Abstract
Peripheral T-cell lymphomas (PTCLs) comprise a heterogeneous group of mature T-cell neoplasms with a poor prognosis. Recently, mutations in TET2 and other epigenetic modifiers as well as RHOA have been identified in these diseases, particularly in angioimmunoblastic T-cell lymphoma (AITL). CD28 is the major co-stimulatory receptor in T-cells which, upon binding ligand, induces sustained T-cell proliferation and cytokine production when combined with T-cell receptor stimulation. We have identified recurrent mutations in CD28 in PTCLs. Two residues – D124 and T195 – were recurrently mutated in 11.3% of cases of AITL and in one case of PTCL, not otherwise specified (PTCL-NOS). Surface plasmon resonance analysis of mutations at these residues with predicted differential partner interactions showed increased affinity for ligand CD86 (residue D124) and increased affinity for intracellular adaptor proteins GRB2 and GADS/GRAP2 (residue T195). Molecular modeling studies on each of these mutations suggested how these mutants result in increased affinities. We found increased transcription of the CD28-responsive genes CD226 and TNFA in cells expressing the T195P mutant in response to CD3 and CD86 co-stimulation and increased downstream activation of NF-κB by both D124V and T195P mutants, suggesting a potential therapeutic target in CD28-mutated PTCLs.
Introduction
Peripheral T-cell lymphomas (PTCLs) comprise approximately 10% of all non-Hodgkin lymphomas in the Western world and generally have a poor prognosis1. Novel therapeutic regimens have largely failed to improve patient outcomes, likely due to the poor understanding of PTCL pathogenesis. We and others have generated extensive gene expression profiling (GEP) data on PTCL entities, including angioimmunoblastic T-cell lymphoma (AITL) and other PTCL subtypes2. Many of these studies support the hypothesis that AITL is derived from follicular helper T-cells (TFH cells)3–5 and that this lymphoma constitutively activates the NF-κB pathway. However, the pathogenic mechanisms for AITL and the other common PTCL category, “not otherwise specified” (PTCL-NOS), have only recently been explored. We and others have reported several common recurrent AITL-associated genetic aberrations affecting TET26, IDH27,8, DNMT3A9, and RHOA10–12. The first three mutations likely lead to global epigenetic perturbations. Other than in FYN, few somatic mutations affecting the T-cell receptor (TCR) activation pathway have been observed in PTCLs, despite our GEP studies pointing to frequent activation of this pathway13. Here, we report recurrent mutations of CD28, a member of the immunoglobulin V-set subfamily and the major co-stimulatory molecule for TCR-mediated activation. By binding B7 family members CD80 (B7.1) and CD86 (B7.2)14, CD28 promotes T-cell activation and prevents anergy after TCR stimulation15. The intracellular domain is necessary for signal transduction after ligand binding16 and includes a YMNM motif, which, when phosphorylated, binds the SH2 domains of Src family kinases, PI3K regulatory subunits, and GADS/GRAP2 and GRB2. The CD28 intracellular tail also contains two proline-rich motifs that bind the SH3 domains of various signaling proteins, including ITK, GRAP2, and LCK17. Together, CD28-mediated PI3K, GRB2, and VAV1 signaling increase NF-κB and NFAT nuclear translocation, augmenting T-cell survival, production of the proliferative cytokine IL-2, and cell cycling.
We performed whole-transcriptome sequencing (WTS) on 20 AITL cases and identified two recurrent sites of mutation in CD28, at D124 and T195. We then performed targeted sequencing of CD28 in 90 PTCL cases, including five of the AITL cases with WTS, and studied the functional alterations associated with these two most frequent mutation sites. We observed that they augment ligand/receptor interactions or signal transduction adaptor binding affinities, both of which could contribute to T-cell activation in PTCLs. This was supported by increased downstream signaling of CD28 mutants upon ligand binding.
Materials and Methods
Patient specimens
The clinical and pathological characteristics of the patients included in the study have been published13 and are included in Supplemental Table 1. Informed consent was obtained from all patients. We included 20 molecularly diagnosed AITL lymphomas for transcriptome sequencing in our study and 85 additional cases of molecularly and/or histologically diagnosed AITL, PTCL-NOS, and ALK-negative ALCL. This study was approved by the Institutional Review Boards of the University of Nebraska Medical Center and City of Hope National Medical Center.
Patient RNA sequencing and targeted sequencing platform
Total RNA from 20 AITLs was extracted using the QIAgen RNEasy kit, and RNAseq was performed using the Illumina GIIx sequencer. TopHat18,19 was used for alignment, and Cufflinks20 was used for gene expression analysis. For the targeted sequencing platform, we used the TruSeq Custom Amplicon (TSCA) platform (Illumina, Inc.) to interrogate all four exons of CD28, all eleven exons of TET2, and all five exons of RHOA in PTCL specimens. Alignment was performed with Mutascope21. VarScan222 was used to call variants.
Fusion transcript validation and sequencing
TopHat-Fusion23 was used to analyze aberrant transcripts. The ICOS-CD28 fusion mRNA discovered on whole transcriptome sequencing was verified by PCR and Sanger sequencing on patient cDNA using the following primers: ICOS (forward): 5’-TGAACACTGAACGCGAGGAC-3’; CD28 (reverse): 5’-CATTGGTGGCCCAACAGG-3’.
Survival analysis
The Kaplan-Meier curve was constructed, and difference in survival was tested by the log-rank method, using the survival package in R24.
Surface plasmon resonance (SPR) spectrometry
Details of the SPR studies are available in the supplement. Briefly, binding experiments were carried out at 37° using surface plasmon resonance as implemented in the BiacoreTM T200 (GE Healthcare). CD28-Fc fusion protein at 0.1 mg/ml was directly immobilized on Research Grade CM5 sensor chips (GE Healthcare) by amine coupling, giving immobilization levels of 1000–2500 RU. Biotinylated tyrosyl phosphopeptides or a “universal” control peptide were indirectly immobilized via streptavidin to ~250 RU as described25. For both, equilibrium binding analysis was undertaken as described26,27. Data were analyzed using BIAevaluation Software (GE Healthcare) and Origin version 5.0 (MicroCal Software Inc., Northampton, MA).
Molecular modeling of novel CD28 mutants
Full details of the modeling procedure are available in the supplement. Briefly, to analyze the effects of the D124V mutation in CD28, PDB files 1YJD (CD28)28, 1I85 (CD86 and CTLA4)29, 1I8L (CD80 and CTLA4)30, and 3WA4 (GRB2 and CD28)31 were used and analyzed as previously described32–34.
