Abstract
Alternative polyadenylation (APA) is a mechanism that generates multiple mRNA isoforms with different 3′UTRs and/or coding sequences from a single gene. Here, using 3′ region extraction and deep sequencing (3′READS), we have systematically mapped cleavage and polyadenylation sites (PASs) in Drosophila melanogaster, expanding the total repertoire of PASs previously identified for the species, especially those located in A-rich genomic sequences. Cis-element analysis revealed distinct sequence motifs around fly PASs when compared to mammalian ones, including the greater enrichment of upstream UAUA elements and the less prominent presence of downstream UGUG elements. We found that over 75% of mRNA genes in Drosophila melanogaster undergo APA. The head tissue tends to use distal PASs when compared to the body, leading to preferential expression of APA isoforms with long 3′UTRs as well as with distal terminal exons. The distance between the APA sites and intron location of PAS are important parameters for APA difference between body and head, suggesting distinct PAS selection contexts. APA analysis of the RpII215C4 mutant strain, which harbors a mutant RNA polymerase II (RNAPII) with a slower elongation rate, revealed that a 50% decrease in transcriptional elongation rate leads to a mild trend of more usage of proximal, weaker PASs, both in 3′UTRs and in introns, consistent with the “first come, first served” model of APA regulation. However, this trend was not observed in the head, suggesting a different regulatory context in neuronal cells. Together, our data expand the PAS collection for Drosophila melanogaster and reveal a tissue-specific effect of APA regulation by RNAPII elongation rate.
Keywords: Drosophila, alternative polyadenylation, RNA polymerase II, transcription elongation rate, 3′READS
INTRODUCTION
The 3′ end maturation of almost all eukaryotic mRNAs involves endonucleolytic cleavage of the nascent RNA and subsequent poly(A) tail synthesis, two coupled reactions that are also linked to transcription termination (Proudfoot 2016). Cleavage and polyadenylation (C/P) are controlled by cis-elements located upstream and downstream from the cleavage and polyadenylation site (PAS) that bind specific protein factors (Zhao et al. 1999; Proudfoot 2011; Tian and Graber 2012; Shi and Manley 2015). In vertebrates, upstream elements include the hexamer A[A/U]UAAA or other close variants, U-rich elements, and UGUA elements (Hu et al. 2005; Darmon and Lutz 2012). Downstream elements include U- and GU-rich elements. In addition, upstream UAUA elements and downstream G-rich sequences are frequently present near PASs (Hu et al. 2005). Cis-elements around the PAS in lower species differ from those in mammals. For example, downstream GU-rich elements are absent from the nematode PASs (Jan et al. 2011), and substantial differences can be found with the PASs in yeast and plants (Graber et al. 2002; Xing and Li 2011; Mata 2013).
It is now well established that most eukaryotic genes harbor multiple PASs, giving rise to transcript isoforms, a process called alternative polyadenylation (APA) (Mangone et al. 2010; Ozsolak et al. 2010; Jan et al. 2011; Wu et al. 2011; Derti et al. 2012; Haenni et al. 2012; Sherstnev et al. 2012; Smibert et al. 2012; Ulitsky et al. 2012; Hoque et al. 2013; Mata 2013; Schlackow et al. 2013; Wilkening et al. 2013; Tian and Manley 2017). APA has increasingly been found to play important roles in health and diseases (Di Giammartino et al. 2011; Lutz and Moreira 2011; Pinto et al. 2011; Lianoglou et al. 2013). APA sites located in the 3′ untranslated region (3′UTR) of mRNA result in transcripts with different 3′UTR sizes. Because the 3′UTR contains various elements that regulate mRNA metabolism, including mRNA stability, translation, and localization, 3′UTR-APA can play important roles in post-transcriptional regulation of gene expression (Lutz and Moreira 2011; Elkon et al. 2013; Tian and Manley 2017). In addition, APA sites located upstream of the last exon affect both the coding sequence and the 3′UTR of transcripts. For simplicity, we refer to these as upstream region (UR) APA events. Several studies indicate that APA regulation is highly tissue-specific. For example, some human tissues have a global tendency favoring certain isoform types, such as promoter-distal PASs in neuronal tissues and proximal PASs in the blood and testis (Zhang et al. 2005; Liu et al. 2007; Smibert et al. 2012). The APA profiles in different tissues are well conserved across species (Derti et al. 2012) and ubiquitously expressed genes are more likely to have APA isoforms than those genes with restricted expression breadth (Lianoglou et al. 2013). APA has also been shown to be dynamically regulated in cell proliferation (Sandberg et al. 2008), differentiation (Ji and Tian 2009; Shepard et al. 2011), cancer transformation (Mayr and Bartel 2009; Singh et al. 2009; Fu et al. 2011; Morris et al. 2012), and in response to extracellular cues (Flavell et al. 2008; Chang et al. 2015).
A growing number of mechanisms have been found to regulate APA, including core cleavage and polyadenylation factors, splicing features, transcriptional dynamics, and many RNA-binding proteins (RBPs) (Di Giammartino et al. 2011; Lutz and Moreira 2011; Shi 2012; Elkon et al. 2013; Zheng and Tian 2014; Mayr 2016; Tian and Manley 2017). Here, using 3′ region extraction and deep sequencing (3′READS), we systematically map PASs in the Drosophila melanogaster genome and examine APA profiles in the fly body and head. In addition, we compare APA in wild-type and RpII215C4 flies, which express a mutant RNA polymerase II (RNAPII) with a slower elongation rate. We report a context-specific impact of RNAPII elongation rate on APA in different tissues.
RESULTS
Genome-wide PAS mapping in Drosophila melanogaster
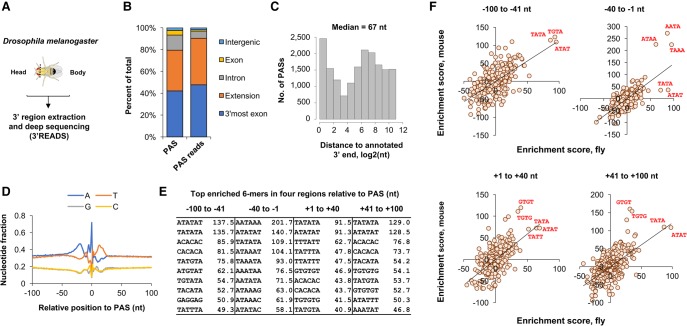
We were interested in understanding APA determinants in Drosophila melanogaster. To comprehensively identify PASs in the fly genome, we isolated RNA from the body and the head of young adult male flies and subjected it to 3′ region extraction and deep sequencing (3′READS) (Fig. 1A), a sequencing method for 3′-end identification and APA analysis (Hoque et al. 2013; Zheng et al. 2016). We generated ∼47 million reads containing PASs (Supplemental Table S1), leading to the identification of 52,844 PASs in the fly genome (Fig. 1B). About half of the PASs reads were aligned to annotated 3′-most exons of genes (Fig. 1B). However, ∼37% of PASs (corresponding to ∼42.8% PAS-containing reads) were found to be in the downstream region of the 3′ ends annotated by the Ensembl database (Fig. 1B), indicating poor annotation of the mRNA 3′ ends in the database. We thus reannotated 3′ ends of genes using our data (see Materials and Methods for details), resulting in extension of the 3′ end for 10,112 genes, with a median extension size of 67 nucleotides (nt) (Fig. 1C). Overall, 44,169 PASs were mapped to 12,890 mRNA genes.
