Abstract
The RNA helicase Prp2 facilitates the remodeling of the spliceosomal Bact complex to the catalytically activated B* complex just before step one of splicing. As a high-resolution cryo-EM structure of the B* complex is currently lacking, the precise spliceosome remodeling events mediated by Prp2 remain poorly understood. To investigate the latter, we used chemical structure probing to compare the RNA structure of purified yeast Bact and B* complexes. Our studies reveal deviations from conventional RNA helices in the functionally important U6 snRNA internal stem–loop and U2/U6 helix Ib in the activated Bact complex, and to a lesser extent in B*. Interestingly, the N7 of U6–G60 of the catalytic triad becomes accessible to DMS modification in the B* complex, suggesting that the Hoogsteen interaction with U6–A52 is destabilized in B*. Our data show that Prp2 action does not unwind double-stranded RNA, but enhances the flexibility of the first step reactants, the pre-mRNA's 5′ splice site and branch site adenosine. Prp2 therefore appears to act primarily as an RNPase to achieve catalytic activation by liberating the first step reactants in preparation for catalysis of the first step of splicing.
Keywords: activated spliceosome, catalytic activation, RNA structure, Prp2 RNA helicase
INTRODUCTION
The removal of introns from nuclear pre-mRNA proceeds by way of two phosphoester transfer reactions and is catalyzed by the spliceosome, a large ribonucleoprotein (RNP) complex composed of the snRNPs U1, U2, U4/U6, and U5 and numerous proteins (Wahl et al. 2009). The spliceosome is a highly dynamic RNP machine that undergoes many changes in composition and conformation during its work cycle, which are driven by at least eight conserved DEAH/D-box ATPases or RNA helicases (Staley and Guthrie 1998; Wahl et al. 2009). The stepwise assembly of the spliceosome is initiated by the base-pairing of U1 snRNP with the 5′ splice site (ss) and U2 snRNP with the downstream branch site (BS) sequence, yielding the A complex. The U4/U6.U5 tri-snRNP, in which the U4 and U6 snRNAs are extensively base paired, associates with the A complex, and a short helix between the 3′ end of U6 and the 5′ end of U2 (U2/U6 helix II) is formed. The U1/5′ ss interaction is disrupted by the action of the DEAD-box ATPase Prp28, allowing base-pairing of the 5′ ss with the conserved ACAGA sequence of U6 snRNA, thus forming the spliceosomal B complex (Staley and Guthrie 1999). Subsequently, the activated spliceosome (Bact) and then the catalytically activated B* complex are generated. Step 1 of splicing then takes place, transforming the B* into the C complex, which contains the cleaved 5′ exon and the intron lariat 3′ exon splicing intermediates. Complex C catalyzes step 2 of splicing, during which the lariat intron is excised and the 5′ and 3′ exons are ligated.
The transformation of the precatalytic B complex into a catalytically active machine is a multistep process that requires extensive structural rearrangements. To generate the Bact complex, structural activation of the B complex is achieved by the Brr2 RNA helicase-mediated dissociation of the U4/U6 base-pairing interaction (Raghunathan and Guthrie 1998). This allows U6 snRNA to form the U6 internal stem–loop (ISL) and to engage in a new base pair formation with U2 snRNA, generating the U2/U6 helices Ia and Ib (Staley and Guthrie 1998; Wahl et al. 2009). These RNA structural elements are at the heart of the catalytic core of the spliceosome (Nilsen 1998). Biochemical and genetic studies in yeast provided strong evidence that within the Bact complex a catalytic U2/U6 RNA–RNA interaction network is assembled that is highly similar to the catalytic RNA network of group II self-splicing introns (Fica et al. 2013), and this was recently further confirmed by cryo-EM structures of yeast and human spliceosomes (Hang et al. 2015; Yan et al. 2015, 2016, 2017; Galej et al. 2016; Rauhut et al. 2016; Wan et al. 2016a,b; Bertram et al. 2017; Fica et al. 2017; Zhang et al. 2017). Thus, similarly to the catalytic domain V of group II introns, the U2/U6 helix Ib and the U6 ISL adopt a secondary structure in which a conserved AGC triad is situated 5 bp away from a conserved bulged nucleotide in U6 (U6–U80 in S. cerevisiae) (Keating et al. 2010). Moreover, the U6 catalytic metal ligands, situated in the triad and the bulge, correspond one-to-one to the domain V ligands. Finally, similarly to the group II intron catalytic core, a group II-like triple helix also appears to form in U6 snRNA to juxtapose the two metal binding sites at the U6–U80 bulge loop and the AGC triad. In yeast, this spliceosomal triplex is comprised of Hoogsteen interactions of the terminal G52 and A53 of the conserved U6 ACAGA box with nucleotides G60 and A59, and of U6–U80 with C61 of the AGC triad, respectively (Fig. 1A,B).
FIGURE 1.
