Abstract
RNA contains over 100 modified nucleotides that are created post-transcriptionally, among which pseudouridine (Ψ) is one of the most abundant. Although it was one of the first modifications discovered, the biological role of this modification is still not fully understood. Recently, we reported that a pseudouridine synthase (TgPUS1) is necessary for differentiation of the single-celled eukaryotic parasite Toxoplasma gondii from active to chronic infection. To better understand the biological role of pseudouridylation, we report here gel-based and deep-sequencing methods to identify TgPUS1-dependent Ψ’s in Toxoplasma RNA, and the use of TgPUS1 mutants to examine the effect of this modification on mRNAs. In addition to identifying conserved sites of pseudouridylation in Toxoplasma rRNA, tRNA, and snRNA, we also report extensive pseudouridylation of Toxoplasma mRNAs, with the Ψ’s being relatively depleted in the 3′-UTR but enriched at position 1 of codons. We show that many Ψ’s in tRNA and mRNA are dependent on the action of TgPUS1 and that TgPUS1-dependent mRNA Ψ’s are enriched in developmentally regulated transcripts. RNA-seq data obtained from wild-type and TgPUS1-mutant parasites shows that genes containing a TgPUS1-dependent Ψ are relatively more abundant in mutant parasites, while pulse/chase labeling of RNA with 4-thiouracil shows that mRNAs containing TgPUS1-dependent Ψ have a modest but statistically significant increase in half-life in the mutant parasites. These data are some of the first evidence suggesting that mRNA Ψ’s play an important biological role.
Keywords: pseudouridine, Toxoplasma gondii
INTRODUCTION
Although more than 100 RNA modifications have been discovered to date, the biological significance of the majority remains elusive (Machnicka et al. 2013). One of the most abundant, ubiquitous, and highly conserved modifications is pseudouridine (psi or Ψ), an isomerization of uridine in which the C5 position, rather than the N1, is incorporated into the glycosidic bond (Cohn 1959, 1960; Scannell et al. 1959; Yu and Allen 1959). This modification is found at high levels in rRNA in multiple organisms and is highly evolutionarily conserved at multiple sites in rRNA, tRNA, and spliceosomal RNA (Volkin and Cohn 1951; Spenkuch et al. 2014; Li et al. 2016). Most recently, with the advent of high-throughput sequencing tools, Ψ has also been found in mRNAs (Carlile et al. 2014; Lovejoy et al. 2014; Schwartz et al. 2014; Li et al. 2015).
This extensive pseudouridylation is enabled by a large family of enzymes, called pseudouridine synthases (or PUSs), which catalyze the isomerization. These enzymes can be divided into RNA-independent PUSs, which recognize their targets based on a combination of sequence and secondary structure, and RNA-dependent PUSs, which recognize their targets with the help of a protein complex containing an H/ACA guide RNA that is complementary to the target sequence (Ofengand 2002; Hamma and Ferré-D'Amaré 2006). While RNA-independent PUSs target tRNAs, snRNAs, mRNAs, and prokaryotic rRNA, RNA-dependent PUSs target snRNAs, mRNAs, and eukaryotic rRNA (Spenkuch et al. 2014; Safra et al. 2017).
The role of Ψ remains poorly understood, although it is known that Ψ can affect the secondary structure and base-pairing of RNA (Arnez and Steitz 1994; Davis 1995; Newby and Greenbaum 2002a; Hudson et al. 2013; Kierzek et al. 2014). In this way, Ψ’s can stabilize tRNA folding and increase the fidelity of codon–anticodon interactions, stabilize certain stem–loop conformations of snRNAs, and enable ribosome assembly (Yarian et al. 1999; Newby and Greenbaum 2001, 2002b; Nobles et al. 2002; Cabello-Villegas and Nikonowicz 2005; Denmon et al. 2011; Jack et al. 2011; Penzo et al. 2015). The majority of indications that pseudouridylation plays an important biological role stems indirectly from the observation that Ψ’s are modulated in response to different cellular conditions. Changes in the extent of rRNA pseudouridylation at specific sites changes with temperature and development, while entirely novel sites of pseudouridylation appear in snRNAs and mRNAs with stresses such as heat shock, starvation, oxidative stress, and immune stimulation (Wu et al. 2011; Basak and Query 2014; Carlile et al. 2014; Schwartz et al. 2014; Li et al. 2015; Chikne et al. 2016).
The functional consequences of changes in pseudouridylation have only been characterized in a small number of cases. First, mutations in the human rRNA PUS (called DKC1) can lead to impaired binding affinity to both IRESs and tRNAs, leading to reduced translation of specific anti-tumor factors and reduced translational fidelity (Jack et al. 2011; Penzo et al. 2015). These mutations manifest as the disease X-linked dyskeratosis congenital (X-DC), which ultimately manifests as cancer (Marrone and Mason 2003). Second, Basak and Query (2014) showed that a Ψ in U6 snRNA, which is induced when yeast transition to filamentous growth, can alter spliceosome function.
We previously reported that a PUS is necessary for differentiation of the parasite Toxoplasma gondii (Anderson et al. 2009). This single-celled eukaryote can exist within its human host as fast-growing tachyzoites or as slower-growing encysted bradyzoites. In two independent screens for Toxoplasma mutants that are defective in differentiation from tachyzoites to bradyzoites, we found mutants with disruption of a predicted PUS (TGME49_202640) (Singh et al. 2002; Anderson et al. 2009). This protein was named TgPUS1, as it was the first PUS investigated in Toxoplasma, and not due to any specific homology between TgPUS1 and yeast PUS1. Anderson (2011) went on to show that a point mutant in the predicted key catalytic residue of TgPUS1 phenocopied the knockout and was unable to differentiate, strongly suggesting that the PUS activity of this protein is necessary for its role in differentiation. Given the paucity of phenotypes associated with PUSs, we exploited this finding to determine the role of pseudouridylation in a tightly regulated biological process. To this end, we have used a deep-sequencing method to identify TgPUS1-dependent Ψ’s in Toxoplasma RNA from multiple developmental stages and taken advantage of the mutant parasites to examine the effect of pseudouridylation on RNA biology. We show that the extensive pseudouridylation of Toxoplasma mRNAs is unequally distributed throughout transcripts and that these mRNA Ψ’s have a modest but significant impact on RNA expression.
RESULTS
CMCT treatment combined with deep sequencing can be used to identify putative Ψ’s
In a series of preliminary experiments we validated our ability to detect Ψ’s using the chemical CMCT (which specifically labels Ψ’s) using low- and high-throughput methods that were previously published (Ofengand et al. 2001; Lovejoy et al. 2014). Primer extensions of CMCT-treated human RNA targeting a region known to contain five Ψ’s were used to identify conditions that produce the most efficient tagging of Ψ’s (data not shown). Next we profiled pseudouridylation using the deep-sequencing method PSI-seq (pseudouridine site identification sequencing) (Lovejoy et al. 2014). Briefly, after CMCT-treating RNA and reverse-transcribing it, a gel-based size-selection was used to enrich for aborted cDNAs (i.e., sites where the RT stopped prematurely). After mapping, each base in the transcriptome was analyzed to determine the number of sequencing reads that had a 3′-end one base downstream, indicative of a RT stop. Analysis of human rRNA showed that a subset of U residues enriched for known sites of human large subunit (LSU) rRNA Ψ’s tended to show more stops in the CMCT+ condition (data not shown). Additionally while a comparable number of reads stopped at the 4 nucleotides (nt) in the CMCT– condition, 58% of reads in the CMCT+ condition stopped at a U residue (the only residue at which we would expect to see pseudouridylation) (data not shown). Taken together these results indicate that deep sequencing with CMCT treatment can be used to detect Ψ’s in bulk RNA.
Human rRNA data can be used to establish criteria for identification of Ψ’s with high confidence
To define the hallmarks of a Ψ in our deep-sequencing data, we examined five parameters (adapted from Schwartz et al. 2014): signal, penetrance, penetrance ratio, identity of the initially transcribed nucleotide, and reproducibility, as defined below. Each of these parameters was optimized to reduce the rate of false positives due to experimental noise.
The “signal” parameter required that a certain minimum number of reads stopped at the residue of interest in the CMCT+ condition. The “penetrance” parameter required that a certain fraction of reads covering each nucleotide position stop at that nucleotide instead of reading through it. “Penetrance” may be affected by several factors including completeness of the CMCT treatment and extent of pseudouridylation of a site within the total cellular pool of a given gene's transcripts. “Penetrance ratio” was defined as the penetrance in the CMCT+ condition divided by the penetrance in the CMCT– condition (used to ensure that the RT did not stop constitutively at the site). The fourth parameter required that all hits be at positions corresponding to U in the initial transcript, since U is the only residue that can be pseudouridylated. Finally, the “reproducibility” parameter required that a residue meet these requirements in two independent sequencing runs. Although some non-U residues met the signal, penetrance, penetrance ratio, and reproducibility requirements that were ultimately chosen, the vast majority (89%) of the residues meeting these requirements were U (data not shown).
To determine where to set these various parameters to identify Ψ’s while minimizing false-discovery (FDR) and false-omission rates (FOR), we examined the CMCT– and CMCT+ data from human LSU rRNA (which has well-defined Ψ’s). After first setting the signal requirement to ≥5 reads, the penetrance and penetrance-ratio parameters were adjusted and the FDR and FOR calculated at various cutoffs (data not shown). Requiring that the penetrance in the CMCT+ condition be ≥0.12 (i.e., at least 12% of reads covering that nucleotide stopped there) and the penetrance ratio be ≥3.5 (i.e., penetrance was ≥3.5-fold higher in CMCT+ vs. CMCT– conditions) yielded the optimal FDR and FOR. Using these criteria, 41/55 known Ψ’s in human LSU rRNA were correctly identified with six false positives (Table 1; Supplemental Table 1). Interestingly, we note that three of these six “false positives” were identified as Ψ’s in two of the six other data sets we generated (from duplicates of tachyzoite-infected, bradyzoite-infected or stressed cells) and the other three were found in three, four, and five of the data sets, respectively. Thus it is possible that some or all of these six positions may be novel sites of pseudouridylation in HFFs, a cell line not previously examined, and the true FDR for identification of Ψ’s in RNA from the uninfected cells may be lower than 0.13. When applied to the other conditions examined, these criteria produced similar or lower FDRs for human rRNA Ψ’s (Table 1; Supplemental Table 1).
