Abstract
Tristetraprolin (TTP) is a nonclassical zinc finger (ZF) protein that plays a key role in regulating inflammatory response. TTP regulates cytokines at the mRNA level by binding to AU-rich sequences present at the 3′-untranslated region, forming a complex that is then degraded. TTP contains two conserved CCCH domains with the sequence Cys-X8CysX5CysX3His that are activated to bind RNA when zinc is coordinated. During inflammation, copper levels are elevated, which is associated with increased inflammatory response. A potential target for Cu(I) during inflammation is TTP. To determine whether Cu(I) binds to TTP and how Cu(I) can affect TTP/RNA binding, two TTP constructs were prepared. One construct contained just the first CCCH domain (TTP-1D) and serves as a peptide model for a CCCH domain; the second construct contains both CCCH domains (TTP-2D) and is functional (binds RNA) when Zn(II) is coordinated. Cu(I) binding to TTP-1D was assessed via electronic absorption spectroscopy titrations, and Cu(I) binding to TTP-2D was assessed via both absorption spectroscopy and a spin filter/inductively coupled plasma mass spectrometry (ICP-MS) assay. Cu(I) binds to TTP-1D with a 1:1 stoichiometry and to TTP-2D with a 3:1 stoichiometry. The CD spectrum of Cu(I)-TTP-2D did not exhibit any secondary structure, matching that of apo-TTP-2D, while Zn(II)-TTP-2D exhibited a secondary structure. Measurement of RNA binding via fluorescence anisotropy revealed that Cu(I)-TTP-2D does not bind to the TTP-2D RNA target sequence UUUAUUUAUUU with any measurable affinity, while Zn(II)-TTP-2D binds to this site with nanomolar affinity. Similarly, addition of Cu(I) to the Zn(II)-TTP-2D/RNA complex resulted in inhibition of RNA binding. Together, these data indicate that, while Cu(I) binds to TTP-2D, it does not result in a folded or functional protein and that Cu(I) inhibits Zn(II)-TTP-2D/RNA binding.
Graphical Abstract
INTRODUCTION
Zinc finger (ZF) proteins are eukaryotic proteins that use zinc as a structural cofactor to fold and function.1,2 The common feature of all ZF proteins is that they contain modular domains with conserved cysteine and/or histidine residues, which serve as zinc coordinating ligands.1–4 Upon zinc coordination, ZFs adopt a secondary structure that allows for function. ZFs are involved in a range of biological processes including neuronal development, inflammatory response, viral development, and oncogenesis.1,2,5–13 Biochemical roles for ZFs include serving as transcription factors via DNA binding, regulating translation via RNA binding, and regulating signaling upon binding to other proteins.1,2,14–16
The best-studied ZF is the “classical” ZF, which was the first ZF to be identified.17 Classical ZFs contain Cys2His2 sequence repeats, adopt an β β α fold upon zinc coordination, and function primarily in transcriptional regulation by binding to specific DNA sequences.1,2 In addition to the classical ZFs, at least 14 other classes of ZFs are known.1,2,4,14,18 Collectively these are called nonclassical ZFs, and they are grouped on the basis of the number of cysteine and histidine residues, the spacing between the residues, and the structure that is formed upon zinc coordination (if known).1–3,14,15,18 For many nonclassical ZFs, experimental evidence for Zn coordination has not yet been obtained, and they have been annotated as ZFs on the basis of their amino acid sequence.19
Although zinc is the presumed metal cofactor for ZF proteins, the presence of mixed sulfur and nitrogen donor ligands within ZF motifs allows for ZFs to coordinate other metal ions. There are several reports of other divalent, monovalent, and trivalent metal ions binding to ZF sites. These include Sb(III), Au(III), As(III), Fe(II), Cd(II), Pb(II), Cu(II), Ni(II), Au(I), and Cu(I).20–53 In some instances, the alternative metal ions are toxic (e.g., Pb) and their coordination to ZFs has been proposed to be part of the mechanism of toxicity.23–26,42,54 In other cases, alternate metal ions have been proposed to substitute for zinc under conditions of zinc depletion (e.g., Fe(II)).28,31,32,44
One metal of particular interest is copper. Copper is the third most abundant transition metal ion present in eukaryotic cells. Copper serves as a cofactor for several key enzymes, including superoxide dismutase and cytochrome c oxidase.55–57 In the reducing environment of the cell, copper is present in the cuprous state Cu(I). Free Cu(I) can be toxic, because it can undergo Fenton chemistry with reactive oxygen species (ROS) leading to cellular damage, including targeting metalloprotein sites and disrupting their function.58–61 As a result, cellular Cu levels must be tightly controlled and there is limited labile or “free” Cu present in cells.55,62–64 There is some interesting recent evidence that pools of exchangeable copper can be present under certain conditions, suggesting that Cu may be more bioavailable than initially described.65,66
Cu(I) is thiophilic, and thiol-rich proteins such as ZFs are potential cellular targets for Cu(I) when they are available (e.g., from the exchangeable pool or under conditions of over-load). 37,65,67 Recent work by one of our laboratories examined Cu(I) binding to several ZF peptides.37 These previous studies involved measuring Cu(I) coordination to the ZF consensus peptides CP-CCHH, CP-CCHC, and CP-CCCC that all form the classical ββα fold upon Zn(II) coordination and differ only in the metal binding residues, and the C-terminal domain of the HIV nucleocapsid protein (NCp7), a nonclassical ZF with a Cys2HisCys (or CCHC) ligand set, often referred to as a zinc knuckle peptide.37 Cu(I) bound to all four peptides studied and was thermodynamically favored over both Co(II) and Zn(II). However, unlike the Zn(II)-bound species, addition of Cu(I) to selected peptides did not induce the formation of any secondary structure observable via circular dichroism spectroscopy. The lack of folding for these Cu(I)-ZF single peptides suggests that, when Cu(I) binds to a functional ZF (i.e., with two or more domains, single ZF peptides are not functional), DNA or RNA binding will be significantly compromised for ZF proteins based upon these two specific folds.
The functional effects of Cu(I) coordination to any ZF have, to our knowledge, not been determined. Here, we sought to investigate how Cu(I) interacts with a functional ZF construct to determine how Cu(I) coordination to a ZF affects function. The protein chosen for these studies was TTP, which is a Cys3His (CCCH) type ZF (Figure 1).1 TTP plays a key role in regulating inflammation by controlling levels of cytokines (inflammatory proteins), at the mRNA level.68,69 Specifically, the two CCCH domains of TTP bind to a specific AU-rich sequence found in cytokine mRNA, forming a complex that is then degraded.68,70–72 The two CCCH domains must be bound to zinc (or iron) and folded in order for the protein to bind to mRNA.1,44 There is evidence that Cu(I) levels are elevated during inflammation,73,74 suggesting that Cu(I) may target TTP and affect its function, making it a particularly relevant ZF to utilize for studies aimed at understanding how Cu(I) can target ZF sites. We prepared a single- and double-domain construct of TTP, called TTP-1D and TTP-2D, and measured the structural and, for TTP-2D, the functional affects of Cu(I) binding. We report that Cu(I) binds to the CCCH domains of TTP but does not induce any secondary structure. We developed a novel spin-filter/inductively coupled plasma mass spectrometry assay to measure metal stoichiometry for TTP-2D and determined that Cu(I) binds in a 3:1 ratio while Zn(II) binds in a 2:1 ratio. Cu(I)-TTP-2D does not recognize the physiologically relevant RNA sequence UUUAUUUAUUU, while Zn(II)-TTP-2D does. When Cu(I) is titrated with Zn(II)-TTP-2D/RNA, RNA binding is disrupted. These results are discussed in the context of a functional role for Cu and TTP in modulating the inflammatory response.
Figure 1.
Sequence of TTP-2D with the CCCH domain in purple and the additional histidine residues in yellow.
METHODS AND MATERIALS
Co(II), Zn(II), and Cu(I) Binding Titrations
CuCl (Sigma-Aldrich) stock solutions were prepared in a Coy anaerobic chamber by dissolving CuCl in 0.01 M HCl/1.0 M NaCl. The concentrations of the resulting solutions were determined upon the addition of small aliquots of the Cu(I) stock to 1 mM bicinchoninate (BCA) in 200 mM HEPES buffer at pH 7.5 to form Cu(BCA)2. The [Cu(I)] in the resulting solution was quantitated via published data for the Cu(BCA)2 complex (A562; ε = 7900 M−1 cm−1 for Cu(BCA)2).75 The addition of Cu(II) to excess BCA will result in the reduction of Cu(II) to Cu(I) over time and a concomitant increase in absorbance at 562 nm. Therefore, we monitored the absorbance of Cu(BCA)2 over time during our concentration determinations to ensure that our Cu(I) stock solutions remained fully reduced. During this process, no increase in absorbance was observed over time, nor did the addition of ascorbate as a reducing agent produce additional absorbance increases at 562 nm, indicating that the copper stock solution is stabilized as Cu(I). ZnCl2 (Sigma-Aldrich) and CoCl2 (EM Science) stock solutions were also prepared in a Coy anaerobic chamber by dissolving in degassed Milli-Q water. Stock solutions used for Co(II), Zn(II), and Cu(I) titrations for TTP-1D and TTP-2D were buffered in 200 mM HEPES, 100 mM NaCl at pH 7.5. Titrations of TTP-1D and TTP-2D with Co(II), Zn(II), and Cu(I) were performed in screw-capped quartz cuvettes (Starna Cells) maintained in a Coy anaerobic chamber (97% nitrogen/3% hydrogen atmosphere), and spectra were measured on a PerkinElmer Lambda 25 spectrometer. The buffer used for all titrations was 200 mM HEPES, 100 mM NaCl at pH 7.5. The buffers were prepared using metal-free reagents and water that had been purified via a MillQ purification system and Sigma Chelex resin, to prevent adventitious metal binding. All experiments were performed in triplicate.