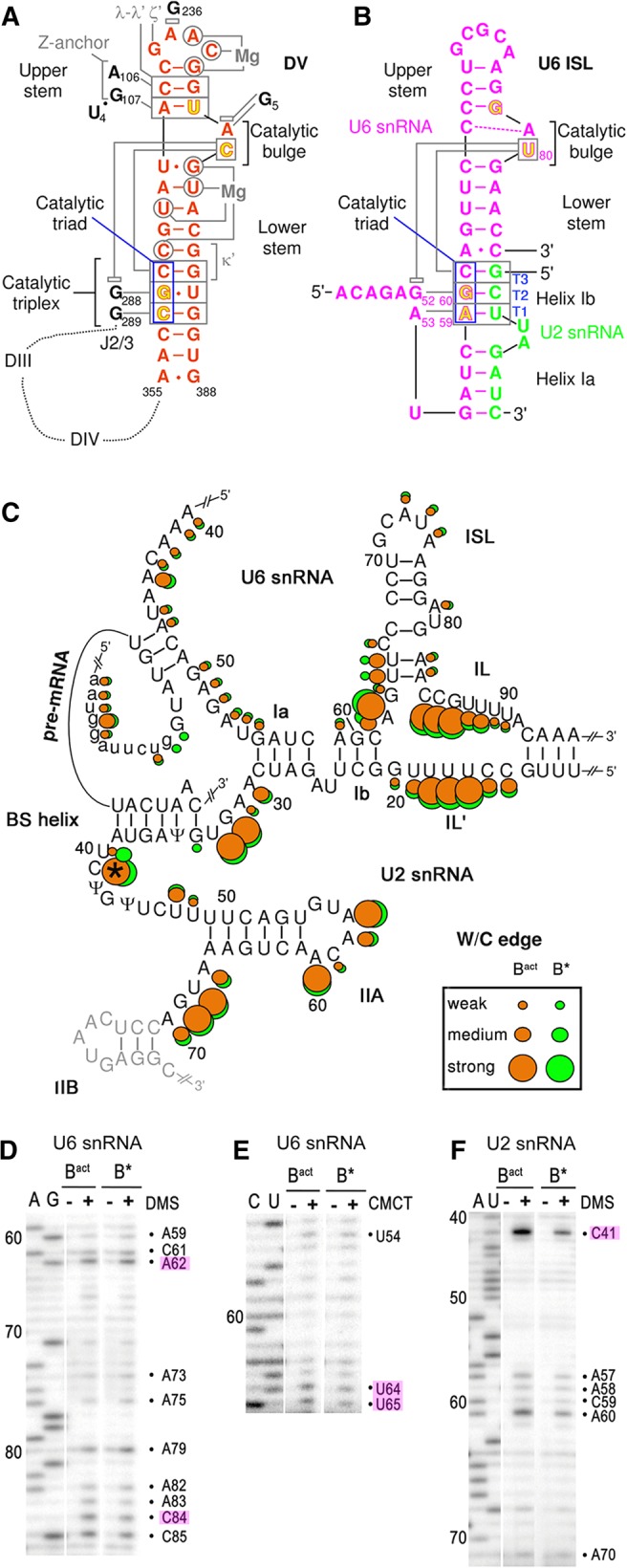
Structure of the core RNA–RNA network in yeast activated spliceosomes. (A) Secondary structure of the group II intron domain V (DV; adapted from Pyle 2010 with permission from Taylor and Francis © 2010). Essential RNA–RNA interactions with other intron elements are denoted with small Greek letters and nonstructural magnesium ions are indicated. The J2/3 linker is indicated and positions of intron domains III and IV are indicated schematically. Tertiary interactions are shown as gray lines, and stacking interactions as gray open rectangles. (B) Secondary structure of the yeast U6 ISL and U6/U2 helices Ia and Ib (adapted from Fica et al. 2014 with permission from Nature Publishing Group © 2014). Phosphates of nucleotides (colored in yellow with red outlines) that coordinate the two essential magnesium ions. Tertiary interactions of the AGC triad are shown as gray lines. Stacking of U6–G52 and U6–U80 is indicated (Fica et al. 2014). (C) Watson/Crick accessibilities of the nucleotides in the core of the spliceosomal RNA network in purified Bact (orange) and B* complexes (green). The size of the dots corresponds to the degree of accessibility to chemical modification. The asterisk denotes nucleotide U2–C41 that was hyperreactive toward DMS in the Bact complex. 5′ exon nucleotides are shown in lowercase letters. The last nucleotides of the 5′ exon are base-pairing to U5 loop 1 nucleotides. (D–F) Representative gels of the DMS (D) and CMCT (E) chemical modification of U6 snRNA, as well as DMS modification of U2 snRNA (F) in Bact and B* complexes (as indicated above the lanes), analyzed by primer extension. Key nucleotides are highlighted. See Supplemental Figures S1–S3 for the full data set of all RNAs and chemical probes. B* complexes were reconstituted by complementing purified Bact complexes with recombinant Prp2 and its cofactor Spp2 and ATP.
Cryo-EM 3D structures of the S. cerevisiae Bact complex (Rauhut et al. 2016; Yan et al. 2016) demonstrated that this intricate catalytic U2/U6 RNA network is already in place in the activated spliceosome. Moreover, the cryo-EM 3D structures of the yeast C complex (Galej et al. 2016; Wan et al. 2016a), the yeast and human C* complexes, which are activated for catalysis of step II of splicing (Bertram et al. 2017; Fica et al. 2017; Yan et al. 2017; Zhang et al. 2017), and of the S. pombe intron lariat spliceosome (ILS) (Hang et al. 2015; Yan et al. 2015), revealed that the catalytic U2/U6 RNA core exhibits a similar overall 3D structure during the two catalytic phases of the spliceosome. These cryo-EM structures also revealed how the 3D structure of the catalytic U2/U6 RNA core is stabilized by numerous protein–RNA interactions, in particular by Prp8 and members of the nineteen complex (NTC).
Despite the fact that the catalytic U2/U6 RNA network is fully established in the Bact spliceosome, this complex is still not catalytically active, as the 2′ hydroxyl of the BS-adenosine (BS-A) cannot attack the phosphodiester bond at the 5′ ss (Warkocki et al. 2009; Liu and Cheng 2012; Schneider et al. 2015). The molecular basis for this was revealed by the cryo-EM structures of the yeast Bact complex (Rauhut et al. 2016; Yan et al. 2016). First, the BS/U2 helix is clamped between the terminal HEAT repeats of the U2 SF3b protein Hsh155, and the BS-A is sequestered in a protein pocket formed by Hsh155 and the SF3b protein Rds3. The BS/U2 helix is oriented in such a way that the 5′ terminal end of U2 in the BS/U2 helix is ∼27 Å above the 3′ terminal end of U2 in the U2/U6 helix Ia and both helices are connected by the 4-nt-long U2 linker (Fig. 1C). The 2′ hydroxyl of the bulged BS-A is thus spatially separated from the scissile bond of the 5′ ss by ∼50 Å. In addition, the 5′ ss, although positioned close to the catalytic center, is shielded by proteins, including residues of Cwc24 and Prp11 (Rauhut et al. 2016; Yan et al. 2016). This molecular organization ensures that the first catalytic step of splicing does not occur prematurely during the B to Bact complex transition.
The final step of catalytic activation, that is, the conversion of Bact into the B* complex, is mediated by the DEAH-box ATPase Prp2 in conjunction with its G patch cofactor Spp2 (King and Beggs 1990; Kim and Lin 1996; Warkocki et al. 2009, 2015). Due to the lack of a cryo-EM 3D structure of the B* complex, the structural rearrangements that occur during this step are poorly understood. Biochemical studies revealed that the strength of interaction of a number of proteins with the spliceosome is affected during B* formation. For example, the Bact proteins Cwc24 and Cwc27 are largely displaced, and the interaction of the RES complex protein Bud13 is less stable, in affinity-purified B* complexes (Warkocki et al. 2009; Ohrt et al. 2012). Moreover, the U2 snRNP SF3a and SF3b protein complexes, which are known to bind to the intron upstream and downstream from the BS, are also destabilized (Warkocki et al. 2009; Lardelli et al. 2010; Schneider et al. 2015). The remodeling and destabilization of these proteins by Prp2/Spp2 suggests that the structure of the catalytic core, in particular in the vicinity of the BS and the 5′ ss, is also remodeled. Genetic data suggest that Prp2 might destabilize the catalytic RNA network and remodel U2/U6 helix I (Wlodaver and Staley 2014). However, it is presently not known whether Prp2, like the RNA helicases Prp28 and Brr2, facilitates changes in the spliceosomal RNA–RNA base-pairing pattern, or whether Prp2 instead acts primarily as an RNPase, that is, by remodeling protein–RNA interactions (Fairman et al. 2004).