FIGURE 1.
PASs in the Drosophila melanogaster genome. (A) Experimental design. RNA isolated from Drosophila head or body was subjected to 3′ region extraction and deep sequencing (3′READS) analysis for identification of PAS and analysis of APA isoform abundance. (B) Distribution of PASs and PAS reads in the Drosophila genome. (C) Extension of annotated 3′ ends by 3′READS data (see Materials and Methods for details). A total of 16,908 PASs were found to be in the extended region. The median extension size is indicated. (D) Nucleotide profile around fly PASs. (E) Top 10 enriched 6-mers around the fly PAS. Four regions around the PAS were analyzed, as indicated. Numbers are enrichment scores based on comparison of observed sequences with expected sequences (see Materials and Methods for details). (F) Comparison of 4-mer enrichment between mouse and fly in four regions surrounding the PAS. Values are enrichment score as in E. Several top 4-mers in each graph are highlighted to indicate consistency or distinction between PAS cis-elements in fly and mouse.
Notably, our PAS collection is substantially larger than that from a recent study (Supplemental Fig. S1; Smibert et al., 2012). As such, for mRNA genes, while the two studies overlap on 12,957 PASs, we report 31,212 unique sites whereas Smibert et al. (2012) had 1540 sites not found in our collection. One important difference between the two studies is that our collection contains significantly more PASs located in A-rich sequences, i.e., more than six consecutive A's or seven A's in a 10-nt window around the PAS (Supplemental Fig. S1; see Materials and Methods for details). These PASs are typically considered by other studies as internal priming cases, i.e., internal A-rich sequences falsely identified as the poly(A) tail, and are eliminated from the final result. However, because our 3′READS method does not utilize oligo(dT) for priming from the poly(A) tail and each PAS-containing read contains a few nucleotides from the poly(A) tail as evidence of the tail, PASs within A-rich sequences can be confidently identified (Zheng et al. 2016). Indeed, based on our data, ∼20% of fly PASs are located near A-rich sequences, a frequency much higher than the ∼8% reported in the mouse genome (Hoque et al. 2013), attesting to the importance of using a method unaffected by internal priming for accurate identification of fly PASs.
The nucleotide profile around the fly PASs is similar to that of mammalian PASs (Tian et al. 2005), containing an upstream A-rich peak and a downstream U-rich peak (Fig. 1D). Consistently, comparisons of k-mer (6-mer or 4-mer) frequencies in four surrounding regions (−100 to −41 nt, −40 to −1 nt, +1 to +40 nt, and +41 to +100 nt, with PAS set to position 0) with randomized sequences (based on dinucleotide frequencies, see Materials and Methods for details) showed that, as in mammals, AATAAA is the top hexamer in the −40- to −1-nt region of fly PASs, and TGTA and TATA elements are highly enriched in the upstream region (Fig. 1E). However, TATA-containing motifs are prominent in all four analyzed regions around the PAS (Fig. 1E), a feature distinct from mammalian PASs (Hu et al. 2005).
To directly compare cis-elements in the vicinity of fly PASs with those around mammalian PASs, we used the same method to calculate k-mer enrichment scores with mouse PASs, which we previously mapped also using 3′READS (Hoque et al. 2013). Comparisons of enrichment scores indicate that while most enriched 4-mers were similar between mouse and fly, TATA and ATAT sequences were conspicuously different in the −40- to −1-nt region between the two species (Fig. 1F). The enrichment scores of these motifs were much higher in the fly when compared to the mouse. In contrast, 4-mers related to AATAAA were slightly more enriched for mouse PASs than for fly PASs in the −40- to −1-nt region (Fig. 1F). In addition, downstream GU-rich elements (GTGT and TGTG sequences) were significantly more enriched for mouse PASs than in flies in both +1- to +40-nt and +41- to +100-nt downstream regions (Fig. 1F). This result indicates distinct upstream and downstream cis-elements around fly PASs when compared to those in mammals.
