Abstract
The early macrophage response to biomaterials has been shown to be a critical and predictive determinant of downstream outcomes. When properly prepared, bioscaffolds composed of mammalian extracellular matrix (ECM) have been shown to promote a transition in macrophage behavior from a proinflammatory to a regulatory/anti-inflammatory phenotype, which in turn has been associated with constructive and functional tissue repair. The mechanism by which ECM bioscaffolds promote this phenotypic transition, however, is poorly understood. The present study shows that matrix-bound nanovesicles (MBV), a component of ECM bioscaffolds, are capable of recapitulating the macrophage activation effects of the ECM bioscaffold from which they are derived. MBV isolated from two different source tissues, porcine urinary bladder and small intestinal submucosa, were found to be enriched in miRNA125b-5p, 143-3p, and 145-5p. Inhibition of these miRNAs within macrophages was associated with a gene and protein expression profile more consistent with a proinflammatory rather than an anti-inflammatory/regulatory phenotype. MBV and their associated miRNA cargo appear to play a significant role in mediating the effects of ECM bioscaffolds on macrophage phenotype.
Keywords: : matrix-bound nanovesicles, extracellular matrix, macrophage, phenotype
Introduction
Biologic scaffolds composed of mammalian extracellular matrix (ECM) have been used in a variety of anatomic sites, including the gastrointestinal tract,1 body wall,2 and cardiovascular system,3 among others, to promote the formation of site-appropriate, functional tissue following injury. These bioscaffold materials influence the default tissue healing response by mitigating inflammation and scar tissue formation,1,4 promoting an accumulation of endogenous stem/progenitor cells at the site of scaffold placement,5,6 and perhaps most importantly, modulating the local innate and adaptive immune response.7–9 Specifically, implantation of ECM bioscaffolds has been shown to enhance the ratio of anti-inflammatory/regulatory (M2-like) macrophages to proinflammatory (M1-like) macrophages at the site of implantation.8 This effect on macrophage phenotype has been positively correlated with constructive and functional tissue remodeling outcomes in animal models of soft tissue repair.8,10
Macrophage phenotype plasticity is well established11 and the importance of, in fact necessity of, a transition from a proinflammatory to an anti-inflammatory/regulatory phenotype for normal functional tissue repair has been shown in many different body systems.12–15 However, the intercellular and intracellular signaling mechanisms responsible for macrophage phenotype transition in normal wound healing, tissue homeostasis and development, and biomaterial-mediated tissue repair are not fully understood. Reasonable explanations for cell–matrix interactions and the associated effects on cell behavior include integrin-mediated responses to the topographical ligand landscape,16,17 mechanical cues,18,19 and release of embedded growth factors/cytokines/cryptic peptides from the bioscaffold.20,21 While these factors are all suspected to contribute to the bioactivity attributed to the ECM, a specific mechanism by which ECM promotes a constructive macrophage phenotype has not been established. Recently, the presence of matrix-bound nanovesicles (MBV) within ECM bioscaffolds has been reported,22 and the miRNA cargo within these MBV has been associated with essential biologic processes such as normal tissue and organ development, inflammation, and immune cell regulation, among others.23–29 Three miRNAs that are preferentially overexpressed in M2-like macrophages30 were chosen as targets in the present study and these miRNAs have been shown to play a role in macrophage functions such as phagocytosis, nitric oxide (NO) production, and alteration of their secretome.31 The present study investigates the role of MBV in ECM bioscaffold-mediated macrophage activation.
Materials and Methods
Chemicals and reagents
Pepsin (MP Biomedicals, Santa Ana, CA), collagenase from Clostridium histolyticum (Sigma-Aldrich, St. Louis, Missouri), proteinase-K, and RNase A (Thermo Fisher Scientific, Waltham, MA) were confirmed by transmission electron microscopy (TEM) to be free of contaminating extracellular vesicles.
ECM bioscaffold production
Urinary bladder matrix
Urinary bladder matrix (UBM) was prepared from market-weight pigs (Tissue Source; LLC, Lafayette, IN) as previously described.7 Briefly, the tunica serosa, muscularis externa, submucosa, and muscularis mucosa were removed by mechanical delamination, and the urothelial cells of the tunica mucosa were dissociated from the basement membrane by washing with deionized water. The remaining basement membrane and the lamina propria (collectively referred to as UBM) were decellularized by agitation in 0.1% peracetic acid with 4% ethanol for 2 h at 300 rpm followed by phosphate-buffered saline (PBS) and type 1 water washes. UBM was then lyophilized and milled using a Wiley Mill with a #60 mesh screen.
Small intestinal submucosa
Preparation of small intestinal submucosa (SIS) bioscaffold has been previously described.32 Briefly, jejunum was harvested from market-weight pigs (Tissue Source; LLC). The superficial layers of the tunica mucosa, the tunica serosa, and tunica muscularis externa were mechanically removed after the jejunum was split longitudinally. The tunica submucosa, muscularis mucosa, and basilar portion of the tunica mucosa (stratum compactum) remain intact (collectively referred to as SIS). The tissue was agitated in 0.1% peracetic acid with 4% ethanol for 2 h at 300 rpm and then extensively rinsed with PBS and sterile water. The SIS was then lyophilized and milled using a Wiley Mill with a #60 mesh screen.