Here, using a purified splicing system (Warkocki et al. 2009), we have performed chemical structure probing to investigate possible changes in the secondary and tertiary structure of the RNA network of the S. cerevisiae spliceosome during the Prp2-mediated transformation of the Bact complex into the catalytically active B* complex. We show that the U2/U6 snRNA duplex forms the conserved three-way junction in both complexes, but that there are deviations from conventional RNA helices at the lower stem of U6 ISL. Unexpectedly, N7 of U6–G60 in the catalytic triad becomes fully accessible in the B* complex. The major effect of Prp2 action is a significant increase in the flexibility of the first step reactants upon catalytic activation, without any evidence for changes in Watson–Crick RNA base-pairing. Prp2 thus appears to remodel RNA–protein units of the spliceosome in order to achieve liberation of the first step reactants for the first step of splicing.
RESULTS AND DISCUSSION
RNA structure probing of purified yeast Bact and B* complexes
S. cerevisiae Bact spliceosomes were assembled on an actin pre-mRNA containing MS2 aptamers, in heat-inactivated splicing extracts from a temperature-sensitive prp2-1 yeast strain, and were purified by glycerol-gradient centrifugation, followed by MS2-MBP affinity chromatography. To generate B* complexes, purified Bact complexes were complemented with ATP and recombinant Prp2 and its cofactor Spp2; under these conditions ∼90% of Bact complexes are converted to B* (Warkocki et al. 2009). To determine the RNA structure of the Bact and B* complexes, the Watson/Crick (W/C) edges of RNA were probed with dimethylsulfate (DMS, A-, and C-specific), β-ethoxy-α-ketobutyraldehyde (kethoxal, G-specific), or 1-cyclohexyl1-3-(2-morpholinoethyl) carbodiimide metho-p-toluene (CMCT, U-specific with low reactivity toward G). With these reagents, reactivity is only observed with unpaired nucleotides, providing information about RNA secondary structure (Ehresmann et al. 1987). Single-stranded nucleotides protected by protein are unreactive. The flexibility of the 2′-OH groups was probed by selective 2′-hydroxyl acylation followed by primer extension (SHAPE) (Merino et al. 2005). With SHAPE, 2′-hydroxyl groups of helical or otherwise constrained nucleotides show decreased reactivity (Merino et al. 2005; Wilkinson et al. 2005; Gherghe et al. 2010), while those of flexible nucleotides exhibit greater reactivity (Weeks 2010). SHAPE thus directly measures RNA motion and is largely independent of solvent accessibility (McGinnis et al. 2012). To monitor tertiary RNA interactions, we probed the accessibility of the N7 position of purines with diethylpyrocarbonate (DEPC; specific for N7-A) or DMS (specific for N7-G), followed by aniline-induced strand scission (Ehresmann et al. 1987). All modifications were analyzed by primer extension of the RNAs at the catalytic center of the spliceosome (i.e., the pre-mRNA, U2, and U6 snRNAs). In all cases, the primer extension analysis leads to a stop 1 nt before the chemically modified nucleotide. The RNA structure in the Bact and B* spliceosomal complexes was analyzed in parallel, and the results are summarized in Figures 1–3 with representative data sets (see Supplemental Figs. S1–S3 for the full data set).
FIGURE 3.
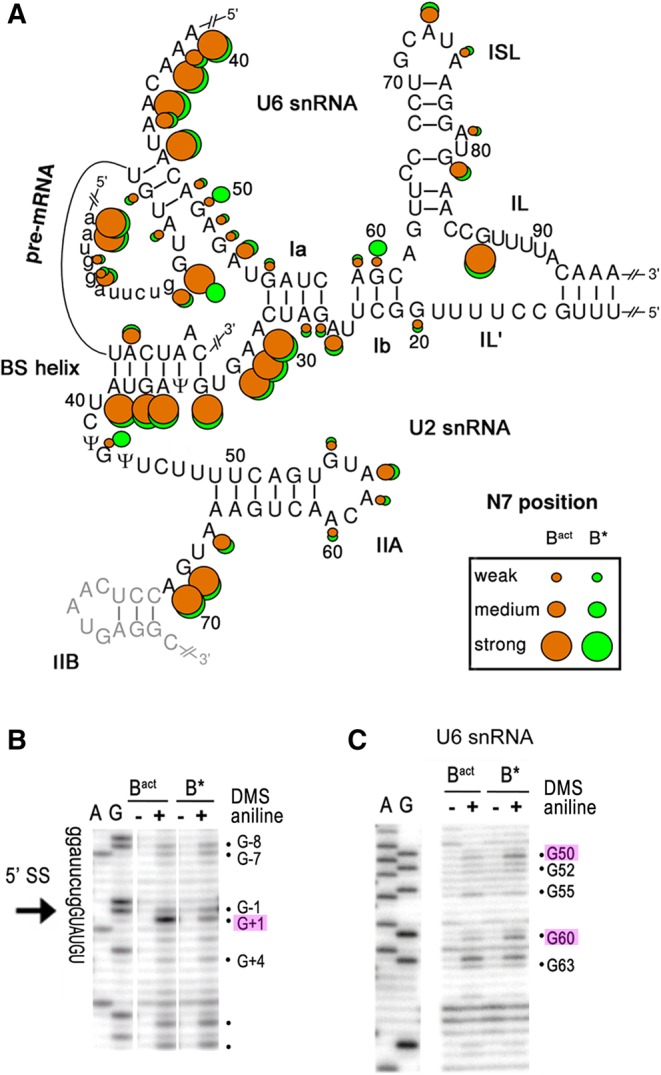
Analysis of tertiary RNA interactions in the catalytic core of the yeast Bact and B* spliceosomes. (A) N7-purine accessibilities of the nucleotides of the spliceosomal RNAs in the Bact and B* complexes as indicated in Figure 1C. (B,C) Representative gels of the chemical modification of the pre-mRNA around the 5′ ss (B) and of the U6 snRNA (C) in purified Bact and B* complexes (as indicated above the lanes) analyzed by primer extension. Key nucleotides are highlighted.
