Abstract
Background
The interplay between host genetics, immunity, and microbiota is central to the pathogenesis of inflammatory bowel disease (IBD). Previous population-based studies suggested a link between antibiotic use and increased IBD risk, but the mechanisms are unknown. The purpose of this study was to determine the long-term effects of antibiotic administration on microbiota composition, innate immunity, and susceptibility to colitis, as well as the mechanism by which antibiotics alter host colitogenicity.
Methods
Wild-type mice were given broad-spectrum antibiotics or no antibiotics for two weeks, and subsequent immunophenotyping and 16S rRNA gene sequencing-based analysis of the fecal microbiome were performed six weeks later. In a separate experiment, control and antibiotic-treated mice were given seven days of DSS, six weeks after completing antibiotic treatment, and the severity of colitis scored histologically. Fecal transfer was performed from control or antibiotic-treated mice to recipient mice whose endogenous microbiota had been cleared with antibiotics, and the susceptibility of the recipients to DSS-induced colitis was analyzed. Naïve CD4+ T cells were transferred from control and antibiotic-treated mice to immunodeficient Rag-1-/- recipients and the severity of colitis compared.
Results
Antibiotics led to sustained dysbiosis and changes in T-cell subpopulations, including reductions in colonic lamina propria total T cells and CD4+ T cells. Antibiotics conferred protection against DSS colitis, and this effect was transferable by fecal transplant but not by naïve T cells.
Conclusions
Antibiotic exposure protects against colitis, and this effect is transferable with fecal microbiota from antibiotic-treated mice, supporting a protective effect of the microbial community.
Keywords: antibiotics, colitis, microbiome, immunity
Introduction
We live in symbiosis with our intestinal microbiota, which comprises a vast population of microbes that confer numerous physiologic benefits vital to our health. Maintenance of a normal microbiota is essential for the normal development and maintenance of fundamental processes, including gut organogenesis, intestinal immunity, barrier integrity, nutrient absorption, and prevention of pathogenic infections 1–4. Our relationship with commensal bacteria is thus essential to our health, and forces that alter that relationship contribute to disease, including inflammatory bowel disease (IBD), necrotizing enterocolitis, enteric infections, obesity, allergy, and asthma 5–7. The “hygiene hypothesis” is based on the fundamental fact that we evolved in a microbe-rich environment that has become gradually sanitized in modern, industrialized societies. The resulting decreased exposure to microbial stimuli, particularly early in life, whether due to “hygiene” or to antibiotic use, is believed to adversely affect the developing host immune system, leading to an increased incidence of immune-mediated diseases later in life 8. Establishment of a normal microbiome early in life is critical, since microbial exposure in infants has a lasting effect into adulthood 9,10. One of the principal determinants of microbiome composition is antibiotic use. Antibiotics for any indication lead to major disruptions in microbiome composition, with Clostridium difficile colitis serving as a common and significant example of the major consequences that can result.
Although the ill effects of antibiotics have generally been considered to be temporary, recent evidence suggests otherwise, with antibiotics producing sustained alterations in microbiome composition lasting at least months after completing a brief course 11. It is estimated that children in the U.S. receive an average of 10-20 courses of antibiotics before reaching 18 years of age 12. Nearly 50% of women receive antibiotics during pregnancy, which alters their microbiome and consequently changes the bacteria acquired by the newborn at birth 12. Several studies support an association between peripartum and early life exposure to antibiotics and changes in mucosal and systemic immunity 9, but how these changes alter long-term susceptibility to disease remains largely unknown. A prospective study in Denmark demonstrated a link between antibiotic use and development of Crohn's disease in childhood 13, an observation consistent with evidence that interactions between the gut flora and host immunity are involved in the pathogenesis of IBD 14. Another recent study analyzed a large IBD database and found that subjects with either Crohn's disease or ulcerative colitis were more likely to have received antibiotics 2-5 years prior to their diagnosis 15. While these epidemiologic studies support a connection between antibiotics and IBD, the underlying mechanisms are not known. Yet, previous work indicates important relationships between microbiomes and host immunity. Germ-free mice accumulate invariant natural killer T cells in the colonic lamina propria and exhibit an increased susceptibility to oxazolone-induced colitis 16. Antibiotic administration in mice also leads to elevated serum IgE and increased numbers of circulating basophils, resulting in an exaggerated inflammatory response in the lung 17. Commensal bacteria, acting via Toll-like receptor signaling, have been shown to protect against colitis 18, and several studies demonstrate that commensal metabolites may be protective as well 19–21.
While the intestinal microbiota may have a protective effect against mucosal inflammation, they are also essential for the development of colitis. Commensal bacteria play a critical role in the development and maintenance of intestinal inflammation 22, and genetically susceptible rodents do not develop chronic colitis in germ-free conditions 23. Additional studies indicate that commensal bacteria contribute to colitis pathogenesis, with Proteus species identified as one potential culprit 24,25. These seemingly contradictory observations emphasize that commensal bacteria can be both protective and pathogenic in the context of colitis 26, highlighting the need for studies on the effects of antibiotics on the pathogenesis of gut inflammation, especially given the increasingly prevalent use of antibiotics. The primary goal of this study was to determine the long-term effects of antibiotics on susceptibility to colitis. We found that antibiotic treatment led to sustained microbiota changes that were protective against experimentally-induced colitis, and that this protective effect was transferable by fecal transplantation. Sequence analysis of microbiota composition in control and antibiotic-treated mice identified potential candidate organisms that may contribute to this protective effect.
Materials and Methods
Antibiotic treatment
(subsequently referred to as the “observational study”) C57BL/6 mice (3-4 weeks of age) received a 14-day course of broad-spectrum antibiotics as described by 18, consisting of ampicillin (1g/l) vancomycin (0.5g/l), neomycin (1g/l) and metronidazole (1g/l) added to the drinking water. They were then maintained on antibiotic-free water for 6 weeks (Fig. S1). Immunophenotyping was performed at the end of Weeks 2 and 8, and stool collected at Week 8 for pyrosequencing. Colitis was induced starting at Week 8 by adding 3% dextran sulfate sodium (DSS; MP Biomedicals, Santa Ana, CA) to the drinking water for 7 days. Mice were sacrificed at the completion of DSS treatment.
Colitis scoring
The severity of colitis was determined histologically in a blinded fashion by examination of H&E-stained slides and scoring the extent of inflammatory infiltration and tissue damage using an established scale 27. A total of 3-4 representative fields (10x objective) in each sample were examined to derive the score. The total colon pathology score is the sum of the inflammation score (0: normal cell pattern, 1: scattered inflammatory cells in the lamina propria, 2: increased number of inflammatory cells in the lamina propria, 3: confluence of inflammatory cells extending into the submucosa, 4: transmural extension of the infiltrating inflammatory cells) and the tissue damage score (0: normal tissue pattern, 2: mild colonic crypt hyperplasia +/- focal invasion of epithelium, 3: obvious colonic crypt hyperplasia, invasion of epithelium and goblet cell depletion, 4: extensive mucosal damage and extension through deeper structures of the bowel wall). The final score represents the mean +/- standard deviation. Significance was calculated by the Mann-Whitney test using Prism (v6.0, GraphPad Software, Inc.). Colon length was also recorded separately as an additional indicator of inflammation.
Fecal transfer
(subsequently referred to as the “transfer experiment”. Fecal donor mice were either treated with the 4-antibiotic regimen for 14 days as above or were maintained without antibiotic treatment as controls. Six weeks after completing antibiotics, stool was collected for fecal transfer. Recipient mice (3-4 week-old male C57BL/6) received the 4-antibiotic regimen for 14 days and, 2 days later, fecal transfer was performed as previously described 28. Colonic and cecal feces were pooled from 5 antibiotic-treated donor mice or from 5 untreated control donor mice. A fecal slurry (200 microliter volume), made by homogenizing stool pellets in PBS, was delivered to the recipients by orogastric gavage. At 3 weeks after the fecal transfer, stool was collected from the recipient mice for pyrosequencing and they were then given 3% DSS for 7 days, after which mice were sacrificed and the colon analyzed histologically to determine the presence and severity of colitis, as described above.