TTP-1D
A peptide corresponding to the first ZF domain of TTP (called TTP-1D) was purchased from Biosynthesis (Lewisville, TX) at >75% purity. TTP-1D has the sequence TSSRYKTELCRTYSESGRCRYGAKCQFAHGLGELRQ, (cysteine and histidine ligands are underlined). To ensure reduction of cysteine thiols, 10 equiv of dithiothreitol was added to TTP-1D and the peptide was heated at 55 °C for 2 h. Apo-TTP-1D was then purified using a Symmetry-C18 reversed phase HPLC column on a metal-free, nonmetallic HPLC (Waters 626 LC). An H2O/CH3CN/TFA solvent gradient was utilized for the separation, and the peptide eluted as a single peak at 65% H2O/35% CH3CN/0.1% TFA. The TTP-1D peptide was then transferred to a Coy anaerobic chamber (97% nitrogen/3% hydrogen atmosphere), where it was lyophilized in a Savant SpeedVac concentrator. All further manipulations were performed anaerobically.
TTP-2D Overexpression and Purification
A construct of TTP, called TTP-2D, which encodes for the two ZF domains of TTP with the amino acid sequence of MSRYKTELCRTYSESGRCRYGAKCQFAHGLGELRQANRHPKYKTELCHKFYLQGRCPYGSRCHFIHNPTEDLAL was overexpressed and purified. The expression vector, previously reported by our laboratory, was based on the pET-15b vector to which the gene that encoded for the two-domain construct of TTP-2D was ligated.44 The expression vector was transformed into BL21 (DE3)-competent cells (Novagen), and the cells were grown in Luria–Bertani (LB) medium containing 100 μg/ mL ampicillin and 100 mM ZnCl2 at 37 °C until mid log phase (~OD600 of 0.6–0.8). At this point, protein expression was induced via addition of 1 mM IPTG (isopropyl β-D-1-thiogalactopyranoside). At 4 h post induction, the cells were harvested by centrifugation at 7800g for 15 min at 4 °C. A solution of 8 M urea, 10 mM MES buffer at pH 6 and EDTA-free protease inhibitor mini-tablet (Roche) was then utilized to resuspend the cells. The cells were then lysed by sonication (Fisher Scientific Sonic Dismembrator Model 100) on ice and and centrifuged at 12100 rpm for 15 min at 4 °C to remove cellular debris. The supernatant was then applied to an SP-Sepharose column at room temperature and equilibrated via rocking for 60 min. Separation was accomplished by performing a step gradient from 0 to 2 M NaCl in 4 M urea, 10 mM MES pH 6; TTP-2D eluted at 600 mM NaCl. To ensure that the cysteine thiols of the isolated TTP-2D were fully reduced, 25 mM DTT was added to the peptide and the peptide was heated at 56 °C for 2 h. A second purification step was then performed. The peptide was applied to a C18-reverse phase HPLC column on a Waters 626 bioinert LC and a H2O/CH3CN/TFA gradient was applied. TTP-2D eluted at 32% CH3CN. Purified TTP-2D was then transferred to a Coy anaerobic chamber (97% nitrogen/ 3% hydrogen atmosphere), where it was lyophilized to dryness (Savant SpeedVac concentrator). The purity of the peptide was verified by SDS-PAGE and MALDI-MS (calculated 8581.8 Da; observed 8581.7 Da). All subsequent handling of TTP-2D was performed anaerobically, to prevent cysteine oxidation.
Co(II) and Zn(II) Binding
To verify that the isolated apo-TTP-1D and apo-TTP-2D bound Zn(II), as expected, Co(II) was used as a spectroscopic probe following the protocol we previously reported.44 In a typical experiment, either apo-TTP-1D or apo-TTP-2D (40–50 μM) was titrated with CoCl2 past saturation and the data were fit to a 1:1 binding equilibrium, by plotting data in the d–d regions (e.g., at 655 nm). A cotitration with ZnCl2 was then performed, and the relative affinities of Zn for TTP-1D and TTP-2D were determined.
Direct Titrations of TTP-1D and TTP-2D with Cu(I)
CuCl was added to apo-TTP-1D or apo-TTP-2D in a stepwise fashion. For apo-TTP-1D, the following additions were made: 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0, 1.25, 1.5, 1.75, 2.0, 2.5, 3.0 equiv of Cu(I). After the addition of 3 equiv, the baseline drifted upward, suggesting precipitation or aggregation. For apo-TTP-2D the following additions were made: 0.25, 0.5, 0.75, 1.0, 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 5.0, 6.0 equiv of Cu(I). After the addition of 6 equiv, the baseline drifted upward, suggesting precipitation or aggregation.
Job Plot
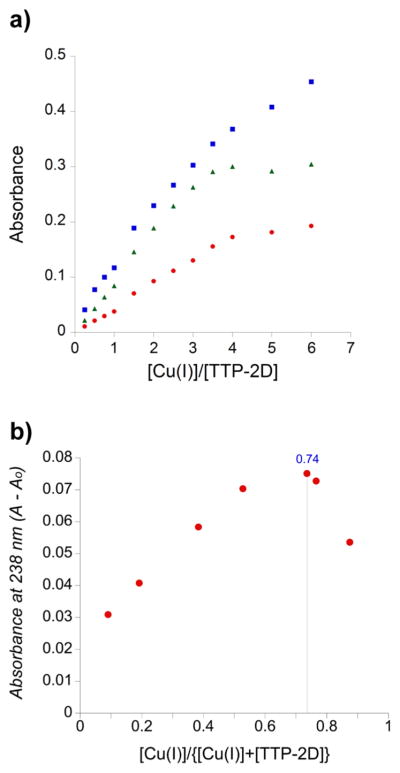
Solutions of apo-TTP-2D and CuCl at a constant concentration of 10 μM were prepared in screw-capped cuvettes maintained in a Coy anaerobic box. The mole fraction of Cu(I) in each sample was varied as 0.09, 0.19, 0.38, 0.53, 0.7, 0.77, and 0.87. A Job plot was generated from the data by plotting the corrected absorbance (A – A0) at 238 nm versus mole fraction [Cu(I)]/{[Cu(I)] + [apo-TTP-2D]}. The maximum absorbance in the Job plot of [Cu(I)]/ {[Cu(I)] + [apo-TTP-2D]} was reached at 0.74, indicating a 2.8:1 binding stoichiometry.
Cu(I)TTP-2D Affinity and Stoichiometry Measured via Cu(BCA)2 Competition Assays
Apo-TTP-2D was added stepwise into a solution of Cu(BCA)2 prepared at a defined molecular ratio (either 9.5 μM Cu(I)/0.5 mM BCA or 9 μM Cu(I)/5 mM BCA)S. The loss of the Cu(BCA)2 signal at 562 nm was monitored. The data were analyzed according to methods described by Xiao et al., resulting in an approximate KD value for the Cu(I)-TTP-2D interaction.76
Indirect Titrations of Co(II)-TTP-2D with Cu(I) or Zn(II)
Co(II)-TTP-2D was prepared by addition of 2.4 equiv of CoCl2 to apo-TTP-2D, followed by addition of CuCl or ZnCl2. CuCl and ZnCl2 in 0.6, 1.2, 1.7, 2.3, 2.8, and 3.8 equiv portions were added, resulting in a diminution of the d–d bands. A 2.8 equiv amount of Cu and 2.3 equiv of Zn were required for complete loss of the d–d bands, which was accompanied by some baseline drift indicative of precipitation or aggregation.
Preparation of ICP-MS Samples
Cu(I)-TTP-2D and Zn(II)-TTP-2D samples for ICP-MS analysis were prepared by addition of 5.0 equiv of CuCl or ZnCl2 to 40 μM of apo-TTP-2D in 200 mM HEPES, 100 mM NaCl at pH 7.5. Control samples were 200 μM CuCl, 200 μM ZnCl2, and 40 μM apo-TTP-2D. All samples were incubated via shaking at 300 rpm for 30 min at room temperature. After incubation, the samples were applied to 3KDa MWCO centrifugal filters (Amicon Ultra −0.5 mL) and spun for 14000g for 30 min on a 40° angle fixed table-top centrifuge (Denville 260D). The flow-through was then diluted in 200 mM HEPES, 100 mM NaCl at pH 7.5 to 500 μL, and the spin filter step was repeated (3× total). The samples were split in half. One half was used to measure metal content: 6% nitric acid was added, and ICP-MS was performed. The other half of the sample was utilized to measure peptide concentration. The peptide was unfolded via addition of HCl and the peptide concentration measured via absorbance spectroscopy (A276; ε = 8520 M−1 cm−1). All samples were prepared under anaerobic conditions (3% H2, 97% N2, anaerobic chamber, Coy Laboratories). Measurements were performed in triplicate.