Widespread APA in the fly genome
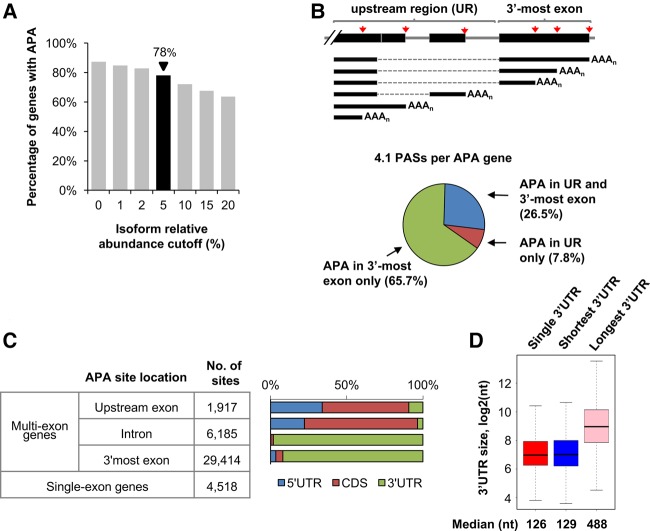
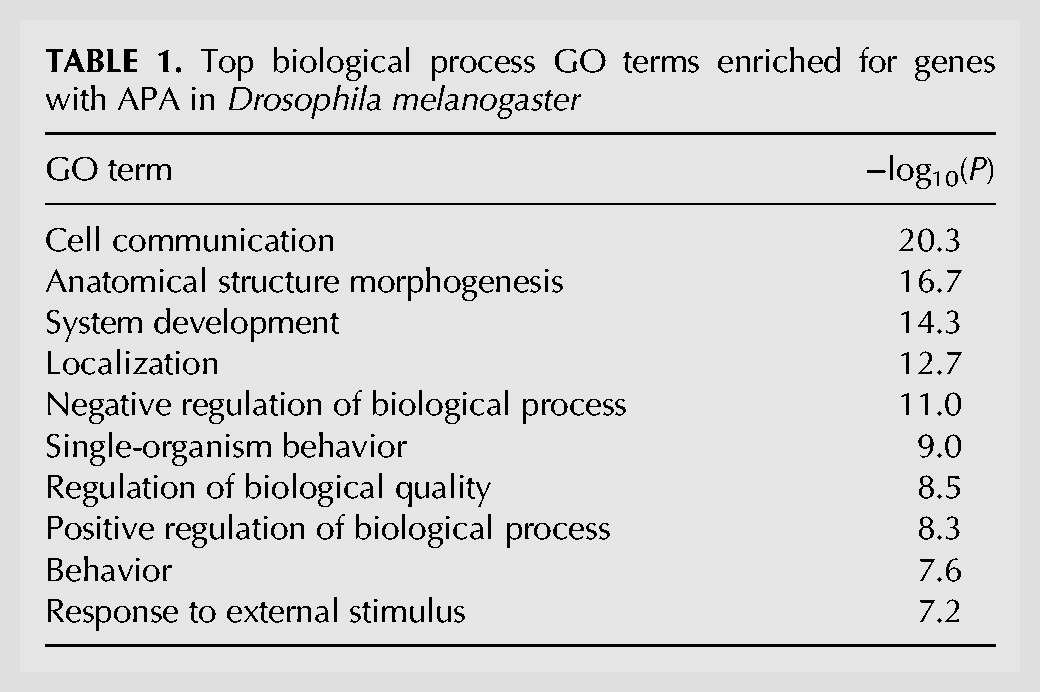
The frequency of genes with APA isoforms varied depending upon the cutoff used to call an isoform (Fig. 2A). With the relative abundance of 5% being the cutoff, 78% of mRNA genes were found to have APA isoform expression (Fig. 2A). Gene Ontology (GO) analysis showed that genes with APA isoforms tend to be associated with a set of GO terms, including “cell communication,” “anatomical structure morphogenesis,” “system development,” “localization,” and “negative regulation of biological process” (Table 1), suggesting that APA may play a role in organism complexity and may be dynamically regulated in development as previously reported in mouse (Ji and Tian 2009). On average, genes with detectable APA isoforms have 4.1 PASs per gene (Fig. 2A). Most fly genes have APA sites in the 3′-most exon only (∼65%, Fig. 2B), affecting the 3′UTR length (Fig. 2C). About one-third of all fly genes have APA sites in the upstream region (Fig. 2B), leading to transcripts with different 5′UTRs and CDSs (Fig. 2C). For genes with multiple PASs in the 3′UTR, the shortest 3′UTR size had a median of 129 nt, similar to genes without APA (126 nt, Fig. 2D), and the longest 3′UTR was about 3.8-fold longer, with a median of 488 nt. These values are significantly smaller than mouse 3′UTR isoforms, which have ∼250 nt and ∼1.7 kb for the shortest and longest 3′UTR isoforms, respectively (Hoque et al. 2013), presumably due to the compact size of the fly genome when compared to the mouse genome.
FIGURE 2.
APA in Drosophila melanogaster. (A) Percentage of protein-coding genes in the fly genome found to express APA mRNA isoforms using isoform expression cutoffs. At 5% relative abundance cutoff, 78% of fly mRNA genes displayed APA with 4.1 PASs per gene. (B) (Top) Scheme of the different APA types. APA sites are divided into two groups, i.e., 3′-most exon and upstream region (UR). (Bottom) Percentage of genes with APA sites in upstream regions (UR) and/or 3′-most exons. Only genes with APA sites were included. (C) mRNA regions affected by APA. Genes were divided into multiexon or single exon groups. The former was further divided into upstream exon (non-3′-most), intron, and 3′-most exon groups. The number of PASs in each group is indicated. mRNA regions were separated into 5′UTR, coding sequence (CDS), and 3′UTR. For intronic PASs, the mRNA region affected was defined by the exon immediately upstream of the PAS. (D) The 3′UTR size of transcripts from genes without APA sites (single 3′UTR) or with APA sites (shortest and longest isoforms are shown).
TABLE 1.
Top biological process GO terms enriched for genes with APA in Drosophila melanogaster
3′UTR-APA in fly body and head
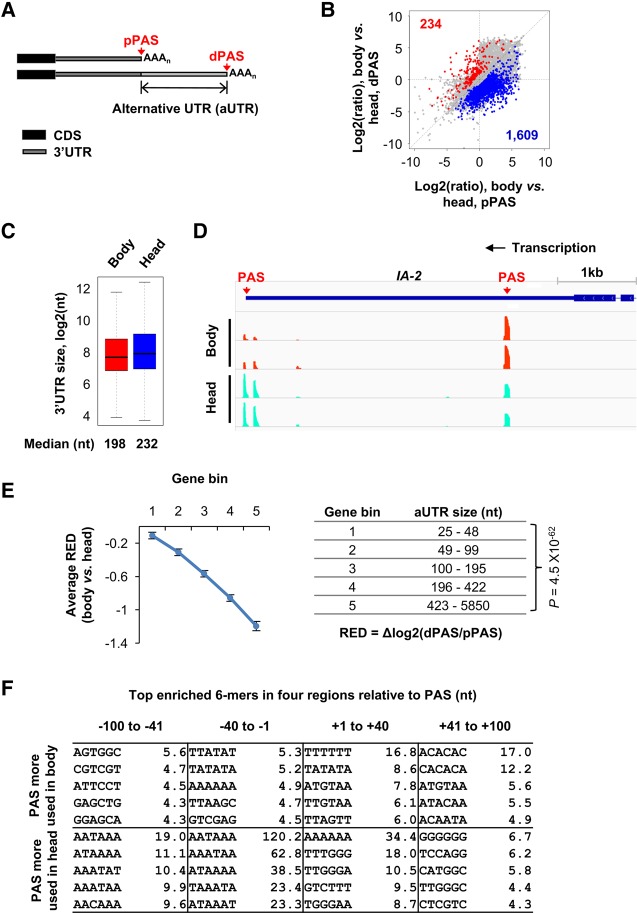
APA is highly tissue-specific (Zhang et al. 2005; Wang et al. 2008; Derti et al. 2012; Smibert et al. 2012; Lianoglou et al. 2013) and the brain and the neural system have been found to express long 3′UTR isoforms in mammals (Zhang et al. 2005) and in fly (Hilgers et al. 2012; Smibert et al. 2012). Therefore, we compared APA profiles in the fly body and head starting by PASs in 3′UTRs. To simplify the analysis, we focused on the top two PASs based on isoform abundance that we named proximal PAS (pPAS) and distal PAS (dPAS), respectively, based on the relative location to the 5′ end of the gene (Fig. 3A). The distance between the two PASs was named alternative 3′UTR (aUTR). Consistent with previous reports (Smibert et al. 2012), we found that isoforms using distal PASs tend to have a higher abundance in the head compared to the body (Fig. 3B). Consequently, the median sizes of the 3′UTR per gene based on all APA isoforms were 198 and 232 nt for body and head samples, respectively (Fig. 3C). An example gene, IA-2, which codes for a protein tyrosine phosphatase whose mammalian ortholog is a major autoantigen in type I diabetes (Genovese et al. 1996; Cai et al. 2001; Williams et al. 2008), is shown in Figure 3D. IA-2 expresses higher levels of dPAS mRNA in the head than in the body (Fig. 3D).