Cell lines, transduction, and luciferase assay
The entire coding region of CD28 with or without D124V or T195P mutations was cloned into GFP-containing pMIG vectors (Promega, Madison, WI). Retrovirus production in 293T cells and infection of ~500,000 Jurkat cells, clone E6.1 (ATCC) were performed essentially as described35. Cells were confirmed free of mycoplasma with the Universal Mycoplasma Contamination Kit (ATCC). Transduced cells stably maintaining GFP expression after one week were sorted for equivalent GFP expression (Supplemental Table 4) using flow cytometry. Cells were cultured in RPMI-1640, 10% FBS (Omega Scientific, Tarzana, CA), 1% Pen/Strep, 1% 1M HEPES and frozen or used in the activation assay. All cell culture reagents were obtained from Life Technologies (Waltham, MA) unless otherwise stated. The in vitro assays for T-cell stimulation and NF-κB luciferase assay are described in detail in the supplemental section.
Nanostring nCounter assay
To measure gene expression over time36, one million Jurkat cells stably transduced with either CD28 mutants or WT were stimulated with one million of the indicated beads for the indicated time, then immediately washed in PBS, lysed in Qiagen buffer RLT, and placed at −80°C until use. Aliquots were thawed, and 10,000 cells per reaction were prepared for nCounter expression analysis per the manufacturer’s instructions (NanoString Technologies, Inc. Seattle, WA). We designed a customized codeset panel for 29 genes and two housekeeping genes (Supplemental Table 2). Cell lysate was hybridized to the custom codeset at 65° overnight. The reaction was processed on the nCounter prepstation and gene expression data were acquired on the nCounter Digital Analyzer on the “high resolution” setting. Standard quality control by the nSolver analysis software was employed.
Results
Detection of a CD28 fusion transcript and CD28 mutations through whole transcriptome sequencing
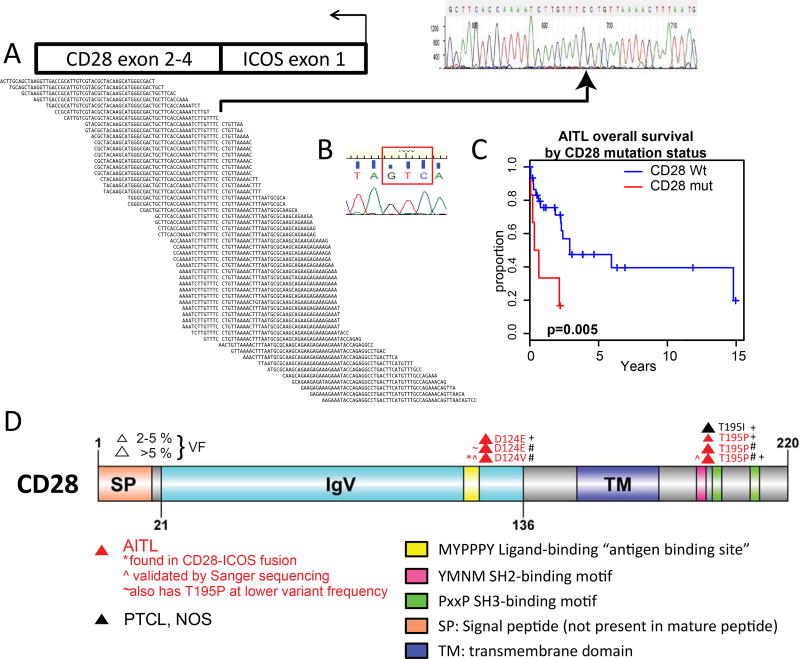
Our analysis of WTS data from 20 AITL cases revealed a single case showing an in-frame ICOS-CD28 fusion transcript. This fusion transcript was confirmed by Sanger sequencing of cDNA from the case showing a fusion of ICOS exon 1 (forward) with CD28 exon 2 (Fig. 1A). ICOS exon 1 encodes the membrane signal sequence, but this is cleaved from the protein; thus, ICOS promoter-mediated expression of the transcript would be expected but with no change in the final product. ICOS is highly expressed in AITL and in TFH cells4. The fusion transcript was found to contain a mutation of aspartate 124 of CD28 to valine (D124V; Fig. 1B). This AITL case was found to express CD28 at a level higher than most other cases in this study; additionally, the AITL cases expressed CD28 at a level higher than normal T-cell subsets when normalized for T-cell content37 (Supplemental Fig. 1A). Three cases with the CD28 residue threonine 195 mutated to proline (T195P) with varying variant frequencies (VFs) were also identified. Additionally, we also found TET2 single-nucleotide variants and indels as reported previously6 as well as RHOA mutations8. Based on these findings, we performed targeted whole-exon sequencing for CD28, TET2, and RHOA on 90 T-cell lymphoma cases (38 AITL – including five also having WTS, 40 PTCL-NOS, and 12 ALK- ALCL). In 88 cases, subtype was assigned molecularly4. For PTCL-NOS cases, GEP previously performed13 classified the cases into TBX21 or GATA3 subtypes; cases that did not fit into either category were considered “unclassified” (Table 1). Clinical information for all cases in this study is available in Supplemental Table 1.
Figure 1. Distribution of CD28 mutations discovered on transcriptome and targeted sequencing in T-cell lymphoma subtypes.
A: Alignment of ICOS-CD28 fusion transcript and identification of breakpoint, verified by Sanger sequencing. B: Demonstration by Sanger sequencing of D124V mutant in ICOS-CD28 transcript, GAC>GTC (red box). C: Kaplan-Meier survival analysis of AITL cases with CD28 mutations (red) versus AITL cases with no CD28 mutations (black). CD28-mutant cases had inferior survival after diagnosis (p=0.005). D: CD28 map and mutations found in 20 AITL cases with whole transcriptome sequencing plus 38 AITL (including five cases overlapping with transcriptome sequencing; red), 40 PTCL-NOS (black), and 12 ALK-ALCL cases. SP: signal peptide; IgV: Ig variable region-like domain CD28 and CTLA4; TM: transmembrane domain. Yellow: “antigen-binding” site required for interaction with ligand within IgV domain; pink: SH2-binding motif; green: SH3-binding motifs. #: identified in whole-transcriptome sequencing; +: identified in targeted sequencing platform. The diagram was built using DOG, version 2.060.
Table 1. Recurrent mutations in CD28 in screened T-cell lymphoma cases.
Summary of mutations in CD28 found on targeted sequencing of PTCL cases, grouped by resultant mutated residue and diagnosis. Seven of 105 total cases (6.7%) had mutations at recurrent residues within CD28.
| Number (% of screened) | Total by diagnosis (%) |
|||
|---|---|---|---|---|
| T195 | D124 | |||
| AITL (n=53)° | 3 (5.7) | 3 (5.7)* | 6 (11.3) | |
| PTCL, NOS (n=40) | TBX21 (n=19) | 1 (5.3) | 0 | 1 (5.3) |
| GATA3 (n=12) | 0 | 0 | 0 | |
| Unclassified (n=9)° | 0 | 0 | 0 | |
| ALCL, ALK- (n=12) | 0 | 0 | 0 | |
| Total by residue | 4 | 3 | 7 (6.7) | |
: this D124E case also has a T195P mutation at lower variant frequency; this case is only tabulated in the D124 column.