Copper and Zinc Detection by ICP-MS
The concentrations of copper and zinc in protein samples were determined by injecting samples into an Agilent 7700x ICP-MS instrument (Agilent Technologies, Santa Clara, CA, USA). Metal levels were detected using an Octopole Reaction System cell (ORS) in He mode. The ICP-MS parameters used for the analysis were an RF power of 1550 W, an argon carrier gas flow of 0.99 L/min, a helium gas flow of 4.3 mL/min, an octopole RF of 190 V, and an OctP bias of −18 V. Samples were directly infused using the 7700x peristaltic pump with a speed of 0.1 rps and a micromist nebulizer. Copper and zinc concentrations in the samples were derived from a calibration curve generated by a series of dilutions of atomic absorption standard (Fluka Analytical) prepared in the same matrix as the samples. Data analysis was performed using Agilent’s Mass Hunter software.
Circular Dichroism (CD) Studies
To investigate changes in secondary structure due to Zn(II) and Cu(I) coordination to TTP-2D, the far-UV circular dichroism (CD) spectra were obtained using a JASCO-810 spectropolarimeter. A 50 μM solution of apo-TTP-2D was prepared in 300 μL of 10 mM sodium phosphate, pH 7.5 (prepared using Chelex-treated water and degassed). Either 2 mol equiv of ZnCl2 or 3 mol equiv of CuCl was added to the cuvette, and scans of either Zn(II)-TTP-2D or Cu(I)-TTP-2D were performed. CD data were collected over the wavelength range of 180–280 nm, with a scan rate of 100 nm/min, at 25 °C in a 1 mm path length quartz rectangular cell (Starna Cells). A total of five scans were obtained for each point, and the average was determined. Experiments were performed in triplicate.
RNA Binding Studies
Fluorescence anisotropy (FA) was performed to determine how Cu(I) coordination to TTP-2D affects RNA recognition and binding. The 3′-fluorescein (F)-labeled RNA oligonucleotide with the sequence UUUAUUUAUUU-F (Dharmacon Research Inc., PAGE-purified, deprotected, and desalted and dissolved in DEPC treated water) was utilized for these studies. An ISS PC-1 spectrofluorometer, configured in the L format, was used for all FA measures. FA experiments were performed with the excitation wavelength/band pass at 495 nm/2 nm and the emission wavelength/ band pass at 517 nm/2 nm, identified from a full excitation/ emission scan of the RNA probe. The buffer system for the experiments was 200 mM HEPES, 100 mM NaCl, 0.05 mg/mL bovine serum albumin (to prevent protein adherence to the cuvette) at pH 7.5, and the cuvettes utilized were Spectrosil far-UV quartz window fluorescence cuvettes (Starna Cells). Three experiments were performed: (1) 10 nM of UUUAUUUAUUU-F was titrated with the Zn(II)-TTP-2D to verify RNA binding, as previously reported by one of our laboratories.44 10 nM of UUUAUUUAUUU-F was titrated with Cu(I)-TTP-2D to determine if Cu(I)-TTP-2D binds RNA, and (3) 10 nM of UUUAUUUAUUU-F was titrated with Zn(II)-TTP-2D to saturation followed by addition of CuICl. In all experiments, the anisotropy (r) was monitored and the data were analyzed. Each data point is the average of 31 readings obtained over 100 s. All titrations were carried out in triplicate.
RESULTS AND DISCUSSION
Cu(I) Binding to Single- and Double-Domain Constructs of TTP
To determine whether Cu(I) bound to each construct, direct titrations with CuCl under anaerobic conditions were performed.37 For the titration of the single ZF domain with Cu(I), TTP-1D, peaks at 238 and 262 nm with a shoulder at 300 nm grew in as Cu(I) was added (Figure 2). The peaks are proposed to be Cu–S charge transfer bands, as such bands have been observed for metallothionein and Cu(I)-ZF-peptides.37 A plot of absorbance versus concentration of Cu(I) (Figure 3) revealed a clear break point upon the addition of 1 equiv of Cu(I), indicating that Cu(I) binding initially yields a 1:1 stoichiometry. We note that the absorption at 238 and 262 nm does not perfectly saturate. This may be due to Cu(I)–buffer (Hepes) interactions, as previously reported by Splan et al., or to either nonspecific binding to the peptide or aggregation, as has been reported previously by LeBrun, Giedroc, and Merchant.37,40,77
Figure 2.
Plot of the change in the absorption spectrum, between 230 and 500 nm, as CuCl is added to apo-TTP-1D. The apo-TTP-1D spectrum has been subtracted, and the titration was performed in 200 mM HEPES, 100 mM NaCl, at pH 7.5.
Figure 3.
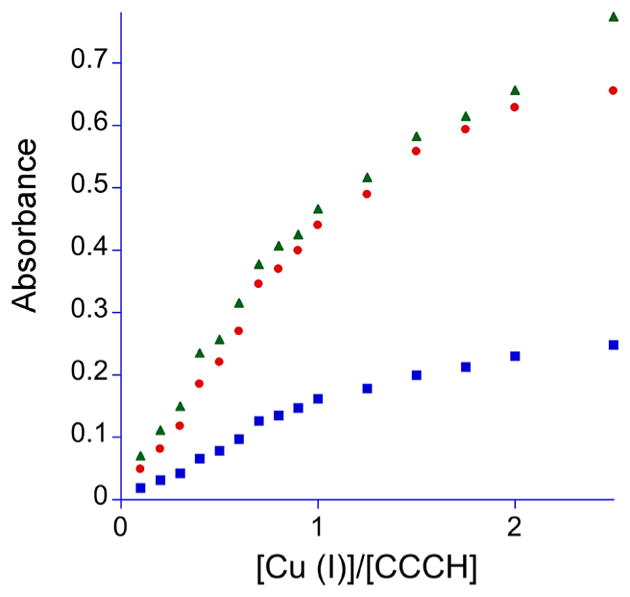
Plot of the increase in absorbance at 238 nm (green triangles), 262 nm (red circles), and 300 nm (blue squares), as CuCl is added to apo-TTP-1D.
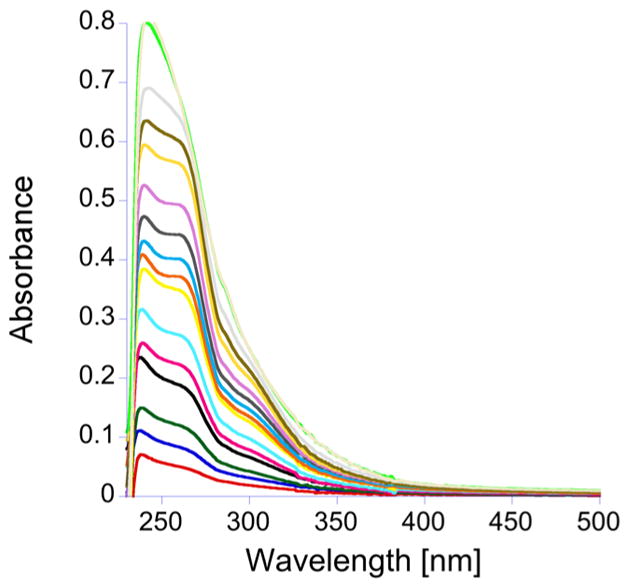
For the titration of the double-ZF domain, TTP-2D, with Cu(I) absorbance peaks at 238, 262, and 300 nm, again appeared (Figure 4). A plot of Cu(I) concentration versus absorbance showed a break point at 3 equiv of Cu(I)/TTP-2D (3:1) (Figure 5a). These data also show absorption at 238 and 262 nm that do not saturate, like the data observed for TTP-1D. In addition, a slight shift is observed in the data after the addition of 1 equiv of Cu(I), which suggests sequential metal binding wherein the second metal exhibits a slightly different spectral signature.78 To further investigate the Cu:TTP-2D stoichiometry, we used Job’s method.79,80 As shown in Figure 5b, the Job plot indicates a 2.8:1 binding stoichiometry. These data are consistent with the direct Cu binding titration data.
Figure 4.
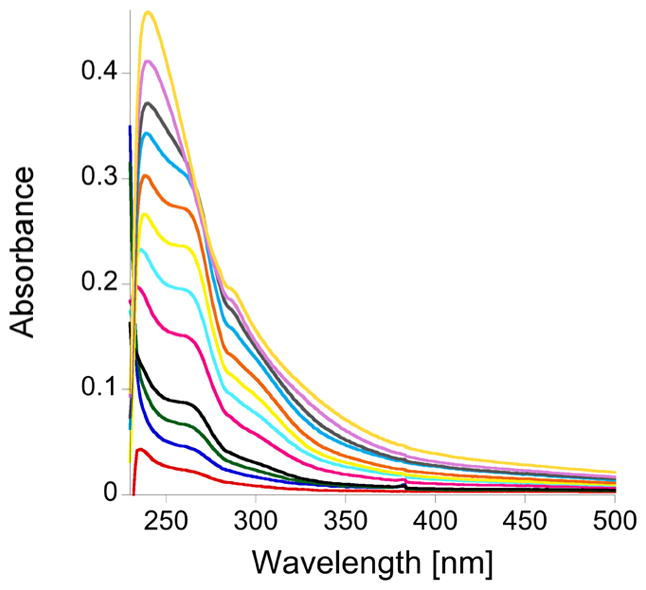
Plot of the change in the absorption spectrum, between 230 and 500 nm, as CuCl is is added to apo-TTP-2D. The apo-TTP-2D spectrum was subtracted, and the titration was performed in 200 mM HEPES, 100 mM NaCl, at pH 7.5.
Figure 5.
(a) Plot of the increase in absorbance at 238 nm (blue squares), 262 nm (green triangles), and 300 nm (red circles), as CuCl was added to apo-TTP-2D. (b) Job plot (X axis, mole fraction [Cu(I)]/{[Cu(I)] + [apo-TTP-2D]}; Y axis, A – A0 at 238 nm).