Enzymatic digestion of ECM samples
Enzymatic digestion was performed by treating each sample (100 mg dry weight) with either proteinase K or collagenase (0.1 mg/mL) for 24 h at room temperature in 50 mM Tris (pH 8), 5 mM CaCl2, and 200 mM NaCl buffer. Pepsin (1 mg/mL) digestion was performed in 0.01 M HCl solution for 24 h in room temperature. Before addition of ECM, all enzymatic solutions were passed through a 0.22-μm filter (Millipore, Oak Brook, IL).
MBV isolation
MBV were isolated as previously described.22 Collagenase was used for isolating MBV that were used for treating cells. Proteinase-K was used for isolating MBV that were used for RNA isolation or for visualization by TEM. Enzymatically digested ECM was subjected to successive centrifugations at 500 g (10 min), 2500 g (20 min), and 10,000 g (30 min). Supernatant was then centrifuged at 100,000 g (Beckman Coulter Optima L-90K ultracentrifuge, Brea, CA) at 4°C for 70 min. Pellets were washed and suspended in 500 μL of PBS and passed through a 0.22-μm filter (Millipore).
MBV imaging
TEM imaging was conducted on MBV loaded on carbon-coated grids and fixed in 1% uranyl acetate. Grids were imaged at 80 kV with a JEOL 1210 microscope.
RNA isolation
RNA was isolated from 2 × 106 cells using the miRNeasy Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. Reverse transcription of 500 ng of RNA to cDNA was performed via a high-capacity RT kit (ABI, Foster City, CA) according to the manufacturer's instructions. RNA was isolated from, at a minimum, 50 μL of MBV using the SeraMir Kit (System Biosciences) according to the manufacturer's instructions. Before RNA isolation, MBV samples were treated with RNase A (10 μg/mL) (Applied Biosystems, Palo Alto, CA) at 37°C for 30 min to degrade any contaminating free RNA that may have remained as a result of the tissue decellularization process. RNA concentration was determined using a NanoDrop spectrophotometer (NanoDrop, Wilmington, DE).
SYBR Green gene expression assays (premade sequences, ABI) were used to determine the relative expression levels of the following genes from DNA harvested from murine bone marrow-derived macrophages (BMDM): inos, tnf-α, stat1, stat2, stat5, irf3, irf4, irf5, il1rn, cd206, tgm2, stat3, stat6, klf4, klf6, fizz-1, arg1, bfkbf3, glut1, hif1α, hk3, pgk1, pdk4, rpia, ldha, pck2, g6pc3, and pparδ. Results were analyzed by the ΔΔCt method using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) to normalize the results. Fold change was calculated using untreated macrophages (M0) as the baseline. Results are displayed in a heat map format created by Java Treeview (Oracle, Redwood City, California).
TaqMan MicroRNA assays (ABI) were used to determine the relative levels of mmu-miR-145-5p, mmu-miR-143-3p, and mmu-miR-125b-5p inhibitors. These three miRNAs were found to be enriched in MBV and have been suggested as mediators of macrophage activation.23,29,31,33,34 mmu-SNO-55 was used to normalize the results.
RNA sequencing and data analysis
RNA sequencing and data analysis were performed as previously described.22 Briefly, small RNA libraries were prepared using Ion Total RNA-Seq Kit version 2, according to the manufacturer's instructions. Following bead-based size selection of RNA (10- to 20-nt range), cDNA was created. Amplified library was again size-selected using a bead-based method. Library size distribution was verified using Agilent Bioanalyzer (Agilent, Santa Clara, CA). The Ion One Touch 2 System was used to perform automated emulsion polymerase chain reaction of the prepared libraries and templated Ion Sphere Particle enrichment. Sequencing was performed on the Ion Proton platform using a single P1 sequencing chip. Obtained data were imported into CLC Genomics Workbench 8 (Qiagen). The adaptors were trimmed, and all reads that had two ambiguous nucleotides, had a Phred score <30, or were lower than 15 nt, or above 100 nt were removed. Conserved reads were then aligned to the human genome (hg38) to verify valid reads. Reads were extracted, counted, and then annotated on miRBase v.21 (human genome reference); a 2-nt mismatch was allowed per read. Only sequences that matched a mature miRNA were used for downstream analysis.
MBV fluorescent labeling
MBV nucleic acid cargo was labeled using Exo-Glow (System Biosciences), according to the manufacturer's instructions. Briefly, 500 μL of resuspended MBV was labeled with Exo-Glow and incubated at 37°C for 10 min. ExoQuick-TC (100 μL) was added to stop the reaction, and samples were placed on ice for 30 min. Samples were then centrifuged for 10 min at 14,000 g. The supernatant was removed and the pellet was resuspended with 500 μL of 1 × PBS, and 50 μL of this MBV suspension was added to BMDM cell culture. The cells were cultured for 4 h, and the transfer of the MBV cargo to the cells was determined by imaging using a 100 × objective and Axio Observer Z1 microscope.