: one case of AITL and one case of PTCL-Unclassified has only a histological diagnosis.
CD28 mutations are relatively frequent in AITL and correlate with poor survival
The frequencies of CD28 mutations are presented in Table 1 and Fig. 1D, and the TET2 and RHOA mutation status and molecular diagnosis for individual cases are in Table 2. The average depth in our targeted sequencing platform was >1000-fold, and variant frequencies for a single nucleotide polymorphism (SNP; rs3116496) within the targeted region reveal a SNP proportion of approximately fifty percent (Supplemental Table 3). Paired-end reads filtered out likely misreads, and only recurrent mutations affecting the same codon were identified to reduce the possibility of artifact or random calls. Mutations at D124 and T195 were identified by these criteria, and seven of 105 (6.7%) PTCL cases had a mutation at one of these residues, including 6/53 (11.3%) AITLs. Two mutations, both at residue T195, had VFs greater than 10% in the targeted platform data, and one mutation at D124 had a VF greater than 5%. There was no difference in relative expression of the T-cell signature (as defined in ref. 13; Supplemental Fig. 1B) between CD28 wild-type and mutant cases in the transcriptome data. There was also no significant difference in average TET2 or RHOA VF based on CD28 mutation status in either the RNAseq or the targeted sequencing platforms (Supp. Fig. 1C–F). AITL cases with CD28 mutations have inferior survival to CD28-WT cases (p=0.005; Fig. 1C). We also found apparent mutations at residue phenylalanine 51 (F51) as previously reported in T-cell lymphomas12,38, but these were all found at extremely low apparent VF (<2%) and could not be verified by droplet digital PCR (ddPCR)39. We also subjected published PTCL sequencing datasets10–12 to the same analysis pipeline and found two T195P and three F51 mutations, ranging from 2.29% to 41.12% variant frequency (Supplemental Table 4). A case of a fusion of CD28 with family member CTLA4 has recently been reported in Sézary syndrome40, and a recent report on adult T-cell leukemia/lymphoma shows several mutations and fusions of CD28 (ref. 41). We examined our WTS data for additional CD28 fusion transcripts but none were found. We also examined our NK/γδTCL data42 and found no CD28 mutations.
Table 2. Mutated residue in CD28 by diagnosis, variant frequency, and TET2 and RHOA mutation status and frequency.
The CD28 mutation reads are grouped by diagnosis. Because TET2 mutations are varied, and individual cases often have more than one single-nucleotide variant (SNV) or indel, only the presence of SNVs or indels and the highest variant frequency is shown. Variant frequencies are either from transcriptome (Tr) or targeted sequencing data as indicated.
| Codon | Disease | Residue change |
Variant frequency |
TET2 mutation variant frequency |
RHOA mutation variant frequency |
|---|---|---|---|---|---|
| D124 | AITL | D124V | 21.9 (Tr) | 16.0 (Tr) | 11.9 (Tr) |
| D124E* | 49.1 (Tr) | 45.6 (Tr) | 28.5 (Tr) | ||
| D124E | 5.9 | 11.57 | 9.75 | ||
| T195 | AITL | T195P | 64.2 (Tr) | 50.0 (Tr) | 11.2 (Tr) |
| 10.58 | 17.73 | 3.72 | |||
| T195P | 42.7 (Tr) | 28.12 (Tr) | 20.6 (Tr) | ||
| T195P | 2.17 | 44.98 | 21.1 | ||
| PTCL-TBX21 | T195I | 12.3 | 13.09 | 14.4 |
: this case with D124E also had a T195P mutation at 6.6% variant frequency.
CD28 mutant D124V has a greater affinity for CD86 than CD28 WT
The D124V mutant found in the ICOS-CD28 fusion protein in an AITL case exchanges a negatively charged amino acid for an uncharged, hydrophobic one, and D124E (AITL) substitutes it for a slightly larger but equally charged R-group. These recurrent D124 mutations are immediately C-terminal to the core of the highly conserved ligand-binding site of CD28 (refs. 30, 43) (see Fig. 1C).
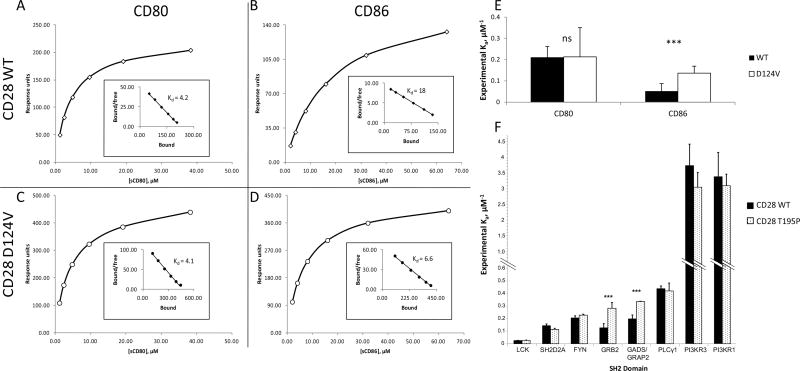
To assess whether D124 mutations affect the affinity of CD28 for its physiological ligands, we selected the CD28 D124V mutant for surface plasmon resonance (SPR)-based analysis of the binding affinity of CD28 for its ligands. Representative binding curves and Scatchard plots for determination of experimental Kd and Ka are shown in Fig. 2A–D. The affinity of CD28 D124V for CD86 was an average 2.6-fold higher than that of CD28 WT (Fig. 2E; p < 1×10−5 for 6 replicates), whereas CD80 binding affinity was not significantly different.
Figure 2. Differential binding affinities in CD28 WT and mutants.
A–D: Binding affinities to B7 family ligands CD80 and CD86 were compared between CD28 WT and the D124V mutant. Representative curves of CD28 WT (A, B) and D124V (C, D) binding soluble CD80 (A, C) or CD86 (B, D). The inset Scatchard plot shows the regression for the dissociation constant Kd calculation. E: The values for the association constant Ka were experimentally determined six different times; the average and standard deviation are shown. ***: p < 1×10−5 for 6 replicates; ns, not significant. F: CD28 tail motif phosphopeptides were indirectly immobilized in a flow cell and introduced to the SH2 domains of the indicated proteins. Relative affinities (mean ± standard deviation) for CD28 species experimentally determined for the SH2 domains from the indicated proteins. ***: p < 1×10−5 for 8 replicates.
