longitudinal. VO2max tested; body composition; pre & post intervention; dietary stabilisation (American Heart Association). | N = 46 M & 55 F Caucasians (50–75 years); inactive; no major health concerns; BP < 160/99; non-smokers; BMI < 37 kg/m2; no meds for BP, cholesterol or glucose control; at least one lipid abnormality. | Unknown | TaqMan allelic discrimination assay using qPCR | 24 weeks; supervised; multi-modal MICT; progressive; 3×/wk.; 20–40 min; up to 70% VO2max reached; 60 min walk home included post 12 wks. |
| Alves, 2009, Brazil | ACE & Angiotensin | Single group, longitudinal. VO2max and echocardiography of left ventricle pre and post intervention. | N = 83 Brazilian policemen; age 26 years ±4.5; BMI 24 kg/m2 ± 1; healthy; normotensive. | Unknown | Polymerase chain reaction protocol. | 17 weeks; supervised MICT; 50–80%VO2peak; 60 min × 3/week. |
| He, 2008a, China | NRF-1 | Single group, longitudinal; VO2max, VT and RE tested pre & post intervention. | N = 102 Chinese male soldiers; no health concerns; age 18.8 ± 0.9 yrs.; wt 60.3 ± 6.5 kg; ht. 1.71 ± 5.8 m; no medications; non-smokers. | Peripheral blood leucocytes | PCR-RFLP assay | 18 wks; supervised; 3×5000m running sessions/wk.; 95%–105% VT. |
| He, 2008b, China | PPARGC1 | Single group, longitudinal; VO2max, VT and RE tested pre & post intervention. | N = 102 Chinese male soldiers; no health concerns; age 18.8 ± 0.9 yrs.; wt 60.3 ± 6.5 kg; ht. 1.71 ± 5.8 m; no medications; non-smokers. | Peripheral blood leucocytes | PCR-RFLP assay | 18 wks; supervised; 3×5000m running sessions/wk.; 95%–105% VT. |
| He, 2007a, China | TFAM | Single group, longitudinal. VO2max, VT and RE tested pre & post intervention. | N = 102 Chinese male soldiers; no health concerns; age 18.8 ± 0.9 yrs.; wt 60.3 ± 6.5 kg; ht. 1.71 ± 5.8 m; no medications; non-smokers. | Peripheral blood leucocytes | PCR-RFLP assay | 18 wks; supervised; 3×5000m running sessions/wk.; 95%–105% VT. |
| He, 2007b, China | NRF-2/NFE2L2 | Single group, longitudinal. VO2max, VT and RE tested pre & post intervention. | N = 102 Chinese male soldiers; no health concerns; age 18.8 ± 0.9 yrs.; wt 60.3 ± 6.5 kg; ht. 1.71 ± 5.8 m; no medications; non-smokers. | Peripheral blood leucocytes | PCR-RFLP assay | 18 wks; supervised; 3×5000m running sessions/wk.; 95%–105% VT. |
| Hautala, 2007, USA | PPARD | Retrospective, single group, longitudinal. VO2max, body composition and lipids tested pre & post intervention. |
N = 477 from HERITAGE Caucasian study (183 female) N = 264 from HERITAGE African-American study (247 female) |
Unknown | SNP scorer genotyping software | 20 wks; supervised; progressive, MICT; 3×/wk.; 55–75% VO2max; 30–50 min. |
| Defoor, 2006a, Belgium | ADRB1 | Retrospective, single group, longitudinal. VO2peak tested pre & post intervention. | N = 935 coronary artery disease patients (CAD); 76 females; Caucasian; age 56 ± 0.3 yrs.; BMI 25.8 ± 0.1 kg/m2; 5% smokers; 85% cardiac medications; 5% diabetes; 27% hypertension. | Peripheral blood leucocytes | Invader assay | CARAGENE: 3 mths; supervised; 2-3×/wk.; 80% HRmax; 90 mins/session. |
| Defoor, 2006b, Belgium | ACE | Retrospective, single group, longitudinal. VO2peak tested pre & post intervention. | N = 935 coronary artery disease patients (CAD); 76 females; Caucasian; age 56 ± 0.3 yrs.; BMI 25.8 ± 0.1 kg/m2; 5% smokers; 85% cardiac medications; 5% diabetes; 27% hypertension. | Peripheral blood leucocytes | Invader assay | CARAGENE: 3 mths; supervised; 2-3×/wk.; 80% HRmax; 90 mins/session. |
| He, 2006, China | HBB | Retrospective, single group, longitudinal. VO2max, VT and RE tested pre & post intervention. | N = 102 Chinese male soldiers; no health concerns; age 18.8 ± 0.9 yrs.; wt 60.3 ± 6.5 kg; ht. 1.71 ± 5.8 m; no medications; non-smokers | Peripheral blood leucocytes | PCR-RFLP assay | 18 wks; supervised; 3x5000m running sessions/wk.; 95%–105% VT |
| Defoor, 2005 | CKMM | Retrospective, single group, longitudinal. VO2peak tested pre & post intervention. | N = 935 coronary artery disease patients (CAD); 76 females; Caucasian; age 56 yrs. ± 0.3; BMI 25.8 kg/m2 ± 0.1; 5% smokers; 85% cardiac medications; 5% diabetes; 27% hypertension. | Peripheral blood leucocytes | Invader assay | CARAGENE: 3 mths; supervised; 2-3×/wk.; 80% HRmax; 90 mins/session. |
| Leon, 2004, USA | APOE | Retrospective, single group, longitudinal. VO2max, blood lipids tested pre & post intervention; counselled not to alter health habits. | N = 241 male and 89 female HERTIAGE Caucasians; 17–65 years; inactive; no major health concerns | Lymphoblastoid cell lines from venous blood | PCR-RFLP assay | HERTIAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min. |
| Thompson, 2004, USA | APOE | Single group, longitudinal. VO2max, anthropometric data and lipid levels collected pre & post intervention; dietary control. | N = 170 Caucasians (120 completed program – 60 M and F); 18–70 years (39 ± 11 years); consumed less than 2 drinks/day; physically inactive; BMI <31; no major health concerns. | Peripheral blood leucocytes | PCR-RFLP assay | 6 months supervised progressive training; 60–80% of VO2max; increasing from 15 to 40 mins during first 4 wks. Once at 40 mins, maintained this for 4 sessions each week for 5–6 months. Multimodal but treadmill primary aerobic activity. |
| Rico-Sanz, 2003, Canada | AMPD1 | Retrospective, single group, longitudinal. VO2max, submax and submax to maximal tested pre & post intervention. | N = 329 HERTAGE Caucasians and 90 HERITGAE African-Americans measured for training response; 17–65 years; inactive; no major health concerns. | Unknown | PCR protocol + separation on agarose gels | HERITAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min |
| Prior, 2003, USA | HIF1A | Single group, longitudinal. VO2max tested pre & post intervention. | N = 101 Caucasian and 22 African-Americans in good health; age 57.7 ± 0.91 yrs.; BMI 29.2 ± 0.64 kg/m2 | Peripheral blood lymphocytes | PCR-RFLP assay | 24 weeks; supervised; progressive MICT; 3×/wk.; 20–40 min; 50–70% VO2max |
| Woods, 2002, UK | ACE | Single group, longitudinal. VO2max, and HR/VO2 relationship tested pre & post intervention. | N = 59 Caucasians with ACE II allele and 29 without ACE DD allele; ~age 18.9 yrs.; ~ht. 1.78 m; ~ wt 73.4 kg; military camp. | Peripheral blood leucocytes | PCR protocol + polyacrylamide gel separation | 11 weeks; supervised aerobic training; 75% squads; 35% adventurous training; 25% running and circuit training. |
| Murakami, 2001, Japan | MtDNA | Single group, longitudinal. VO2max tested pre & post intervention | N = 41 Japanese M (age 20.6 ± 2.2 yrs), inactive; no major health concerns; wt 62.8 ± 7.5 kg; ht. 171.8 ± 6.7 cm. | Peripheral blood leucocytes | PCR-RFLP assay | 8 weeks; supervised 1×/week out of 3.5; 60 min/session; 70% VO2max |
| Sonna, 2001, USA | ACE | Double-blind study. VO2peak, anthropometrics physical fitness assessment for active duty personnel tested pre and post intervention. | N = 85 F and 62 M; age 21.7 ± 3.6 yrs.; 84 Caucasian, 20 Hispanic, 1 Native Americans, 5 Asian and 37 African-American; no major health concerns; BMI 23.1 ± 3.1 kg/m2; BF% 27.9 ± 6.1 F and 16.4 ± 5.7 M. | Peripheral blood leucocytes | PCR-RFLP assay | 8 weeks supervised; 6 days/week; 2 x aerobic (sprints & 3–5 miles) & 2 x strength. Participants place in 1 of 4 ability groups so all running for same duration. Participants also completed road marches and other drills. |
| Rankinen, 2000a, USA | Na + −K + ATPaseα | Retrospective, single group, longitudinal. VO2max and max power output tested pre & post intervention. | HERITAGE WHITES: 472 Caucasians; 17–65 years; inactive; no major health concerns. | Lympohblastoid cell lines | PCR protocol + agarose gel separation | HERTIAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min |
| Rankinen,2000b, USA | ACE | Retrospective, single group, longitudinal. V02max, VE, VT, blood lactate, oxygen, stroke volume, carbon dioxide, HR, tested pre & post intervention (submax VO2 test for older patients). | HERITAGE WHITES AND BLACKS: 476 Caucasian & 248 Blacks; 17–65 years; inactive; no major health concerns. | Lympohblastoid cell lines | PCR protocol + agarose gel separation | HERTIAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min |
| Hagberg, USA, 1999 | APOE | Retrospective, single group, longitudinal. VO2max and lipid levels tested pre and post; stabilised on American Heart Association diet 8 weeks prior to intervention. | N = 51; 40–80-year-old sedentary men (61 ± 3 yrs); overweight with ~BF% 30 ± 3; BP < 160/95 mmHg; no major health concerns or medications for blood lipids or glucose. | Peripheral blood leucocytes | PCR-RFLP assay | 9 months’ endurance training; multimodal; 5–7 months supervised and last 2–4 months used heart rate monitor to ensure 70–80% VO2max intensity and 3 days/week for 45 min was complied with. |
| Rivera, 1999, Canada | CKMM | Retrospective, single group, longitudinal. VO2max tested pre & post intervention. | HERITAGE WHITES: 495 Caucasians from 98 families; 17–65 years; inactive; no major health concerns. | Lympohblastoid cell lines | PCR-RFLP assay | HERTIAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min |
| Rivera, 1997, Canada | CKMM | Retrospective, single group, longitudinal. VO2max tested pre & post intervention. | HERITAGE WHITES: 160 Caucasian parents and 80 offspring; 17–65 years; inactive; no major health concerns. | Lympohblastoid cell lines | PCR-RFLP assay | HERTIAGE: 20 wks; supervised; progressive MICT; 3×/wk.; 55–75% VO2max; 30–50 min |
| Dionne, 1991, Canada | mtDNA | Single group, longitudinal. VO2max tested pre & post intervention. | N = 46 M from Quebec (17–27 yrs) & 27 M from Tempe (24–29 yrs); inactive | Peripheral blood leucocytes | PCR-RFLP assay | Quebec: 20 weeks; supervised; progressive training; Max 85% HRR; max 45 min/session; 3×/wk. Tempe: 12 weeks; supervised; progressive training; max 70–77% VO2max; max 40 min/session; 3×/wk |
| Bouchard, 1989, Canada |
AK1M
CKM |
RCT. VO2max, total power output tested pre & post intervention. | N = 295 M 7 F (18–30 years); healthy Caucasians | Muscle biopsy and peripheral blood leucocytes | Formazan technique? | Group 1: 15 weeks; supervised; progressive MICT; 30–45 min/session; 3-5×/wk.; 60–85% HRR Group 2: 15 weeks; supervised; progressive interval training; 1-2×/week; 80–85% HRR separated by 5 min recovery. |
M male, F female, wks weeks, mths months, wt weight, ht. height, yrs. years, BMI body mas index, BF % body fat percentage, VO 2max maximal oxygen uptake/cardiorespiratory fitness, PCR polymerase chain reaction protocol, RFLP restriction fragment length polymorphism, qPCR Quantatitive Polymerase Chain Reaction, RCT randomised controlled trial, GWAS genome wide association study, HRT hormone replacement therapy, SNP single nucleotide polymorphism, AT anaerobic threshold, MICT moderate intensity interval training, HR heart rate, HRR heart rate reserve, HR max heart rate maximum, P max maximal aerobic power, Submax submaximal, Cal/kcal calories, mtDNA mitochondrial DNA, BP blood pressure
A summary of key findings from the included articles is provided in Tables 2 and 3. Limitations were assessed by two authors (CW and JC) based on the intervention, genotyping method used, study design and sample used. Table 4 was developed to highlight which predictor genes for VO2max trainability merited further exploration. A third author (MW) examined Tables 1, 2, 3 and 4 to ensure all genetic variants, genomic coordinates and genotypes, were described with a consistent annotation.
Table 2.
