Abstract
Noncanonical RNA nucleotidyltransferases (NTases), including poly(A), poly(U) polymerases (PAPs/PUPs), and C/U-adding enzymes, modify 3′-ends of different transcripts affecting their functionality and stability. They contain PAP/OAS1 substrate-binding domain (SBD) with inserted NTase domain. Aspergillus nidulans CutA (AnCutA), synthesizes C/U-rich 3′-terminal extensions in vivo. Here, using high-throughput sequencing of the 3′-RACE products for tails generated by CutA proteins in vitro in the presence of all four NTPs, we show that even upon physiological ATP excess synthesized tails indeed contain an unprecedented number of cytidines interrupted by uridines and stretches of adenosines, and that the majority end with two cytidines. Strikingly, processivity assays documented that in the presence of CTP as a sole nucleotide, the enzyme terminates after adding two cytidines only. Comparison of our CutA 3D model to selected noncanonical NTases of known structures revealed substantial differences in the nucleotide recognition motif (NRM) within PAP/OAS1 SBD. We demonstrate that CutA specificity toward CTP can be partially changed to PAP or PUP by rational mutagenesis within NRM and, analogously, Cid1 PUP can be converted into a C/U-adding enzyme. Collectively, we suggest that a short cluster of amino acids within NRM is a determinant of NTases’ substrate preference, which may allow us to predict their specificity.
Keywords: CutA, nucleotidyltransferase (NTase), noncanonical poly(A) polymerase, NTP specificity, 3′-RACE-seq
INTRODUCTION
3′-End processing and modification contribute to determination of stability of various RNA molecules. The most widespread post-transcriptional modification of the RNA 3′-end is a template-independent addition of nucleotides. This process is catalyzed by enzymes belonging to the DNA polymerase β-like nucleotidyltransferase (NTase) superfamily (Aravind and Koonin 1999; Martin and Keller 2007) (also known as NTase fold superfamily [Kuchta et al. 2009]), which display variable substrate specificity. The best characterized representatives of these NTases are (i) canonical poly(A) polymerases (PAPs), responsible for polyadenylation of mRNA and (ii) CCA-adding enzymes, synthesizing CCA triplet at the 3′-end of tRNAs (Yue et al. 1996; Martin and Keller 2007). Furthermore, there is an expanding group of noncanonical NTases, encompassing not only atypical PAPs, but also 2′-5′-oligoadenylate synthetases (OAS), poly(U) polymerases (PUPs), and terminal uridyltransferases (TUTases) (Aphasizhev 2005; Stevenson and Norbury 2006; Kwak and Wickens 2007; Martin and Keller 2007; Rissland and Norbury 2008; Torralba et al. 2008; Schmidt and Norbury 2010).
Shortening or removal of poly(A) tails from the 3′-ends of mRNAs has been long considered as a rate-limiting step of their degradation, which may then proceed in a 5′–3′ direction (through concerted action of decapping enzymes and Xrn1 exoribonuclease) or in a 3′–5′ direction (catalyzed by the RNA exosome complex) (for review, see Labno et al. 2016a). Studies in Schizosaccharomyces pombe revealed that deadenylation is not an absolute prerequisite for degradation, and uridylation by noncanonical cytoplasmic NTase Cid1 stimulates decapping instead (Rissland and Norbury 2009). Importantly, both oligo(A) tails arising after deadenylation and 3′-uridylation tags are recognized by the Lsm1-7/Pat1 complex, which recruits decapping machinery and is therefore crucial for both deadenylation-dependent and -independent decay (Chowdhury et al. 2007; Song and Kiledjian 2007).
mRNA uridylation and the responsible PUPs gained significant attention because they are conserved among eukaryotes (except Saccharomyces cerevisiae, which genome does not encode noncanonical NTase with PUP activity), although some species-specific differences exist (Scott and Norbury 2013; Munoz-Tello et al. 2015; Viegas et al. 2015; Scheer et al. 2016). For instance, uridylation of oligo(A) tails in Arabidopsis thaliana by URT1 PUP does not seem to influence the mRNA degradation rates globally, but rather protects 3′-ends from trimming to ensure a 5′–3′ directionality of cotranslational mRNA decay (Sement et al. 2013; Scheer et al. 2016; Zuber et al. 2016). In mammals, uridylation catalyzed by Cid1 homologs—TUT4 (ZCCHC11) or TUT7 (ZCCHC6)—occurs on deadenylated mRNAs with oligo(A) tails shorter than 25 nt (Chang et al. 2014; Lim et al. 2014). Furthermore, all above-mentioned TUTases regulate small RNA biogenesis and activity. In human stem cells or certain cancer cells, TUT4/7 are recruited to pre-miRNAs by Lin28 protein, which leads to 3′-uridylation and degradation by a Perlman syndrome exonuclease DIS3L2 (Heo et al. 2009; Chang et al. 2013; Ustianenko et al. 2013; Faehnle et al. 2014; Kim et al. 2015). Notably, nucleolytic activity of the latter, stimulated by TUTase-mediated oligo(U) tail synthesis, has been recently documented to control quality of multiple noncoding cytoplasmic transcripts in different species (Labno et al. 2016b; Pirouz et al. 2016; Reimao-Pinto et al. 2016; Ustianenko et al. 2016).
In addition to uridylation, a substantial fraction of eukaryotic poly(A)+ mRNAs is subject to guanylation, performed by as yet unidentified NTases (Chang et al. 2014; Zuber et al. 2016). The function of guanosine-rich extensions remains unknown—it was proposed that they may protect poly(A) tails from deadenylation (Chang et al. 2014).
Another interesting example of posttranscriptional mRNA 3′-end modification was discovered in Aspergillus nidulans. It involves addition of short (1–7 nt) C/U-rich tails by a Cid1-related enzyme, CutA (Morozov et al. 2010). Similar to S. pombe, such heteropolymeric C/U extensions are added to poly(A) tails, circumventing the need for their shortening prior to decapping (Morozov et al. 2010).
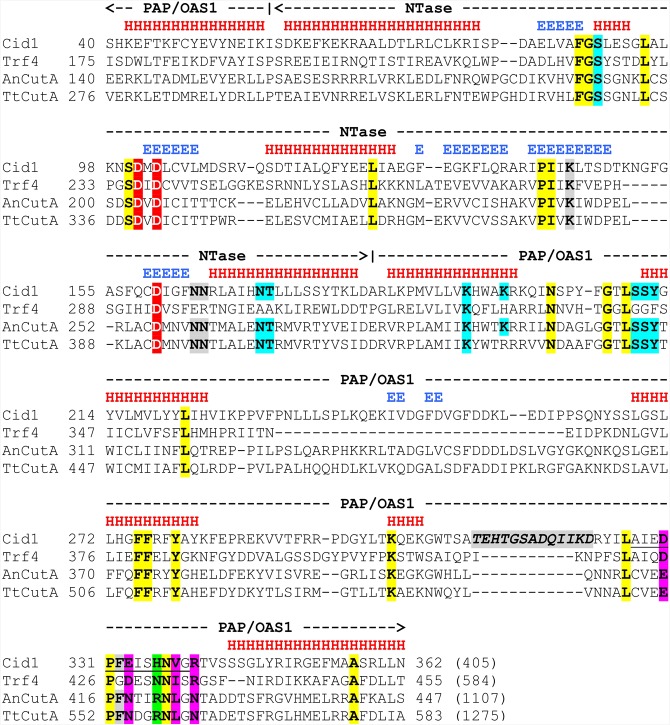
Several crystal structures of Cid1 provided insights into the catalytic mechanism of uridylation (Lunde et al. 2012; Munoz-Tello et al. 2012, 2014; Yates et al. 2012, 2015). The structural basis for addition of other nucleotides, like cytidine in the case of CutA, has not been explained. Noncanonical NTases generally comprise catalytic and central domains (the latter also known as PAP/OAS1 substrate binding domain [SBD]) (Fig. 1; Aravind and Koonin 1999; Stevenson and Norbury 2006; Martin and Keller 2007; Kuchta et al. 2009), and some of them may additionally contain other functionally important motifs, like zinc fingers (Hagan et al. 2009; Heo et al. 2009; Rajappa-Titu et al. 2016). The catalytic domain of Cid1 is built from a five-stranded β-sheet flanked by α-helices (Lunde et al. 2012; Yates et al. 2012). The second β-strand contains a DxD motif between positions 101–103, which, together with the third D in the fifth β-strand (D160) (Fig. 1), interacts with the incoming RNA molecule and two metal ions, thus creating the active site, located at the bottom of the cleft separating catalytic and central domains (Lunde et al. 2012; Munoz-Tello et al. 2012; Yates et al. 2012). The three catalytic aspartates also face β- and γ-phosphates of the UTP substrate (Munoz-Tello et al. 2012). The α-helical central domain contains a nucleotide recognition motif (NRM)—it corresponds to a ∼10–15 aa-long loop (located between positions 327–339 in Cid1) (Fig. 1), forming one end of the NTP-binding pocket (Lunde et al. 2012; Munoz-Tello et al. 2012). The residues in the NRM, together with amino acids from other parts of the protein (Fig. 1) stabilize the base of the NTP substrate via water-mediated and/or direct hydrogen bonds with their side chain atoms (Munoz-Tello et al. 2012). The hydrogen bond crucial for UTP selectivity of Cid1 is formed by the unique histidine (H336) in the NRM (Fig. 1), and the nucleobase (Lunde et al. 2012; Munoz-Tello et al. 2012; Yates et al. 2012). The affinity of Cid1 toward UTP is 30-fold higher than toward ATP and substituting H336 with asparagine or alanine significantly increases Cid1 ability to utilize ATP as a substrate (Lunde et al. 2012; Yates et al. 2012). UTP recognition by Cid1 is supported by water-mediated interactions between D330 and E333 (Fig. 1) and uridine (Lunde et al. 2012; Yates et al. 2012). The preference for UTP over CTP is partially due to the protonation state of H336, which disfavors formation of the hydrogen bond with the latter (Lunde et al. 2012). Cid1 interaction with UTP is further stabilized by several other hydrogen bonds, involving particularly N171 and T172 (Fig. 1), which contact the ribose directly or via water bridge, and K193/K197, interacting with phosphate groups with some contribution from S90, S211, and Y212 (Fig. 1; Lunde et al. 2012; Munoz-Tello et al. 2012; Yates et al. 2012).
FIGURE 1.
Multiple sequence alignment of the catalytic (NTase) and central (PAP/OAS1 SBD) domains from S. pombe Cid1 poly(U) polymerase, S. cerevisiae Trf4 poly(A) polymerase, and CutA proteins from A. nidulans (AnCutA) and T. terrestris (TtCutA). Domain ranges are shown above the alignment with dashed lines ending with arrowheads. Locations of observed secondary structure elements in Cid1: α-helices and β-strands are marked with red “H” and blue “E” letters, respectively, above its amino acid sequence. Three conserved aspartates in the active site are highlighted in red. H336, primarily responsible for UTP selectivity of Cid1, and corresponding amino acids in other proteins are marked with green. Other residues within or in the vicinity of the nucleotide recognition motif (NRM), including those that were subjected to mutations in our study, are highlighted in magenta. Amino acids from other parts of the protein, involved in NTP recognition, are marked with blue. Remaining residues important for nucleotidyltransferase activity are highlighted in gray (β-trapdoor loop, unique for Cid1, is additionally marked with italics). Cid1 NRM (residues 327–339) is underlined. Other highly evolutionary conserved amino acids are indicated in yellow.
In this study, we investigated the molecular basis for the ability of A. nidulans CutA and its homolog from Thielavia terrestris—a thermophilic fungus species—to add both cytidine and uridine to the 3′-ends of RNA. To assess preference of these enzymes toward different NTPs, products of in vitro nucleotidylation were subjected to the 3′-RACE analysis coupled with high-throughput sequencing (3′-RACE-seq), a methodology never previously used to study specificity of NTases. We noted that the wild-type versions of CutA proteins have an unprecedented preference toward cytidine, even in the excess of ATP over the other nucleotides, mimicking physiological conditions. Furthermore, most of the tails ended with two cytidines. This was in agreement with results of biochemical assays, which demonstrated low CutA processivity in the presence of CTP as the sole nucleotide and termination of the reaction after addition of only two cytidine residues. In silico structure modelling revealed substantial differences in the NTP-binding site, which likely explain distinct substrate specificity of CutA in comparison to other noncanonical NTases. Corroborating our presumptions, we were able to partially convert CutA into PAP or PUP by changing several amino acids critical for NTP recognition. Similarly, we successfully transformed Cid1 PUP into a variant with increased specificity toward CTP. Our data provide new mechanistic insight into the catalytic mechanism of RNA nucleotidylation.
RESULTS
Fungal CutA proteins are noncanonical NTases with enzymatic activity dependent on the intact conserved aspartate residues in the active site
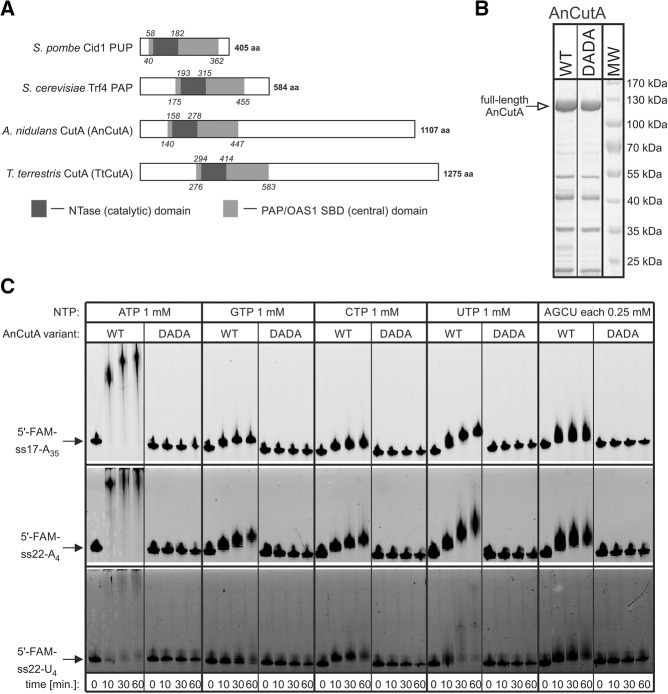
In this study, we focused on CutA protein from A. nidulans (AnCutA), which was previously shown to synthesize C/U-rich 3′-terminal tails in vivo (Morozov et al. 2010), and on its close homolog from T. terrestris (TtCutA). CutA proteins contain both catalytic (NTase) and central (PAP/OAS1 SBD) domains and show significant sequence similarity to several NTases of known structure, including poly(U) polymerase Cid1 and noncanonical poly(A) polymerase Trf4 (Figs. 1, 2A; Lunde et al. 2012; Munoz-Tello et al. 2012, 2014; Yates et al. 2012, 2015). In CutA enzymes, NTase and PAP/OAS1 SBD domains are flanked with a relatively short upstream fragment and C-terminal region, considerably extended in comparison to Cid1 and Trf4 (Fig. 2A).