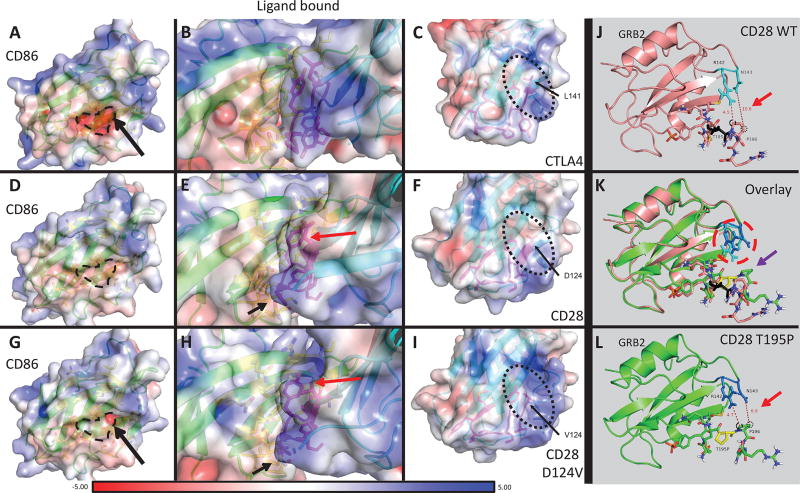
To understand why CD86 has a greater affinity for the CD28 D124V mutant than for CD28 WT, we modeled their interaction (Fig. 3). Because a complete crystal structure of CD28 bound to ligand is not available, we used the crystal structures of the CD28 extracellular domain28 and the complex between CD86 and CD28 family member CTLA4 (ref. 29). In this model, the protein surface in the vicinity of the D124V mutation has a more positive overall charge compared to CD28 WT and more closely resembles the surface charge of CTLA4 (Fig. 3C, F, I), which binds CD86 with a 20- to 100-fold greater affinity than CD28 (refs. 44–46). This is possibly because the negatively charged D124 counters any surrounding positive charges, leading to a neutral surface on CD28 WT. The D124V mutant is therefore expected to interact more strongly with the negatively charged pocket of CD86 (Fig. 3A, D, G, black dashed outline). Thus, the increased affinity of the mutant may result from improved charge complementarity. Furthermore, several residues, notably in the ligand-binding site, are rotated compared to CD28 WT. This is likely a result of the improved electrostatic interactions with the mutant CD28, leading to a better packing of the interface. We also performed molecular modeling on the CD80-CD28 interaction (Supplemental Figure 3; Supplemental Table 5) in the same manner using the crystal structure of CTLA4 in complex with CD80 (ref. 30).
Figure 3. Protein modeling of CD28 WT and D124V interacting with CD86, and CD28 WT and T195P interacting with GRB2.
A–I: PyMOL models of CD86 bound to receptor CTLA4, CD28 WT, or CD28 D124V with APBS-generated electrostatic surfaces. A–C: CD86-CTLA4 interaction. The residue corresponding to D124 in CD28 (L141) is indicated in C and the protein surface in the vicinity of this residue has an overall positive charge (also apparent in B). The negatively charged binding surface of CD86 can be seen in A (dashed line and surrounding area). D–F: CD86-CD28 WT interaction. The surface around D124 is more neutral compared to CTLA4. G–I: CD86-CD28 D124V interaction. Replacement of the negatively charged aspartate residue leads to a more positively charged surface, similar to CTLA4. The orientation of several residues is changed (Y118 – red arrow, P121 – black arrow) between CD28 WT and CD28 D124V in these models. J-L: PyMOL models of the CD28 cytoplasmic tail with or without the T195P mutant binding adaptor protein GRB2. J, L: CD28 WT (J) and T195P (L) cytoplasmic tail binding GRB2 (J, salmon; L, chartreuse). T195 is colored black (J); T195P is colored yellow (L). Note the distances between CD28 P196 carbons Cβ (black dotted circle) or Cδ (black dashed circle) and the closest non-hydrogen atoms on GRB2 (red arrows). CD28 P196 Cβ moves from 10.6Å away from GRB2 N143 in the CD28 WT to 6.9Å in CD28 T195P. K: Overlay of CD28 WT versus T195P mutant binding of GRB2. There is a significant change in orientation of several CD28 residues: proximal SH3-domain P196 (purple arrow) rotates and approaches GRB2 N143 in the CD28 T195P mutant. GRB2 R142 and N143 have a strikingly different rotation in the CD28 T195P mutant compared to WT (red dashed circle).
CD28 mutant T195P has a higher affinity for GRB2 and GADS/GRAP2 than the CD28 WT
The T195P mutation changes a polar amino acid to a hydrophobic one at the residue between the YMNM-containing SH2-binding motif and proximal PxxP-containing SH3-binding motif, essential mediators of adaptor protein binding during downstream signaling47. This mutant of CD28 is predicted to have a higher affinity for GRB2 than WT according to ScanSite48. To examine experimentally whether T195P alters the affinity of CD28 for adaptor proteins, we determined the affinity of this form of CD28 for several SH2 domains in known binding partners (Fig. 2). For two replicates, only GADS/GRAP2 and GRB2 showed significant differences, so each was assayed six more times. The CD28 T195P mutant had an average 1.7-fold greater affinity for GADS/GRAP2 and a 2.0-fold greater affinity for GRB2 than the CD28 WT (p < 1×10−5 for both).
No complete crystal structure of the CD28 cytoplasmic domain is available. However, there is one model of a synthesized (non-native) CD28 phosphotyrosine motif interacting with the SH2 domain of the adaptor protein GRB231, which we employed to build our model. The CD28 T195P mutation, which alters the lone residue between the SH2- and proximal SH3-binding motifs, was predicted to modify the conformation of the cytoplasmic tail C-terminal to the mutation (Fig. 3J–L). Interestingly, the HADDOCK model49 predicts that the interaction with GRB2’s SH2 domain is not directly altered. Rather, P196, within the adjacent SH3-binding motif, contorts significantly, potentially increasing the interaction between the CD28 P196 Cβ and Cδ atoms and the side chains of GRB2 R142 and N143, respectively. Comparing wild-type CD28 to the T195P mutant, the distance between P196 Cβ and the closest guanidino [−NHC(=NH)NH2] group of GRB2 R142 is essentially unchanged (4.5 vs. 4.7Å). However, the distance between P196 Cδ and the closest amino group on GRB2 N143 decreases from 10.6 to 6.9Å, drawing it close enough to interact. Given the lack of constraints in this model that would be imposed by the remainder of the CD28 tail, these results must be interpreted with caution; however, they do provide a plausible explanation for the altered affinity.
CD28 mutants alter transcription and induce higher NF-κB pathway activation than CD28 WT
To determine whether these mutations alter the kinetics or magnitude of CD28 signaling, we examined the expression of transcripts previously determined to be regulated upon CD28 ligand binding in Jurkat and/or CD4+ T-cells50. Jurkat cells transduced with CD28 WT or mutants with similar levels of expression (Supplemental Figure 4) were incubated for the indicated times with beads ligated with anti-CD3 antibody and CD86. CD28-positive Jurkat cells were chosen instead of CD28-null for this experiment to best model the heterozygous-mutant tumor as only one allele in the tumor cells is expected to be mutated.