Overall similarity between RNA structure probing and the 3D cryo-EM structure of the Bact complex
Reactivities and protections of the W/C positions in the Bact complex (orange dots in Fig. 1C) were in good overall agreement with the three-way junction model of the U2/U6 snRNA duplex as proposed previously (Madhani and Guthrie 1992). Thus, most of the nucleotides in U2/U6 helices Ia and Ib, in the upper part of the U6-ISL and in U2/U6 helix II, were well protected from chemical modification (Fig. 1C). In contrast, U6 nts 84–90 and U2 nts 14–19 exhibited strong W/C accessibilities (Fig. 1D; Supplemental Figs. S1A,C, S2A–C) and/or high flexibilities of their 2′-OH groups (Fig. 2A,B), indicating that both internal loop regions are single-stranded, consistent with the structural organization of the U2 and U6 snRNAs in the cryo-EM structure of the Bact complex (Rauhut et al. 2016; Yan et al. 2016).
FIGURE 2.
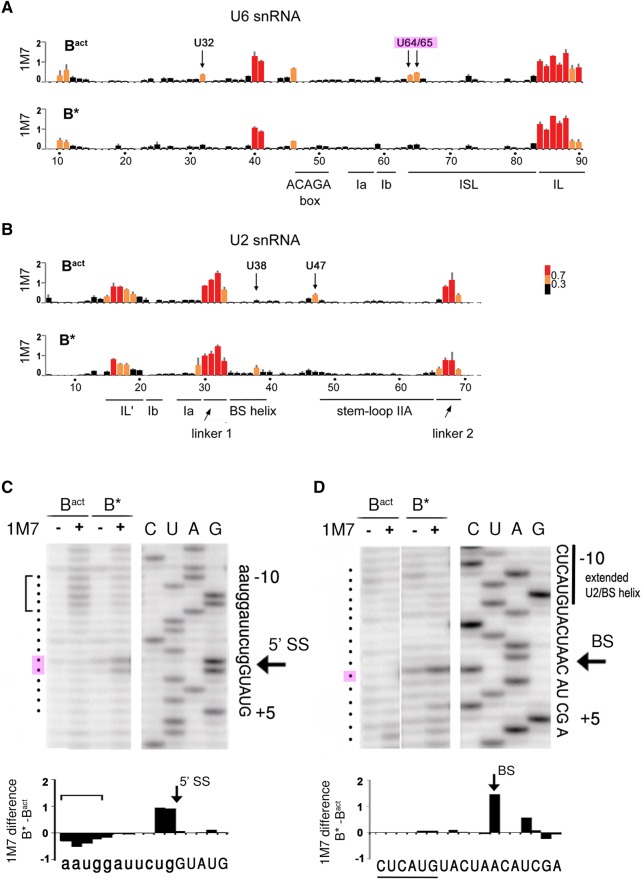
SHAPE analysis of the RNA–RNA network in yeast Bact and B* spliceosomes. (A,B) Shape analysis of U6 snRNA (A) and U2 snRNA (B) in Bact and B* complexes, as indicated. The data represent the mean of three independent experiments. Normalized reactivities are color-coded (inset) into three different categories of flexibility as previously described (Mortimer and Weeks 2007): black (0–0.3; not flexible, helically constrained), orange (0.3–0.7; intermediate flexibility, no internal Watson/Crick base pairs), and red (>0.7; fully unconstrained, single stranded). Key nucleotides are labeled above the top graph in each panel. Nucleotide numbers and the positions of structural elements are shown below the bottom graph in each panel. (C,D) Representative gels of the 1M7 reactivities of the pre-mRNA in Bact and B* around the 5′ splice site (C) and the branch site sequence (D) analyzed by primer extension. Primer extension will stop 1 nt before the 2′-O-acylated nucleotide relative to the dideoxy sequence. The bracket in C denotes the stretch of nucleotides with reduced 2′-OH flexibility in the B* complex. Quantifications of the autoradiographs in C and D were used for the difference plots (B* − Bact) shown below the respective autoradiographs. The sequences shown below the difference plots correspond to the actual nucleotide involved in attack or protection from 2′-O-acylation by 1M7.
Most of the U6 ACAGA box nucleotides and their complementary nucleotides near the 5′ end of the intron were not, or only very weakly, accessible to all chemicals, consistent with their involvement in helix formation (Fig. 1C). In addition, three single-stranded nucleotides at the 5′ ss of the pre-mRNA and six at the 3′ end of exon 1 were protected at their W/C edges (Fig. 1C; Supplemental Fig. S3A) and were not flexible based on SHAPE analyses (Fig. 2C), indicating protection by protein. This is consistent with the situation found in the cryo-EM structure of the Bact complex, where the G of the 5′ ss GU dinucleotide is caged by the Cwc24 and Prp11 proteins (Rauhut et al. 2016; Yan et al. 2016), with the latter extending to the U5 loop 1 that interacts with the 3′-most nucleotides of the 5′ exon.
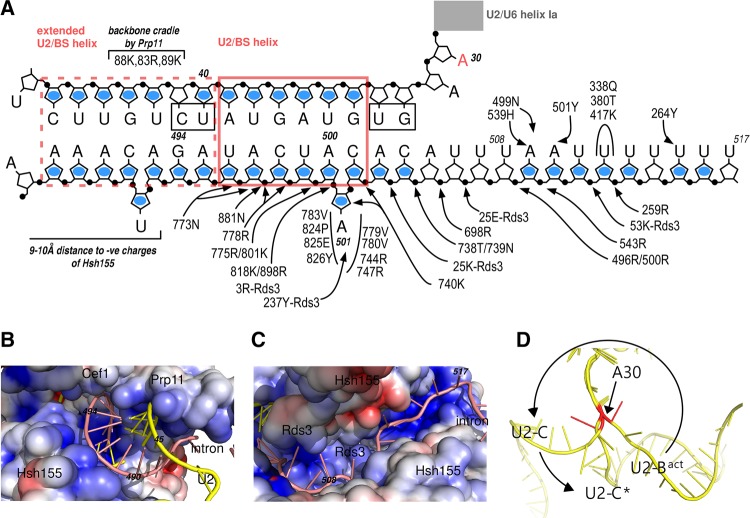
As expected for a helical structure, the W/C edges of the nucleotides of U2 snRNA and the intron that form the BS/U2 helix were not accessible (Fig. 1C; Supplemental Figs. S2A–C, S3E). The N7-purine positions of U2 snRNA nucleotides of the BS/U2 helix showed strong to medium accessibility, whereas the complementary nucleotides of the intron were not, or only weakly, reactive (Fig. 3A; Supplemental Figs. S2A,D, S3F,G). These observations suggest that the BS/U2 helix is distorted, with an accessible major groove on the U2 face (Weeks and Crothers 1993). This is most likely due to the binding of U2 SF3a and SF3b proteins to the region upstream of the BS region of the intron (McPheeters et al. 2000; Schneider et al. 2015; Cretu et al. 2016). Twelve intron nucleotides immediately upstream of the BS were not reactive at their W/C edges or were not flexible, or both (Figs. 2D, 4A; Supplemental Figs. S3E, S4). This is consistent with the cryo-EM structure of the yeast Bact complex (Yan et al. 2016), which shows that these intron nucleotides are indeed base paired and/or bound by the Prp11 and Hsh155 proteins (Fig. 4A–C). The RNA nucleotides immediately upstream of the BS constitute an extended BS/U2 helix (Fig. 4A), which may be particular to the pre-mRNA substrate used. However, a cavity is created by the proteins (Fig. 4B) that would allow for protection by protein–RNA interactions in the absence of a perfect RNA helical structure (Fig. 4C). Downstream from the BS, three patches of protection of the WC/edges were observed, which partially overlapped with regions of low flexibility (Supplemental Fig. S4). The cryo-EM structure of the Bact complex revealed that this region of the intron runs from the BS in a single-stranded fashion, across the superhelical α-solenoid-like structure of the Hsh155 protein (Fig. 4C). Strikingly, the W/C protections observed roughly align to regions where the RNA bases in the cryo-EM structure are contacted by the Hsh155 protein when they cross and exit from the α-solenoid formed by the Hsh155 HEAT repeats (Fig. 4A,C).