TABLE 1.
False-discovery rate of human LSU rRNA Ψ’s in tachyzoite-infected, bradyzoite-infected, uninfected, and stressed sequencing runs 1 and 2
PSI-seq identifies evolutionarily conserved sites of pseudouridylation in Toxoplasma rRNAs
To begin the identification of Ψ’s in Toxoplasma, i.e., its predicted “pseudouridylome,” we performed PSI-seq on tachyzoite and bradyzoite RNA isolated from wild-type parasites (specifically C32 parasites, a clone derived from a Type II Pru strain [Singh et al. 2002]). Tachyzoites were grown in neutral pH and serum-rich conditions, while bradyzoite differentiation was induced using starvation (1% FBS) and high pH (8.0). Two independent samples were prepared for each condition and sequenced independently (these samples will be referred to as “Tachy 1,” “Tachy 2,” “Brady 1,” and “Brady 2” for tachyzoite and bradyzoite, sequencing runs 1 and 2, respectively). All samples except Tachy 1 were sequenced on an Illumina NextSeq 500, while Tachy 1 was sequenced on an Illumina GAII. We calculated the number of reads for which we had 3′-end information as a reflection of sequencing depth. The results showed that the sequencing depth varied significantly between samples, particularly in relation to the number of reads consumed by human RNA, although this was generally not a problem since all our analyses are based on relative stop frequencies.
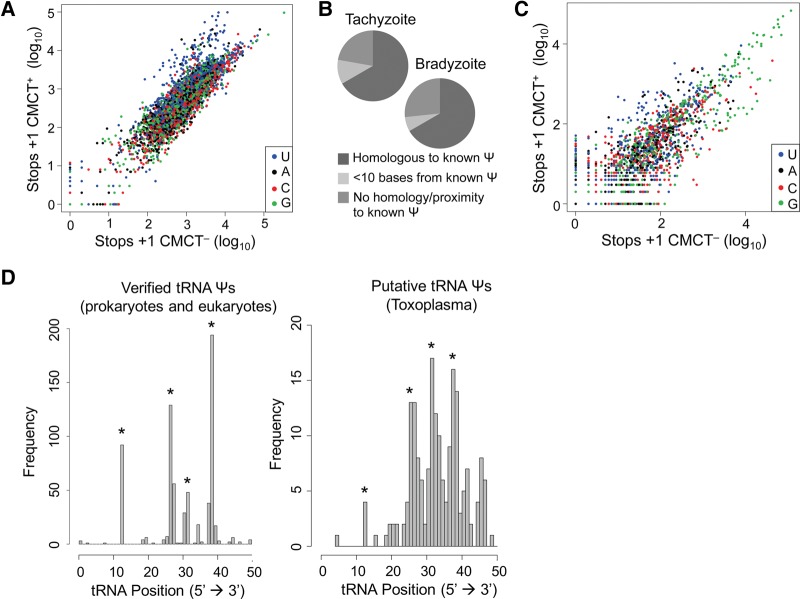
To confirm that the approach successfully identified Ψ’s in Toxoplasma RNA, we examined strongly predicted sites of pseudouridylation in rRNA and tRNA based on evolutionarily conserved positions found throughout eukaryotes. First, the RT stops seen in CMCT– and CMCT+ conditions were plotted for each nucleotide in Toxoplasma LSU rRNA. The results showed that the residues with more RT stops in the CMCT+ sample are predominantly U (Fig. 1A). This suggests that CMCT treatment of rRNA was effective. Next we applied the criteria developed with the human rRNA to these Toxoplasma rRNA sequencing data. The results showed that in Toxoplasma LSU rRNA, 18 U's are predicted to be Ψ’s in tachyzoites, while 15 were predicted in bradyzoites (Supplemental Table 1). Of the 18 putative Ψ’s identified in tachyzoites, 12 correspond to known Ψ’s in human LSU rRNA, two were within 10 nt of a known human LSU Ψ, and four showed no homology with human LSU Ψ’s (Fig. 1B). Of the 15 putative bradyzoite Ψ’s, 10 showed direct correspondence, one was within 10 nt of a human LSU Ψ, and four showed no homology with known human LSU Ψ’s (Fig. 1B). We note that of the four “false positives” found in each condition, two were identified as putative Ψ’s in all four sequencing runs (Tachy 1, Tachy 2, Brady 1, and Brady 2); therefore, these are likely to be true Ψ’s that are at positions that are not evolutionarily conserved. In addition to the sites in Toxoplasma LSU rRNA, the data also predict the location of nine and 11 Ψ’s in small subunit (SSU) rRNA of tachyzoites and bradyzoites, respectively (Supplemental Table 1).
FIGURE 1.
PSI-seq successfully identifies predicted pseudouridines in Toxoplasma rRNAs and tRNAs. (A) Scatterplot depicting the number of 3′-ends mapping to each rRNA residue in CMCT– conditions (x-axis) and CMCT+ conditions (y-axis). One read was added to each read number to allow the axes to be log-transformed. Residues are colored by nucleotide, as indicated. (B) Proportion of Toxoplasma rRNA Ψ’s showing homology with known Ψ’s in the tachyzoite and bradyzoite samples. (C) As in A, but depicting the number of 3′-ends mapping to each tRNA residue. (D) (Left) Histogram showing frequency of Ψ’s verified at each tRNA position across the total population of tRNAs in studied eukaryotes and prokaryotes (Auffinger and Westhof 1998). Asterisks indicate the most frequently pseudouridylated sites. (Right) Histogram showing frequency of a Ψ hit at each position in the total population of Toxoplasma tRNAs, with the asterisked sites being the same positions indicated in the left panel. Only tRNA regions accessible by PSI-seq are plotted (i.e., ≤50 nt from the 5′ end).
To determine whether there are any Ψ’s in Toxoplasma rRNA that are dependent on TgPUS1, we compared predicted Ψ’s in wild-type vs. TgPUS1-deficient parasites in both tachyzoite and bradyzoite conditions. We note that these TgPUS1-deficient parasites are insertional mutants (TBD5, isolated by Singh et al. 2002 and characterized by Anderson et al. 2009), rather than TgPUS1 knockouts. Furthermore TgPUS1-deficient parasites incubated under bradyzoite conditions for 84 h are not able to differentiate; therefore, TgPUS1-deficient “bradyzoites” express subsets of both tachyzoite and bradyzoite markers (Anderson et al. 2009 and data not shown). Because of this any change in pseudouridylation observed between wild-type and TgPUS1-deficient parasites grown in bradyzoite conditions could be attributed to either the loss of TgPUS1 or the change in their development.
At each residue in the wild-type RNA, the probability of an RT stop in the CMCT+ condition was modeled using a binomial distribution based on the proportion of reads overlapping the position that stopped at the position. This model was used to calculate a P-value for the proportion of reads stopping in the TgPUS1-deficient parasites, allowing us to estimate the probability that a given residue represents a TgPUS1-dependent RT stop. To gain statistical power, the reads from both sequencing runs were pooled for these analyses. We required that the P-value be <0.05 to define a residue as “missing” in the mutant condition. To ensure that any change in pseudouridylation was both statistically and biologically significant, we also calculated a new parameter, the pooled penetrance. This is defined as the average penetrance for a given position in all combined sequencing runs. We required that the pooled penetrance in the wild-type CMCT+ sample be at least twofold greater than the pooled penetrance for that position in the TgPUS1-deficient CMCT+ sample. When these criteria were applied to the rRNA sequencing data from both tachyzoites and bradyzoites, we did not find any rRNA residues that were pseudouridylated in wild-type parasites but not in TgPUS1-deficient parasites. This is consistent with the fact that BLAST analysis places TgPUS1 as similar to members of the yeast RluA family of PUSs, PUS6, PUS8, and PUS9 (data not shown), all of which are known to modify tRNAs, not rRNAs (Hamma and Ferré-D'Amaré 2006).
PSI-seq identifies predicted and TgPUS1-dependent Ψ’s in Toxoplasma tRNAs
Given the results above, we next examined the tRNA sequencing data to look for conserved Ψ’s. Again, to determine whether CMCT treatment was effective, the RT stops seen in CMCT– and CMCT+ conditions were plotted for each nucleotide. The results show that the residues that show more RT stops in the CMCT+ sample are predominantly U's, consistent with CMCT treatment being effective (Fig. 1C). To determine whether PSI-seq identified Ψ’s corresponding to known sites of pseudouridylation in tRNAs from other eukaryotes, the criteria above were applied to the sequencing data. We note that PSI-seq is optimized to detect Ψ’s in RNAs that do not have other RT stops due to secondary structure or other modifications, whereas tRNAs are heavily modified and contain extensive secondary structure. A number of the CMCT-independent stops we observed were found in the region between positions 35 and 45 of the tRNAs, which are known to contain RT-blocking modifications (data not shown; Cozen et al. 2015). Because the majority of Ψ’s in tRNAs are typically located 5′ of positions 35–45, these blocks significantly lowered our power to detect tRNA Ψ’s. The frequency of putative Ψ’s predicted by this approach at each position of tachyzoite tRNAs (in both or either of the sequencing runs) was then plotted. The results showed putative Ψ’s that cluster at positions 13, 26–27, 32, and 39 of tRNAs (Fig. 1D, right; Supplemental Table 2). These positions are known to be pseudouridylated in multiple tRNAs across many organisms (Fig. 1D, left). We note that we could not detect Ψ’s at the 3′-end of tRNAs, including the most highly conserved site of pseudouridylation, site 55, due to the size selections that were used during PSI-seq.