Cell culture
Murine BMDM were isolated and characterized as previously described.7 Briefly, bone marrow was harvested from 6- to 8-week-old C57bl/6 mice. Harvested cells from the bone marrow were washed and plated at 1 × 106 cells/mL and were allowed to differentiate into macrophages for 7 days in the presence of macrophage colony-stimulating factor (MCSF) with complete medium changes every 48 h.
Macrophage activation
Macrophages were activated for 24 h with one of the following: (1) 20 ng/mL interferon-γ (IFNγ) and 100 ng/mL lipopolysaccharide (LPS) (Affymetrix eBioscience, Santa Clara, CA; Sigma Aldrich) to promote an MIFNγ+LPS phenotype (M1-like), (2) 20 ng/mL interleukin (IL)-4 (Invitrogen) to promote an MIL-4 phenotype (M2-like), (3) 250 ug/mL of UBM-ECM, or SIS-ECM to promote an MECM phenotype, or (4) 25 μg/mL of UBM-MBV or SIS-MBV to promote an MMBV phenotype. The concentration of MBV was determined by bicinchoninic acid assay. Pepsin (1 mg/mL) and collagenase (0.1 mg/mL) were used as baseline controls for ECM and MBV, respectively. After the incubation period at 37°C, cells were washed with sterile PBS and fixed with 2% paraformaldehyde (PFA) for immunolabeling, or harvested with TRIzol lysis reagent (Thermo Fisher) for protein/RNA assessment, respectively. Macrophage function was evaluated by assays for phagocytosis and NO production.
Transfection
BMDM were transfected with 50 nM mmu-miR-154-5p, mmu-miR-143-3p, and mmu-miR-125b-5p inhibitor or a cocktail mix of the inhibitors (Thermo Fisher), as well as a scrambled negative control (Thermo Fisher), using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer's instructions. miRNA inhibitors were transfected for 4 h in Opti-MEM media. Cells were then washed and incubated overnight with Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (P/S).
NO quantification
Following 24 h of treatment with test articles, macrophage supernatants were transferred to a 96-well plate and frozen at −80°C. Fifty microliters of samples or internal assay standards consisting of sodium nitrite from 100 to 1.56 μM in a 1:2 serial dilution was added to the plate. The wells were treated with 50 μL of 1% sulfanilamide in 5% phosphoric acid for 10 min, followed by addition of 50 μL of 0.1% N-1-napthylethylenediamine dihydrochloride in water for 10 min. The wells were then read at 540 nm and compared to the standard curve. Cells were counted using Cell Profiler software using 4’6-diamidino-2-phenylindole (DAPI) nuclear staining. NO values were normalized to the number of cells per well.
Phagocytosis assay
Following 24 h of treatment with the test articles, macrophages were incubated with the Vybrant Phagocytosis Kit (Thermo Fisher) FITC-labeled Escherichia coli beads for 2 h. Wells were then washed once with 1 × PBS and fixed with 2% PFA for 30 min. Wells were washed three times with PBS, then stained with DAPI for 10 min, and washed again three times with PBS. Wells were imaged using an automated Live Cell Scope and quantified for mean fluorescence intensity of the cells using Cell Profiler software.
Macrophage immunolabeling
Cells were fixed with 2% PFA for 45 min at room temperature, then washed with PBS, followed by immunolabeling to determine surface marker expression. To prevent nonspecific binding, the cells were incubated in a blocking solution composed of PBS, 0.1% Triton-X, 0.1% Tween-20, 4% goat serum, and 2% bovine serum albumin for 1 h at room temperature. The blocking buffer was then removed and cells were incubated in a solution of one of the following primary antibodies: (1) monoclonal anti-F4/80 (Abcam, Cambridge, MA) at 1:200 dilution as a pan-macrophage marker, (2,3) polyclonal anti-inducible nitric oxide synthase (iNOS) and anti-tumor necrosis factor-α (TNF-α) (Abcam, Cambridge, MA) at 1:100 dilution, each as M1-like markers, and (4,5) polyclonal anti-Fizz1 (Peprotech, Rocky Hill, NJ) and anti-Arginase1 (Abcam, Cambridge, MA) at 1:200 dilution, each as M2-like markers. The cells were incubated at 4°C for 16 h, the primary antibody was removed, and the cells washed with PBS. A solution of fluorophore-conjugated secondary antibody (Alexa donkey anti-rabbit 488 or donkey anti-rat 488; Invitrogen, Carlsbad, CA) was added to the appropriate well for 1 h at room temperature. The antibody was then removed, the cells washed with PBS, and the nuclei were counterstained using DAPI. Cytokine-activated macrophages were used to establish standardized exposure times (positive control), which were held constant throughout groups thereafter. CellProfiler (Broad Institute, Cambridge, MA) was used to quantify images.
Statistical analysis
Data were analyzed for statistical significance using either an unpaired Student's t-test, through which treated macrophages were compared to the appropriate M0 media control, or a one-way analysis of variance with Tukey's post-hoc test for multiple comparisons. Data are reported as mean ± standard deviation with a minimum of N = 3. p-Values of <0.05 were considered to be statistically significant. For gene expression data represented as a heat map, the p-values that were generated by Student's t-tests comparing treated macrophages with M0 media control, and results are displayed in Supplementary Tables S1 and S3 (Supplementary Data are available online at www.liebertpub.com/tea).