FIGURE 4.
RNA–protein interactions at/near the pre-mRNA branch site in the yeast Bact complex and the pivotal role of A30 of U2 snRNA. (A) Schematic of the region upstream and downstream from the branch site (A501), as found in the cryo-EM structure of the yeast Bact complex (pdb: 5GM6; EM density: EMD-9524), containing an ensemble of natural pre-mRNA (Yan et al. 2016). (Black dots) 5′ phosphates. (Blue dots) The homologous nucleotides of the actin pre-mRNA found protected or weakly accessible on their W/C positions (this manuscript, Supplemental Fig. S4). (Boxed nucleotides) Suboptimal base-pairing due to the large distance between the pairing partners. Amino acid side chain interactions are shown with arrows, with the protein only indicated when it is not Hsh155. The branch site is nucleotide A501 of the pre-mRNA; the U2/BS helix and the extended U2/BS helix are indicated. (B) Path of the extended U2/BS helix. The electrostatic surface of the cavity harboring the extended U2/BS helix and formed by Prp11 (U2 snRNA contacts) and Hsh155 (intron contacts). Intron sequences are not conserved and hence the backbone contacts are the major determinants for the paths of the RNAs. Charges are shown as heat maps, with blue for positive and red for negative potential (±5 kT). (C) The path of the single-stranded polypyrimidine tract across the superhelical α-solenoid structure of the Hsh155 HEAT repeats. Charges are as in B. (D) A close-up view of the counter-clockwise rotational movement of the region of U2 snRNA downstream from U2–A30 (red) encompassing the U2/BS helix and extended U2/BS helix. The coordinates from the cryo-EM structures from the Bact (5GM6, Yan et al. 2016) to C (5GMK, Wan et al. 2016a) to C* (5WSG, Yan et al. 2017) were aligned on U5 snRNA and only U2 snRNA (yellow) is shown. The U2/U6 helix Ia is behind the plane of the drawing. The pivotal U2–A30 is in red and is part of the flexible linker 1, AAGU (nts 30–33 of yeast U2 snRNA; Fig. 2B).
The conserved AAGU (nucleotides 30–33) U2 linker sequence (linker 1; nts 30–33) that connects the BS/U2 helix to helix Ia was fully accessible at the W/C edges and the N7-purine positions, and showed high flexibility (Figs. 1C, 2B, 3A; Supplemental Fig. S2). This region is thus single-stranded and its properties make it a perfect candidate to act as a flexible hinge during RNA helicase-mediated remodeling of the spliceosome for catalytic activation. In fact, such a rotation was instrumental in the initial modeling of the RNA network of the human C complex (Anokhina et al. 2013). A comparison of the yeast Bact (Rauhut et al. 2016; Yan et al. 2016), C (Galej et al. 2016; Wan et al. 2016a), and C* (Fica et al. 2017; Yan et al. 2017) complexes reveals that the major rotational movement of the U2 snRNA hinges on this flexible linker (Fig. 4D; Supplemental Fig. S6).
Deviations from conventional RNA helices in the lower stem of the U6 ISL and U2/U6 helix Ib in the Bact complex
The U6 ISL in the structures of the Bact and C complexes is generally depicted as composed of a lower and upper stem separated by a bulge structure (see Fig. 1B). Our structural probing however revealed subtle differences. First, U6–C84, at the base of the lower U6 ISL, showed strong W/C accessibility and high flexibility (Figs. 1C,D, 2A; Supplemental Fig. S1A), while U6–G63 was slightly accessible (Fig. 1C; Supplemental Fig. S1C) and not flexible (Fig. 2A), indicating that the two nucleotides do not form a canonical base pair. Second, we find that the two U nts following U6–G63 (U64 and U65) exhibited medium W/C reactivity (Fig. 1E; Supplemental Fig. S1B) and some flexibility (Fig. 2A; Supplemental Fig. 1A), but their base-pairing partners, A82 and A83 of U6 snRNA, showed low W/C reactivity (Fig. 1D; Supplemental Fig. S1A) and no flexibility (Fig. 2A). This suggests an inherent potential for breathing of the lower U6 ISL. Consistent with these findings, the W/C edges of U6–G63 and U6–C84 are ∼3.9 Å apart in the Bact structure (Rauhut et al. 2016; Yan et al. 2016). Further, U6–A62, which connects the lower U6 ISL with the U2/U6 helix Ib, was fully accessible at the W/C edge (Fig. 1D), but not flexible at all (Fig. 2A). This demonstrates that U6–G63 is not base-paired, but the nucleotide must be constrained otherwise. The likely cause is the single-stranded stack between U6 bases C61 and A62 to U65 that was observed in the cryo-EM structure of the yeast Bact complex (Rauhut et al. 2016). Finally, helix Ib also showed some asymmetric reactivities. For example, U6–C61 exhibited medium reactivity at its W/C edge (Fig. 1D; Supplemental Fig. S1A) but no flexibility (Fig. 2A); in contrast, its presumed base-pair partner, U2–G21, was only weakly modified (Supplemental Fig. 2A,C). Moreover, U6–A59 also showed weak W/C reactivity (Fig. 1D), while U2–U23 did not (Supplemental Fig. S2B). In summary, these asymmetric reactivities of the nucleotides in the lower stem of the U6 ISL and the U2/U6 helix Ib indicate significant deviations from perfect helices. These nucleotides (with the exception of U6–G63 and U6–C84) appear to be base paired in the cryo-EM structures of the yeast Bact complex (Rauhut et al. 2016; Yan et al. 2016), but they may breathe when in solution.