FIGURE 2.
AnCutA displays a nucleotidyltransferase activity, dependent on the intact aspartates in the active site. (A) Domain organization of different fungal noncanonical nucleotidyltransferases: S. pombe Cid1 poly(U) polymerase, S. cerevisiae Trf4 poly(A) polymerase, and CutA proteins from A. nidulans and T. terrestris (AnCutA and TtCutA, respectively). All proteins contain NTase domain (dark gray rectangle), which is inserted into PAP/OAS1 SBD (light gray rectangle), formed by protein fragments separated in the primary sequence, but located in proximity in the three-dimensional structure. Italicized numbering refers to amino acids located at the borders of NTase (above) and PAP/OAS1 SBD (below) domains. (B) SDS-PAGE analysis of the recombinant full-length AnCutA WT and DADA mutant (with aspartates at positions 203 and 205 substituted with alanines) variants purified by nickel-column affinity chromatography. Protein preparations were analyzed together with PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) as a molecular weight marker (lane MW). Masses of individual bands are indicated on the right. Position of band corresponding to the full-length protein is marked with an open arrow. (C) Analysis of nucleotidyltransferase activity, performed for AnCutA WT and DADA variants with three different RNA oligonucleotides, labeled with fluorescein amidite (FAM) at the 5′-end. Reactions were programmed with individual NTPs or a mixture, as indicated at the top, and terminated after the time points indicated below the lanes.
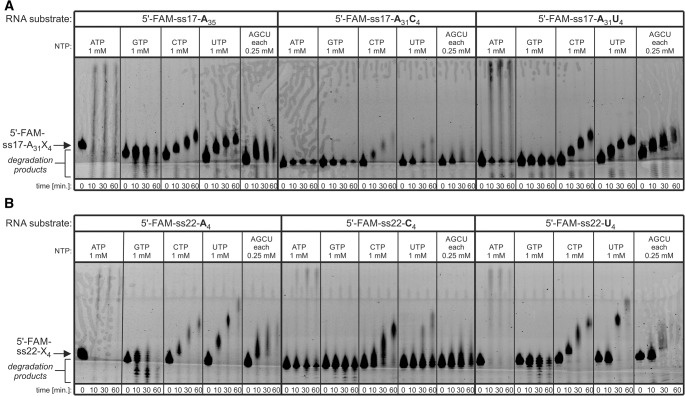
To analyze the specificity of CutA enzymes, we first produced recombinant AnCutA (Fig. 2B), which displayed a nucleotidyltransferase activity, but its efficiency varied depending on the NTP utilized (ATP > CTP ≈ UTP > GTP) (Fig. 2C). These results unveil differences in the enzyme processivity rather than specificity. Notably, the length of tails synthesized in the presence of ATP + CTP + GTP + UTP (AGCU) combinations was comparable to those generated in the presence of individual CTP or UTP (Fig. 2C), suggesting that AnCutA may indeed have an unusual substrate specificity, which would agree with in vivo data (Morozov et al. 2010). Importantly, the AnCutAD203A/D205A catalytic mutant (DADA variant) was inactive in vitro, proving that the observed activity was indeed attributable to the analyzed protein and demonstrating the importance of the conserved aspartic acid residues (Fig. 2C). Finally, the enzyme displayed a preference toward RNA substrates with terminal A or U, while it had low activity for terminal C (Fig. 3). The activity toward substrate with terminal G could not be analyzed, because such RNA precipitated in the gel wells. Therefore, only substrates terminating with adenosine or uridine stretches were utilized in subsequent experiments.
FIGURE 3.
AnCutA efficiently extends 3′-ends of RNA substrates terminating with adenosines or uridines, but not cytidines. Nucleotidyltransferase activity assays were performed for the full-length AnCutA WT protein with two sets of synthetic oligoribonucleotides (ss17-A31 or ss22 in panels A and B, respectively), labeled with fluorescein amidite (FAM) at the 5′-end, having 4-nucleotide-long stretches of adenosines, cytidines, or uridines at their 3′-ends. Reactions were programmed with individual NTPs or a mixture and terminated after the time points indicated below the lanes. Position of the substrates is marked with an arrow on the left. Products of unspecific degradation are indicated.
Identification of the enzymatically active core of a fungal CutA enzyme
Recombinant full-length AnCutA and TtCutA proteins were prone to degradation and their enzymatic activity successively decreased after consecutive stages of purification (Figs. 2B, 4A and data not shown). Thus, in further experiments, we aimed at using more stable, highly active enzyme fragments. To this end, TtCutA was subjected to limited proteolysis (Fig. 4). Protein obtained after nickel affinity chromatography (Fig. 4A) was digested using different concentrations of trypsin, and the samples were analyzed in SDS-PAGE (Fig. 4B). We observed that the full-length TtCutA (lane ND) got degraded to ∼38- to 44-kDa-long fragments (lanes B1–B7), of which the longest, relatively stable one was present in the lane B3 (Fig. 4B). Since all samples collected after limited proteolysis comprised some common contaminant migrating at ∼60 kDa, we further separated proteins present in the sample B3 by gel filtration (Fig. 4C). The band corresponding to the TtCutA proteolytic digestion product was analyzed by Edman degradation, which revealed that the four most N-terminal residues of this fragment have the following sequence: SAYS (Fig. 4C). This allowed us to localize the N-terminal boundary of this fragment to position 216 in the TtCutA amino acid sequence.
FIGURE 4.
Identification of the N terminus of enzymatically active TtCutA proteolytic degradation fragment. (A) SDS-PAGE analysis of protein fractions obtained after purification of the full-length TtCutA by affinity chromatography on the nickel resin. Fraction A1 was subjected to limited proteolysis with trypsin. (B) SDS-PAGE analysis of TtCutA sample, which was not treated with trypsin (lane ND) and samples subjected to proteolytic digestion with different concentrations of trypsin (increasing from lane B1 to lane B7, as indicated above the lanes). Fraction B3 was further purified by gel filtration. (C) SDS-PAGE analysis of protein fractions obtained after separation of the TtCutA limited proteolysis products by gel filtration. Band marked with a white rectangle in lane C6 was subjected to Edman degradation analysis, which demonstrated that the N terminus of the proteolysis-derived fragment begins with S at position 216 of TtCutA, as indicated below the gel. Protein fractions were loaded on SDS-PAGE gels shown in A–C together with PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) as a molecular weight marker (lanes MW). Masses of individual bands are indicated on the left. Positions of bands corresponding to the full-length protein are marked with an open arrow in A and B. Fractions highlighted in bold were tested for nucleotidyltransferase activity in the corresponding assays presented in panels D–F. (D–F) Nucleotidyltransferase activity assays were performed for protein fractions highlighted in bold in panels A–C, using 5′-FAM-ss17-A35 RNA oligonucleotide as a substrate (position indicated with an arrow). Reactions were programmed with individual NTPs or a mixture and terminated after the time points indicated below the lanes.
In parallel to SDS-PAGE analyses, the nucleotidyltransferase activity of the fractions obtained in the course of limited proteolysis and gel filtration was monitored by in vitro assays. Notably, fractions resulting from limited proteolysis retained nucleotidyltransferase activity, but it was profoundly reduced in comparison with the full-length protein, particularly in the case of CTP as an NTP substrate (Fig. 4D,E). This was most likely related to significantly lower protein concentration (cf. lanes B3 and A1 in Fig. 4B and 4A, respectively). The decrease of the enzymatic activity correlated with the increased trypsin concentration and thus, with the reduced length of the proteolytic fragments (Fig. 4E). Analysis of nucleotidyltransferase activity for fractions obtained after gel filtration revealed that the faster-migrating band (see fractions C4–C6 in Fig. 4C) indeed represented the proteolytic fragment of TtCutA, at least partially retaining its catalytic properties (Fig. 4F), while the slower-migrating protein (see fractions C1–C3 in Fig. 4C) was an unrelated contamination, lacking enzymatic activity (Fig. 4F).
To summarize, a limited proteolysis approach and Edman degradation analysis, in conjunction with biochemical investigation of nucleotidyltransferase activity allowed us to identify a stable, enzymatically active fragment of TtCutA protein.
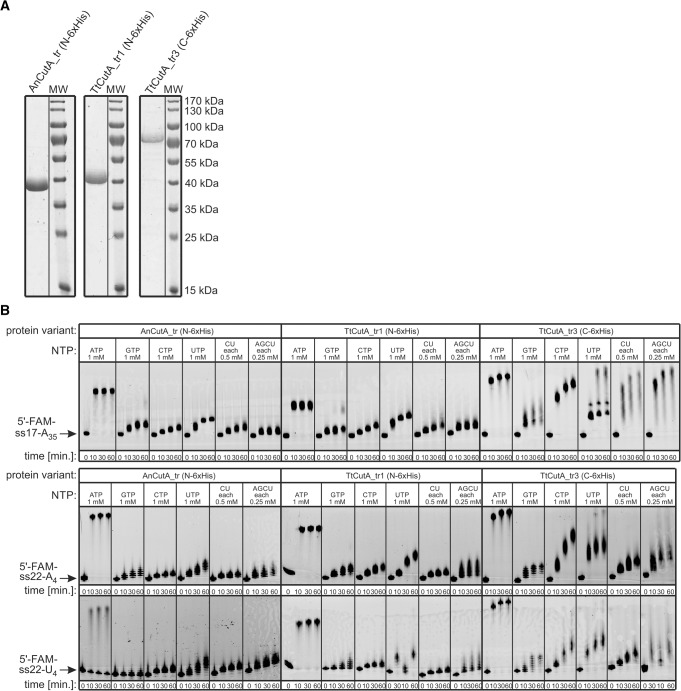
Based on the results of limited proteolysis and secondary structure predictions, we designed truncated variants of TtCutA and AnCutA: TtCutA216-617 and AnCutA84-487, as well as a longer version of TtCutA: TtCutA127-827, further referred to herein as TtCutA_tr1, AnCutA_tr, and TtCutA_tr3, respectively. All three truncated proteins were successfully purified (Fig. 5A), and biochemical analyses confirmed their ability to synthesize 3′-terminal extensions in the presence of each of the four individual NTPs as well as CU and AGCU mixtures, for each of the RNA substrates tested (Fig. 5B). 3′-Tails synthesized by TtCutA_tr3 were considerably longer than in the case of TtCutA_tr1, irrespective of the NTPs utilized. Furthermore, TtCutA_tr3 was able to generate 3′-terminal tails with individual NTPs in the following order of decreasing efficiency: ATP > CTP > UTP > GTP (Fig. 5B). On the contrary, for TtCutA_tr1, as well as its counterpart from A. nidulans—AnCutA_tr—reaction efficiency was comparable in the presence of CTP and UTP (Fig. 5B). This indicates that the lateral extensions present in TtCutA_tr3, when compared to TtCutA_tr1, may influence enzyme processivity. Importantly, there were no significant differences in the pattern of nucleotidylation between substrates having stretches of either adenosine (ss17-A35, ss22-A4) or uridine (ss22-U4) residues of different lengths at their 3′-termini for any of the examined protein variants (Fig. 5B).
FIGURE 5.
Truncated AnCutA and TtCutA variants are enzymatically active. (A) SDS-PAGE analysis of the recombinant (wild-type) AnCutA_tr, TtCutA_tr1, and TtCutA_tr3 variants, purified by nickel-column affinity chromatography followed by gel filtration. Protein preparations were analyzed together with PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) as a molecular weight marker (lanes MW). Masses of individual bands are indicated on the right. (B) Analysis of nucleotidyltransferase activity, performed for truncated CutA protein variants from panel A with three different RNA oligonucleotides, labeled with FAM at the 5′-end, as substrates (positions indicated with arrows on the left). Reactions were programmed with individual NTPs or mixtures (CU or AGCU), as indicated at the top, and terminated after the time points indicated below the lanes.
Full-length and truncated wild-type CutA proteins display a strong preference toward CTP as an NTP substrate in vitro
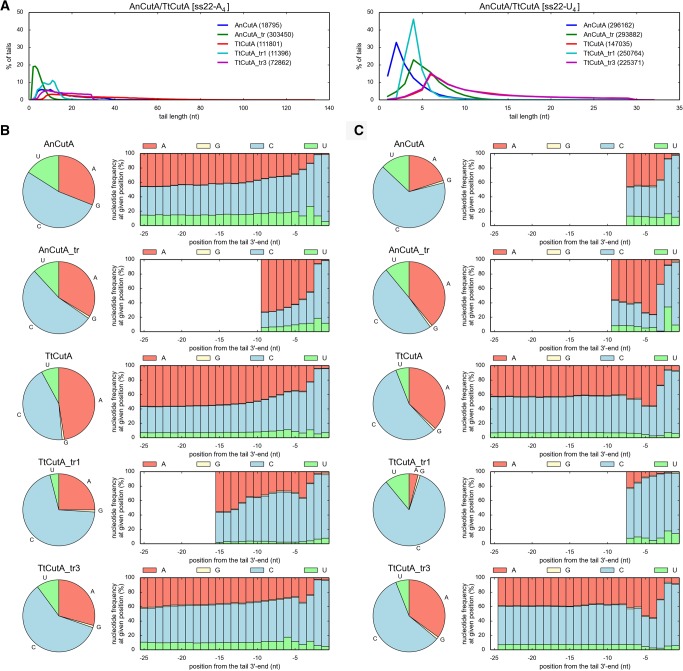
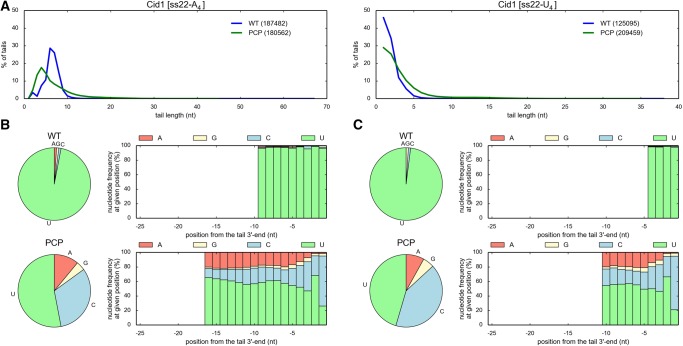
In vitro assays for nucleotidyltransferase activity based on incubation of the recombinant proteins in the presence of labeled RNA substrates and individual NTPs, combined with an in-gel analysis of the products may be misleading in terms of the evaluation of the NTP preference. Therefore, to analyze NTP-specificity of the wild-type full-length and truncated AnCutA and TtCutA protein variants in a more reliable way, we devised a 3′-RACE procedure combined with high-throughput sequencing (3′-RACE-seq). Libraries were prepared for products of reactions catalyzed by AnCutA, AnCutA_tr, TtCutA, TtCutA_tr1, and TtCutA_tr3 in the presence of either ss22-A4 or ss22-U4 substrate and an equimolar AGCU mixture.