Immunophenotyping
Single cells were harvested from spleen, mesenteric lymph nodes, and colonic lamina propria (LP). LP isolation was performed as previously described 29. Cells then underwent flow cytometric analysis for markers of T cells and T-cell subsets (TCR-b, CD4, CD8, Foxp3). Antibodies include TCR-b clone #H57-59, CD4-FITC clone #RM4.4, and CD8-PercP clone #53-6.7, and Foxp3-PE clone #150D, all purchased from Biolegend (San Diego, CA). Foxp3 staining was performed according to manufacturer's protocol.
T-Cell Transfer Colitis
This was performed as previously described 29. Naïve CD4+ T cells (4x105), sorted for CD4+CD25-CD45RBhi by flow cytometry, were isolated from the mesenteric and peripheral lymph nodes and spleens of mice treated with either PBS or antibiotics and transferred into RAG-1-deficient mice by intraperitoneal injection. Weights were followed weekly and colons were harvested for assessment at week 8. Histologic score was measured in a blinded fashion on a scale of 0-8: colonic wall thickening (0-3), lamina propria infiltration (0-3), and presence of crypt abscesses (0-2) as described 29.
16S ribosomal (16S rRNA) gene pyrosequencing
Tag-encoded FLX amplicon pyrosequencing was performed as described 30,31 using Gray28F 5′TTTGATCNTGGCTCAG and Gray519r 5′ GTNTTACNGCGGCKGCTG, with primers numbered in relation to the primary sequence of E. coli 16S rRNA 32. Tag-encoded FLX amplicon pyrosequencing extending from 28F utilized a Roche 454 FLX instrument with Titanium reagents, and Titanium procedures performed at the Research and Testing Laboratory (Lubbock, TX).
Processing of pyrosequencing data
Sequence data were processed using Research and Testing Laboratory's in-house pipeline, described at http://www.researchandtesting.com/docs/Data_Analysis_Methodology.pdf. Briefly, sequences were grouped using their barcodes and any sequence that contained a low quality barcode or that failed to be at least half the expected amplicon length (or 250 bp, whichever was shortest) was removed from the data pool. All sequencing reads then were denoised using an algorithm based on USEARCH 33 and then checked for chimeras using UCHIME 34. Finally, sequence data were separated into operational taxonomic units (OTUs) and annotated using USEARCH 33. GreenGenes v. 12.10 35 was used as the reference database for taxonomic assignment.
Diversity estimates
Two measures of alpha diversity (microbiota diversity within samples) were calculated from the final OTU table. The first, Chao1, provided an estimate of true species richness by incorporating information about the number of species observed as singletons and doubletons 36. The second metric, Shannon, provided an estimate of ecological diversity by considering both species evenness and richness 37. To quantify microbiota compositional distances among samples the UniFrac metric was employed 38. The UniFrac metric incorporates branch length and evolutionary relationship information from the bacterial phylogenetic tree when estimating distances among samples. The unweighted UniFrac metric is calculated from a bacterial presence/absence matrix, whereas the weighted UniFrac metric is calculated from a bacterial matrix of relative abundances. Using UniFrac distance matrices relationships among samples were visualized using Principal Coordinates Analysis (PCoA).
Statistical analyses of pyrosequencing data
Data were expressed as mean ± standard error (SEM). Statistical analysis was performed using SPSS (version 21 for Mac OS; SPSS, Inc., Chicago, IL) and R Software (R Development Core Team, 2011). The results were considered as statistically significant at p<0.05. Analysis of variance (ANOVA) was used to test for group differences in alpha diversity and an alpha value of 0.05 was employed as the significance threshold. Statistical testing for multivariate differences among groups was performed using a permutational multivariate ANOVA using the UniFrac distance matrices 39. Bacterial lineages classified at the generic and OTU levels were screened for statistically significant differences in abundances among groups. For bacterial genera arcsin-transformed relative abundances were analyzed using ANOVA. P-values from individual ANOVAs were adjusted to maintain a false discovery rate (FDR) of 5%. OTU count data were analyzed using a generalized linear model with a negative binomial distribution 40. All statistical analyses were conducted in R (R Development Core Team, 2011) using the vegan 39, ladbsv 41, and DESeq 40 packages.
Ethical Considerations
All animal studies were approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital.
Results
Antibiotics lead to sustained changes in microbiota composition
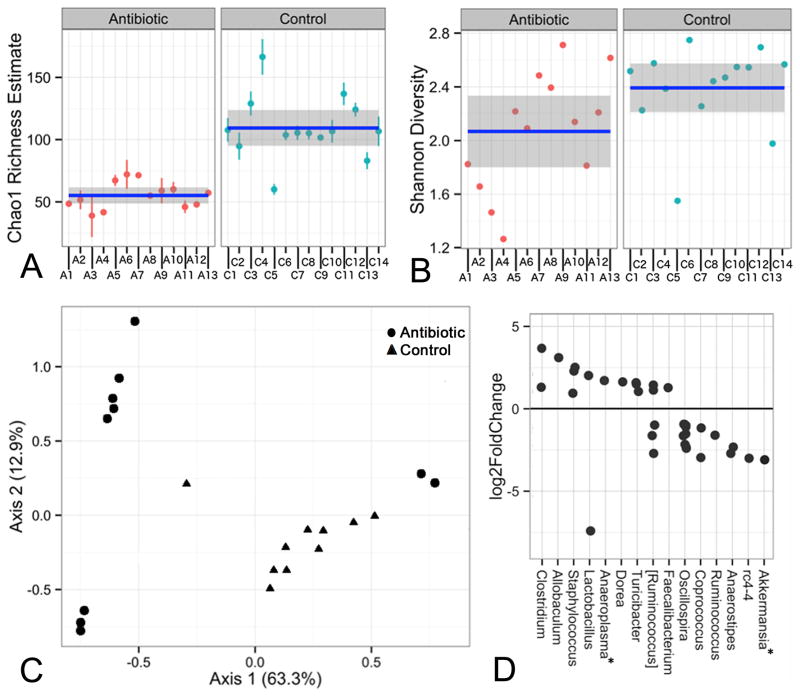
Fecal microbiota analysis was performed six weeks after completing antibiotic treatment. Antibiotic exposure was associated with a sustained reduction in microbial diversity, as demonstrated by statistically supported differences between antibiotic and control groups for both Chao1 (Fig. 1A) and Shannon (Fig. 1B) indices. There was corresponding support (p<0.05) for general treatment-based differences in overall microbiome composition, revealed by PCoA exploration of weighted UniFrac distances (Fig. 1C). Differences between treatment groups were also observed when unweighted distances were used (data not shown). The antibiotic-treated mice were housed in three cages (A1-4, A5-8, A9-13), and the control mice in four cages (C1-5, C6-9, C10-12, C13-14). An apparent cage effect was observed for this treatment, as demonstrated by additional segregation of these data points along both axes, resulting in two distinct groups of data points for antibiotic-treated samples. These segregations were more pronounced with the use of weighted vs unweighted distances (data not shown), and may be indicative of stochastic community assembly processes following antibiotic treatment (see Discussion).
Figure 1. Antibiotics lead to sustained changes in fecal microbiota composition.
Species richness was measured using the Chao (A) and Shannon (B) indices. The mean value and confidence interval for each group are illustrated. Standard errors around each Chao1 estimator value are also shown. The differences are statistically significant (p<0.05) in both A and B. A1-13 = antibiotic-treated; C1-14 = control (C) Plot of the PCoA results, based on weighted UniFrac for control (triangles) and antibiotic (circles) groups. The percent of overall variability accounted for by each axis is denoted in the axis labels. (D) OTUs that exhibited a significant difference in abundance between the antibiotic and control treatments are shown. Each dot represents an individual OTU. Positive values indicate an increase in abundance of a given OTU in the antibiotic group relative to control. OTUs are grouped by the genus (x-axis) to which they belong. All OTUs shown belong to phylum Firmicutes, with the exception of those (denoted with an asterisk) assigned to Anaeroplasma (phylum Tenericutes) and Akkermansia (phylum Verrucomicrobia). Square brackets around the genus name Ruminococcus indicate that the genus affiliation of the species-level taxa within this group is disputed, and in the future they may be assigned to a genus other than Ruminococcus.