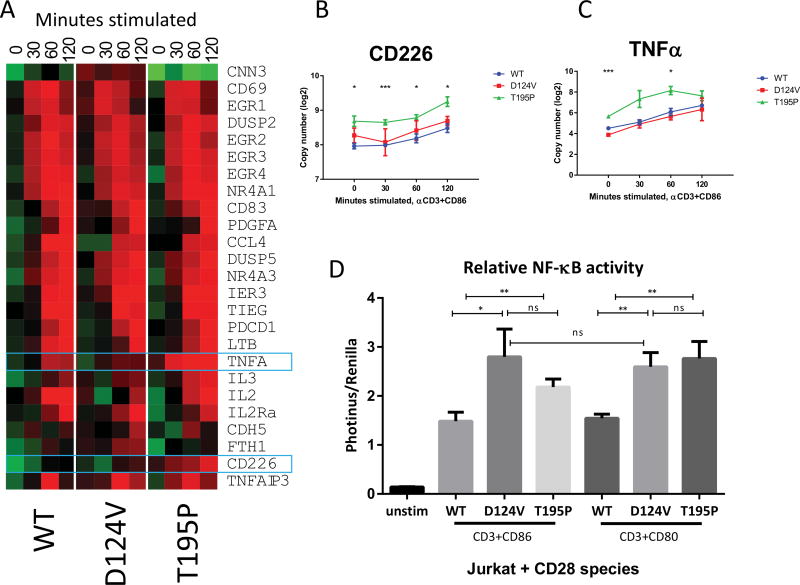
Expression profiles were similar among the samples (Fig. 4A); the Nanostring method, which directly counts the number of transcripts in a given sample, was chosen because it has greater reproducibility than qPCR36. Two of the assayed genes, CD226 and TNFA, showed upregulation in CD28 T195P over WT (4B, 4C). No assessed transcript was significantly different between CD28 D124V and WT.
Figure 4. CD28 D124V and T195P mutants alter transcriptional profiles and increase NF-κB signaling in response to CD28 ligation.
A: Jurkat cells stably transduced with GFP-containing retroviral constructs expressing CD28 WT or mutants were stimulated with beads ligated with anti-CD3+CD86 for the indicated time, and specific transcripts were quantitated using the Nanostring nCounter. The heat map was constructed from comparative expression profiling of the indicated genes. B–C: Two transcripts, CD226 and TNFA, showed significantly increased expression in cells expressing the CD28 T195P mutant. *: p<0.05 between T195P and WT; ***: p<0.005 between T195P and WT. There is no significant difference between the D124V and WT. D: Luciferase reporters of NF-κB activation were transduced into Jurkat cells expressing the indicated CD28 transgene and stimulated with the indicated beads for four hours. Diagram averages three replicates ± standard deviation. *, p<0.05; **, p<0.01; ns, not significant. Both CD28 mutants activated NF-κB more strongly than WT upon ligation of CD80 or CD86 with CD3 stimulation.
An NF-κB reporter vector was transfected into these Jurkat cells, and the cells were exposed to beads coated with anti-CD3 and either CD80 or CD86 (Fig. 4D). Compared to Jurkat cells transduced with CD28 WT, D124V showed a 1.9-fold greater NF-κB induction in response to CD80 and a 1.7-fold greater induction to CD86. T195P showed 1.5-fold and 1.8-fold higher NF-κB induction to CD80 and CD86, respectively. The increase in activation by the D124V mutant did not significantly differ between CD80 and CD86, an unexpected finding given the difference seen in the SPR (Fig. 2). We also compared the GEPs of the six CD28-mutant AITL cases to those of CD28 WT AITL cases and found 178 differentially-regulated genes (Supplemental Fig. 5A). Gene set enrichment analysis (GSEA) showed several differentially regulated pathways, most notably an increased T-cell signal transduction signature in CD28 mutant cases and a higher B-cell development signature in CD28 WT cases (Supplemental Fig. 5B–I).
Discussion
Recent sequencing studies on PTCLs, particularly AITL, have shown frequent mutations in RHOA and in the epigenetic modifiers TET2, DNMT3A, and IDH2, as well as less frequent mutations affecting TCR signaling proteins, including FYN. Our study finds similar frequencies and locations of TET2 mutations in AITL and other types of PTCL as previously determined, and we show that the less frequent CD28 mutations may play a role in promoting TCR and NF-κB signaling.
CD28 stimulation by ligand CD80 (B7.1) or CD86 (B7.2) is required for normal activation of T cells. Via multiple adaptor proteins, CD28 stimulation increases NFAT and NF-κB signaling, leading to proliferation, cytokine production, and the prevention of anergy. Thus, enhanced CD28 activation by mutation could lead to abnormal T-cell activation and contribute to the development of PTCL. Interestingly, CD28 expression in multiple myeloma has been shown to alter myeloma proliferation and survival51, and to be predictive of disease progression and relapse in conjunction with CD86, but not CD80, expression52,53.
In the current study, we found multiple CD28 mutations and a fusion transcript involving CD28 in PTCLs. One such mutation, T195P, was observed in AITL in previous studies12,54, including the demonstration of mildly enhanced CD28 signaling. Here, we report a number of novel findings regarding the CD28 mutations. In particular, our study suggests a survival disadvantage for AITL patients with tumors containing CD28 D124 and T195 mutations, demonstrates increased affinity of the CD28 mutants for ligand and interacting proteins, visualizes these interactions through molecular modeling, and quantifies the mutants’ influence on downstream signaling.
The fusion transcript between ICOS and CD28 is driven by the ICOS promoter. Because ICOS is highly expressed in AITL, it is expected that this fusion can drive higher CD28 expression (Supplementary Figure 1). However, the signal peptide, encoded by the first exon of ICOS, is cleaved from the final protein, such that the product only contains CD28 with the mutation. A recently reported case of Sézary syndrome had a CTLA4-CD28 fusion40, whose expression is expected to be driven by CTLA4; in this case, the extracellular domain of CD28 is replaced by CTLA4, which augments CD28 intracellular signaling through the higher-affinity CTLA4 extracellular domain. A study on adult T-cell leukemia/lymphoma41 found rare D124, T195, and F51 mutations, as well as both ICOS-CD28 and CTLA4-CD28 fusion transcripts. Although we found no CTLA4-CD28 fusions, we show that enhanced CD28 pathway activation can be achieved also through mutation. The D124V mutant, which occurs adjacent to the extracellular ligand-binding site, has an increased affinity for ligand CD86 shown by modeling and SPR analysis (Fig. 2–3). Our modeling of the interaction between CD86 and CD28 indicates that the increased affinity of the D124V mutant is likely due to improved electrostatic interactions as opposed to steric or hydrophobic factors. There is also evidence of enhanced downstream target activation. The fact that the D124V mutant has the same NF-κB activation with either CD80 or CD86 stimulation is unexpected based on the binding analysis (Fig. 2) which shows CD86, but not CD80, having a higher affinity for the D124V mutant. The model of the CD80-CD28 interaction shows several interactions that could explain these findings (Supplemental Fig. 3). On the cell surface, the orientation of CD28 homodimers is highly constrained, whereas in the SPR flow cell, they are three-dimensionally unconstrained. Three-dimensional Ka measurements by SPR of TCR/MHC interactions are approximately 1000-fold lower than the same analysis in two dimensions55. Thus, it is possible that a difference in affinity for CD80 between WT and mutant CD28 when confined to the plasma membrane may not be apparent by SPR analysis. Also, crystal structures of the modeled complexes would be useful, as they may reveal changes not predicted by the molecular modeling.