Summary of findings from candidate gene studies
| Gene | Variant | Chromosome | Author & Date | Race | Age | Sex | Health concerns | (+/−/0)* Genotype & VO2max training response | P-value (x) | Highest training intensity | Sessions/week | Duration per session (min) | Training period | Training modality |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPARGC1 | Intron 7G/C | 22 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | GG, CG, CC (0) | 0.51 | 80% HRmax | 2–3 | 90 | 3 months | Ambulatory |
| He, 2008b | 102 Chinese | ~19 | M | N | All variants (0) | > 0.05 | 95–105% VT | 3 | Time to finish. | 18 weeks | 5000 m running | |||
| APOE | E2: rs7412 (c.526C > T; p.Arg176Cys) E3: WT E4: rs429358 (c.388 T > C; p.Cys130Arg) E3/E3: WT/WT E2/E3: p.Arg176Cys/WT E4/E3: p.Cys130Arg/WT E2/E2: p.Arg176Cys/p.Arg176Cys E2/E4: p.Arg176Cys/p.Cys130Arg E4/E4: p.Cys130Arg/p.Cys130Arg |
19 | Yu, 2014 | 360 Chinese | 18–40 | M F M F M&F |
N | E2/E3 in M (+) n = 20 E2/E3 F (+) n = 25 E3/E4 M (+) n = 31 E3/E4 F (+) n = 29 E2/E2; E2/E4; E3/E3; E4/E4 in M&F (0) |
0.04 0.03 0.02 0.02 > 0.05 |
60–85% VO2max | ‘Progressive’ but details NA | ‘Progressive’ but details NA | 6 months | Treadmill |
| Leon, 2004 | 265 Caucasian | 17–65 | M&F | N | All variants (0) | > 0.05 | 75% VO2max | 3 | 30–50 | 20 weeks | Cycle ergo | |||
| Thompson, 2004 | 170 Unknown | ~39 | M&F | N | E3/E3 (−) n = 43 E2/E3 (0) n = 40 E3/E4 (0) n = 41 |
< 0.01 | 60–85% VO2max | 4 | Up to 50 min | 6 months | Treadmill | |||
| CKM | 1170 & 985 + 185 | 19 | Defoor, 2005 | 935 Caucasian | ~56 | M&F | Y (CAD) | AA; GG; A/G (0) | > 0.05 | 80% HRmax | 2–3 | 90 | 3 months | Ambulatory |
| Rivera, 1999 | 240 Caucasian | 17–65 | M&F | N | CKM locus (n = 227) | < 0.01 | 75% VO2max | 3 | 30–50 | 20 weeks | Cycle ergo | |||
| Rivera, 1997 | 495 Caucasian | 17–65 | M&F | N | Homozygotes 1170bpa allele (−) n = 12 | < 0.05 | 75% VO2max | 3 | 30–50 | 20 weeks | Cycle ergo | |||
| Bouchard, 1989 | 295 Caucasian | 18–30 | M&F | N | All variants (0) | > 0.05 | 1. 60–85% HRR 2: 80–85% HRR |
1: 1–2 2: 3–5 |
1: Intervals 2: 30–45 |
1: 15 2: 15 |
1: Cycling 2: Cycling |
|||
| ACE | Insertion (I) or Deletion (D) | 17 | Alves, 2009 | 83 Brazilian | ~26 | M | N | All variants (0) | > 0.05 | 50–80% VO2peak | 2–3 | 60 min | 17 weeks | Running |
| Rankinen, 2000b | 476 Caucasian 248 AA |
17–65 | M&F | N | DD Caucasian offspring (+) n = 81 |
0.042 | 75% VO2max | 3 | 30–50 | 20 weeks | Ergo cycle | |||
| Defoor, 2006 | 935 Caucasian | ~56 | M&F | Y (CAD) | II (+) (frequency of 0.3 M and 0.36 F) | Entire group: 0.047 No Ace inhibitors: 0.013 |
80% HRmax | 2–3 | 90 | 3 months | Ambulatory | |||
| Woods, 2002 | 59 Caucasian | ~19 | M | N | II; I/D; DD (0) | >0.22 | NA | NA | NA | 11 weeks | Squads, adventure training, running, circuits | |||
| Sonna, 2001 | 147 Caucasian, 37 AA, 26 other | 19–24 | M&F | N | II, DD (0) | >0.05 | NA | 4–6 | 90 min | 8 weeks | Military training | |||
| CYBA; P22Phox | A24G – 640A > G | 16 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | AA, AG, GG (0) CC, CT, TT (0) |
0.78 0.94 |
80% HRmax | 2–3 | 90 | 3 months | Ambulatory |
| PLIN | PLIN1 (6209 T > C) – rs2289487 15:g.90217096C > T PLIN4 (11482G > A) – rs894160 15:g.90211823C > T PLIN5 (13041A > G – rs2304795 15:g.90210263A > G PLIN6 (149954A > T – rs1052700 15:g.90208310A > T |
15 | Jenkins, 2010 | 101 Caucasian | NA | M&F | N | Genotypes and haplotypes (0) | p > 0.05 | Up to 70% VO2max | 3 | 20–40 min | 24 weeks | Multi-modal |
| AKT | rs1130214 (4:g.105259734C > A) | 14 | McKenzie, 2011 | 109 Caucasian | 50–75 | M F |
Elevated BP, cholesterol, menopause | All genotypes sig. Increased, but GT/TT men (+) n = 22 | 0.037 | 50–70%HRR | 3 | 20–40 min | 24 weeks | Multi-modal |
| HIF1A | T + 140C (rs11549465) A-2500 T |
Ch 14 | Prior, 2003 | 101 Caucasian 22 AA |
>60 <60 |
M&F | N | CT & TT in Caucasian over 60 (−) n = 37 All other ages, race and genotypes (0) |
0.03 >0.05 >0.05 |
50–70% VO2max | 3 | 20–40 min | 24 weeks | ‘Aerobic training’ |
| Na + −K + −ATPase α2 | Alpha2 exon 1 Alpha2 exon 21–22 |
13 | Rankinen, 2000a | 472 Caucasian | 17–65 | M&F | N | 3.3/3.3 (−) n = 5 10.5/10.5 offspring (+) n = 14 |
0.018 0.017 |
55–75% VO2max | 3 | 30–50 | 20 weeks | Cycle ergo |
| HBB | -551C/T – no rs ID 11:g.5248801 T > C +16, intron 2 - rs10768683 11:g.5247791C > G +340 – no rs ID 11:g.5246488 T > A |
11 | He, 2006 | 102 Chinese | ~19 | M | N | CC, CT, TT (0) CC, CG, GG (0) AA, AT, TT (0) |
>0.05 | 95–105% VT | 3 | Time to finish. | 18 weeks | 5000 m running |
| CNTF | rs1800169 (11:g.58391501G > A) | 11 | Thomaes, 2011 | 935 Caucasian | ~56 | M&F | N | AA (+) n = 21 | 0.002 | 80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| CAT | -262C > T | 11 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | TT (−) n = 342 | 0.02 | 80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| GSTP1 | rs1695 (11:g67352689A > G c.313A > G p.Ile105Val) |
11 | Zabreska, 2014 | 66 Polish | 19–24 | F | N | GG & AG (+) n = 30 | Absolute: 0.029 Relative: 0.025 |
50–75% HR max | 3 | 60 | 3 months | ‘Aerobic routine’ |
| ADRB1 | Pos. 145 Pos. 1165 |
10 | Defoor, 2006 | 935 Caucasian | ~56 | M&F | Y(CAD) | Ser49Gly49, Ser49Ser49, | 80% HR max | 2–3 | 90 | 3 months | Ambulatory | |
| Gly49Gly49 (0) GLy389Gly389, |
0.18 | |||||||||||||
| Gly389Arg389, Arg389Arg389 (0) | 0.75 | |||||||||||||
| TFAM | rs1937 (10:g.60145342G > C c.35G > C p.Ser12Thr) rs2306604 (10:g.60148692A > G) rs1049432 (10:g.60155120G > T) |
10 | He, 2007b | 102 Chinese | ~19 | M | N | GG, CG, CC (0) AA, AG, GG (0) GG, GT, TT (0) |
>0.05 | 95–105% VT | 3 | Time to finish. | 18 weeks | 5000 m running |
| NOS3 | T-1495A – No rs ID 7:g.150689397A > T A-949G – rs1800779 7:g.150689943G > A -786 T > C– rs41322052 7:g150690106C > T G298A – rs1799983 7:g.150696111 T > G c.894 T > G (p.Asp298Glu)) |
7 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | TT, TA, AA (0) AA, AG, GG (0) TT, TC, CC (0) TT, CT, C (0) CC, CT, TT (0) GG, GA, AA (0) |
0.54 0.76 0.69 0.69 1.88 1.04 |
80% HRmax | 2–3 | 90 | 3 months | Ambulatory |
| -786 T > C– rs41322052 7:g150690106C > T Intron 4 – rs61722009 VNTR (repeat) 7:g.150694276_150694302AGGGGTG 894G > T – rs1799983 7:g.150696111 T > G c.894 T > G (p.Asp298Glu)) |
7 | Silva, 2011 | 80 Portuguese | 20–35 | M | N | TT, CC, TC (0) 4b4b, 4ba4c, 4a4a (0) GG, GT, TT (0) *All genotypes sig. Increased. fitness, thus no difference between groups |
0.001 | Graded to VT HR | 3 | 80 min | 18 weeks | Running | |
| NRF-1 | C&T - rs2402970 7:g.80647382G > T A & G - rs10500120 7:g.129393341A > G rs6949152 7:g129286436A > G |
7 | He, 2008a | 102 Chinese | ~19 | M | N | CC, CT, TT (0) AA, AG, GG (0) AA, AG, GG (0) |
0.38 0.110 0.094 |
95–105% VT | 3 | Time to finish. | 18 weeks | 5000 m running |
| AK1M | common and rare variants | 7 | Bouchard, 1989 | 295 Caucasian | 18–30 | M&F | N | (0) | > 0.05 | 1. 85% HRR 2: 85% HRR |
1: 1–2 2: 3–5 |
1: Intervals 2: 30–45 |
1: 15 2: 15 |
1: Cycling 2: Cycling |
| PPARD | Exon 4 + 15 Exon 7 + 65 |
Ch 6 | Hautala, 2007 | Caucasian AA | 17–65 | M&F | N | CC genotype in AA of Exon 4 + 15 (−) n = 19 | 0.005 | 75% VO2max | 3 | 30–50 | 20 weeks | Cycle ergo |
| VEGF | 405 460 |
6 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | GG, GC, CC (0) CC, CT, TT (0) |
0.52 0.52 |
80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| GR/NR3C1 | rs6190 (5:g.142780337C > T c.68G > A p.Arg23Lys) |
5 | Thomaes, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | G/A (+) n = 55 | <0.01 | 80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| PPARα | Gly482Ser | 4 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | GG, G, SS (0) | 0.59 0.8 |
80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| SOD3 | C760G | 4 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y (CAD) | CC (0) G carrier (0) |
0.12 0.18 |
80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| GPX | 197P > L | 3 | Onkelinx, 2011 | 935 Caucasian | ~56 | M&F | Y(CAD) | Pro197Pro (0) Leu-carrier (0) |
0.18 0.78 |
80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| NFE2L2 | Rs125949 Rs8031031 Rs718186 |
2 | He, 2007b | 102 Chinese | ~19 | M | N | CC, CA, AA (0) CT, TT, AA (0) AG, GG (0) |
> 0.05 | 95–105% VT | 3 | Time to finish. | 18 weeks | 5000 m running |
| AMPD1 | AMPD1:c.133C (rs17602729) | 1 | Thomaes, 2011 | 935 Caucasian | ~56 | M&F | N | CC (+) n = 652 | < 0.05 | 80% HR max | 2–3 | 90 | 3 months | Ambulatory |
| Rico-Sanz, 2003 | 329 Caucasian 90 AA |
17–65 | M&F | N | TT (−) in Caucasians (n = 6) | < 0.006 | 75% VO2max | 3 | 30–50 | 20 weeks | Cycling | |||
| mtDNA | MTND5 m.13470A > C or A > G m.12406G > A m.13365C > T |
mtDNA SNP via restriction enzyme | Murakami, 2001 | 21 Japanese | 20.6 | M | N | All variants (0) | > 0.05 | 70% VO2max | 3–4 | 60 min | 8 weeks | Ergo Cycle |
| mtDNA | Within mitochondria | Dionne, 1991 | 53 Quebec, Tempe | 17–27 | M | N | mtDNA subunit 5 N5 (−) n = 3 | 0.05 | Quebec: 85% HRR Tempe:77% VO2max |
Quebec: 3 Tempe: 3–5 |
Quebec:45 min Tempe:40 min |
Quebec: 20 wks Tempe: 12 wks |
Ergo Cycle | |
| ALAS2 | ≤166 bp | Mitochondria | Xu, 2015 | 72 Chinese | 18–22 | M | N | ≤166 bp (+) n = 25 | < 0.05 | ‘High/Low training’ | 3 | 30 min | 4 weeks | Ergo Cycle |
where possible, gene variants were annotated using the references sequence (GRCh37/hg19)
CAD coronary artery disease, wks weeks, mths months, VO 2max maximal oxygen uptake/cardiorespiratory fitness, AT anaerobic threshold, HRR heart rate reserve, HRmax heart rate maximum, Pmax maximal aerobic power, Cauc Caucasian, AA African-American, M male, F female
**(+) = high training response, (−) = low training response, (0) = neutral training response
(x) = p-value has been adjusted for covariates except for article by Xu et al. (2015) where it wasn’t clear if p-value had been adjusted (ALAS2)
Table 3.