Next we used the binomial distribution to compare these predicted Ψ’s in wild-type and TgPUS1-deficient parasites. The results revealed three TgPUS1-dependent Ψ’s, all in the 5′ half of the anticodon arm, specifically at positions 27 and 34 of tRNA Val and Asn (TGME49_251700 and TGME49_293750) in tachyzoites and position 28 of tRNA Ile (TGME49_262568) in bradyzoites. The lack of further TgPUS1-dependent residues may be a result of the low number of Ψ’s that we could reliably identify in tRNAs due to the technical limitations described above. Nevertheless, together with the rRNA data, these results show that PSI-seq correctly identified conserved sites of pseudouridylation in rRNA and tRNA and that TgPUS1 modifies tRNAs not rRNA.
Pseudouridylation of spliceosomal RNAs is TgPUS1-independent
Having identified Ψ’s in rRNA and tRNA, we next sought to characterize spliceosomal RNAs (snRNAs), which are known to contain Ψ’s in other systems. Because these RNAs are not expressed as highly as rRNAs or tRNAs, they were not present in sufficient quantities after two rounds of poly(A) purification to be profiled using PSI-seq. Thus, to identify such Ψ’s we performed primer extension reactions with snRNA-specific primers. Primers were designed to specifically target predicted Toxoplasma U2, U4, U5, and U6 snRNAs (see Materials and Methods for the gene numbers for each), end-labeled with 32P, and used to perform primer extensions on 1–2 µg of total RNA isolated from wild-type and TgPUS1-deficient tachyzoites and bradyzoites. RNA from uninfected host cells was also used as a control to ensure that the primers were specific to Toxoplasma snRNAs.
Primer extension on the predicted Toxoplasma U6 snRNA showed a single CMCT-dependent stop near the 5′ end of the molecule (Fig. 2A, arrow). This extension product was only seen with Toxoplasma RNA (cf. lanes 1–2 and lanes 3–10), and was not TgPUS1-dependent (arrow, lanes 4 and 6, 8 and 10). This putative Ψ corresponds to the single Ψ identified at position 28 in S. cerevisiae U6 snRNA.
FIGURE 2.
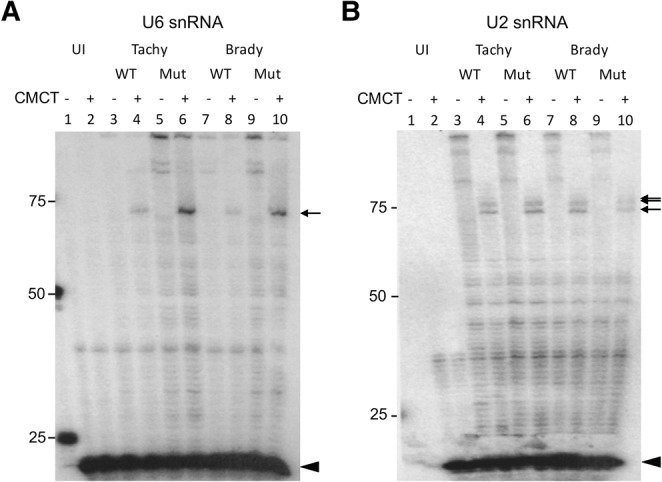
Pseudouridines in Toxoplasma snRNAs are homologous to those in Saccharomyces cerevisiae and human snRNAs, and are not TgPUS1-dependent. (A) Primer extension of Toxoplasma U6 snRNA using a primer targeting the 3′-end of the U6 transcript. RNA from uninfected HFF cultures (UI) and HFF cultures infected with wild-type (WT) or TgPUS1-deficient (Mut) tachyzoites and bradyzoites was analyzed following treatment with CMCT (+) or mock treatment (−), as indicated. The 3′-end of the primer is located 25 residues from the putative 3′-end of the U6 snRNA. Radiolabeled cDNAs were resolved on an 8% polyacrylamide gel. Unextended primer is seen at the bottom of the gel (black arrowhead) while the location of the putative Ψ is indicated with an arrow. The migration of size markers (given in nucleotides) are indicated on the left. (B) As in A, but showing primer extension of Toxoplasma U2 snRNA. The 3′-end of the primer is located 87 residues from the putative 3′-end of the U2 snRNA.
Primer extension on the predicted U2 snRNA showed three CMCT-dependent stops specific to Toxoplasma RNA (Fig. 2B, arrow, cf. lanes 1–2 and 3–10) and, as for U6, these stops were not dependent onTgPUS1 (arrows, lanes 4 and 6, 8 and 10). These putative Ψ’s are found in a region of U2 that is pseudouridylated in S. cerevisiae and humans (Spenkuch et al. 2014).
Primer extension on the remaining predicted snRNAs did not conclusively identify any Toxoplasma-specific Ψ’s. While U4 snRNA showed three CMCT-dependent stops, the same patterns of stops were observed in the uninfected samples, making it unclear whether these correspond to Toxoplasma-specific Ψ’s or the known Ψ’s at these positions in host U4 snRNA (data not shown). Although S. cerevisiae and human U1 and U5 snRNAs contain 1–3 Ψ’s each, primer extension of the putative Toxoplasma U5 and U1 snRNAs revealed no CMCT-dependent stops (data not shown). The lack of putative Ψ’s identified in U5 and U1 snRNAs may indicate that these snRNAs are not pseudouridylated in Toxoplasma, although we cannot exclude the possibility that the putative U1 and U5 snRNAs are incorrectly annotated and other sequences are the true U5 and U1 genes (these two snRNAs are less conserved than the other snRNAs and so less easily identified unambiguously). Regardless, these results indicate that at least some Toxoplasma spliceosomal RNAs contain sites of pseudouridylation corresponding to those seen in S. cerevisiae and human snRNAs, and that TgPUS1 is not required for the Toxoplasma snRNA Ψ’s identified here.
There is extensive, regulated pseudouridylation of Toxoplasma mRNA
To determine whether Toxoplasma mRNAs contain pseudouridines, we applied the five criteria for identification of Ψ’s to mRNA sequencing data. The results showed evidence for 1669 and 394 sites of pseudouridylation in tachyzoite and bradyzoite mRNAs, respectively (Supplemental Table 3). These results indicate that there is extensive pseudouridylation of parasite mRNA in both tachyzoite and bradyzoite conditions, supporting data recently published for yeast and mammalian mRNA (Carlile et al. 2014; Schwartz et al. 2014; Li et al. 2015).
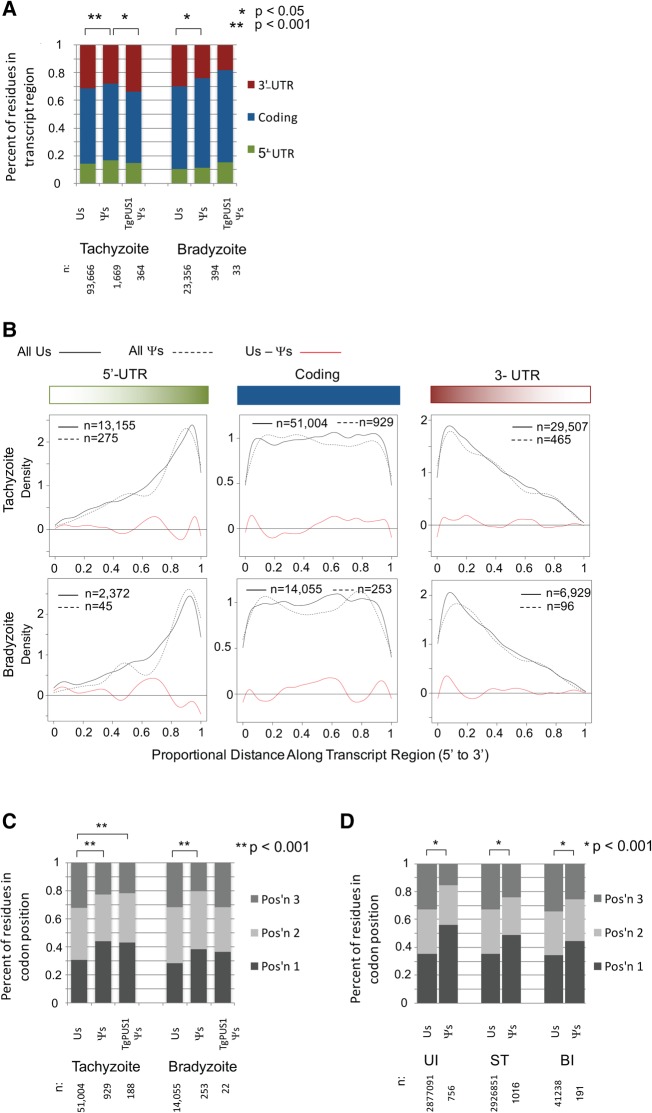
To determine whether these Ψ’s are enriched in particular regions of the mRNAs, the Ψ’s were divided into those found in the 5′-UTR, the coding region, and the 3′-UTR. The results showed that Ψ’s are present in all regions of the transcript (Fig. 3A, Tachyzoite Ψ’s and Bradyzoite Ψ’s). A χ2 test was used to determine whether the distribution of Ψ’s differed from the distribution of all transcribed U's in these mRNAs. The results showed that the distribution of U's and Ψ’s is significantly different between the three regions of the transcript (Fig. 3A, Tachyzoite and Bradyzoite U's vs. Ψ’s). The decrease in Ψ’s relative to U's in the 3′-UTR is similar to what has been observed in yeast, where it was also found that pseudouridylation is underrepresented in the 3′-UTR (Carlile et al. 2014). In contrast to what has been reported for human mRNAs, however, our data do not show underrepresentation of Ψ’s in the 5′-UTR of Toxoplasma mRNAs (Li et al. 2015). Together, these data indicate that Ψ’s are distributed nonrandomly in Toxoplasma mRNAs.