A dramatic increase in flexibility of first step reactants induced by Prp2-mediated catalytic activation
Next, we investigated the structural changes occurring in the RNA–RNA interaction network of the spliceosome during the Prp2-induced catalytic activation of the Bact complex to B*. First, our data indicated that none of the double-helical elements in the RNA catalytic core are disrupted in B* (Figs. 1–3; Supplemental Figs. S1–S3). This includes the three-way junction, the U6 ACAGA box region, and the extended BS helix (Fig. 4). At the bottom of the U6 ISL, U6–U64 and U6–U65 showed a decrease in W/C reactivity (Fig. 1C,E; Supplemental Fig. S1A,B) and a strong decrease in flexibility (Fig. 2A). This likely indicates consolidation/stabilization of the helical structure of the lower U6 ISL in preparation for catalysis. However, U6–C84 is still fully accessible in the B* complex, showing that U6–G63 is not base paired to U6–C84, not even directly prior to catalysis.
We did not detect any changes in the structure of the U2 snRNA during the Bact to B* transition (Fig. 1F; Supplemental Fig. 2A–C). Previous genetic data suggested that U2 snRNA toggles during catalytic activation between a conformation containing stem–loop IIA and a conformation where stem–loop IIA is completely unwound and instead U2 intramolecular helix IIC is formed (Hilliker et al. 2007; Perriman and Ares 2007). As cryo-EM structural data showed that helix IIC is likely present in the C (Galej et al. 2016; Wan et al. 2016a) and C* (Bertram et al. 2017; Fica et al. 2017; Yan et al. 2017) complexes, but we observe stem–loop IIA in both Bact and B*, our data indicate that U2 helix IIC first forms after catalytic activation by Prp2. This helix most likely forms as a result of the large-scale movement/repositioning of U2 snRNP, which occurs during step one of splicing (Bertram et al. 2017; Fica et al. 2017; Yan et al. 2017).
SHAPE analysis revealed a significant increase in the flexibility of the first step reactants upon Prp2 action. First, the 3′-most nucleotides of the 5′ exon, G-1 and U-2, showed a strong reaction toward the 1-methyl-7-nitroisatoic anhydride (1M7) acylation reagent only in the B* complex (Fig. 2C). As the 3′ position of G-1 is linked to the reactive phosphate of the first step of splicing, this implies that the observed increase in flexibility, i.e., structural disorder or RNA motion (McGinnis et al. 2012), involves the phosphate directly involved in catalytic step 1. The first nucleotide of the intron, G+1, bearing this phosphate at the 5′ position, is sequestered by the Cwc24 and Prp11 protein in the Bact complex (see above). The expulsion or destabilization of Cwc24 and Prp11 upon Prp2 action (Ohrt et al. 2012) results in a decrease in N7 accessibility in B* compared to Bact (Fig. 3B), suggesting that the base of G+1 forms new hydrogen bond interactions just before catalysis.
The second major change upon Prp2 activation that was detected by SHAPE is a significant increase in the 2′-OH flexibility of the BS-adenosine, as indicated by 1M7 acylation of its 2′-OH only in the B* complex (Fig. 2D). Considering that the BS is sequestered by the Hsh155/Rds3 proteins in the Bact complex (see above), the data imply that this sequestration is abolished by Prp2 action, allowing the increased flexibility of the first step nucleophile, the 2′-OH of the BS-adenosine. However, the situation appears to be more complicated as the BS and intron nucleotides upstream and downstream from it show very similar chemical accessibilities in the Bact and B* complexes (Supplemental Fig. S4), suggesting that protein contacts with the BS region do not change appreciably. Likely, the SF3a/b protein complexes are nonetheless repositioned within the spliceosome during Prp2 action and thus exhibit altered binding affinities with other spliceosomal proteins; this could lead to their partial loss upon affinity purification of the spliceosome after Prp2 action (Bessonov et al. 2008; Lardelli et al. 2010). In summary, our data provide direct evidence for the liberation of the two reacting partners of the first step of splicing by Prp2-mediated catalytic activation.
The structural properties of the BS/U2 helix, based on its overall W/C and N7 accessibilities, do not change substantially upon B* formation. In particular, the BS-adenosine remains protected after activation. However, subtle structural differences in the immediate vicinity of the BS/U2 helix are detected (Figs. 1C,F, 3A; Supplemental Fig. S2). There are minor increases in W/C accessibility of U2–G34 and in N7 accessibility of G43 in B* compared to Bact. In addition, the extreme W/C reactivity of U2–C41 is decreased while the W/C reactivity of the neighboring U40 is increased (Supplemental Fig. S2), and flexibility changes are detected by SHAPE at U33 in the linker region of U2, at U38 in the BS/U2 helix, and at the downstream U47 (Fig. 2B). These changes at the borders of the BS/U2 helix suggest that Prp2 acts on the RNP structure comprised of the Hsh155 and Rds3 proteins and the BS/U2 helix. Moreover, the data show that the structure of the BS/U2 helix does not change while the BS moves the large distance (∼50 Å) toward the catalytic center (Yan et al. 2015; Rauhut et al. 2016).
SHAPE analysis further demonstrated that four nucleotides of the 5′ exon (AAUG, located 5 nt upstream of the 5′ ss) lose much of their flexibility upon Prp2 action (Fig. 2C, bracketed nucleotides). In the Bact cryo-EM structure (Rauhut et al. 2016; Yan et al. 2016), this region of the 5′ exon lies in a cleft formed by the Cwc22 protein and part of the N-terminal domain of Prp8, with the two proteins forming the so-called exon entry channel (Supplemental Fig. S5). The observed decrease in flexibility suggests that the N-terminal domains of Prp8 and/or Cwc22 strengthen their grip on the 5′ exon in the B* complex. Together with the RNA–RNA interactions between U5 loop 1 and the 5′ exon, these protein–RNA interactions would ensure that the 3′-OH of the 5′ exon remains at the catalytic center after 5′ ss cleavage.
Two further changes were observed upon Prp2 action. First, U6–G50 in the ACAGA box showed increased N7-purine accessibility in B* compared to Bact (Fig. 3C), suggesting a repositioning of the unpaired base, perhaps by interacting with the Cus1 protein that is located nearby (Yan et al. 2016). The 5′ phosphate of U6–G50 was previously implicated in metal binding (Fabrizio and Abelson 1992), but at a stage before catalysis (Fica et al. 2013), and base repositioning may thus be related to the metal requirement for splicing catalysis. Second, U6–G60 of the catalytic triad showed an unanticipated increase in N7 accessibility (Fig. 3C; Supplemental Fig. S1D). U6–G60 is base paired with U2–C22 and is thought to form a base triple by Hoogsteen interaction with U6–G52 (Fig. 1B). However, our data show that in the catalytically activated B* complex this central base triple does not form or is substantially destabilized, suggesting deviations from this triple interaction in the B* complex. Similarly, the low levels of W/C reactivities found at U6 A59 and C61 in both the Bact and B* complex (Fig. 1C) suggest that base-pairing to U2 at these positions is relaxed.
