Abstract
Recent work has identified cancer-associated U2AF35 missense mutations in two zinc-finger (ZnF) domains, but little is known about Q157R/P substitutions within the second ZnF. Surprisingly, we find that the c.470A>G mutation not only leads to the Q157R substitution, but also creates an alternative 5′ splice site (ss) resulting in the deletion of four amino acids (Q157Rdel). Q157P, Q157R, and Q157Rdel control alternative splicing of distinct groups of exons in cell culture and in human patients, suggesting that missplicing of different targets may contribute to cellular aberrations. Our data emphasize the importance to explore missense mutations beyond altered protein sequence.
Keywords: U2AF, U2AF mutations and cancer, alternative splicing
INTRODUCTION
In higher eukaryotes, most genes contain noncoding introns in the pre-mRNA that have to be removed to yield the mature mRNA with a continuous open reading frame. Removal of introns and joining of exons is a two-step transesterification reaction catalyzed by the spliceosome. The spliceosome assembles de novo on every intron, which is enabled by the recognition of several sequence elements within the pre-mRNA (Wahl et al. 2009). For example, the major class of intron boundaries is marked by GU and AG dinucleotides, which are essential for splicing to occur. U2AF35 and its heterodimerization partner U2AF65 recognize the 3′ splice site and the polypyrimidine tract, respectively, in an early step during spliceosome assembly (Zamore et al. 1992; Merendino et al. 1999; Wu et al. 1999; Zorio and Blumenthal 1999). U2AF35 consists of two zinc-finger (ZnF) domains, an RNA-recognition motif (RRM), and a C-terminal arginine/serine-rich-domain (Zhang et al. 1992; Webb and Wise 2004). This architecture is highly conserved across species and several paralogs are found in mouse and human (Wu and Fu 2015). In a recent publication the crystal structure of the yeast U2AF35 homolog (U2AF23) was solved and, somewhat surprisingly, showed that RNA binding of U2AF23 is mainly mediated through the ZnF domains, whereas the “RRM” serves as a scaffold for the ZnF domains and as a protein–protein interaction surface (Yoshida et al. 2015). The ZnF domains of the human U2AF35 have received further attention since the recent identification of cancer-associated mutations within these domains. The first report connecting mutations of spliceosome components with oncogenesis was published in 2011 and was followed by many studies confirming and extending the initial observations (Yoshida et al. 2011; Graubert et al. 2012). In particular, heterozygous somatic mutations in two proteins required for early steps in spliceosome assembly, SF3B1 and U2AF35, have been identified in a variety of hematological malignancies such as myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), and chronic myelomonocytic leukemia (CMML). Furthermore, mutations in SF3B1 and U2AF35 occur in solid tumors like uveal melanoma, breast cancer, and lung adenocarcinoma (Hahn et al. 2015; Inoue et al. 2016). Myeloid neoplasm-associated mutations in U2AF35 include the more frequent S34F/Y substitutions (4.3% of all cases analyzed) and the rare Q157R (1.2%) and Q157P (0.7%) substitutions that fall within the first and second ZnF domain, respectively (Yoshida et al. 2011). While several studies have analyzed the more frequent S34F/Y mutations, little is known about the molecular consequences of the Q157R/P alleles; one study that included data from both Q157R and Q157P patients has actually combined the data of the two genotypes and treated them as equivalent (Qiu et al. 2016). It thus remains an open question whether there is a functional difference between the Q157R and Q157P variants affecting the same position within U2AF35. In contrast, expression of the S34F U2AF35 mutant was shown to alter pre-mRNA splicing in a variety of cell types and mice, leading, among other phenotypes, to altered hematopoiesis (Shirai et al. 2015). In cell culture experiments, the S34F mutant leads to reduced cell growth and increased cell death, which makes its precise role in malignant transformation less clear (Yoshida et al. 2011; Shirai et al. 2015). At the molecular level, the S34F mutant was shown to promote splicing of exons with a C or A preceding the AG of the 3′ss and to repress those with a T at this position (Przychodzen et al. 2013; Ilagan et al. 2014; Okeyo-Owuor et al. 2015; Shirai et al. 2015). The S34F mutant has also been suggested to control 3′ end processing of Atg7 pre-mRNA, leading to a defect in autophagy that provides a further link between the mutation and oncogenesis (Park et al. 2016). An additional connection between the spliceosome and oncogenesis has been shown for Myc-driven cancers, where the spliceosome has been suggested to become rate limiting for gene expression due to increased pre-mRNA synthesis. Targeting the core spliceosome using pharmacological inhibitors thus led to increased intron retention in the tumor but not in healthy tissue, where the splicing machinery can cope with reduced spliceosome activity (Hsu et al. 2015; Koh et al. 2015). These findings together make the spliceosome an interesting target to investigate molecular mechanisms of malignant transformation and to explore new therapeutic concepts.
In the present work, we have analyzed consequences of the Q157R and Q157P mutations in detail, in cell culture using a knockdown complementation assay with quantitative readout, as well as in patient data. We find that the c.470A>G mutation in addition to causing the Q157R substitution generates an alternative 5′ splice site that leads to the deletion of four amino acids within the second ZnF (Q157Rdel). The Q157Rdel variant is also present in c.470A>G patients, suggesting a contribution of this variant in disease development. We have quantified the effect of the Q157P, Q157R, and Q157Rdel mutants in our cell culture assay and show considerable differences between the different mutants. Importantly, these differences are also observed in RNA-seq data from patients carrying the respective mutations, confirming mutation-specific missplicing of different targets in vivo. Exons preferentially affected in Q157P-, Q157R-, and Q157Rdel-expressing cells show distinct 3′ splice site sequences, suggesting that the mutations alter the binding specificity of the respective proteins. In summary, we present the surprising finding that the c.470A>G mutation, considered to result in the Q157R amino acid change, also creates an alternative 5′ss that leads to a protein with altered functionality. More such cases are likely to be discovered if further disease-associated missense mutations are investigated beyond their role in changing the coding sequence. In addition, we define distinct splicing signatures in c.470A>G (Q157R, Q157del) and c.470A>C (Q157P) patients, thus contributing to a molecular understanding of these cancer-associated mutations.
