Abstract
X inactive-specific transcript (Xist) is a long noncoding RNA that plays an essential role in X chromosome inactivation. Although Xist RNA, like common protein-coding mRNAs, is transcribed by RNA polymerase II, spliced and polyadenylated, it is retained in the nucleus and associates with the X chromosome it originates from. It has been assumed that Xist RNA recruits proteins involved in epigenetic modifications and chromatin compaction to the X chromosome. One of the major proteins constituting the nuclear matrix, hnRNP U, has been shown to be required for the association of Xist RNA with the inactive X chromosome (Xi). In this study, we found that the first 950-nt sequence of Xist RNA had the potential to associate with chromatin in a manner independent of hnRNP U. Furthermore, its chromatin association is apparently dependent on the presence of an intact A-repeat sequence, which is one of the repeats in Xist/XIST RNA conserved among many mammalian species, and has been shown to be important for Xist RNA-mediated silencing. Taking this unexpected finding and a previous study demonstrating the effect of Xist RNA lacking the A-repeat on the formation of the silent heterochromatin domain together, we suggest that the A-repeat captures chromatin near the initial loading site of Xist RNA and relocates it into the core of the heterochromatin domain.
Keywords: Xist, A-repeat, chromatin, X chromosome inactivation
INTRODUCTION
X chromosome inactivation (XCI) is a mechanism to compensate for the dosage difference of X-linked genes between XX females and XY males (Lyon 1961). An X-linked noncoding RNA, X-inactive specific transcript (Xist), is a key regulator of XCI, and an X chromosome deficient for Xist fails to initiate XCI (Borsani et al. 1991; Brockdorff et al. 1991; Brown et al. 1991, 1992; Penny et al. 1996; Marahrens et al. 1997). Although Xist RNA, like common protein-coding mRNAs, undergoes splicing and polyadenylation, which facilitate nuclear export of mRNA, Xist RNA escapes nuclear export and stays in the nucleus (Brockdorff et al. 1992; Brown et al. 1992). In addition, Xist RNA spreads along the X chromosome from which it is transcribed, and associates with the entire chromosome in a cis-limited manner at the onset of XCI. Recent studies demonstrated that one of the nuclear matrix proteins, heterogeneous nuclear ribonucleoprotein U (hnRNP U; also known as SAF-A) mediates the association of Xist RNA with the inactive X (Xi) (Hasegawa et al. 2010; Sakaguchi et al. 2016), and hnRNP U depletion results in dissociation of Xist RNA from the X chromosome. In addition, hnRNP U was not only detected as one of the proteins associated with Xist RNA by mass spectrometry analysis following a pull-down of Xist RNA–protein complexes (Chu et al. 2015; McHugh et al. 2015; Minajigi et al. 2015), but it was also found in close spatial proximity to Xist RNA by 3D-SIM (Smeets et al. 2014), suggesting a physical interaction between Xist RNA and hnRNP U. Xist RNA anchored by hnRNP U on the X chromosome subsequently induces a series of changes in the epigenetic state and causes compaction of the chromosome for heterochromatinization. It has been suggested that a human ortholog of structural maintenance of chromosomes hinges on the domain containing 1 (SmcHD1), which is a protein whose loss causes female-specific lethality during embryonic development in the mouse, and its binding partner, HBiX1, are involved in compaction of Xi (Blewitt et al. 2008; Nozawa et al. 2013). Their depletion results in decompaction of Xi in human cultured cells even though XIST RNA stays associated with Xi. This observation suggests that the chromosomal association of XIST/Xist RNA on its own cannot induce compaction of the X chromosome, and that instead, compaction requires additional factors such as SMCHD1/SmcHD1 and HBiX1. While the analysis of a series of deletions in Xist RNA has identified the A-repeat as a domain essential for chromosome silencing (Wutz et al. 2002), it is largely unknown whether any sequences of Xist RNA are involved in compaction of the chromosome.
In this study, we showed that a 950-nucleotide (nt) sequence near the 5′ end of Xist RNA containing the A-repeat possessed the potential to associate with chromatin in a manner independent of hnRNP U. We reexamined previous CLIP-seq data (Huelga et al. 2012) and found that HNRNP U in human cells bound poorly to the 5′ region of XIST RNA. This finding led us to overexpress the corresponding 950-nt region in the mouse Xist sequence and examine its behavior. The results demonstrated that the 950-nt RNA formed dispersed signals in the nucleus, as was the case for full-length Xist RNA in those cells depleted for hnRNP U. In addition, a substantial portion of the 950-nt RNA was found in the chromatin fraction, but not in the nucleoplasmic or cytoplasmic fraction. Interestingly, mutation in the conserved A-repeat sequence altered the distribution of the mutated RNA, that is, the RNA did not form dispersed signals but single pinpoint signals at the integration sites. Given the functional importance of the A-repeat, chromatin binding of the 950-nt RNA might be involved in XCI. The functional implications of this finding are discussed.