Results
MBV imaging and gene expression signature of MBV-treated cells
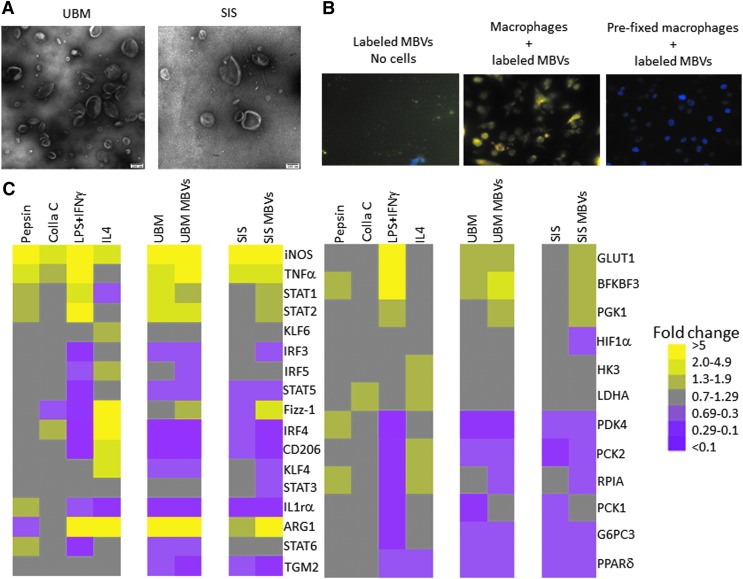
Particulate UBM-ECM or SIS-ECM was enzymatically digested for 16 h at room temperature with proteinase-K or 0.1% collagenase solution. The solubilized ECM samples were then subjected to centrifugation at increasing g forces to isolate MBV. MBV were visualized at 100,000 × magnification using TEM (Fig. 1A). Cellular uptake of MBV was determined by labeling MBV with acridine orange. Labeled MBV were visible within BMDM 2 h after their addition to the culture media (Fig. 1B). The effect of MBV on macrophage activation was evaluated by quantitative polymerase chain reaction (qPCR) analysis of more than 25 commonly used markers of macrophage activation, including surface markers, cytokines, transcription factors, and metabolic markers. The gene expression profile of macrophages treated with MBV was qualitatively very similar to the gene expression profile of macrophages treated with ECM (Fig. 1C). Little to no effect on gene expression was observed after exposing the macrophages to pepsin or collagenase indicating that these enzymes, which are used to digest ECM before extraction of MBV, are not responsible for macrophage activation (Supplementary Table S1). Exposure of BMDM to IFNγ+LPS (MIFNγ+LPS) or IL-4 (MIL-4) led to distinct gene expression profiles consistent with previous studies.11,35–37
FIG. 1.
MBV imaging and gene expression signature of MBV-treated macrophages. (A) MBV isolated using proteinase-K digestion were visualized by transmission electron microscopy at 100,000 × magnification. (B) Following exposure of BMDMs (counterstained with DAPI) to MBVs whose nucleic acid content was labeled with acridine orange, the cells were visualized using fluorescence microscopy at 200 × magnification. MBV are seen within the cytosol of cells after a 2-h incubation. (C) Gene expression analysis of cells exposed to either ECM or their respective MBVs was evaluated using qPCR. Results are presented in a heatmap form that was generated using Treeview software; all fold changes are with respect to media control (N = 3). Scale bar scoring system is demonstrated as follows: less than 0.1-fold change (darkest purple), 0.1–0.29-fold change (intermediate purple), 0.3–0.69-fold change (light purple), 0.7–1.29-fold change (gray), 1.3–1.9-fold change (light yellow), 2.0–4.9-fold change (intermediate yellow), greater than 5.0-fold change (bright yellow). Supplementary Table S1 indicates significant differences between media control and treated macrophages as determined using t-tests with p < 0.05 considered significant. BMDM, bone marrow-derived macrophages; ECM, extracellular matrix; DAPI, 4’6-diamidino-2-phenylindole; MBV, matrix-bound nanovesicles.