RESULTS AND DISCUSSION
The cancer-associated U2AF35 c.470A>G (Q157R) mutation creates an alternative 5′ss
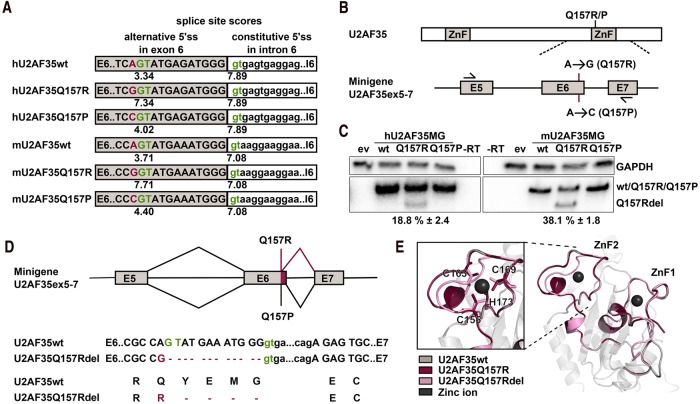
In human patients, the Q157R and Q157P mutations in U2AF35 are less frequent than the S34F allele, and therefore most studies have focused on the latter mutant. The mutations in the second ZnF are thus less well understood; in particular, it remains an open question whether they induce missplicing by the same molecular mechanism and whether the same set of exons is affected in Q157R and Q157P patients. To address these questions, we first analyzed the consequences of the two point mutants in more detail. By using splice site predictions, we noticed that the Q157R mutant in addition to altering the coding sequence also generates an alternative 5′ splice site with a splice site score only slightly below the score of the annotated splice site (MaxEntScan scores 7.34 vs 7.89) (Fig. 1A). To investigate whether the mutation leads to missplicing of U2AF35, we constructed a minigene comprising exons 5–7 of U2AF35, either as wt or with the Q157P or Q157R mutations, only the latter generating a potential alternative 5′ splice site in exon 6 (Fig. 1B). Transfecting these minigenes into HEK293T cells indeed revealed that the alternative 5′ss present in the Q157R minigene is used, leading to significant accumulation of the alternatively spliced, shortened product (Fig. 1C). This was not a cell type-specific effect, as we observed similar splicing patterns also in HeLa cells (Supplemental Fig. 1A). Consistent with a low splice site score in the Q157P mutant, this alternatively spliced product was specific for the Q157R mutation. To confirm that formation of the shorter product in the Q157R mutant was not due to aberrant splicing of this particular minigene, we introduced the mutation in the context of the mouse U2AF35 gene and observed a similar effect, which was consistent with a higher splice site score, even stronger than in the human minigene (Fig. 1A,C). Sequencing of the shortened product confirmed the usage of the alternative 5′ss, which deletes four amino acids within the second ZnF domain (Fig. 1D). The Q157Rdel mRNA did not show substantially altered stability, and as it is an in-frame deletion does not represent an NMD target (Supplemental Fig. 1B). At the amino acid level this deletion does not involve the zinc complexing cysteine and histidine residues, and structure predictions suggest the structure of the ZnF domain to remain functional (Fig. 1E). The structure prediction points to the loss of a short helix in Q157Rdel, which leads to a slightly altered orientation and distance between the Zn complexing residues (Supplemental Fig. 1C) but leaves the overall structure intact. However, since the ZnF domains within U2AF35 have been shown to make direct contact with the 3′ss of target exons (Yoshida et al. 2015), a changed structure of the ZnF may lead to altered binding specificity and thus altered target exons. Together, these data provide a compelling example for a missense mutation that has an additional, so far overlooked, effect on splicing regulation; such a connection has been suggested before (Cartegni et al. 2002), but only a few cases have been analyzed in detail (Ito et al. 2017), and a broader role for missense mutations that additionally control alternative splicing is only beginning to be appreciated (Soemedi et al. 2017).
FIGURE 1.
The U2AF35 Q157R mutation creates an alternative 5′ splice site. (A) Sequences of the exon 6 to intron 6 junction of human and mouse U2AF35. Constitutive and potential alternative donor splice sites are shown in green and nucleotides encoding AML-associated substitutions Q157R/P are shown in red. The numbers below the donor sites show the splice site scores. (B) Schematic depiction of minigene constructs spanning introns 4–8, including exons 5–7, and the introduced point mutations. (C) Minigene-specific, radioactive splicing-sensitive RT-PCR and quantification using HEK293T cells transfected with human and mouse U2AF35 minigenes or the empty vector (ev). GAPDH-specific radioactive RT-PCR was used as the loading control. (D) Schematic depiction of the splicing pattern of minigenes with the Q157R and Q157P mutation. Sequences of the exon 6 to exon 7 junction of wt U2AF35 and U2AF35 Q157Rdel and the translated proteins are shown. (E) Structure prediction of U2AF35wt, Q157R, and Q157Rdel with the coordinated zinc ions. The zoom on ZnF2 shows the zinc-coordinating cysteine and histidine residues as sticks.
U2AF35 Q157P, Q157R, and Q157Rdel control alternative splicing of different sets of exons
Since a potential difference in target exons of the Q157R and Q157P mutants has not been addressed in a quantitative manner and to directly test the role of the Q157Rdel mutant in splicing regulation, we set up a knockdown system in which missplicing of exons sensitive to loss of endogenous U2AF35 can be rescued by expression of U2AF35 variants. Endogenous U2AF35 mRNA was knocked down to around 10% using U2AF35-specific siRNA (Supplemental Fig. 2A) and replaced by the respective ectopically expressed, siRNA-resistant mutants. Western blot confirmed knockdown of endogenous U2AF35 and equal amounts of ectopically expressed U2AF35 variants (Fig. 2A). Knockdown of U2AF35 led to reduced cell growth and slightly altered cell cycle distribution, which was rescued by overexpression of wt but not the mutated U2AF35 versions (Supplemental Fig. 2B,C); this is consistent with previous observations showing that knockdown of U2AF35 inhibits cell proliferation and leads to accumulation of cells in G2/M phase (Pacheco et al. 2006). Furthermore, it was shown that overexpression of the Q157R/P variants leads to reduced proliferation (Shao et al. 2014). These results indicate that mutations in U2AF35 contribute to similar cellular aberrations, although how reduced proliferation can be linked to malignant transformation has still to be analyzed. To analyze the impact of the U2AF35 mutants on splicing, we interrogated splicing of 16 alternative exons. We selected these exons either as they showed a substantial decrease in inclusion upon U2AF35 knockdown or as they are misregulated in patients with the U2AF35 S34F mutation (RNF10, CHEK2, EIF4A2, THYN1, CEP164, TPD52L2, ASUN, RIPK2, FXR1, and SETX; Przychodzen et al. 2013; Shao et al. 2014). We also selected targets that were misregulated in Q157R or Q157P patients based on our subsequent analysis (see below; DLG1, FNTB, CEP57, DICER1, BCL2L12, and CHD3). In contrast to previous studies, we have used a well-established, radioactive RT-PCR protocol that allows a precise quantification of isoform ratios (Preußner et al. 2014; Schultz et al. 2016; Wilhelmi et al. 2016; example in Fig. 2B). The use of a radioactively labeled primer allows a low-cycle PCR, as detection is much more sensitive than with other methods, thus avoiding problems with saturating PCR conditions; in addition, exclusion and inclusion products are each labeled with one primer, i.e., the same amount of radioactivity, which is not the case in ethidium bromide stained gels, and Phosphorimager analysis allows accurate quantification of signal strength over several orders of magnitude.