Comparison of the average tail lengths revealed that truncated protein variants usually produced shorter 3′-terminal extensions than full-length CutA enzymes, possibly due to the lack of some additional interactions between the synthesized tail and the C-terminal unstructured CutA region, absent in shorter versions (Fig. 6A). Inspection of the percentage distribution of synthesized tails’ lengths showed that most of the examined wild-type proteins and their variants synthesized shorter tails for the ss22-U4 substrate than for the ss22-A4 substrate (Fig. 6A). This might be the consequence of the fact that the frequency of cytidine addition to the ss22-U4 substrate is high, and the resulting tail, ending with uridines and cytidines, interacts most probably very weakly with the substrate binding site (see below).
FIGURE 6.
Full-length and truncated recombinant AnCutA and TtCutA proteins synthesize 3′-terminal tails rich in cytidines, most frequently ending with two C residues. (A) Length distribution for tails added by different AnCutA/TtCutA variants to ss22-A4 (left) and ss22-U4 (right) oligoribonucleotide substrates. Numbers of analyzed sequences, obtained after 3′-RACE-seq high-throughput analysis for each protein variant, are indicated in parentheses. (B,C) Analysis of nucleotide composition for tails added by different AnCutA/TtCutA variants to ss22-A4 (panel B) and ss22-U4 (panel C) RNA substrates. Pie charts demonstrate an overall frequency of incorporation of individual nucleotides into the 3′-terminal extensions. Histograms show the percentage of nucleotide types incorporated at the given position of the tail (the ultimate, 3′-terminal position is on the right). Only those positions, which were covered by at least 5% of the 3′-RACE-seq reads, are presented in the histograms. Furthermore, a limit of 25 nucleotides (nt) from the 3′-end of the tail was arbitrarily set for simplicity.
Importantly, for virtually all tested AnCutA and TtCutA protein variants, cytidine was the predominant incorporated nucleotide, usually followed by adenosine, while the incorporation of uridine and—particularly—guanosine, was significantly less frequent (Fig. 6B,C).
We also realized that the nucleotide incorporation is nonrandom. Thus, to analyze the distribution of nucleotides in the added tails, we have aligned all the sequences starting from the 3′-end. Most importantly, almost all tails synthesized by AnCutA and TtCutA ended with at least two cytidines (Fig. 6B,C), which seems to be a unique feature of CutA enzymes. Furthermore, we observed that the addition of the particular nucleotide strongly depends on the previously added nucleotide. For instance, AnCutA and TtCutA apparently incorporated adenosine more frequently when the antecedent nucleotide of the substrate was also adenosine. For ss22-A4 oligonucleotide, the percentage of added adenosines is increased in such case to 42% and 59%, compared to the average frequency of adenosine incorporation, which is 31% and 48% for AnCutA and TtCutA, respectively (Supplemental Table S1). The situation was different when the last nucleotide of the primer was cytidine or uridine. In such an instance, for the same RNA substrate, we observed: (i) decrease of the adenosine incorporation, (ii) increase of the cytidine incorporation (from 53% or 44% on average to 77% or 76% for AnCutA or TtCutA, respectively) when the preceding nucleotide was uridine, and (iii) increase of the uridine incorporation (from 16% and 8% on average to 22% and 11% for AnCutA and TtCutA, respectively) when the previous nucleotide of the substrate was cytidine (Supplemental Table S1). It is worth noting that the described effect was slightly enhanced when the primer ended with the CU doublet (increasing the percentage of added cytidines to 79% and 78% for AnCutA and TtCutA, respectively) or UC (increasing the percentage of added uridines to 14% for TtCutA) (Supplemental Table S1). This is important in the context of findings reported by Morozov et al. (2010), who suggested that AnCutA is a CUCU-adding enzyme that participates in mRNA modification, which promotes decapping and transcript degradation in A. nidulans. The results of our analyses are partially consistent with their observation, as we noticed that the presence of uridine at the 3′-end of the RNA substrate enhances the possibility of cytidine incorporation, and vice versa, by CutA proteins. Consequently, it is evident that nucleotide selectivity of these enzymes depends on preceding added nucleotides and is determined by protein–substrate interactions. It must be pointed out, however, that AnCutA and TtCutA protein variants do not terminate the nucleotides incorporation process after adding a single (CU) or double (CUCU) nucleotide doublet, in contrast to CCA-adding enzymes, which add a single (CCA) or double (CCACCA) nucleotide triplet to the 3′end of tRNAs (Kuhn et al. 2015).
To sum up, high-throughput 3′-RACE-seq analysis unambiguously demonstrated that CutA enzymes from two fungal species display strong preference toward CTP as an NTP substrate in vitro, which has not been observed for noncanonical nucleotidyltransferases studied so far.
3D model of T. terrestris CutA proteins reveals the location of the potential NTP specificity determinant region
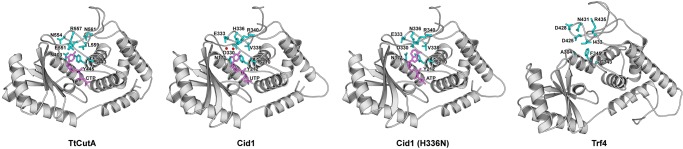
We next asked what could be the structural basis for the observed in vitro CutA enzymes’ preference toward CTP. To answer this question, we generated a 3D homology model of TtCutA and compared it to the experimentally solved structures of closely related enzymes: wild-type and H336N mutant of poly(U) polymerase Cid1 from S. pombe (Lunde et al. 2012), and noncanonical poly(A) polymerase Trf4 from S. cerevisiae (Hamill et al. 2010) (Fig. 7; see Fig. 1 for alignment of amino acid sequences of these NTases).
FIGURE 7.
Comparison of the 3D model of TtCutA with CTP, and available crystal structures of poly(U) polymerase Cid1 with UTP (pdb|4fh5), Cid1 (H336N) mutant with ATP (pdb|4fhx), and noncanonical poly(A) polymerase Trf4 (pdb|3nyb). Key amino acids forming a nucleobase binding pocket are shown in blue. Nucleotides are shown in magenta. Position of CTP in TtCutA was taken from pdb|4fhv. Selected water molecules responsible for interactions between Cid1 and UTP are denoted by red spheres.
In addition, we investigated our 3D model of TtCutA in complex with RNA [oligo(A)], which was taken from S. cerevisiae canonical poly(A) polymerase (pdb|2q66) (Balbo and Bohm 2007) after superposition of the structures. We noticed that the analyzed CutA variants might interact mainly with the last three nucleotides of the RNA substrate. The interaction of the third nucleotide with the protein is the weakest, while the second nucleotide is most probably interacting mainly with the C-terminal, unstructured extension of the enzyme. As both AnCutA and TtCutA modify mRNA in vivo, they are evolutionarily designed to bind to RNAs ending with adenosines. Consequently, substrates with two pyrimidines at the 3′-end probably could not be strongly bound in their active sites. This observation, combined with the ability of these proteins to incorporate their preferred nucleotide—cytidine, may fully explain the phenomenon that the final product is almost always terminated with two cytidines (see above).
The Cid1 PUP incorporates mostly uridines, while Trf4 and H336N mutants of Cid1 are effective in adding poly(A) tails. TtCutA, similarly to Cid1 and Trf4, possesses the two following domains: a PAP/OAS1 SBD with an inserted NTase domain right after the first core PAP/OAS1 SBD α-helix (Figs. 1, 2A). All these proteins share highly conserved NTase domain motifs ([DE]h[DE]h, h[DE]h, and hG[GS]) (Kuchta et al. 2009) and, in addition, they also retain three residues critical for NTP binding (T404, K425, Y445 in TtCutA), which are conserved in proteins possessing both NTase domain and PAP/OAS1 SBD (Fig. 1; Kuchta et al. 2016). The PAP/OAS1 SBD specifically binds a nucleobase of the incoming NTP mainly by amino acids that provide Watson–Crick hydrogen bonds, either directly or indirectly (via water molecules). Those amino acids are located mostly in the NRM region, between the fourth and fifth core α-helixes of PAP/OAS1 SBD (Kuchta et al. 2016). In Cid1, residues from this region, which are responsible for UTP selectivity (D330, E333, and H336; see Fig. 1), cluster around the N3 amine group of uridine and interact with uridine directly or indirectly via one or more water molecules (Fig. 7; Yates et al. 2012, 2015). Another residue, N171 (Fig. 1), binds the uridine O2 carbonyl group, while O4 carbonyl group forms a water-mediated contact to S210 (Figs. 1, 7). In addition, V338 makes a hydrophobic interaction with the nucleobase (Figs. 1, 7; Lunde et al. 2012). The H336N mutant of Cid1 is effective in adding poly(A) tails mainly through the direct interaction of arginine R340 with the N1 position of the incoming adenosine (Figs. 1, 7; Lunde et al. 2012). Position R340 is stabilized by N336, which also forms a hydrogen bond to the side chain of D330 (Figs. 1, 7), probably having a discriminating effect on the uridine selection in the H336N mutant (Lunde et al. 2012). On the contrary, larger H336 in wild-type Cid1 precludes such unique positioning of the R340 side chain, which adopts a different conformation (Fig. 7). It should be noted that the key residues (D330, N336, and R340) present in the active site of the H336N mutant of Cid1 are also conserved in the noncanonical poly(A) polymerase Trf4 from S. cerevisiae (D425, N431 and R435) (Figs. 1, 7; Hamill et al. 2010), which also utilizes ATP in the reaction. We thus concluded that the TtCutA NRM region located between the fourth and fifth core α-helices of the PAP/OAS1 SBD (mostly the fragment encompassing amino acids 551–559—see Fig. 1) remains likely the most important NTP-specificity determinant (NSD) of the analyzed enzymes.
Deciphering the mechanism of CutA preference toward CTP through site-directed mutagenesis in the NRM region
Having identified the NSD of CutA enzymes, we focused on verifying whether particular amino acids within NRM indeed play a role in the CTP specificity, and rationally designed mutations in this region of TtCutA.
We noticed that R557, substituted by H336 in Cid1, is unique for both TtCutA and AnCutA (Fig. 1). It allowed us to speculate that changing this residue may strongly influence substrate specificity. We thus constructed the R557A mutant (PAP1) following the H336A mutation in Cid1 proposed by Yates and coworkers (Yates et al. 2012, 2015) to strengthen the production of poly(A) tails. We also proposed R557Q (PAP2) mutation following the study of Lunde and coworkers, who demonstrated that the Cid1H336N mutant catalyzes robust adenosine incorporation (Lunde et al. 2012). We chose the mutation R557Q over R557N, because the Q557 side chain in the PAP2 mutant could probably be better stabilized by longer E551, compared to a shorter D330 in the H336N mutant of Cid1. We also wondered how to enhance the poly(A) polymerase activity by additional changes in the NRM, apart from substituting a R557 residue. Therefore, we created a PAP3 mutant (E551D/N554E/R557A), in which we reconstructed other amino acids present in the active site of Cid1H336A, which has proven PAP activity (Yates et al. 2012). Additionally, we constructed another mutant—PAP4 (E551T/R557A/L559S), with the aim of enlarging the TtCutA active site and eliminating some (in)direct interactions between the protein and its preferred incoming nucleotide—CTP. The construction of PAP4 variant was guided by the idea that a canonical poly(A) polymerase (e.g., PAP from S. cerevisiae) binds the incoming NTP mainly through nonpolar interactions above and below the plane of the adenosine base and to the sugar, and water mediated interactions to polar groups in the plane of the base, and to the sugar hydroxyl groups (Balbo and Bohm 2007).
We also proposed three mutants: PUP1 (R557H), PUP2 (E551D/R557H), and PUP3 (E551D/N554E/R557H), which we assumed should allow us to gradually construct a modified nucleobase recognition site in TtCutA, resembling the one observed in the Cid1 poly(U) polymerase.