RESULTS
Xist RNA is retained in the nucleus by a mechanism independent of hnRNP U-mediated chromosomal accumulation
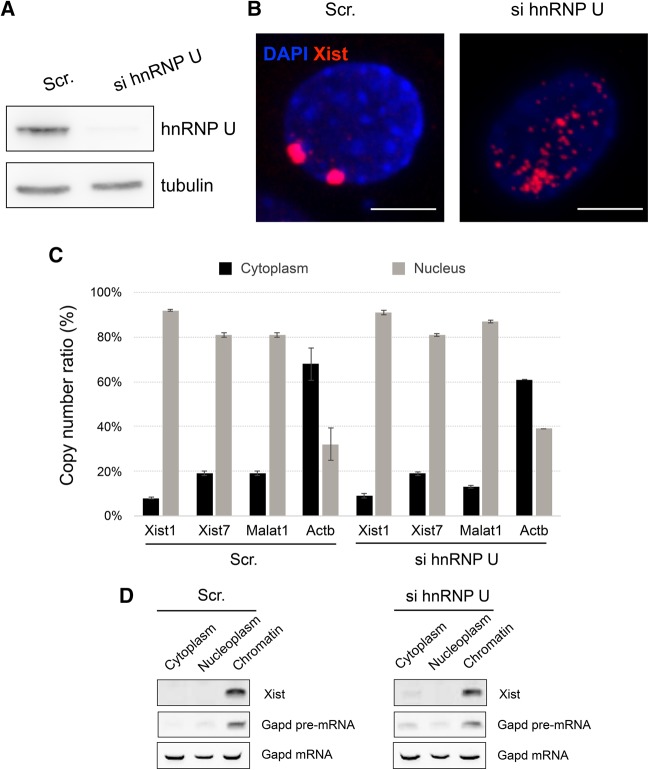
A previous study showed that depletion of hnRNP U in female somatic cells leads to dissociation of Xist RNA from the inactive X chromosome (Xi) (Hasegawa et al. 2010), suggesting that association of Xist RNA with an Xi is mediated by hnRNP U. However, in that study, diffuse Xist RNA signals detected by RNA-FISH were still retained in the nucleus and apparently associated with chromatin. This implies that the nuclear retention and chromosomal association of Xist RNA are not necessarily a result of its association with Xi mediated by hnRNP U, and that a distinct mechanism also operates not only to prevent the nuclear export of Xist RNA but also to facilitate its association with chromatin. To investigate this hypothesis, we knocked down hnRNP U in female immortalized mouse embryonic fibroblasts (iMEFs) by siRNA (Fig. 1A) and analyzed the subcellular localization of Xist RNA by not only RNA-FISH but also RT-qPCR on cDNA prepared from nuclear and cytoplasmic RNA (Fig. 1B,C). Upon knockdown of hnRNP U, the hybridization signal of Xist RNA detected by RNA-FISH became dispersed in the nucleus, consistent with a previous report (Fig. 1B; Hasegawa et al. 2010). RT-qPCR also demonstrated that Xist RNA was still substantially enriched in the nucleus (at the same level of enrichment as Malat1 nuclear lncRNA) (Tripathi et al. 2010), contrasting subcellular localization of protein coding mRNA such as β-actin (Fig. 1C). To further examine if the Xist RNA delocalized from Xi upon knockdown of hnRNA U still associated with chromatin, we isolated RNA from the chromatin and nucleoplasmic fractions prepared using urea buffer (Pandya-Jones and Black 2009) for RT-PCR. As shown in Figure 1D, Xist RNA was substantially enriched in the chromatin fraction. These results suggested that Xist RNA delocalized from Xi still stayed in the nucleus and associated with chromatin at ectopic sites, raising the possibility that factors other than hnRNP U interact with Xist RNA and allow it to associate with chromatin.
FIGURE 1.
Effect of knockdown using siRNA against hnRNP U. (A) Efficiency and specificity of knockdown in female iMEFs were confirmed by Western blotting using cells treated with siRNA against hnRNP U (si hnRNP U) in comparison with cells treated with scrambled siRNA against hnRNP U (Scr.). (B) Xist RNA was detected by RNA-FISH in female iMEFs upon siRNA treatment. Scale bar, 10 µm. (C) Quantitative RT-PCR comparing absolute copy number of each transcript between the cytoplasmic and nuclear fractions after knockdown of hnRNP U. Error bars, standard deviation (SD). (D) Subcellular localization of Xist RNA delocalized from Xi upon knockdown of hnRNP U in iMEFs was examined in comparison with pre-mRNA and mRNA encoding Gapd by RT-PCR.
Partial fragments of Xist RNA accumulated on the chromosome in cis
To explore which part of Xist RNA delocalized from Xi was involved in the apparent chromatin association, we employed an inducible overexpression approach. We first prepared NIH3T3 cells stably expressing rtTA, a tetracycline-inducible transactivator, and subsequently introduced a test fragment under the control of a tetracycline-inducible promoter via a piggyBac transposon system to facilitate efficient integration into the genome (Supplemental Fig. S1A). When a sequence encoding either EGFP or full-length Xist cDNA was used as a test fragment, RNA transcribed from the inducible promoter upon administration of doxycycline (dox) behaved in the manner expected, that is, mRNA encoding EGFP was exported to the cytoplasm, whereas a full-length Xist RNA stayed in the nucleus to form a so-called Xist cloud representing its chromosomal accumulation (Supplemental Fig. S1B–E). This confirmed that overexpression of Xist RNA did not perturb its localization property. Expression of these test fragments was observed upon administration of dox in almost all cells recovered after drug selection, indicating efficient integration and dox-induction of our system.