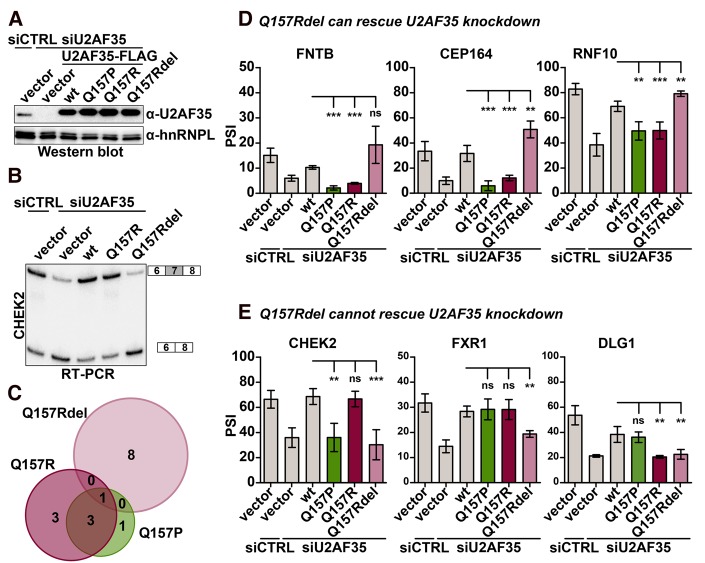
FIGURE 2.
The U2AF35 Q157R and Q157P alleles create three proteins that affect different target exons. (A) Western blot of endogenous U2AF35 in the presence and absence of siU2AF35 and rescue with ectopically expressed Flag-tagged U2AF35wt, Q157R, Q157Rdel, and Q157P variants in HEK293T cells. hnRNPL was used as the loading control. (B) Example of a splicing-sensitive radioactive RT-PCR for CHEK2 in HEK293T cells with a knockdown of endogenous U2AF35 and rescue with ectopically expressed U2AF35 mutants. (C) Venn diagram showing overlaps of Q157P, Q157R, and Q157Rdel rescued U2AF35-dependent splicing targets analyzed via radioactive RT-PCR. In total, splicing of 16 alternative exons was analyzed. (D,E) Quantifications of splicing-sensitive radioactive RT-PCRs as in B for targets where Q157Rdel can (D) or cannot (E) rescue U2AF35 knockdown. n ≥ 3; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Knockdown of U2AF35 resulted in decreased exon inclusion for 14 targets and increased inclusion for two targets (DICER1 and EIF4A2). Overexpression of U2AF35wt reversed this effect and led to a complete rescue of the splicing change for CEP164, CHEK2, FXR1, CHD3, EIF4A2, BCL2L12, CEP57, TPD52L2, and to a significant change compared to knockdown for the other eight targets exons, thus validating our experimental system (Fig. 2B,D,E; Supplemental Fig. 2D,E). As some cells may not be transfected with the expression construct, the actual rescue effect may be underestimated in this setup. Quantifying the effect of the Q157R, Q157P, and Q157Rdel variants revealed that Q157Rdel was able to rescue the knockdown of endogenous U2AF35 in nine out of 16 targets (FNTB, CEP164, RNF10 in Fig. 2C and DICER1, RIPK2, CHD3, EIF4A2, BCL2L12, CEP57 in Supplemental Fig. 2D); importantly, eight out of these nine targets were not rescued by the Q157R or Q157P mutants, showing that the Q157Rdel variant controls alternative splicing of a distinct set of target exons (Fig. 2C). Out of seven exons that were not rescued by the Q157Rdel variant, six could be rescued by the Q157R protein (CHEK2, FXR1, TPD52L2, SETX, ASUN, THYN1), further suggesting that the Q157R and Q157Rdel mutants affect largely nonoverlapping groups of exons (Fig. 2C–E; Supplemental Fig. 2D,E). As the Q157Rdel mutant is able to actively rescue the missplicing of specific targets upon U2AF35 knockdown, these results further indicate that the mutation does not abolish RNA binding but rather changes the sequence specificity. When coexpressed with wt U2AF35, the Q157Rdel variant did not act in a dominant-negative manner (Supplemental Fig. 2F), again supporting a sequence-specific role in controlling the alternative splicing of a specific set of target exons. Many examples for CCCH ZnF domains deviating from the classical C–X7/8–C–X5–C–X3–H arrangement are known, also with shorter linkers between the first two cysteine residues (Wang et al. 2008), suggesting that a C–X3–C–X5–C–X3–H structure, which is found in the Q157Rdel mutant, can remain functional. Whether the shorter ZnF domain shows altered RNA-binding affinity remains an open question that may be addressed once recombinant proteins are available. Together with the structure prediction, the functionality of the Q157Rdel mutant in regulating alternative splicing of specific exons strongly suggests that the second ZnF of U2AF35 tolerates the deletion of four amino acids, but that this leads to changes in binding specificity.