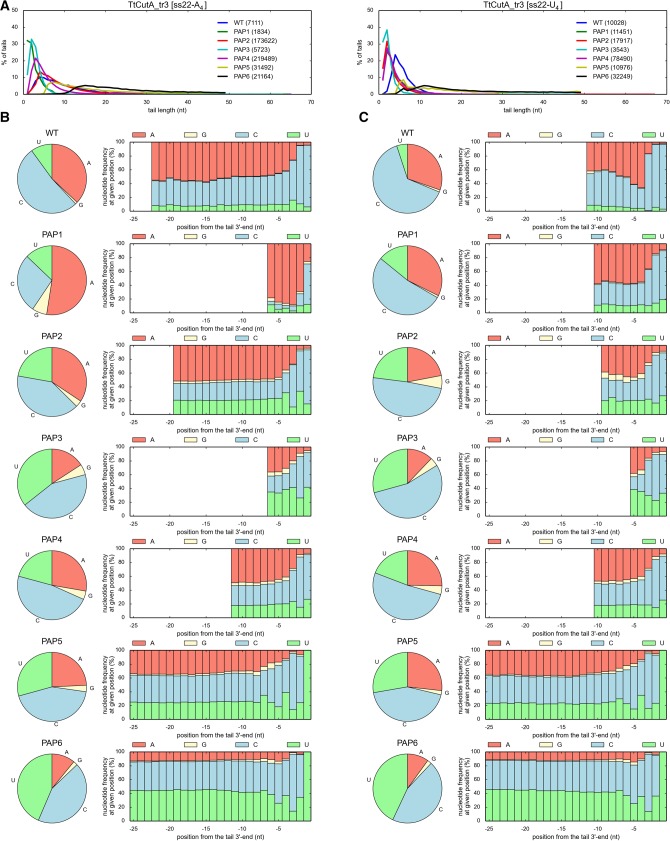
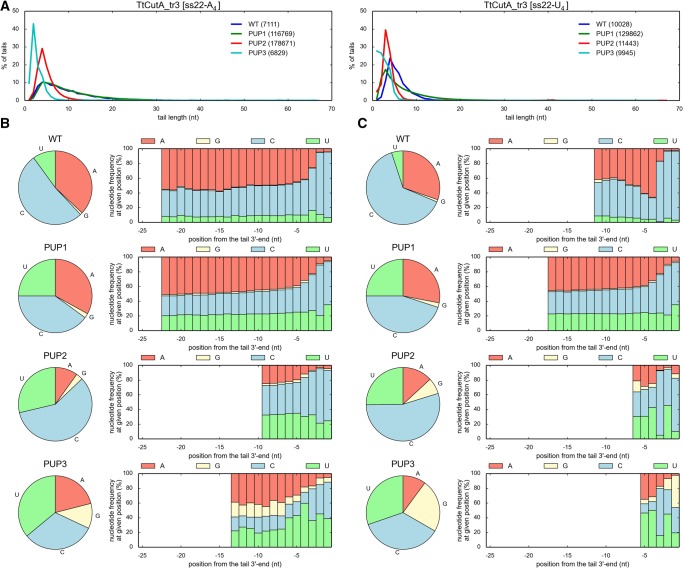
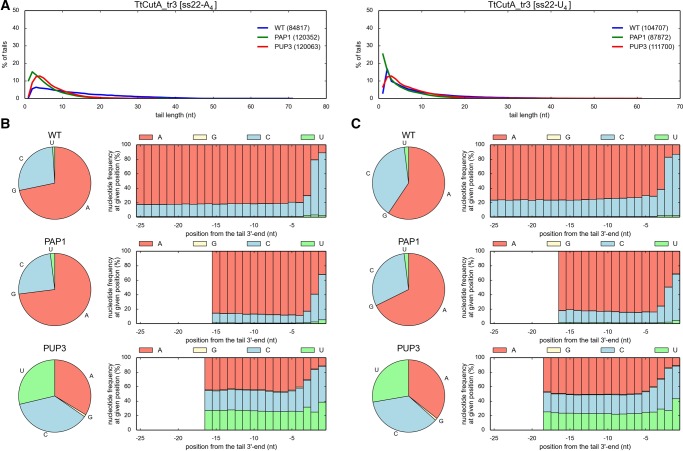
To experimentally analyze the influence of the designed mutations on the NTP specificity, we expressed and purified TtCutA_tr3 variants containing appropriate amino acid substitutions within NRM. All purified TtCutA_tr3 versions still displayed nucleotidyltransferase activity, but based on the biochemical data it was impossible to assess whether the NTP substrate preference was definitely changed upon a given mutation (data not shown). Thus, to analyze the specificity, 3′-RACE-seq for both ss22-A4 and ss22-U4 substrates was performed (Figs. 8 and 9). In concordance with previously obtained results, the most predominant nucleotide added by the wild-type TtCutA_tr3 in the presence of an equimolar AGCU mixture, was cytidine, followed by adenosine and uridine (Figs. 8 and 9). The frequency of guanosine incorporation fluctuated around 0.5%–2%, corroborating biochemical data indicating that it is definitely the least often utilized nucleotide (Figs. 8 and 9).
FIGURE 8.
Effect of mutations designed to change TtCutA specificity into PAP. (A) Length distribution for tails added by wild-type (WT) and mutant (PAP1-PAP6) TtCutA_tr3 variants to ss22-A4 (left) and ss22-U4 (right) oligoribonucleotide substrates. Numbers of analyzed sequences, obtained after 3′-RACE-seq high-throughput analysis for each protein variant, are indicated in parentheses. (B,C) Analysis of nucleotide composition for tails added by different TtCutA_tr3 variants to ss22-A4 (panel B) and ss22-U4 (panel C) RNA substrates. Results are presented as in Figure 6.
FIGURE 9.
Effect of mutations designed to change TtCutA specificity into PUP. (A) Length distribution for tails added by wild-type (WT) and mutant (PUP1-PUP3) TtCutA_tr3 variants to ss22-A4 (left) and ss22-U4 (right) oligoribonucleotide substrates. Numbers of analyzed sequences, obtained after 3′-RACE-seq high-throughput analysis for each protein variant, are indicated in parentheses. (B,C) Analysis of nucleotide composition for tails added by different TtCutA_tr3 variants to ss22-A4 (panel B) and ss22-U4 (panel C) RNA substrates. Results are presented as in Figure 6.
Importantly, in the case of the TtCutA_tr3R557A (PAP1) mutant, the frequency of adenosine incorporation was increased, which was particularly visible for tails added to ss22-A4 substrate (Fig. 8), indicating that R557A substitution partially converted the enzyme specificity from PCP to PAP. This implied that R557 is an important residue for selection of CTP over other nucleotides, but not as crucial as its Cid1 counterpart (H336) for the UTP selectivity of the latter (Yates et al. 2012). The R557A mutation weakened the CTP binding affinity, probably because of destroying one of the key contacts between the protein and this preferred nucleotide. Additionally, replacing arginine with much smaller alanine enlarged the TtCutA nucleobase binding site. As a result, the R557A mutation, by diminishing the cytidine-adding activity of the enzyme, increased the presence of adenosines in synthesized tails, as we expected.
The influence of PAP2, PAP3, and PAP4 mutations on the NTP specificity of TtCutA_tr3 was less obvious (Fig. 8). Similar to other mutant protein variants, the number of cytidines built into the 3′-terminal tails by TtCutA_tr3R557Q (PAP2), TtCutA_tr3E551D/N554E/R557A (PAP3), and TtCutA_tr3E551T/R557A/L559S (PAP4) was reduced in comparison to the wild-type enzyme (Fig. 8). The percentage of adenosines was also lower for these protein variants than in the wild-type control—such an effect was most remarkable for the PAP3 triple mutant (Fig. 8). On the other hand, the parallel increase in the frequency of uridine incorporation, observed for all these protein variants (again—most clearly in the case of PAP3 mutant) (Fig. 8), suggested that R557Q single mutation and E551D/N554E/R557A or E551T/R557A/L559S triple substitutions change NTP preference more toward UTP than ATP. It thus seems that PAP2, PAP3, and PAP4 mutants gained additional interactions with uridine, in the case of PAP2 and PAP3 probably through increase of similarity to Cid1 in the NRM region. Specifically, Q557 in PAP2 corresponds to H336 in Cid1 and thus seems to replace it functionally to perform a similar role in UTP recognition. In addition, we did not observe the increased amount of adenosines in tails synthesized by PAP2 (Fig. 8), probably because TtCutA possesses asparagine (N561) instead of key arginine, which can be found at this position in the noncanonical poly(A) polymerase Trf4 (R435) and Cid1H336N mutant, specific toward adenosine (R340). On the other hand, according to the 3D modeling, R557 may be a structural equivalent of this key arginine responsible for ATP recognition, which may explain substantial adenosine-adding activity of wild-type TtCutA. Consequently, we introduced the N561R mutation and gradually changed TtCutA to make it more similar to both the Cid1H336N mutant and Trf4, by generation of PAP5 (R557N/N561R) and PAP6 (E551D/R557N/N561R) variants. Surprisingly, these mutants had significantly decreased frequency of adenosine incorporation and largely increased specificity toward uridines (particularly PAP6) (Fig. 8). This suggests that in TtCutA—probably due to slight differences in the geometry of the NTP binding site (e.g., NRM conformation)—the role of arginine at position 557 in adenosine recognition cannot be substituted by moving this arginine to position 561 (where it is present in Trf4 poly(A) polymerase).
When trying to re-engineer poly(U) polymerase from TtCutA, we noticed other apparent changes in the substrate preference in the case of TtCutA_tr3R557H (PUP1), TtCutA_tr3E551D/R557H (PUP2), and TtCutA_tr3E551D/N554E/R557H (PUP3) mutants. First of all, they all synthesized 3′-tails with fewer adenosines than the wild-type counterpart (Fig. 9). At the same time, the percentage of uridines increased considerably (three- to ninefold, depending on the substrate and the length of analyzed tails), which suggested that for these variants, the specificity toward uridines was increased (Fig. 9). Furthermore, the amount of uridines in the 3′-terminal extensions generated by TtCutA_tr3E551D/N554E/R557H was higher than for TtCutA_tr3E551D/R557H, and simultaneously the frequency of cytidine incorporation decreased by ∼20%–25% (Fig. 9). This implicated that while E551D and R557H substitutions are partially responsible for PCP→PUP modification of specificity, N554E amino acid change may additionally increase enzyme preference toward UTP. These results collectively indicate that while replacing TtCutA E551 and R557 residues with their Cid1 counterparts (aspartic acid and histidine, respectively) apparently failed to reconstruct all water-mediated contacts between protein and UTP, an additional N554E substitution caused UTP to be the preferred nucleotide. This also implies that N554, together with R557 is a very important residue responsible for CTP selectivity of TtCutA.
In sum, results of our high-throughput 3′-RACE-seq analysis, combined with structural inspection, demonstrated the critical importance of arginine (R), glutamic acid (E), and asparagine (N) at positions 557, 551, and 554 of TtCutA (Fig. 1), located within NRM, for CTP-specificity of the examined enzyme. Furthermore, R557A substitution enabled partial conversion of TtCutA nucleotidyltransferase activity from PCP to PAP, whereas R557H, E551D, and N554E substitutions changed the enzyme specificity from PCP to PUP. Notably, however, while we successfully engineered proteins with significantly reduced or increased specific nucleotide selectivity, we were not able to construct the enzymes, which would use only one preferred nucleotide for the polymerization process. This may be caused by the character of the CutA enzymes’ NRM region, which allows binding of all NTPs in the active site without a stringent preference for a single nucleotide.
Nucleotide specificity of TtCutA variants is preserved at a physiologically relevant NTP ratio
Since the 3′-RACE-seq analyses described above were performed for products of reactions with equimolar AGCU mixtures, the increased frequency of cytidine incorporation in vitro by TtCutA wild-type protein could potentially result from artificially high CTP concentration. To test this possibility, assays for TtCutA_tr3 WT, as well as for PAP1 and PUP3 variants, which displayed the most obvious changes in NTP specificity, were repeated at more physiologically accurate 10:1:1:1 ratio of ATP:GTP:CTP:UTP, assuming that ATP is in approximately 10-fold excess over other NTPs in vivo.
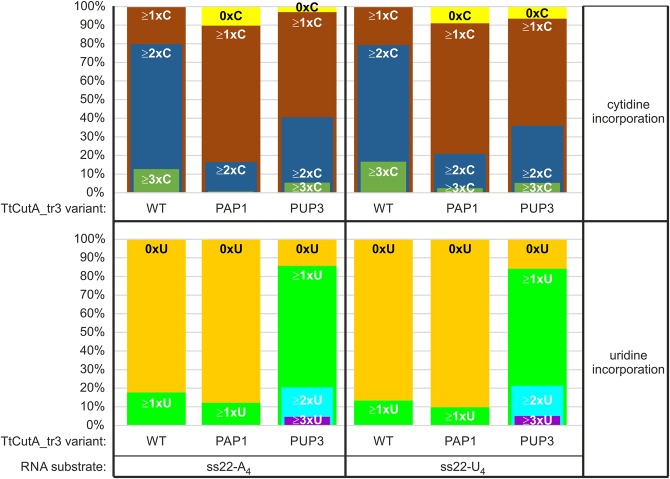
Analysis of the 3′-RACE-seq data derived from assays conducted in such modified conditions (Fig. 10) revealed for the TtCutA_tr3 wild-type version that although the overall percentage of adenosines increased to 60%–70% (from 30% to 40% for equimolar NTP ratio), depending on the substrate, the level of cytidine incorporation decreased at the same time from 50%–60% to 30%–40% (cf. Fig. 10B,C with Fig. 8B,C). This demonstrates that even when CTP is underrepresented, the enzyme retains high specificity toward this nucleotide. Furthermore, decreasing CTP concentration did not preclude predominant incorporation of cytidine at the 3′-terminal and penultimate positions of the extension (Fig. 10B,C and data not shown). In addition, we noticed that more than 99%, ∼80%, and ∼15% of tails contained at least one, two, and three cytidines, respectively (Fig. 11).
FIGURE 10.
Specificity of TtCutA with an excess of ATP over other nucleotides. (A) Length distribution for tails added by wild-type (WT) and mutant (PAP1 and PUP3) TtCutA_tr3 variants to ss22-A4 (left) and ss22-U4 (right) oligoribonucleotide substrates in the presence of 10:1:1:1 ATP:GTP:CTP:UTP molar ratio. Numbers of analyzed sequences, obtained after 3′-RACE-seq high-throughput analysis for each protein variant, are indicated in parentheses. (B,C) Analysis of nucleotide composition for tails added by different TtCutA_tr3 variants to ss22-A4 (panel B) and ss22-U4 (panel C) RNA substrates at physiologically relevant NTP ratio. Results are presented as in Figure 6.
FIGURE 11.
Different fractions of tails synthesized by WT, PAP1, and PUP3 variants of TtCutA_tr3 contain a given number of cytidines or uridines. Upper graph shows percentage of tails derived from the 3′-RACE-seq data presented in Figure 10, which entirely lack cytidine (0xC), or contain at least one single cytidine or at least one 2- or 3-nt-long string of consecutive cytidines. Lower graph demonstrates analogous results for uridines.