The effects of antibiotic treatment on microbiome composition were supported by a classification-dependent approach based on the relative abundance of dominant genera. As shown in Fig. S2, samples from the control cages clustered together to the exclusion of those from the antibiotic-treated cages. A cage effect for the antibiotic-treated mice was again observed, as seen in the UniFrac data (Fig. 1C). Samples from one antibiotic-treated cage (A5-A8) clustered with the control samples, to the exclusion of the other antibiotic-treated cages, albeit with a short branch length which likely indicates an unstable relationship (Fig. S2).
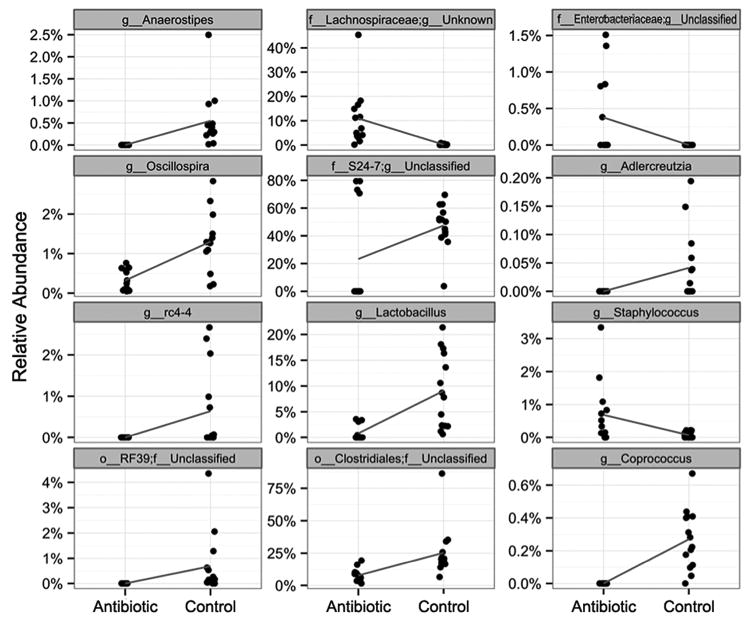
The taxonomic affiliations of sequences contributing to treatment-based differences in microbiome composition were further explored through visual representation of their abundance (Fig. S3) and statistical analysis (Fig. 2). Visually striking differences were observed in the abundance of four genus-level groups that represented the dominant populations, accounting for 20-80% of the sequences obtained (Fig. S3). These differences were also supported by the statistical analysis, as shown in Fig. 2 (center column). These four groups include the genus Lactobacillus, as well as three genus-level groups belonging to the order Clostridiales, the family-level group S24-7, and the family Lachnospiraceae. The first three taxa were less abundant in antibiotic-treated mice relative to controls, while the Lachnospiraceae genus-level group showed the opposite relationship. Statistical analysis also revealed significant treatment-based differences in eight other genera or genus-level groupings that formed minority components of the microbial populations identified (Fig. 2).
Figure 2. Sequencing analysis reveals genus-level taxa with significant sustained differences in relative abundance associated with antibiotic treatment (observational study).
Relative abundances from each sample are shown as single points, and the data are jittered along the x-axis to alleviate overplotting. The blue line connects the means in each group and indicates the change between groups.
The genus-level composition of the control samples was highly consistent (Fig. S3), and dominated by the genera Turicibacter and Lactobacillus, as well as genus-level groups within the order Clostridiales and the family-level group S24-7. One group of antibiotic-treated mice (samples A5-A8) resembled this control mouse profile, albeit with greater dominance of genus-level groups within S24.7 and the family Rikenellaceae, as well as fewer groups within the order Clostridiales. The other antibiotic-treated mice showed a more striking compositional difference relative to controls, with replacement of S24.7 by the genus Turicibacter, as well as genus-level groups within the families Clostridiaceae and Lachnospiraceae. One cage of antibiotic-treated mice (samples A9-A13) show proliferation of the genus Clostridium as well as genera within the family Ruminococcaceae (Fig. S3).
Both visual representation and statistical analysis of treatment-based differences in relative abundance at the species level (data not shown) closely resembled the analysis performed at the genus level (Figs. 2,S3). Within the genus-level groups, only one of the most dominant genera (Ruminococcus, Fig. S3) was further resolved to the species level (Ruminococcus gnavus, data not shown), but treatment-based differences for this species were not supported by statistical analysis.
Additional analysis of treatment-based effects was performed at the OTU level (Fig. 1D), revealing multiple significant differences, with most falling within a 5-fold level of change. While many of these OTUs could not be classified to a specific genus, either because that genus is unknown or unclassified, some were assigned to genera that displayed significant treatment-based differences, as described above (Figs. S2,S3). Of those genera or genus-level groups showing statistically supported differences in relative abundance (Fig. 2), the following also contained significant differences at the OTU level: Anaerostipes, Oscillospira, rc4-4, Lactobacillus, Staphylococcus, and Coprococcus. In all but one case, the treatment effect (enrichment or depletion in antibiotic-treated mice relative to controls) at the OTU level was the same as at the genus level. The exception was genus Lactobacillus, which at the genus level was depleted in antibiotic-treated mice whereas it contained one OTU that was enriched and another that was depleted. In addition, OTUs within the genera Clostridium, Allobaculum, Anaeroplasma, Dorea, Turicibacter, and Faecalibacterium were over-represented in antibiotic-treated mice relative to controls, whereas Ruminococcus, and Akkermansia OTUs were depleted, and different OTUs within [Ruminococcus] displayed either enrichment or depletion (Fig. 1D). Note that square brackets around the genus name indicate that the genus affiliation of the species-level taxa within this group is disputed, and in the future they may be assigned to a genus other than Ruminococcus.
Antibiotic treatment leads to early and sustained changes in immunity
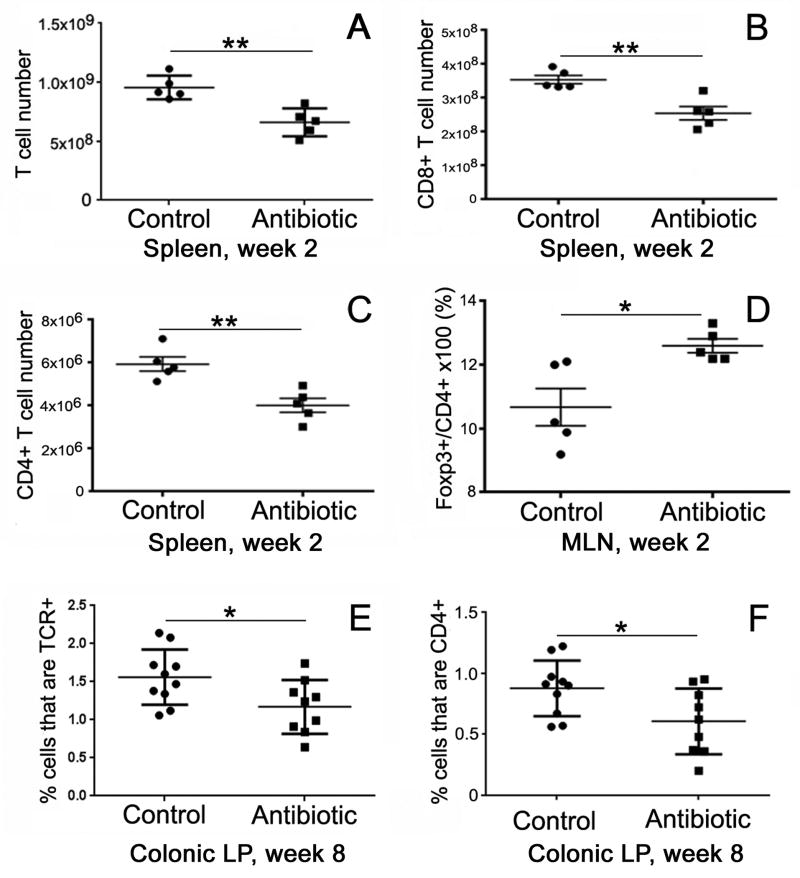
In order to test how antibiotics impact immunity, we analyzed various immune compartments following antibiotic treatment, including quantitative analysis of total cellularity as well as absolute counts of total T cells and T-cell subpopulations in the colonic lamina propria (LP), mesenteric lymph nodes (MLN), and spleen at Week 2 and Week 8, as shown in Fig. 3. With respect to spleen cellularity, a significant decrease in cell numbers was observed at Week 2, immediately after completion of the antibiotic treatment. This included decreases in total cell number as well as in absolute numbers of total T cells, CD8+ T cells, and CD4+ T cells (Fig. 3A-C). By Week 8, however, six weeks after completing antibiotics, cell numbers in each of these categories returned to normal and were not significantly different from untreated controls (data not shown).