Target exons of U2AF35 Q157 mutants show distinct 3′ splice site signatures
Although the targeted analysis of 16 exons already suggests different rescue abilities of the three U2AF35 Q157 mutants, we used RNA-seq for a more comprehensive analysis. Knockdown of endogenous U2AF35 led to missplicing of around 940 cassette exons, 600 of which were rescued by expression of wt U2AF35 (see Materials and Methods for details). This result validates our cell culture system, because despite overexpression of U2AF35, splicing of ∼70% of the target exons can be rescued to control levels. In addition, splicing patterns of control siRNA transfected and wt U2AF35 rescued samples clustered together (Supplemental Fig. 5A), further supporting our experimental setup. Importantly, despite stringent filter criteria, nine out of the 16 exons analyzed in Figure 2 (EIF4A2, ASUN, THYN1, RIPK2, RNF10, CEP164, TPD52L2, CEP57, and BCL2L12; Supplemental Fig. 3A) are among the RNA-seq targets, validating the sequencing and analysis pipeline. Two additional RT-PCR targets (DICER1 and CHEK2) show the same tendency in the RNA-seq data but have slightly too large confidence intervals. As expected, of the 600 exons, the majority (90%) shows increased skipping upon knockdown of U2AF35, which was rescued by ectopic U2AF35 expression. For further analysis, we focused on this group of 535 exons. When comparing the rescue abilities of the different U2AF35 mutants, we found that the Q157R mutant rescues 354 targets whereas this number was reduced for Q157P (286) and Q157Rdel (238; Fig. 3A). The highest overlap was found between the Q157R and the Q157P mutants (157 exons) followed by exons that were rescued by all three mutants (98 exons). However, we also found exons that were uniquely rescued by either one of the three mutants: The largest group was found for the Q157Rdel mutant (111), followed by 82 (Q157R) and 19 (Q157P) unique target exons. We then used this transcriptome-wide data set of uniquely rescued targets to define 3′ splice site signatures for the individual mutants. This analysis revealed a striking difference with the Q157Rdel, showing a strong preference for an A in position +1 (105 out of 111 unique targets), a G in position +1 for the Q157P mutant (17 out of 19), and a C in the same position for the Q157R mutant (68 out of 82; Fig. 3A). Analysis of all 535 wt U2AF35 targets showed no preference at the +1 position at all, and the preference for one nucleotide in the respective mutants was strongly reduced when looking at all rescued targets and not just the unique ones (Supplemental Fig. 3B). Exons not rescued by the Q157R/P mutants showed an enrichment of A in the +1 position, which is consistent with previous observations (Ilagan et al. 2014), further validating our approach. Interestingly, 207 out of 287 cassette exons not rescued by wt U2AF35 were responsive to either one of the different mutants, with sequence preferences similar to the one described above (Supplemental Fig. 3C); these exons may react more sensitively to the precise expression level of U2AF35 and binding affinities of the different mutants. Altogether, these data confirm and expand the conclusion that the three Q157 mutants control alternative splicing of a distinct set of target exons and strongly suggest that the respective specificity is controlled through distinct 3′ splice site sequences. This analysis also suggests that the wt U2AF35 has flexibility in recognizing 3′ splice sites and that the Q157 mutants show sequence constraints that select for and against certain nucleotides at the +1 position (Fig. 3; Supplemental Fig. 3B).
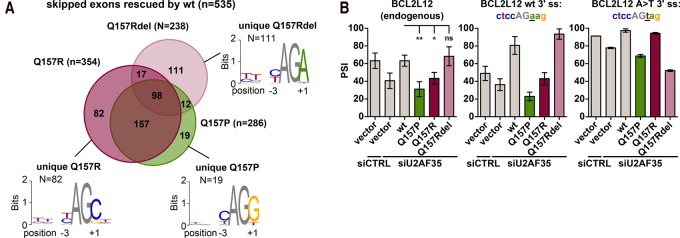
FIGURE 3.
Q157P, Q157R, and Q157Rdel have distinct 3′ splice site sequence preferences. (A) Identification of U2AF35 target exons responding to Q157P, Q157R, and Q157Rdel variant expression in RNA sequencing data. Cassette exons with increased skipping upon U2AF35 knockdown and rescue by wt U2AF35 expression were defined as U2AF35 target exons. Overlap of Q157P, Q157R, and Q157Rdel responsive U2AF35 target exons is shown together with the 3′ splice site consensus motifs of unique Q157P, Q157R, and Q157Rdel targets. (B) Splicing of BCL2L12 alternative exon 3 in response to U2AF35 knockdown and rescue by U2AF35 variants. Splicing of endogenous BCL2L12 (left, n = 3) was compared to the wild-type and mutated minigenes of BCL2L12 containing exons 2, 3, and 4. Minigenes were transfected in duplicates in two independent experiments.
To further confirm differential 3′ splice site preferences experimentally, we focused on the Q157Rdel mutant and cloned a BCL2L12 minigene, which, in the wt gene, contains an adenosine at position +1. As for the endogenous gene, missplicing of the BCL2L12 minigene upon knockdown of endogenous U2AF35 was rescued by wt and the Q157Rdel U2AF35, but not by overexpression of the Q157R or Q157P point mutants (Fig. 3B; Supplemental Fig. 3D). This is in agreement with an enrichment of an adenosine at the +1 position for Q157Rdel rescued targets and for nonrescued targets of Q157P and Q157R (Fig. 3A; Supplemental Fig. 2B). However, when mutating the adenosine in position +1 to a thymidine, the Q157Rdel mutant did not rescue missplicing upon the U2AF35 knockdown anymore, which is consistent with the motif analysis. In contrast, the Q157R mutant was now able to compensate for the loss of endogenous U2AF35, which was not the case for the Q157P mutant. Although this Q157R/P difference for a T in the +1 position was not predicted by our motif analysis, it further confirms that the ability of the different mutants to rescue splicing of different exons is based on the +1 position of the 3′ splice site sequence. Introducing a C in the +1 position of this minigene leads to complete inclusion of the alternative exon, which was not responsive to U2AF35 knockdown anymore and was thus not further analyzed (Supplemental Fig. 3D).
In addition to the characterization of the Q157Rdel mutant, its unique targets and their 3′ splice site signature, our results for the first time show quantitative differences in target exons between Q157P and Q157R mutations and extend previous qualitative observations (Ilagan et al. 2014; Shao et al. 2014). As the mechanistic basis we suggest different preferences for a G or a C in the +1 position of the 3′ splice site of the respective target exons.