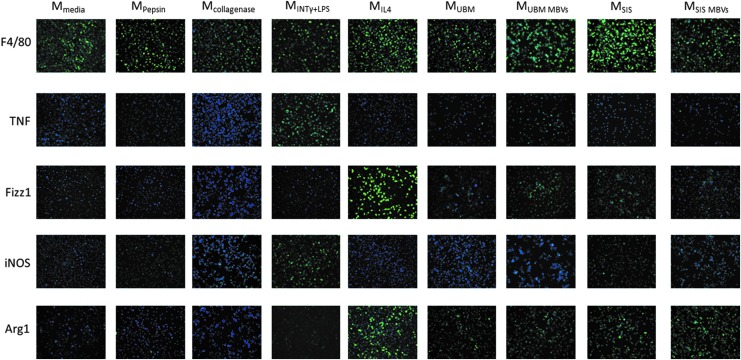
MBV treatment increases M2-like protein expression
Immunolabeling was performed to evaluate protein expression of BMDM (Fig. 2). Similar to the gene expression results, MBV-treated groups had a qualitatively similar protein expression profile as the ECM-treated groups. Both ECM groups, as well as their corresponding MBV groups, had protein expression profiles similar to MIL4 cells. Macrophage treatment with SIS-ECM and UBM-ECM and their corresponding MBV resulted in expression of Fizz-1 and Arg-1 (markers that are associated with the MIL-4 phenotype). Low levels of iNOS expression were detected in the SIS-MBV group. TNF-α was detectable only in the UBM-MBV group. Both TNF-α and iNOS are markers associated with the MIFNγ+LPS phenotype. No expression of these proteins was noted in the control groups. The majority of cells expressed F4/80, confirming their macrophage differentiation state. Quantification of immunolabeling images using CellProfiler software supported the qualitative interpretation of the results (Supplementary Fig. S1A).
FIG. 2.
MBV treatment increases M2-like protein expression. BMDM were exposed for 24 h to media control, 1 mg/mL pepsin or 0.1 mg/mL collagenase controls, 250 μg/mL ECM, 25 μg/mL MBVs, or the cytokine controls IFNγ+LPS or interleukin (IL)-4. Cells were then fixed with 4% PFA .The cells were then incubated with anti-murine antibody for markers of the M1-like phenotype TNFα and iNOS, or markers of the M2-like phenotype Fizz1 and Arginase1. All images for the same antibody were taken at the same exposure time normalized to the positive control. Cell nuclei were stained with DAPI. Images were taken at 200 × magnification (N = 3). The percentage of cells positive for each stain was quantified using Cell Profiler software and is presented in Supplementary Figure S1A. PFA, paraformaldehyde.
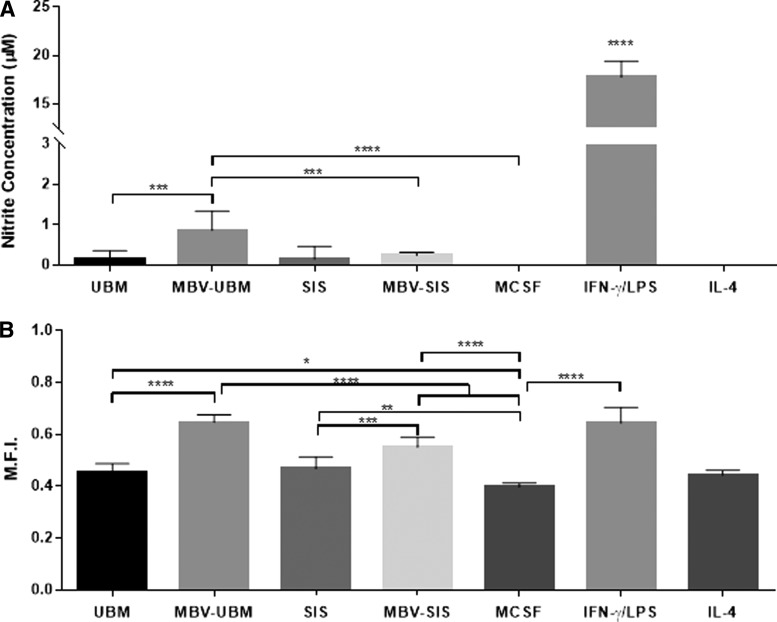
MBV treatment affects BMDM function more than ECM treatment
To determine the effect of MBV on macrophage function, NO production and phagocytosis were measured in macrophages exposed to MBV, ECM, or cytokines (Fig. 3).
FIG. 3.
MBV treatment affects BMDM function more than ECM treatment. Macrophages were exposed for 24 h to MCSF control, 250 μg/mL ECM, 25 μg/mL MBVs, or the cytokine controls IFNγ+LPS or IL-4. (A) Macrophage supernatants were mixed with 1% sulfanilamide in 5% phosphoric acid for 10 min, followed by addition of 0.1% N-1-napthylethylenediamine dihydrochloride in water. The solutions were read in a spectrophotometer at 540 nm and compared to the standard curve of sodium nitrite to assess nitric oxide production levels. Values: mean ± standard deviation, N = 6, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way analysis of variance (ANOVA) with Tukey's post-hoc test. (B) Treated macrophages were incubated with Vybrant Phagocytosis Kit FITC-labeled Escherichia coli beads for 2 h. Cells were fixed and stained with DAPI. Using fluorescence microscopy, the cells were visualized and quantified for mean fluorescence intensity of the cells using Cell Profiler software. Values: mean ± standard deviation, N = 6, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by one-way ANOVA with Tukey's post-hoc test. MCSF, macrophage colony-stimulating factor.
Nitric oxide production
NO was assessed in MBV-exposed BMDM. NO production was not detectable in naive and MIL-4 macrophages, consistent with the findings of previous studies.38,39 MIFN-γ+LPS macrophages produced a significant increase in NO. BMDM treated with MBV derived from UBM was the only treatment group to increase NO production (Fig. 3A).