Summary of hypothesis-free studies
| Gene | Variant | Chromosome | MapPosition | Minor allele frequency (MAF) frequency | Race | Gender | Age | Training period | Sessions/wk | Session duration | Sessions intensity | (+/−/0)** genotype/expression and VO2max response to training | P-value | Author, Date |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ^*CAMTA1 intronic | rs884736 | 1 | 6,937,692 | 0.41 |
1. 473 Caucasian
2. 259 African-American |
M&F
M&F |
17–65
17–65 |
20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | AA (−) |
1. 1.49 × 10-
4
2. 0.03 3. 1.54 × 10 −4 |
Bouchard, 2011 (1&2)
Ghosh, 2013 (3) |
| +ID3 | rs11574 (1:g.23559007 T > C c.313A > G p. Thr105Ala) | 1 | 23,758,085 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 2.1 × 10−3 | Timmons, 2010 |
|
*RGS18
5′ upstream of gene (non-coding) |
rs10921078 (1:g.192059022G > A) | 1 | 190,325,645 | 0.15 |
1. 483 Caucasian
2. 259 African-American |
M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | GG (−) n = 567 |
1. 7.17 × 10~
5
2. 0.032 |
Bouchard, 2011 |
|
^RYR2
intronic |
rs7531957 (1:g.237789656 T > G) | 1 | 235,856,279 | 0.08 | 473 Caucasian) | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA |
1:6.42 × 10–
5
2:1.18 × 10 −4 |
Bouchard, 2011 (1)
Ghosh, 2013 (2) |
| #SCLC45A1 | NA | 1 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | #89.1 | Ghosh, 2013 |
| MAST2 | rs2236560 | 1 | 46,268,021 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| SYPL2 | rs12049330 | 1 | 109,832,711 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2.12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| #ACVR1C | NA | 2 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #85.8 | Ghosh, 2013 |
| SLC4A5 | rs828902 | 2 | 74,323,642 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| KCNF1/NLGN1 | rs2003298 (2:g.11086150 T > C) | 2 | 11,003,601 | 0.42 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.21 × 10~4 | Bouchard, 2011 |
| * FLJ44450 | rs4952535 (2:g.42131523G > A) | 2 | 41,985,027 | 0.41 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | G (+) | 1.01 × 10-4 | Bouchard, 2011 |
| ++TTN | rs10497520 (2:g.179644855 T > C c3601A > G p.Lys1201Glu) | 2 | 175,353,100 | 0.50 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 2.5 × 10−3 | Timmons, 2010 |
| ++NRP2 intronic |
rs3770991 (2:g.206655739A > G) | 2 | 206,363,984 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.4 × 10−3 | Timmons, 2010 |
| CREB1 | rs2709356 | 2 | 208,120,337 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| SCN3A | rs7574918 | 2 | 165,647,425 | NA | 473 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| ^HCG22 | rs2517512 (6:g.31029685C > T) | 6 | NA | 0.18 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 3.09 × 10−5 | Ghosh, 2013 |
| *KCNH8 (268 kb) | rs4973706 (3:g.18921772 T > C) | 3 | 18,896,776 | 0.24 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 5.31 × 10~5 | Bouchard, 2011 |
| *ZIC4 (146 kb) intronic | rs11715829 | 3 | 148,439,856 | 0.08 |
1. 473 Caucasian
2. 183 Caucasian |
M&F
M&F |
17–65
40–65 |
20 wks
6 mths |
3×/wk.
3-4×/wk |
30–50 min
4-8 kcal/kg/week |
55–75% VO
2
max
+50%VO 2 max |
AA (−) n = 4 |
8.68 × 10-
6
0.032 |
Bouchard, 2011 |
| *NLGN1 (110 kb) intronic |
rs2030398 (3:g.173005973G > A) | 3 | 174,488,667 | 0.20 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 1.32 × 10~4 | Bouchard, 2011 |
| ^ADCY | NA | 3 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #86.1 | Ghosh, 2013 |
| AMOTL2 | rs13322269 | 3 | 135,569,834 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2.12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| CSN1S2B intronic |
rs2272040 (4:g71007047A > G) | 4 | 71,041,636 | 0.13 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 5.05 × 10-5 | Bouchard, 2011 |
| *LOC100289626 (134 kb) | rs2053896 (4:g137154796G > A) | 4 | 137,374,246 | 0.10 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 6.62 × 10~5 | Bouchard, 2011 |
| ^*ACSL1 | rs6552828 (4:g.185725416A > G) | 4 | 185,962,410 | 0.37 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | AA (−) |
1:1.31 × 10–
6
2:3.8 × 10 −6 |
Bouchard, 2011 (1)
Ghosh, 2013 (2) |
| ^SLED1 | rs6552828 | 4 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 3.8 × 10−6 | Ghosh, 2013 |
| ^C4orf40 | rs3775758 (4:g.71008910C > T) | 4 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.09 × 10−4 | Ghosh, 2013 |
| ^TEC | rs13117386 (4:g.48252763G > C) | 4 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.97 × 10−5 | Ghosh, 2013 |
| #NLN | NA | 5 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #88 | Ghosh, 2013 |
| FAABP6 | rs7734683 | 5 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.44 × 10−4 | Ghosh, 2013 |
| TTC1 | rs2176830 | 5 | 159,380,714 | 0.13 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.42 × 10~4 | Bouchard, 2011 |
| BTNL9 | rs888949 | 5 | 180,425,011 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2.12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| RTN4IP1/QRSL1 | rs898896 | 6 | 107,169,855 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2.12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| HCG22 | rs2523849 | 6 | 31,133,030 | 0.17 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.53 × 10-5 | Bouchard, 2011 |
| HCG22 | rs2523848 | 6 | 31,133,083 | 0.17 | 473 Caucasian | M & F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.53 × 10~5 | Bouchard, 2011 |
| HCG22 | rs2428514 | 6 | 31,135,495 | 0.15 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 8.22 × 10-5 | Bouchard, 2011 |
| HCG22 | rs2517518 | 6 | 31,136,324 | 0.17 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.53 × 10~5 | Bouchard, 2011 |
| HCG22 | rs2523840 | 6 | 31,138,404 | 0.17 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.53 × 10-5 | Bouchard, 2011 |
| HCG22 | rs2517506 | 6 | 31,139,659 | 0.17 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.53 × 10~5 | Bouchard, 2011 |
| *PRDM1 (287 kb) | rs10499043 | 6 | 106,353,830 | 0.13 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 3.93 × 10-6 | Bouchard, 2011 |
| *ENPP3 (17 kb) | rs10452621 | 6 | 132,127,094 | 0.12 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 1.23 × 10~4 | Bouchard, 2011 |
| +SLC22A3 | rs2457571 | 6 | 160,754,818 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | Downregulated in high responders | 3.0 × 10−3 | Timmons, 2010 |
| ^TMEM181 | NA | 6 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #84.5 | Ghosh, 2013 |
| ^PARK2 | NA | 6 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #84.8 | Ghosh, 2013 |
| ^SNX14 | NA | 6 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #86.7 | Ghosh, 2013 |
| ^BTBD9 | NA | 6 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #86 | Ghosh, 2013 |
| ^KCNQ5 | NA | 6 | NA | NA | 473 Caucasian | 1.M&F 2. M |
1.17–65 2. young adults |
1.20 wks 2. 6–12 wks |
1. 3×/wk. 2. 3–4/wk |
1. 30–50 min 2. 45 min vs progressive |
1. 55–75% VO2max 2. 70% vs progressive |
NA NA |
1:#85.9 2:NA |
Ghosh, 2013 (1), Timmons, 2010 (2) |
| PPARD | rs2076167 | 6 | 35,499,765 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| HDAC9 | rs3814991 | 7 | 18,601,428 | 0.11 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.46 × 10-4 | Bouchard, 2011 |
| WBSCR17 (35 kb) | rs12538806 | 7 | 70,200,777 | 0.30 | 473 Caucasian | M & F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.47 × 10~4 | Bouchard, 2011 |
| WBSCR17 (33 kb) | rs13235325 | 7 | 70,202,943 | 0.30 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.47 × 10-4 | Bouchard, 2011 |
| ++CPVL | rs4257918 | 7 | 29,020,374 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | Upregulated in high responders | 3.1 × 10−3 | Timmons, 2010 |
| ^ITGB8 | rs10265149 | 7 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.04 × 10−5 | Timmons, 2010 |
| LHFPL3 | NA | 7 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 84.34 | Ghosh, 2013 |
| PILRB | rs13228694 | 7 | 99,778,243 | NA | 41 Caucasian | Young adults | 17–65 | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| +DEPDC6 | rs7386139 | 8 | 121,096,600 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.85×10−2 | Timmons, 2010 |
| #PINX1 | N/A | 8 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | 88.2 | Ghosh, 2013 |
| *GRIN3A (516 kb) | rs1535628 | 9 | 104,056,570 | 0.09 | 473 Caucasian | M & F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 6.81 × 10~6 | Bouchard, 2011 |
| GRIN3A (540 kb) | rs959066 | 9 | 104,081,084 | 0.27 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.35 × 10-4 | Bouchard, 2011 |
| *C9orf27 (33 kb) | rs12115454 | 9 | 117,759,871 | 0.11 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | G (+) | 7.74 × 10~5 | Bouchard, 2011 |
| ^TTLL11 | rs7022103 | 9 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.08 × 10−4 | Ghosh, 2013 |
| KCNT1 | N/A | 9 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #86.5 | Ghosh, 2013 |
| KLF4 | rs4631527 | 9 | 109,309,857 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| TET1 | rs12413410 | 10 | 70,055,236 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| PRKG1 | N/A | 10 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #87.3 | Ghosh, 2013 |
| ^+SVIL | rs6481619 | 10 | 30,022,960 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.0 × 10−3 | Timmons, 2010 |
| +BTAF1 | rs2792022 | 10 | 93.730,409 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.2 × 10−2 | Timmons, 2010 |
| CASC2 | rs1413184 | 10 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.65 × 10−4 | Ghosh, 2013 |
| KIF5B | rs806819 | 10 | 32,403,990 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| +H19 | rs22551375 | 11 | 1,976,072 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | Upregulated in high responders | 4.0 × 10−4 | Timmons, 2010 |
| ACTN3 | rs1815739 | 10 | 66,084,671 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| BTAF1 | rs2792022 | 10 | 93,730,409 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| *LOC100130460 | rs2198009 | 11 | 10,360,153 | 0.50 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 2.28 × 10-5 | Bouchard, 2011 |
| *DBX1 (64 kb) | rs10500872 | 11 | 20,202,299 | 0.15 | 473 Caucasian | M & F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 6.49 × 10~6 | Bouchard, 2011 |
| ^*CD44 | rs353625 | 11 | 35,125,122 | 0.32 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA |
1:1.12 × 10–
4
2:1.64 × 10 −4 |
Bouchard, 2011 (1)
Ghosh, 2013 (2) |
| CXCR5 (36 kb) | rs4938561 | 11 | 118,223,695 | 0.23 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 9.29 × 10~5 | Bouchard, 2011 |
| * CXCR5 (24 kb/) BLR1 | rs7933007 | 11 | 118,235,879 | 0.23 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 7.35 × 10-5 | Bouchard, 2011 |
| ^CD6 | rs175098 | 11 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.11 × 10−4 | Ghosh, 2013 |
| ^SHANK2 | rs10751308 | 11 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | 8.11 × 10 −5 | Ghosh, 2013 |
| #GRIK4 | N/A | 11 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | 88.32 | Ghosh, 2013 |
| H19 | rs2251375 | 11 | 1,976,076 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| FAM19A2 | rs2168452 | 12 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.34 × 10−4 | Ghosh, 2013 |
| ^C12orf36 (14 kb) | rs12580476 | 12 | 13,435,330 | 0.14 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.08 × 10~4
2. 1.45 × 10−4 |
Bouchard, 2011 (1) Ghosh, 2013 (2) |
| ^NALCN | N/A | 13 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | #85 | Ghosh, 2013 |
| +MIPEP | rs7324557 | 13 | 23,194,862 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 5.1 × 10−3 | Timmons, 2010 |
| ^EEF1DP3 | rs2773968 | 13 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 3.67 × 10−6 | Ghosh, 2013 |
| ^CLYBL | N/A | 13 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #85.4 | Ghosh, 2013 |
| *TTC6 | rs12896790 | 14 | 37,343,673 | 0.09 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 3.59 × 10-5 | Bouchard, 2011 |
| METTL3 | rs1263809 | 14 | 21,058,740 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| TTC6 | rs8018889 | 14 | 37,353,342 | 0.09 | 473 Caucasian | M & F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 5.25 × 10~5 | Bouchard, 2011 |
| *DAAM1 | rs1956197 (14:g.59477414C > T) | 14 | 58,547,167 | 0.16 |
1. 473 Caucasian
2. 464 Caucasian |
1.M
2. F |
17–65
Post menopause |
20 wks
6 mths |
3×/wk.
120-170 min/wk |
30–50 min
120–170 min/wk |
55–75% VO
2
max
+50%VO 2 max |
AA (−) n = 84 | 1.43 × 10- 5 | Bouchard, 2011 |
|
*NDN (75 kb)
Downstream of NDN |
rs824205 | 15 | 21,559,164 | 0.15 |
1. 473 Caucasian
2. 464 Caucasian |
1.M
2.F |
17–65
Post menopause |
20 wks
9 mths |
3×/wk.
120-170 min/wk |
30–50 min
120-170 m in/wk |
55–75% VO
2
max
40–85%VO 2 max |
GG (−) n = 521 |
3.45 × 10~
5
0.05 |
Bouchard, 2011 |
| +DIS3L | rs1546570 | 15 | 64,382,829 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 2.3 × 10−2 | Timmons, 2010 |
| UNKL | rs3751894 | 16 | 1,426,876 | NA | 473 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| IL32 | rs13335 | 16 | 3,052,198 | NA | 473 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| #RPTOR | N/A | 17 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | #89 | Ghosh, 2013 |
| #VPS53 | N/A | 17 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #84 | Ghosh, 2013 |
| ACE | DI | 17 | 58,919,622 | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | NA | Timmons, 2010 |
| SMTNL2 | rs7217556 | 17 | 4,425,585 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| ZSWIM7 | R21 | 17 | 15,825,286 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| ENOSF1 | rs3786355 | 18 | 671,962 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| EMR4 | rs7256163 | 19 | 6,909,134 | 0.31 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.13 × 10-4 | Bouchard, 2011 |
| IER2 | rs892020 | 19 | 13,8185 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| DNAJB1 | rs4926222 | 19 | 14,488,050 | NA | 41 Caucasian | M | Young adults | 1.6 wks 2. 12 wks |
1. 4×/wk. 2. 3×/wk |
1. 45 min 2. Progressive |
1. 70% VO2max 2. Progressive |
NA | NA | Timmons, 2010 |
| g.63226200G > A | rs6090314 | 20 | 61,327,997 | 0.16 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 1:6.48 × 10~5
2:6.24 × 10−5 |
Bouchard, 2011 (1) Ghosh, 2013 (2) |
| ^YTHDF1 | rs6122403 | 20 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 6.24 × 10−5 | Ghosh, 2013 |
| ^MACROD2 | N/A | 20 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO 2 max | NA | #86.6 | Ghosh, 2013 |
| ^HLS21 | N/A | 21 | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | #84.7 | Ghosh, 2013 |
| *MN1 (14 kb) | rs738353 | 22 | 26,460,072 | 0.35 | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | A (+) | 1.23 × 10–4 | Bouchard, 2011 |
| LOC731789 | rs11015207 | NA | NA | NA | 473 Caucasian | M&F | 17–65 | 20 wks | 3×/wk | 30–50 min | 55–75% VO2max | NA | 1.61 × 10−4 | Ghosh, 2013 |
There were no other possible mediators (such as medications, health concerns) or other significant findings noted in the above three studies. Where possible, gene variants were annotated using the references sequence (GRCh37/hg19)
*Out of the 39 SNPs identified via GWAS, 21 (*) explained 49% of the VO2 max trainability variance (after regression analysis). The 15 most significant were then examined using data from the following studies: HERITAGE African-Americans, DREW study, STRRIDE study. The variants replicated are in italics
+11 SNPs from a regression analysis explained ~23% of the estimated VO2 max variance. 90% RNA expression remained unchanged by exercise training. (++) were found in study by Bouchard (2011) but weren’t included in the regression analysis because they weren’t considered significant at the 0.00015 level
^Top 20 GWAS associated genes based on second-best SNP-P values
#Candidate genes identified through CANDID software based on literature search; GWAS association data; sequence conversion & gene expression. This equates to a ‘final score’ rather than p-value. Bolded text indicates moderate-strong related biological mechanisms that influence VO2 max trainability
**(+) = significantly higher training response
(0) = no significant difference in training response between genotypes
(−) = significantly lower training response
Table 4.