FIGURE 3.
3′UTR-APA difference between Drosophila melanogaster body and head. (A) Scheme showing 3′UTR-APA and its analysis. Top two most abundant PAS isoforms per gene were selected for comparison, which are named proximal PAS (pPAS) and distal PAS (dPAS) isoforms, respectively. The distance between the two PASs is considered alternative 3′UTR (aUTR). (B) Scatterplot showing pPAS and dPAS isoform abundance differences between head and body. Two biological replicates were used. Genes with significantly (FDR < 0.05, DEXseq analysis) higher abundance of pPAS isoforms in the body vs. head are shown in blue (1609 genes), and those with higher abundance of dPAS isoforms are in red (234 genes). Total numbers of blue and red genes are shown. (C) Box plot of the 3′UTR length for genes with expression in body and head samples. The weighted mean based on multiple APA isoforms was used to calculate the 3′UTR size of expressed transcripts of each gene. The median value is indicated. (D) UCSC snapshot of 3′READS data for IA-2, showing that pPASs are preferentially expressed in fly bodies while dPASs are in heads. Two replicates are shown. (E) Relationship between the extent of 3′UTR-APA difference and aUTR size. Expressed genes with 3′UTR-APA were evenly divided into five bins based on the aUTR size (distance between pPAS and dPAS), resulting in ∼1300 genes in each bin. The aUTR size range for each bin is shown in the table next to the graph. The extent of 3′UTR-APA difference is represented by relative expression difference (RED), which is the difference in log2(ratio) of dPAS isoform abundance to pPAS isoform abundance between body and head. Error bars are SEM. Values for genes in bin #1 were compared with those in bin #5 by the Wilcoxon rank sum test, and the P-value is shown. Only PASs with ≥5 reads were used for analysis. (F) Top enriched 6-mers in four regions around the PASs up-regulated in body (top) or in head (bottom). Values are −log10(P), where P is based on the Fisher's exact test.
Several GO terms, including “system development,” “organelle organization,” and “macromolecule localization” were the highest statistically enriched terms for genes with longer 3′UTRs in the head (Supplemental Table S3). This result is largely consistent with a previous report (Smibert et al. 2012), in which biological processes such as neural development or function were also found to be highly enriched for neural-extended transcripts.
The degree of 3′UTR lengthening was indicated by relative expression difference (RED), which was calculated by the difference in distal vs. proximal PAS usage between two samples (see Materials and Methods for details). We found that the RED values between the head and body of genes correlated with the aUTR size (Fig. 3E). Because the aUTR size reflects the time lag between the transcription of proximal PAS and that of distal PAS, this result suggests that the difference in the kinetic competition between proximal and distal PAS usage leads to APA changes in head vs. body.
We next identified enriched sequence motifs around the PASs preferentially used in the body or in the head (Fig. 3F). Most significantly, U-rich downstream elements (TTTTTT) in the +1- to +40-nt region were highly enriched for PASs up-regulated in the body, whereas AAUAAA and related A-rich motifs in the upstream region as well as G-rich motifs, for example, UUUGGG, in the downstream region were highly enriched for PASs up-regulated in the head. This result suggests that the PASs preferentially used in the head are stronger than those in the body, which is consistent with the notion that proximal PASs, which are generally up-regulated in the body, are typically weaker than distal PASs, which are up-regulated in the head.
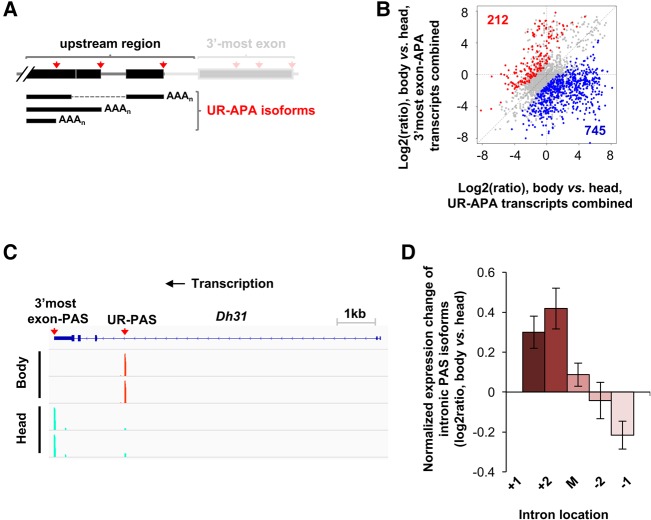
UR-APA in fly body and head
We found that about one-third of APA sites are located in upstream regions of the 3′-most exon. These sites are collectively called upstream region (UR)-APA sites (Fig. 4A). Comparing UR-APA isoform abundance with that of transcripts using 3′-most exon PASs, we observed a global trend in which genes expressed in the head tended to use PAS in the 3′-most exon, whereas those expressed in the body had the opposite trend (Fig. 4B). The genes with up-regulated UR-APA in the body outnumbered the genes with up-regulated UR-APA in the head by ∼3.5-fold (Fig. 4B). GO analysis indicated that several biological processes, such as “signaling,” “anatomical structure morphogenesis,” and “system development,” were enriched in genes with down-regulated UR-APA in the head (Supplemental Table S4). Interestingly, some development-related GO terms, such as “system development,” were also enriched for genes with long 3′UTRs in the head, indicating that the transcripts of these genes are generally lengthened by both 3′UTR-APA and UR-APA mechanisms. Most UR-APA sites are located in introns as exemplified by Dh31, which encodes the fly ortholog of the vertebrate neuropeptide calcitonin gene-related peptide (Fig. 4C). Separating APA sites in different introns based on their locations, we found that PASs in 5′ introns tend to be more suppressed in the head than those in 3′ introns (Fig. 4D), indicating polarity in intronic PAS regulation in head vs. body. This trend is in line with the result from aUTR vs. RED analysis for 3′UTR-APA sites, suggesting a common mechanism underlying differential 3′UTR- and UR-APA events between head and body.
FIGURE 4.