Phagocytosis
A basal level of phagocytosis as measured by uptake of FITC-E. coli particles was shown by all macrophages. Treatment with IFN-γ+LPS resulted in a significant increase in phagocytic activity compared to MCSF (M0). ECM and MBV treatment resulted in a significant increase in phagocytic uptake compared to MCSF (M0). When compared to their parent ECM bioscaffolds, exposure to MBV alone led to greater phagocytosis. Whereas there was no significant difference in phagocytosis between macrophages treated with UBM or SIS, UBM-MBV caused an increase in macrophage phagocytosis compared to macrophages treated with SIS-MBV (Fig. 3B).
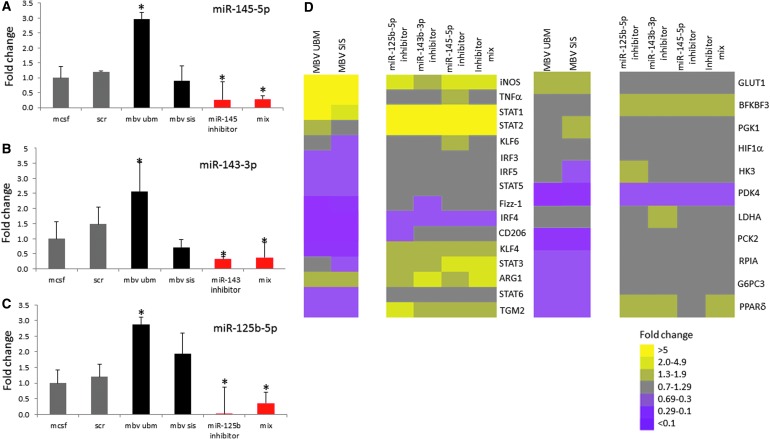
miRNA inhibition reverses gene expression patterns compared to MBV-exposed BMDM
Sequencing results revealed expression of 240 different miRNAs in UBM-MBV and 53 in SIS-MBV (Fig. 4).22 miRNAs with the greatest expression in UBM-MBV and SIS-MBV are listed in Supplementary Table S2. miRNAs highlighted in red were selected for downstream analysis based on previous studies that showed their involvement in macrophage activation.23,30,31
FIG. 4.
miRNA inhibition reverses gene expression patterns compared to MBV-exposed BMDM. Macrophage miRNA inhibition. (A–C) Selective inhibition of specific miRNAs, miR-145-5p, miR-145-3p, and miR-125-b-5p using 50 nM of inhibitor for each. Relative abundance of miRNA levels following inhibition was determined by TaqMan miRNA qPCR assays. Values: mean ± standard deviation, N = 3, *p < 0.05 by Student's t-test. Data represents fold change in comparison to M0. (D) Gene expression analysis of cells exposed to MBVs, or transfected with scrambled control miRNA inhibitor, mmu-miR-154-5p inhibitor, mmu-miR-143-3p inhibitor, mmu-miR-125b-5p inhibitor, or a combination of all three inhibitors was evaluated using qPCR. Results are presented in a heatmap form that was generated using Treeview software; all fold changes are with respect to media control. Scale bar scoring system is demonstrated as follows: less than 0.1-fold change (darkest purple), 0.1–0.29-fold change (intermediate purple), 0.3–0.69-fold change (light purple), 0.7–1.29-fold change (gray), 1.3–1.9-fold change (light yellow), 2.0–4.9-fold change (intermediate yellow), and greater than 5.0-fold change (bright yellow). Supplementary Table S3 indicates significant differences between media control and treated macrophages as determined using t-tests with p < 0.05 considered significant.
miR-145-5p, miR-143-3p, and miR-125b-5p were inhibited in BMDM to determine their role in macrophage expression of activation markers. The degree of inhibition of each miRNA was determined by qPCR (Fig. 4A–C). miR-145 expression was reduced by more than 70%, miR-143 expression was reduced by 65%, and miR-125b expression was reduced by more than 95%. qPCR analysis of MBV-treated and miRNA inhibitor-treated cells showed that 6 of 27 genes had a markedly different expression pattern (Fig. 4D and Supplementary Table S3). Interestingly, the six genes whose expression was increased by miRNA inhibition were the same as those that were decreased by MBV treatment. Importantly, KLF4, which is a transcription factor associated with macrophage activation and also a known target of miR-145-5p, shows an opposite expression pattern for all the inhibited miRNA treatment groups compared to both UBM-MBV- and SIS-MBV-treated cells. These results suggest that inhibition of KLF4 may be a target of the miRNA cargo within MBV.