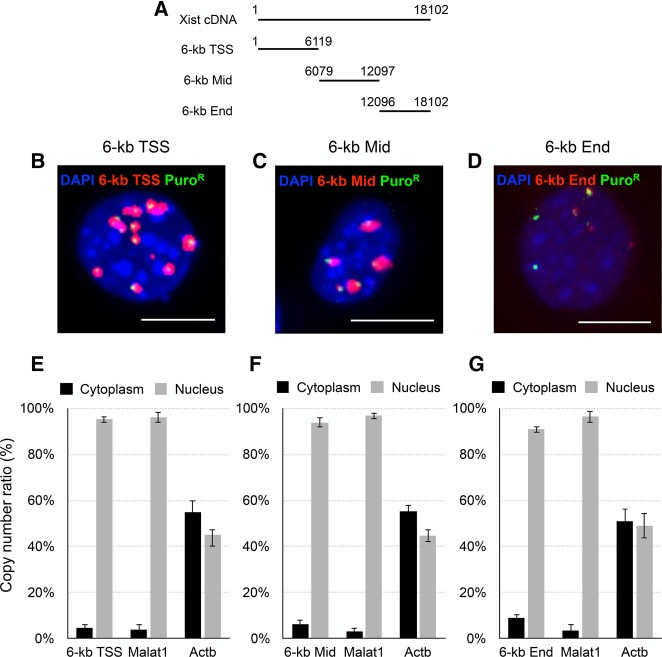
We then divided the full-length Xist cDNA into three pieces, each of which consisted of a 6-kb sequence (TSS, Mid, and End), and examined the localization of their transcripts upon induction by doxycycline (Fig. 2A). While one of the three fragments (End) failed to produce a prominent signal, the other two fragments (TSS and Mid) were still able to produce large signals resembling Xist clouds (Fig. 2B–D). Quantitative RT-PCR revealed, however, that all three of these kinds of transcripts were retained in the nucleus (Fig. 2E–G). The abundance of the transcripts derived from the TSS and Mid fragments in the nucleus was much higher than that from the End fragment (Supplemental Fig. S2), implying that either the expression level and/or stability of the latter transcripts was lower than that of the former two transcripts. In addition, these results suggested that hnRNP U, which has been shown to associate with broad regions of Xist RNA (Hasegawa et al. 2010), still interacted with the transcripts of the 6-kb TSS and 6-kb Mid fragments and anchored them onto chromatin, and therefore these fragments would not be suitable for exploring hnRNP U-independent chromatin association of Xist RNA.
FIGURE 2.
Overexpression of partial Xist RNA fragments. (A) Xist cDNA fragments used for overexpression assay. (B–D) Each of the fragments overexpressed in NIH3T3 cells was detected by RNA-FISH. Scale bar, 10 µm. (E–G) Distribution of overexpressed Xist fragments and Malat1 was compared between the cytoplasm and nucleus by RT-qPCR. Error bars, standard deviation (SD).
A 0.9-kb truncated Xist RNA fragment could associate with chromatin in an hnRNP U-independent manner
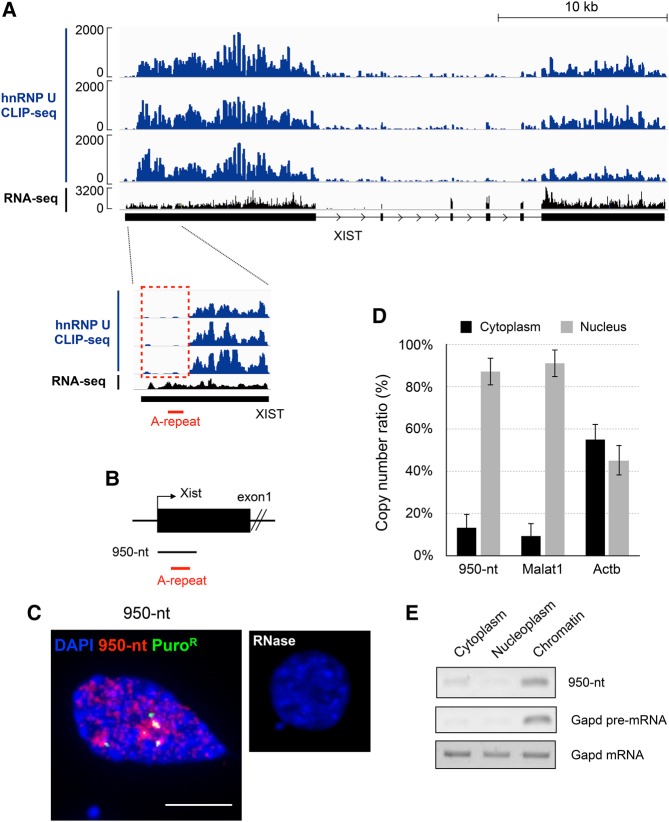
A recent study demonstrated by RIP-qPCR that hnRNP U binds to almost the entire span of mouse and human Xist/XIST RNA (Yamada et al. 2015). We therefore next investigated more precisely the binding sites of hnRNP U with XIST RNA in human by reanalyzing CLIP-seq data (Huelga et al. 2012). This analysis revealed that although hnRNP U bound to XIST RNA essentially along its entire length, as previously reported, the 5′ region of XIST RNA containing the A-repeat was largely devoid of hnRNP U (Fig. 3A). The presence of transcripts containing this region was evident from the RNA-seq data. In addition, more recent study by fRIP-seq also revealed poor binding of hnRNP U at the 5′ region of Xist containing the A-repeat in the mouse (Hendrickson et al. 2016). Since this region is conserved among many species and required for Xist RNA-mediated silencing in the mouse, it was of interest to examine the behavior of this fragment when it was expressed on its own. Accordingly, we introduced the corresponding mouse 950-nucleotide (nt) sequence as a test fragment and induced its expression in NIH3T3 cells containing rtTA for the analysis of subcellular localization (Fig. 3B). Interestingly, RNA derived from the 950-nt sequence formed dispersed signals in the nucleus, like delocalized full-length Xist RNA (Figs. 