Upstream region APA difference between fly body and head. (A) Scheme showing various upstream region (UR)-APA isoforms. (B) Scatter plot comparing abundance of UR-APA isoforms and 3′-most exon APA isoform in body vs. head. Genes with significantly higher abundance of UR-APA isoforms in body compared to head are shown in blue (745 genes). Those with higher abundance of 3′-most exon PAS isoforms in the body vs. head are shown in red (212 genes). Two biological replicates were used. Significance of APA was based on the DEXseq analysis (FDR < 0.05). (C) UCSC snapshot of 3′READS data for Dh31, showing that an UR-PAS isoform is preferentially expressed in bodies while 3′-most exon PAS expression is biased to heads. (D) Introns were divided into first (+1), second (+2), last (−1), second to last (−2), and middle (between +2 and −2 introns) groups. Only genes with ≥4 introns were analyzed, and only PASs with ≥5 reads were selected. Expression changes are log2(ratio) of PAS reads in test sample vs. control sample. Values for five intron groups were normalized by mean-centering. Error bars are SEM.
Impact of slower RNAPII elongation on APA
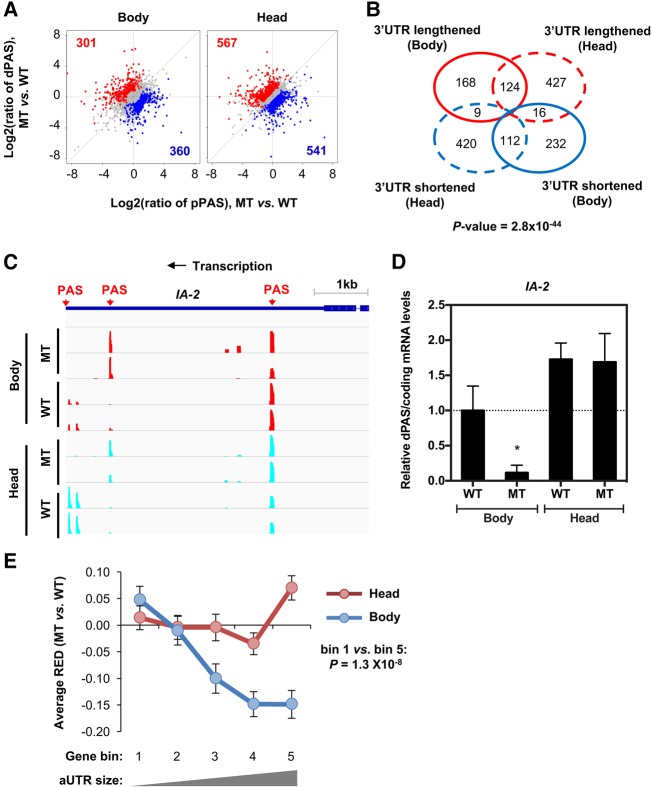
One of the mechanisms that can regulate PAS usage is the elongation rate of RNAPII. In theory, a slower elongation rate could favor the usage of proximal PAS due to the “first come, first served” kinetics (de la Mata et al. 2010; Pinto et al. 2011; Fong et al. 2015; Laitem et al. 2015). We thus applied 3′READS to examine the APA profiles in the Drosophila mutant strain RpII215C4, which has a 50% reduction in RNAPII elongation rate (Chen et al. 1996), again using RNA samples from bodies and heads of male flies. Overall, the APA differences between body and head in the mutant strain were similar to those in the wild type, for both 3′UTR- and UR-APA events (Supplemental Fig. S2). In line with the “first come, first served” model, the slower RNAPII altered PAS usage in the 3′-most exon with a mild preference to proximal PASs in the body (301 genes with dPAS up-regulated vs. 360 genes with pPAS up-regulated, Fig. 5A). In contrast, this trend was much subdued with head samples (541 genes with dPAS up-regulated vs. 567 genes with pPAS up-regulated, Fig. 5B). Consistently, genes with regulated 3′UTR-APA events in the body did not have a substantial overlap with those in the head (Fig. 5B).
FIGURE 5.
3′UTR-APA regulation in a fly mutant with a slower RNAPII elongation. (A) Scatter plots comparing 3′UTR-APA isoform abundance between wild-type (WT; w1118) and mutant (MT; RpII215) in body (left) and head (right), as in Figure 3B. (B) Venn diagram comparing 3′UTR-APA changes between WT and MT in body vs. in head. (C) UCSC snapshot of 3′READS for IA-2, showing that dPAS expression is decreased in mutant body in comparison to the wild-type body. Two replicates are shown. (D) Ratio of dPAS/coding relative mRNA expression levels for IA-2 in wild-type (WT) and mutant (MT) bodies and heads, quantified by RT-qPCR. Data show the mean ± SD normalized to WT body, for at least three independent experiments. Comparisons were performed against WT body and head using an unpaired two-tailed t-test ([*] P < 0.01). (E) Relationship between the extent of 3′UTR-APA difference (relative expression difference, or RED) and aUTR size, as in Figure 3E.
A case in point is APA regulation of IA-2. Levels of IA-2 dPAS mRNA isoform were strongly decreased in the body of the mutant strain in comparison to the wild type while this difference was not observed in the head, as determined by 3′READS (Fig. 5C) and validated by RT-qPCR using two primer pairs specific for a common coding region and a region specific for the long 3′UTR isoform (Fig. 5D). We also validated APA of rassf8 (ras-associated domain family 8) (Supplemental Fig. S3) and slmo (Reeve et al. 2007) by the same method (Supplemental Fig. S3). Both showed similar APA regulation in both body and head, but with different directions. The 3′UTR was shortened in rassf8, but lengthened in slmo (Supplemental Fig. S3), highlighting the gene-specific impact of RNAPII elongation rate on APA.
We next examined the aUTR size vs. RED for regulated APA events. Consistent with the overall trend of 3′UTR shortening, genes in all aUTR size bins showed negative RED values in the body of mutant fly (Fig. 5E). Importantly, there was an inverse correlation between aUTR size and the extent of regulation (more negative RED values), consistent with the “first come, first served” model. In contrast, the correlation in the head was not obvious and genes with large aUTR sizes (bin 5) in fact displayed slight 3′UTR lengthening (Fig. 5E).