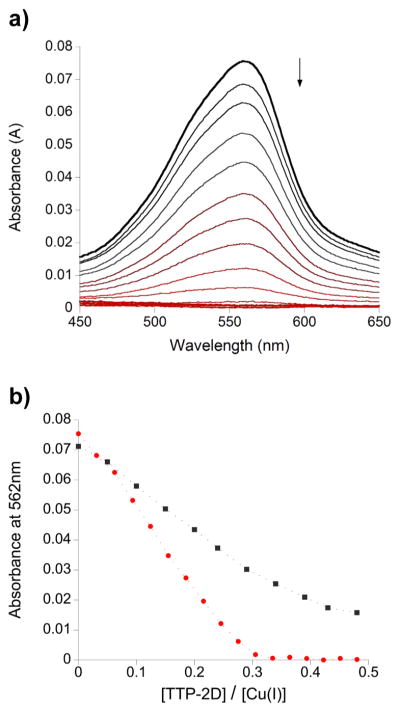
To examine binding under copper limiting conditions wherein weak and/or nonspecific binding is prohibited, further experiments were conducted in the presence of the copper-specific chelator BCA. Addition of Cu(I) to excess BCA results in a highly stable, colored 1:2 complex ((A562; ε = 7900 M−1 cm−1 for Cu(BCA)2; log β2 = 17.2)75 As shown in Figure 6a, addition of increasing amounts of TTP-2D resulted in the loss of absorbance signal owing to Cu(BCA)2, indicating that TTP-2D effectively competes with BCA for the available copper. Analysis of the decrease in absorbance as a function of protein added revealed that, even in the presence of 500 μM BCA for which stoichiometric titration data are observed, TTP-2D binds Cu(I) with a stoichiometry of ~3:1, consistent with our data presented above (Figure 6b). While the direct titration data presented in Figures 2–5 do not fully saturate and leave the question of stoichiometry uncertain, this observed 3:1 stoichiometry under limited copper availability that more closely approximates intracellular conditions strongly supports the conclusion that TTP-2D binds three Cu(I) ions. When the amount of BCA present in the experiment was increased to 5 mM, equilibrium binding data were observed and were used to estimate an average dissociation constant (KD) for the binding of Cu(I) to TTP-2D per the method described by Xiao et al.75,76 Therein, the average KD (per copper ion) for a protein or peptide that binds multiple Cu(I) ions can be calculated from eq 1:
| (1) |
where n equals the number of Cu(I) ions bound to the protein. The fractional occupancy and [Cu]f were calculated from the total concentrations of Cu(I), TTP-2D, and BCA. Assuming n = 3, a dissociation constant KD ≈ 10−18 M for the Cu(I)-TTP-2D interaction was estimated, which is within the range of several Cu(I)–protein interactions previously reported.75 The upper limit KD for Zn(II) binding to TTP-2D has previously been reported.44 This affinity is 6.2 × 10−11 M, which is weaker than the affinity of Cu(I) for TTP-2D.
Figure 6.
(a) Plot of the change in the absorption spectrum between 450 and 650 nm as apo-TTP-2D was added to Cu(BCA)2 (9.5 μM Cu/0.5 mM BCA). (b) Plot of the absorption spectrum at 592 nm as apo-TTP-2D was added to Cu(BCA)2 (9.5 μM Cu/0.5 mM BCA (red) and 9 μM Cu/5 mM BCA (black)). Experiments were performed in 200 mM HEPES, 100 mM NaCl, at pH 7.5.
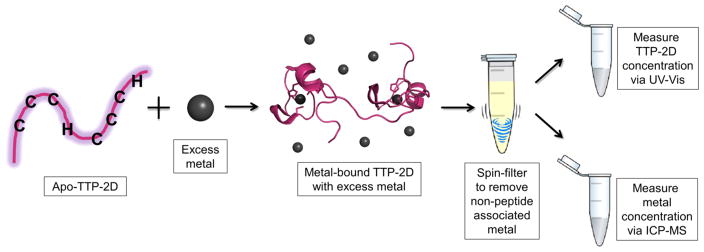
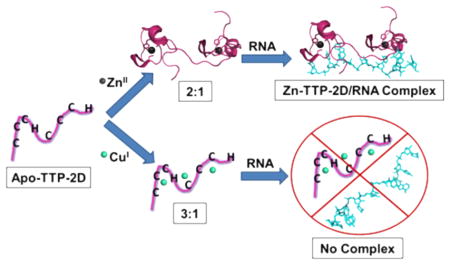
TTP-2D contains two CCCH domains; therefore, a 2:1, not a 3:1, stoichiometry is expected if each Cu(I) binds to a single CCCH site. Therefore, an assay based upon inductively coupled plasma mass spectrometry (ICP-MS) was developed. In this assay, outlined in Scheme 1, M-TTP-2D complexes (M = Cu(I), Zn(II)) were prepared by addition of 5 equiv (excess) of the metal ion to apo-TTP-2D followed by incubation to form the M-TTP-2D complex. The complex was then applied to a 3 kDa molecular weight cutoff spin filter and washed three times to remove any adventitiously bound metal ions. This approach should isolate the fully metal bound TTP-2D species. The resultant M:TTP-2D or metal:TTP-2D stoichiometry was determined by measuring the total metal content via inductively coupled plasma mass spectrometry (ICP-MS). The total protein content was determined by denaturing the protein and using the determined extinction coefficient of ε = 8520 M−1 cm−1 at 276 nm.29 The metal:TTP-2D stoichiometry was then determined from this ratio. As shown in Table 1, the Cu:TTP-2D stoichiometry was 3.1:1, matching the UV–visible data. The Zn-TTP-2D stoichiometry was 2.3:1, which was expected on the basis of previously reported NMR data.34 The consistency seen in the Zn and Cu data, between ICP-MS and other measures of stoichiometry, provides strong support for the viability of this spin filtration/ICP-MS approach.
Scheme 1.
Table 1.
Measurement of Metal/TTP-2D Stoichiometry Using ICP-MS
| Cu stoichiometry
|
Zn stoichiometry
|
||
|---|---|---|---|
| Cu-TTP-2D | 3.1 (±0.27a, ± 0.13b) | Zn-TTP-2D | 2.3 (±0.41,a 0.10b) |
| apo-TTP-2D | 0.034 (±0.013,a ±0.004b) | apo-TTP-2D | 0.004 (±0.005,a 0.001b) |
| bufferc | 0.057 (±0.097,a ±0.028b) | bufferc | 0.027 (±0.010,a 0.002b) |
SD, standard deviation.
SEM, estimated standard error of the mean (n = 12).
Conditions: buffer 200 mM HEPES, 100 mM NaCl, pH 7.5 with 5 equiv of Cu or Zn.
TTP-2D has two CCCH domains. When zinc is bound, each CCCH domain provides four ligands—all three cysteines and the histidine—resulting in four-coordinate, tetrahedral complexes. The 3:1 Cu:TTP-2D binding stoichiometry observed here suggests that the coordination number and geometry differ when Cu(I) is bound instead of Zn(II). Cu(I) typically binds with a coordination number of 2–3 and is often planar.81 Therefore, the three Cu(I) ions may bind to the two CCCH domains by utilizing 2–3 ligands per Cu(I). We also note that, within the amino acid sequence of TTP-2D, there are three additional histidine residues that are not part of the CCCH domains. These may also serve as ligands for Cu(I).
Competitive Metal Titrations: Displacement of Co(II) by Cu(I) and Zn(II)
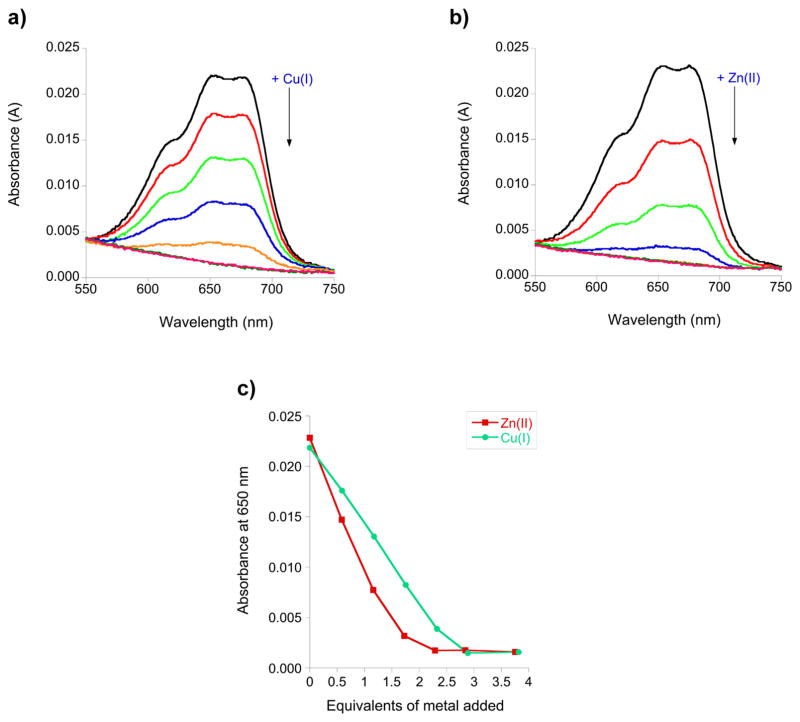
To determine if Cu(I) binds to the CCCH sites, like Zn(II), competition titrations were performed. In these experiments, the TTP-2D peptide was loaded with a slight excess of Co(II) (2.4 equiv), and then aliquots of either Zn(II) or Cu(I) were added (Figure 7a, b). Co(II) is a surrogate for Zn(II)—it binds to the CCCH domains in a tetrahedral geometry and exhibits distinct d–d bands between 550 and 750 nm indicative of this coordination; however, it can be displaced by Zn(II), which binds more tightly to tetrahedral sites in comparison to Co(II), due to ligand field stabilization energy preferences.1,2,44 Additions of 2.3 equiv of Zn and 2.8 equiv of Cu were sufficient to fully replace the Co, as evidenced by the loss of the Co d–d bands (Figure 7c). The finding that slightly more Cu is required to replace Co in comparison to the amount for Zn is consistent with our observation of a 3:1 Cu:TTP-2D stoichiometry from both the direct titrations of apo-TTP-2D with Cu(I) and ICP-MS analysis. Cu(I) typically binds with a lower coordination number than Zn(II) or Co(II), and we propose that the third Cu(I) binds to some of the ligands within the CCCH domains and possibly to other ligands within the TTP-2D sequence. The property of metal ions displacing Co and binding to the CCCH domains of TTP appears to be generalizable for TTP: we have previously shown that Fe(II), Fe(III), and Cd(II) all displace Co from Co-TTP and that Cu(I) will displace Co(II) from the ZF consensus peptides and zinc knuckle peptides.34,37,44
Figure 7.