1B, 3C) and was retained in the nucleus (Fig. 3D). RNase A treatment prior to RNA-FISH completely abolished the dispersed signals, indicating that they represented RNA hybridized with the probe (Fig. 3C). We further examined if the RNA derived from the 950-nt fragment was associated with chromatin by RT-PCR using RNA isolated from the chromatin and nucleoplasmic fractions. As Figure 3E shows, the RNA derived from the 950-nt fragment was enriched in the chromatin fraction, but not detected in the nucleoplasmic or cytoplasmic fraction. If the dispersed signals represented RNA that was not associated with chromatin, this RNA should have been detected in the nucleoplasmic fraction. The results therefore suggested the association of this RNA with chromatin in a manner independent of hnRNP U.
FIGURE 3.
Analysis of binding regions of hnRNP U on Xist RNA. (A) The data set of previous hnRNP U CLIP-seq and RNA-seq in human HEK293 cells (Huelga et al. 2012) was analyzed. The 5′ region of Xist RNA was found to be poorly bound by hnRNP U (rectangle drawn with dashed red line). (B) A 950-nt test fragment derived from the 5′ region of Xist RNA overexpressed in NIH3T3 cells. (C) RNA-FISH detecting the 950-nt RNA fragment (left). RNA-FISH was carried out in the same manner, but using cells treated with RNase A (right). Scale bar, 10 µm. (D) Comparison of absolute copy number of the 950-nt RNA fragment between cytoplasmic and nuclear fractions. Error bars, standard deviation (SD). (E) Subcellular localization of the 950-nt RNA fragment was examined by RT-PCR.
Dispersed chromatin association of 950-nt transcripts depended on the presence of an intact A-repeat sequence
Since the A-repeat, which is present in the RNA derived from the 950-nt sequence, has been shown to be essential for the proper silencing function of Xist RNA (Wutz et al. 2002; Sakata et al. 2017), we were interested in the functional relationship between A-repeat-mediated silencing and the chromatin association of 950-nt RNA. It has been suggested that the A-repeat could form multiple stem–loop structures, and if the formation of such structures was disrupted by point mutations or inversions, the silencing function of Xist RNA was weakened (Wutz et al. 2002; Duszczyk et al. 2011). Accordingly, we attempted to destroy the stem–loop structure by introducing two base substitutions (C to G and G to A) in the first part of the two stem–loop structures in each repeat unit of the 950-nt sequence, as previously described (Fig. 4A 950-nt mut and 950-nt as, respectively, and Fig. 4B; Wutz et al. 2002) and induced expression of the respective mutated RNAs in NIH3T3 cells carrying rtTA. Interestingly, both mutant RNAs failed to form dispersed signals and instead formed pinpoint signals at the integration sites (Fig. 4C,D), suggesting a loss of the ability to associate with chromatin shown by 950-nt RNA. It is therefore likely that the hnRNP U-independent chromatin association of Xist RNA requires an intact A-repeat sequence that forms multiple stem–loop structures.
FIGURE 4.