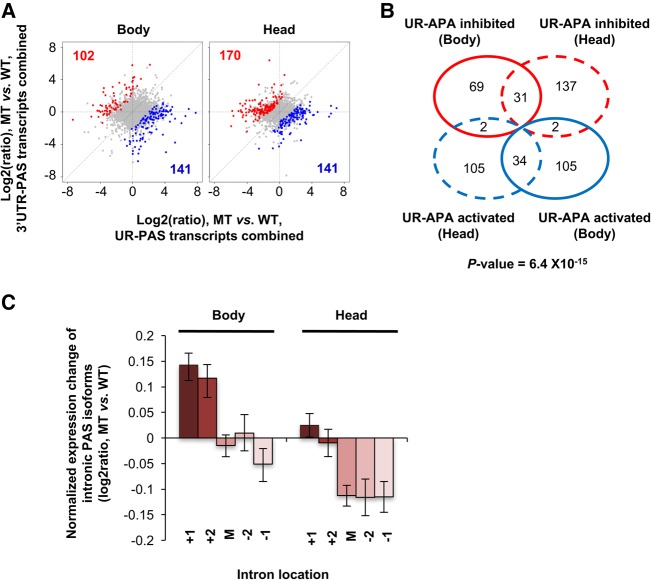
Like 3′UTR-APA events, UR-APA isoforms in the body of mutant fly showed a modest general trend of up-regulation (141 genes with UR-APA isoforms up-regulated vs. 102 genes with UR-APA isoforms down-regulated, Fig. 6A), whereas a slightly inverse trend could be discerned in the head (141 genes with UR-APA isoforms up-regulated vs. 170 genes with UR-APA isoforms down-regulated). Again, only a small set of genes showed UR-APA regulation in both body and head (Fig. 6B). For regulated UR-APA events, PASs located in 5′ introns tended to be more up-regulated than those in downstream introns in the body of mutant fly (Fig. 6C). However, PASs in all intron groups were slightly down-regulated in the head (Fig. 6C).
FIGURE 6.
UR-APA regulation in a fly mutant with slower RNAPII elongation. (A) Scatterplot comparing UR-APA and 3′-most exon APA isoform abundance between wild-type (WT) and mutant (MT) in body (left) and head (right), as in Figure 4B. (B) Venn diagram comparing UR-APA changes between WT and MT in body vs. head. Two biological replicates were used for the analysis. (C) Expression difference in intronic APA isoforms between body and head in different intron groups, as in Figure 4D.
We also found that PASs associated with the AAUAAA motif were less frequently used in the mutant body (Supplemental Fig. S4A), suggesting that weaker PAS usage is generally preferred when the RNAPII elongation rate is decreased. In contrast, this was not observed in the mutant head (Supplemental Fig. S4B). Intriguingly, PASs associated with the upstream A-rich motif tended to be more used in the mutant head (Supplemental Fig. S4B). Taken together, our results indicate that a slower RNAPII elongation mildly enhances proximal PAS usage in the body, in line with the “first come, first served” mechanism. Presumably, slower transcriptional elongation provides proximal, relatively weaker PAS more time for processing before the distal, stronger PAS is transcribed. However, this model does not apply to the head, suggesting that the “first come, first served” mechanism is context-specific and is overwritten in the neuronal system.
DISCUSSION
In this work, we systematically mapped PASs in Drosophila melanogaster, uncovering a substantial number of PASs previously overlooked, including ∼20% of fly PASs located in A-rich sequence regions. We identified distinct sequence motifs between fly and mouse PASs. While fly PASs tend to have higher frequencies of UA-rich motifs upstream of the PAS when compared to mammalian ones, they are associated with GU-rich downstream elements to a much lesser extent, suggesting different evolutionary paths for these elements. This result is in general agreement with a previous study, which reported different downstream elements in fly and human PASs (Retelska et al. 2006). However, using a vastly greater number of PASs (fewer than 3000 PASs in the previous study), we were able to clearly define this difference and reveal new differences in UA-rich elements.
Consistent with a previous study of the fly transcriptomes in different tissues (Smibert et al. 2012), we found that the fly head preferentially uses distal PASs while the fly body preferentially uses proximal PASs in the 3′-most exon. We additionally extended this global APA trend to upstream regions, which affects both the coding sequences and 3′UTRs. The consistent directions of 3′UTR-APA and UR-APA in favoring promoter-distal PASs in the head indicate general transcript lengthening in the neural system of Drosophila, similar to mammals (Zhang et al. 2005; Wang et al. 2008; Derti et al. 2012). Intriguingly, we also observed that most genes encoding cleavage/polyadenylation, elongation, and termination factors showed increased expression levels in Drosophila head as compared to the body (Supplemental Table S5).
The RpII215C4 mutant strain was previously shown to have alterations in alternative splicing (de la Mata et al. 2003; Fong et al. 2014), in transcription termination (Fong et al. 2015), and in APA (Pinto et al. 2011). Our analysis of the RpII215C4 mutant strain by 3′READS reveals that a 50% decrease in RNAPII elongation rate (Chen et al. 1993) mildly enhances proximal PAS usage in the fly body, which is consistent with the “first come, first served” model for APA regulation. However, this is not the case in the head, indicating that different contexts in body and head have different impacts on APA regulation. One potential source of this difference may be the differential expression of proteins with functions in pre-mRNA cleavage/polyadenylation, such as core cleavage/polyadenylation factors, regulators of RNAPII elongation and termination and/or RBPs. Many of these genes are differentially expressed in the mutant in comparison to the wild type (Supplemental Table S5), and it is possible that they play a role in the tissue-specific APA observed. Our results indicate that neuronal cells respond to decreased RNAPII elongation rate differently than the body, masking the “first come, first served” mechanism. Taken together, our data indicate a tissue-specific effect of the transcriptional elongation rate on APA.
MATERIALS AND METHODS
Drosophila melanogaster samples
Fly strains were obtained from the Bloomington Stock Centre and grown at 25°C using standard culture conditions and media. w1118 flies were used as a wild-type strain and the RpII215 flies, carrying the RNA polymerase II C4 allele (Chen et al. 1993), as the mutant fly strain. Heads and bodies were dissected from w1118 and RpII215C4 1- to 5-d-old adult male flies using standard procedures. Total RNA was extracted and purified using TRIzol (Invitrogen) according to the manufacturer's protocol.
3′READS and PAS identification
The 3′ region extraction and deep sequencing (3′READS) and 3′READS+ methods were described in Hoque et al. (2014) and in Zheng et al. (2016), respectively. Data processing was based on the methods previously described (Zheng et al. 2016). Briefly, reads from 3′READS were mapped to the fly genome (BDGP5, Ensembl v70) using Bowtie 2 (Langmead and Salzberg 2012). Uniquely mapped reads (with MAPQ score >10) that had at least two nongenomic T's at the 5′ end were considered as PAS-containing reads. PASs located within 24 nt from each other were clustered together, as previously described. PASs mapped to the genome were further assigned to genes using gene models defined by the Ensembl database. The 3′ ends of the gene models were extended by 2 kb to include downstream PASs, but the extension did not go beyond the transcription start site of the downstream gene. To eliminate spurious PASs, we further required that the number of PAS reads for a PAS was ≥5% of all PAS reads for the gene in at least two samples. Statistics of samples are shown in Supplemental Table S1.