(a) Plot of the change in the absorption spectrum, between 550 and 750 nm, as 0.6, 1.2, 1.7, 2.3, 2.8, and 3.8 equiv of CuCl was added to Co(II)-TTP-2D. (b) Plot of the change in the absorption spectrum, between 550 and 750 nm, as 0.6, 1.2, 1.7, 2.3, 2.8, and 3.8 equiv of ZnCl2 was added to Co(II)-TTP-2D. (c) Plot of the absorption spectrum at 650 nm as a function of either added Cu(I) or Zn(II). Titrations were performed with 16.5 μM Apo-TTP-2D in 200 mM HEPES, 100 mM NaCl, at pH 7.5.
Circular Dichroism (CD) of apo-TTP-2D, Zn(II)-TTP-2D, and Cu(I)-TTP-2D
Circular dichroism (CD) spectroscopy was employed to measure the secondary structure of TTP-2D as a function of metal coordination. As shown in Figure 8, apo-TTP-2D adopts a random coil conformation while Zn(II)-TTP-2D shows some evidence of secondary structure, mostly α helical character. This is in keeping with the NMR structures of TTP and a close homologue Tis11d—both of which exhibit some α helical structure upon Zn binding.82,83 The Cu(I)-TTP-2D spectrum lacked any secondary structure and instead is similar to the spectrum of apo-TTP-2D. These results indicate that while Cu(I) binds to TTP-2D, as evidenced by changes in the UV–visible spectrum including the appearance of charge transfer bands between Cu(I) and sulfur, that the metal coordination is not accompanied by adoption of any secondary structure.
Figure 8.
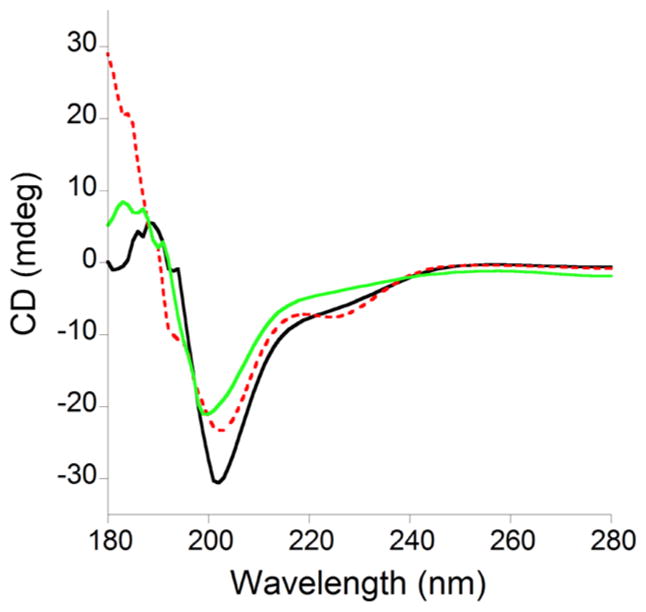
Overlay of the CD spectra of 50 μM apo-TTP-2D (black line), Zn(II)-TTP-2D (red dotted line), and Cu(I)-TTP-2D (green line). All experiments were performed in 10 mM sodium phosphate, at pH 7.5.
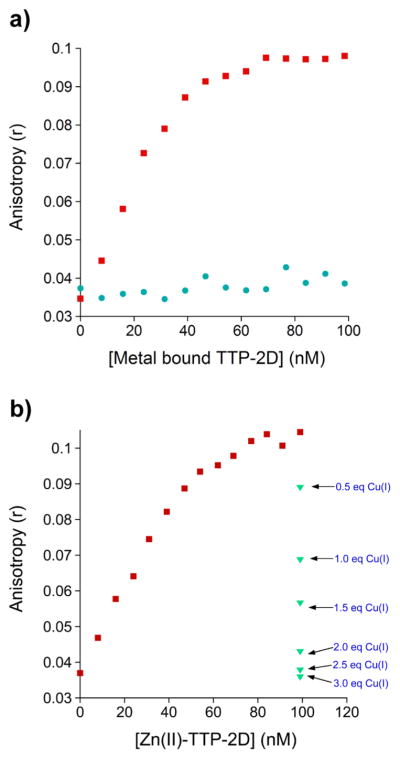
Cu(I)-TTP-2D/RNA Binding
TTP-2D binds specifically to the mRNA sequence UUUAUUUAUUU, which is found at the 3′-end of cytokine mRNAs. When Zn(II) is bound to TTP-2D, high-affinity binding to this RNA target is observed.1,44,82,84 Similar high-affinity RNA binding is observed when Fe(II), Fe(III), or Cd(II) is coordinated to TTP-2D.34,44 To determine how Cu(I)-TTP-2D binds to RNA, a fluorescence anisotropy assay, developed by one of our laboratories, was utilized.34,44 In the experiment, Cu(I)-TTP-2D was added to a fluorescently labeled RNA target sequence, UUUAUUUAUUU-F (F = fluorescein), and binding was monitored via a change in anisotropy (r). No binding to RNA was observed (Figure 9a) in the presence of Cu(I)-TTP-2D. This is in contrast with Zn(II)-TTP-2D, Fe(II)-TTP-2D, Fe(III)-TTP-2D, and Cd(II)-TTP-2D, all of which bind to the AU-rich RNA sequence with high affinities (Kd = 16 ± 1, 12 ± 1, 25 ± 3, 2.4 ± 0.2, respectively).34,44 This result suggests that when Cu(I) binds to TTP-2D, in lieu of Zn(II) or other divalent or trivalent metal ions, the protein is not functional. This lack of function is likely due to the effect of Cu(I) on folding. While Cu(I) binds to TTP-2D, as evidenced by the appearance of charge transfer bands in the UV–visible spectrum (Figures 2 and 4), the protein does not exhibit any secondary structure, as indicated by the CD spectrum of Cu(I)-TTP-2D, which resembles that of apo-TTP-2D. Moreover, the observation that 3 equiv of Cu(I) binds to TTP-2D provides further support for the altered/ disrupted structure of TTP-2D.
Figure 9.
(a) Comparison of the change in anisotropy upon the addition of Zn(II)-TTP-2D (red) or Cu(I)-TTP-2D (teal green) to the RNA oligonucleotide UUUAUUUAUUU-F (F = fluorescein). (b) Plot of the change in anisotropy upon the addition of Zn(II)-TTP-2D (red) to the RNA oligonucleotide UUUAUUUAUUU-F (F = fluorescein) followed by addition of CuCl. All FA experiments were performed in a 200 mM HEPES, 100 mM NaCl, at pH 7.5.
A second fluorescence anisotropy experiment, in which Cu(I) was added to Zn(II)-TTP-2D bound to RNA, was also performed. The goal was to determine if Cu(I) disrupts the protein/RNA binding interaction. We observe that addition of Cu(I), starting with 0.5 equiv (vs 1 equiv of Zn(II)-TTP2D/ RNA), resulted in a diminution of anisotropy. After the addition of 3.5 equiv of Cu(I), the anisotropy equaled that of free RNA, indicating that Zn(II)-TTP binding to RNA was completely inhibited (Figure 9b). These data suggest a potential role for Cu(I) in turning off TTP function by disrupting the protein/RNA binding interaction.
CONCLUSIONS
The work described in this paper is the first study of how Cu(I) can affect the function of a ZF protein. Cu(I)-coordinated TTP-2D does not bind to RNA, and Cu(I) inhibits Zn(II)-TTP-2D/RNA binding. The lack of RNA binding when Cu(I) is coordinated to TTP-2D is likely due to the lack of folding of the CCCH domains. Although TTP-2D has limited fold when the native Zn(II) metal is bound, this limited fold is clearly important for RNA binding. The inhibition of Zn(II)-TTP-2D/ RNA by Cu(I) suggests that Cu(I) can also associate with the Zn(II)-TTP-2D/RNA complex and disrupt function. Whether Cu(I) disrupts the function of other types of ZF proteins, classical or nonclassical, remains to be seen. We note that the only other study of Cu(I) binding to a ZF site, in addition to that of one reported by one of our laboratories that examined Cu(I) binding to short ZF peptides,37 examined Cu(I) binding to the zinc binding domain, SBP, from the copper response regulator 1 (CRR1) protein from Chlamydomonas reinhardtii.40 Cu(I)-SBP adopted the same structure as Zn(II)-SBP, as measured by CD, in contrast to TTP (vide supra), CP-CCHC, and NCp7-C, all of which exhibited no secondary structure when Cu(I) was coordinated. This finding suggests that the type of ZF (i.e., types of ligands at each ZF domain) must play a role in whether Cu(I) leads to a structured domain. The functional effects of Cu(I)-SBP were not investigated, and it would be interesting to learn how this protein’s ability to bind to DNA is affected by Cu(I) coordination.