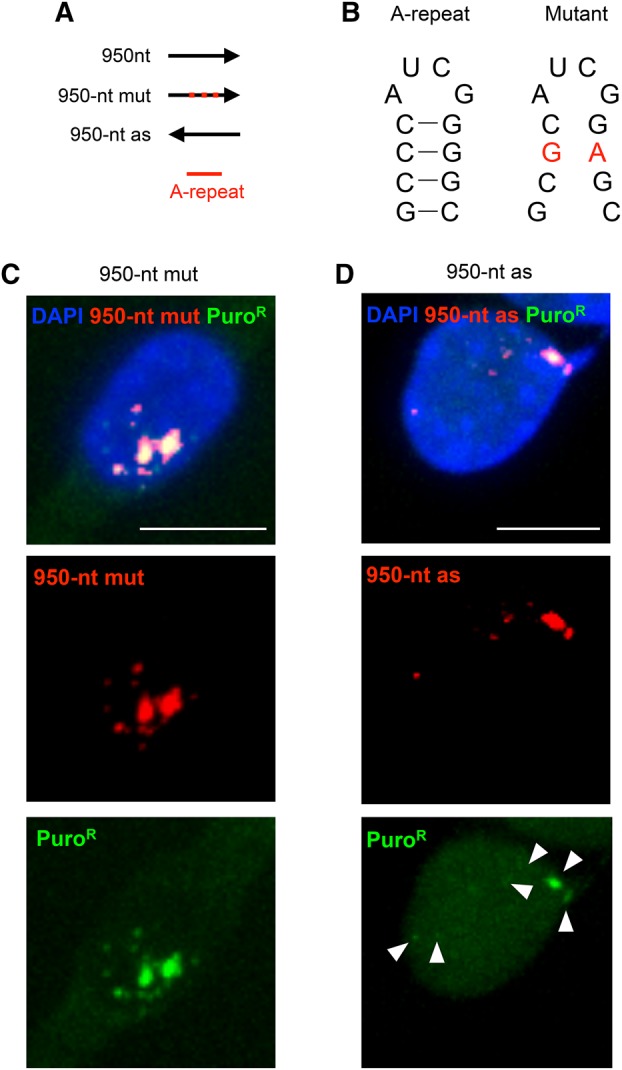
Mutation of the A-repeat affected the behavior of the 950-nt RNA fragment. (A) Test fragments used for the analysis. Red dots indicate mutations introduced in the A-repeat. (B) Possible secondary structure, which might be formed by the consensus sequence of the A-repeat (Wutz et al. 2002), and positions of two point mutations introduced according to Wutz et al. (2002) in the 950-nt mut RNA fragment are shown in red. (C,D) RNA-FISH was carried out to detect each of these test fragments. Scale bar, 10 µm.
950-nt RNA forms dispersed signals regardless of its abundance
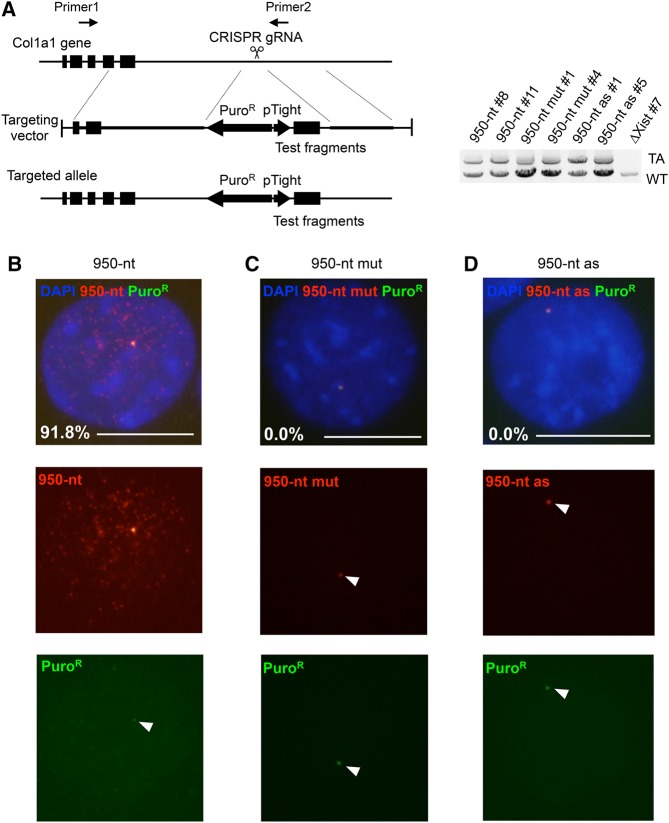
Although the behavior of 950-nt RNA described above was in accord with the A-repeat-dependency of the chromatin association of Xist RNA, this conclusion was based on observations of these transcripts expressed at variable levels from transgenes randomly integrated at multiple sites in the genome. Accordingly, we attempted to examine the behavior of each of these RNAs when they were expressed from a single genomic locus. A single copy of each of the three transgenes was introduced at the Col1a1 locus by homologous recombination (Fig. 5A,B) in male ES cells expressing nlsrtTA (Supplemental Fig. S3A; Wutz and Jaenisch 2000), in which the 950-nt sequence present in the endogenous Xist locus had previously been removed using the CRISPR-Cas9 system (Supplemental Fig. S3B,C). We successfully isolated cells in which each of the three transgenes, 950-nt, 950-nt mut, and 950-nt as, had been introduced into one of the two Col1a1 alleles (Fig. 5A). RNA-FISH was carried out in these cells expressing one of the three forms of the RNA. As shown in Figure 5B–D, while the two mutant forms of the RNA formed a single pinpoint signal, 950-nt RNA was detected as many signals dispersed in the nucleus of undifferentiated ES cells, as was seen in NIH3T3 cells. These results demonstrated that the 950-nt RNA carrying the intact A-repeat sequence, but not the two forms of A-repeat mutant RNA, formed dispersed signals regardless of its abundance. This indicated that the chromatin association is a unique property of 950-nt RNA in both differentiated and undifferentiated cells.
FIGURE 5.