Human Q157P and Q157R patients show distinct splicing signatures
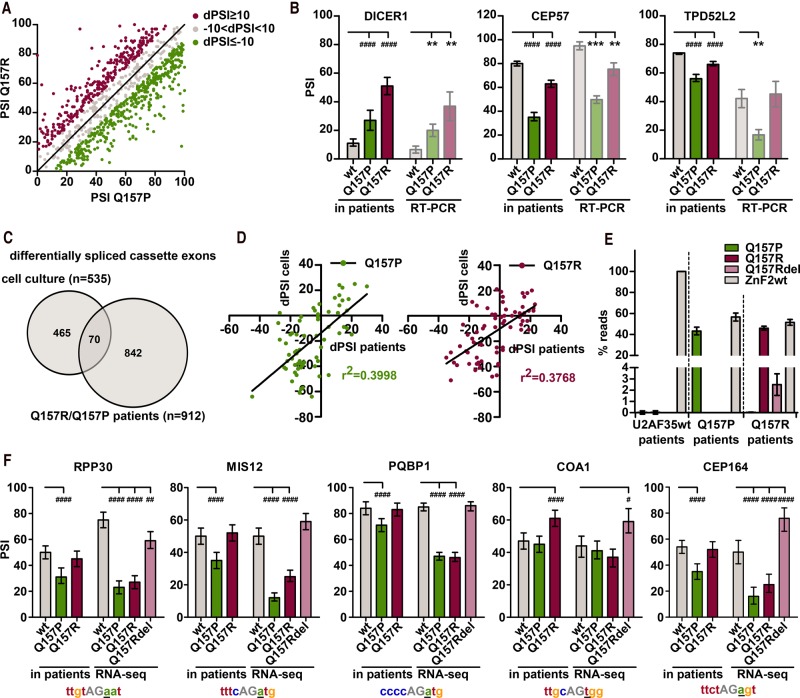
Considering the results of our knockdown and complementation assay, we suggest that Q157P and Q157R mutations have distinct groups of target exons, which we aimed to confirm in human patients. A previous study that addressed U2AF35 mutations in vivo combined Q157P/Q157R patients with S34 mutations in the first ZnF and thus missed mutation-specific targets (Qiu et al. 2016); therefore, in vivo analysis of Q157R and Q157P splicing defects is still missing. To identify Q157P- and Q157R-specific targets in vivo, we analyzed publicly available RNA sequencing data of 533 AML patients (GSE67040) (Lavallée et al. 2015; Pabst et al. 2016) and searched for U2AF35 transcripts carrying either the c.470A>G (Q157R) or c.470A>C (Q157P) mutation. In the analyzed cohort, we identified three Q157R and two Q157P patients. Comparison of these patients with 10 patients with wt U2AF35 identified 1874 (Q157P) and 1849 (Q157R) differentially regulated alternative splicing events, with a delta percent spliced in (dPSI) above 10%, a Bayes factor ≥5, and confidence intervals that do not diverge by more than 10% from the calculated PSI value (see Materials and Methods for a detailed description of the analysis). For further analysis we focused on skipped exons, as they represent the largest group of differentially regulated events (Supplemental Fig. 4A), and only maintained exons where the above criteria were met for all three genotypes. This analysis identified 912 distinct alternatively spliced cassette exons misspliced in either Q157R or Q157P patients when compared to U2AF35 wt patients. Of these 912 events, 22% (n = 201) were similarly affected by Q157R and Q157P because the observed inclusion values differed by <10% between Q157R and Q157P. However, 32% (n = 289) and 46% (n = 422) of the identified exons showed a dPSI value of at least 10% when compared between Q157R or Q157P patients (Fig. 4A). To confirm these results, we first used our set of 16 exons analyzed by RT-PCR (Fig. 2) and indeed found that splicing in patients is almost perfectly recapitulated in the knockdown complementation cell culture system for seven exons (ASUN, THYN1, BCL2L12, FXR1; DICER1, CEP57, and TPD52L2 as examples in Fig. 4B). To more broadly analyze a correlation between patients and cell culture, we used our RNA-seq data and looked for events that are Q157R/P targets in patients as well as U2AF35-dependent target exons in cell culture. The overlap consists of only 70 events, likely due to largely different gene expression profiles of the hematopoietic primary cells and the HEK293T cells used in culture (Fig. 4C). However, when correlating dPSI (wt-mutant) values of these 70 exons of patients versus cell culture, we obtained reasonable r2 values of close to 0.4 for both the Q157R and Q157P pairs (Fig. 4D). Furthermore, alternative splicing patterns of Q157R and Q157P patients clustered together (Supplemental Fig. 5B, see Materials and Methods for details), suggesting that the observed differences are primarily due to the mutation and not due to random tumor heterogeneity. These data validate the differences observed between Q157R and Q157P patients and strongly suggest that these mutants are not equivalent but rather cause missplicing of a distinct set of exons. An interesting case is missplicing of DICER1 in both Q157R and Q157P patients (Fig. 4B), as this could lead to aberrant processing of miRNAs, thereby contributing to oncogenesis (Lin and Gregory 2015).
FIGURE 4.
Splicing signature and expression of the Q157Rdel variant in Q157R and Q157P patients. (A) The diagram shows 912 cassette exons with a dPSI value above 10% between Q157R/Q157P and U2AF35 wt patients with a Bayes factor ≥5 and confidence intervals not diverging more than 10% from the calculated PSI value. Plotted are the PSI values of these 912 cassette exons in patients with Q157R and Q157P mutation. Differentially spliced exons with dPSI higher or equal to 10% are shown in green for Q157P and red for Q157R. (dPSI) delta percent spliced in. (B) Comparison of PSI values of cassette exons obtained in cell culture (Fig. 2) by RT-PCR to PSI values in U2AF35 wt, Q157P and Q157R patients. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (####) Bayes factor >100. (C) Overlap between U2AF35-dependent cassette exons in RNA sequencing of HEK293T cells (Fig. 3) and differentially spliced exons in Q157P/Q157R patients. (D) Comparison of alternative splicing in cell culture experiments to splicing observed in patients. dPSI values between wt and either Q157P or Q157R were calculated for the 70 overlapping targets (C) between cell culture and patient sequencing and plotted against each other. Correlation coefficients were calculated. (E) Identification of the Q157Rdel isoform in AML patients. Depicted is the number of reads of the Q157P, Q157R, Q157Rdel, and wt variants in U2AF35wt, Q157R, and Q157P patients as percent of total reads at that position. (F) Examples of differentially spliced exons in patients and cell culture where the Q157Rdel rather than the Q157R mutant might regulate splicing in patients. 3′ splice sites of the alternative exons are depicted below the graphs. (#) Bayes factor >3, (##) Bayes factor >20, (####) Bayes factor >100.
Having identified Q157P- and Q157R-responsive exons in patients, we asked whether the respective 3′ splice sites also show a nucleotide preference at the +1 position. Comparison of all exons rescued by either the Q157R or the Q157P variant in cell culture to all exons with increased inclusion in the respective patients and all nonrescued exons in cell culture to all exons with less inclusion in patients revealed the same enrichment patterns, although less prominent in patients (Supplemental Figs. 3B, 4B). For example, for exons not rescued by the Q157P mutant in cells and with less inclusion in Q157P patients, we observe an enrichment of an A and C at the +1 position. The less prominent enrichment in patients compared to our cell culture model is likely due to the presence of 50% wild-type U2AF35 in patients, as the mutations occur heterozygously.