Conclusions
Our investigation of the structural dynamics of the RNA–RNA network in the spliceosome during the poorly understood catalytic activation phase yielded three major novel insights. First, the structure probing data concerning the Bact complex were in full agreement with the cryo-EM structures (Rauhut et al. 2016; Yan et al. 2016), but provided additional information not readily apparent from the cryo-EM structures. These were the formation of an incomplete helix in the lower part of the U6 ISL, the complete lack of base-pairing involving U6–A62 and G63, and the breathing of U2/U6 helix Ib. These observations suggest that the junction between helix Ib and the U6 ISL is less rigid than previously thought and suggested by the homology with group II introns (Toor et al. 2008; Fica et al. 2013). Second, an additional effect of Prp2 during catalytic activation, i.e., B* complex formation, is the apparent loss of the triple interaction at the central position of the AGC catalytic triad. While this requires additional experimental investigation, the data suggest that this interaction is not essential for splicing. It is conceivable that the embedding of the RNA network in the protein scaffold within the spliceosome helps to stabilize Mg coordination at this stage and thereby can substitute for this triple interaction. Third, Prp2 action does not lead to substantial changes in RNA secondary structure. However, our data show a dramatic increase in flexibility of the first step reactants (the 5′ss and BS-A) upon catalytic activation and subtle structural dynamics around and within the U2/BS helix. In the Bact complex, the 5′ ss is already close to the catalytic center by virtue of its interaction with the U6 ACAGA box (Fica et al. 2014), but it is caged by the Cwc24 and Prp11 proteins (Yan et al. 2016), whereas the BS is sequestered in a sandwich between Hsh155 and Rds3 and is located ∼50 Å from the catalytic center (Rauhut et al. 2016; Yan et al. 2016). For catalytic activation, the sequestration of the first step reactants has to be relieved and the BS has to be moved into the catalytic center. As Prp2 is located on the periphery of the Bact spliceosome on the outer side of the Hsh155 HEAT repeats (Rauhut et al. 2016; Yan et al. 2016), catalytic activation must entail Prp2 action from a distance, i.e., from beyond the Hsh155 superhelical ring (Rauhut et al. 2016; Yan et al. 2016). Our observations suggest that the target of Prp2 action to achieve correct positioning of the branch site in the catalytic center is an RNP structure consisting of the SF3a and SF3b, the BS/U2 helix, and the intron fragment downstream from the BS. In addition, the highly flexible 4-nt-long linker connecting the BS/U2 helix to U2/U6 helix Ia allows for rotation of the entire downstream region of U2 snRNP around U2–A30 (Fig. 4D; Supplemental Fig. S6). This is accompanied by subtle local structural changes in nucleotides around and within the BS/U2 helix, as well as other important RNA elements, such as the 5′ ss and the catalytic triad regions. Prp2 thus remodels RNA–protein units of the spliceosome in order to liberate the reactants of the first step of splicing from their molecular embrace in preparation for catalysis of the first step of splicing. We will have to await further structural studies of the spliceosome, combined with biophysical investigations, before we understand the molecular details and molecular mechanism governing the mechanics of this evolutionary ancient ribonucleoprotein machine.
MATERIALS AND METHODS
Materials
Oligodeoxynucleotides were obtained from Eurofins Genomics. Synthesis of 1M7 was from 4-nitroisatoic anhydride (ABCR) as previously described (Mortimer and Weeks 2008).
Spliceosome preparation for chemical probing
Yeast Bact spliceosomes were assembled on actin pre-mRNA bound by MS2-MBP at the 5′ end (Warkocki et al. 2009), using whole-cell extracts from the yeast strain 3.2.AID (α, prp2-1, ade2, his3, lys2-801, ura3, carrying the G360D mutation in Prp2 [Kim and Lin 1996]). After MS2-MBP affinity purification on an amylose matrix, eluted Bact spliceosomes were further purified on glycerol gradients containing 150 mM KCl, 20 mM HEPES-KOH (pH 7.9), and 1.5 mM MgCl2. Bact spliceosomes from the peak fractions were further incubated with recombinant Prp2, Spp2, and ATP for 30 min at 23°C to generate the B* complex (Warkocki et al. 2009).
Chemical modification and analysis
Chemical modifications were performed essentially as previously described (Hartmuth et al. 1999; Anokhina et al. 2013), except for using 0.1–0.2 pmol of the respective complexes in a total volume of 200 µL containing 40 mM potassium phosphate buffer (pH 7.9), 75 mM KCl, 1.5 mM MgCl2, 5% (v/v) glycerol, and 10 µg tRNA were used. For SHAPE, spliceosomes were treated with 6.5 mM 1M7 at 25°C for 5 min. Modifications were analyzed by primer extension as previously described (Hartmuth et al. 1999) using 5′-end labeled oligodeoxynucleotides. For U6 snRNA, primer U6_94–112 (5′-AAAACGAAATAAATCTCTT-3′) was generally used except U6_68–84 (5′-GTTCATCCTTATGCAGG-3′) for Supplemental Figure S1B. For U2 snRNA, U2_74–91 (5′-AGGTAATGAGCCTCATTG-3′) was used. Pre-mRNA was analyzed by primers 5′-GTGCAATTCTTCTTACAGTT-3′ (Supplemental Fig. S3A), 5′-GCAATTGGGACCGTCG-3′ (Supplemental Fig. S3B–D), 5′-TACAATAACCAAAGCAGCAAC-3′ (Supplemental Fig. S3E–G). Data were quantified as previously described (Anokhina et al. 2013) using Quantity ONE (Bio-Rad). Signals were divided into three classes according to the fold increase observed over background: 1.1- to 2.4-fold increase, weak; 2.5- to threefold increase, medium; greater than threefold increase, strong. SHAPE data were additionally quantified using SAFA (Das et al. 2005) and normalized as previously described (Mortimer and Weeks 2007). The pymol program (http://www.pymol.com) was used to analyze and draw the molecular structures.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Zbigniew Warkocki for helpful discussions and protocols for spliceosome reconstitution, and Cindy L. Will for helpful comments on the manuscript. This work was supported by a grant from the Deutsche Forschungs Gemeinschaft (SFB860 to R.L.).