Behavior of the test RNA fragments expressed from the Col1a1 locus. (A) Targeting scheme to introduce expression cassette of the test fragments from the Col1a1 locus (pTight, Doxycycline-inducible promoter) (left). The test fragments used for the assay encode 950-nt, 950-mut, and 950-as sequences. Cells harboring the expected targeted recombination were identified by genomic PCR (right). (TA) Targeted allele, (WT) wild-type allele. ΔXist#7 is the parental cell line used for targeting (Supplemental Fig. S3). Positions of each primer are shown on the left. (B–D) Representative images of RNA-FISH detecting the test RNA fragments expressed from the Col1a1 locus. Prevalence of the nuclei with indicated expression pattern of respective RNA are shown.
DISCUSSION
In this study, we found that a 950-nt sequence of the 5′ region of Xist RNA was the only region devoid of hnRNP U in the Xist RNA sequence, and that upon overexpression, this 950-nt sequence formed dispersed signals, which most likely represented association with chromatin, in the nucleus. Interestingly, this chromatin association was apparently dependent on the presence of the intact A-repeat sequence. The importance of the A-repeat in chromosome silencing has been shown by the induced expression of the mutated Xist RNA lacking the A-repeat and its failure to induce gene silencing on the chromosome it coats in ES cells (Wutz et al. 2002; Chaumeil et al. 2006; Engreitz et al. 2013) and in vivo (Sakata et al. 2017). Detailed microscopy analysis has revealed that genes that fail to be silenced by the mutated Xist RNA lacking the A-repeat are located outside or at the periphery of the mutated Xist RNA cloud. This finding led to the idea that the A-repeat is required for relocation of X-linked genes, which are subjected to inactivation, into the core of the Xist cloud constituting a heterochromatin compartment (Chaumeil et al. 2006). A more recent study using RAP-seq analysis further demonstrated that Xist RNA does not necessarily continuously propagate along the chromosome from the Xist locus, but rather associates with chromatin in three-dimensionally close proximity to the Xist locus, from which Xist RNA spreads locally along the chromosome (Engreitz et al. 2013). Interestingly, the local spreading of the mutated Xist RNA lacking the A-repeat appears to be less efficient than that of wild-type Xist RNA, especially into regions where actively transcribed genes are densely located. Our finding that the A-repeat on its own has the property of associating with chromatin in a manner independent of hnRNP U might account for the difference in behavior between Xist RNA with and without the A-repeat. It is tempting to speculate that wild-type Xist RNA associated with the chromatin sites spatially juxtaposed to the Xist locus by hnRNP U would be able to efficiently spread along the chromosome and capture local chromatin nearby. If this is the case, the mutated Xist RNA would still be able to associate with such initial chromatin sites via hnRNP U but would not be able to capture chromatin nearby due to the lack of the A-repeat. This could result in the failure of the mutated Xist RNA to relocate such a chromatin region into the core of the heterochromatin domain. Lamin B receptor and SHARP, which have been identified as proteins that can interact with the A-repeat (Chu et al. 2015; McHugh et al. 2015; Chen et al. 2016), might be involved in the proposed chromatin capture mediated by the A-repeat. Previous RAP-seq analysis could not reveal differences in the distribution of Xist RNA with and without the A-repeat at high resolution, most probably due to limited recovery of chromatin associated with the respective RNA (Engreitz et al. 2013). It will be of interest to know if there are any common features in the chromatin regions associated with the 950-nt RNA overexpressed in our assay system. This would provide further insights into how the A-repeat exerts its effect on chromosome silencing.
MATERIALS AND METHODS
Construction of plasmids
To construct pEFrtTAIB containing an rtTA expression cassette, an EF1α promoter fragment in CSII-EF-RfA (gift from Kunitada Shimotono), an IRES sequence in pSA-IRES-EGFP (Sado et al. 2005), and a blasticidin S deaminase (BSD)-pA fragment amplified by PCR from pcDNA6TR (Thermo) were serially cloned into pBluescript II SK (+) to generate pEFIB. To convert tTA to rtTA, a tTA fragment amplified from ptTA-bleo (a gift from Dr. Tatsuo Fukagawa) was cloned into pBluescript II SK (+), and subsequently mutations D95N, L101S, G102D, and E71K were introduced according to Gossen et al. (1995) by sequential inverse PCR using primers containing respective mutations. The resultant rtTA fragment was cloned into pEFIB to produce pEFrtTAIB.
A basic construct for overexpression of respective test fragments, pPBTight, which contains the tetracycline-inducible promoter and a CAG-IRES-Puromycin resistance gene (PuroR) cassette between piggybac transposon terminal repeats, was generated using pPBCAG-BstXI-IP (a gift from Hitoshi Niwa) and pTRE-Tight (Clontech). A unique NotI site was used for the cloning of respective test fragments (6-kb TSS, Mid, and End, 950-nt and 950-nt as). To make 950-nt mut, pBluescript II SK (+) containing the 950-nt fragment was used to replace the A-repeat region with a synthetic 469-bp A-repeat mutant oligo sequence (IDT) by inverse PCR in combination with In-Fusion cloning technology (Clontech). Full-length Xist cDNA was cloned into pPBTight by combining partial genomic fragments and fragments amplified by PCR.
To disrupt the Xist locus by CRISPR-Cas9-mediated genome editing, two gRNA sequences were designed using CRISPRdirect (https://crispr.dbcls.jp) (Naito et al. 2015). Double-stranded gRNA oligos were cloned into pX330 vector to generate pXPR-Xist(-20) and pXPR(0.9k).