APA analysis
APA analysis was carried out largely following the methods previously described (Hoque et al. 2013; Li et al. 2015, 2016). Briefly, relative expression (RE) of two PAS isoforms in the 3′-most exon, e.g., pPAS and dPAS, was calculated by log2(RPM) of dPAS vs. pPAS, where RPM was reads per million PAS-containing reads. Relative expression difference (RED) of two isoforms in two comparing samples was based on the difference in RE of the two isoforms between the two samples. DEXSeq was used to derive statistically significant APA events (FDR < 0.05) (Anders et al. 2012).
Cis-element analysis
We used the PROBE method, which we previously used for human PAS analysis, to examine cis-elements around fly PASs (Hu et al. 2005). Briefly, for each k-mer (6-mer or 4-mer), we enumerated its frequency (observed value) in a defined region of PAS and compared it with the frequency (expected value) using the randomized sequences of the region. The randomization was based on the one-order Markov chain model. The enrichment score was calculated as a Z-score based on the difference between the observed and expected frequencies. Four regions around the PAS were analyzed, including −100 to −41 nt, −40 to −1 nt, +1 to +40, and +41 to +100 nt, with the PAS position set at zero. Comparisons between two sets of PASs (body vs. head) were based on the Fisher's exact test.
Intron analysis
The intron location was based on the RefSeq database, considering all RefSeq-supported splicing isoforms. Introns were divided into first (+1), second (+2), last (−1), second to last (−2), and middle (between +2 and −2 introns) groups. Only genes with ≥4 introns were analyzed. Expression changes are log2(ratio) of PAS reads in test sample vs. control sample. Values for five intron groups were normalized by mean-centering to reveal bias of intron location.
Gene Ontology analysis
Gene Ontology (GO) annotation of genes was obtained from the Ensembl database. GO entries were tested for bias to significantly regulated APA genes using the hypergeometric test. GO terms associated with more than 2000 genes were considered too generic and were discarded. To remove redundancy, each reported GO term was required to have at least 30% of genes that are not associated with another term with a more significant P-value.
RT-qPCR
A minimum of 10 heads and bodies was collected from w1118 and RpII215C4 1- to 5-d-old adult male flies, and total RNA was purified using TRIzol (Invitrogen). cDNA synthesis was performed using random hexamers (Sigma-Aldrich) with SuperScript IV (Invitrogen) following the manufacturer's instructions and quantified in an Applied Biosystems 7500 Fast Real-Time PCR System with SYBR Select Master Mix (Applied Biosystems). rp49 was used for normalization in all assays. Oligonucleotide sequences used are shown in Supplemental Table S2.
DATA DEPOSITION
All data can be obtained from the NCBI GEO database (GSE85368).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a National Institutes of Health grant (GM084089) to B.T. and by Norte-01-0145-FEDER-000008–Porto Neurosciences and Neurologic Disease Research Initiative at I3S, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement through the European Regional Development Fund (ERDF) to A.M.; by FCT–Fundação para a Ciência e Tecnologia under the project FCOMP-01-0124-FEDER-028252 (PTDC/BEX-BCM/0468/2012) to J.F.; and FCT PhD fellowship SFRH/BD/102002/2014 to M.S.O. We thank members of A.M. and B.T. laboratories for helpful discussions.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062661.117.
REFERENCES
- Anders S, Reyes A, Huber W. 2012. Detecting differential usage of exons from RNA-seq data. Genome Res 22: 2008–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai T, Krause MW, Odenwald WF, Toyama R, Notkins AL. 2001. The IA-2 gene family: homologs in Caenorhabditis elegans, Drosophila and zebrafish. Diabetologia 44: 81–88. [DOI] [PubMed] [Google Scholar]
- Chang JW, Zhang W, Yeh HS, de Jong EP, Jun S, Kim KH, Bae SS, Beckman K, Hwang TH, Kim KS, et al. 2015. mRNA 3′-UTR shortening is a molecular signature of mTORC1 activation. Nat Commun 6: 7218. [DOI] [PubMed] [Google Scholar]
- Chen Y, Weeks J, Mortin MA, Greenleaf AL. 1993. Mapping mutations in genes encoding the two large subunits of Drosophila RNA polymerase II defines domains essential for basic transcription functions and for proper expression of developmental genes. Mol Cell Biol 13: 4214–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Chafin D, Price DH, Greenleaf AL. 1996. Drosophila RNA polymerase II mutants that affect transcription elongation. J Biol Chem 271: 5993–5999. [PubMed] [Google Scholar]
- Darmon SK, Lutz CS. 2012. Novel upstream and downstream sequence elements contribute to polyadenylation efficiency. RNA Biol 9: 1255–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. 2003. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell 12: 525–532. [DOI] [PubMed] [Google Scholar]
- de la Mata M, Lafaille C, Kornblihtt AR. 2010. First come, first served revisited: factors affecting the same alternative splicing event have different effects on the relative rates of intron removal. RNA 16: 904–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, Rohl CA, Johnson JM, Babak T. 2012. A quantitative atlas of polyadenylation in five mammals. Genome Res 22: 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giammartino DC, Nishida K, Manley JL. 2011. Mechanisms and consequences of alternative polyadenylation. Mol Cell 43: 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat Rev Genet 14: 496–506. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. 2008. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron 60: 1022–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Kim H, Zhou Y, Ji X, Qiu J, Saldi T, Diener K, Jones K, Fu XD, Bentley DL. 2014. Pre-mRNA splicing is facilitated by an optimal RNA polymerase II elongation rate. Genes Dev 28: 2663–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong N, Brannan K, Erickson B, Kim H, Cortazar MA, Sheridan RM, Nguyen T, Karp S, Bentley DL. 2015. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol Cell 60: 256–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genovese S, Bonfanti R, Bazzigaluppi E, Lampasona V, Benazzi E, Bosi E, Chiumello G, Bonifacio E. 1996. Association of IA-2 autoantibodies with HLA DR4 phenotypes in IDDM. Diabetologia 39: 1223–1226. [DOI] [PubMed] [Google Scholar]
- Graber JH, McAllister GD, Smith TF. 2002. Probabilistic prediction of Saccharomyces cerevisiae mRNA 3′-processing sites. Nucleic Acids Res 30: 1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenni S, Ji Z, Hoque M, Rust N, Sharpe H, Eberhard R, Browne C, Hengartner MO, Mellor J, Tian B, et al. 2012. Analysis of C. elegans intestinal gene expression and polyadenylation by fluorescence-activated nuclei sorting and 3′-end-seq. Nucleic Acids Res 40: 6304–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers V, Lemke SB, Levine M. 2012. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev 26: 2259–2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Ji Z, Zheng D, Luo W, Li W, You B, Park JY, Yehia G, Tian B. 2013. Analysis of alternative cleavage and polyadenylation by 3′ region extraction and deep sequencing. Nat Methods 10: 133–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque M, Li W, Tian B. 2014. Accurate mapping of cleavage and polyadenylation sites by 3′ region extraction and deep sequencing. Methods Mol Biol 1125: 119–129. [DOI] [PubMed] [Google Scholar]