Expression of the Q157Rdel variant in Q157R patients
We next asked whether the Q157Rdel mutant is expressed in Q157R patients. We therefore searched for U2AF35 transcripts carrying the Q157Rdel, Q157R, or Q157P sequence in the identified Q157R, Q157P, and U2AF35 wt patients. Consistent with the heterozygosity of these mutations, slightly below 50% of the reads that mapped to the second ZnF of U2AF35 encode either the Q157R or the Q157P mutation, and the other 50% encode the wild-type sequence, indicating a marginally reduced stability of the mutant mRNA (Fig. 4E). Interestingly, as predicted by our minigene experiments, this analysis indeed confirmed the presence of the Q157Rdel mutant resulting from the usage of the alternative 5′ss in Q157R but not in Q157P patients. Although we find only 2.5% of all U2AF35 reads at that position to encode the Q157Rdel variant, the average RPKM value in the three patients is 1.25, which places the Q157Rdel mutant in the group of highly expressed genes, which has been suggested to have an RPKM value above one (Hebenstreit et al. 2011, also see Supplemental Fig. 4B). To investigate a potential contribution of the Q157Rdel variant to the splicing pattern in Q157R patients, we asked whether for some exons the splicing pattern in Q157R patients is more closely mimicked by the Q157Rdel variant than the Q157R mutant in cell culture. Sixteen out of 70 exons indeed showed such a pattern (Fig. 4F for five examples), pointing to a role of the Q157Rdel variant in splicing regulation in Q157R patients. This idea is further supported by the finding that the correlation between cell culture and patient dPSI values is strongly improved when Q157R cell culture values are corrected by better fitting Q157Rdel values [Δr2 (patient-cell culture; patient-cell culture corrected) = 0.1586], whereas the improvement is less good for the Q157P pair (Δr2 = 0.0822) serving as control. Although this analysis does not take into account the coexpression of wt, Q157R and Q157Rdel U2AF35 variants and the different expression levels of the respective proteins in “Q157R” patients, it points to an active role of the Q157Rdel mutant in regulating splicing in vivo. Together these data show that the U2AF35 Q157R and Q157P alleles create three proteins that affect different target exons based on the 3′ splice site sequence and likely differentially contribute to missplicing and potentially disease formation in human patients.
In summary, we provide evidence for differential splicing regulation by the cancer-associated Q157R and Q157P mutations and, based on our analysis of target splice sites, suggest differential RNA-binding preferences as the underlying cause. In addition, our data further illustrate the importance of analyzing exonic missense mutations beside their role in altering one amino acid. We expect that more examples will be discovered where a point mutant affects, in addition to changing one amino acid, additional RNA processing events such as splicing, mRNA stability, or translation that control protein expression and thereby contribute to disease development.
MATERIALS AND METHODS
Cell culture, transfection, and treatments
HEK293T and HeLa cells were cultured in DMEM High glucose (Biowest) containing 10% FCS (Biochrom) and 1% penicillin/streptomycin (Biowest). Transfections of plasmid and siRNA were performed using RotiFect (Carl Roth) following manufacturer's instructions. For minigene analysis, 1.8 × 105 HEK293T and 1.0 × 105 HeLa cells were seeded in 12-well plates and were transfected with 0.8 µg human and mouse U2AF35 Exon5-7 minigenes. For rescue experiments, knockdown of U2AF35 was performed by transfecting 0.5 × 105 HEK293T cells in a 12-well plate with 20 pmol siRNAs (siU2AF35: GAAAGUGUUGUAGUUGAUUGA; siCTRL: UUCUCCGAACGUGUCACGU). After 24 h, knockdown was rescued by transfection of 0.8 µg expression vectors for Flag-tagged U2AF35 expression constructs for an additional 48 h.
Constructs
Cloning of minigenes was performed by amplifying the human and mouse genomic region of U2AF35 spanning exon 5 to exon 7 and the human genomic region of BCL2L12 spanning exon 2 to exon 4. AML-associated point mutations were introduced into the U2AF35 and the 3′ss mutations into the BCL2L12 minigene. Minigenes were cloned in the CMV promoter-driven vector pcDNA3.1(+). U2AF35 protein expression constructs were cloned by amplifying the full-length mouse U2AF35 from cDNA followed by insertion of point mutations and deletions in the second zinc finger. Human and mouse U2AF35 are 97% conserved. PCR products were cloned in a CMV promoter-driven vector with a C-terminal Flag-tag. Primer sequences are provided in the Supplemental Table 1G.
Cell proliferation and cell cycle assay
For cell proliferation and cycle analysis, HEK293T cells were seeded at a density of 0.3 × 105 into 48-well plates. Twenty hours after seeding, cells were transfected with 7.5 pmol siU2AF35 or siCTRL and after 10 h with 0.2 µg expression vectors for Flag-tagged U2AF35 expression constructs. For cell proliferation analysis, cells were washed twice in PBS with 5.6 mM d-glucose, stained with 1 µM CFSE (carboxyfluorescein succinimidyl ester) in PBS with 5.6 mM d-glucose for 10 min light protected at room temperature, and washed twice in DMEM containing 10% FCS prior to seeding. Twenty-four, 48, 72, and 96 h after the second transfection, cells were trypsinized and the remaining CFSE signal was measured with a guava easyCyte 8 cytometer. The median CFSE signal was calculated and normalized to 24 h after transfection. For cell cycle analysis, cells were trypsinized 72 h after the second transfection, fixed in 70% ethanol, treated with 2.5 µg/µL RNase A in PBS for 1 h at 37°C and stained with 12.5 µg/mL PI (propidium iodide) in 0.2 mL PBS. PI signal was measured with a guava easyCyte 8 cytometer and percentage of cells in the G0/G1, S, and G2/M phase was calculated.
RNA, RT-PCR, and RT-qPCR
RNA extraction, RT-PCR, Phosphor imager quantification, and RT-qPCR were done as previously described (Preußner et al. 2014; Wilhelmi et al. 2016). Results of endogenous alternative splicing represent the mean value of at least three independent experiments with the corresponding standard deviation. Significance was calculated by Student's unpaired t-test: (*) P < 0.05, (**) P < 0.01, (***) P < 0.001. For minigene analysis, total RNA was isolated and DNaseI treated prior to radioactive RT-PCR. Primer sequences are provided in the Supplemental Table 1G.
Immunoblotting and antibodies
Cells were boiled in 2× SDS-loading dye; proteins were separated on an SDS-page and immunoblotted to a PVDF membrane according to standard protocols. Expression of U2AF35 proteins was analyzed using α-U2AF35 antibody (a kind gift from Angus Lamond; Chusainow et al. 2005) and α-hnRNPL antibody (Santa Cruz, sc-32317) as a loading control.
Structure prediction
Structure predictions of the U2AF35wt, Q157R, and Q157Rdel proteins were performed with the Protein Homology/analog Recognition Engine V 2.0 (Phyre2) (Kelly et al. 2015) and visualized with PyMOL (by DeLano [http://www.pymol.org]). Distances between the zinc-coordinating residues within the second zinc finger were measured with Coot (Emsley and Cowtan 2004).