The T195P mutant has an increased affinity for adaptor proteins GRB2 and GADS/GRAP2 by SPR (Fig. 4). Our limited molecular modeling supports these SPR results and suggests that interactions between GRB2 residues R142 and N143 and the mutant CD28 T195P may stabilize the docking of GRB2 at the CD28 SH2-binding motif (Fig. 3).
There are several possible mechanisms by which increased ligand affinity may augment CD28 signaling. First, the increased affinity for SH2-containing adaptor proteins may directly increase CD28 signaling. Another possibility is through reduced receptor-mediated endocytosis. The PI3K regulatory subunit’s SH2 domain binds to the phosphotyrosine motif of the CD28 cytoplasmic tail and effects a wide range of changes, including CD28 receptor internalization56. Our binding assays show no significant difference between the CD28 WT and CD28 T195P binding of PI3K regulatory subunits (Fig. 2D). Because GRB2 and GADS/GRAP2 have a significantly higher affinity for the T195P mutant tail over WT, perhaps these adaptor proteins outcompete PI3K for binding to the CD28 SH2-binding motif and thereby compromise CD28 endocytosis. A decreased downregulation of CD28 signaling would have the same effect as direct over-activation, e.g. to augment cell cycling, proliferation, and cytokine production.
Analysis of gene expression using nCounter (Fig. 4) showed enhanced upregulation of two CD28-responsive genes, CD226 and TNFA, by CD28 T195P compared to WT. CD226 is crucial for TFH differentiation57 and can significantly modulate T-cell function by outcompeting its inhibitor TIGIT to bind receptor CD15558. TNFA is a potent cytokine that enhances proliferation, in part by inducing IκB-kinase phosphorylation and NF-κB activation59. It is also interesting that CD28-mutant AITL cases showed upregulation of the T-cell signal transduction signature over CD28 WT AITL cases (Supplemental Fig. 5B). One major consequence of TCR/CD28 signaling is the activation of the NF-κB signaling pathway, and both mutants demonstrate enhanced NF-κB activity using a luciferase reporter assay. Taken together, these data support the notion that the two CD28 mutations explored in this study may impart a functional advantage to CD28-mutant T-cells.
We also observed other recurrent possible CD28 mutations with variant frequencies below the standard cutoff for inclusion. One such mutation at F51 was observed in other T-cell lymphomas12,38, and we also detected mutations at this residue through our bioinformatics pipeline in the PTCL datasets of others (Supplemental Table 4), but we could not verify them in our own samples by ddPCR.
TET2 mutations tend to have the highest allelic frequencies among the common mutations in PTCL. CD28 mutations tend to have lower variant frequencies and may represent a late-stage mutation, giving a subclone a selective advantage. Alternatively, the low VF of CD28 mutations may represent not a subclone that has acquired a proliferative advantage but one that supports other cells in a paracrine fashion, such as through cytokine secretion. It would be highly interesting to determine the allelic frequency of CD28 mutants in refractory or relapsed disease to assess their contributions in these settings. The presence of CD28 mutations in PTCLs supports the importance of TCR/CD28 signaling from our GEP data. The CTLA4-CD28 fusions reported in Sézary syndrome as well as the other infrequent fusions and CD28 mutations found in adult T-cell leukemia/lymphoma40,41 underscore the relevance of T-cell activation via enhanced CD28 signaling as important for the pathogenesis of T-cell lymphomas and suggests that targeted treatments against these activation steps may yield novel methods to treat these cancers.
Supplementary Material
Acknowledgments
We thank R. Redder and Drs. A. Dhar and J. Eudy of the UNMC sequencing core; V. Smith and Dr. P Hexley of the UNMC flow cytometry core; and L. Brown, N. Feng, and S. Hsueh of the COH Analytical Cytometry Core. We would like to thank Elizabeth Chavez at BCCA for performing the TSCA experiment. Thanks to Dr. Françoise Berger of Université Lyon 1 for contributing several samples to this study. WCC is supported by NCI SPECS II 5 UO1 CA157581-01, NCI SPORE 1P50CA 136411-01 01A1 PP-4, and City of Hope internal funds; JI is supported by the Lymphoma Research Foundation (F-263549), the Leukemia and Lymphoma Society (TRP-6129-04), the UNMC Clinical-Translational Research Scholars Program, and the NCI Eppley Cancer Center Support Grant P30CA036727. AM is supported by the Dr. Mildred-Scheel Cancer Foundation and the Michael Smith Foundation for Health Research. Partial support is from NCI Eppley Cancer Center Support Grant P30CA036727, National Center for Research Resources 5P20RR016469, and National Institute for General Medical Science 8P20GM103427 to GB. The University of Nebraska DNA Sequencing Core receives partial support from the National Center for Research Resources (1S10RR027754-01, 5P20RR016469, RR018788-08) and the National Institute for General Medical Science (8P20GM103427, GM103471-09). Research reported in this publication included work performed in the City of Hope Analytical Cytometry Core supported by the NCI and NIH under award number P30CA33572. This publication’s contents are the sole responsibility of the authors and do not necessarily represent the official views of the NIH, FDA, or NIGMS.
Footnotes
Conflict of interest disclosure
K.F. is a potential inventor on a patent application using Nanostring technology for the Lymph2Cx assay, which has been licensed from the NIH by Nanostring Patents & Royalties
Authorship
Contributions
J.R. designed and performed research, analyzed data, and wrote the manuscript.
S.G. designed and performed research
Y.L. and C.S. designed research.
J.H. and S.D. designed and performed SPR analysis.
A.B., C.L., A.M., T.H., and A.C. performed research.
P.S. and G.B. designed and performed molecular modeling analysis.
W.Z., Q.G., C.W., S.H., and W.X. analyzed sequencing data.
A.R., R.G., K.F., T.G., D.W., and J.V. provided clinical samples and edited the manuscript.
T.M. and J.I. designed research, analyzed data, and edited and approved the final version of the manuscript.
W.C.C. supervised and designed research, and edited and approved the final version of the manuscript.