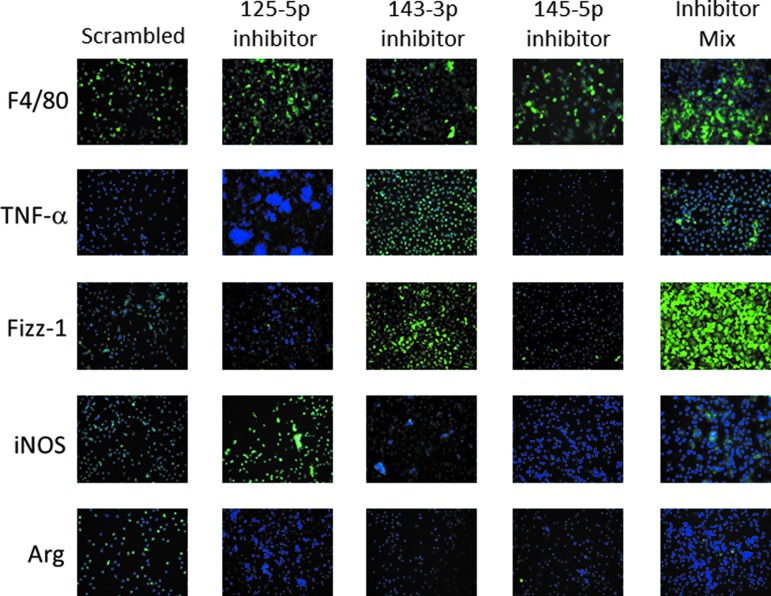
miRNA inhibition shows opposite protein expression in BMDM
Immunolabeling was performed to evaluate protein expression of BMDM in which miRNA had been selectively inhibited. Similar to the gene expression results, treatment with miRNA inhibitors led to an opposite protein expression profile compared to MBV-treated groups (Fig. 5). Inhibition of miRNA-125b-5p led to an increase in iNOS expression, whereas inhibition of miRNA-143-3p led to an increase in the expression of TNF-α and Fizz1 compared to uninhibited controls. The majority of cells expressed F4/80 consistent with a macrophage differentiation state. Quantification of immunolabeling images using CellProfiler software supported the qualitative interpretation of the results (Supplementary Fig. S1B).
FIG. 5.
miRNA inhibition shows opposite protein expression compared to MBV. BMDM were exposed for 4 h to 50 nM of one of the following: scrambled control, mmu-miR-125b-5p inhibitor, mmu-miR-143-3p inhibitor, or miR-145-5p. Treatment media were then changed to normal growth media for an additional 18 h. Cells were then fixed with 4% PFA. Cells were then incubated with anti-murine antibody for markers of the M1-like phenotype TNFα and iNOS, or markers of the M2-like phenotype Fizz1 and Arginase1. Exposure times were established based on a negative isotype control and cytokine-treated controls and kept constant for each marker tested. Cell nuclei were stained with DAPI. Images were taken at 200 × magnification. The percentage of cells positive for each stain was quantified using Cell Profiler software and is presented in Supplementary Figure S1B. The results show that miRNA inhibition is capable of impacting the expression of several probed proteins, implicating the role of miR-125b-5p, miR-143-3p, and miR-145-5p in the formation of the MMBV phenotype.
Discussion
The results of the present study show that MBV can largely recapitulate the effects of ECM on macrophage phenotype. MBV were rapidly internalized by macrophages and direct inhibition of specific miRNA cargo (found in high abundance in MBV) within these macrophages notably affected the macrophage phenotype. Macrophage gene and protein expression, cell surface markers, and functional capacity as determined by phagocytic activity, NO production, and antimicrobial activity were most representative of a regulatory/anti-inflammatory phenotype following treatment with MBV, which is consistent with previous reports describing the effects of ECM bioscaffolds on macrophage phenotype.40–43 These findings provide a plausible mechanism by which MBV embedded within ECM bioscaffolds can regulate the macrophage component of the innate immune response during matrix remodeling events.
Macrophages have been shown to be important, in fact necessary, regulators of normal healing following injury, and/or in normal tissue development.44 Specifically, a transition in macrophage activation state from a proinflammatory to an anti-inflammatory and pro-remodeling phenotype is required for initiation and resolution of the healing process and a return to homeostasis.7,43,45,46 Failure of this transition is associated with chronic inflammation, impaired wound healing, and dysregulation of the microenvironmental niche,47–49 and has been suggested to play a causal role in conditions such as inflammatory bowel disease, muscular dystrophy, and kidney disease.47,50,51 While the importance of this phenotypic switch is recognized, the specific endogenous signals regulating spatiotemporal patterns of macrophage phenotype are poorly understood. Phagocytosis of neutrophils as the cause of the phenotype switching in macrophages has been suggested,52 but definitive studies have been lacking.
ECM harvested from many different tissues and commercially available biomaterials composed of ECM consistently and reproducibly promote an M2-like regulatory macrophage phenotype,4,8,40,53 a process that drives downstream constructive and functional tissue remodeling. ECM bioscaffolds have thus far been shown to promote this phenotype activation by at least two mechanisms: (1) a direct transmembrane process involving intracellular pathways such as COX1/254 and (2) an indirect effect through stem/progenitor cell paracrine signaling.4 Macrophages have been shown to be necessary for the degradation of ECM bioscaffolds in vivo.10 It is plausible that during degradation of the ECM, MBV are released, internalized by the local macrophage population, and facilitate transition to an anti-inflammatory/regulatory phenotype. It is also logical to hypothesize that these events occur not only in the presence of ECM bioscaffolds but also during normal wound healing events; however, additional work is required to test such a hypothesis.