RNA sequencing
Total RNAs from control siRNA and U2AF35 siRNA transfected cells, as well as Q157R, Q157Rdel, and Q157P rescue cell lines, were purified using RNA-Tri (Bio&SELL) and further purified using the RNeasy mini kit (Qiagen) in combination with a DNase I (Qiagen) treatment to prevent genomic DNA contamination. RNA sequencing libraries were prepared by using the TruSeq mRNA Library Preparation kit (Illumina). Of note, 125-bp paired-end reads were generated by using a HiSeq 2500 sequencer (Illumina) with V4 sequencing chemistry. Biological duplicates were sequenced for all conditions.
Accession numbers, annotations, and read mapping
Initial RNA-seq data analysis was performed similarly to that described in Preußner et al. (2017). In short, reads were mapped to the hg18 genome using TopHat (Trapnell et al. 2012), version 2.0.13 (Bowtie2 version 2.2.5). Numbers of mapped reads are listed in Supplemental Table 1A. For analysis of alternative splicing events, a mixture of isoforms Bayesian inference model (MISO) (Katz et al. 2010), version 0.5.3, was used. Alternative splicing events were considered differential when they showed a minimal dPSI of 10% with a Bayes factor higher than five. Events with confidence intervals diverging more than 10% of the calculated PSI value were eliminated. Coverage tracks with junction reads were visualized using the Integrative Genomics Viewer (Thorvaldsdóttir et al. 2013).
To find RNA-seq data from AML patients with mutations in U2AF35, a publicly available data set of the Sauvageau laboratory (GSE67040) was used (Lavallée et al. 2015; Pabst et al. 2016). The data set was mapped to the U2AF35 genomic region. To identify patients with the relevant point mutations c.470A>G (Q157R) and c.470A>C (Q157P), SAMtools (Li 2011) version 0.1.19 and custom UNIX scripts were used. To assess the question whether missplicing of U2AF35 occurs in Q157R patients, the abundance of the Q157Rdel isoform was quantified. Reads containing nonamers of either wt, 470A>G mutation or deletion sequences at the position of the potential new splice site were counted to this end. The number of reads found for wt, the mutation and the deletion isoform are shown as percent of total reads in that position. The data set included two patients with a c.470A>C (Q157P) mutation and three patients with a c.470A>G (Q157R) mutation, which were selected for further analysis together with 10 randomly selected patients with U2AF35 wt.
To obtain single values for each alternative splicing event per condition (a prerequisite for the analysis with MISO), data from duplicate cell culture samples or all patients with the same mutation were concatenated after mapping. To confirm that merging of alignments is appropriate, hierarchical clustering was performed with the individual data sets. To this end, the Euclidean distances between PSI values of events identified in our analysis were calculated and cluster dendrograms were computed using Ward's method (Ward 1963). This analysis was performed using a custom R script. For our sequencing data, the heatmap and dendrogram visualizes that the duplicates cluster closely together as expected. Additionally, the siCtrl-vector and wild-type rescue samples form a large cluster, which shows that our system recapitulates endogenous splicing levels. For the patient data, we observe that the patients with the same mutation cluster together. The wild-type samples form two clusters, which in our view can be tolerated considering the larger number of 10 wild-type patients (heatmaps and dendrograms in Supplemental Fig. 5).
For analysis of the cell sequencing, custom Python scripts were used. We defined an event to be affected by knockdown of U2AF35, when the dPSI between the siCtrl and knockdown sample was at least 10% and the confidence intervals did not touch. The event was defined as rescued, when expression of U2AF35 restored the PSI value at least to the mean between endogenous and knockdown level, again considering confidence intervals. This resulted in 535 events with a knockdown effect that leads to more skipping of the target and that can be rescued by expression of full-length U2AF35. These events were used for the splice site analysis and for clustering. Raw sequencing data are available as BioProject ID PRJNA401377.
To analyze the patient data, differential events between wt and Q157P as well as wt and Q157R patients were computed. All events that were contained in any of these two lists were used for the comparison of Q157P and Q157R patients as well as for the clustering.
To compare effects between cell lines and patients, we focused on the 535 knockdown-rescue targets in the cell culture defined above and exons that were differentially spliced between U2AF35 wild-type and mutant patients. To account for the variation in basal splicing level between patient blood cells and HEK cells, we calculated dPSI values between mutant versus wild-type in patients and wild-type rescue versus mutant rescue in HEK cells, respectively. We then correlated values for these events for the same genotypes. To find events potentially controlled by Q157Rdel in patients, events were identified for which splicing in patients is better mimicked by the Q157Rdel mutant in cell culture (increased PSI correlation of at least 5%). The Q157Rdel corrected cell culture values were than correlated with patient data and an improvement was calculated (Δr2). Improvement of the Q157P (in which Q157Rdel is not expressed) served as control.
Splice site analysis
Splice site sequences were retrieved using custom scripts and the REST API of http://TogoWS.org (Katayama et al. 2010). Splice site scores were obtained using the first-order Markov model of the MaxEntScan algorithm (Yeo and Burge 2004). Visualization of the splice sites was obtained by using the web-based application WebLogo (Crooks et al. 2004).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the HPC Service of ZEDAT, Freie Universität Berlin, for computing time. We also thank members of the Heyd laboratory for discussion and comments on the manuscript and Markus Wahl and laboratory members for help with structure predictions. Work in the Heyd laboratory was funded through an Emmy-Noether-Fellowship by the Deutsche Forschungsgemeinschaft (He5398/3); additional support from the Deutsche Forschungsgemeinschaft came from the SFB958/A21 to F.H.
Author contributions: O.H. performed experiments. A.N. performed bioinformatics analysis. B.T. performed RNA sequencing. F.H. and O.H. designed the study, planned experiments, analyzed data, and wrote the manuscript. F.H. initiated and designed the study and supervised the work.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061432.117.