We then checked whether changes in the NTP specificity that we observed for TtCutA_tr3 PAP1 and PUP3 mutant variants could be due to the excess of ATP. Surprisingly, the frequency of overall ATP incorporation increased very modestly for PAP1 variant, when compared to WT (Fig. 10B,C), but this could be partly due to a shorter average length of the tails generated by the PAP1 mutant relative to the wild-type protein (Fig. 10A and data not shown). Nevertheless, the R557A mutation present in TtCutA_tr3 PAP1 variant led to other apparent changes, e.g., the increased presence of adenosine at two ultimate tail positions (more tails contained AA, AC, or CA 3′-terminal dinucleotides) (Fig. 10B,C and data not shown), appearance of a fraction of tails (∼10%) lacking cytidine, and concomitant reduction in the percentage of tails containing at least two (∼20%) consecutive cytidines (Fig. 11). This confirms that the partial change of specificity from CTP toward ATP has been achieved in the case of the TtCutA_tr3 PAP1 variant.
More pronounced changes were noted at the physiologically relevant NTP ratio for the TtCutA_tr3 PUP3 mutant (Fig. 10B,C). First, despite significant ATP excess, uridine incorporation into tails increased to ∼30% (compared to ∼2% for the WT variant). Second, numerous tails contained CU, UC, UU, UA, and AU dinucleotides at the 3′-end, instead of CC (Fig. 10B,C and data not shown). Finally, fractions of tails lacking cytidines and containing two or more cytidines in a row were more and less abundant, respectively, than for TtCutA_tr3 WT (Fig. 11). At the same time, the percentage of extensions encompassing at least one uridine was as high as ∼85%, compared to only 10%–20% for WT and PAP1 versions (Fig. 11). Furthermore, longer (2–5 nt) stretches of uridines were also evident in some tails generated by the PUP3 mutant (Fig. 11 and data not shown), which virtually did not occur in the extensions synthesized by TtCutA_tr3 WT and PAP1.
In summary, the relatively high in vitro preference of TtCutA_tr3 WT toward CTP is maintained even in conditions where the availability of this nucleotide is considerably reduced with respect to ATP, which likely reflects the situation in vivo. Moreover, changes of NTP specificity toward adenosine or uridine due to mutations within NRM introduced in PAP1 or PUP3 variant, respectively, could be observed under the same conditions, which further supports our conclusion about the importance of E, N, and R amino acids at TtCutA positions 551, 554, and 557, respectively, for CTP selectivity.
Distinct affinities and variable enzyme processivity in the presence of various NTPs account for differences in the nucleotide composition of extensions generated by different TtCutA variants
We next sought to determine whether differential nucleotide composition of tails synthesized by different variants of TtCutA_tr3 could result from distinct affinities toward various NTPs. To this end, we first attempted to monitor polymerization efficiency in the presence of individual NTPs by fluorescence-based coupled activity assays, which rely on indirect measuring of pyrophosphate release with the use of inorganic pyrophosphatase (PPase) and purine nucleoside phosphorylase (PNP), able to cleave a fluorescent 7-methylguanosine derivative in the presence of generated phosphate. Similar approach proved previously to be successful for calculation of kinetic parameters for S. pombe Cid1 (Lunde et al. 2012). However, we found it inappropriate in the case of TtCutA, since the reaction conditions optimized for PPase and PNP were totally incompatible with requirements of TtCutA polymerization activity.
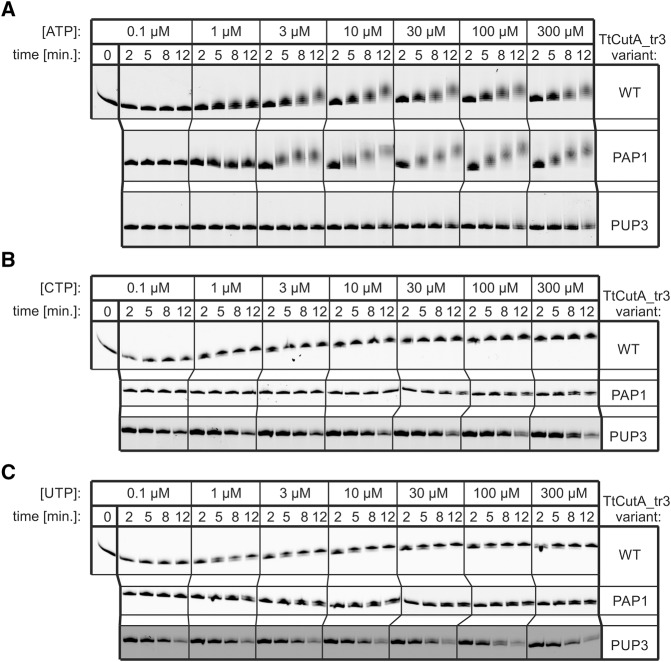
Therefore, to roughly compare affinities of TtCutA_tr3 WT, PAP1, and PUP3 variants toward different NTPs, a set of gel-based assays was performed under conditions where the RNA:protein molar ratio was 40:1 and individual ATP, CTP, or UTP were utilized at various concentrations, ranging from 0.1 to 300 µM and reactions were stopped after much shorter time-points than in previous biochemical analyses. Results of these experiments are presented in Figure 12.
FIGURE 12.
Different affinity of TtCutA_tr3 variants toward individual NTPs. (A–C) Nucleotidyltransferase activity assays were performed for WT, PAP1, and PUP3 TtCutA_tr3 protein variants, with 40-fold molar excess of the 5′-FAM-labeled ss22-A4 RNA substrate. Reactions were programmed with individual ATP (A), CTP (B), or UTP (C) at different concentrations, as indicated above the lanes and terminated after short time points (2–12 min) following start of incubation.
While basing on data obtained in this manner we were unable to provide quantitative measurements of kinetic constants (which is also due to substantial differences in enzyme processivity in the presence of ATP relative to CTP or UTP, as discussed below), some relative comparisons could be made. First, both WT and PAP1 variants displayed a similar affinity toward ATP, starting polymerization at 1 µM ATP concentration (Fig. 12A). In contrast, the PUP3 mutant was unable to begin ATP addition unless its concentration was higher than 10 µM. Moreover, the reaction progressed much more slowly and resulting tails were significantly shorter (Fig. 12A). This, together with other observations discussed below, may partially explain why the stretches of adenosines present in tails generated by TtCutA_tr3 PUP3 variant are more often interrupted with other residues, particularly uridine.
In the case of CTP, PUP3 and PAP1 were able to start polymerization at a much higher concentration of this nucleotide (10 and 30 µM, respectively), when compared to the wild-type TtCutA_tr3 version (Fig. 12B). This indicates that the affinity of PAP1 and PUP3 mutants toward CTP might be indeed somewhat lower, which is reflected by a lower incidence of cytidine in tails generated by these protein variants in comparison to TtCutA_tr3 WT (see Fig. 11). On the contrary, affinity of all three tested TtCutA_tr3 versions toward UTP did not seem to differ dramatically, since all of them started adding uridine already at 1 µM UTP concentration.
In addition, we noted that when the RNA substrate was in excess over protein, reflecting an in vivo situation, all TtCutA_tr3 variants synthesized very short tails with CTP and UTP, when compared to ATP, indicating lower enzyme processivity in the presence of pyrimidines. This difference was less obvious in previous gel-based assays, where a slight excess of protein over RNA (2.5:1) was applied.
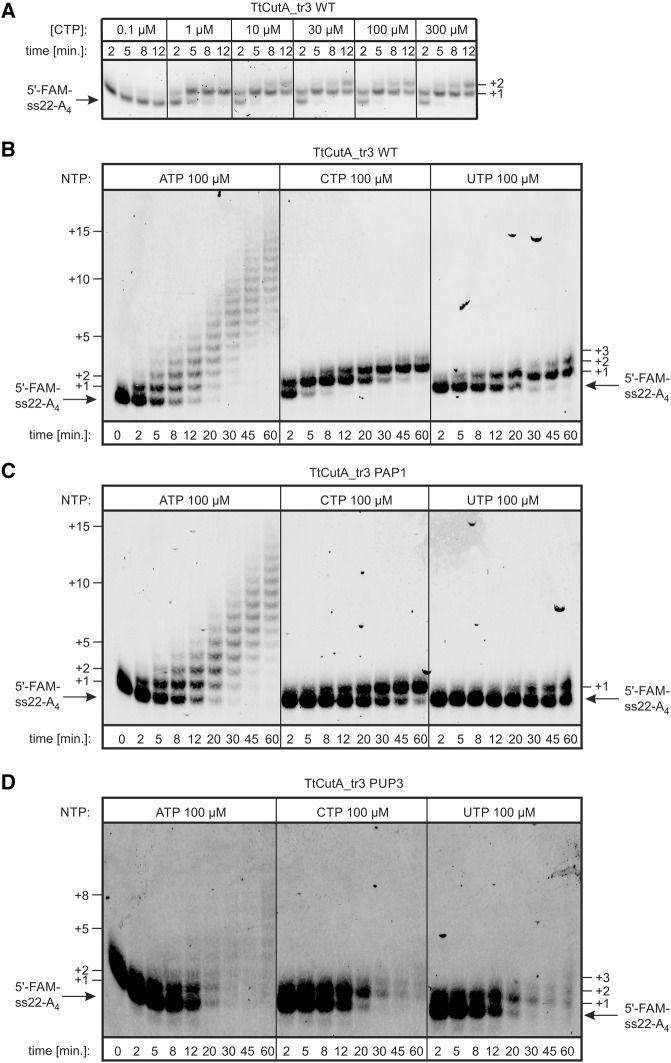
To investigate differences in processivity more systematically, reactions were carried out individually for each of the three TtCutA_tr3 variants with either ATP, CTP, or UTP at a fixed, nonlimiting (100 µM) concentration in a time-course extended to 60 min, and the products were analyzed in a sequencing gel to obtain single-nucleotide resolution (Fig. 13). Initial analysis revealed that at 1 µM CTP concentration, TtCutA_tr3 WT was able to add only one cytidine, whereas at higher concentrations of this nucleotide maximum length of the extension achieved only two residues, demonstrating that the enzyme tends to terminate after adding two cytidines (at least by the end of a short reaction time-course) (Fig. 13A). Further experiments demonstrated that all protein variants were processive with ATP and acted distributively in the presence of CTP or UTP (Fig. 13B–D). Processivity of TtCutA_tr3 PUP3 variant with ATP was a bit lower than that observed for WT or PAP1 versions, but this conclusion may be obscured due to RNA degradation at later time points. On the contrary, both WT and PUP3 variants stopped after adding maximally two cytidines or uridines, whereas in the case of the PAP1 mutant termination occurred even after the addition of one pyrimidine.
FIGURE 13.
TtCutA_tr3 variants act processively with ATP and distributively in the presence of CTP or UTP. (A) Products of reactions shown in Figure 12B for TtCutA_tr3 WT protein were resolved in 10% high-resolution sequencing gel, showing that the substrate 3′-end can be extended by not more than two cytidines, as indicated on the right (B–D). Nucleotidyltransferase activity assays were performed for WT (B), PAP1 (C), and PUP3 (D) TtCutA_tr3 protein variants, with 40-fold molar excess of the 5′-FAM-labeled ss22-A4 RNA substrate (position marked with arrows). Reactions were programmed with 100 µM ATP, CTP, or UTP, as indicated above the lanes and terminated after 2–60 min following start of incubation. Lengths of the synthesized tails are indicated on both sides of the gel images.
These observations corroborate our 3′-RACE-seq data, demonstrating for instance that in the presence of AGCU mixture, TtCutA_tr3 wild-type protein usually processively adds a stretch of adenosines, before switching at some point to cytidine (or sometimes uridine), but is able to distributively add only one or two such residues. Then, depending on the nature of the next incoming nucleotide, the enzyme likely dissociates from the substrate, resulting in the frequent generation of tails terminating with two cytidines, or continues synthesis of longer adenosine stretches if the subsequent nucleotide is ATP. Furthermore, the majority of tails generated by PAP1 variant contain only one cytidine, since addition of the second one seems to be inefficient in this case.
In conclusion, nucleotide composition of tails synthesized by different TtCutA variants is determined by a combination of several interdependent factors, such as NTP specificity determined by the character of the NRM (affinity toward particular NTPs), enzyme processivity and the nature of preceding nucleotide in the RNA substrate. It should be also noted that the analyzed mutations (e.g., substitution of arginine) in the NRM region may also have some effect on the electrostatic environment of the nucleotide binding pocket, which could influence interaction with the RNA 3′-end near the active site.
Mutations in the Cid1 NRM region enable its partial conversion from PUP to PCP
To further confirm our conclusions about the importance of particular amino acids within the NRM of CutA enzymes, and to extend our findings to other fungal noncanonical NTases, we attempted to construct the mutant of S. pombe Cid1 PUP with increased preference for cytidine. To this end, we expressed in E. coli a full-length wild-type Cid1 (Cid1WT), as well as its mutated variant, in which aspartic acid, glutamic acid, and histidine, at positions 330, 333, and 336, equivalent to critical TtCutA positions 551, 554, and 557 (Fig. 1), were changed into arginine, glutamate, and asparagine present in the latter protein, respectively. Such amino acid changes, designed to convert Cid1 PUP into PCP were rationalized not only by 3D structure modeling, but also justified by data obtained for TtCutA, demonstrating that the reciprocal substitutions within its NRM partially alter NTP specificity of this enzyme from CTP toward UTP (see above). We will further refer to this Cid1 triple mutant as Cid1D330E/E333N/H336R or Cid1 PCP variant. Both Cid1WT and Cid1D330E/E333N/H336R proteins were successfully purified following heterologous expression, and biochemical assays confirmed that they have nucleotidyltransferase activity (data not shown).