Copper levels are upregulated during inflammation, and consequently cytokine levels are elevated.73,85,86 The mechanisms by which increased copper leads to inflammation are not clearly understood. One current hypothesis is that copper is released from the protein Ceruloplasmin, a multicopper oxidase, that stores most of the circulating copper in the plasma.69 Once released, the copper can bind to other proteins, and inflammatory mediators such as NFκB have been proposed to be targets. TTP is another inflammatory mediator and participates in the same signaling pathway as NFκB, making it a potential target. Our biochemical data revealed that TTP does not bind to RNA when Cu(I)-loaded, suggesting that in cells Cu(I)-TTP may inhibit TTP’s function to shut off cytokine production (and therefore the inflammatory response), leading to overinflammation. Work is now underway to examine how TTP function is modulated by metal ions in an in vivo model.
More broadly, the ability of Cu(I) to disrupt Zn-metalloproteins may have clear implications in the context of copper toxicity and/or metallo-regulation. While under most cellular conditions Cu(I) levels are tightly controlled and free Cu(I) concentrations are very low,62 several disease states are characterized by misregulation of cellular copper.56,73,87–91 A clear understanding of Cu(I) binding to Zn(II)-metalloproteins is important in identifying putative targets for disruption by Cu(I) under conditions of copper misregulation. Previous work has shown that the ZF-based Sp1 transcription factor regulates the expression of multiple copper-binding proteins in response to fluctuations in copper concentrations, and it is hypothesized that the ZFs in Sp1 function as intracellular copper sensors via copper interaction at the zinc binding sites.92–95 The disruption of TTP-2D function by Cu(I) observed here suggests that Cu(I) may also play a regulatory role in the inflammatory response and may be another example of metalloprotein substitution for regulatory purposes. Finally, recent work suggests that fluctuations of intracellular transition metal ion concentrations, including Cu(I), may play important roles in signal transduction, providing support for the notion that transition metal ion–protein interactions are more dynamic than previously believed.65,66
A final contribution of this work is the utilization of a rapid ICP-MS approach to measure metal:protein stoichiometry. Conventionally, UV–visible monitored titrations are used to determine stoichiometry; however, this approach relies on the presence of clear absorbance bands (often charge transfer bands), which can be obscured or modulated by protein bands, making it difficult to definitively ascribe stoichiometry.44 Our approach directly measures the metal stoichiometry via a rapid spin filter/ICP-MS approach, and we envision its use in confirming metal stoichiometry for other metalloprotein systems.
Acknowledgments
S.L.J.M. is grateful to the NSF (CHE1306208) for support of this research. G.D.S. is grateful to the NIH CBI Training Grant T32GM066706-13; K.E.S. is grateful to the NSF (CHE1308675) for support of this research.
Footnotes
Notes
The authors declare no competing financial interest.
References
- 1.Lee SJ, Michel SL. Structural metal sites in nonclassical zinc finger proteins involved in transcriptional and translational regulation. Acc Chem Res. 2014;47:2643–2650. doi: 10.1021/ar500182d. [DOI] [PubMed] [Google Scholar]
- 2.Michalek JL, Besold AN, Michel SL. Cysteine and histidine shuffling: mixing and matching cysteine and histidine residues in zinc finger proteins to afford different folds and function. Dalton Trans. 2011;40:12619–12632. doi: 10.1039/c1dt11071c. [DOI] [PubMed] [Google Scholar]
- 3.Decaria L, Bertini I, Williams RJ. Zinc proteomes, phylogenetics and evolution. Metallomics. 2010;2:706–709. doi: 10.1039/c0mt00024h. [DOI] [PubMed] [Google Scholar]
- 4.Bertini I, Decaria L, Rosato A. The annotation of full zinc proteomes. JBIC, J Biol Inorg Chem. 2010;15:1071–1078. doi: 10.1007/s00775-010-0666-6. [DOI] [PubMed] [Google Scholar]
- 5.Gower-Winter SD, Levenson CW. Zinc in the central nervous system: From molecules to behavior. Biofactors. 2012;38:186–193. doi: 10.1002/biof.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swamynathan SK. Kruppel-like factors: three fingers in control. Hum Genomics. 2010;4:263–270. doi: 10.1186/1479-7364-4-4-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shifera AS. The zinc finger domain of IKKgamma (NEMO) protein in health and disease. J Cell Mol Med. 2010;14:2404–2414. doi: 10.1111/j.1582-4934.2010.01054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heyninck K, Beyaert R. A20 inhibits NF-kappaB activation by dual ubiquitin-editing functions. Trends Biochem Sci. 2005;30:1–4. doi: 10.1016/j.tibs.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Todorova T, Bock FJ, Chang P. Poly(ADP-ribose) polymerase-13 and RNA regulation in immunity and cancer. Trends Mol Med. 2015;21:373–384. doi: 10.1016/j.molmed.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang C, Huang S, Wang X, Gu Y. Emerging Roles of CCCH-Type Zinc Finger Proteins in Destabilizing mRNA Encoding Inflammatory Factors and Regulating Immune Responses. Crit Rev Eukaryotic Gene Expression. 2015;25:77–89. doi: 10.1615/critreveukaryotgeneexpr.2015013022. [DOI] [PubMed] [Google Scholar]
- 11.Mori M, Kovalenko L, Lyonnais S, Antaki D, Torbett BE, Botta M, Mirambeau G, Mely Y. Nucleocapsid Protein: A Desirable Target for Future Therapies Against HIV-1. Curr Top Microbiol Immunol. 2015;389:53–92. doi: 10.1007/82_2015_433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mani RS. The emerging role of speckle-type POZ protein (SPOP) in cancer development. Drug Discovery Today. 2014;19:1498–1502. doi: 10.1016/j.drudis.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jen J, Wang YC. Zinc finger proteins in cancer progression. J Biomed Sci. 2016;23:53. doi: 10.1186/s12929-016-0269-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 15.Klug A. The discovery of zinc fingers and their development for practical applications in gene regulation and genome manipulation. Q Rev Biophys. 2010;43:1–21. doi: 10.1017/S0033583510000089. [DOI] [PubMed] [Google Scholar]
- 16.Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- 17.Berg JM. Potential metal-binding domains in nucleic acid binding proteins. Science. 1986;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 18.Andreini C, Banci L, Bertini I, Rosato A. Counting the zinc-proteins encoded in the human genome. J Proteome Res. 2006;5:196–201. doi: 10.1021/pr050361j. [DOI] [PubMed] [Google Scholar]
- 19.Andreini C, Bertini I, Cavallaro G. Minimal functional sites allow a classification of zinc sites in proteins. PLoS One. 2011;6:e26325. doi: 10.1371/journal.pone.0026325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laskay UA, Garino C, Tsybin YO, Salassa L, Casini A. Gold finger formation studied by high-resolution mass spectrometry and in silico methods. Chem Commun. 2015;51:1612–1615. doi: 10.1039/c4cc07490d. [DOI] [PubMed] [Google Scholar]
- 21.Bertrand B, Spreckelmeyer S, Bodio E, Cocco F, Picquet M, Richard P, Le Gendre P, Orvig C, Cinellu MA, Casini A. Exploring the potential of gold(III) cyclometallated compounds as cytotoxic agents: variations on the C∧N theme. Dalton Trans. 2015;44:11911–11918. doi: 10.1039/c5dt01023c. [DOI] [PubMed] [Google Scholar]
- 22.Jacques A, Lebrun C, Casini A, Kieffer I, Proux O, Latour JM, Seneque O. Reactivity of Cys4 zinc finger domains with gold(III) complexes: insights into the formation of “gold fingers”. Inorg Chem. 2015;54:4104–4113. doi: 10.1021/acs.inorgchem.5b00360. [DOI] [PubMed] [Google Scholar]
- 23.Ordemann JM, Austin RN. Lead neurotoxicity: exploring the potential impact of lead substitution in zinc-finger proteins on mental health. Metallomics. 2016;8:579–588. doi: 10.1039/c5mt00300h. [DOI] [PubMed] [Google Scholar]
- 24.Petering DH, Huang M, Moteki S, Shaw CF., III Cadmium and lead interactions with transcription factor IIIA from Xenopus laevis: a model for zinc finger protein reactions with toxic metal ions and metallothionein. Mar Environ Res. 2000;50:89–92. doi: 10.1016/s0141-1136(00)00099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zawia NH, Crumpton T, Brydie M, Reddy GR, Razmiafshari M. Disruption of the zinc finger domain: a common target that underlies many of the effects of lead. Neurotoxicology. 2000;21:1069–1080. [PubMed] [Google Scholar]