Figure 3. Antibiotic treatment alters immune cell compartments.
Immediately following antibiotic treatment, total numbers of T cells (A), including both CD8+ (B) and CD4+ (C) T cells, are decreased in the spleen. These numbers returned to normal six weeks later (not shown). The proportion of Tregs (Foxp3+/CD4+ T cells) was significantly increased in the mesenteric lymph nodes at Week 2 following antibiotic treatment (D). By Week 8, the proportion of T cells (E) and CD4+ cells (F) were both significantly reduced in the colonic lamina propria. TCE, T cell receptor; *p<0.05, **p<0.01. This is representative of two experiments, n = 5 each group. LP, lamina propria; MLN, mesenteric lymph nodes
With respect to cellularity in the MLN and colonic LP, antibiotics did not produce any significant change in cell numbers (total cells, total T cells, CD8+ T cells, or CD4+ T cells) at Week 2 (data not shown). Interestingly, however, the proportion of CD4+ cells that are Foxp3+, a marker of Tregs, was increased in the MLN at Week 2 in the antibiotic group (Fig. 3D), and this increase normalized by week 8 (data not shown). In contrast, antibiotic treatment did not affect Treg numbers in the spleen or LP at Week 2 or Week 8. In the colonic LP, the proportions of total T cells and CD4+ T cells were both significantly reduced in antibiotic-treated mice at Week 8 (Fig. 3E,F).
Antibiotic treatment confers a sustained protection against DSS-induced colitis
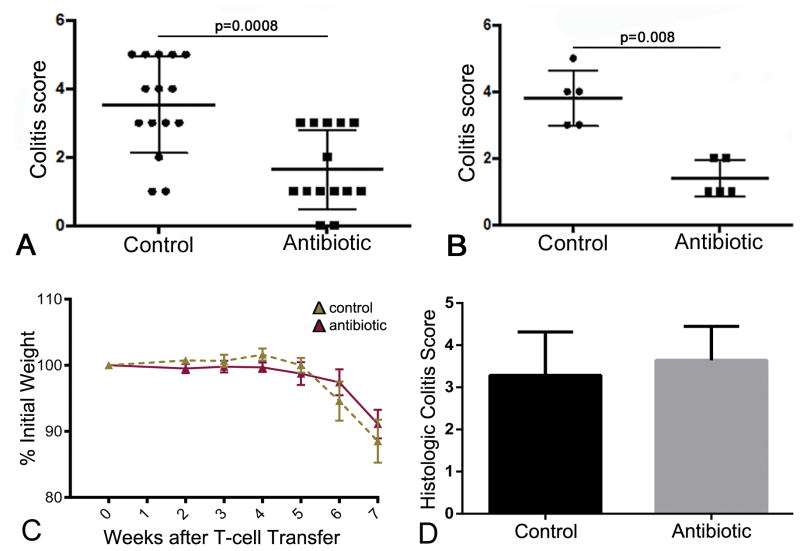
Given the sustained microbiota and immune changes observed following antibiotic treatment, we asked whether these alterations were associated with any change in the susceptibility to colitis. Mice were treated with the 4-antibiotic regimen for 2 weeks and, after a 6-week period off antibiotics, were given 3% DSS for 7 days, as shown in Fig. S1. Control mice, which did not receive antibiotics, had an average colitis score of 3.53±1.41 (n=15). In contrast, the colitis score in the antibiotic-treated group was 1.64±1.15 (n=14), representing a 54% decrease in colitis severity (Fig. 4A; p=0.0008).
Figure 4. Antibiotics protect against DSS colitis by promoting a less colitogenic microbiota and not via changes to T cell populations.
(A) Mice (n=15) were treated with antibiotics for 2 weeks. Six weeks later, DSS was administered for 7 days. Colitis severity was significantly lower in these mice than in control mice (n=14) that did not receive antibiotics (1.64±1.15 v. 3.53±1.41, p=0.0008). (B) Feces were collected from antibiotic-treated mice 6 weeks after completing treatment and transferred to normal mice 48 hours after completing antibiotic treatment to deplete their microbiota. DSS was administered for 7 days to the recipient mice 3 weeks later and colitis severity scored. Recipients of fecal transfer from antibiotic-treated mice (n=5) had a significantly lower colitis score (1.40±0.55) than recipients of stool from untreated controls (n=5; 3.80±0.84; p=0.008). (C,D) Naïve CD4+ T cells were isolated from control and antibiotic-treated mice and transferred intraperitoneally to immunodeficient Rag-1-/- mice to induce colitis. Eight weeks later, the degree of weight loss (C) and histologic colitis scores (D) were no different between the two groups of recipient mice. N = 7 in each group.
Antibiotic treatment does not permanently imprint T cells to become less colitogenic
Antibiotic-induced changes were also seen in the lymphocyte subpopulations. We hypothesized that antibiotic treatment may lead to a reduction in the colitogenicity of T cells. To test this, a naïve CD4+ T-cell transfer mode of colitis was performed as previously described 29. CD4+CD25-CD45RBhi cells were isolated from the mesenteric and peripheral lymph nodes and spleens of control and antibiotic-treated mice at Week 8. These cells were subsequently injected intraperitoneally into Rag-1-/- mice and the development of colitis was assessed 8 weeks later. Normally, mice receiving naive CD4+ T cells develop colitis by this time point. Recipient mice were weighed weekly and the weights decreased to about 90% of baseline in both groups (Fig. 4C). Histologic colitis scores were calculated in all mice and showed no significant difference between control and antibiotic-treated groups (Fig. 4D).
The sustained protective effect of antibiotics is attributed to a less colitogenic microbiota
To determine whether the change in colitogenic susceptibility, as shown in Fig. 4, could be attributed to the microbiota, a fecal transfer experiment was performed. Stool was collected from normal mice or from antibiotic-treated mice 6 weeks after completing their 2-week antibiotic course and transferred by gavage to recipient wild-type mice whose endogenous microbiota had been depleted with the same antibiotic regimen. Three weeks later, colitis was induced with a 7-day course of 3% DSS. As shown in Fig. 4B, recipients of stool from antibiotic-treated mice had a significantly lower colitis score that those receiving stool from untreated controls (1.40±0.55 v. 3.80±0.84; n=5 per group; p=0.008). Colitis severity was also evaluated by measuring the length of the colon, since inflammation is associated with colonic shortening. Consistent with the histologic results, mice receiving control stool had an average colon length of 5.60±0.29cm, whereas recipients of antibiotic-treated stool had colons measuring 6.98±0.40cm (p = 0.008, Mann-Whitney test).
Microbiota composition is altered following fecal transfer
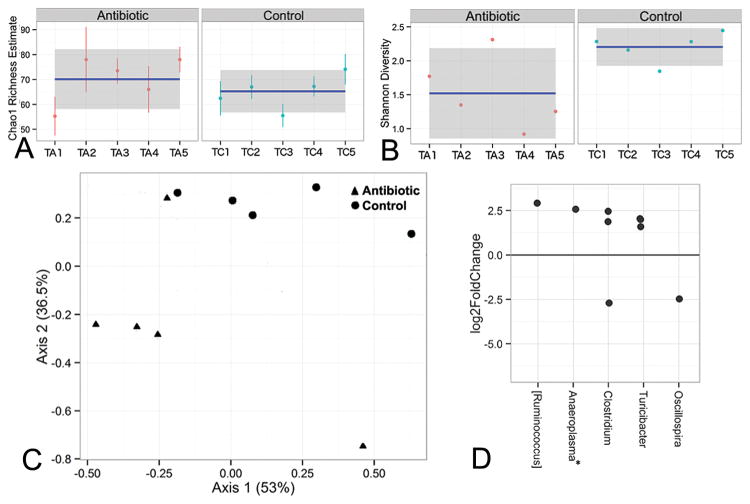
The protective effect of the fecal transplant from antibiotic–treated donor mice (Fig. 4) suggests that the antibiotics either lead to the development of a microbial population that is protective against colitis or to the elimination of a colitogenic microbiome. To characterize this protective effect further, we determined the diversity and composition of the fecal microbiota in recipient mice 3 weeks after the fecal transfer from antibiotic-treated (TA1-TA5) or control (TC1-TC5) donors. Exposure to an antibiotic-treated donor microbiota was associated with a significantly reduced diversity, although statistically supported differences (p<0.05) between antibiotic and control groups were observed only for the Shannon index (Fig. 5A,B). There was a significant difference in overall microbiome composition for both weighted (F1,25 = 5.44, p = 0.01) and unweighted (F1,25 = 9.34, p < 0.001) UniFrac distances, further illustrated by PCoA exploration (Fig. 5C shows weighted analysis).