FIGURE 3.
Toxoplasma mRNA pseudouridines are distributed unequally within the RNA transcript as well as within the codons. (A) The proportion of tachyzoite and bradyzoite U's, Ψ’s, and TgPUS1-dependent Ψ’s found in each region of the transcript (5′-UTR, coding region, and 3′-UTR) based on gene models available through ToxoDB.org (release 27). The absolute number of residues in each category is displayed below the graph (n). Asterisks indicate statistical significance as determined using a χ2 test. All differences that were statistically significant are indicated. (B) Density plot showing the distribution of U's (solid line) and Ψ’s (dashed line) within each region of the transcript. X-axis shows the proportional distance along the transcript region from 5′ to 3′. (Left) 5′-UTR; (center) coding region; (right) 3′-UTR. Red line shows the differential between the density plots (U's–Ψ’s). The absolute number of residues is displayed on the graph (n). (C) The proportion of tachyzoite and bradyzoite U's, Ψ’s, and TgPUS1- dependent Ψ’s found in each position of the codon. Details as in A. (D) The proportion of human U's and Ψ’s found in each position of the codon in Uninfected (UI), Stressed (ST), and Bradyzoite-infected (BI) conditions. Details as in A. (E) DNA sequence motif surrounding all positions of pseudouridylation identified in tachyzoites and bradyzoites (left), non-U positions that met Ψ criteria of signal, penetrance, penetrance ratio, and reproducibility (middle), and TgPUS1-dependent sites of pseudouridylation identified in tachyzoites and bradyzoites (right). The position being analyzed is site 0; sites −4 through −1 are found 5′ of the residue, while sites 1–4 are found 3′ of the residue. The height of each letter reflects the degree of conservation (maximum possible value is 2). (F) Cumulative distribution function of the ratio of (expression in tachyzoites):(expression in bradyzoites) for genes containing a TgPUS1-independent Ψ (red line) or a TgPUS1-dependent Ψ (blue line) in tachyzoite conditions. Expression values were normalized based on total reads in each sequencing run. Kolmogorov–Smirnov test was performed on the data to determine statistical significance. (G) As in F, but for Ψ’s identified in bradyzoite conditions.
To further refine the analysis, we also mapped the density of pseudouridylation within each region of the transcripts. Because sequencing depths vary along the length of a transcript, the density of all U's in these mRNAs was also plotted as a control. Figure 3B shows the density of U's and Ψ’s within each region of the transcript in tachyzoites (top) and bradyzoites (bottom). These data show that the density of U's is essentially constant throughout the coding region (center, solid lines). Within the UTRs, the density of U's decreases with distance from the translation start/stop sites (right and left plots, solid lines). This is almost certainly because the annotated UTRs are based on algorithms rather than empirical data; our data as well as RNA-seq data available through ToxoDB show that the UTR lengths are frequently overestimated. Consistent with this, the density of Ψ’s is similar to that of U's, with no clustering at start or stop codons, or at any other site within the transcript regions (Fig. 3B, dotted lines). To more accurately quantify the differences, the density of Ψ’s was subtracted from the density of U's. The results show that there is no apparent difference in the distribution of Ψ’s compared to U's within any of the transcript regions (Fig. 3B, red lines). Together with the results above this suggests that although Ψ’s are underrepresented in the 3′-UTR compared to U's, this is not restricted to any particular subregion within the 3′-UTR.
Within the coding region, we also analyzed the distribution of Ψ’s relative to U's at each of the three codon positions. A χ2 test was again used to determine whether the distribution of Ψ’s differed from that of all U's in codons within these mRNAs. The results showed that there was a statistically significant difference in the distribution of Ψ’s compared to U's within the codon, with a higher rate of pseudouridylation seen at position 1 of the codon relative to positions 2 and 3 (Fig. 3C, Tachyzoite and Bradyzoite U's vs. Ψ’s). We note that this bias was also seen when comparing the codon position of Ψ’s to all U's in the host mRNA data (Fig. 3D). These results are consistent with the observation that in human mRNA, positions 1 and 2 of the UUU codon are found to be pseudouridylated, but rarely position 3 (Li et al. 2015). Together, these results indicate that pseudouridylation of mRNAs is nonrandom, both with respect to overall region and specific codon position.
PUS1 catalyzes pseudouridylation events in tachyzoite-specific mRNAs in a sequence-independent manner
To determine whether any mRNA Ψ’s were TgPUS1-dependent we again compared pseudouridylation of residues in wild-type and TgPUS1-deficient parasites using the binomial probability distribution. As with the rRNA and tRNA analyses, we required that the P-value be <0.05 to define a residue as “missing” in the mutant condition, and that the pooled penetrance in the wild-type CMCT+ sample be at least twofold greater than the pooled penetrance of the mutant CMCT+ sample. These criteria identified 364 TgPUS1-dependent Ψ’s in tachyzoite mRNA, and 33 TgPUS1-dependent Ψ’s in bradyzoite mRNA (Supplemental Table 4).
To determine whether TgPUS1 preferentially pseudouridylates a particular region of the mRNAs, we also examined the distribution of TgPUS1-dependent Ψ’s between the UTRs and coding region of RNAs. The data show that TgPUS1-dependent Ψ’s are abundant in all regions of the mRNA (Fig. 3A, Tachyzoite and Bradyzoite TgPUS1 Ψ’s). Notably, the lower extent of pseudouridylation in the 3′-UTR observed above with all Ψ’s was not seen with TgPUS1-dependent Ψs in the Tachyzoite sample, and the distribution of TgPUS1-dependent Ψ’s did not differ significantly from the distribution of all U's by χ2 analysis. Density plots showed that, as with all Ψ’s, tachyzoite TgPUS1-dependent Ψ’s were not found to cluster at any subregion within these coding and untranslated regions (data not shown). Note that because of the low number of TgPUS1-dependent Ψ’s found in the bradyzoite sample, no statistically significant conclusions could be drawn about the distribution of these residues within those RNA transcripts. Overall, however, these data suggest that TgPUS1 does not target a particular region of mRNA for pseudouridylation.
To determine whether TgPUS1-dependent Ψ’s favor a specific codon position, we analyzed the distribution of TgPUS1-dependent Ψ’s in the first, second, and third positions of codons. χ2 analysis shows that the distribution of TgPUS1-dependent Ψ’s in tachyzoites differs significantly from the distribution of all U's within the codon, but does not differ significantly from the distribution of TgPUS1-independent Ψ’s (Fig. 3C, Tachyzoite TgPUS1 Ψ’s). Thus the increase in pseudouridylation at position 1 of the codon described above for all Ψ’s is recapitulated in the subpopulation of tachyzoite Ψ’s that are TgPUS1-dependent. No conclusions could be drawn about the distribution of bradyzoite TgPUS1-dependent Ψ’s due to the low number (22) identified. These results suggest that at least in tachyzoites, TgPUS1-dependent Ψ’s are distributed within the codon comparably toTgPUS1-independent Ψ’s.
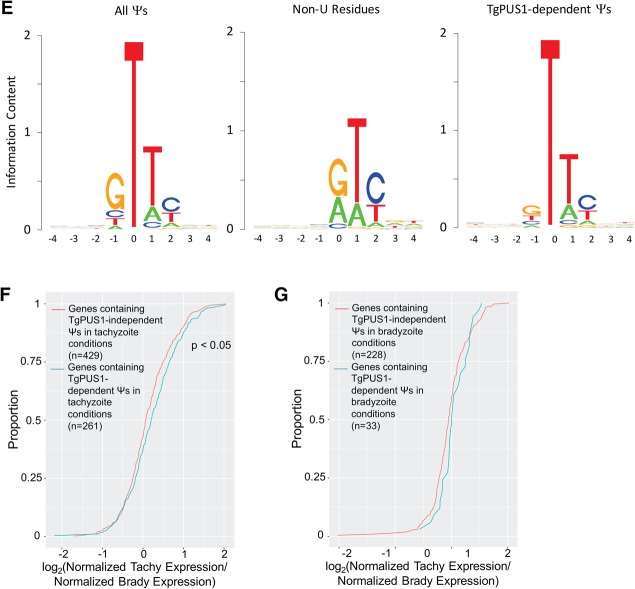
Some tRNA pseudouridine synthases recognize their targets partially based on a motif present in the surrounding sequence, while others seem to rely on secondary structure (Hamma and Ferré-D'Amaré 2006). To determine whether TgPUS1-dependent Ψ’s are found in a particular motif, we analyzed the sequence surrounding such sites. All tachyzoite and bradyzoite TgPUS1-dependent Ψ’s were pooled for this analysis, and the motif surrounding TgPUS1-dependent Ψ’s was compared to that surrounding all tachyzoite and bradyzoite Ψ’s. Sequence motifs were analyzed as previously described (Schneider and Stephens 1990) using the seqLogo package in R (note that this software displays the DNA sequence, and so positions of U's and Ψ’s are represented as “T”). Although TgPUS1-independent Ψ’s were identified in a strong GΨUY motif, the XUY motif (where X is the index residue and Y is a pyrimidine) was also seen when we examined non-U residues that met the Ψ criteria (Fig. 3E, left and center). These results suggest that there is some artifact (potentially due to RT stop bias) that is responsible for the UY motif after the index residue, but that the 5′ G is specific to Ψ residues. Analysis of the sequence surrounding TgPUS1-dependent Ψ’s revealed the same 3′ “UY” bias, although the G bias seen 5′ of TgPUS1-independent Ψ’s was not seen, suggesting that TgPUS1 does not recognize its targets based on sequence context (Fig. 3E, right). This is consistent with TgPUS1's homology with yeast and human tRNA PUSs that also do not rely on a surrounding motif, although the G bias seen 5′ of TgPUS1-independent Ψ’s suggests that the other PUS(s) acting on Toxoplasma mRNA do show some sequence bias.