Predictor genes that may influence VO2max training response
| Number | Chromosome | Gene | Variant | Race | Genotype/expression and VO2max training response (+/−/0)** | Author, Date (x = candidate gene study) |
|---|---|---|---|---|---|---|
| 1 | 1 | AMPD1 | rs17602729 | Caucasian | TT and CT (−) | Thomaes, 2011 (x); Rico-Sanz, 2003 (x) |
| 2 | 1 | CAMTA1 | rs884736 |
Caucasian
African-American |
AA (−) | Bouchard, 2011; Ghosh, 2013 |
| 3 | 1 | ID3 | rs11574 | Caucasian | TBC | Timmons, 2010 |
| 4 | 1 | RGS18 | rs10921078 |
Caucasian
African-American |
GG (−) | Bouchard, 2011 |
| 5 | 1 | RYR2 | rs7531957 | Caucasian | TBC | Bouchard, 2011; Ghosh, 2013 |
| 6 | 1 | SLC45A1 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 7 | 2 | ACVR1C | TBC | Caucasian | TBC | Ghosh, 2013 |
| 8 | 2 | KCNF1 | rs2003298 | Caucasian | TBC | Bouchard, 2011 |
| 9 | 2 | FLJ44450 | rs4952535 | Caucasian | G (+) | Bouchard, 2011 |
| 10 | 2 | TTN | rs10497520 | Caucasian | TBC | Timmons, 2010 |
| 11 | 2 | NRP2 | rs3770991 | Caucasian | TBC | Timmons, 2010 |
| 12 | 3 | KCNH8 | rs4973706 | Caucasian | A (+) | Bouchard, 2011 |
| 13 | 3 | ZIC4 | rs11715829 | Caucasian | AA (−) | Bouchard, 2011 |
| 14 | 3 | NLGN1 | rs2030398 | Caucasian | A (+) | Bouchard, 2011 |
| 15 | 3 | ADCY5 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 16 | 4 | CSN1S2B | rs2272040 | Caucasian | TBC | Bouchard, 2011 |
| 17 | 4 | LOC100289626 | rs2053896 | Caucasian | A (+) | Bouchard, 2011 |
| 18 | 4 | ACSL1 | rs6552828 | Caucasian | AA (−) | Bouchard, 2011; Ghosh, 2013 |
| 19 | 4 | SLED1 | rs6552828 | Caucasian | TBC | Ghosh, 2013 |
| 20 | 4 | PRR27; C4orf40 | rs3775758 | Caucasian | TBC | Ghosh, 2013 |
| 21 | 4 | TEC | rs13117386 | Caucasian | TBC | Ghosh, 2013 |
| 22 | 5 | NR3C1 | rs6190 | Caucasian | GG (−) | Thomaes, 2011 |
| 23 | 5 | NLN | TBC | Caucasian | TBC | Ghosh, 2013 |
| 24 | 5 | FABP6 | rs7734683 | Caucasian | TBC | Ghosh, 2013 |
| 25 | 5 | TTC1 | rs2176830 | Caucasian | TBC | Bouchard, 2011 |
| 26 | 6 | PPARD | Exon 4 + 15 Exon 7 + 65 |
African-American | CC (−) | Hautala, 2007 (x) |
| 27 | 6 | HCG22 | rs2517512 | Caucasian | TBC | Ghosh, 2013 |
| 28 | 6 | HCG22 | rs2523849 | Caucasian | TBC | Bouchard, 2011 |
| 29 | 6 | HCG22 | rs2523848 | Caucasian | TBC | Bouchard, 2011 |
| 30 | 6 | HCG22 | rs2428514 | Caucasian | TBC | Bouchard, 2011 |
| 31 | 6 | HCG22 | rs2517518 | Caucasian | TBC | Bouchard, 2011 |
| 32 | 6 | HCG22 | rs2523840 | Caucasian | TBC | Bouchard, 2011 |
| 33 | 6 | HCG22 | rs2517506 | Caucasian | TBC | Bouchard, 2011 |
| 34 | 6 | PRDM1 | rs10499043 | Caucasian | A (+) | Bouchard, 2011 |
| 35 | 6 | ENPP3 | rs10452621 | Caucasian | A (+) | Bouchard, 2011 |
| 36 | 6 | SLC22A3 | rs2457571 | Caucasian | Downregulated in high responders | Timmons, 2010 |
| 37 | 6 | TMEM181 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 38 | 6 | PARK2 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 39 | 6 | SNX14 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 40 | 6 | BTBD9 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 41 | 6 | KCNQ5 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 42 | 7 | HDAC9 | rs3814991 | Caucasian | TBC | Bouchard, 2011 |
| 43 | 7 | WBSCR17 | rs12538806 | Caucasian | TBC | Bouchard, 2011 |
| 44 | 7 | WBSCR17 | rs13235325 | Caucasian | TBC | Bouchard, 2011 |
| 45 | 7 | CPVL | rs4257918 | Caucasian | TBC | Timmons, 2010 |
| 46 | 7 | ITGB8 | rs10265149 | Caucasian | TBC | Ghosh, 2013 |
| 47 | 7 | LHFPL3 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 48 | 8 | DEPDC6 | rs7386139 | Caucasian | TBC | Timmons, 2010 |
| 49 | 8 | PINX1 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 50 | 9 | GRIN3A | rs1535628 | Caucasian | TBC | Bouchard, 2011 |
| 51 | 9 | GRIN3A | rs959066 | Caucasian | TBC | Bouchard, 2011 |
| 52 | 9 | C9orf27 | rs12115454 | Caucasian | G (+) | Bouchard, 2011 |
| 53 | 9 | TTLL11 | rs7022103 | Caucasian | TBC | Ghosh, 2013 |
| 54 | 9 | KCNT1 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 55 | 10 | FAM238B; LOC731789 | rs11015207 | Caucasian | TBC | Ghosh, 2013 |
| 56 | 10 | PRKG1 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 57 | 10 | SVIL | rs6481619 | Caucasian | TBC | Timmons, 2010 |
| 58 | 10 | BTAF1 | rs2792022 | Caucasian | TBC | Timmons, 2010 |
| 59 | 10 | CASC2 | rs1413184 | Caucasian | TBC | Ghosh, 2013 |
| 60 | 11 | H19 | rs22551375 | Caucasian | Upregulated in high responders | Timmons, 2010 |
| 61 | 11 | LOC100130460 | rs2198009 | Caucasian | A (+) | Bouchard, 2011 |
| 62 | 11 | DBX1 | rs10500872 | Caucasian | A (+) | Bouchard, 2011 |
| 63 | 11 | CD44 | rs353625 | Caucasian | TBC | Bouchard, 2011; Ghosh, 2013 |
| 64 | 11 | CXCR5 (36 kb) | rs4938561 | Caucasian | TBC | Bouchard, 2011 |
| 65 | 11 | CXCR5 (24 kb)/BLR1 | rs7933007 | Caucasian | TBC | Bouchard, 2011 |
| 66 | 11 | CD6 | rs175098 | Caucasian | TBC | Ghosh, 2013 |
| 67 | 11 | SHANK2 | rs10751308 | Caucasian | TBC | Ghosh, 2013 |
| 68 | 11 | GRIK4 | TBC | Caucasian | TBC | Ghosh, 2013 |
| 69 | 11 | CNTF | rs1800169 | Caucasian | AA (+) | Thomaes, 2011 (x) |
| 70 | 11 | CAT | -262C > T | Caucasian | TT (−) | Onkelinx, 2011 (x) |
| 71 | 11 | GSTP1 | c.313A > G (rs1695) | Caucasian | GG & AG (+) | Zarebska, 2014 (x) |
| 72 | 12 | FAM19A2 | rs2168452 | Caucasian | TBC | Ghosh, 2013 |
| 73 | 12 | C12orf36 | rs12580476 | Caucasian | TBC | Bouchard, 2011 Ghosh, 2013 |
| 74 | 13 | NALCN | TBC | Caucasian | TBC | Ghosh, 2013 |
| 75 | 13 | MIPEP | rs7324557 | Caucasian | TBC | Timmons, 2010 |
| 76 | 13 | EEF1DP3 | rs2773968 | Caucasian | TBC | Ghosh, 2013 |
| 77 | 13 | CLYBL | NA | Caucasian | TBC | Ghosh, 2013 |
| 78 | 13 | Na + −K + −ATPase α2 | Alpha2 exon 1 Alpha2 exon 21–22 |
Caucasian | 3.3/3.3 (−) 10.5/10.5 (+) |
Rankinen, 2000a (x) |
| 79 | 14 | HIF1A | T + 140C | Caucasian (60+ years) | C/T (−) | Prior, 2003 (x) |
| 80 | 14 | AKT1 | G205 T (RS1130214) | Caucasian men | GT & TT (+) | McKenzie, 2011 (x) |
| 81 | 14 | TTC6 | rs12896790 | Caucasian | C (+) | Bouchard, 2011 |
| 82 | 14 | DAAM1 | rs1956197 | Caucasian | AA (−) | Bouchard, 2011 |
| 83 | 15 | NDN | rs824205 | Caucasian | GG (−) | Bouchard, 2011 |
| 84 | 15 | DIS3L | Rs1546570 | Caucasian | TBC | Timmons, 2010 |
| 85 | 17 | ACE | Intron 16 | Caucasian |
DD (+)
II (+) |
Rankinen, 2000b (x); Defoor, 2006 (x) |
| 86 | 17 | RPTOR | NA | Caucasian | TBC | Ghosh, 2013 |
| 87 | 17 | VPS53 | NA | Caucasian | TBC | Ghosh, 2013 |
| 88 | 19 | ADGRE3P; EMR4 | rs7256163 | Caucasian | TBC | Bouchard, 2011 |
| 89 | 19 | APOE | TBC | Chinese & unknown |
E2/E3 (+)
E2/E3 (+) E3/E4 (+) E3/E4 (+) E3/E3 (−) |
Yu, 2014 (x); Thompson, 2004 (x) |
| 90 | 19 | CKM | Ncol | Caucasian | Homozygous 1170 bp (−); CKM locus (+/−) | Rivera, 1999(x); Rivera 1997 (x) |
| 91 | 20 | BIRC7 and YTHDF1 | rs6090314 | Caucasian | A (+) |
Bouchard, 2011
Ghosh, 2013 |
| 92 | 20 | YTHDF1 | rs6122403 | Caucasian | TBC | Ghosh, 2013 |
| 93 | 20 | MACROD2 | NA | Caucasian | TBC | Ghosh, 2013 |
| 94 | 21 | HLCS | NA | Caucasian | TBC | Ghosh, 2013 |
| 95 | 22 | MN1 | rs738353 | Caucasian | A (+) | Bouchard, 2011 |
| 96 | Mitochondria | ALAS2 | </=166 bp | Chinese | </=166 bp (+) | Xu, 2015 (x) |
| 97 | Mitochondria | mtDNA | TBC | Quebec, Tempe | mtDNA subunit 5 N5 (−) | Dionne, 1991 (x) |
Where possible, gene variants were annotated using the references sequence (GRCh37/hg19)
Bolded = genes that have been replicated between or within studies
**(+) = high training response, (−) = low training response, (0) = neutral training response, TBC to be confirmed whether variant contributes to a high or low training response
Results
Of the 1635 articles identified, 35 met the inclusion criteria (see Fig. 1). A summary of these articles is provided in Tables 1, 2 and 3. From the 35 articles, 97 genetic variants were identified as being significantly associated with VO2max trainability (Table 4).
Fig. 1.
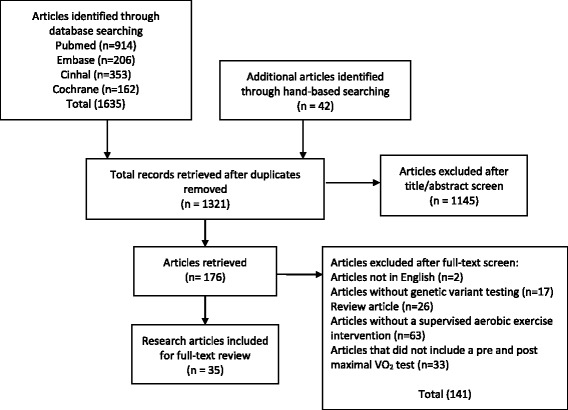
PRISMA flow chart of article selection process
Study characteristics
Across the studies DNA samples from 4212 individuals were used. Tissue sources were predominantly blood leucocytes, lymphoblastoid cell lines and buccal cells. Genotype was primarily identified through PCR-RFLP (polymerase chain reaction restriction fragment length polymorphism based analysis) for candidate genes and Illumina Human CV370-Quad Bead Chips for GWAS analysis (which can capture over 370,000 SNPs per participant).