Figure 5. Fecal transfer from antibiotic-treated mice leads to an altered microbiota compared to transfer from control donors.
Diversity and composition of fecal microbiota was determined in recipient mice three weeks after fecal transfer from control or antibiotic-treated donors. Species richness was measured using the Chao (A) and Shannon (B) indices. The mean value and confidence interval for each group are illustrated. Standard errors around each Chao1 estimator value are also shown. TA1-5 = recipients after transfer from antibiotic-treated donor; TC1-5 = recipients after transfer from control donor. (C) Plot of the PCoA results, based on weighted UniFrac for antibiotic (triangles) and control (circles) groups. The percent of overall variability accounted for by each axis is denoted in the axis labels. (D) OTUs that exhibited a significant difference in abundance between the antibiotic and control treatments. Here, each dot represents an individual OTU. Positive values indicate an increase, and negative values indicate a decrease in the antibiotic group, relative to the control. OTUs are grouped by the genus (x-axis) to which they belong. All OTUs shown belong to phylum Firmicutes, with the exception of those (denoted with an asterisk) assigned to Anaeroplasma (phylum Tenericutes).
These treatment-based effects on microbiome composition were supported by a classification-dependent approach based on the relative abundance of dominant genera (Fig. S4). One antibiotic-treatment recipient (TA1) grouped with controls, albeit basal to the control samples. In this analysis, we also examined the relationship between donor and recipient microbiomes. The antibiotic-treated donor (DA) and control donor (DC) microbiomes clustered with their respective recipient groups, supporting successful engraftment of the donor populations. Additionally, all but one of the pre-transfer recipient microbiomes (P1-P6) clustered together to the exclusion of the post-transfer recipient microbiomes. The outlier pre-transfer microbiome (P4) grouped most closely with the antibiotic-treated donor microbiome, within the larger cluster associated with antibiotic treatment.
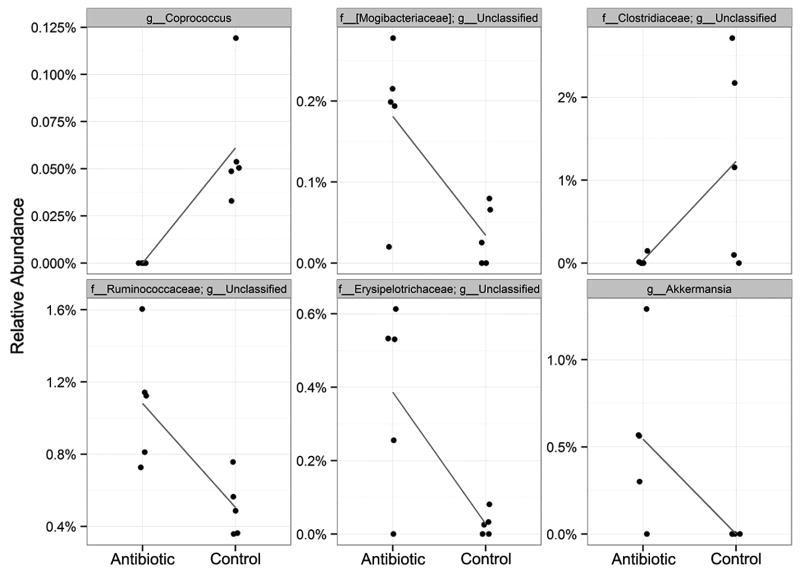
At first glance, the genus-level composition of antibiotic-treatment recipients (TA1-5) was not obviously different from that of controls (TC1-4) (Fig. S5), as was observed for the antibiotic-treated microbiomes from the observational studies described above. One apparent difference was that antibiotic-treated microbiomes generally contained fewer members of a genus-level group within the order Clostridiales (colored aqua) than most control recipients. Further analysis showed statistically supported differences in the abundance of six genera (Fig. 6) that formed minority microbiome components in recipients of both antibiotic-treated and control microbiomes (Fig. 6,S5), with relative abundances ranging from 0 to 3%. Four of the six genera were more abundant in antibiotic treatment recipients than in controls.
Figure 6. Relative abundance of genus-level taxa reveals significant differences between recipients of antibiotic-treated versus control fecal donors (transfer experiment).
Arcsin-transformed observations from each sample are shown as single points, and the data are jittered along the x-axis to alleviate overplotting. The blue line connects the means in each group and indicates the change between groups.
The composition of control microbiome recipients resembled that of the control donor, albeit with expansion of a genus-level group within the family-level group S24-7. In contrast, the recipients of antibiotic-treated microbiomes showed this S24-7 group as the dominant taxon, despite its absence in the donor mouse (Fig. S5). As was seen in the hierarchical clustering analysis (Fig. S4), the recipient mice contained a very different microbiome profile prior to transfer. In all but one mouse (P5), the S24-7 group was absent, and the majority components were members of a genus-level group within the family Clostridiales, as well as the genera Pseudomonas, Oscillospira, and (in variable numbers of mice) Herbaspirillum, Curtobacterium, and Candidatus Phytoplasma.
As was the case for the observational studies described above, both visual representation and statistical analysis of treatment-based differences in relative abundance at the species level (data not shown) closely resembled analysis performed at the genus level for post-transfer recipient microbiomes (Fig. 6,S5). Within the pre-transfer microbiomes, the dominant members of genus Pseudomonas were further resolved to the species Pseudomonas veronii. Analysis of treatment-based differences at the OTU level (Fig. 5D) revealed significant differences, with most differences falling within a 3-fold change, and many OTUs unclassified at the genus level. The majority of OTUs were assigned to the phylum Firmicutes, with a small number belonging to phylum Tenericutes (see legend to Fig. 5). Eight OTUs, classified within the genera Oscillospira, [Ruminococcus], Anaeroplasma, Clostridium, and Turicibacter, showed treatment-based differences. Of these, all but two (OTUs assigned to Clostridium and Oscillospira) were enriched in recipients of antibiotic-treated microbiomes relative to controls.
Sequencing analysis identifies candidate organisms that may explain the antibiotic-induced protection against colitis
Twenty-five OTUs showed consistent statistically supported differences in relative abundance in both the observational study and the fecal transfer experiment (Table S1). The majority of OTUs (19) were enriched in antibiotic-treated mice or in the recipients of stool from antibiotic-treated donor mice relative to controls. Both the enriched and depleted OTU sets were overwhelmingly dominated by members of the order Clostridiales. Where taxonomy could be further resolved, dominant Clostridiales families were Ruminococcaceae and Lachnospiraceae, with two additional OTUs assigned to Clostridiaceae and Mogibactericeae. Within the minority Bacteroidales and Anaeroplasmales components, OTUs were assigned to S24-7 and Anaeroplasmaceae. Six OTUs could be confidently assigned to the genus level, and were affiliated with Ruminococcus, Coprococcus, Oscillospira, Clostridium, and Anaeroplasma.