To determine whether TgPUS1-dependent pseudouridines are found in a functionally related subset of transcripts, we performed GO-term analysis on the set of mRNAs containing TgPUS1-independent Ψ’s as well as those containing TgPUS1-dependent Ψ’s. Because of the stringent requirements imposed for our analyses, the majority of Ψ’s identified were in highly expressed transcripts. This strongly confounded the GO-term enrichment analysis with both sets of genes being dominated by biological function encoded on the most abundant mRNAs (data not shown). The question of whether there is biological relatedness to these different subsets, therefore, cannot be addressed here.
To determine whether TgPUS1 preferentially pseudouridylates developmentally regulated transcripts, we compared the expression of transcripts containing TgPUS1-dependent Ψ’s in the tachyzoite vs. bradyzoite samples. Transcripts containing TgPUS1-independent Ψ’s were used as a control. Although traditional RNA-seq was not performed on these parasites, the control samples from the PSI-seq experiment (which were not chemically treated) could be used to estimate transcript expression levels. First, the number of reads mapped to each transcript in the tachyzoite and bradyzoite samples was normalized to the total number of reads in each sequencing run (reads per million reads mapped, or RPM), and the RPM of each mRNA species was averaged across the two biological replicates performed in each developmental stage. The ratio of (average tachyzoite RPM: average bradyzoite RPM) was then calculated for each gene. The results show that control transcripts containing TgPUS1-independent Ψ’s are, on average, not expressed at different levels in tachyzoites and bradyzoites (Fig. 3F, red line; note that the 0.5 proportion has a log2 value of 0.065; fold-change = 1.04). In contrast, transcripts containing tachyzoite TgPUS1-dependent Ψ’s are expressed ∼11% more abundantly in tachyzoites than bradyzoites (Fig. 3F, blue line; 0.5 proportion has a log2 value of 0.203). This difference was statistically significant when examining genes pseudouridylated in the tachyzoite condition (P = 0.036); while this pattern also existed when we examined transcripts that contain Ψ’s in bradyzoite conditions the difference was not statistically significant (P = 0.115) (Fig. 3G).
Pseudouridylation of the 5′-UTR and coding regions has a modest effect on steady-state mRNA levels
Having identified a number of transcripts containing TgPUS1-dependent Ψ’s, we next used these to study the impact of losing Ψ’s from mRNAs that normally contain them. Because transcripts containing TgPUS1-dependent Ψ’s are pseudouridylated in only the wild-type parasites, the mutant parasites can be used to examine RNA metabolism in the absence of these Ψ’s. Note that this analysis could only be done for transcripts in the tachyzoite stage: Mutant parasites do not fully differentiate in bradyzoite conditions and so differences in RNA metabolism of transcripts in mutant vs. wild-type bradyzoites will be substantially confounded by developmental differences.
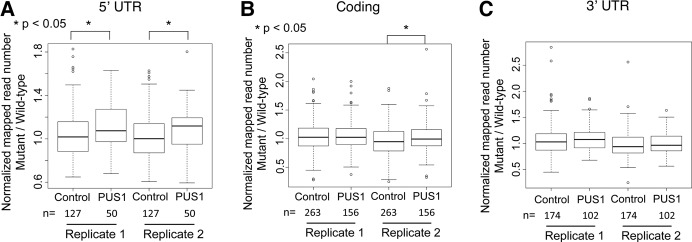
First, to determine whether pseudouridylation might affect the steady-state levels of mRNAs, we compared the RPM of transcripts containing a TgPUS1-dependent Ψ in TgPUS1-deficient and wild-type parasites. Transcripts containing a TgPUS1-independent Ψ were used as a control, as the absence of TgPUS1 would not be expected to have an impact on their abundance. This was also used to account for any biases that may be associated with our stringent criteria for declaring a RNA as Ψ-containing, such as a certain minimum number of reads. Because modifications in different regions of a transcript can affect RNA differently we divided transcripts into groups based on the region of the transcript in which the Ψ was found (5′-UTR, coding region, and 3′-UTR). First, transcripts containing TgPUS1-dependent and -independent Ψ’s in the 5′-UTR were compared. The results showed that in both biological replicates the mutant:wild-type RPM ratio was ∼10% higher for transcripts containing a TgPUS1-dependent Ψ than the control transcripts in both replicates (Fig. 4A). This difference was statistically significant in both cases. The mutant:wild-type RPM ratio was also calculated for transcripts containing TgPUS1-dependent and TgPUS1-independent Ψ’s in the coding region and the 3′-UTR (Fig. 4B,C). The results showed that no significant difference was observed for mRNAs containing Ψ’s in the 3′-UTR, although a difference was observed in one biological replicate for mRNAs with Ψ’s in the coding region. These results suggest that TgPUS1-dependent pseudouridylation leads to lower steady-state levels of substrate mRNAs.
FIGURE 4.
Transcripts containing TgPUS1-dependent Ψ’s show increased expression in mutant parasites. (A) Boxplot showing the distribution of the ratio of (reads mapped to transcripts in TgPUS1-mutant parasites):(reads mapped to transcripts in wild-type parasites). The box indicates the 75th, 50th, and 25th percentiles while tails show the most extreme value within the Q3 to Q3 + 1.5(IQR) range (top) or Q1 to Q1 − 1.5(IQR) range (bottom). Control indicates genes containing a TgPUS1-independent Ψ in the 5′-UTR; PUS1 indicates genes containing a TgPUS1-dependent Ψ in the 5′-UTR. Number of genes in each group of transcripts is displayed below the graph. Two independent biological replicates are displayed. Wilcoxon sum rank test was used to determine statistical significance. (B) As in A, but Ψ’s were identified in the coding region. (C) As in A, but Ψ’s were identified in the 3′-UTR.
4TU can be used to specifically label and purify parasite RNA
To explore the possibility that pseudouridylation of mRNA affects the stability of the RNA in which it is found, a method for pulse–chase labeling of mRNA was needed. The thio-substituted nucleoside 4-thiouracil (4TU) is a substrate for the Toxoplasma salvage enzyme uracil phosphoribosyltransferase (UPRT), which is missing in mammalian cells; this allows the parasite but not the host to incorporate the 4TU into newly synthesized RNAs (Cleary et al. 2005; Zeiner et al. 2008). Thiol-reactive biotin can then be conjugated to the 4TU-containing RNA, and the biotinylated RNA can be purified from the total RNA pool using streptavidin beads and sequenced (Rabani et al. 2011; Schwanhäusser et al. 2011; Windhager et al. 2012).
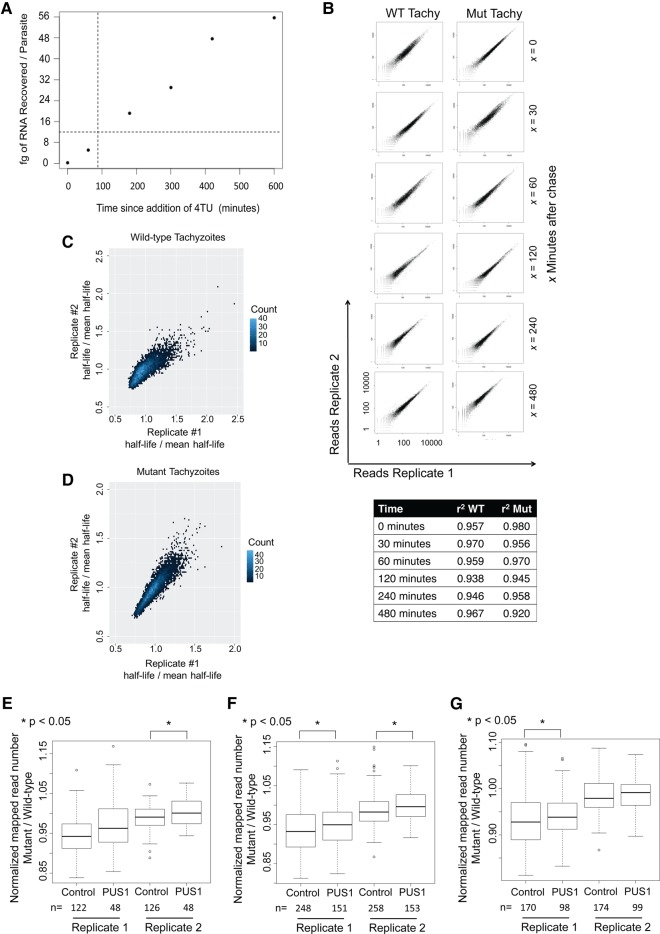
To determine the appropriate pulse length for 4TU labeling, we performed a time course examining 4TU recovery over an extended period. While ideally this pulse would have been under 10 min, the incorporation and subsequent level of RNA recovered from such pulses was not reliably above background (∼1 fg/parasite, data not shown). To be at or above background levels of RNA recovery 4 h into the chase (chosen because detection at this time point should allow examination of both long- and short-lived transcripts), and given that Toxoplasma transcripts were previously reported to have an average half-life of about 1 h (Cleary et al. 2005), it was necessary to start with ∼12 fg/parasite of labeled RNA at the start of the chase. To determine when the parasites were labeled to this extent, HFF cells were infected at an MOI of 2, and 36 h later 4TU was added to the cultures to a concentration of 2 µM. The dishes were collected 0, 60, 180, 300, 420, and 600 min after addition of the 4TU. These data showed that a pulse of 90 min produced the requisite 12 fg/parasite of recovered 4TU-containing RNA (Fig. 5A). Furthermore, there was little if any detectable lag and a linear increase in RNA recovery throughout the time course, indicating that that the UTP pool is rapidly saturated and that the 4TU is present in vast excess.
FIGURE 5.