To verify whether Cid1D330E/E333N/H336R triple mutant indeed acquired specificity toward CTP, reactions for Cid1WT and Cid1D330E/E333N/H336R proteins were carried out for both ss22-A4 and ss22-U4 substrates in the presence of equimolar AGCU combination, followed by 3′-RACE-seq. Results of this experiment are summarized in Figure 14. The 3′-terminal tails synthesized by Cid1WT were composed almost exclusively of uridines (specificity >97%), as expected. On the contrary, Cid1D330E/E333N/H336R variant generated 3′ extensions containing only ∼50% of uridines. In parallel, incorporation of remaining nucleotides increased significantly, in the following order: C > A > G. Importantly, fraction of cytidines built into the tails achieved 30%–40%, indicating that the mutations that we designed indeed significantly increased cytidine and decreased uridine incorporation by Cid1 enzyme.
FIGURE 14.
Substitution of several amino acids within Cid1 NRM increases its specificity toward CTP. (A) Length distribution for tails added by wild-type (WT) and D330E/E333N/H336R triple mutant (PCP) Cid1 variants to ss22-A4 (left) and ss22-U4 (right) oligoribonucleotide substrates. Numbers of analyzed sequences, obtained after 3′-RACE-seq high-throughput analysis for each protein variant, are indicated in parentheses. (B,C) Analysis of nucleotide composition for tails added by Cid1 WT and PCP variants to ss22-A4 (panel B) and ss22-U4 (panel C) RNA substrates. Results are presented as in Figure 6.
In conclusion, investigation of Cid1 specificity allowed us to complement data obtained for TtCutA, demonstrating that the ExxNxxR and DxxExxH clusters present at the equivalent positions of NRM, are primarily responsible for the preference of noncanonical NTases toward CTP or UTP, respectively. Unfortunately, we were unable (via proposed mutations) to convert TtCutA into highly effective poly(A) or poly(U) polymerase, as we probably failed to reconstruct all Watson–Crick hydrogen bonds between protein and the desired nucleotide. It seems that TtCutA has a unique geometry and environment of the nucleotide binding site, which makes it impossible to achieve specificity toward a single nucleotide by only a few mutations.
DISCUSSION
It has been more than 10 years from the discovery of noncanonical PAPs (Saitoh et al. 2002; Wang et al. 2002). Multiple follow-up studies have demonstrated that several enzymes from this group synthesize polyuridine, rather than polyadenosine tails, which classified them as PUPs or TUTases (Kwak and Wickens 2007; Rissland et al. 2007). Since then, cytoplasmic uridylation was demonstrated to play a crucial role in the maintenance of cellular homeostasis by regulating stability of both coding and various noncoding transcripts in virtually all model organisms analyzed (Scott and Norbury 2013; Munoz-Tello et al. 2015; Viegas et al. 2015; Scheer et al. 2016). S. pombe Cid1 has been the most extensively investigated PUP, for which several crystal structures were solved (both apo- and NTP-bound) (Lunde et al. 2012; Munoz-Tello et al. 2012, 2014; Yates et al. 2012, 2015), which laid the ground for the attempts to explain differences in the substrate specificity between noncanonical PAPs and PUPs. What made the situation even more complicated was the discovery that Cid1 homolog in A. nidulans—CutA—does not act as a PUP in vivo, but rather adds C/U-rich tails to the 3′-ends of mRNAs in this fungal species (Morozov et al. 2010). Subsequent global transcriptomic analyses demonstrated that untemplated incorporation of cytidines (and also guanosines) into the 3′-terminal extensions is not an uncommon event (Chang et al. 2014; Zuber et al. 2016); however, the noncanonical NTases responsible for such modifications occurring in mammalian or plant cells have not been identified yet. Nevertheless, these findings unequivocally show that the repertoire of substrate specificity among this class of enzymes is not limited to ATP/UTP and some of the group members display distinct NTP preferences. This warrants further investigations of the molecular basis for the observed variability in the substrate specificities of noncanonical NTases.
Previous structural inspections, confirmed by biochemical studies, revealed that H336 in the Cid1 NRM is a major determinant of its specificity toward UTP (Lunde et al. 2012; Munoz-Tello et al. 2012; Yates et al. 2012). UTP recognition by Cid1 is supported by interactions involving other amino acids from the NRM, particularly D330 and E333, which together with H336 constitute DxxExxH cluster (Table 1). Importantly, very similar clusters are present in the human TUTases ZCCHC6 and ZCCHC11 and proteins from other species, for which PUP activity has been confirmed (Ren et al. 2012; Zhao et al. 2012; Sement et al. 2013; Chang et al. 2014; Lim et al. 2014; Tu et al. 2015; Zuber et al. 2016), although A. thaliana HESO-1 TUTase possesses valine instead of otherwise conserved histidine (Table 1). It would be interesting to check whether differences between these NRM clusters present in A. thaliana HESO-1 and URT1 TUTases (Table 1) contribute somehow to variable preference of these two enzymes toward small RNA substrates terminating with adenosines or uridines, as has been demonstrated recently (Tu et al. 2015).
TABLE 1.
Comparison of cluster sequences present in NRMs of noncanonical NTases with distinct NTP specificities

On the contrary, S. cerevisiae Trf4 and Trf5 proteins, displaying PAP activity (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; Houseley and Tollervey 2006; Hamill et al. 2010), contain DxxDxxN cluster at the equivalent position and the same is true for S. pombe nuclear noncanonical PAP, Cid14 (Table 1; Win et al. 2006). Importantly, asparagine seems to be invariantly present in the last position of the NRM cluster in all noncanonical NTases, for which PAP activity has been experimentally demonstrated, including recently studied human GLD-2 (TUT2) which contains ExxDxxN cluster (Table 1; Chung et al. 2016). In concordance, the Cid1 H336N mutation (creating DxxExxN cluster) changed Cid1 specificity from PUP into PAP (Lunde et al. 2012). The identity of the remaining residues within the cluster seems to be less critical—the first one is always acidic (D or E), whereas the middle one is most often aspartate, but other amino acids also occur at this position, as exemplified by well-characterized C. elegans PAPs: Gld-2 and Gld-4 and Drosophila melanogaster Trf4 homolog, DmTrf4-1 (Table 1; Kwak et al. 2004; Nakamura et al. 2008; Schmid et al. 2009).
These examples indicate that the presence of H or N at the last position of the cluster largely determines whether the noncanonical NTase acts as PUP or PAP, respectively. The only significant exception from this rule regards mitochondrial PAPs (mtPAPs) in animals (Tomecki et al. 2004), which contain a completely different, but highly conserved, NxxExxL cluster (Table 1). Notably, substitution of the asparagine with aspartate results in the compromised PAP activity, both in vitro and in vivo, which leads to development of progressive neurodegenerative disorder—spastic ataxia 4 (Wilson et al. 2014; Lapkouski and Hallberg 2015). Structural studies revealed that this asparagine may play a role in a correct spatial positioning of the mRNA 3′-end and the bound ATP substrate, rather than in discrimination of mtPAP specificity toward NTP (Lapkouski and Hallberg 2015). Therefore, the molecular mechanisms ensuring the same NTP preference of nuclear/cytoplasmic and mitochondrial noncanonical PAPs may be distinct, and further structural insights are clearly required to understand these differences. It would be particularly interesting to determine whether there is any link between the divergence of NRM clusters present in nuclear/cytoplasmic and mitochondrial PAPs and the fact that the former act as homodimers, which is a unique feature among noncanonical NTases, obligatory for enzymatic activity of mtPAPs (Bai et al. 2011; Lapkouski and Hallberg 2015).
In this paper, we investigated the basis of unusual properties of the fungal CutA enzymes. Unlike other authors, we refrained from drawing conclusions based on gel analysis of the in vitro reaction products generated by recombinant proteins in the presence of individual NTPs. We reasoned that such, nota bene commonly used, approach provides information about differences in enzyme processivity toward particular NTPs (see Fig. 13 herein as an example), rather than illustrates genuine NTase specificity. Instead, we took advantage of the high-throughput methodology and subjected 3′-RACE products generated for tails synthesized by recombinant proteins in the presence of AGCU mixture to next-generation sequencing and data analysis. This strategy allowed us to quantify percentage of individual nucleotides in tails generated by analyzed NTases and to analyze distribution of nucleotides at every position of the synthesized extensions. Owing to this approach, we were able to confirm that S. pombe Cid1 is extremely specific toward uridines, whereas CutA enzymes from two different fungal species display less strict specificity, but tend to predominantly incorporate cytidines. Importantly, the most striking feature of these enzymes is a frequent incorporation of two cytidine residues as the last added nucleotides, evident even at a physiologically relevant NTP ratio. Moreover, biochemical experiments with CTP as the only nucleotide present in the reaction mixture revealed that if RNA is in excess over protein, mimicking the in vivo situation, CutA is not able to efficiently extend the RNA substrate with more than two cytidines.
As expected, a 10-fold increase in the ATP concentration over CTP resulted in the appearance of longer adenosine stretches in the tails synthesized by the wild-type CutA, but the overall incorporation of cytidines did not decrease dramatically, indicating that the enzyme retains a high degree of specificity toward CTP, even if the latter is underrepresented. The pattern of tail composition, wherein strings of adenosines are interrupted by one or two cytidines and frequently terminate with two C residues, is due to several factors, as demonstrated herein, namely: character of the nucleotide recognition motif (NRM) and variable affinity of the enzyme toward different NTP substrates, much higher processivity of CutA with ATP than with CTP or UTP, as well as a tendency to poorly bind RNA molecules with two cytidines at their 3′-ends. It should be emphasized that these findings may be relevant to CutA activity in vivo. In A. nidulans, the enzyme was implicated in addition of C/U-rich extensions to polyadenylated mRNAs. However, it was not directly demonstrated that these unique tails are added to nascent poly(A) tails. It could be envisioned that after normal mRNA deadenylation, CutA synthesizes mixed extensions, containing several processively added adenosines, followed by distributively added cytidines and uridines. Interestingly, in addition to CutA's previously described CU-tagging activity (Morozov et al. 2010), it has recently been found that CutA is able to adenylate 3′-ends of some mRNAs in vivo (N Dube, IY Morozov, and MX Caddick, pers. comm.) Thus, although our work was performed on recombinant proteins, concordance with in vivo observations suggests that the nucleotide specificity of CutA was preserved.
Comparison of the 3D model of CutA with solved crystal structures of Cid1 and Trf4, combined with site-directed mutagenesis led to the conclusion that CutA NTases have an unusual nucleotide-binding site, in which the unique ExxNxxR cluster, at position equivalent to NRM clusters present in the noncanonical PAPs and PUPs, is mainly responsible for the observed preference of CutA toward cytidine. To support this conclusion, we demonstrated that introduction of ExxNxxR cluster into Cid1 NRM significantly increased cytidine incorporation at the expense of uridine inclusion into the 3′-terminal tails. Importantly, replacing CutA ExxNxxR with Cid1-like DxxExxH resulted in generation of the tails containing less cytidines and more uridines, but such a triple mutation did not lead to the complete conversion of CutA into PUP enzyme. Similarly, we were not able to convert CutA into highly efficient PAP, probably due to a unique geometry and environment of the nucleotide binding site, which makes it impossible to achieve specificity toward a single nucleotide by only a few mutations. Further structural studies—particularly analysis of CutA cocrystals with different NTP substrates, supported by mutational and biochemical studies are clearly needed to fully understand the complete network of protein-nucleotide interactions responsible for the specificity of NTP selection for these noncanonical NTases.
We believe that based on the NRM cluster sequence one may now try to predict the specificity of the putative noncanonical NTases in the majority of the cases. For instance, there are three yet-uncharacterized enzymes from C. elegans, which all contain DxxD/ExxH NRM cluster and several other proteins from different species with D/ExxXxxN cluster, which are most likely PUPs and PAPs, respectively (Table 1). Most of the NTases from the latter group also possess arginine located four amino acids downstream from the NRM cluster, which is known to stabilize interaction with ATP, provided that the NRM cluster terminates with asparagine (Table 1).
Furthermore, inspection of NRM clusters present in enzymes from several model species suggests that there are enzymes which may display an NTP preference similar to CutA, namely: Cid11 and Cid13 from S. pombe, Mkg-p from D. melanogaster and PUP-3 from C. elegans (Table 1). Neither of these proteins has been thoroughly characterized in terms of NTP substrate specificity, however it was shown that Cid13 utilizes CTP more readily than UTP or GTP (Saitoh et al. 2002). Therefore, it would be worth checking if the enzymes mentioned above are able to synthesize 3′-terminal tails containing mainly cytidines in vitro using the 3′-RACE-seq approach described herein, as well as to perform in parallel global TAIL-seq experiments to analyze whether RNA molecules with C/U-rich tails exist in vivo in fission yeast, fruitfly and worm. Strikingly, it seems that mammalian genomes do not encode a noncanonical PAP with NRM cluster that would suggest such specificity toward CTP, which is in agreement with TAIL-seq analysis of human mRNA 3′-ends, in which tails containing cytidine were very rarely detected, when compared to uridine-rich extensions (Chang et al. 2014). All of that suggests that various species may utilize different enzymes and mechanisms to modify mRNA fate by addition of nontemplated nucleotides to the 3′ ends of RNA molecules, however further research is needed to uncover physiological functions of various noncanonical PAPs with distinct nucleotide specificities.
MATERIALS AND METHODS
Bacterial strains
The following Escherichia coli strains were used in this study:
MH1 (E. coli araD lacX74 galU hsdR hsdM rpsL),
BL21-CodonPlus-RIL [Stratagene; E. coli B F– ompT hsdS(rB− mB−) dcm+ Tetr gal endA Hte (argU ileY leuW Camr)]
Oligonucleotides, plasmids, and cloning
Plasmids and oligonucleotides used in this study are listed in Supplemental Tables S2 and S3, respectively. All restriction enzymes and Phusion DNA polymerase for amplification of inserts were from Thermo Fisher Scientific. DNA purification kits (DNA Plasmid Mini and Gel-Out) were from A&A Biotechnology.
All plasmids for heterologous expression of fungal proteins in E. coli were generated by a sequence and ligation independent cloning (SLIC) approach (Li and Elledge 2007).
Plasmids pUC57-AnCutA and pUC57-TtCutA contained recoded sequences (see Supplemental Fig. S1) of CutA enzymes from A. nidulans and T. terrestris, which were synthesized by Genscript with the aim of codon optimization for heterologous expression in E. coli, and precloned into pUC57 vector.