Discussion
Recent studies have highlighted the integral relationship between the structure of the microbiota and host immune factors in the pathogenesis of IBD 42. The immune system of the host and the composition of its microbiota have co-evolved to maintain a careful balance in order to ensure tolerance of commensal organisms and rejection of pathogens. When that balance is perturbed, inflammatory intestinal diseases, such as IBD, can occur. Many examples of the microbiota-immunity relationship exist. For example, certain colonic microbes metabolize dietary fiber to generate short-chain fatty acids, like butyrate, which induce regulatory T cells (Tregs) and can ameliorate the severity of experimental colitis 19. Other gut bacteria, particularly pathogenic fecal organisms, utilize bile to generate hydrogen sulfide (H2S), which induces proinflammatory T helper type 1 (Th1) cells and increases the incidence of colitis in genetically susceptible mice 43. High levels of IgA coating of bacteria have been shown to identify especially colitogenic species in both mouse and human 44. In contrast, commensal organisms activate toll-like receptors (TLRs) that induce protective cytokines that lessen the severity of experimental colitis 18.
Antibiotics represent a common cause of microbial perturbation 45 and have been associated with increased susceptibility to colitis. Mice treated with a four-antibiotic regimen, similar to that used in the current study, demonstrated markedly increased mortality following DSS treatment immediately after completing antibiotics, as compared to mice receiving only selective or no antibiotics 18. In human studies, antibiotic exposure during childhood has been associated with an increase in the long-term risk for developing IBD 13,15,46,47. However, the specific pathogens contributing to this colitogenic effect are unknown, and the role of antibiotic-induced changes in intestinal innate and adaptive immunity are also unclear. The present study aimed to establish whether antibiotic treatment leads to a sustained alteration in microbiota composition that increases susceptibility to experimental colitis, and whether that effect is mediated directly by intestinal microbes or by resulting abnormalities in gut immunity. We found that a two-week course of a four-antibiotic broad-spectrum regimen, including vancomycin, neomycin, metronidazole, and ampicillin, led to a sustained perturbation in microbial composition and diversity, as previously suggested by others 48,49. Interestingly, rather than making the host more susceptible to colitis, the antibiotic-treated mice exhibited a significant protection against DSS-induced colitis six weeks after completion of antibiotic treatment. Moreover, transfer of feces obtained six weeks after antibiotic treatment to mice depleted of commensals with the same four antibiotics, led to transfer of the protective effect, supporting the presence of a persistent and beneficial alteration in microbiota composition. Despite minor changes in T-cell numbers and proportions in the different immune compartments described, there was no major shift in the T-cell population to explain for the change in DSS colitis phenotype. Although there may have been changes in the innate immune cell subpopulations, which were not examined, the lack of significant T-cell changes suggests that microbes may have exerted a direct colitis-protective effect.
Several factors should be considered in evaluating the contrast between our findings of an antibiotic-associated protective effect, and the results of previous studies. It is likely that the starting microbiota composition of both donors and recipients, the selection of antibiotics, and the read-outs used to assess functional consequences of antibiotic treatment (i.e. indicators of colitis) affect the experimental outcomes. Additionally, the timing of interventions (i.e. DSS administration relative to antibiotic therapy), and interactions between all of these factors may further influence host-microbiome relationships. Further studies, in which each factor is independently investigated and potential interactions are explored, are needed to evaluate the effect of context on the outcomes. For example, it seems possible, even probable, that the antibiotic treatment of the recipients favored the growth of those organisms that had been selected to survive in the antibiotic-treated donors. If we had used a different set of antibiotics to condition the recipients (or used germ-free recipients), the outcome may have been different. Lastly, connections between the persistence of antibiotic-associated microbiota changes, and the colitis-protective effect, must be elucidated. At present this connection constitutes a “black box” that most likely contains taxon-specific microbial metabolism and physiology. Understanding this connection will most likely require study of candidate taxa in isolation.
The antibiotic-associated protective effect led us to perform deeper comparative analysis of data from the two studies (observational and transfer) in order to identify potential candidates for this anti-colitogenic effect. Our observational study showed that antibiotic treatment depleted overall microbial diversity (Fig. 1A,B ), and affected microbiome composition (Fig. 1C, Fig. S2). Mouse fecal transfer experiments have demonstrated the contributions of the microbiome to host phenotypes for several disease states. We therefore applied this approach to determine the microbial contribution to the observed colitis-protective effect of antibiotic therapy. The microbial communities of mice receiving microbiota from antibiotic-treated donors were less diverse than seen for recipients of control microbiota (Fig. 5A,B). Treatment-based differences in microbiota composition were also observed (Fig. 5C, Fig. S3). In this way, the outcomes of the transfer experiment mimicked those of the observational experiment. Additionally, results of both experiments align well with previously published work on the effects of antibiotics on gut microbes, where both reduction in diversity, and compositional shifts have often been reported 50,11,51,52.
The taxonomic affiliation of sequences contributing to microbiome compositional differences in both studies was determined to the lowest level of taxonomy for which a high-confidence assignment could be made. While some contributing taxa could be assigned to the genus level or (even fewer) to species level, the majority represented undescribed taxa within established families or orders. Within the observational study, treatment-based differences in microbiome composition were primarily driven by changes in the abundance of four numerically dominant genera or genus-level taxa (Fig. 2, Fig. S4). For the transfer experiments, our ability to resolve the taxonomy of taxa showing treatment effects in abundance was similarly limited at the genus and, even more so, at the species levels. However, six genera or genus-equivalent taxa displayed treatment-based differences in relative abundance (Fig. 3, Fig. S5).
In the observational study, several minority components also showed treatment-based differences in abundance (Fig. 2). Several of these occurred at or below the 0.1% relative abundance threshold considered to define the “rare” biosphere 53. Data generated from this study do not allow us to predict functional contributions. However, although it is intuitive to assume that only majority components are functionally relevant, low-abundance taxa are known to contribute unique and/or essential ecological functions, including temporal variability and resilience, to their communities 53. Our results may indicate that it isn't the dominant members that convey a colitis- protective effect, but rather some of the lower-abundance members for which a statistically supported treatment-based difference was found.
We observed a cage effect for the antibiotic-treatment groups in both the observational study and transfer experiments. One possible interpretation of this effect is when antibiotics perturb the microbiota, the perturbation is also subject to stochastic effects that are dependent on the identity of early colonizers. These stochastic effects are then amplified as the microbiota develops and matures, through successive colonization events. Including several families of mice allowed us to observe colitis-protective effects associated with very different community composition. That is, although cage-specific effects are consistent with stochastic effects for community dominance, colitis protection was conveyed regardless.
For both the observational study and transfer experiment, we identified multiple OTUs (the finest level of resolution) within several genera that exhibited treatment-based differences in relative abundance (Fig. 1D, 5D). This set of genera partially overlapped with those identified by genus-level comparisons (Fig. 2, 6). Strong evidence for a cohort of 19 colitis-protective species was obtained by identification of OTUs showing treatment effects on abundance across both studies (Table S1; enriched in antibiotic-treated mice, or mice receiving fecal transfers from antibiotic-treated donors, relative to control mice). This cohort was dominated by members of the order Clostridiales, with the remaining OTUs assigned to the orders Bacteroidales and Anaeroplasmatales. Only a few of the OTUs could be classified to a family, and even fewer to the genus level. Here we limit our discussion of these results, in the context of previous literature concerning associations between microbial taxa and colitis, to the three genera to which OTUS were assigned with confidence: Oscillospira, Ruminococcus, and Coprococcus.
Oscillospira guillermondii, and some Ruminococcus species, were reported to be enriched in the mucosa of a patient whose antibiotic-induced colitis was resolved by fecal transplantation; some of these changes were also observed in fecal samples 54. However, Ruminococcus gnavus (apparently colitis-protective in our study) was depleted in the patient after fecal transfer and return to gut homeostasis. This is consistent with reports that R. gnavus is associated with dysbiosis in Crohn's Disease 55–58, and can promote colitis through induction of proinflammatory cytokines and effector TH1 and TH17 immune responses 59. Other species of Ruminococcus are frequently reported to differ in abundance between IBD patients and controls, with both IBD-associated enrichment 56 and depletion 60,61. Coprococcus is reported to be reduced in IBD 60 and is associated with successful treatment of C. difficile infection via fecal transfer 62, although antibodies to certain Coprococcus comes strains have been reported in Crohn's disease and ulcerative colitis patients 63–66.