Transcripts containing PUS1-dependent Ψ’s show increased stability in mutant parasites. (A) Amount of RNA recovered/parasite throughout 10-h time course. Dotted lines indicate the time at which ∼12 fg of RNA were recovered/parasite. (B, top) Absolute number of reads mapped to each gene in the two replicates of each condition at each time point (top to bottom: 0, 30, 60, 120, 240, and 480 min after chase was started). Columns (left to right): wild-type (WT) tachyzoites, TgPUS1-deficient (Mut) tachyzoites. (Bottom) R2 values for each set of replicates. (C) Relative half-life of each gene (normalized to mean half-life in the total data set) in each wild-type tachyzoite replicate are plotted against one another to show reproducibility of the data. Color reflects density of points at each coordinate. (D) As in C, but replicates are shown for mutant tachyzoites. (E) Boxplot showing the distribution of the ratio of (mRNA half-life in TgPUS1-mutant parasites):(mRNA half-life in wild-type parasites) for mRNAs with a Ψ identified in the 5′-UTR. Two independent biological replicates are displayed. All other details as in Figure 4A. (F) As in E, but mRNAs shown are those with Ψ’s in the coding region. (G) As in E, but mRNAs shown are those with Ψ’s in the 3′-UTR.
The recovery of 4TU-containing RNAs can be used to calculate a relative half-life for each transcript
To determine RNA turnover transcriptome-wide we examined each species in the mRNA pool throughout the pulse-chase using deep-sequencing. Briefly, replicate cultures infected with wild-type and mutant parasites, grown under tachyzoite conditions, were pulsed with 4TU for 90 min before a chase with excess uracil was started. Cultures were harvested 30, 60, 120, 240, and 480 min after the chase was started (T30, T60, T120, T240, and T480) and 4TU-containing RNA was harvested. Libraries were made using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech/Takara), sequenced on a HiSeq 500 machine, and the reads were mapped to the Toxoplasma genome.
To determine if any of the samples had produced low-quality data, we examined the correlation between replicates. For each time point in wild-type and mutant parasites, the number of reads mapping to each gene in the first replicate was plotted against mapped reads in the second and the R2 value was calculated (Fig. 5B). The results show that for all time points there was good correlation between the replicates, with R2 values ≥0.92.
Next we set out to calculate the half-life of mRNA transcripts. Relative half-lives were calculated by log-transforming reads and fitting a linear model to the data; the best-fit line was used to calculate a relative half-life for each mRNA. Only mRNAs with at least five reads mapped at each time point and a R2 of the log2 model ≥0.80 were included in the analysis to ensure confidence in the results obtained. Of note, 5405 mRNAs met these requirements in both of the wild-type tachyzoite replicates, while 5751 mRNAs met them in both of the TgPUS1-deficient tachyzoite replicates. To ensure that the half-lives calculated were reproducible we plotted the relative half-life of each mRNA in the two replicates. The results show that in tachyzoite replicates there was good correlation between the relative half-lives of each gene between the wild-type replicates and, separately, between the TgPUS1-deficient replicates (Fig. 5C,D).
mRNA stability is modestly affected by TgPUS1-dependent pseudouridylation
To determine whether pseudouridylation affects mRNA turnover we compared the half-life of mRNAs containing TgPUS1-dependent Ψ’s in wild-type and TgPUS1-deficient parasites. As with the data looking at RNA steady-state levels, transcripts were divided into coding and UTR regions, and transcripts containing TgPUS1-independent Ψ’s were used as a control. For each transcript, the ratio of half-lifemutant:half-lifewild-type was calculated and the distribution of this ratio was represented as a boxplot. The results showed that for all comparisons, transcripts containing TgPUS1-dependent Ψ’s in any region of the transcript show a modest increase in half-life in the mutants compared to wild-type parasites (Fig. 5E–G). This difference was statistically significant in both replicates when examining the coding region (Fig. 5F), but only statistically significant in replicate 2 examining the 5′-UTR and replicate 1 examining the 3′-UTR (Fig. 5E,G). These results indicate that TgPUS1-dependent pseudouridylation in at least the coding region has a modest effect on mRNA stability.
DISCUSSION
The results presented here show that pseudouridylation is dynamically regulated in Toxoplasma mRNAs, and that TgPUS1, a PUS homolog that is necessary for Toxoplasma differentiation, appears to be mediating pseudouridylation of mRNA and tRNA. As expected, the results also indicate that rRNAs, tRNAs, and snRNAs in Toxoplasma are pseudouridylated by other PUSs at the usual, highly conserved sites.
The parameters used in all analyses were chosen to minimize the false-discovery rate rather than false-omission rate. Importantly, this means that only a subset of Ψ’s have been identified here. We used the evolutionary conservation of Ψ’s in rRNA to validate our approach using tachyzoite rRNA; we further attempted to validate the Ψ’s identified with CMCT using SCARLET (Liu et al. 2013) and were able to do this for some of the highly conserved sites in the very abundant rRNA, but the amount of material we could generate for the mRNAs was not sufficient to use this method to validate Ψ’s in this latter class of RNA (data not shown). Among the false negatives will be those that do not meet our Ψ criteria due to a relatively low level of pseudouridylation of a position in the total cellular pool of that transcript. Therefore, the Ψ’s identified in these data sets are biased toward positions that show highly penetrant pseudouridylation. Additionally, the method used here relies on an mRNA being of sufficient abundance that the read numbers are high across the length of the mRNA. This necessarily excludes mRNAs that are low abundance, therefore our data do not address whether low abundance mRNAs are more or less likely to be pseudouridylated than high abundance mRNAs. Overall, while our data provide the first look into the pseudouridylome of Toxoplasma, it is likely that only a fraction of such sites have been identified here.
Despite these drawbacks, our results show that pseudouridylation of mRNAs changes with cellular stresses and development. Although extensive changes in pseudouridylation upon starvation and heat shock in yeast and human cells have also been reported (Carlile et al. 2014; Schwartz et al. 2014; Li et al. 2015), our data provide the first evidence suggesting developmentally regulated Ψ’s in mRNAs. Because the differentiation process for Toxoplasma is induced by stress conditions, however, we cannot exclude the possibility that these effects are mediated by the stress rather than differentiation, per se.
Interestingly, we found that Ψ’s are unequally distributed within the transcript. In both human and Toxoplasma mRNAs, Ψ is disproportionately present at position 1 of the codon, and relatively absent at position 3. This is particularly intriguing in light of the structural and stabilizing role that Ψ can play in base interactions (Hudson et al. 2013; Kierzek et al. 2014). Because a Ψ–A base pair may be stronger than a U–A base pair, modification of position 1 of the codon could increase the fidelity of translation. The reason for the underrepresentation of Ψ’s at position 3 is less easy to propose explanations for, although it could be to facilitate “wobble” at this position by decreasing the strength of the base-pairing interaction here. The observed underrepresentation of Ψ’s in 3′-UTRs is consistent with the results seen by Carlile et al. (2014) when profiling yeast mRNAs. Pseudouridylation of this region is intriguing because Ψ’s are known to affect the secondary structure of RNAs in which they are found (Arnez and Steitz 1994; Davis 1995; Newby and Greenbaum 2001, 2002a). By changing the secondary structure of RNA in the 3′-UTR, Ψ’s could potentially recruit novel regulatory proteins to the transcript.
Direct evidence that mRNA Ψ’s may have a biological consequence in vivo previously came only from Schwartz et al. (2014), who found that when PUS7 was knocked out, the transcripts containing PUS7-dependent Ψ’s were present at a lower level. In contrast, the data presented here point to a model in which some tachyzoite-specific transcripts are pseudouridylated by TgPUS1, marking them for destabilization under bradyzoite conditions. This model is based on (i) the location of TgPUS1-dependent Ψ’s in tachyzoite-specific transcripts; (ii) the decreased abundance of such transcripts in wild-type compared to TgPUS1-deficient parasites; and (iii) the increased half-life of mRNAs that normally harbor TgPUS1-dependent Ψ’s in TgPUS1-deficient parasites. The differences between the results of Schwartz et al. and our own could be due to many factors, including the organisms being examined (yeast vs. Toxoplasma), the conditions being tested (starvation vs. differentiation) and the fact that our analysis compared abundance of the TgPUS1-dependent to TgPUS1-independent pseudouridylated mRNAs whereas Schwartz et al. (2014) compared PUS7-dependent pseudouridylated mRNAs with nonpseudouridylated mRNAs.
While statistically significant, the effect of lacking TgPUS1 was relatively modest. This could be due to a number of factors. First, because TgPUS1-deficient parasites do not differentiate efficiently, we were able to assess TgPUS1-dependent differences in mRNA turnover in wild type vs. mutant tachyzoites but not bradyzoites. While TgPUS1 mRNA is expressed at a low level throughout all developmental stages, it is possible that its major impact is during differentiation and/or in fully differentiated bradyzoites.
Second, for each of the transcripts in which we identified Ψ’s, it is unclear what percentage of that mRNA species is pseudouridylated. Recent work has shown that pseudouridylation of a given position is not binary; instead a specific site can be more or less pseudouridylated in different conditions (Chikne et al. 2016). If pseudouridylation does affect RNA turnover, the modest change we observed could be the result of only 25% of the pool of an RNA being pseudouridylated and showing differential stability, while the other 75% shows no change in stability, thereby dampening the signal.
Third, when identifying TgPUS1-dependent Ψ’s in parasites, we were unable to fully profile tRNAs due to poor processivity of reverse transcriptase on these molecules. It is possible, therefore, that the TgPUS1-dependent Ψ’s affecting differentiation are found in tRNA(s). If changes in pseudouridylation change expression or processing of tRNAs, tRNA cleavage products that act as regulatory molecules could indirectly affect the cellular pool of mRNAs, producing the changes we observed in TgPUS1-deficient parasites (Phizicky and Hopper 2010; Galizi et al. 2013; Hopper 2013).