Plasmids pCC1 and pCC4 were generated by inserting open-reading frames corresponding to the full-length AnCutA and TtCutA, respectively, amplified using respective primer pairs (278F–278R and 280F–280R) and templates (pUC57-AnCutA and pUC57-TtCutA), into NcoI–XhoI sites of pET28 C-6xHisTag vector.
Plasmid pCC2 was generated by site-directed mutagenesis with oligonucleotide pair CUCUmutF–CUCUmutR, using pCC1 as a template.
Plasmid pCC3 was constructed by inserting AnCutA ORF fragment corresponding to amino acids 84–487, amplified with primer pair 278SumoF–278Sumo2 and pCC1 as a template, into BamHI–XhoI sites of pET28 N-6xHis-SUMOTag vector. Plasmid pCC5, containing TtCutA ORF fragment corresponding to amino acids 216–617, was generated similarly, but using primer pair 280SumoF–280Sumo2 and pCC4 as a template in the amplification step.
Plasmids pCC6 and pCC7, containing TtCutA ORF fragments corresponding to amino acids 216–557 or 127–827, respectively, were constructed by inserting amplicons obtained with the respective primer pairs (12.280 S_F–12.280 sI_R or 280sIIIF–280sIIIR), into NcoI–XhoI sites of pET28 C-6xHisTag vector.
Plasmids pCC8, pCC9, pCC12, pCC14, and pCC15 were generated by site-directed mutagenesis with oligonucleotide pairs 280PAP1F–280PAP1R, 280PAP2F–280PAP2R, 280PAP5F–280PAP5R, 280PUP1F–280PUP1R, and 280PUP2F–280PUP2R, respectively, using pCC7 as a template. Plasmids pCC10 and pCC16 were constructed by site-directed mutagenesis with oligonucleotide pairs 280PAP3F–280PAP3R or 280PUP3F–280PUP3R, using pCC8 or pCC15 as templates, respectively. Plasmid pCC11 was generated by site-directed mutagenesis with oligonucleotide pair 280PAP4F–280PAP4R, using pCC8 as a template. Plasmid pCC13 was constructed by site-directed mutagenesis with oligonucleotide pair 280PAP6F–280PAP6R, using pCC15 as a template.
Plasmid pCC17 was constructed by inserting Cid1 ORF, amplified with primer pair Cid1SumoF–Cid1SumoR, using cDNA obtained following oligo(dT)-primed reverse transcription on total S. pombe RNA as a template, into BamHI–XhoI sites of pET28 N-6xHis-SUMOTag vector. Plasmid pCC18 was generated by site-directed mutagenesis with oligonucleotide pair Cid1PCPF–Cid1PCPR, using pCC17 as a template.
The presence of introduced mutations was verified by digestion with relevant restriction enzymes and correctness of sequences was checked by sequencing the inserts with appropriate primers, listed in Supplemental Table S3.
Heterologous expression of proteins in E. coli
For expression of different variants of fungal nucleotidyltransferases, Escherichia coli BL21-CodonPlus-RIL strain was transformed with appropriate plasmids (pCC1–pCC18). Transformants were grown at 18°C for 48 h in 1 L or 50 mL (for large-scale and small-scale purification, respectively) of Auto Induction Media Super Broth Base Including Trace Elements (Formedium), supplemented with 2% glycerol, kanamycin (50 µg/mL), and chloramphenicol (34 µg/mL), following inoculation from a preculture, grown in standard Luria-Broth (LB) medium containing both antibiotics.
Small-scale protein purification
A bacterial pellet from 50 mL of liquid culture was resuspended in 5 mL of the lysis buffer (20 mM Na-phosphate, pH = 4.5 or 8.0/20 mM Tris–HCl, pH = 8.0; 300 mM NaCl; 10 mM imidazole; 1 mM PMSF; 10 mM β-mercaptoethanol; 50 µg/mL lysozyme) and subjected to 60 cycles of sonication (30 sec of full power sonication : 30 sec pause) in a Bioruptor XL device (Diagenode). Homogenate was centrifuged at 10,000g for 45 min at 4°C. The supernatant was then loaded onto a Protino 96 Ni-IDA plate (Macherey-Nagel), pre-equilibrated with the lysis buffer. Following protein binding, the resin was washed sequentially with 2 mL of low-salt (LS) buffer (20 mM Na-phosphate/Tris–HCl, pH = 8.0; 150–300 mM NaCl; 10 mM imidazole), 1 mL of high-salt (HS) buffer (20 mM Na-phosphate/Tris–HCl, pH = 8.0; 1 mM NaCl; 10 mM imidazole) and 2 mL of LS buffer. Bound proteins were recovered by three consecutive rounds of elution with 0.25 mL of buffer E (20 mM Na-phosphate/Tris–HCl, pH = 8.0; 150–300 mM NaCl; 300 mM imidazole). Purified proteins were analyzed by standard SDS-PAGE.
Large-scale protein purification
Following centrifugation of 1 L of the liquid culture at 4500 rpm in a Sorvall H6000A/HBB6 swinging bucket rotor for 15 min at 4°C, a bacterial pellet was resuspended in 70 mL of the lysis buffer, incubated with lysozyme (50 µg/mL; Roth) for 20 min in a cold room, and then broken in the EmulsiFlex-C3 High Pressure homogenizer at 1500 Bar. Homogenate was centrifuged in a Sorvall WX ULTRA SERIES ultracentrifuge (F37L rotor) at 32,000 rpm for 45 min at 4°C. The supernatant (extract) was then used for protein purification with the ÄKTA Xpress system (GE Healthcare).
Depending on the proteins’ stability and contamination level, several various methods were used in purification procedures. The basic step of purification was nickel affinity chromatography on the 5 mL column compatible with ÄKTA, filled with Ni-NTA Superflow resin (Qiagen). The column was equilibrated with 25 mL of LS buffer prior to the extract loading. After protein binding, the resin was washed sequentially with 35–60 mL of LS buffer, 25 mL of HS buffer (to reduce contaminations), and 20 mL of LS buffer. Proteins with a C-terminal HisTag were simply recovered by elution with 25 mL of buffer E. For proteins fused with an N-terminal HisTag-SUMO, purification involving SUMO protease cleavage (see Supplemental Table S4) included additional on-column digestion with 100 µg of the home-made SUMO protease resuspended in 5 mL of buffer E (8 h), followed by buffer exchange to LS on the desalting column and final separation of the desired protein from the cut out HisTag-SUMO epitope and SUMO protease on the Ni-NTA column.
Furthermore, size-exclusion chromatography on a Hiload 16/60 Superdex S200 column (GE Healthcare), was carried out in the majority of cases by applying 5 mL of eluate from the affinity chromatography step, with the use of 1.2 column volumes of gel-filtration (GF) buffer (20 mM Na-phosphate/Tris–HCl, pH = 8.0; 150 mM NaCl), to increase protein purity. Purified proteins were analyzed by standard SDS-PAGE. See Supplemental Table S4 for more details on purification protocols applied to particular CutA variants.
Protein concentration
Following purification, recombinant proteins were concentrated with the use of Vivaspin (500, 2, 4, or 15, depending on the initial volume of protein solution) ultrafiltration devices (Sartorius Stedim), according to the manufacturer's instructions.
Limited proteolysis
After small-scale purification of the full-length TtCutA, 15 µg of the protein was subjected to digestion with various trypsin concentrations (ranging from 0.0005 to 0.125 µg/µL) in 20 µL of the buffer containing 10 mM Tris–HCl, pH = 7.4 and 100 mM NaCl for 30 min at room temperature. Of note, 1 mM PMSF was then added to terminate reactions and standard SDS-PAGE was used for products’ analysis.
Edman degradation analysis
Three hundred micrograms of TtCutA was digested as above in a total volume of 0.8 mL with trypsin (0.001 µg/mL) for Edman degradation analysis. Reaction products were separated by gel filtration on a Hiload 16/60 Superdex 75 column (GE Healthcare) in a buffer containing 10 mM Na-phosphate, pH = 8.0 and 100 mM NaCl. The fraction containing properly sized protein (about 40 kDa, as verified by SDS-PAGE) was concentrated approximately 10-fold. Fifteen microliters of the concentrated protein preparation was then mixed with 5 µL of denaturation buffer (150 mM Tris–HCl, pH = 6.8; 12% SDS; 30% glycerol; 0.05% Coomassie blue G-250; 6% β-mercaptoethanol) and separated in Tricine–SDS-PAGE with the use of anode buffer (0.2 M Tris–HCl, pH = 8.9) and cathode buffer (0.1 M Tris; 0.1 M tricine; 0.1% SDS). The protein was then subjected to wet electrotransfer onto methanol preactivated PVDF membrane (Thermo Fisher Scientific) in CAPS buffer (0.22% CAPS, pH = 11.0; 10% methanol). Following transfer, the membrane was stained with 0.1% Coomassie Brillant Blue R-250 in 40% methanol; 1% acetic acid, then washed four times in 50% methanol and air-dried. The PVDF membrane fragment containing a protein of interest was cut out and an N-terminal sequence was analyzed by chemical Edman degradation with a fully automated Procise 491 Sequencer (Applied Biosystems) at the Selvita company.
Protein secondary structure analysis
CFSSP (Chou&Fasman Secondary Structure Prediction Server, available at https://sourceforge.net/projects/cfpred/files/latest/download) was used to predict AnCutA and TtCutA secondary structures.
3D protein model
Homologs of TtCutA and AnCutA proteins were identified with BLAST (Altschul et al. 1997) searches performed against the NCBI nonredundant protein sequence database using sequences of TtCutA and AnCutA as queries. Multiple sequence alignment was derived using PCMA program (Pei et al. 2003) followed by some manual adjustments. Secondary structures were predicted with PSIPRED (McGuffin et al. 2000). A 3D model of TtCutA protein was constructed with MODELLER (Fiser and Sali 2003) using S. pombe Cid1 poly(U) polymerase (pdb|4fh5) (Lunde et al. 2012) as a template (Ginalski and Rychlewski 2003). The structures were visualized with Pymol (pymol.org).
In vitro biochemical assays
Nucleotidyltransferase activity of different AnCutA, TtCutA, and Cid1 protein variants was assessed by in vitro assays. Reactions were performed in the volume of 20 µL, containing 10 mM Tris–HCl, pH = 8.0; 50 mM NaCl; 1 mM MnCl2; 1 mM DTT; and either individual NTP (ATP/GTP/CTP/UTP; 1 mM) or mixtures of two (0.5 mM each) or four (0.25 mM each, or 0.77 mM ATP:0.077 mM GTP:0.077 CTP:0.077 UTP, when a physiologically relevant NTP ration was desirable) different NTPs. In most experiments, each reaction was programmed with 5 µL of the purified protein (0.5 µM), and started by adding 4 µL of 1 µM 5′-FAM-labeled RNA substrate solution (see Supplemental Table S3 for sequences). For estimation of enzyme affinity towards various nucleotides and analysis of processivity, reactions were performed similarly, but using an excess of RNA substrate (1 µM) over the enzyme (0.025 µM), and individual NTPs (ATP, CTP, or UTP) at final concentrations ranging from 0.1 to 300 µM. Reactions were carried out at 37°C and terminated at various time-points after the start by collecting 4 µL of reaction mixture and adding it to 5 µL of formamide loading dye (90% formamide; 20 mM EDTA; 0.03% bromophenol blue; 0.03% xylene cyanol in 1× Tris–borate–EDTA [TBE]), followed by snap-freezing in liquid nitrogen. After thermal denaturation at 90°C for 5 min, reaction products were resolved either in denaturing 20% polyacrylamide/8 M urea/1× TBE gels, or—if single-nucleotide resolution was required—in 10% sequencing gels. A Typhoon FLA 9000 laser scanner (GE Healthcare) was used to visualize fluorescently labeled RNA molecules.
3′-RACE-seq
Nucleotide composition of the 3′-extensions added by different variants of fungal nucleotidyltransferases in the in vitro assays was analyzed by 3′-RACE and sequencing. Sequences of oligonucleotides used in the 3′-RACE are listed in the Supplemental Table S3.
RNA purification
Following RNA visualization, a gel fragment containing reaction products was cut out, frozen in liquid nitrogen and crushed with a pipette tip. RNA was then eluted from acrylamide with 350 µL of the gel elution buffer (100 mM Tris–HCl, pH = 8.0; 150 mM NaCl; 12.5 mM EDTA; 1% SDS) in the presence of 800 µL of phenol:chloroform (1:1, v:v) on a rotating device at room temperature for 16 h. Following centrifugation at 13,500g for 15 min at room temperature, the aqueous layer was transferred to a new tube and RNA was precipitated with isopropanol in the presence of 1 µL of glycogen (20 mg/mL) at −20°C for 16 h. RNA was pelleted by centrifugation at 21,000g for 30 min at 4°C and washed twice with 80% ethanol. Eventually, the RNA pellet was suspended in 11 µL of RNase-free water.
Adapter ligation
Of note, 10.7 µL of purified RNA was mixed with 2 µL of T4 RNA Ligase Reaction buffer (New England Biolabs), 1 µL of 30 µM RA3 adapter, 4.8 µL of 50% PEG 8000 (New England Biolabs) and 0.5 µL of RiboLock RNase Inhibitor (40 U/µL; Thermo Fisher Scientific). After 2 min of incubation at 70°C, the tube was placed on ice, and 1 µL of T4 RNA Ligase 2, Truncated (200 U/µL; New England Biolabs) was added. The reaction was carried out for 1 h at 25°C.
Reverse transcription
Following ligation of the 3′-adapter, 1.5 µL of 20 µM reverse transcription (RTP) primer was annealed by heating the mixture at 75°C for 5 min, followed by incubation at 37°C for 30 min and at 25°C for 15 min. Ten microliters aliquot of the mixture was transferred to a new tube, mixed with 4 µL of the 5× First Strand Buffer (Invitrogen), 1 µL of 10 mM dNTP Mix, 0.5 µL of RiboLock RNase Inhibitor, 2 µL of 100 mM DTT, 1.5 µL of RNase-free H2O and heated at 42°C for 2 min. Afterwards, 1 µL of the SuperScript III Reverse Transcriptase (Invitrogen) was added, and RT-PCR was carried out at 42°C for 1 h. Enzyme inactivation was performed by heating the reaction mixture at 70°C for 30 min.