- 26.Hanas JS, Rodgers JS, Bantle JA, Cheng YG. Lead inhibition of DNA-binding mechanism of Cys(2)His(2) zinc finger proteins. Mol Pharmacol. 1999;56:982–988. doi: 10.1124/mol.56.5.982. [DOI] [PubMed] [Google Scholar]
- 27.Nagaoka M, Kuwahara J, Sugiura Y. Alteration of DNA binding specificity by nickel (II) substitution in three zinc (II) fingers of transcription factor Sp1. Biochem Biophys Res Commun. 1993;194:1515–1520. doi: 10.1006/bbrc.1993.1996. [DOI] [PubMed] [Google Scholar]
- 28.Besold AN, Lee SJ, Michel SL, Sue NL, Cymet HJ. Functional characterization of iron-substituted neural zinc finger factor 1: metal and DNA binding. JBIC, J Biol Inorg Chem. 2010;15:583–590. doi: 10.1007/s00775-010-0626-1. [DOI] [PubMed] [Google Scholar]
- 29.Lee SJ, Michel SL. Cysteine oxidation enhanced by iron in tristetraprolin, a zinc finger peptide. Inorg Chem. 2010;49:1211–1219. doi: 10.1021/ic9024298. [DOI] [PubMed] [Google Scholar]
- 30.Hider RC, Bittel D, Andrews GK. Competition between iron(III)-selective chelators and zinc-finger domains for zinc(II) Biochem Pharmacol. 1999;57:1031–1035. doi: 10.1016/s0006-2952(99)00014-3. [DOI] [PubMed] [Google Scholar]
- 31.Conte D, Narindrasorasak S, Sarkar B. In vivo and in vitro iron-replaced zinc finger generates free radicals and causes DNA damage. J Biol Chem. 1996;271:5125–5130. doi: 10.1074/jbc.271.9.5125. [DOI] [PubMed] [Google Scholar]
- 32.Omichinski JG, Trainor C, Evans T, Gronenborn AM, Clore GM, Felsenfeld G. A small single-”finger” peptide from the erythroid transcription factor GATA-1 binds specifically to DNA as a zinc or iron complex. Proc Natl Acad Sci U S A. 1993;90:1676–1680. doi: 10.1073/pnas.90.5.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malgieri G, Palmieri M, Esposito S, Maione V, Russo L, Baglivo I, de Paola I, Milardi D, Diana D, Zaccaro L, Pedone PV, Fattorusso R, Isernia C. Zinc to cadmium replacement in the prokaryotic zinc-finger domain. Metallomics. 2014;6:96–104. doi: 10.1039/c3mt00208j. [DOI] [PubMed] [Google Scholar]
- 34.Michalek JL, Lee SJ, Michel SL. Cadmium coordination to the zinc binding domains of the non-classical zinc finger protein Tristetraprolin affects RNA binding selectivity. J Inorg Biochem. 2012;112:32–38. doi: 10.1016/j.jinorgbio.2012.02.023. [DOI] [PubMed] [Google Scholar]
- 35.Malgieri G, Zaccaro L, Leone M, Bucci E, Esposito S, Baglivo I, Del Gatto A, Russo L, Scandurra R, Pedone PV, Fattorusso R, Isernia C. Zinc to cadmium replacement in the A. thaliana SUPERMAN Cys(2) His(2) zinc finger induces structural rearrangements of typical DNA base determinant positions. Biopolymers. 2011;95:801–810. doi: 10.1002/bip.21680. [DOI] [PubMed] [Google Scholar]
- 36.Kothinti RK, Blodgett AB, Petering DH, Tabatabai NM. Cadmium down-regulation of kidney Sp1 binding to mouse SGLT1 and SGLT2 gene promoters: possible reaction of cadmium with the zinc finger domain of Sp1. Toxicol Appl Pharmacol. 2010;244:254–262. doi: 10.1016/j.taap.2009.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doku RT, Park G, Wheeler KE, Splan KE. Spectroscopic characterization of copper(I) binding to apo and metal-reconstituted zinc finger peptides. JBIC, J Biol Inorg Chem. 2013;18:669–678. doi: 10.1007/s00775-013-1012-6. [DOI] [PubMed] [Google Scholar]
- 38.Hutchens TW, Allen MH, Li CM, Yip TT. Occupancy of a C2-C2 type ‘zinc-finger’ protein domain by copper. Direct observation by electrospray ionization mass spectrometry. FEBS Lett. 1992;309:170–174. doi: 10.1016/0014-5793(92)81088-4. [DOI] [PubMed] [Google Scholar]
- 39.Huthens TW, Allen MH. Differences in the conformational state of a zinc-finger DNA-binding protein domain occupied by zinc and copper revealed by electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 1992;6:469–473. doi: 10.1002/rcm.1290060713. [DOI] [PubMed] [Google Scholar]
- 40.Sommer F, Kropat J, Malasarn D, Grossoehme NE, Chen X, Giedroc DP, Merchant SS. The CRR1 nutritional copper sensor in Chlamydomonas contains two distinct metal-responsive domains. Plant Cell. 2010;22:4098–4113. doi: 10.1105/tpc.110.080069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bal W, Schwerdtle T, Hartwig A. Mechanism of nickel assault on the zinc finger of DNA repair protein XPA. Chem Res Toxicol. 2003;16:242–248. doi: 10.1021/tx025639q. [DOI] [PubMed] [Google Scholar]
- 42.Magyar JS, Weng TC, Stern CM, Dye DF, Rous BW, Payne JC, Bridgewater BM, Mijovilovich A, Parkin G, Zaleski JM, Penner-Hahn JE, Godwin HA. Reexamination of lead(II) coordination preferences in sulfur-rich sites: implications for a critical mechanism of lead poisoning. J Am Chem Soc. 2005;127:9495–9505. doi: 10.1021/ja0424530. [DOI] [PubMed] [Google Scholar]
- 43.Ghering AB, Jenkins LM, Schenck BL, Deo S, Mayer RA, Pikaart MJ, Omichinski JG, Godwin HA. Spectroscopic and functional determination of the interaction of Pb2+ with GATA proteins. J Am Chem Soc. 2005;127:3751–3759. doi: 10.1021/ja0464544. [DOI] [PubMed] [Google Scholar]
- 44.diTargiani RC, Lee SJ, Wassink S, Michel SL. Functional characterization of iron-substituted tristetraprolin-2D (TTP-2D, NUP475–2D): RNA binding affinity and selectivity. Biochemistry. 2006;45:13641–13649. doi: 10.1021/bi060747n. [DOI] [PubMed] [Google Scholar]
- 45.Bernardes VH, Qu Y, Du Z, Beaton J, Vargas MD, Farrell NP. Interaction of the HIV NCp7 Protein with Platinum(II) and Gold(III) Complexes Containing Tridentate Ligands. Inorg Chem. 2016;55:11396–11407. doi: 10.1021/acs.inorgchem.6b01925. [DOI] [PubMed] [Google Scholar]
- 46.Abbehausen C, Peterson EJ, de Paiva RE, Corbi PP, Formiga AL, Qu Y, Farrell NP. Gold(I)-phosphine-N-heterocycles: biological activity and specific (ligand) interactions on the C-terminal HIVNCp7 zinc finger. Inorg Chem. 2013;52:11280–11287. doi: 10.1021/ic401535s. [DOI] [PubMed] [Google Scholar]
- 47.Spell SR, Farrell NP. [Au(dien)(N-heterocycle)](3+): reactivity with biomolecules zinc finger peptides. Inorg Chem. 2015;54:79–86. doi: 10.1021/ic501784n. [DOI] [PubMed] [Google Scholar]
- 48.de Paula QA, Mangrum JB, Farrell NP. Zinc finger proteins as templates for metal ion exchange: Substitution effects on the C-finger of HIV nucleocapsid NCp7 using M(chelate) species (M = Pt, Pd, Au) J Inorg Biochem. 2009;103:1347–1354. doi: 10.1016/j.jinorgbio.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 49.Demicheli C, Frezard F, Mangrum JB, Farrell NP. Interaction of trivalent antimony with a CCHC zinc finger domain: potential relevance to the mechanism of action of antimonial drugs. Chem Commun. 2008:4828–4830. doi: 10.1039/b809186b. [DOI] [PubMed] [Google Scholar]
- 50.Demicheli C, Frezard F, Pereira FA, Santos DM, Mangrum JB, Farrell NP. Interaction of arsenite with a zinc finger CCHC peptide: evidence for formation of an As-Zn-peptide mixed complex. J Inorg Biochem. 2011;105:1753–1758. doi: 10.1016/j.jinorgbio.2011.09.023. [DOI] [PubMed] [Google Scholar]
- 51.Frezard F, Silva H, Pimenta AM, Farrell N, Demicheli C. Greater binding affinity of trivalent antimony to a CCCH zinc finger domain compared to a CCHC domain of kinetoplastid proteins. Metallomics. 2012;4:433–440. doi: 10.1039/c2mt00176d. [DOI] [PubMed] [Google Scholar]
- 52.Spuches AM, Wilcox DE. Monomethylarsenite competes with Zn2+ for binding sites in the glucocorticoid receptor. J Am Chem Soc. 2008;130:8148–8149. doi: 10.1021/ja802179p. [DOI] [PubMed] [Google Scholar]
- 53.Franzman MA, Barrios AM. Spectroscopic evidence for the formation of goldfingers. Inorg Chem. 2008;47:3928–3930. doi: 10.1021/ic800157t. [DOI] [PubMed] [Google Scholar]
- 54.Godwin HA. The biological chemistry of lead. Curr Opin Chem Biol. 2001;5:223–227. doi: 10.1016/s1367-5931(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 55.Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. doi: 10.1146/annurev-biochem-030409-143539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Besold AN, Culbertson EM, Culotta VC. The Yin and Yang of copper during infection. JBIC, J Biol Inorg Chem. 2016;21:137–144. doi: 10.1007/s00775-016-1335-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sheng Y, Abreu IA, Cabelli DE, Maroney MJ, Miller AF, Teixeira M, Valentine JS. Superoxide dismutases and superoxide reductases. Chem Rev. 2014;114:3854–3918. doi: 10.1021/cr4005296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Valko M, Jomova K, Rhodes CJ, Kuca K, Musilek K. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch Toxicol. 2016;90:1–37. doi: 10.1007/s00204-015-1579-5. [DOI] [PubMed] [Google Scholar]