For targeted introduction of respective test fragments at the Col1a1 locus, a targeting vector containing the pTight promoter and a pA cassette as well as a Puromycin cassette was generated by serial PCR and In-Fusion cloning. Respective test fragments were cloned at the unique NotI site. One gRNA was designed against the Col1a1 locus and cloned into pX330 (pCRISPR-Col1a1), and was used to increase targeting efficiency. All primers and oligos used for constructions are shown in Supplemental Table 1.
Cell culture and generation of transgenic cell lines
Mouse embryonic fibroblasts immortalized by SV40 large T antigen (iMEFs) and NIH3T3 cells were cultured in DMEM supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. To generate the rtTAIB cell lines, NIH3T3 cells transfected with 1.1 µg of pEFrtTAIB using Fugene HD reagent were selected for 7 d from 24 h after transfection in medium containing 1 µg/mL of Blasticidin S HCl (InvivoGen), and subsequently cloning was done by limited dilution. One of the rtTAIB cell lines thus generated was used for overexpression of respective test fragments. Cells lipofected with 1.1 µg of each overexpression construct in combination with 1.1 µg of piggyBac transposase expression vector (a gift from Dr. Hitoshi Niwa) were selected for 7 d from 24 h after transfection in medium containing 1 µg/mL of each of Blasticidin S HCL and puromycin (Invitrogen).
Mouse embryonic stem cells (ESCs) expressing nls-rtTA (J1rtTA/N, a gift from Anton Wutz [Wutz and Jaenisch 2000]) were cultured in DMEM supplemented with 15% KSR, 1% non-essential amino acids, 1% penicillin–streptomycin, 500 U of LIF and 2i inhibitors (1 µM PD0325901 and 3 µM CHIR99021). To disrupt the Xist locus, lipofection was carried out using 1.1 µg each of pXPR-Xist(-20), pXPR-Xist(0.9k), and a pEFIB containing a BSD gene under the control of the human EF1α promoter. Following selection with 4 µg/mL of Blasticidin S for 2 d, cells were seeded at low density, so that single cell-derived colonies could be isolated. For gene targeting in ESCs, 30 µg of the targeting construct was used for electroporation in combination with 10 µg of pCRISPR-Col1a1, and subsequent selection was carried out using puromycin at 2 µg/mL. Induction by doxycycline was performed at concentrations of 1 µg/mL and 2 µg/mL for NIH3T3 cells and ES cells, respectively.
Knockdown of hnRNP U
Knockdown of hnRNP U was carried out as previously described (Hasegawa et al. 2010) using siRNA with the same sequence. One hundred thousand female iMEFs plated on a 35-mm dish at 24 h prior to transfection were treated with siRNA using RNAiMAX (Invitrogen). Forty-eight hours later, one quarter of the cells were reseeded and treated with siRNA again after an additional 24 h. Sampling for Western blotting and RNA-FISH was done at 24 h after the second transfection of siRNA. Western blotting was carried out using standard procedures with an antibody against hnRNP U (Abcam #20666).
Subcellular fractionation
Cells were lysed in 10 mM Tris–Cl pH 7.5, 10 mM NaCl, 3 mM MgCl2, 0.5% Nonidet P-40 on ice, and centrifuged to precipitate nuclei. To prepare cytoplasmic RNA, the supernatant was treated with 200 µg/mL of proteinase K in 0.1 M Tris–Cl pH 7.5, 0.22 M NaCl, 1% SDS, 12.5 mM EDTA for 60 min at 37°C and subsequent extraction with phenol/chloroform and ethanol precipitation. Nuclear RNA was prepared using TRIzol (Ambion). Chromatin fractionation assays were performed using urea buffer as previously described (Pandya-Jones and Black 2009).
RT-qPCR
First-strand cDNA was prepared from 1 µg of total RNA using SuperscriptIII (Invitrogen) and random primers. One-fiftieth of the cDNA thus prepared was subjected to quantitative PCR using a KAPA SYBR FAST qPCR kit (Kapa Biosystems) according to the manufacturer's instructions. Quantification was performed by comparison with a series of standard plasmids (from 103 to 106 copies) amplified in parallel.
RNA fluorescent in situ hybridization (FISH)
RNA-FISH was carried out as described previously (Sado et al. 2001) using probes prepared by nick translation using plasmids containing each of the test fragments and the puromycin resistance gene.
High-throughput sequencing analysis
To identify the HNRNP U binding region in human XIST RNA, a previous hnRNP U CLIP-seq data set (Huelga et al. 2012) was analyzed to align the reads along the reference genome (Human hg19) using TopHat v2.0.12 (Kim et al. 2013). RNA-seq data in the same data set (Huelga et al. 2012) were analyzed in the same way to confirm the presence of transcripts derived from the 5′ region of XIST. Data visualization was performed with Integrative Genomics Viewer (IGV) v2.3.47 (Robinson et al. 2011; Thorvaldsdóttir et al. 2013).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kunitada Shimotono, Hitoshi Niwa, and Anton Wutz for providing pCSII-EF-RfA, pPBCAG-BstXI-IP plasmid, and the J1rtTA/N ES cell line, respectively. We also thank Motoko Unoki for technical advice about a procedure for knockdown of hnRNP U. This work was supported by Grants-in-Aid for Scientific Research on Innovative Areas (16H01320 and 17H05606 to T.S.) from the Ministry of Education, Science, Sports, and Culture of Japan (MEXT).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.062158.117.