- Hu J, Lutz CS, Wilusz J, Tian B. 2005. Bioinformatic identification of candidate cis-regulatory elements involved in human mRNA polyadenylation. RNA 11: 1485–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan CH, Friedman RC, Ruby JG, Bartel DP. 2011. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 469: 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Tian B. 2009. Reprogramming of 3′ untranslated regions of mRNAs by alternative polyadenylation in generation of pluripotent stem cells from different cell types. PLoS One 4: e8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laitem C, Zaborowska J, Isa NF, Kufs J, Dienstbier M, Murphy S. 2015. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat Struct Mol Biol 22: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, You B, Hoque M, Zheng D, Luo W, Ji Z, Park JY, Gunderson SI, Kalsotra A, Manley JL, et al. 2015. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLoS Genet 11: e1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Park JY, Zheng D, Hoque M, Yehia G, Tian B. 2016. Alternative cleavage and polyadenylation in spermatogenesis connects chromatin regulation with post-transcriptional control. BMC Biol 14: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. 2013. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 27: 2380–2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Brockman JM, Dass B, Hutchins LN, Singh P, McCarrey JR, MacDonald CC, Graber JH. 2007. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res 35: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz CS, Moreira A. 2011. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip Rev RNA 2: 22–31. [DOI] [PubMed] [Google Scholar]
- Mangone M, Manoharan AP, Thierry-Mieg D, Thierry-Mieg J, Han T, Mackowiak SD, Mis E, Zegar C, Gutwein MR, Khivansara V, et al. 2010. The landscape of C. elegans 3′UTRs. Science 329: 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J. 2013. Genome-wide mapping of polyadenylation sites in fission yeast reveals widespread alternative polyadenylation. RNA Biol 10: 1407–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C. 2016. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol 26: 227–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris AR, Bos A, Diosdado B, Rooijers K, Elkon R, Bolijn AS, Carvalho B, Meijer GA, Agami R. 2012. Alternative cleavage and polyadenylation during colorectal cancer development. Clin Cancer Res 18: 5256–5266. [DOI] [PubMed] [Google Scholar]
- Ozsolak F, Kapranov P, Foissac S, Kim SW, Fishilevich E, Monaghan AP, John B, Milos PM. 2010. Comprehensive polyadenylation site maps in yeast and human reveal pervasive alternative polyadenylation. Cell 143: 1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto PA, Henriques T, Freitas MO, Martins T, Domingues RG, Wyrzykowska PS, Coelho PA, Carmo AM, Sunkel CE, Proudfoot NJ, et al. 2011. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J 30: 2431–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2011. Ending the message: poly(A) signals then and now. Genes Dev 25: 1770–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot NJ. 2016. Transcriptional termination in mammals: stopping the RNA polymerase II juggernaut. Science 352: aad9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve S, Carhan A, Dee CT, Moffat KG. 2007. slowmo is required for Drosophila germline proliferation. Genesis 45: 66–75. [DOI] [PubMed] [Google Scholar]
- Retelska D, Iseli C, Bucher P, Jongeneel CV, Naef F. 2006. Similarities and differences of polyadenylation signals in human and fly. BMC Genomics 7: 176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. 2008. Proliferating cells express mRNAs with shortened 3′ untranslated regions and fewer microRNA target sites. Science 320: 1643–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Marguerat S, Proudfoot NJ, Bähler J, Erban R, Gullerova M. 2013. Genome-wide analysis of poly(A) site selection in Schizosaccharomyces pombe. RNA 19: 1617–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard PJ, Choi EA, Lu J, Flanagan LA, Hertel KJ, Shi Y. 2011. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA 17: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherstnev A, Duc C, Cole C, Zacharaki V, Hornyik C, Ozsolak F, Milos PM, Barton GJ, Simpson GG. 2012. Direct sequencing of Arabidopsis thaliana RNA reveals patterns of cleavage and polyadenylation. Nat Struct Mol Biol 19: 845–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2012. Alternative polyadenylation: new insights from global analyses. RNA 18: 2105–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manley JL. 2015. The end of the message: multiple protein–RNA interactions define the mRNA polyadenylation site. Genes Dev 29: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Alley TL, Wright SM, Kamdar S, Schott W, Wilpan RY, Mills KD, Graber JH. 2009. Global changes in processing of mRNA 3′ untranslated regions characterize clinically distinct cancer subtypes. Cancer Res 69: 9422–9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Graber JH. 2012. Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip Rev RNA 3: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Manley JL. 2017. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 18: 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Hu J, Zhang H, Lutz CS. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res 33: 201–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Subtelny AO, Koppstein D, Bell GW, Sive H, Bartel DP. 2012. Extensive alternative polyadenylation during zebrafish development. Genome Res 22: 2054–2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S, Pelechano V, Jarvelin AI, Tekkedil MM, Anders S, Benes V, Steinmetz LM. 2013. An efficient method for genome-wide polyadenylation site mapping and RNA quantification. Nucleic Acids Res 41: e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AJ, Aitken RJ, Chandler MA, Gillespie KM, Lampasona V, Bingley PJ. 2008. Autoantibodies to islet antigen-2 are associated with HLA-DRB1*07 and DRB1*09 haplotypes as well as DRB1*04 at onset of type 1 diabetes: the possible role of HLA-DQA in autoimmunity to IA-2. Diabetologia 51: 1444–1448. [DOI] [PubMed] [Google Scholar]
- Wu X, Liu M, Downie B, Liang C, Ji G, Li QQ, Hunt AG. 2011. Genome-wide landscape of polyadenylation in Arabidopsis provides evidence for extensive alternative polyadenylation. Proc Natl Acad Sci 108: 12533–12538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing D, Li QQ. 2011. Alternative polyadenylation and gene expression regulation in plants. Wiley Interdiscip Rev RNA 2: 445–458. [DOI] [PubMed] [Google Scholar]
- Zhang H, Lee JY, Tian B. 2005. Biased alternative polyadenylation in human tissues. Genome Biol 6: R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Hyman L, Moore C. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev 63: 405–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Tian B. 2014. RNA-binding proteins in regulation of alternative cleavage and polyadenylation. Adv Exp Med Biol 825: 97–127. [DOI] [PubMed] [Google Scholar]
- Zheng D, Liu X, Tian B. 2016. 3′READS+, a sensitive and accurate method for 3′ end sequencing of polyadenylated RNA. RNA 22: 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.