Author contributions: P.B., K.H., and R.L. designed the experiments. P.B. prepared spliceosomes and performed RNA structure probing and C.H. synthesized 1M7. P.B. and K.H. analyzed data and prepared figures. K.H., P.B., and R.L. wrote the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063115.117.
REFERENCES
- Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K, Lührmann R. 2013. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J 32: 2804–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram K, Agafonov DE, Liu WT, Dybkov O, Will CL, Hartmuth K, Urlaub H, Kastner B, Stark H, Lührmann R. 2017. Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature 542: 318–323. [DOI] [PubMed] [Google Scholar]
- Bessonov S, Anokhina M, Will CL, Urlaub H, Lührmann R. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452: 846–850. [DOI] [PubMed] [Google Scholar]
- Cretu C, Schmitzová J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, Will CL, Urlaub H, Lührmann R, Pena V. 2016. Molecular architecture of SF3b and structural consequences of its cancer-related mutations. Mol Cell 64: 307–319. [DOI] [PubMed] [Google Scholar]
- Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. 2005. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA 11: 344–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res 15: 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Abelson J. 1992. Thiophosphates in yeast U6 snRNA specifically affect pre-mRNA splicing in vitro. Nucleic Acids Res 20: 3659–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. 2004. Protein displacement by DExH/D “RNA helicases” without duplex unwinding. Science 304: 730–734. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. 2013. RNA catalyses nuclear pre-mRNA splicing. Nature 503: 229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Mefford MA, Piccirilli JA, Staley JP. 2014. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol 21: 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fica SM, Oubridge C, Galej WP, Wilkinson ME, Bai XC, Newman AJ, Nagai K. 2017. Structure of a spliceosome remodelled for exon ligation. Nature 542: 377–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Wilkinson ME, Fica SM, Oubridge C, Newman AJ, Nagai K. 2016. Cryo-EM structure of the spliceosome immediately after branching. Nature 537: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW Jr, Gorelick RJ, Rein A, Weeks KM. 2010. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc Natl Acad Sci 107: 19248–19253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J, Wan R, Yan C, Shi Y. 2015. Structural basis of pre-mRNA splicing. Science 349: 1191–1198. [DOI] [PubMed] [Google Scholar]
- Hartmuth K, Raker VA, Huber J, Branlant C, Lührmann R. 1999. An unusual chemical reactivity of Sm site adenosines strongly correlates with proper assembly of core U snRNP particles. J Mol Biol 285: 133–147. [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP. 2007. U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating KS, Toor N, Perlman PS, Pyle AM. 2010. A structural analysis of the group II intron active site and implications for the spliceosome. RNA 16: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin RJ. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 16: 6810–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DS, Beggs JD. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res 18: 6559–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HL, Cheng SC. 2012. The interaction of Prp2 with a defined region of the intron is required for the first splicing reaction. Mol Cell Biol 32: 5056–5066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. 1992. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817. [DOI] [PubMed] [Google Scholar]
- McGinnis JL, Dunkle JA, Cate JH, Weeks KM. 2012. The mechanisms of RNA SHAPE chemistry. J Am Chem Soc 134: 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Schwer B, Muhlenkamp P. 2000. Interaction of the yeast DExH-box RNA helicase Prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res 28: 1313–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J Am Chem Soc 129: 4144–4145. [DOI] [PubMed] [Google Scholar]
- Mortimer SA, Weeks KM. 2008. Time-resolved RNA SHAPE chemistry. J Am Chem Soc 130: 16178–16180. [DOI] [PubMed] [Google Scholar]
- Nilsen TW. 1998. RNA–RNA interactions in nuclear pre-mRNA splicing. In RNA structure and function (ed. Simons RW, Grunberg-Manago M), pp. 279–307. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, et al. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18: 1244–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman RJ, Ares M Jr. 2007. Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM. 2010. The tertiary structure of group II introns: implications for biological function and evolution. Crit Rev Biochem Mol Biol 45: 215–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. 1998. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol 8: 847–855. [DOI] [PubMed] [Google Scholar]
- Rauhut R, Fabrizio P, Dybkov O, Hartmuth K, Pena V, Chari A, Kumar V, Lee CT, Urlaub H, Kastner B, et al. 2016. Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science 353: 1399–1405. [DOI] [PubMed] [Google Scholar]
- Schneider C, Agafonov DE, Schmitzová J, Hartmuth K, Fabrizio P, Lührmann R. 2015. Dynamic contacts of U2, RES, Cwc25, Prp8 and Prp45 proteins with the pre-mRNA branch-site and 3′ splice site during catalytic activation and step 1 catalysis in yeast spliceosomes. PLoS Genet 11: e1005539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92: 315–326. [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. 1999. An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell 3: 55–64. [DOI] [PubMed] [Google Scholar]
- Toor N, Keating KS, Taylor SD, Pyle AM. 2008. Crystal structure of a self-spliced group II intron. Science 320: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Huang G, Shi Y. 2016a. Structure of a yeast catalytic step I spliceosome at 3.4 Å resolution. Science 353: 895–904. [DOI] [PubMed] [Google Scholar]
- Wan R, Yan C, Bai R, Wang L, Huang M, Wong CC, Shi Y. 2016b. The 3.8 Å structure of the U4/U6.U5 tri-snRNP: insights into spliceosome assembly and catalysis. Science 351: 466–475. [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243. [DOI] [PubMed] [Google Scholar]
- Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzová J, Höbartner C, Fabrizio P, Lührmann R. 2015. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev 29: 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM. 2010. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 20: 295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks KM, Crothers DM. 1993. Major groove accessibility of RNA. Science 261: 1574–1577. [DOI] [PubMed] [Google Scholar]
- Wilkinson KA, Merino EJ, Weeks KM. 2005. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J Am Chem Soc 127: 4659–4667. [DOI] [PubMed] [Google Scholar]
- Wlodaver AM, Staley JP. 2014. The DExD/H-box ATPase Prp2p destabilizes and proofreads the catalytic RNA core of the spliceosome. RNA 20: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Hang J, Wan R, Huang M, Wong CCL, Shi Y. 2015. Structure of a yeast spliceosome at 3.6-angstrom resolution. Science 349: 1182–1191. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2016. Structure of a yeast activated spliceosome at 3.5 Å resolution. Science 353: 904–911. [DOI] [PubMed] [Google Scholar]
- Yan C, Wan R, Bai R, Huang G, Shi Y. 2017. Structure of a yeast step II catalytically activated spliceosome. Science 355: 149–155. [DOI] [PubMed] [Google Scholar]
- Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y. 2017. An atomic structure of the human spliceosome. Cell 169: 918–929.e14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.