- 59.Jomova K, Valko M. Advances in metal-induced oxidative stress and human disease. Toxicology. 2011;283:65–87. doi: 10.1016/j.tox.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 60.Brewer GJ. Iron and copper toxicity in diseases of aging, particularly atherosclerosis and Alzheimer’s disease. Exp Biol Med. 2007;232:323–335. [PubMed] [Google Scholar]
- 61.Gaetke LM, Chow-Johnson HS, Chow CK. Copper: toxicological relevance and mechanisms. Arch Toxicol. 2014;88:1929–1938. doi: 10.1007/s00204-014-1355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 63.Boal AK, Rosenzweig AC. Structural biology of copper trafficking. Chem Rev. 2009;109:4760–4779. doi: 10.1021/cr900104z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nevitt T, Ohrvik H, Thiele DJ. Charting the travels of copper in eukaryotes from yeast to mammals. Biochim Biophys Acta, Mol Cell Res. 2012;1823:1580–1593. doi: 10.1016/j.bbamcr.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dodani SC, Firl A, Chan J, Nam CI, Aron AT, Onak CS, Ramos-Torres KM, Paek J, Webster CM, Feller MB, Chang CJ. Copper is an endogenous modulator of neural circuit spontaneous activity. Proc Natl Acad Sci U S A. 2014;111:16280–16285. doi: 10.1073/pnas.1409796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Krishnamoorthy L, Cotruvo JA, Jr, Chan J, Kaluarachchi H, Muchenditsi A, Pendyala VS, Jia S, Aron AT, Ackerman CM, Wal MN, Guan T, Smaga LP, Farhi SL, New EJ, Lutsenko S, Chang CJ. Copper regulates cyclic-AMP-dependent lipolysis. Nat Chem Biol. 2016;12:586–592. doi: 10.1038/nchembio.2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McElwee MK, Song MO, Freedman JH. Copper activation of NF-kappaB signaling in HepG2 cells. J Mol Biol. 2009;393:1013–1021. doi: 10.1016/j.jmb.2009.08.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blackshear PJ. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem Soc Trans. 2002;30:945–952. doi: 10.1042/bst0300945. [DOI] [PubMed] [Google Scholar]
- 69.Brooks SA, Blackshear PJ. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta, Gene Regul Mech. 2013;1829:666–679. doi: 10.1016/j.bbagrm.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carballo E, Lai WS, Blackshear PJ. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 71.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 72.Carrick DM, Lai WS, Blackshear PJ. The tandem CCCH zinc finger protein tristetraprolin and its relevance to cytokine mRNA turnover and arthritis. Arthritis Res Ther. 2004;6:248–264. doi: 10.1186/ar1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pereira TC, Campos MM, Bogo MR. Copper toxicology, oxidative stress and inflammation using zebrafish as experimental model. J Appl Toxicol. 2016;36:876–885. doi: 10.1002/jat.3303. [DOI] [PubMed] [Google Scholar]
- 74.Malavolta M, Giacconi R, Piacenza F, Santarelli L, Cipriano C, Costarelli L, Tesei S, Pierpaoli S, Basso A, Galeazzi R, Lattanzio F, Mocchegiani E. Plasma copper/zinc ratio: an inflammatory/nutritional biomarker as predictor of all-cause mortality in elderly population. Biogerontology. 2010;11:309–319. doi: 10.1007/s10522-009-9251-1. [DOI] [PubMed] [Google Scholar]
- 75.Xiao Z, Brose J, Schimo S, Ackland SM, La Fontaine S, Wedd AG. Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1, and related proteins: detection probes and affinity standards. J Biol Chem. 2011;286:11047–11055. doi: 10.1074/jbc.M110.213074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xiao Z, Loughlin F, George GN, Howlett GJ, Wedd AG. C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J Am Chem Soc. 2004;126:3081–3090. doi: 10.1021/ja0390350. [DOI] [PubMed] [Google Scholar]
- 77.Kihlken MA, Leech AP, Le Brun NE. Copper-mediated dimerization of CopZ, a predicted copper chaperone from Bacillus subtilis. Biochem J. 2002;368:729–739. doi: 10.1042/BJ20021036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roehm PC, Berg JM. Sequential metal binding by the RING finger domain of BRCA1. Biochemistry. 1997;36:10240–10245. doi: 10.1021/bi970863d. [DOI] [PubMed] [Google Scholar]
- 79.Gil VMS, Oliveira NC. On the use of the method of continuous variations. J Chem Educ. 1990;67:473–478. [Google Scholar]
- 80.Leeladee P, Baglia RA, Prokop KA, Latifi R, de Visser SP, Goldberg DP. Valence tautomerism in a high-valent manganese-oxo porphyrinoid complex induced by a Lewis acid. J Am Chem Soc. 2012;134:10397–10400. doi: 10.1021/ja304609n. [DOI] [PubMed] [Google Scholar]
- 81.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. Advanced Inorganic Chemistry. Wiley; New York: 1999. [Google Scholar]
- 82.Hudson BP, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the mRNA AU-rich element by the zinc finger domain of TIS11d. Nat Struct Mol Biol. 2004;11:257–264. doi: 10.1038/nsmb738. [DOI] [PubMed] [Google Scholar]
- 83.Amann BT, Worthington MT, Berg JM. A Cys3His zinc-binding domain from Nup475/tristetraprolin: a novel fold with a disklike structure. Biochemistry. 2003;42:217–221. doi: 10.1021/bi026988m. [DOI] [PubMed] [Google Scholar]
- 84.Brewer BY, Malicka J, Blackshear PJ, Wilson GM. RNA sequence elements required for high affinity binding by the zinc finger domain of tristetraprolin: conformational changes coupled to the bipartite nature of Au-rich MRNA-destabilizing motifs. J Biol Chem. 2004;279:27870–27877. doi: 10.1074/jbc.M402551200. [DOI] [PubMed] [Google Scholar]
- 85.Saghiri MA, Asatourian A, Orangi J, Sorenson CM, Sheibani N. Functional role of inorganic trace elements in angiogenesis-Part II: Cr, Si, Zn, Cu, and S. Crit Rev Oncol Hematol. 2015;96:143–155. doi: 10.1016/j.critrevonc.2015.05.011. [DOI] [PubMed] [Google Scholar]
- 86.Brewer GJ. Alzheimer’s disease causation by copper toxicity and treatment with zinc. Front Aging Neurosci. 2014;6:92. doi: 10.3389/fnagi.2014.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mercer JF. The molecular basis of copper-transport diseases. Trends Mol Med. 2001;7:64–69. doi: 10.1016/s1471-4914(01)01920-7. [DOI] [PubMed] [Google Scholar]
- 88.Marchetti C. Interaction of metal ions with neurotransmitter receptors and potential role in neurodiseases. BioMetals. 2014;27:1097–1113. doi: 10.1007/s10534-014-9791-y. [DOI] [PubMed] [Google Scholar]
- 89.Gaggelli E, Kozlowski H, Valensin D, Valensin G. Copper homeostasis and neurodegenerative disorders (Alzheimer’s, prion, and Parkinson’s diseases and amyotrophic lateral sclerosis) Chem Rev. 2006;106:1995–2044. doi: 10.1021/cr040410w. [DOI] [PubMed] [Google Scholar]
- 90.Sheng Y, Chattopadhyay M, Whitelegge J, Valentine JS. SOD1 aggregation and ALS: role of metallation states and disulfide status. Curr Top Med Chem. 2013;12:2560–2572. doi: 10.2174/1568026611212220010. [DOI] [PubMed] [Google Scholar]
- 91.Culotta VC, Yang M, O’Halloran TV. Activation of superoxide dismutases: putting the metal to the pedal. Biochim Biophys Acta, Mol Cell Res. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Song IS, Chen HH, Aiba I, Hossain A, Liang ZD, Klomp LW, Kuo MT. Transcription factor Sp1 plays an important role in the regulation of copper homeostasis in mammalian cells. Mol Pharmacol. 2008;74:705–713. doi: 10.1124/mol.108.046771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liang ZD, Tsai WB, Lee MY, Savaraj N, Kuo MT. Specificity protein 1 (sp1) oscillation is involved in copper homeostasis maintenance by regulating human high-affinity copper transporter 1 expression. Mol Pharmacol. 2012;81:455–464. doi: 10.1124/mol.111.076422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ogra Y, Suzuki K, Gong P, Otsuka F, Koizumi S. Negative regulatory role of Sp1 in metal responsive element-mediated transcriptional activation. J Biol Chem. 2001;276:16534–16539. doi: 10.1074/jbc.M100570200. [DOI] [PubMed] [Google Scholar]
- 95.Bellingham SA, Coleman LA, Masters CL, Camakaris J, Hill AF. Regulation of prion gene expression by transcription factors SP1 and metal transcription factor-1. J Biol Chem. 2009;284:1291–1301. doi: 10.1074/jbc.M804755200. [DOI] [PubMed] [Google Scholar]