Overall, 68% of participants in the reviewed studies were men, and ages ranged from 17 to 75 years. The average BMI of participants was 25.3 kg/m2 (SD 2.36). Where detailed, DNA samples were taken from a variety of ethnicities, including Caucasian (74.5%), Asian (13.5%), African-American (7.5%), Hispanic (4.3%) and Native American (0.2%).
The 35 included articles described 15 cohorts, with three cohorts providing subject data for 19 articles (see Table 1 for details). Nine articles [20–28] used data from the HERITAGE study and five [29–33] reviewed Caucasian participant data from the Cardiac Rehabilitation and Genetics of Exercise Performance and Training Effect (CARAGENE) study. Five studies examined clinical data from 102 young male and apparently healthy police recruits in China [34–38]. The remaining samples came from independent clinical studies focusing on apparently healthy but sedentary adults from a variety of ethnicities including Caucasians, Asians, African-Americans, Native American and Hispanics [13, 39–53].
Most reviewed studies (n = 32) used a single-group longitudinal design. However, one study compared three groups using a longitudinal design [28]. One study used retrospective data from two Randomized Controlled Trials (RCT) [20]; and one was a double-blind study [39].
Twenty-eight studies examined a MICT intervention. Two studies examined protocols using High Intensity Interval Training (HIIT) [28, 40]. The 5 remaining studies trained participants by running at Ventilatory Threshold (VT) [34–38]. Training intensity was measured using a percentage of VO2max, Heart Rate Reserve (HRR), VT, Maximal Power (Pmax) or Maximum Heart Rate (HRmax). Intensities varied between 50 and 85% VO2max, 95% -105% VT, 50–85% Pmax, 80–85% HRR and 50–80% HRmax. Training volume varied between 20 to 90 min per session (2-4×/week). The period of interventions ranged from 4 weeks to 9 months. Training modalities consisted primarily of cycle ergometers and treadmills.
Only six studies incorporated a standardized diet prior to and during the intervention period [23, 41–45]. Three articles included strength training [20, 39, 47] and two studies included military training [39, 47] as the intervention.
Genotyping findings
Candidate gene studies
The candidate gene association approach requires a prior hypothesis that the genetic polymorphisms of interest are causal variants or in strong linkage disequilibrium (LD) with a causal variant, and would be associated with a particular exercise-related phenotype at a significantly different rate than predicted by chance alone (may be higher or lower). This approach is effective in detecting genetic variants that are either directly causative, or belong to a shared haplotype that is causative [54]. Thirty-two candidate gene studies were based on the gene’s molecular function and possible association with VO2max trainability (Table 2).
Genes associated with muscular subsystems
VO2peak can be influenced by muscle efficiency and it has been hypothesized that genes encoding muscular subsystems may contribute to the genetic variability in VO2peak training response [33]. Twelve genes and 21 genetic variants related to muscular phenotypes were investigated in 935 (76 female) cardiac patients from the CARAGENE study [33]. Three out of the 21 genetic variants were significantly associated (p < 0.05) with an increase in VO2peak following 3 months of MICT (2–3 × 90-min sessions per week at 80% HRmax; p < 0.05). These variants included GR:c.68 > A (G/A genotype, number of people with genotype; n = 55) in the glucocorticoid receptor gene (GR; rs6190), CNTF:c.115-6G > A (AA genotype, n = 21) in the ciliary neurotrophic factor gene (CNTF; rs1800169) and the AMPD1:c.133C wild type (CC genotype, n = 652) of the adenosine monophosphate deaminase gene (AMPD1; rs17602729). Furthermore, a larger change in relative VO2peak was reported in patients with a greater number of these variants described (Area Under the Curve (AUC): 0.63; 95% Confidence Interval (CI): 0.56–0.7; p < 0.01). More specifically, those with a gene predictor score (GPS) of one or less positive response alleles had an average increase in VO2peak of 16.7%. Those with four or more positive response alleles had an average increase of 25%, with each positive response allele contributing approximately 1% (13.5 mL/min) to the increase in VO2peak.
Caucasians aged between 17 and 65 years from the HERITAGE study who were homozygous (TT genotype) for the AMPD1:c.133C > T (p.(Gln45*)) (rs17602729) variant (n = 6), had a lower VO2max training response (<121 mL/min; p = 0.006), compared to the CT and CC genotypes (n = 497) following 20 weeks of MICT (3 × 50 min per week at 55–75% HRmax) [46].
The serine/threonine protein kinase 1 (AKT1) gene has been linked to growth and skeletal muscle differentiation [44]. In a study of 109 Caucasians (50–75 years old), men (n = 22) with the AKT1:c.-350G > T (rs1130214) variant (TT/GT genotype) significantly increased their VO2max compared to men (n = 29) with the GG genotype (fold increase of 1.2 ± 0.02 vs 1.1 ± 0.02, p = 0.037) following 24 weeks of MICT (3 × 20–40 min per week at 50–75% HRR) [44].
The glutathione S-transferase P1 (GSTP1) c.313A > G variant has been associated with an impaired ability to remove excess reactive oxygen species. This is hypothesised to increase the exercise training response by better activation of cell signalling pathways resulting in positive muscle adaptations [45]. While investigating 62 Polish females’ (19–24 years-old) response to 12 weeks of MICT (3 × 60 min per week at 50–75% HRmax), participants (n = 30) with the GSTP1:c.313A > G (GG + GA genotype) demonstrated a 2 mL/kg/min greater improvement in VO2max compared to AA genotypes (n = 5) following training (absolute p = 0.029, relative p = 0.026, effect size = 0.06) [45].
Genes associated with electrolyte balance
The electrogenic transmembrane ATPase (NA+/K + −ATPase) gene may contribute to VO2max trainability by affecting the electrolyte balance and membrane excitability in working muscles [24]. Examining Caucasian data from the HERITAGE study, it was found that those homozygous for a recurrent 3.3-kb deletion in the exon 1 of the ATP1A2 gene (n = 5) had a 41% (45 mL/min) lower training response compared to heterozygotes (n = 87) [24]. This exon encodes on part (alpha-2-subunit) of the Na+/K + ATPase protein. This genotype also had a 48% (197 mL/min) lower VO2max training response than homozygotes (n = 380) for a repeated 8.8-kb in the exon 1 of the ATP1A2 gene following 20 weeks of MICT (p = 0.018) [24]. VO2max gains were 29% (130 mL/min) and 39% (160 mL/min) greater in offspring homozygous for a 10.5-kb deletion in exon 21–22 (n = 14) compared to heterozygotes (n = 93) and homozygotes (n = 187) respectively (p = 0.017) [24].
The angiotensin-converting enzyme (ACE) gene contributes to blood pressure, fluid and salt balance [55]. Elite endurance athletes are more likely to have the Insertion (I) allele [56] which relates to lower ACE activity and reduced blood pressure response during exercise, whereas sprint/power athletes are more likely to have the Deletion (D) allele and the DD genotype [57] and subsequently higher ACE activity. Caucasians from the CARAGENE study with the homozygous II genotype (frequency of 0.23 and 0.18 for men and women respectively) had a 2.1% greater VO2max training response (p = 0.047) compared to the DD genotype (frequency of 0.3 and 0.36 for men and women respectively) [31]. When eliminating those on ACE inhibitors, the improvement increased by 3% (p = 0.013) [31]. On the other hand, VO2max trainability was 14–38% greater (p = 0.042) in HERITAGE Caucasian offspring with the DD genotype (n = 81) [25]. Three studies found no association with ACE or angiotensinogen genetic variants and VO2max training response in 53 Caucasians (average age 19 years) following 12 weeks of military training [47]; 147 multi-ethnic 19–24 year-old adults following 8 weeks of military training [39]; and 83 Brazilian policemen (average age 26 years) following 17 weeks of MICT (3 × 60 min per week at 50–85% VO2peak) [48].
Genes associated with lipid metabolism
Genotypes of the perilipin (PLIN1) gene may influence training response via intracellular lipolysis and energy production [43]. In 101 Caucasians (50–75 years old), there were no significant differences between carriers and non-carriers of the PLIN1:c.504 T > A variant (rs1052700) after 24 weeks of MICT (20–40 min, 3 × per week) [43].
The peroxisome proliferator activated receptor delta (PPARD) gene affects fatty acid oxidation and energy production [22]. African-Americans (n = 19) from the HERITAGE study with the PPARD exon 4 + 15 (CC genotype) had a significantly lower VO2max training response (> 50 mL/min lower; p = 0.028) and power output (> 15 W lower; p = 0.005) compared to the C/T and TT genotypes (n = 230) [22].
Apolipoprotein E (APOE) variants affect the level of lipids in the blood, cell lipid uptake and endothelial vascular dilation [23]. APOE has 3 common alleles: E2 (TT/TT), E3 (TT/CC), E4 (CC/CC) at two SNPs (rs429358, rs7412), which can create six possible genotypes (E2/E2, E3/E3, E4/E4, E2/E3, E2/E4, E3/E4) [58]. The APOE E4 allele has been associated with Alzheimer’s disease [59], higher levels of low density cholesterol (LDL-C) and a greater risk of coronary heart disease compared to E3 (wild-type) and E2 carriers [23]. Chinese men (18–40 years) with the APOE E2/E3 (n = 20) and E3/E4 (n = 31) genotypes had a significantly higher VO2max training response (Odds Ratio (OR) = 0.68 (95% CI (0.04, 1.32); p = 0.04 and OR = 0.60 (95% CI (0.09, 1.11); p = 0.02 respectively) compared to other APOE genotypes following 6 months of progressive MICT (3 x per week at 60–85% VO2max) [13]. Similarly, Chinese women (18–40 years) with the APOE E2/E3 (n = 25) and E3/E4 (n = 29) genotypes had significantly higher VO2max training responses compared to other APOE genotypes (OR = 0.62 (95% CI = 0.05, 1.18); p = 0.03 and OR = 0.62(95% CI = 0.09,1.15); p = 0.02 respectively) [13]. Men and women (ethnicity unknown) with the E3/E3 APOE genotype (n = 43) had an 8% lower training response compared to the E2/E3 (n = 40) and E3/E4 genotypes (n = 37) (p < 0.01, Bonferroni-corrected) following 6 months of MICT (4 × 50 min per week at 60–85% VO2max) [42]. However, there was no significant difference in the VO2max training response between APOE genotypes in men and women from the HERITAGE study (n = 766) [23]. Similarly, in 51 males (40–80 years old, ethnicity not confirmed) there was no difference in VO2max training response between genotypes [41].
Genes associated with oxidative phosphorylation and energy production
Mitochondrial DNA (mtDNA) encodes several enzyme subunits involved in oxidative phosphorylation, and may be a key factor in endurance and cardiorespiratory fitness [56]. Research of mtDNA variants in 41 inactive Japanese men (mean age 20.6) failed to find a significant difference in trainability after 8 weeks of MICT (3–4 × 60 min per week at 70% VO2max) [49]. On the contrary, 3 men (17–25 years) with the mtDNA variant in subunit 5 of ND5 had a lower VO2max training response compared to other mtDNA variants (~ gain 0.22 L/min less, p < 0.05) following 12-weeks of MICT (3–5 × 45 min per week at 85%HRRmax) [50].
The creatine kinase muscle (CKM) gene has been associated with reduced fatigue from increased adenosine triphosphate (ADP) production [26, 27]. Using data from the HERITAGE study, parents and offspring homozygote for the 1170 bp allele (n = 12) had a lower VO2max training response (3 times and 1.5 times lower respectively; p < 0.05) compared to other CKM genotypes (n = 148). This explained 9 and 10% of the inter-individual variation in VO2max change respectively [26]. A nominal genetic linkage was identified in siblings (n = 277) who shared two alleles (1170 base pairs or 985 + 185 base pairs) at the CKM locus identical by descent (IBD), with these siblings having similar changes in VO2max compared to siblings with fewer alleles IBD (p = 0.04) [27]. In an earlier study focusing on muscle specific inherited variations, no association was found in 295 Caucasians (18–30 years old) between CKM or adenylate kinase (AK1) variants after a randomized control trial that included 15 weeks of endurance training versus maximal power contraction interval training [40]. Similarly, no association was found with the CKM gene and VO2max trainability in 937 Caucasian patients with coronary artery disease following 3 months of MICT (2–3 × 90 min aerobic sessions per week at 80% HRmax) [29].
Nuclear respiratory factor 1 (NRF1) and nuclear factor (erythroid-derived 2)-like 2 (NFE2L2) [36, 37], contribute to mitochondrial biogenesis and oxidative phosphorylation [60]. In a study involving 102 physically active Chinese male soldiers (average age 19 years), there was no association between NRF1 and NFE2L2 genotypes or haplotypes and VO2max trainability after 18 weeks of 3 × 5000 m runs per week at 95–105% VT [36, 37].
Genes associated with oxygen delivery
Nitric oxide causes coronary and arterial vasodilation, contributing to oxygen delivery regulation [32]. Data from the CARAGENE study was used to investigate genes associated with nitric oxide bioavailability [32]. These included nitric oxide synthase 3 (NOS3), cytochrome b-245 alpha chain (CYBA, also known as p22-PHOX), glutathione peroxidase (GPX1), catalase (CAT), superoxide dismutase 3 (SOD3), vascular endothelial growth factor A (VEGFA), peroxisome proliferator-activated receptor alpha (PPARα) and peroxisome proliferator-activated receptor gamma coactivator-related 1 (PPARC1) [32]. Participants carrying the C allele of the CAT:c.262 T > C variant (n = 342) had up to 3.1% greater improvements in VO2max training response compared to participants with the TT genotype (n = 521) following MICT (f = 3.6; p = 0.02). Participants with the NOS3 1.4 haplotype combinations (n = 36) had a 6.4% lower training response compared to the 3.3. haplotype combinations (n = 133) (p < 0.05). However, these associations were not significant after Bonferroni correction. No other associations were found with other genes or haplotypes related to nitric oxide availability and endothelial function [32]. Similarly, in a cohort of 80 Portuguese (20–35 years old) police recruits, there was no association between NOS3 genotypes (−786 TT/TC/CC, 894 GT/TT/GG) and VO2peak response following 18 weeks of 3 × 80-min per week of graded running training [59]. Additionally, no association was found with PPARGC1 and VO2max trainability in 102 Chinese male polices recruits following MICT [36].