Amplification
Two-step amplification was performed for high-throughput RNA-seq of the 3′-RACE products. The first round PCR reaction mixture contained 20 µL of the first-strand cDNA, 0.2 mM dNTP Mix, 20 µM primer pair RA5_ss22-RPI_X (with Illumina index for RNA-seq), 5× Phusion HF Buffer, Phusion DNA polymerase, and sterile water to 40 µL. The following PCR profile was utilized: initial denaturation at 98°C for 30 sec, 25 cycles of amplification (with denaturation at 98°C, annealing at 55°C, and elongation at 72°C carried out for 10, 10, and 30 sec, respectively) and final elongation at 72°C for 5 min. The second round PCR reaction mixture comprised 1 µL of the first-round PCR products, 0.2 mM dNTP Mix, 20 µM primer pair RP1-RPI_X, 5× Phusion HF Buffer, Phusion DNA polymerase, and sterile water to 50 µL. The PCR profile described above was essentially the same as in the first round, but with only five cycles of amplification. PCR products were analyzed by electrophoresis in 2% agarose gels.
Purification of the 3′-RACE-seq libraries
PCR products obtained following amplification were purified with the use of Agencourt AMPure XP magnetic beads (Beckman Coulter). Briefly, 48 µL of PCR products were mixed with 48 µL of the beads, incubated for 15 min at room temperature, and then placed on a magnetic stand. After 5 min, supernatant was discarded, and the beads were washed twice with 100 µL of 80% ethanol and air-dried for 15 min. Elution was done by suspending the beads in 20 µL of sterile water and 5 min of incubation at room temperature. Magnetic stand was used to separate eluted DNA from the beads.
High-throughput sequencing of the 3′-RACE products (3′-RACE-seq)
Concentration of the 3′-RACE-seq libraries was measured with the KAPA Library Quantification Universal Kit (Kapa Biosystems). Sequencing was performed using an Illumina NextSeq 500 System and NextSeq 500 v2 High Output Kit (2 × 75 cycles), applying standard procedures of a pair-end run.
Analysis of the 3′-RACE-seq results
Initially, the strands encompassing sequences of RNA substrates in the sense orientation were subjected to filtering, consisting in a selection of sequences containing 26 nt of the respective RNA substrate (ss22-A4 or ss22-U4) at the 5′-end, with no more than two erroneous bases, and an entirely correct sequence of the RA3 oligonucleotide (21 nt) at the 3′-end. Detailed analyses for all filtered sequences were carried out with in-house scripts using Python, NumPy, SciPy, and Matplotlib.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was mainly supported by a grant from The National Centre for Research and Development (NCBR LIDER/35/46/L-3/11/NCBR/2012 to R.T.) and co-supported by the European Research Council (ERC Starting Grant 309419-PAP&PUPs to A.D.), the Polish National Science Centre (2011/02/A/NZ2/00014, 2014/15/B/NZ1/03357 to K.G.), and the Foundation for Polish Science (TEAM/2010-6, TEAM/2016-2 to K.G.). Experiments were carried out with the use of CePT infrastructure that was financed by the European Union via the European Regional Development Fund (Innovative economy 2007–13, Agreement POIG.02.02.00-14-024/08-00). We acknowledge Dorota Adamska and Paweł Krawczyk for assistance with 3′-RACE-seq and data analysis. We thank Karolina Drazkowska for reading the manuscript.
Author contributions: K. Kobyłecki prepared plasmid constructs, expressed and purified all proteins, performed the majority of biochemical experiments, including generation of the 3′-RACE-seq libraries, and carried out initial analysis of high-throughput sequencing data under the supervision of R.T. and A.D. K. Kuchta generated the 3D TtCutA model, performed structural analyses, and thoroughly analyzed 3′-RACE-seq results under the supervision of K.G., with contributions from R.T. A.D. participated in the design of the experiments. R.T. conceived and directed the studies with the help of K.G., performed biochemical experiments at the stage of revision, and wrote the manuscript, with contributions from K.G., K. Kuchta, and A.D.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.061010.117.
REFERENCES
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev R. 2005. RNA uridylyltransferases. Cell Mol Life Sci 62: 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. 1999. DNA polymerase β-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res 27: 1609–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y, Srivastava SK, Chang JH, Manley JL, Tong L. 2011. Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol Cell 41: 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo PB, Bohm A. 2007. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure 15: 1117–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HM, Triboulet R, Thornton JE, Gregory RI. 2013. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature 497: 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Mukhopadhyay J, Tharun S. 2007. The decapping activator Lsm1p-7p-Pat1p complex has the intrinsic ability to distinguish between oligoadenylated and polyadenylated RNAs. RNA 13: 998–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung CZ, Jo DH, Heinemann IU. 2016. Nucleotide specificity of the human terminal nucleotidyltransferase Gld2 (TUT2). RNA 22: 1239–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Walleshauser J, Joshua-Tor L. 2014. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature 514: 252–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiser A, Sali A. 2003. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 374: 461–491. [DOI] [PubMed] [Google Scholar]
- Ginalski K, Rychlewski L. 2003. Protein structure prediction of CASP5 comparative modeling and fold recognition targets using consensus alignment approach and 3D assessment. Proteins 53 (Suppl 6: 410–417. [DOI] [PubMed] [Google Scholar]
- Hagan JP, Piskounova E, Gregory RI. 2009. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol 16: 1021–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamill S, Wolin SL, Reinisch KM. 2010. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci 107: 15045–15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN. 2009. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell 138: 696–708. [DOI] [PubMed] [Google Scholar]
- Houseley J, Tollervey D. 2006. Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Rep 7: 205–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Ha M, Loeff L, Chang H, Simanshu DK, Li S, Fareh M, Patel DJ, Joo C, Kim VN. 2015. TUT7 controls the fate of precursor microRNAs by using three different uridylation mechanisms. EMBO J 34: 1801–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K, Knizewski L, Wyrwicz LS, Rychlewski L, Ginalski K. 2009. Comprehensive classification of nucleotidyltransferase fold proteins: identification of novel families and their representatives in human. Nucleic Acids Res 37: 7701–7714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta K, Muszewska A, Knizewski L, Steczkiewicz K, Wyrwicz LS, Pawlowski K, Rychlewski L, Ginalski K. 2016. FAM46 proteins are novel eukaryotic non-canonical poly(A) polymerases. Nucleic Acids Res 44: 3534–3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, Wilusz JE, Zheng Y, Beal PA, Joshua-Tor L. 2015. On-enzyme refolding permits small RNA and tRNA surveillance by the CCA-adding enzyme. Cell 160: 644–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wickens M. 2007. A family of poly(U) polymerases. RNA 13: 860–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak JE, Wang L, Ballantyne S, Kimble J, Wickens M. 2004. Mammalian GLD-2 homologs are poly(A) polymerases. Proc Natl Acad Sci 101: 4407–4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno A, Tomecki R, Dziembowski A. 2016a. Cytoplasmic RNA decay pathways - enzymes and mechanisms. Biochim Biophys Acta 1863: 3125–3147. [DOI] [PubMed] [Google Scholar]
- Labno A, Warkocki Z, Kulinski T, Krawczyk PS, Bijata K, Tomecki R, Dziembowski A. 2016b. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res 44: 10437–10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. [DOI] [PubMed] [Google Scholar]
- Lapkouski M, Hallberg BM. 2015. Structure of mitochondrial poly(A) RNA polymerase reveals the structural basis for dimerization, ATP selectivity and the SPAX4 disease phenotype. Nucleic Acids Res 43: 9065–9075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. 2007. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nat Methods 4: 251–256. [DOI] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Magler I, Meinhart A. 2012. Crystal structures of the Cid1 poly (U) polymerase reveal the mechanism for UTP selectivity. Nucleic Acids Res 40: 9815–9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W. 2007. RNA-specific ribonucleotidyl transferases. RNA 13: 1834–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16: 404–405. [DOI] [PubMed] [Google Scholar]
- Morozov IY, Jones MG, Razak AA, Rigden DJ, Caddick MX. 2010. CUCU modification of mRNA promotes decapping and transcript degradation in Aspergillus nidulans. Mol Cell Biol 30: 460–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Tello P, Gabus C, Thore S. 2012. Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure 20: 977–986. [DOI] [PubMed] [Google Scholar]
- Munoz-Tello P, Gabus C, Thore S. 2014. A critical switch in the enzymatic properties of the Cid1 protein deciphered from its product-bound crystal structure. Nucleic Acids Res 42: 3372–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Tello P, Rajappa L, Coquille S, Thore S. 2015. Polyuridylation in eukaryotes: a 3′-end modification regulating RNA life. Biomed Res Int 2015: 968127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura R, Takeuchi R, Takata K, Shimanouchi K, Abe Y, Kanai Y, Ruike T, Ihara A, Sakaguchi K. 2008. TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster. Mol Cell Biol 28: 6620–6631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Sadreyev R, Grishin NV. 2003. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics 19: 427–428. [DOI] [PubMed] [Google Scholar]
- Pirouz M, Du P, Munafo M, Gregory RI. 2016. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep 16: 1861–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajappa-Titu L, Suematsu T, Munoz-Tello P, Long M, Demir O, Cheng KJ, Stagno JR, Luecke H, Amaro RE, Aphasizheva I, et al. 2016. RNA editing TUTase 1: structural foundation of substrate recognition, complex interactions and drug targeting. Nucleic Acids Res 44: 10862–10878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimao-Pinto MM, Manzenreither RA, Burkard TR, Sledz P, Jinek M, Mechtler K, Ameres SL. 2016. Molecular basis for cytoplasmic RNA surveillance by uridylation-triggered decay in Drosophila. EMBO J 35: 2417–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren G, Chen X, Yu B. 2012. Uridylation of miRNAs by HEN1 SUPPRESSOR1 in Arabidopsis. Curr Biol 22: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2008. The Cid1 poly(U) polymerase. Biochim Biophys Acta 1779: 286–294. [DOI] [PubMed] [Google Scholar]
- Rissland OS, Norbury CJ. 2009. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol 16: 616–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Mikulasova A, Norbury CJ. 2007. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol Cell Biol 27: 3612–3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Chabes A, McDonald WH, Thelander L, Yates JR, Russell P. 2002. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell 109: 563–573. [DOI] [PubMed] [Google Scholar]
- Scheer H, Zuber H, De Almeida C, Gagliardi D. 2016. Uridylation earmarks mRNAs for degradation… and more. Trends Genet 32: 607–619. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Norbury CJ. 2010. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. Wiley Interdiscip Rev RNA 1: 142–151. [DOI] [PubMed] [Google Scholar]
- Schmid M, Kuchler B, Eckmann CR. 2009. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev 23: 824–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DD, Norbury CJ. 2013. RNA decay via 3′ uridylation. Biochim Biophys Acta 1829: 654–665. [DOI] [PubMed] [Google Scholar]
- Sement FM, Ferrier E, Zuber H, Merret R, Alioua M, Deragon JM, Bousquet-Antonelli C, Lange H, Gagliardi D. 2013. Uridylation prevents 3′ trimming of oligoadenylated mRNAs. Nucleic Acids Res 41: 7115–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song MG, Kiledjian M. 2007. 3′ Terminal oligo U-tract-mediated stimulation of decapping. RNA 13: 2356–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson AL, Norbury CJ. 2006. The Cid1 family of non-canonical poly(A) polymerases. Yeast 23: 991–1000. [DOI] [PubMed] [Google Scholar]
- Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. 2004. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res 32: 6001–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torralba S, Sojat J, Hartmann R. 2008. 2′-5′ oligoadenylate synthetase shares active site architecture with the archaeal CCA-adding enzyme. Cell Mol Life Sci 65: 2613–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu B, Liu L, Xu C, Zhai J, Li S, Lopez MA, Zhao Y, Yu Y, Ramachandran V, Ren G, et al. 2015. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet 11: e1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Hrossova D, Potesil D, Chalupnikova K, Hrazdilova K, Pachernik J, Cetkovska K, Uldrijan S, Zdrahal Z, Vanacova S. 2013. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA 19: 1632–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko D, Pasulka J, Feketova Z, Bednarik L, Zigackova D, Fortova A, Zavolan M, Vanacova S. 2016. TUT-DIS3L2 is a mammalian surveillance pathway for aberrant structured non-coding RNAs. EMBO J 35: 2179–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. 2005. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3: e189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Silva IJ, Apura P, Matos RG, Arraiano CM. 2015. Surprises in the 3′-end: ‘U’ can decide too! FEBS J 282: 3489–3499. [DOI] [PubMed] [Google Scholar]
- Wang L, Eckmann CR, Kadyk LC, Wickens M, Kimble J. 2002. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature 419: 312–316. [DOI] [PubMed] [Google Scholar]
- Wilson WC, Hornig-Do HT, Bruni F, Chang JH, Jourdain AA, Martinou JC, Falkenberg M, Spahr H, Larsson NG, Lewis RJ, et al. 2014. A human mitochondrial poly(A) polymerase mutation reveals the complexities of post-transcriptional mitochondrial gene expression. Hum Mol Genet 23: 6345–6355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win TZ, Draper S, Read RL, Pearce J, Norbury CJ, Wang SW. 2006. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol Cell Biol 26: 1710–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. [DOI] [PubMed] [Google Scholar]
- Yates LA, Fleurdepine S, Rissland OS, De Colibus L, Harlos K, Norbury CJ, Gilbert RJ. 2012. Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nat Struct Mol Biol 19: 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates LA, Durrant BP, Fleurdepine S, Harlos K, Norbury CJ, Gilbert RJ. 2015. Structural plasticity of Cid1 provides a basis for its distributive RNA terminal uridylyl transferase activity. Nucleic Acids Res 43: 2968–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue D, Maizels N, Weiner AM. 1996. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae. RNA 2: 895–908. [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Yu Y, Zhai J, Ramachandran V, Dinh TT, Meyers BC, Mo B, Chen X. 2012. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr Biol 22: 689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuber H, Scheer H, Ferrier E, Sement FM, Mercier P, Stupfler B, Gagliardi D. 2016. Uridylation and PABP cooperate to repair mRNA deadenylated ends in Arabidopsis. Cell Rep 14: 2707–2717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.