Despite these limitations, our data shed light on the possible mechanistic basis for how loss of TgPUS1 disrupts Toxoplasma differentiation. Further work will be needed to determine how pseudouridylation impacts mRNA stability and which specific pseudouridylation events, either within the mRNAs or even in tRNAs, are necessary for efficient differentiation.
MATERIALS AND METHODS
Toxoplasma culture and harvest
HFFs (human foreskin fibroblasts) were maintained in complete DMEM (Gibco) supplemented with 10% FBS, 1% PenStrep, and 1% l-glutamine. Parasite strains C32 (a clone derived from a type II Pru strain of Toxoplasma expressing GFP under the bradyzoite-specific LDH2 promoter) and TBD5 (a mutant made in the C32 strain that contains an insertion in the PUS1 gene [TgME49_ 202640]) were originally isolated as described in Singh et al. (2002). Toxoplasma gondii tachyzoites were maintained by serial passage in confluent HFFs and kept at 37°C with 5% CO2. For serial passage, parasites were lysed out of host cells by mechanical forcing through a 27-gauge needle every 3–4 d and passed onto new host cells. Throughout experiments the parasites were regularly monitored to ensure an absence of Mycoplasma infection.
For PSI-seq experiments, parasites were passed onto new HFFs at an MOI of 2 and allowed to grow for 48 h to full vacuoles. Infected cells and free parasites were scraped from the container, transferred to conicals, and spun at 2000 RPM (770g) for 5 min at room temperature. Supernatant was removed, pellets were resuspended in TRIzol (Invitrogen), and samples were stored at −20°C until the samples were ready for RNA isolation.
Bradyzoite generation
Bradyzoite differentiation was induced using nutrient and pH stress (Weiss and Kim 2013; Weiss et al. 1995). Tachyzoites were lysed out of host cells by passage through a syringe, passed onto new HFFs at an MOI of 2, and allowed to invade for 3.5 h. Uninvaded parasites were then washed off the monolayer with PBS prewarmed to 37°C, and media were replaced with differentiation media (RPMI 1640 medium lacking sodium bicarbonate, supplemented with 1% FBS, HEPES, and 1% PenStrep, and adjusted to a pH of 8.0). Cultures were moved to an incubator kept at 37°C with ambient CO2 levels. After ∼84 h, parasites were checked to confirm GFP expression and harvested in the same fashion as tachyzoites.
Human RNA harvest
“Uninfected” HFF cells were grown in identical conditions to tachyzoite-infected HFFs. Cells were grown for ∼1 wk, then were harvested in the same way as infected cultures.
For experimental harvest of “stressed” HFF cells, media on cells at least 1 wk old was replaced with differentiation media. Cells were then moved to an incubator kept at 37°C with ambient CO2 levels. Approximately eighty-four hours later, cells and parasites were harvested as above.
Primer extensions
RNA was isolated from heavily infected HFFs using phenol chloroform extraction. CMCT treatment was performed as described in Ofengand et al. (2001). We note that this buffer was used at a pH of 11.4 rather than 10.4, as a pH of 10.4 led to significant U laddering in primer extensions (data not shown), potentially reflecting incomplete removal of CMCT from uridines. Samples were ultimately resuspended to a concentration of 0.25 µg/µL.
Primers complementary to rRNA (3′ end at residue 3741) or snRNAs were synthesized by IDT. Four hundred picomoles of primer were labeled in a 50 µL reaction containing 10 units of PNK in 1× PNK buffer (NEB) as well as 10 μCi of 32P ATP (3 µCi/pmol) (PerkinElmer); labeled primers were cleaned up using QiaQuick Nucleotide Kit (QIAGEN) and resuspended in 50 µL of water.
Of note, 0.25 µg of total RNA (for rRNA) or 1–2 µg of total RNA (for snRNAs) was used as input for the primer extension reactions using 2 µL of labeled primer. Samples were denatured at 80°C for 2 min and cooled before a 30-min reverse transcription using AMV at 42°C. After adding 2× formamide loading buffer (95% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 5 mM EDTA), samples were immediately loaded onto a 7 M urea–acrylamide (19:1) gel and run at 1200 V for ∼1 h. The gel was then transferred to Whatman paper, dried at 80°C for 1 h, and imaged overnight on a phosphor screen.
PSI-seq
The PSI-seq method was adapted from Lovejoy et al. (2014) with changes in initial RNA isolation, CMCT removal, and the sizes of RNA, cDNA, and DNA extracted from gels (Ingolia et al. 2012). Specifically, total RNA was isolated from heavily infected HFFs using phenol chloroform extraction, and mRNA was isolated using Oligo(dT)25 beads (Thermo Fisher). RNA was then treated as detailed in Lovejoy et al. (2014), with the exception that CMCT removal was performed with buffer at pH 11.4 rather than 10.4 (as detailed in Primer Extension). Additionally, RNAs extracted from the gel were split into fragments 100–200 (“small subset”) and 200–300 (“large subset”) nucleotides long; these subsets were kept separate through linker ligation and cDNA isolation, and were recombined before library circularization. Amplified libraries were submitted for sequencing on Illumina GAII and NextSeq platforms as 1 × 50 bp reads.
RNA-seq analysis
Using Bowtie, reads were first aligned to concatenated human and Toxoplasma ribosomal RNA (downloaded from NCBI), then unaligned reads were aligned to concatenated human and Toxoplasma transcripts (downloaded from the UCSC Genome Bioinformatics Website, GRCh38, and ToxoDB, version 9.0, respectively) For alignment we used default Bowtie options, except for the following: “-3 11” was used to trim nucleotides off the 3′ end of the read to ensure adapter was no longer part of the read, “-l 25” was used to set seed length to 25 to reflect the short reads, and the “–best” option was used. Bowtie alignments were analyzed further using SAMtools and R.
Binomial distribution
A binomial distribution was calculated based on the proportion of reads stopping at each putative Ψ in CMCT-treated wild-type parasites. Using the “pbinom” command in R, the probability that the proportion of reads stopping at the site in CMCT-treated TgPUS1-deficient parasites fell under the same distribution was calculated.
Identification of spliceosomal RNAs
We identified spliceosomal RNAs in Toxoplasma using Rfam software (Nawrocki et al. 2015). Genomic locations (multiple listed when there are identical sequences in the genome) and first and last 20 nt of each snRNA (sense strand) are as follows:
U6: TGME49_chrIII:1000000–1000107 plus strand; GGCCUCCUUCGGGAGUGCCA…GAGAAGUGUAACCAAUUUUU
U2: TGME49_chrV:2894399–2894590 plus strand or TGME 49_chrXI:3723892–3724084 minus strand; AUACCUUCUCGGCCUUUUGG…GGAAGAGCGGUGCACCCUUU
U4: TGME49_chrIa:1488531–1488661 plus strand; AUCCUUGCGGUAGGGGCAAU…AUUUCUGGGGCCCCCCGGGG
U5: TGME49_chrIa:1489175–1489284 minus strand; AUCGCAGCGGUUGUCACAGA…UUGACGGCGUCUCGACGCCA
U1: TGME49_chrV:2,898,404–2,898,530 plus strand or TGME 49_chrVIIb:3224162–3224288 plus strand or TGME49_ chrVIIa:371404–371530 minus strand; AACTTACCTGGCAGGCGCTC…GCCAACTGCAGAATTTCTG
4TU labeling of parasite RNA
Heavily infected cells were scraped from 25 cm2 flasks and passed through a 27-gauge needle to release intracellular parasites. The resulting material was used to infect new monolayers growing in 8 × 15 cm dishes at an MOI of 5. 24 h later, 4-thiouracil (4TU) (Sigma-Aldrich) was added at a concentration of 2 µM. Of note, 1.5 h after the 4TU was added, the monolayer was gently washed with prewarmed PBS, and fresh media containing 20 µM of unlabeled uracil and prewarmed to 37°C, were added to the dishes. Plates were harvested at the beginning of the pulse (no 4TU added), at the beginning of the chase, and 30, 60, 120, 240, and 480 min into the chase. Bradyzoite differentiation was induced as above in 8 × 15 cm dishes at an MOI of 5; 4TU was added to bradyzoite cultures 72 h into differentiation; 4TU pulse/chase and harvest were then carried out as with tachyzoites.
Biotinylation and purification of 4TU-containing RNA
4TU labeling and purification were done as described in Cleary et al. (2005) and Rädle et al. (2013). RNA was eluted from the columns by adding 100 µL of 5% 2-mercaptoethanol to the columns; this was repeated 3 min later for a second elution, after which samples were immediately treated to remove DNA using RNeasy kits (QIAGEN).
Library preparations and sequencing
cDNAs were made using the SMART-Seq v4 Ultra Low Input RNA Kit (Clontech/Takara) and amplified for library preparation using the Illumina Nextera XT DNA Library Preparation Kit (Illumina). Notably, all volumes and quantities (including cDNA input) were reduced 2.5× when using the Nextera XT Kit (Combs and Eisen 2015). RNA input for each library was 2.5 ng for the “Tachyzoite 1” and 2 ng for the “Tachyzoite 2” samples. Samples were sequenced using a HiSeq 500 machine, with all samples in one lane. Sequences were aligned to a concatenated human and Toxoplasma genome using CLC Genomics (default settings).
Half-life calculation
Because it was not possible to know how much RNA was lost at each stage of the RNA purification and subsequent stages of library preparation and sequencing, the number of reads mapped to a gene could not be expressed in absolute terms (i.e., number of transcripts of gene x per parasite at each time point). To account for this, we instead calculated a relative half-life for each transcript, adjusting the total number of reads at each time point to fit an exponential decay curve. A half-life of 1 h was chosen for the curve to which the data were adjusted based on previous empirical data from Cleary et al. (2005). Importantly, whether the estimate of an average half-life of 1 h is precisely correct is not important as we used it to determine the relative half-life of an mRNA in the mutant and wild-type conditions (i.e., this number cancels out in the comparison).