The beta-2-adrenergic receptor (ADBR2) gene helps to support oxygen delivery to working muscles via the adrenergic receptors [30]. In participants from the CARAGENE study, there was no association found between ADBR2 genotypes or haplotypes, and VO2max trainability [30].
The hypoxia-inducible factor 1 alpha (HIF1A) gene is a transcriptional regulator that controls angiogenesis (blood vessel development) and metabolism by increasing the expression of hypoxia-induced genes, such as VEGF [52]. Caucasians 60 years and over with the H1F1A:c.1744C > T (rs11549465; C/T genotype; n = 37) had a significantly lower training response (0.3 mL/kg/min; p = 0.03) compared to those with the CC genotype (n = 64) following 24 weeks of MICT (3 × 20–40 min per week at 50–70% VO2max) [52].
The 5′-aminolevulinate synthase 2 (ALAS2) gene is highly expressed in erythroid cells and is imperative for hemoglobin and myoglobin synthesis [53]. Seventy-two Chinese participants (18–22 years old) allocated to one of 13 ALAS2 genotypes with compound dinucleotide repeats lengths (157 bp −184 bp), were placed in a 4-week ‘HiHiLo’ training program (varying between low and high altitude training at 75% VO2max) [53]. Baseline hemoglobin levels and change in VO2max with training was significantly higher in subjects (n = 25) with the dinucleotide repeats ≤ 166 bp (p < 0.05). No significant associations were found between VO2max trainability and other genes related to oxygen transport and utilization genotypes in 102 young Chinese soldiers following 18 weeks of 3 × 5000 m runs per week [35, 37, 38]. These genes include mitochondrial transcription factor A (TFAM) [35] and hemoglobin-beta locus (HBB) [38].
-
2.
Hypotheses free studies
Over the last decade, with the advent of technological advances allowing researchers to genotype millions of genetic variants (e.g. SNPs) in each individual, the investigation of the contribution of common variants to traits is now feasible. Unbiased and hypothesis-free genome wide association studies (GWAS) for exercise/health-related traits have emerged.
Three studies have used GWAS to identify genes associated with the VO2max response to exercise training [20, 21 28]. These are outlined in Table 3.
The first investigated two clinical trials and data from the HERITAGE study [28]. RNA expression profiling and VO2max testing was performed on 24 healthy and inactive Caucasian men (average age 24 years) before and after a 6-week training intervention (4 × 45-min cycling sessions per week at 70% VO2max). Muscle biopsies from the vastus lateralis were collected and the RNA expression of genes was correlated with changes in VO2max by analysing oligonucleotide arrays. Pearson correlations were used to identify the relationships between the median logit normalised probe sets and the number of times they were selected. In the 24 subjects, using a median correlation cut-off greater than 0.3, 29 genes were selected greater than 22 out of 24 times. The sum of expression of these 29 genes were found to have a significant linear relationship with VO2max change following endurance training (r 2 = 0.58, p < 0.00001). Across the group, VO2max changes improved on average by 14% and ranged from −2.8% to 27.5% (p = 0.0001). More than 20% of the group had a response less than 5%. A gene set enrichment analysis found that the oxidative phosphorylation gene was upregulated (False Discovery Rate (FDR) = 1.1%), which was associated with an increased reliance on lipids during training (RER decreased on average by 10% post training, p < 0.0001). To identify if these predictor genes would be similar in a different sample, a 12-week blind study on 17 young and active Caucasian men was conducted. Training consisted of 1-day of testing, 2 sessions of interval training (3 × 3-min intervals at 40–85% Pmax) and 2 × 60–120-min cycle sessions (55–60% Pmax) each week. The 29 predictor genes were also significantly associated with VO2max trainability in this group (p = 0.02). The haplotypes of these predictor genes were then genotyped using candidate genes identified from the HERITAGE study. Six genetic variants were associated with VO2max trainability: SMTNL2, DEPDC6, SLC22A3, METTL3, ID3 and BTNL9 (p < 0.01 each). A stepwise regression model using 25 variants from the predictor set and 10 variants from the HERTIAGE study (Table 3) found that eleven SNPs (included in Table 4) contributed to 23% of the differences seen in residual VO2 max gains, which correlated to approximately 50% of the genetic variability in VO2max trainability (seven variants from the RNA predictor set and four from the HERITAGE project). Reciprocal RNA expression validation found that three of four HERITAGE candidate genes enhanced the original RNA transcript predictor model. Overall, more than 90% of gene expression did not change. However, OCT3 was downregulated in high responders and H19 was upregulated in low responders (FDR <5%). BTNL9, KLF4 and SMTNL2 also had small but inconsistent changes in expression (i.e. dissimilar in high vs low responders) (FDR < 5%).
A GWAS examining 324,611 variants from the HERITAGE study was completed to identify possible predictor genes associated with VO2peak [20]. Based on single-variant analysis, 39 variants (Table 3) were associated with gains in VO2peak although none of these achieved genome-wide or suggestive significance (p = 1.5 × 10−4) [19]. The strongest predictor for training response was found in the Acyl-CoA synthetase long-chain family member 1 (ACSL1) gene (4:g.185725416A > G; rs6552828) which accounted for 7% of the training response (p = 1.31 × 10−6). After a stepwise multiple regression analysis of the thirty-nine variants, 21 were suggested to account for (or at least contribute to) 49% of the variance in VO2max trainability (included in Table 4; p < 0.05). The strongest predictors were found in SNPs associated with: PR domain-containing protein 1 (PRDM1); glutamate receptor, ionotropic, N-methyl-D-aspartate 3A (GRIN3A); N-methyl-D-aspartate receptor (NMDA); potassium voltage-gated channel subfamily H member 8 (KCNH8); zinc finger protein of cerebellum 4 (ZIC4); and, ACSL1. An unweighted ‘predictor score’ based on contribution to VO2max of these 21 variants was created. A score of ‘0’ represented homozygote for the low-response variant; ‘1’ represented heterozygous and ‘2’ represented homozygous for the high-response allele. Individuals with a score equal to or less than 9 (n = 36) had an average VO2max score improvement of 221 mL O2/min. Alternatively, those (n = 52) with a score equal to or greater than 19 had an average VO2max increase of 604 mL/min.
The 15 most significant variants were tested for replication in a sample of African-Americans from the HERITAGE study, women in the Dose Response to Exercise (DREW) study (n = 112), and the men and women in the Study of a Targeted Risk Reduction Intervention through Defined Exercises (STRRIDE) (n = 183) [20]. Variants in the NDN (15:g.24008071 T > C; rs824205) and DAAM1 (14:g.59477414C > T; rs1956197) were replicated in the DREW study, the Z1C4 (3:g.146957166 T > C T; rs11715829) variant was replicated in the STRRIDE study and CAMTA1 (7:g.7015105 T > C; rs884736) and RGS18 (1:g.192059022G > A; rs10921078) variants were replicated in African-Americans from the HERITAGE study. Four variants in the genes supervillin (SVIL), neuropillin 2 (NRP2), titin (TTN) and carbozypeptidase (CPVL) identified by Timmons et al. [28] were also found by Bouchard et al. [20], however, at a significance of 0.008, these variants were not included in the multi-variate regression analysis.
Using the HERITAGE cohort, an extended analysis was performed, with 2.5 million variants analysed [21]. To reduce bias associated with outlier variants, the second most significant variant p-value was used to determine genotype and changes in VO2max. Even with an extended analysis, the ACSL1 gene was shown to have the most significant variant (4:g.185725416A > G; rs6552828), which confirmed findings by Bouchard et al. [20], whom identified the most significant variant at each gene (Table 3). The following genes and their variants were also replicated in both studies: CAMTA1 (rs884736), RYR2 (rs7531957), g.63226200G > A (rs6090314), C12orf36 (rs12580476) and CD44 (rs353625) [20, 21].
The gene prioritisation tool ‘CANDID’ was then used to rank candidate genes for changes in VO2max [21]. This was done via: 1) a weighted analysis based on variant gene expression in targeted tissues; 2) GWAS p-value change in VO2max; 3) literature related to candidate genes; and 4) ‘cross species sequence conservation’ [21]. The top-ranking candidate genes from the GWAS and CANDID tool (Table 1) were then investigated for possible biological mechanisms and changes in VO2max. As a result, variants were allocated into four groups: 1) broad effects on exercise-related processes (such as the electron transport chain, physical fitness, skeletal development and other cardiorespiratory markers); 2) moderately strong scores against selective exercise-related processes; 3) high and low scores across several exercise-related processes; 4) low scores across all exercise-related processes.
Variants and their involvement in pathways related to changes in VO2max response were then examined [21]. Out of the sixteen pathways found, variants related to pantothenate and co-enzyme A (CoA) biosynthesis, PPAR gene signalling and immune function signalling had the highest level of ‘burden’ (variants contributing to trainability). The variants related to long-chain fatty acid transport (including ACSL1) and fatty acid oxidation strongly influence VO2max training response via lipid metabolism process and the tricarboxylic acid cycle, both of which affect the availability of adenosine triphosphate and subsequently training response.
Predictor genes
Out of the 35 articles analysed (candidate genes and GWAS studies), 97 predictor genes were identified as possible contributors to VO2max trainability (Table 4). These genes were based on what authors deemed significant, or the most significant, for their particular study. Thirteen of these predictor genes were replicated between at least two studies (bolded in Table 4). The traits for VO2max trainability (e.g. which genotype was related to the training effect and whether it was a low or high responding genotype) was not outlined for each variant and hence this will require confirmation in future studies.
Discussion
This systematic review aimed to summarize genetic variants that have been identified as influencing VO2max trainability. We have reviewed 35 studies that have reported 97 genes associated with an exercise training-induced improvement in VO2max. It has been estimated that VO2max trainability has a significant heritable component of around 50% [39].
There were several studies that identified the same variant, including: the lipid-related ACSL1:c.-32-716 T > C (rs6552828) [20, 21] and skeletal muscle-related AMPD1:c.133C > T [33, 46]; intra-cellular calcium regulator RYR2:c.6166 + 552 T > G; cellular function-related CD44 (rs3653625), transcriptional activator CAMTA1 (rs884736), non-coding C12orf36 (rs12580476) and apoptotic regulator 20:g.63226200G > A (rs6090314) [20, 21]. Additionally, Bouchard et al. [20] were able to replicate the variants in genes from the HERITAGE study, including: growth suppressor NDN, cell cortex function-related DAAM1, development-related Z1C4 and signal transduction inhibitor RGS18. Numerous identified variants were found in pathways that contribute to training response (e.g. calcium signaling, immune function, angiogenesis, mitochondrial biogenesis) with pathways and associated SNPs possibly influencing each other and overall trainability [21]. Several articles found conflicting results with electrolyte balance, lipid production and energy production genes ACE [25, 31, 47, 48], APOE [13, 23, 41, 42], mtDNA [49, 50] and CKMM variants respectively [26, 27, 29, 40]. All other ‘predictor genes’ identified are yet to be replicated.
While most of the articles examined in this review have focused on one or a few candidate genes/markers (n = 32), it is noted that exercise-related phenotypes are complex traits and are polygenic (i.e. influenced by many genes working together) with each genetic variant likely to be contributing a small percentage (typically less than 1%) to the overall change in VO2max [33, 39, 61]. Thus relying on one variant as a predictor is misguided; rather it has been suggested that a gene predictor score (GPS) based on numerous variants has a greater probability to determine higher and lower responders for VO2max trainability. For example, a score of ‘0’ represents a homozygote for a low-response variant; ‘1’ represents heterozygous and ‘2’ represents homozygous for a high-response variant [20]. A higher score indicates a greater possible VO2max training response (and vice versa). A similar model has been suggested in elite athletes aiming to determine the probability of an individual with a theoretically ‘optimal’ polygenic profile for endurance sports. The ‘optimal’ profile using a so-called ‘total genotype score’ (TGS, ranging from 0 to 100, with ‘0’ and ‘100’ being the worst and best genotype combinations, respectively) was quantified from a simple algorithm resulting from the combination of candidate polymorphisms [62, 63].
These predictor genes, along with muscle RNA and protein expression data provide a sound platform to further explore the cellular mechanisms underlying VO2max trainability. Further research will need to consider several limitations identified from the literature to-date. For example, the lack of replication found between articles and conflicting results with certain variants, may be a result of several main limitations (typically in study design). Firstly, most of the articles used a hypotheses-driven candidate gene approach (n = 32), several articles used retrospective data from similar cohorts (n = 19), and many lacked a control group and randomization (n = 31). While it is understandable that in the past, high-throughput SNP microarray or gene sequencing technology was not available to use, by looking at one or only a few gene variants (whereas it is estimated that the human genome consists of about 40 million common gene variants) it is almost impossible to generate meaningful information. Similarly, a lack of control group makes it challenging to distinguish between individual response to an intervention and within-subject random variation [64]. Secondly, most of the exercise training studies involve a relatively small number of participants (typically n = 20 to 30; with the exception of the HERITAGE and CARAGENE studies), which results in lack of statistical power when associating genotype with a phenotype. Many of the studies also failed to include a robust significance criterion (p < 0.05 occurs approximately 106 times in the genome by chance). Thirdly, a lack of racial diversity (74.5% Caucasian) further reduces the power of variants detected. Finally, many of the training studies were not tightly controlled in terms of nutrition, participant baseline data (study entry), physical activity status and other lifestyle factors.
Future research needs to consider epigenetic variation of gene activity that can occur in reaction to external factors, such as additional physical activity, drugs, diet and environmental toxins [61, 65]. Such epigenetic modifications can affect all adaptions to exercise training [10]. For example, in addition to nutrition and baseline physical activity status, there were many other differences in subjects between articles not taken into consideration including: age, training duration and volume (MICT vs. HIIT), body weight, body fat percentage, medications, clinical versus healthy populations; sleep, psychological status and the gut microbiome. Together, these are potential epigenetic modifiers (e.g. DNA methylation and histone acetylation) that can influence gene expression, molecular function and thereby influence VO2max training response [61, 66]. Whether genes or epigenetic modifiers play a larger percentage role in adaptive variability in a specific situation requires further exploration.