REFERENCES
- Blewitt ME, Gendrel AV, Pang Z, Sparrow DB, Whitelaw N, Craig JM, Apedaile A, Hilton DJ, Dunwoodie SL, Brockdorff N, et al. 2008. SmcHD1, containing a structural-maintenance-of-chromosomes hinge domain, has a critical role in X inactivation. Nat Genet 40: 663–669. [DOI] [PubMed] [Google Scholar]
- Borsani G, Tonlorenzi R, Simmler MC, Dandolo L, Arnaud D, Capra V, Grompe M, Pizzuti A, Muzny D, Lawrence C, et al. 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, Cooper P, Smith S, McCabe VM, Norris DP, Penny GD, Patel D, Rastan S. 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. 1992. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 71: 515–526. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafrenière RG, Grompe M, Tonlorenzi R, Willard HF. 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF. 1992. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542. [DOI] [PubMed] [Google Scholar]
- Chaumeil J, Le Baccon P, Wutz A, Heard E. 2006. A novel role for Xist RNA in the formation of a repressive nuclear compartment into which genes are recruited when silenced. Genes Dev 20: 2223–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Blanco M, Jackson C, Aznauryan E, Ollikainen N, Surka C, Chow A, McDonel P, Cerase A, Guttman M. 2016. Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science 354: 468–472. [DOI] [PubMed] [Google Scholar]
- Chu C, Zhang QC, da Rocha ST, Flynn RA, Bharadwaj M, Calabrese JM, Magnuson T, Heard E, Chang HY. 2015. Systematic discovery of Xist RNA binding proteins. Cell 161: 404–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszczyk MM, Wutz A, Rybin V, Sattler M. 2011. The Xist RNA A-repeat comprises a novel AUCG tetraloop fold and a platform for multimerization. RNA 17: 1973–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. 2013. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341: 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. 2010. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell 19: 469–476. [DOI] [PubMed] [Google Scholar]
- Hendrickson DG, Kelley DR, Tenen D, Bernstein B, Rinn JL. 2016. Widespread RNA binding by chromatin-associated proteins. Genome Biol 17: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelga SC, Vu AQ, Arnold JD, Liang TY, Liu PP, Yan BY, Donohue JP, Shiue L, Hoon S, Brenner S, et al. 2012. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep 1: 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon MF. 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R. 1997. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. 2015. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521: 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minajigi A, Froberg JE, Wei C, Sunwoo H, Kesner B, Colognori D, Lessing D, Payer B, Boukhali M, Haas W, et al. 2015. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H, Ui-Tei K. 2015. CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa RS, Nagao K, Igami KT, Shibata S, Shirai N, Nozaki N, Sado T, Kimura H, Obuse C. 2013. Human inactive X chromosome is compacted through a PRC2-independent SMCHD1-HBiX1 pathway. Nat Struct Mol Biol 20: 566–573. [DOI] [PubMed] [Google Scholar]
- Pandya-Jones A, Black DL. 2009. Co-transcriptional splicing of constitutive and alternative exons. RNA 15: 1896–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penny GD, Kay GF, Sheardown SA, Rastan S, Brockdorff N. 1996. Requirement for Xist in X chromosome inactivation. Nature 379: 131–137. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sado T, Wang Z, Sasaki H, Li E. 2001. Regulation of imprinted X-chromosome inactivation in mice by Tsix. Development 128: 1275–1286. [DOI] [PubMed] [Google Scholar]
- Sado T, Hoki Y, Sasaki H. 2005. Tsix silences Xist through modification of chromatin structure. Dev Cell 9: 159–165. [DOI] [PubMed] [Google Scholar]
- Sakaguchi T, Hasegawa Y, Brockdorff N, Tsutsui K, Tsutsui KM, Sado T, Nakagawa S. 2016. Control of chromosomal localization of Xist by hnRNP U family molecules. Dev Cell 39: 11–12. [DOI] [PubMed] [Google Scholar]
- Sakata Y, Nagao K, Hoki Y, Sasaki H, Obuse C, Sado T. 2017. Defects in dosage compensation impact global gene regulation in the mouse trophoblast. Development 144: 2784–2797. [DOI] [PubMed] [Google Scholar]
- Smeets D, Markaki Y, Schmid VJ, Kraus F, Tattermusch A, Cerase A, Sterr M, Fiedler S, Demmerle J, Popken J, et al. 2014. Three-dimensional super-resolution microscopy of the inactive X chromosome territory reveals a collapse of its active nuclear compartment harboring distinct Xist RNA foci. Epigenetics Chromatin 7: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14: 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A, Jaenisch R. 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol Cell 5: 695–705. [DOI] [PubMed] [Google Scholar]
- Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet 30: 167–174. [DOI] [PubMed] [Google Scholar]
- Yamada N, Hasegawa Y, Yue M, Hamada T, Nakagawa S, Ogawa Y. 2015. Xist exon 7 contributes to the stable localization of Xist RNA on the inactive X-chromosome. PLoS Genet 11: e1005430. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.