Members of the genera Oscillospira, Coprococcus, and Ruminococcus are fiber-degrading SCFA producers, including butyrate. Two of the taxa (Clostridium and Anaeroplasma) found depleted in the colitis-predisposed control group include aerobic fermenters that produce fatty acids 67,68, and several studies report an inverse correlation between abundance of Clostridium, and both total SCFA levels, and butyrate levels 67. It thus seems that fatty acid profile and abundance may be associated with the colitis-protective effects observed in this study. Reduction in SCFAs, and alteration of SCFA profile, are associated with multiple diseases featuring colitis including inflammatory bowel diseases 69 and Hirschsprung's-associated enterocolitis 70.
Summary
Our study found that mice exposed to antibiotics were protected against colitis, and this effect was transferrable to recipient mice with fecal microbiota from antibiotic-treated donors, suggesting that the colitis-protective effect was due to properties of the microbial community. We identified several OTUs associated with colitis protection across independent observational and transfer experiments. While it will be necessary to further examine the effect of experimental design parameters on the antibiotic-induced colitis protection observed here, our results suggest that the effect of antibiotics on susceptibility to colitis may be more complex than previously reported.
Supplementary Material
Figure S1. Study schema. Antibiotics were administered for two weeks, then immunophenotyping performed. Six weeks later, immunophenotyping was repeated and fecal pyrosequencing performed. At Week 8, DSS was administered to control and antibiotic-treated mice for seven days and animals were subsequently scored for the severity of colitis.
Figure S2. Heatmap summarizing the relative abundance of the 20 most dominant genera in all samples for the observational studies. Samples and taxa are sorted based on hierarchical clustering of unweighted UniFrac distances among samples (rows) or Euclidian distances among taxa (columns). A1-13 = antibiotic-treated; C1-14 = control.
Figure S3. Relative abundance of the 19 most dominant genera in all samples for the observational studies. A1-13 = antibiotic-treated; C1-14 = control.
Figure S4. Heatmap summarizing the relative abundance of the 20 most dominant genera in all samples for the transfer experiment. Samples and taxa are sorted based on hierarchical clustering of unweighted UniFrac distances among samples (rows) or Euclidian distances among taxa (columns). DA = antibiotic-treated donor; DC = control donor; P1-6 = recipients prior to transfer; DA = TA1-5 = recipients after transfer from antibiotic-treated donor; TC1-5 = recipients after transfer from control donor.
Figure S5. Relative abundance of the 19 most dominant genera in all samples for the transfer experiment. DA = antibiotic-treated donor; DC = control donor; P1-6 = recipients prior to transfer; DA = TA1-5 = recipients after transfer from antibiotic-treated donor; TC1-5 = recipients after transfer from control donor.
Table S1. Identities of OTUs showing statistically supported changes in abundance across observational studies (Fig. 1D) and transfer experiments (Fig. 5D).
Acknowledgments
We are grateful to Estela Trebicka for technical assistance and to Kyle Bochanski for help with preparation of the manuscript.
Source of Funding: NKNS and BJC were supported by NIH grant R01 AI089700. DDN was supported by NIH grant K08DK083430. AMG was supported by a grant from the CSIBD (Center for the Study of Inflammatory Bowel Disease) at Massachusetts General Hospital.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to report.
Literature cited
- 1.Bartlett JG. Clinical practice. Antibiotic-associated diarrhea. N Engl J Med. 2002;346:334–339. doi: 10.1056/NEJMcp011603. [DOI] [PubMed] [Google Scholar]
- 2.Kaiko GE, Stappenbeck TS. Host-microbe interactions shaping the gastrointestinal environment. Trends Immunol. 2014;35:538–548. doi: 10.1016/j.it.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Possemiers S, Grootaert C, Vermeiren J, et al. The intestinal environment in health and disease - recent insights on the potential of intestinal bacteria to influence human health. Curr Pharm Des. 2009;15:2051–2065. doi: 10.2174/138161209788489159. [DOI] [PubMed] [Google Scholar]
- 4.Tappenden KA, Deutsch AS. The physiological relevance of the intestinal microbiota--contributions to human health. J Am Coll Nutr. 2007;26:679S–83S. doi: 10.1080/07315724.2007.10719647. [DOI] [PubMed] [Google Scholar]
- 5.Carlisle EM, Morowitz MJ. Pediatric surgery and the human microbiome. J Pediatr Surg. 2011;46:577–584. doi: 10.1016/j.jpedsurg.2010.12.018. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y, Blaser MJ. Inverse associations of Helicobacter pylori with asthma and allergy. Arch Intern Med. 2007;167:821–827. doi: 10.1001/archinte.167.8.821. [DOI] [PubMed] [Google Scholar]
- 7.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaplan JL, Shi HN, Walker WA. The role of microbes in developmental immunologic programming. Pediatr Res. 2011;69:465–472. doi: 10.1203/PDR.0b013e318217638a. [DOI] [PubMed] [Google Scholar]
- 9.Bedford Russell AR, Murch SH. Could peripartum antibiotics have delayed health consequences for the infant? BJOG Int J Obstet Gynaecol. 2006;113:758–765. doi: 10.1111/j.1471-0528.2006.00952.x. [DOI] [PubMed] [Google Scholar]
- 10.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blaser M. Antibiotic overuse: Stop the killing of beneficial bacteria. Nature. 2011;476:393–394. doi: 10.1038/476393a. [DOI] [PubMed] [Google Scholar]
- 13.Hviid A, Svanström H, Frisch M. Antibiotic use and inflammatory bowel diseases in childhood. Gut. 2011;60:49–54. doi: 10.1136/gut.2010.219683. [DOI] [PubMed] [Google Scholar]
- 14.Elson CO, Cong Y, McCracken VJ, et al. Experimental models of inflammatory bowel disease reveal innate, adaptive, and regulatory mechanisms of host dialogue with the microbiota. Immunol Rev. 2005;206:260–276. doi: 10.1111/j.0105-2896.2005.00291.x. [DOI] [PubMed] [Google Scholar]
- 15.Shaw SY, Blanchard JF, Bernstein CN. Association between the use of antibiotics and new diagnoses of Crohn's disease and ulcerative colitis. Am J Gastroenterol. 2011;106:2133–2142. doi: 10.1038/ajg.2011.304. [DOI] [PubMed] [Google Scholar]
- 16.Olszak T, An D, Zeissig S, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill DA, Siracusa MC, Abt MC, et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat Med. 2012;18:538–546. doi: 10.1038/nm.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, et al. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Furusawa Y, Obata Y, Fukuda S, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 20.Singh N, Gurav A, Sivaprakasam S, et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity. 2014;40:128–139. doi: 10.1016/j.immuni.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith PM, Howitt MR, Panikov N, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nell S, Suerbaum S, Josenhans C. The impact of the microbiota on the pathogenesis of IBD: lessons from mouse infection models. Nat Rev Microbiol. 2010;8:564–577. doi: 10.1038/nrmicro2403. [DOI] [PubMed] [Google Scholar]
- 23.Sellon RK, Tonkonogy S, Schultz M, et al. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun. 1998;66:5224–5231. doi: 10.1128/iai.66.11.5224-5231.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garrett WS, Lord GM, Punit S, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garrett WS, Gallini CA, Yatsunenko T, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peloquin JM, Nguyen DD. The microbiota and inflammatory bowel disease: insights from animal models. Anaerobe. 2013;24:102–106. doi: 10.1016/j.anaerobe.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen CC, Louie S, McCormick B, et al. Concurrent infection with an intestinal helminth parasite impairs host resistance to enteric Citrobacter rodentium and enhances Citrobacter-induced colitis in mice. Infect Immun. 2005;73:5468–5481. doi: 10.1128/IAI.73.9.5468-5481.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shanmugam NKN, Trebicka E, Fu LL, et al. Intestinal inflammation modulates expression of the iron-regulating hormone hepcidin depending on erythropoietic activity and the commensal microbiota. J Immunol Baltim Md 1950. 2014;193:1398–1407. doi: 10.4049/jimmunol.1400278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen DD, Wurbel MA, Goettel JA, et al. Wiskott-Aldrich syndrome protein deficiency in innate immune cells leads to mucosal immune dysregulation and colitis in mice. Gastroenterology. 2012;143:719–729–2. doi: 10.1053/j.gastro.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Callaway TR, Dowd SE, Edrington TS, et al. Evaluation of bacterial diversity in the rumen and feces of cattle fed different levels of dried distillers grains plus solubles using bacterial tag-encoded FLX amplicon pyrosequencing. J Anim Sci. 2010;88:3977–3983. doi: 10.2527/jas.2010-2900. [DOI] [PubMed] [Google Scholar]