To address these limitations, larger-scale studies are required to ascertain if the 97 predictor genes identified from this review are similar in various cohorts (e.g. several ethnicities, ages, gender). The Athlome Project Consortium, which includes the Gene SMART study, is an example of a current larger-scale investigation examining ‘omic markers’ of training response, elite performance and injury rates/predisposition in variety of populations [67]. Ideally, future studies will complement and expand on this research, and consider alternative forms of exercise training intensity and volume, lifestyle factors, general health, diet, medications and health history when implementing interventions and analyzing data.
Furthermore, the role of the gut microbiome, and its influence on metabolism and physiology, needs to be explored. For example, gut microbiota (which has its own genome) can interact with the tissue cellular environment to regulate gene expression [61]. Poor diet, stress, illness, the use of antibiotics, environmental toxins and poor lifestyle choices can increase inflammation within the gut, causing dysbiosis; this appears to contribute to chronic diseases and other illnesses, irrespective of genotype, age and gender [68, 69]. Interestingly, VO2max was recently shown to be related to gut microbial diversity in a human cross-sectional study [70], suggesting a link between VO2max and gut microbes. Pre- and probiotics, resistant starch and a Mediterranean diet (dietary diversification) can alter the gut microbiome [68]. Investigating how the gut and human genome interact to positively influence VO2max is warranted.
With these points in mind, the analysis of stool samples, in addition to incorporating epigenetic, transcription and proteomic analysis, may help to identify the best aerobic training or lifestyle intervention to upregulate or downregulate certain genes, signaling pathways and molecular responses required for a greater VO2max training response. Implementing tightly-controlled studies examining various mediators (training intervention, diet, lifestyle) and molecular biomarkers across various populations will help to capture accurate information related to ideal traits for VO2max trainability.
Conclusion
In total, 97 genes that predicted VO2max trainability were identified. Phenotype is dependent on several of these genotypes/variants, which may contribute to approximately 50% of an individual’s VO2max trainability. Higher responders to exercise training have more positive response alleles (greater gene predictor score) than lower responders. Whilst these findings are exciting, further randomized-controlled research with larger and diverse cohorts are needed. Additional exploration is required to identify genetic variants and the mediators (training intensity and volume, diet, drugs, other lifestyle factors) that can potentially affect gene expression, molecular function and training response. Findings from this review and future research may assist clinicians to provide precision evidence-based medicine centered on phenotype, contributing to the fight against chronic disease.
Pubmed, embase, cinahl and cochrane search terms
Pubmed search
gene*[ti] OR allele [tiab] OR SNP [tiab] OR genetic profiling[tiab] OR genetic variant*[tiab] OR Genomic predictor*[tiab] OR polymorphism[tiab] OR heritability[tiab] AND (exercise training [tiab] OR VO2peak[tiab] OR ‘cardiorespiratory fitness’[tiab] OR ‘maximal/maximum VO2peak’[tiab] OR maximal/maximum VO2max’[tiab] OR maximal oxygen consumption’[tiab]OR peak oxygen uptake’[tiab] OR interval exercise’[tiab] OR ‘high/low intensity exercise’[tiab] OR peak fitness [tiab] OR endurance*[tiab] OR physical fitness[tiab] OR cardiorespiratory fitness[tiab] OR endurance training [tiab] OR cardiovascular fitness[tiab] OR VO2max[tiab] OR aerobic power[tiab] OR aerobic fitness[tiab] OR exercise capacity[tiab] OR exercise training response[tiab] OR response to exercise training[tiab]) NOT animal*.
Embase
gene:ab,ti OR allele:ab,ti OR snp:ab,ti OR ‘genetic profiling’:ab,ti OR ‘genetic variant’:ab,ti OR ‘genomic predictor’:ab,ti OR heritability:ab,ti AND (vo2peak:ab,ti OR vo2max:ab,ti OR ‘cardiovascular fitness’:ab,ti OR ‘cardiorespiratory fitness’:ab,ti OR ‘aerobic power’:ab,ti OR ‘aerobic fitness’:ab,ti OR ‘exercise training response’:ab,ti OR ‘physical fitness’:ab,ti).
Cinahl
(genes OR ‘genetic variant’ OR ‘Genomic predictor’ OR polymorphism OR ‘genetic profiling’ OR ‘single nucleotide polymorphisms’ OR ‘SNPs’ heritability) AND (‘trainability’ OR’ cardiovascular fitness’ OR ‘interval exercise’ OR ‘maximum O2’ OR maximal oxygen consumption’ OR ‘peak oxygen consumption’ OR maximal aerobic capacity’ OR ‘high/low intensity exercise’ OR ‘cardiorespiratory fitness’ OR ‘aerobic power’ OR ‘response to exercise training’ OR ‘exercise capacity’ OR ‘VO2max’ OR ‘VO2peak’ OR endurance).
Cochrane database for systematic reviews
(genes OR ‘genetic variant’ OR ‘Genomic predictor’ OR polymorphism OR ‘genetic profiling’ OR ‘single nucleotide polymorphisms’ OR ‘SNPs’ OR heritability) AND (‘trainability’ OR’ cardiovascular fitness’ OR ‘interval exercise’ OR ‘maximum O2’ OR maximal oxygen consumption’ OR ‘peak oxygen consumption’ OR maximal aerobic capacity’ OR ‘high/low intensity exercise’ OR ‘cardiorespiratory fitness’ OR ‘aerobic power’ OR ‘response to exercise training’ OR ‘exercise capacity’ OR ‘VO2max’ OR ‘VO2peak’ OR endurance).
Cochrane central register of controlled trial
(genes OR ‘genetic variant’ OR ‘Genomic predictor’ OR polymorphism OR ‘genetic profiling’ OR ‘single nucleotide polymorphisms’ OR ‘SNPs’ heritability) AND (‘trainability’ OR’ cardiovascular fitness’ OR ‘cardiorespiratory fitness’ OR ‘interval exercise’ OR ‘maximum O2’ OR maximal oxygen consumption’ OR ‘peak oxygen consumption’ OR maximal aerobic capacity’ OR ‘high/low intensity exercise’ OR ‘aerobic power’ OR ‘response to exercise training’ OR ‘exercise capacity’ OR ‘VO2max’ OR ‘VO2peak’ OR endurance).
Acknowledgments
Funding
Publication of this manuscript was supported by the Collaborative Research Network for Advancing Exercise and Sport Science (CRN-AESS).
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article.
About this supplement
This article has been published as part of BMC Genomics Volume 18 Supplement 8, 2017: Proceedings of the 34th FIMS World Sports Medicine Congress. The full contents of the supplement are available online at https://bmcgenomics.biomedcentral.com/articles/supplements/volume-18-supplement-8.
Authors’ contributions
CW was the primary author. MW checked the nomenclature of all variants and terminology used. JC, NE, UW, JL and KA provided expert advice and edits to the manuscript. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Ethics approval from Bellberry.
Consent for publication
Written informed consent was obtained from the individuals involved in this study.
Competing interests
The authors declare they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Camilla J. Williams, Email: camilla.williams@uqconnect.edu.au
Mark G. Williams, Email: mark.williams@mater.org.au
Nir Eynon, Phone: +61 399195615, Email: Nir.Eynon@vu.edu.au.
Kevin J. Ashton, Email: keashton@bond.edu.au
Jonathan P. Little, Email: jonathan.little@ubc.ca
Ulrik Wisloff, Email: ulrik.wisloff@ntnu.no.
Jeff S. Coombes, Email: jcoombes@uq.edu.au
References
- 1.World Health Organization. Chronic diseases and health promotion. 2016. http://www.who.int/chp/en/. Accessed 21 Oct 2016.
- 2.Bacon AP, Carter RE, Ogle EA, Joyner MJ. VO2max trainability and high intensity interval training in humans: a meta-analysis. PLoS One. 2013;8:e73182. doi: 10.1371/journal.pone.0073182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denham J, Marques F, O'Brien B, Charchar F. Exercise: putting action into our epigenome. Sports Med. 2014;44:189–209. doi: 10.1007/s40279-013-0114-1. [DOI] [PubMed] [Google Scholar]
- 4.Mann TN, Lamberts RP, Lambert MI. High responders and low responders: factors associated with individual variation in response to standardized training. Sports Med. 2014;44:1113–1124. doi: 10.1007/s40279-014-0197-3. [DOI] [PubMed] [Google Scholar]
- 5.Bouchard C, Antunes-Correa LM, Ashley EA, Franklin N, Hwang PM, Mattsson CM, et al. Personalized preventive medicine: genetics and the response to regular exercise in preventive interventions. Prog Cardiovasc Dis. 2015;57:337–346. doi: 10.1016/j.pcad.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DeFina LHW, Willis B, Barlow C, Finley C, Levine B, et al. Physical activity versus cardiorespiraotry fitness: two (partly) distinct components of cardiovascular health? Prog in Cardiovasc Dis. 2015;57:324–329. doi: 10.1016/j.pcad.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Wilson GM, Ellison G, Cable TN. Basic science behind the cardiovascular benefits of exericse. Br J Sports Med. 2016;50:93–99. doi: 10.1136/bjsports-2014-306596rep. [DOI] [PubMed] [Google Scholar]
- 8.Bouchard C. Health risk extimation, risk reduction and health promotion. Proceedings for the 18th annual meeting of the society of prospective medicine. Ottowa: Canadian Public Health Institution; 1983. Human adaptability may have a genetic basis; pp. 443–476. [Google Scholar]
- 9.Lortie G, Simoneau JA, Hamel P, Boulay MR, Landry F, Bouchard C. Response of maximal aerobic power and capacity to aerobic training. Int J Sports Med. 1984;5:232–236. doi: 10.1055/s-2008-1025911. [DOI] [PubMed] [Google Scholar]
- 10.Mori M, Higuchi K, Sakurai A, Tabara Y, Miki T, Nose H. Genetic basis of inter-individual variability in the effects of exercise on the alleviation of lifestyle-related diseases. J Physiol. 2009;587:5577–5584. doi: 10.1113/jphysiol.2009.179283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rankinen TBC. Genetic predictors of exercise training response. Curr Cardiovasc Risk Rep. 2011;5:368–372. doi: 10.1007/s12170-011-0179-z. [DOI] [Google Scholar]
- 12.Atkinson G, Batterham AM. True and false interindividual differences in the physiological response to an intervention. Exp Physiol. 2015;100:577–588. doi: 10.1113/EP085070. [DOI] [PubMed] [Google Scholar]
- 13.Yu B, Chen W, Wang R, Qi Q, Li K, Zhang W, et al. Association of apolipoprotein E polymorphism with maximal oxygen uptake after exercise training: a study of chinese young adult. Lipids Health Dis. 2014;13:40. doi: 10.1186/1476-511X-13-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klissouras V. Adaption to maximal effort: genetics and age. J Appl Physiol. 1973;35:288–293. doi: 10.1152/jappl.1973.35.2.288. [DOI] [PubMed] [Google Scholar]
- 15.Monotype HJGR. Familial relationships in maximal oxygen uptake. Hum Biol. 1978;50:241–249. [PubMed] [Google Scholar]
- 16.Hamel P, Simoneu J, Gilles L, Boulay M, Bouchard C. Heredity and muscle adaptiation to endurance training. Med Sci Sports Exerc. 1986;18:690–696. doi: 10.1249/00005768-198612000-00015. [DOI] [PubMed] [Google Scholar]
- 17.Prud'homme D, Bouchard C, Leblanc C, Landry F, Fontaine E. Sensitivity of maximal aerobic power to training is genotype-dependent. Med Sci Sports Exerc. 1984;16:489–493. doi: 10.1249/00005768-198410000-00012. [DOI] [PubMed] [Google Scholar]
- 18.Bouchard C, An P, Rice T, Skinner JS, Wilmore JH, Gagnon J, et al. Familial aggregation of VO(2max) response to exercise training: results from the HERITAGE family study. J Appl Physiol. 1999;87:1003–1008. doi: 10.1152/jappl.1999.87.3.1003. [DOI] [PubMed] [Google Scholar]
- 19.Webborn N, Williams A, McNamee M, Bouchard C, Pitsiladis Y, Ahmetov I, et al. Direct-to-consumer genetic testing for predicting sports performance and talent identification: consensus statement. Br J Sports Med. 2015;49:1486–1491. doi: 10.1136/bjsports-2015-095343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchard C, Sarzynski MA, Rice TK, Kraus WE, Church TS, Sung YJ, et al. Genomic predictors of the maximal O(2) uptake response to standardized exercise training programs. J Appl Physiol. 2011;110:1160–1170. doi: 10.1152/japplphysiol.00973.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh S, Vivar JC, Sarzynski MA, Sung YJ, Timmons JA, Bouchard C, et al. Integrative pathway analysis of a genome-wide association study of (V)O(2max) response to exercise training. J Appl Physiol. 2013;115:1343–1359. doi: 10.1152/japplphysiol.01487.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hautala ALA, Skinner JS, Rao DC, Bouchard C, Rankinen T. Peroxisome proliferator-activated receptor-beta polymorphims are associated with physical performance and plasma lipids: the HERITAGE family study. Am J Physiol Heart Circ Physiol. 2007; doi:10.1152/ajpheart.01092.2006. [DOI] [PubMed]