REFERENCES
- Cartegni L, Chew SL, Krainer AR. 2002. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298. [DOI] [PubMed] [Google Scholar]
- Chusainow J, Ajuh PM, Trinkle-Mulcahy L, Sleeman JE, Ellenberg J, Lamond AI. 2005. FRET analyses of the U2AF complex localize the U2AF35/U2AF65 interaction in vivo and reveal a novel self-interaction of U2AF35. RNA 11: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. 2004. WebLogo: a sequence logo generator. Genome Res 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Biol Crystallogr D Biol Crystallogr 60: 2126–2132. [DOI] [PubMed] [Google Scholar]
- Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, Krysiak K, Harris CC, Koboldt DC, Larson DE, et al. 2012. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 44: 53–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn CN, Venugopal P, Scott HS, Hiwase DK. 2015. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev 263: 257–278. [DOI] [PubMed] [Google Scholar]
- Hebenstreit D, Fang M, Gu M, Charoensawan V, van Oudenaarden A, Teichmann SA. 2011. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol Syst Biol 7: 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu TYT, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, Echeverria GV, Sun T, Kurley SJ, Tyagi S, et al. 2015. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525: 384–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, Bradley RK. 2014. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome Res 25: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue D, Bradley RK, Abdel-wahab O. 2016. Spliceosomal gene mutations in myelodysplasia: molecular links to clonal abnormalities of hematopoiesis. Genes Dev 30: 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Patel PN, Gorham JM, Mcdonough B, Depalma SR, Adler EE, Lam L, Macrae CA, Mohiuddin SM, Fatkin D, et al. 2017. Identification of pathogenic gene mutations in LMNA and MYBPC3 that alter RNA splicing. Proc Natl Acad Sci 114: 7689–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama T, Nakao M, Takagi T. 2010. TogoWS: Integrated SOAP and REST APIs for interoperable bioinformatics web services. Nucleic Acids Res 38: W706–W711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Wang ET, Airoldi EM, Burge CB. 2010. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7: 1009–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly LA, Mezulis S, Yates C, Wass M, Sternberg M. 2015. The Phyre2 web portal for protein modelling, prediction, and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh CM, Bezzi M, Low DHP, Ang WX, Teo SX, Gay FPH, Al-Haddawi M, Tan SY, Osato M, Sabò A, et al. 2015. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523: 96–100. [DOI] [PubMed] [Google Scholar]
- Lavallée V-P, Baccelli I, Krosl J, Wilhelm B, Barabé F, Gendron P, Boucher G, Lemieux S, Marinier A, Meloche S, et al. 2015. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet 47: 1030–1037. [DOI] [PubMed] [Google Scholar]
- Li H. 2011. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Gregory RI. 2015. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15: 321–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merendino L, Guth S, Bilbao D, Martínez C, Valcárcel J. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402: 838–841. [DOI] [PubMed] [Google Scholar]
- Okeyo-Owuor T, White BS, Chatrikhi R, Mohan DR, Kim S, Griffith M, Ding L, Ketkar-Kulkarni S, Hundal J, Laird KM, et al. 2015. U2AF1 mutations alter sequence specificity of pre-mRNA binding and splicing. Leukemia 29: 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst C, Bergeron A, Lavallée V-P, Yeh J, Gendron P, Norddahl GL, Krosl J, Boivin I, Deneault E, Simard J, et al. 2016. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood 127: 2018–2027. [DOI] [PubMed] [Google Scholar]
- Pacheco TR, Coelho MB, Desterro JMP, Mollet I, Carmo-Fonseca M. 2006. In vivo requirement of the small subunit of U2AF for recognition of a weak 3′ splice site. Mol Cell Biol 26: 8183–8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius C-M, Ali AM, Zhu LJ, Mukherjee S, Raza A, et al. 2016. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3′ end formation. Mol Cell 62: 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preußner M, Wilhelmi I, Schultz A-S, Finkernagel F, Michel M, Möröy T, Heyd F. 2014. Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol Cell 54: 651–662. [DOI] [PubMed] [Google Scholar]
- Preußner M, Goldammer G, Neumann A, Haltenhof T, Rautenstrauch P, Müller-McNicoll M, Heyd F. 2017. Body temperature cycles control rhythmic alternative splicing in mammals. Mol Cell 67: 433– 446.e4. [DOI] [PubMed] [Google Scholar]
- Przychodzen B, Jerez A, Guinta K, Sekeres MA, Padgett R, Maciejewski JP, Makishima H. 2013. Patterns of missplicing due to somatic U2AF1 mutations in myeloid neoplasms. Blood 122: 999–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Zhou B, Thol F, Zhou Y, Chen L, Shao C, DeBoever C, Hou J, Li H, Chaturvedi A, et al. 2016. Distinct splicing signatures affect converged pathways in myelodysplastic syndrome patients carrying mutations in different splicing regulators. RNA 22: 1535–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz A-S, Preußner M, Bunse M, Karni R, Heyd F. 2016. Activation-dependent TRAF3 exon 8 alternative splicing is controlled by CELF2 and hnRNP C binding to an upstream intronic element. Mol Cell Biol 37: e00488-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. 2014. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 21: 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, Ndonwi M, Wadugu B, Duncavage EJ, Okeyo-Owuor T, et al. 2015. Mutant U2AF1 expression alters hematopoiesis and pre-mRNA splicing in vivo. Cancer Cell 27: 631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Cygan KJ, Rhine CL, Wang J, Bulacan C, Yang J, Bayrak-Toydemir P, McDonald J, Fairbrother WG. 2017. Pathogenic variants that alter protein code often disrupt splicing. Nat Genet 49: 848–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl MC, Will CL, Lührmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136: 701–718. [DOI] [PubMed] [Google Scholar]
- Wang D, Guo Y, Wu C, Yang G, Li Y, Zheng C. 2008. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genomics 9: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JH. 1963. Hierarchical grouping to optimize an objective function. J Am Stat Assoc 58: 236–244. [Google Scholar]
- Webb CJ, Wise JA. 2004. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol 24: 4229–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelmi I, Kanski R, Neumann A, Herdt O, Hoff F, Jacob R, Preußner M, Heyd F. 2016. Sec16 alternative splicing dynamically controls COPII transport efficiency. Nat Commun 7: 12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Fu X-D. 2015. Genomic functions of U2AF in constitutive and regulated splicing. RNA Biol 12: 479–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Romfo CM, Nilsen TW, Green MR. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402: 832–835. [DOI] [PubMed] [Google Scholar]
- Yeo G, Burge CB. 2004. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol 11: 377–394. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, Sato Y, Sato-Otsubo A, Kon A, Nagasaki M, et al. 2011. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 478: 64–69. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Park S, Oda T, Akiyoshi T, Sato M, Shirouzu M, Tsuda K, Kuwasako K, Unzai S, Muto Y, et al. 2015. A novel 3′ splice site recognition by the two zinc fingers in the U2AF small subunit. Genes Dev 29: 1649–1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Patton JG, Green MR. 1992. Cloning and domain structure of the mammalian splicing factor U2AF. Nature 355: 609–614. [DOI] [PubMed] [Google Scholar]
- Zhang M, Zamore PD, Carmo-Fonseca M, Lamond AI, Green MR. 1992. Cloning and intracellular localization of the U2 small nuclear ribonucleoprotein auxiliary factor small subunit. Proc Natl Acad Sci 89: 8769–8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorio DA, Blumenthal T. 1999. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature 402: 835–838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.