Supplementary information is available at Leukemia’s website
References
- 1.A clinical evaluation of the International Lymphoma Study Group classification of non-Hodgkin's lymphoma. The Non-Hodgkin's Lymphoma Classification Project. Blood. 1997 Jun 1;89(11):3909–18. [PubMed] [Google Scholar]
- 2.Mourad N, Mounier N, Briere J, Raffoux E, Delmer A, Feller A, et al. Clinical, biologic, and pathologic features in 157 patients with angioimmunoblastic T-cell lymphoma treated within the Groupe d'Etude des Lymphomes de l'Adulte (GELA) trials. Blood. 2008 May 1;111(9):4463–70. doi: 10.1182/blood-2007-08-105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood. 2007 Jun 1;109(11):4952–63. doi: 10.1182/blood-2006-10-055145. [DOI] [PubMed] [Google Scholar]
- 4.Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, et al. Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood. 2010 Feb 4;115(5):1026–36. doi: 10.1182/blood-2009-06-227579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krenacs L, Schaerli P, Kis G, Bagdi E. Phenotype of neoplastic cells in angioimmunoblastic T-cell lymphoma is consistent with activated follicular B helper T cells. Blood. 2006 Aug 1;108(3):1110–1. doi: 10.1182/blood-2006-01-0394. [DOI] [PubMed] [Google Scholar]
- 6.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011 Jul 12;20(1):25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 7.Cairns RA, Iqbal J, Lemonnier F, Kucuk C, de Leval L, Jais JP, et al. IDH2 mutations are frequent in angioimmunoblastic T-cell lymphoma. Blood. 2012 Feb 23;119(8):1901–3. doi: 10.1182/blood-2011-11-391748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C, McKeithan TW, Gong Q, Zhang W, Bouska A, Rosenwald A, et al. IDH2R172 mutations define a unique subgroup of patients with angioimmunoblastic T-cell lymphoma. Blood. 2015 Oct 8;126(15):1741–52. doi: 10.1182/blood-2015-05-644591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014 Feb 27;123(9):1293–6. doi: 10.1182/blood-2013-10-531509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet. 2014 Feb;46(2):166–70. doi: 10.1038/ng.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet. 2014 Feb;46(2):171–5. doi: 10.1038/ng.2872. [DOI] [PubMed] [Google Scholar]
- 12.Yoo HY, Sung MK, Lee SH, Kim S, Lee H, Park S, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014 Apr;46(4):371–5. doi: 10.1038/ng.2916. [DOI] [PubMed] [Google Scholar]
- 13.Iqbal J, Wright G, Wang C, Rosenwald A, Gascoyne RD, Weisenburger DD, et al. Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood. 2014 May 8;123(19):2915–23. doi: 10.1182/blood-2013-11-536359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greaves P, Gribben JG. The role of B7 family molecules in hematologic malignancy. Blood. 2013 Jan 31;121(5):734–44. doi: 10.1182/blood-2012-10-385591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gimmi CD, Freeman GJ, Gribben JG, Gray G, Nadler LM. Human T-cell clonal anergy is induced by antigen presentation in the absence of B7 costimulation. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6586–90. doi: 10.1073/pnas.90.14.6586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudd CE, Schneider H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat Rev Immunol. 2003 Jul;3(7):544–56. doi: 10.1038/nri1131. [DOI] [PubMed] [Google Scholar]
- 17.Boomer JS, Green JM. An enigmatic tail of CD28 signaling. Cold Spring Harb Perspect Biol. 2010 Aug;2(8):a002436. doi: 10.1101/cshperspect.a002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009 May 1;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010 May;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yost SE, Alakus H, Matsui H, Schwab RB, Jepsen K, Frazer KA, et al. Mutascope: sensitive detection of somatic mutations from deep amplicon sequencing. Bioinformatics. 2013 Aug 1;29(15):1908–9. doi: 10.1093/bioinformatics/btt305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012 Mar;22(3):568–76. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Therneau TM. A Package for Survival Analysis in S. 2015 [Google Scholar]
- 25.Evans EJ, Castro MA, O'Brien R, Kearney A, Walsh H, Sparks LM, et al. Crystal structure and binding properties of the CD2 and CD244 (2B4)-binding protein, CD48. J Biol Chem. 2006 Sep 29;281(39):29309–20. doi: 10.1074/jbc.M601314200. [DOI] [PubMed] [Google Scholar]
- 26.Collins AV, Brodie DW, Gilbert RJ, Iaboni A, Manso-Sancho R, Walse B, et al. The interaction properties of costimulatory molecules revisited. Immunity. 2002 Aug;17(2):201–10. doi: 10.1016/s1074-7613(02)00362-x. [DOI] [PubMed] [Google Scholar]
- 27.van der Merwe PA, Bodian DL, Daenke S, Linsley P, Davis SJ. CD80 (B7-1) binds both CD28 and CTLA-4 with a low affinity and very fast kinetics. J Exp Med. 1997 Feb 3;185(3):393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans EJ, Esnouf RM, Manso-Sancho R, Gilbert RJ, James JR, Yu C, et al. Crystal structure of a soluble CD28-Fab complex. Nat Immunol. 2005 Mar;6(3):271–9. doi: 10.1038/ni1170. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz JC, Zhang X, Fedorov AA, Nathenson SG, Almo SC. Structural basis for co-stimulation by the human CTLA-4/B7-2 complex. Nature. 2001 Mar 29;410(6828):604–8. doi: 10.1038/35069112. [DOI] [PubMed] [Google Scholar]
- 30.Stamper CC, Zhang Y, Tobin JF, Erbe DV, Ikemizu S, Davis SJ, et al. Crystal structure of the B7-1/CTLA-4 complex that inhibits human immune responses. Nature. 2001 Mar 29;410(6828):608–11. doi: 10.1038/35069118. [DOI] [PubMed] [Google Scholar]