- 23.Leon AS, Togashi K, Rankinen T, Despres JP, Rao DC, Skinner JS, et al. Association of apolipoprotein E polymorphism with blood lipids and maximal oxygen uptake in the sedentary state and after exercise training in the HERITAGE family study. Metabolism. 2004;53:108–116. doi: 10.1016/j.metabol.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 24.Rankinen T, Perusse L, Borecki I, Chagnon YC, Gagnon J, Leon AS, et al. The NA(+)-K(+)-ATPase alpha2 gene and trainability of cardiorespiratory endurance: the HERITAGE family study. J Appl Physiol. 2000;88:346–351. doi: 10.1152/jappl.2000.88.1.346. [DOI] [PubMed] [Google Scholar]
- 25.Rankinen T, Perusse L, Gagnon J, Chagnon YC, Leon AS, Skinner JS, et al. Angiotensin-converting enzyme id polymorphism and fitness phenotype in the heritage family study. J Appl Physiol. 2000;88:1029–1035. doi: 10.1152/jappl.2000.88.3.1029. [DOI] [PubMed] [Google Scholar]
- 26.Rivera M, Dionne A, Fance T, Simoneau J, Perusse L, Chagnon M, et al. Muscle-specific creatine kinase gene polymorphism and VO2max in the HERITAGE family study. Med Sci Sports Exerc. 1997;29:1311–1317. doi: 10.1097/00005768-199710000-00006. [DOI] [PubMed] [Google Scholar]
- 27.Rivera MA, Perusse L, Simoneau J, Gagnon J, Dionne FT, Leon AS, et al. Linkage between a muscle-specific ck gene marker and VO2max in the HERITAGE family study. Med Sci Sports Exerc. 1999;31:698–701. doi: 10.1097/00005768-199905000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Timmons JA, Knudsen S, Rankinen T, Koch LG, Sarzynski M, Jensen T, et al. Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J Appl Physiol. 2010;108:1487–1496. doi: 10.1152/japplphysiol.01295.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Defoor J, Martens K, Matthijs G, Zielinska D, Schepers D, Philips T, et al. The CARAGENE study: muscle-specific creatine kinase gene and aerobic power in coronary artery disease. Eur J Cardiovasc Prev Rehabil. 2005;12:415–417. doi: 10.1097/01.hjr.0000170266.30562.59. [DOI] [PubMed] [Google Scholar]
- 30.Defoor J, Martens K, Zielinska D, Matthijs G, Van Nerum H, Schepers D, et al. The CARAGENE study: polymorphisms of the beta1-adrenoceptor gene and aerobic power in coronary artery disease. Eur Heart J. 2006;27:808–816. doi: 10.1093/eurheartj/ehi737. [DOI] [PubMed] [Google Scholar]
- 31.Defoor J, Vanhees L, Martens K, Matthijs G, Van Vlerken A, Zielinska D, et al. The CARAGENE study: ACE gene I/D polymorphism and effect of physical training on aerobic power in coronary artery disease. Heart. 2006;92:527–528. doi: 10.1136/hrt.2004.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onkelinx S, Cornelissen V, Defoor J, Matthijs G, Thomaes T, Coeckelberghs E, et al. The CARAGENE study: genetic variants of the endothelium and aerobic power in patients with coronary artery disease. Acta Cardiol. 2011;66:407–414. doi: 10.1080/AC.66.4.2126586. [DOI] [PubMed] [Google Scholar]
- 33.Thomaes T, Thomis M, Onkelinx S, Fagard R, Matthijs G, Buys R, et al. A genetic predisposition score for muscular endophenotypes predicts the increase in aerobic power after training: the CARAGENE study. BMC Genet. 2011;12:84. doi: 10.1186/1471-2156-12-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He Z, Hu Y, Feng L, Bao D, Wang L, Li Y, et al. Is there an association between PPARGC1a genotypes and endurance capacity in chinese men? Scand J Med Sci Sports. 2008;18:195–204. doi: 10.1111/j.1600-0838.2007.00648.x. [DOI] [PubMed] [Google Scholar]
- 35.He Z, Hu Y, Feng L, Bao D, Xi Y, Wen L, et al. Relationship between TFAM gene polymorphisms and endurance capacity in response to training. Int J Sports Med. 2007;28:1059–1064. doi: 10.1055/s-2007-965064. [DOI] [PubMed] [Google Scholar]
- 36.He Z, Hu Y, Feng L, Li Y, Liu G, Xi Y, et al. NRF-1 genotypes and endurance exercise capacity in young chinese men. Br J Sports Med. 2008;42:361–366. doi: 10.1136/bjsm.2007.042945. [DOI] [PubMed] [Google Scholar]
- 37.He Z, Hu Y, Feng L, Lu Y, Liu G, Xi Y, et al. NRF-2 genotype improves endurance capacity in response to training. Int J Sports Med. 2007;28:717–721. doi: 10.1055/s-2007-964913. [DOI] [PubMed] [Google Scholar]
- 38.He Z, Hu Y, Feng L, Lu Y, Liu G, Xi Y, et al. Polymorphisms in the HBB gene relate to individual cardiorespiratory adaptation in response to endurance training. Br J Sports Med. 2006;40:998–1002. doi: 10.1136/bjsm.2006.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sonna LA, Sharp MA, Knapik J, Cullivan M, Angel KC, Patton JF, Lilly CM. Angiotensin-converting enzyme genotype and physical performance during US army basic training. J Appl Physiol (1985) 2001;91:1355–1363. doi: 10.1152/jappl.2001.91.3.1355. [DOI] [PubMed] [Google Scholar]
- 40.Bouchard C, Chagnon M, Thibault M, Boulay M, Marcotte M, Cote C. Muscle genetic variants and relationship with performance and trainability. Med Sci Sports Exerc. 1989;21:71–77. doi: 10.1249/00005768-198902000-00013. [DOI] [PubMed] [Google Scholar]
- 41.Hagberg J, Katzel DR, Sorkin DR, Dengel JD, Golder AB, Apolipoprotein E. Genotype and exercise training-induced increases in plasma high-density lipoprotein (HDL) and HDL2 cholesterol levels in overweight men. Metabolism. 1999;48:943–945. doi: 10.1016/S0026-0495(99)90185-3. [DOI] [PubMed] [Google Scholar]
- 42.Thompson PD, Tsongalis GJ, Seip RL, Biblie C, Miles M, Zoeller R, et al. Apolipoprotein E genotype and changes in serum lipids and maximal oxygen uptake with exercise training. Metabolism. 2004;53:193–202. doi: 10.1016/j.metabol.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 43.Jenkins NT, McKenzie JA, Damcott CM, Witkowski S, Hagberg JM. Endurance exercise training effects on body fatness, VO2max hdl-c subfractions, and glucose tolerance are influenced by a PLIN haplotype in older caucasians. J Appl Physiol. 2010;108:498–506. doi: 10.1152/japplphysiol.01018.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McKenzie JA, Witkowski S, Ludlow AT, Roth SM, Hagberg JM. AKT1 G205T genotype influences obesity-related metabolic phenotypes and their responses to aerobic exercise training in older caucasians. Exp Physiol. 2011;96:338–347. doi: 10.1113/expphysiol.2010.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zarebska A, Jastrzebski Z, Kaczmarczyk M, Ficek K, Maciejewska-Karlowska A, Sawczuk M, et al. The GSTP1 c.313A>G polymorphism modulates the cardiorespiratory response to aerobic training. Biol Sport. 2014;31:261–266. doi: 10.5604/20831862.1120932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rico-Sanz J, Rankinen T, Joanisse DR, Leon AS, Skinner JS, Wilmore JH, et al. Associations between cardiorespiratory responses to exercise and the C34T AMPD1 gene polymorphism in the HERITAGE family study. Physiol Genomics. 2003;14:161–166. doi: 10.1152/physiolgenomics.00165.2002. [DOI] [PubMed] [Google Scholar]
- 47.Woods DR, World M, Rayson MP, Williams AG, Jubb M, Jamshidi Y, et al. Endurance enhancement related to the human angiotensin I-converting enzyme I-D polymorphism is not due to differences in the cardiorespiratory response to training. Eur J Appl Physiol. 2002;86:240–244. doi: 10.1007/s00421-001-0545-5. [DOI] [PubMed] [Google Scholar]
- 48.Alves BGOE, Alves RC, Rached HRS, Mota GFA, Pereira AC, et al. Influence of angiotensinogen and angiotensin-converting enzyme polymorphisms on cardiac hypertrophy and improvement on maximal aerobic capacity caused by exercise training. Eur J Cardiovasc Prev Rehabil. 2009;16:487–492. doi: 10.1097/HJR.0b013e32832c5a8a. [DOI] [PubMed] [Google Scholar]
- 49.Murakami H, Soma R, Hayashi JI, Katsuta S, Matsuda M, Ajisaka R, et al. Relationship between mitochondrial DNA polymorphism and the individual differences in aerobic performance. Jpn J Physiol. 2001;51:563–568. doi: 10.2170/jjphysiol.51.563. [DOI] [PubMed] [Google Scholar]
- 50.Dionne FT, Turcotte L, Thibault MC, Boulay MR, Skinner JS, Bouchard C. Mitochondrial DNA sequence polymorphism, VO(2max), and response to endurance training. Med Sci Sports Exerc. 1991;23:177–185. doi: 10.1249/00005768-199102000-00006. [DOI] [PubMed] [Google Scholar]
- 51.Silva BM, Neves FJ, Negrao MV, Alves CR, Dias RG, Alves GB, et al. Endothelial nitric oxide synthase polymorphisms and adaptation of parasympathetic modulation to exercise training. Med Sci Sports Exerc. 2011;43:1611–1618. doi: 10.1249/MSS.0b013e3182152197. [DOI] [PubMed] [Google Scholar]
- 52.Prior SJ, Hagberg JM, Phares DA, Brown MD, Fairfull L, Ferrell RE, et al. Sequence variation in hypoxia-inducible factor 1alpha (HIF1a): association with maximal oxygen consumption. Physiol Genomics. 2003;15:20–26. doi: 10.1152/physiolgenomics.00061.2003. [DOI] [PubMed] [Google Scholar]
- 53.Xu Y, Yang H, Ren Z, Yi L. Delta-aminolevulinate synthase 2 polymorphism is associated with maximal oxygen uptake after living-high exercise-high training-low in a male chinese population. Int J Clin Exp Med. 2015;8:21617–21622. [PMC free article] [PubMed] [Google Scholar]
- 54.Wang G, Padmanabhan S, Wolfarth B, Fuku N, Lucia A, Ahmetov II, et al. Genomics of elite sporting performance: what little we know and necessary advances. Adv Genet. 2013;84:123–149. doi: 10.1016/B978-0-12-407703-4.00004-9. [DOI] [PubMed] [Google Scholar]
- 55.Genetics Home Reference. 2016. https://ghr.nlm.nih.gov/gene/ACE. Accessed 1 Nov 2016.
- 56.Woods DR, Pollard AJ, Collier DJ, Jamshidi Y, Vassiliou V, Hawe E, et al. Insertion/deletion polymorphism of the angiotensin i-converting enzyme gene and arterial oxygen saturation at high altitude. Am J Respir Crit Care Med. 2002;166:362–366. doi: 10.1164/rccm.2103060. [DOI] [PubMed] [Google Scholar]
- 57.Papadimitriou ID, Lucia A, Pitsiladis YP, Pushkarev VP, Dyatlov DA, Orekhov EF, et al. ACTN3 r577x and ACE I/D gene variants influence performance in elite sprinters: a multi-cohort study. BMC Genomics. 2016;17:285. doi: 10.1186/s12864-016-2462-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Genetics Home Reference. 2016. https://ghr.nlm.nih.gov/gene/APOE. Accessed 31 Oct 2016.
- 59.Depypere H, Vierin A, Weyers S, Sieben A. Alzheimer's disease, apolipoprotein E and hormone replacement therapy. Maturitas. 2016;94:98–105. doi: 10.1016/j.maturitas.2016.09.009. [DOI] [PubMed] [Google Scholar]
- 60.Bishop DJ, Granata C, Eynon N. Can we optimise the exercise training prescription to maximise improvements in mitochondria function and content? Biochim Biophys Acta. 1840;2014:1266–1275. doi: 10.1016/j.bbagen.2013.10.012. [DOI] [PubMed] [Google Scholar]
- 61.Paul B, Denmark-Wahnefried W, Morrow C, Salvador C, Skibola C, Toolefsbol T. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin Epigenetics. 2015;7:1-11. doi:10.1186/s13148-015-0144-7. [DOI] [PMC free article] [PubMed]
- 62.Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol. 2008;586:113–121. doi: 10.1113/jphysiol.2007.141887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eynon N, Ruiz JR, Meckel Y, Moran M, Lucia A. Mitochondrial biogenesis related endurance genotype score and sports performance in athletes. Mitochondrion. 2011;11:64–69. doi: 10.1016/j.mito.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 64.Aktinson G, Batterham AM. True and false interindividual differences in the physiological resone to an internvention. Exp Physiol. 2015;100(6):577–88. doi: 10.1113/EP085070. [DOI] [PubMed] [Google Scholar]
- 65.Voisin S, Eynon N, Bishop DJ. Exercise training and DNA methylation in humans. Acta Physiol. 2014;213(1):39-59. https://doi.org/10.1111/alpha.12414. [DOI] [PubMed]
- 66.Barres R, Yan J, Egan B, Treebak JT, Rasmussen M, Fritz T, et al. Acute exercise remodels promoter methylation in human skeletal muscle. Cell Metab. 2012;15:405–411. doi: 10.1016/j.cmet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 67.Pitsiladis YP, Tanaka M, Eynon N, Bouchard C, North KN, Williams AG, et al. Athlome project consortium: a concerted effort to discover genomic and other “omic” markers of athletic performance. Physiol Genomics. 2016;48:183–190. doi: 10.1152/physiolgenomics.00105.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shanahan GD. Microbial contributions to chronic inflammation and metabolic disease. Curr Opin lin Nut Metab Care. 2016;19 [DOI] [PubMed]
- 69.Zhang Y, Gan R, Zhou T, Xu D, Li H. Impacts of gut bacteria on human health and diseases. Int J Mol Sci. 2015;16 [DOI] [PMC free article] [PubMed]
- 70.Estaki M, Pither J, Baumeister P, Little JP, Gill SK, Ghosh S, et al. Cardiorespiratory fitness as a predictor of intestinal microbial diversity and distinct metagenomic functions. Microbiome. 2016;4:42. doi: 10.1186/s40168-016-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article.