DATA DEPOSITION
Pseudouridylation and RNA turnover data have been submitted to the NCBI Sequence Reads Archive under accession numbers SAMN07211859 through SAMN07211874 (uninfected host cells); SAMN07211777 through SAMN07211792 (Toxoplasma and infected host cells); and SAMN07224326 through SAMN07224373 (RNA turnover).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kerry Buchholz for experimental help as well as Melanie Espiritu for assistance with tissue culture work; and the Boothroyd laboratory, Pedro Batista, Andrew Fire, and Peter Sarnow for helpful input. This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant R21 AI120476 and the Abbott Laboratories Stanford Graduate Fellowship.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062794.117.
REFERENCES
- Anderson MZ. 2011. Genetics of Toxoplasma gondii tachyzoite to bradyzoite differentiation. PhD thesis, Stanford University. ProQuest, UMI Dissertation Publishing. [Google Scholar]
- Anderson MZ, Brewer J, Singh U, Boothroyd JC. 2009. A pseudouridine synthase homologue is critical to cellular differentiation in Toxoplasma gondii. Eukaryot Cell 8: 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnez JG, Steitz TA. 1994. Crystal structure of unmodified tRNAGln complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 33: 7560–7567. [DOI] [PubMed] [Google Scholar]
- Auffinger P, Westhof E. 1998. Effects of pseudouridylation on tRNA hydration and dynamics: a theoretical approach. In Modification and editing of RNA (ed. Grosjean H, Benne R), pp. 103–112. ASM Press, Washington, DC. [Google Scholar]
- Basak A, Query CC. 2014. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 8: 966–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabello-Villegas J, Nikonowicz EP. 2005. Solution structure of ψ32-modified anticodon stem–loop of Escherichia coli tRNAPhe. Nucleic Acids Res 33: 6961–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. 2014. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 515: 143–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikne V, Doniger T, Rajan KS, Bartok O, Eliaz D, Cohen-Chalamish S, Tschudi C, Unger R, Hashem Y, Kadener S, et al. 2016. A pseudouridylation switch in rRNA is implicated in ribosome function during the life cycle of Trypanosoma brucei. Sci Rep 6: 25296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary MD, Meiering CD, Jan E, Guymon R, Boothroyd JC. 2005. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nat Biotechnol 23: 232–237. [DOI] [PubMed] [Google Scholar]
- Cohn WE. 1959. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim Biophys Acta 32: 569–571. [DOI] [PubMed] [Google Scholar]
- Cohn WE. 1960. Pseudouridine, a carbon-carbon linked ribonucleoside in ribonucleic acids: isolation, structure, and chemical characteristics. J Biol Chem 235: 1488–1498. [PubMed] [Google Scholar]
- Combs PA, Eisen MB. 2015. Low-cost, low-input RNA-seq protocols perform nearly as well as high-input protocols. PeerJ 3: e869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DR. 1995. Stabilization of RNA stacking by pseudouridine. Nucleic Acids Res 23: 5020–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denmon AP, Wang J, Nikonowicz EP. 2011. Conformation effects of base modification on the anticodon stem–loop of Bacillus subtilis tRNATyr. J Mol Biol 412: 285–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizi R, Spano F, Giubilei MA, Capuccini B, Magini A, Urbanelli L, Ogawa T, Dubey JP, Spaccapelo R, Emiliani C, et al. 2013. Evidence of tRNA cleavage in apicomplexan parasites: half-tRNAs as new potential regulatory molecules of Toxoplasma gondii and Plasmodium berghei. Mol Biochem Parasitol 188: 99–108. [DOI] [PubMed] [Google Scholar]
- Hamma T, Ferré-D'Amaré AR. 2006. Pseudouridine synthases. Chem Biol 13: 1125–1135. [DOI] [PubMed] [Google Scholar]
- Hopper AK. 2013. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 194: 43–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson GA, Bloomingdale RJ, Znosko BM. 2013. Thermodynamic contribution and nearest-neighbor parameters of pseudouridine-adenosine base pairs in oligoribonucleotides. RNA 19: 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. 2012. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc 7: 1534–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack KK, Bellodi CC, Landry DMDM, Niederer RORO, Meskauskas AA, Musalgaonkar SS, Kopmar NN, Krasnykh OO, Dean AMAM, Thompson SR, et al. 2011. rRNA pseudouridylation defects affect ribosomal ligand binding and translational fidelity from yeast to human cells. Mol Cell 44: 660–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierzek E, Malgowska M, Lisowiec J, Turner DH, Gdaniec Z, Kierzek R. 2014. The contribution of pseudouridine to stabilities and structure of RNAs. Nucleic Acids Res 42: 3492–3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. 2015. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 11: 592–597. [DOI] [PubMed] [Google Scholar]
- Li X, Ma S, Yi C. 2016. Pseudouridine: the fifth RNA nucleotide with renewed interests. Curr Opin Chem Biol 33: 108–116. [DOI] [PubMed] [Google Scholar]
- Liu N, Parisien M, Dai Q, Zheng G, He C, Pan T. 2013. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 19: 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovejoy AF, Riordan DP, Brown PO. 2014. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 9: e110799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machnicka MA, Milanowska K, Oglou OO, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. 2013. MODOMICS: a database of RNA modification pathways - 2013 update. Nucleic Acids Res 41: D262–D267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrone A, Mason PJ. 2003. Dyskeratosis congenita. Cell Mol life Sci 60: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, et al. 2015. Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43: D130–D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2001. A conserved pseudouridine modification in eukaryotic U2 snRNA induces a change in branch-site architecture. RNA 7: 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2002a. Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci 99: 12697–12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL. 2002b. Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965. [DOI] [PubMed] [Google Scholar]
- Nobles KN, Yarian CS, Liu G, Guenther RH, Agris PF. 2002. Highly conserved modified nucleosides influence Mg2+-dependent tRNA folding. Nucleic Acids Res 30: 4751–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofengand J. 2002. Ribosomal RNA pseudouridines and pseudouridine synthases. FEBS Lett 514: 17–25. [DOI] [PubMed] [Google Scholar]
- Ofengand J, Del Campo M, Kaya Y. 2001. Mapping pseudouridines in RNA molecules. Methods 25: 365–373. [DOI] [PubMed] [Google Scholar]
- Penzo M, Rocchi L, Brugiere S, Carnicelli D, Onofrillo C, Couté Y, Brigotti M, Montanaro L. 2015. Human ribosomes from cells with reduced dyskerin levels are intrinsically altered in translation. FASEB J 29: 3472–3482. [DOI] [PubMed] [Google Scholar]
- Phizicky EM, Hopper AK. 2010. tRNA biology charges to the front. Genes Dev 24: 1832–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabani M, Levin JZ, Fan L, Adiconis X, Raychowdhury R, Garber M, Gnirke A, Nusbaum C, Hacohen N, Friedman N, et al. 2011. Metabolic labeling of RNA uncovers principles of RNA production and degradation dynamics in mammalian cells. Nat Biotechnol 29: 436–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rädle B, Rutkowski AJ, Ruzsics Z, Friedel CC, Koszinowski UH, Dölken L. 2013. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. J Vis Exp. 10.3791/50195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safra M, Nir R, Farouq D, Vainberg Slutskin I, Schwartz S. 2017. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res 27: 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell JP, Crestfield AM, Allen FW. 1959. Methylation studies on various uracil derivatives and on an isomer of uridine isolated from ribonucleic acids. Biochim Biophys Acta 32: 406–412. [DOI] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. 1990. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18: 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473: 337–342. [DOI] [PubMed] [Google Scholar]
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. 2014. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 159: 148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh U, Brewer JL, Boothroyd JC. 2002. Genetic analysis of tachyzoite to bradyzoite differentiation mutants in Toxoplasma gondii reveals a hierarchy of gene induction. Mol Microbiol 44: 721–733. [DOI] [PubMed] [Google Scholar]
- Spenkuch F, Motorin Y, Helm M. 2014. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11: 1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkin E, Cohn WE. 1951. Nucleoside-from ribonucleic 5′-phosphates acid. Nature 167: 483–484. [Google Scholar]
- Weiss LM, Kim K. 2013. Toxoplasma gondii: the model Apicomplexan: perspectives and methods, 2nd ed Elsevier, Amsterdam. [Google Scholar]
- Weiss LM, Laplace D, Takvorian PM, Tanowitz HB, Cali A, Wittner M. 1995. A cell culture system for study of the development of Toxoplasma gondii bradyzoites. J Eukaryot Microbiol 42: 150–157. [DOI] [PubMed] [Google Scholar]
- Windhager L, Bonfert T, Burger K, Ruzsics Z, Krebs S, Kaufmann S, Malterer G, L'Hernault A, Schilhabel M, Schreiber S, et al. 2012. Ultrashort and progressive 4sU-tagging reveals key characteristics of RNA processing at nucleotide resolution. Genome Res 22: 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Xiao M, Yang C, Yu YT. 2011. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 30: 79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarian CS, Basti MM, Cain RJ, Ansari G, Guenther RH, Sochacka E, Czerwinska G, Malkiewicz A, Agris PF. 1999. Structural and functional roles of the N1- and N3-protons of pseudouridine at tRNA's position 39. Nucleic Acids Res 27: 3543–3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu CT, Allen FW. 1959. Studies on an isomer of uridine isolated from ribonucleic acids. Biochim Biophys Acta 32: 393–406. [DOI] [PubMed] [Google Scholar]
- Zeiner GM, Cleary MD, Fouts AE, Meiring CD, Mocarski ES, Boothroyd JC. 2008. RNA analysis by biosynthetic tagging using 4-thiouracil and uracil phosphoribosyltransferase. Methods Mol Biol 419: 135–146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.