- 31.Handl S, Dowd SE, Garcia-Mazcorro JF, et al. Massive parallel 16S rRNA gene pyrosequencing reveals highly diverse fecal bacterial and fungal communities in healthy dogs and cats. FEMS Microbiol Ecol. 2011;76:301–310. doi: 10.1111/j.1574-6941.2011.01058.x. [DOI] [PubMed] [Google Scholar]
- 32.Brosius J, Palmer ML, Kennedy PJ, et al. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinforma Oxf Engl. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinforma Oxf Engl. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McDonald D, Price MN, Goodrich J, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao A. Estimating the population size for capture-recapture data with unequal catchability. Biometrics. 1987;43:783–791. [PubMed] [Google Scholar]
- 37.Shannon C. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423–656. [Google Scholar]
- 38.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oksanen J, Guillaume Blanchet F, Kindt R, et al. vegan: Community Ecology Package. 2011 [Google Scholar]
- 40.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts D. labdsv: Ordination and Multivariate Analysis for Ecology. 2010 [Google Scholar]
- 42.Wlodarska M, Kostic AD, Xavier RJ. An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe. 2015;17:577–591. doi: 10.1016/j.chom.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devkota S, Wang Y, Musch MW, et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palm NW, Zoete MR de, Cullen TW, et al. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 2014;158:1000–1010. doi: 10.1016/j.cell.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jernberg C, Löfmark S, Edlund C, et al. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiol Read Engl. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- 46.Virta L, Auvinen A, Helenius H, et al. Association of repeated exposure to antibiotics with the development of pediatric Crohn's disease--a nationwide, register-based finnish case-control study. Am J Epidemiol. 2012;175:775–784. doi: 10.1093/aje/kwr400. [DOI] [PubMed] [Google Scholar]
- 47.Kronman MP, Zaoutis TE, Haynes K, et al. Antibiotic exposure and IBD development among children: a population-based cohort study. Pediatrics. 2012;130:e794–803. doi: 10.1542/peds.2011-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Antonopoulos DA, Huse SM, Morrison HG, et al. Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect Immun. 2009;77:2367–2375. doi: 10.1128/IAI.01520-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Croswell A, Amir E, Teggatz P, et al. Prolonged impact of antibiotics on intestinal microbial ecology and susceptibility to enteric Salmonella infection. Infect Immun. 2009;77:2741–2753. doi: 10.1128/IAI.00006-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho I, Yamanishi S, Cox L, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6:e280. doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schubert AM, Sinani H, Schloss PD. Antibiotic-Induced Alterations of the Murine Gut Microbiota and Subsequent Effects on Colonization Resistance against Clostridium difficile. mBio. 2015;6:e00974–15. doi: 10.1128/mBio.00974-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lynch MDJ, Neufeld JD. Ecology and exploration of the rare biosphere. Nat Rev Microbiol. 2015;13:217–229. doi: 10.1038/nrmicro3400. [DOI] [PubMed] [Google Scholar]
- 54.Satokari R, Fuentes S, Mattila E, et al. Fecal transplantation treatment of antibiotic-induced, noninfectious colitis and long-term microbiota follow-up. Case Rep Med. 2014;2014:913867. doi: 10.1155/2014/913867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joossens M, Huys G, Cnockaert M, et al. Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut. 2011;60:631–637. doi: 10.1136/gut.2010.223263. [DOI] [PubMed] [Google Scholar]
- 56.Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 57.Prindiville T, Cantrell M, Wilson KH. Ribosomal DNA sequence analysis of mucosa-associated bacteria in Crohn's disease. Inflamm Bowel Dis. 2004;10:824–833. doi: 10.1097/00054725-200411000-00017. [DOI] [PubMed] [Google Scholar]
- 58.Willing BP, Dicksved J, Halfvarson J, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010;139:1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 59.Eun CS, Mishima Y, Wohlgemuth S, et al. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect Immun. 2014;82:2239–2246. doi: 10.1128/IAI.01513-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen L, Wang W, Zhou R, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore) 2014;93:e51. doi: 10.1097/MD.0000000000000051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rajilić-Stojanović M, Shanahan F, Guarner F, et al. Phylogenetic Analysis of Dysbiosis in Ulcerative Colitis During Remission. Inflamm Bowel Dis. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- 62.Shankar V, Hamilton MJ, Khoruts A, et al. Species and genus level resolution analysis of gut microbiota in Clostridium difficile patients following fecal microbiota transplantation. Microbiome. 2014;2:13. doi: 10.1186/2049-2618-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hazenberg MP, van de Merwe JP, Peña AS, et al. Antibodies to Coprococcus comes in sera of patients with Crohn's disease. Isolation and purification of the agglutinating antigen tested with an ELISA technique. J Clin Lab Immunol. 1987;23:143–148. [PubMed] [Google Scholar]
- 64.Bull K, Matthews N, Rhodes J. Antibody response to anaerobic coccoid rods in Crohn's disease. J Clin Pathol. 1986;39:1130–1134. doi: 10.1136/jcp.39.10.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Auer IO, Röder A, Wensinck F, et al. Selected bacterial antibodies in Crohn's disease and ulcerative colitis. Scand J Gastroenterol. 1983;18:217–223. doi: 10.3109/00365528309181586. [DOI] [PubMed] [Google Scholar]
- 66.Wensinck F, van de Merwe JP, Mayberry JF. An international study of agglutinins to Eubacterium, Peptostreptococcus and Coprococcus species in Crohn's disease, ulcerative colitis and control subjects. Digestion. 1983;27:63–69. doi: 10.1159/000198931. [DOI] [PubMed] [Google Scholar]
- 67.Hester CM, Jala VR, Langille MG, et al. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J Gastroenterol. 2015;21:2759–2769. doi: 10.3748/wjg.v21.i9.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rose CS, Pirt SJ. Conversion of glucose to fatty acids and methane: roles of two mycoplasmal agents. J Bacteriol. 1981;147:248–254. doi: 10.1128/jb.147.1.248-254.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumari R, Ahuja V, Paul J. Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J Gastroenterol. 2013;19:3404–3414. doi: 10.3748/wjg.v19.i22.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Demehri FR, Frykman PK, Cheng Z, et al. Altered fecal short chain fatty acid composition in children with a history of Hirschsprung-associated enterocolitis. [Accessed November 30, 2015];J Pediatr Surg. 0 doi: 10.1016/j.jpedsurg.2015.10.012. Available at: http://www.jpedsurg.org/article/S0022346815006211/abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Study schema. Antibiotics were administered for two weeks, then immunophenotyping performed. Six weeks later, immunophenotyping was repeated and fecal pyrosequencing performed. At Week 8, DSS was administered to control and antibiotic-treated mice for seven days and animals were subsequently scored for the severity of colitis.
Figure S2. Heatmap summarizing the relative abundance of the 20 most dominant genera in all samples for the observational studies. Samples and taxa are sorted based on hierarchical clustering of unweighted UniFrac distances among samples (rows) or Euclidian distances among taxa (columns). A1-13 = antibiotic-treated; C1-14 = control.
Figure S3. Relative abundance of the 19 most dominant genera in all samples for the observational studies. A1-13 = antibiotic-treated; C1-14 = control.
Figure S4. Heatmap summarizing the relative abundance of the 20 most dominant genera in all samples for the transfer experiment. Samples and taxa are sorted based on hierarchical clustering of unweighted UniFrac distances among samples (rows) or Euclidian distances among taxa (columns). DA = antibiotic-treated donor; DC = control donor; P1-6 = recipients prior to transfer; DA = TA1-5 = recipients after transfer from antibiotic-treated donor; TC1-5 = recipients after transfer from control donor.
Figure S5. Relative abundance of the 19 most dominant genera in all samples for the transfer experiment. DA = antibiotic-treated donor; DC = control donor; P1-6 = recipients prior to transfer; DA = TA1-5 = recipients after transfer from antibiotic-treated donor; TC1-5 = recipients after transfer from control donor.
Table S1. Identities of OTUs showing statistically supported changes in abundance across observational studies (Fig. 1D) and transfer experiments (Fig. 5D).