- 31.Higo K, Ikura T, Oda M, Morii H, Takahashi J, Abe R, et al. High resolution crystal structure of the Grb2 SH2 domain with a phosphopeptide derived from CD28. PLoS One. 2013;8(9):e74482. doi: 10.1371/journal.pone.0074482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simone PD, Struble LR, Kellezi A, Brown CA, Grabow CE, Khutsishvili I, et al. The human ITPA polymorphic variant P32T is destabilized by the unpacking of the hydrophobic core. J Struct Biol. 2013 Jun;182(3):197–208. doi: 10.1016/j.jsb.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simone LC, Georgesen CJ, Simone PD, Wang X, Solheim JC. Productive association between MHC class I and tapasin requires the tapasin transmembrane/cytosolic region and the tapasin C-terminal Ig-like domain. Mol Immunol. 2013 Jan;49(4):628–39. doi: 10.1016/j.molimm.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krishna SS, Aravind L, Bakolitsa C, Caruthers J, Carlton D, Miller MD, et al. The structure of SSO2064, the first representative of Pfam family PF01796, reveals a novel two-domain zinc-ribbon OB-fold architecture with a potential acyl-CoA-binding role. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Oct 1;66(Pt 10):1160–6. doi: 10.1107/S1744309110002514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kucuk C, Iqbal J, Hu X, Gaulard P, De Leval L, Srivastava G, et al. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc Natl Acad Sci U S A. 2011 Dec 13;108(50):20119–24. doi: 10.1073/pnas.1115128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008 Mar;26(3):317–25. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 37.Weinstein JS, Lezon-Geyda K, Maksimova Y, Craft S, Zhang Y, Su M, et al. Global transcriptome analysis and enhancer landscape of human primary T follicular helper and T effector lymphocytes. Blood. 2014 Dec 11;124(25):3719–29. doi: 10.1182/blood-2014-06-582700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi J, Goh G, Walradt T, Hong BS, Bunick CG, Chen K, et al. Genomic landscape of cutaneous T cell lymphoma. Nat Genet. 2015 Sep;47(9):1011–9. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hindson BJ, Ness KD, Masquelier DA, Belgrader P, Heredia NJ, Makarewicz AJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011 Nov 15;83(22):8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekulic A, Liang WS, Tembe W, Izatt T, Kruglyak S, Kiefer JA, et al. Personalized treatment of Sezary syndrome by targeting a novel CTLA4:CD28 fusion. Mol Genet Genomic Med. 2015 Mar;3(2):130–6. doi: 10.1002/mgg3.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga JI, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015 Oct 5; doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 42.Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025. doi: 10.1038/ncomms7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bajorath J, Metzler WJ, Linsley PS. Molecular modeling of CD28 and three-dimensional analysis of residue conservation in the CD28/CD152 family. J Mol Graph Model. 1997 Apr;15(2):135–9. 08–11. doi: 10.1016/S1093-3263(97)00020-X. [DOI] [PubMed] [Google Scholar]
- 44.Linsley PS, Brady W, Grosmaire L, Aruffo A, Damle NK, Ledbetter JA. Binding of the B cell activation antigen B7 to CD28 costimulates T cell proliferation and interleukin 2 mRNA accumulation. J Exp Med. 1991 Mar 1;173(3):721–30. doi: 10.1084/jem.173.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linsley PS, Brady W, Urnes M, Grosmaire LS, Damle NK, Ledbetter JA. CTLA-4 is a second receptor for the B cell activation antigen B7. J Exp Med. 1991 Sep 1;174(3):561–9. doi: 10.1084/jem.174.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Linsley PS, Greene JL, Brady W, Bajorath J, Ledbetter JA, Peach R. Human B7-1 (CD80) and B7-2 (CD86) bind with similar avidities but distinct kinetics to CD28 and CTLA-4 receptors. Immunity. 1994 Dec;1(9):793–801. doi: 10.1016/s1074-7613(94)80021-9. [DOI] [PubMed] [Google Scholar]
- 47.Watanabe R, Harada Y, Takeda K, Takahashi J, Ohnuki K, Ogawa S, et al. Grb2 and Gads exhibit different interactions with CD28 and play distinct roles in CD28-mediated costimulation. J Immunol. 2006 Jul 15;177(2):1085–91. doi: 10.4049/jimmunol.177.2.1085. [DOI] [PubMed] [Google Scholar]
- 48.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: Proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003 Jul 1;31(13):3635–41. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003 Feb 19;125(7):1731–7. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 50.Riley JL, Mao M, Kobayashi S, Biery M, Burchard J, Cavet G, et al. Modulation of TCR-induced transcriptional profiles by ligation of CD28, ICOS, and CTLA-4 receptors. Proc Natl Acad Sci U S A. 2002 Sep 3;99(18):11790–5. doi: 10.1073/pnas.162359999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bahlis NJ, King AM, Kolonias D, Carlson LM, Liu HY, Hussein MA, et al. CD28-mediated regulation of multiple myeloma cell proliferation and survival. Blood. 2007 Jun 1;109(11):5002–10. doi: 10.1182/blood-2006-03-012542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robillard N, Jego G, Pellat-Deceunynck C, Pineau D, Puthier D, Mellerin MP, et al. CD28, a marker associated with tumoral expansion in multiple myeloma. Clin Cancer Res. 1998 Jun;4(6):1521–6. [PubMed] [Google Scholar]
- 53.Pope B, Brown RD, Gibson J, Yuen E, Joshua D. B7-2-positive myeloma: incidence, clinical characteristics, prognostic significance, and implications for tumor immunotherapy. Blood. 2000 Aug 15;96(4):1274–9. [PubMed] [Google Scholar]
- 54.Lee SH, Kim JS, Kim J, Kim SJ, Kim WS, Lee S, et al. A highly recurrent novel missense mutation in CD28 among angioimmunoblastic T-cell lymphoma patients. Haematologica. 2015 Sep 24; doi: 10.3324/haematol.2015.133074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang J, Zarnitsyna VI, Liu B, Edwards LJ, Jiang N, Evavold BD, et al. The kinetics of two-dimensional TCR and pMHC interactions determine T-cell responsiveness. Nature. 2010 Apr 8;464(7290):932–6. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cefai D, Schneider H, Matangkasombut O, Kang H, Brody J, Rudd CE. CD28 receptor endocytosis is targeted by mutations that disrupt phosphatidylinositol 3-kinase binding and costimulation. J Immunol. 1998 Mar 1;160(5):2223–30. [PubMed] [Google Scholar]
- 57.Seth S, Ravens I, Kremmer E, Maier MK, Hadis U, Hardtke S, et al. Abundance of follicular helper T cells in Peyer's patches is modulated by CD155. Eur J Immunol. 2009 Nov;39(11):3160–70. doi: 10.1002/eji.200939470. [DOI] [PubMed] [Google Scholar]
- 58.Johnston RJ, Comps-Agrar L, Hackney J, Yu X, Huseni M, Yang Y, et al. The immunoreceptor TIGIT regulates antitumor and antiviral CD8(+) T cell effector function. Cancer Cell. 2014 Dec 8;26(6):923–37. doi: 10.1016/j.ccell.2014.10.018. [DOI] [PubMed] [Google Scholar]
- 59.Sakurai H, Suzuki S, Kawasaki N, Nakano H, Okazaki T, Chino A, et al. Tumor necrosis factor-alpha-induced IKK phosphorylation of NF-kappaB p65 on serine 536 is mediated through the TRAF2, TRAF5, and TAK1 signaling pathway. J Biol Chem. 2003 Sep 19;278(38):36916–23. doi: 10.1074/jbc.M301598200. [DOI] [PubMed] [Google Scholar]
- 60.Ren J, Wen L, Gao X, Jin C, Xue Y, Yao X. DOG 1.0: illustrator of protein domain structures. Cell Res. 2009 Feb;19(2):271–3. doi: 10.1038/cr.2009.6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.