The profile of markers used in the present study to characterize macrophage activation state included transcription factors, surface markers, gene and protein expression, and functional assays. Stated differently, the activity of MBV was characterized on many levels, and it was shown that they mimic or even outperform ECM bioscaffolds with regard to macrophage activation in certain assays. These results suggest that MBV are a key bioactive component within ECM.
Sequencing of MBV nucleic acid cargo from UBM-ECM and SIS-ECM showed that specific miRNAs are particularly enriched in these nanovesicles, depending on the ECM tissue source. Three of these miRNAs, 125b-3p, 143-3p, 145-5p, were highly expressed within MBV and these specific miRNAs have been implicated in macrophage activation and phenotypes.23,30,31 These miRNAs were therefore targeted in the present study for closer scrutiny. There is not a consensus in the literature with respect to known functions of the particular miRNA investigated in the present study. It has been reported that, in murine Raw 264.7 macrophages,55 miRNA-125b-5p targets TNF-α and 5-lipoxygenase, negatively regulating the inflammatory response.56 However, in isolated murine peritoneal macrophages, miRNA-125b-5p was shown to enhance the inflammatory response in certain contexts by targeting IRF4.23 In the present study, inhibition of miR-125b-5p did not lead to increases in TNF-α, however, a reduction in IRF4 gene expression levels was noted in both miR-125b-5p inhibition and MBV-treated macrophages. This incongruity suggests that IRF4 regulation is not dependent solely on miR-125-5p and likely represents the net actions of various miRNAs and proteins present in the MBV cargo. Discrepancies observed between the results of the current study and previous reports with respect to miRNA-125b-5p could arise from the use of macrophages derived from different sources.
It has been shown that miRNA-143/145 are elevated in M2-like macrophages and downregulated in M1-like macrophages, implying their role in macrophage activation.30 Moreover, it has been shown that miRNA-145-5p is capable of activating the epigenetic IL-10 gene silencer, HDAC11, in murine macrophage cell lines. The previous report showed that downregulation of miRNA-145-5p expression as a result of IFN-γ signaling directly contributes to proinflammatory macrophage activity and phenotype.57
miRNA-143-3p is less well studied in the context of innate immunity and its targets within macrophages are poorly described. Nonetheless, inhibition of both miRNA-143-3p and miRNA-145-5p in naive macrophages resulted in increased expression of the macrophage surface marker associated with an M1-like phenotype, TNF-α, which is consistent with the findings of the previous report.30 These results are further corroborated by the finding that MBV treatment led to increased expression of M2-like markers, Fizz1 and Arg1, which are associated with a constructive macrophage phenotype; however, there are a multitude of miRNAs within MBV that could similarly contribute to this downregulation of proinflammatory markers. Taken together, the results of the miRNA inhibition and MBV treatments corroborate previous reports that miRNA-145-5p contributes to a downregulation of the inflammatory phenotype and suggest that both miRNA-143/145 contribute to the macrophage phenotype in response to MBV and ECM bioscaffolds.
Further analysis of gene and protein expression assays of naive BMDM in which these miRNAs were inhibited showed that their inhibition resulted in opposite patterns of expression in nearly 25% of the genes that were investigated. Among these genes are members of the KLF, STAT, and IRF families of transcription factors. Interestingly, KLF4 was consistently downregulated by both ECM and MBV treatment. Inhibition of miR-145-5p resulted in an increase of KLF4. The regulation of KLF4 by miR-145-5p has been shown to be an effector of macrophage activation,24,34,58 which together with findings of the present study, suggests its role in this process. Importantly, inhibition of the different miRNAs led to a similar gene expression profile. Using TargetScan software, 18 mutual genes were identified that are predicted to be regulated by miR-145-5p, miR-143-3p, and miR125-5p. Among those genes is QKI, which was previously identified as a macrophage differentiation regulator.59,60 We postulate that mutual target genes are being regulated by these miRNAs, affecting specific pathways.
Overall, these results strongly suggest that MBV and their miRNA cargo are at least partially responsible for the macrophage response that is observed when ECM bioscaffolds facilitate functional tissue repair.
MBV treatment also impacted macrophage function, with effects that were greater in magnitude than the effects of the parent ECMs on all metrics. The rate of phagocytosis in UBM-MBV-exposed macrophages rose to a level indistinguishable from that of MIFNγ+LPS macrophages, significantly greater than all other treatment groups.
Labeling MBV with ExoRed showed that MBV were internalized by macrophages within 2 h. This finding does not differentiate between phagocytosis and endocytosis as the major route of uptake, however, and this represents an area of future study.
While miRNA-125/143/145 appear to modify the macrophage activation state, there are more than 200 miRNAs present within the MBV,22 which have the potential to affect macrophage activation or the behavior of other cell types. In addition, the effect of MBV protein cargo on macrophage activation is yet to be determined. The present work did not utilize an MBV-depleted ECM control due to the inability to remove MBV without destroying all remaining ECM constituents.
Conclusion
The results of the present study clearly show the ability of MBV to recapitulate many effects of ECM on macrophage activation, which is an important bioactive property of ECM bioscaffolds. Furthermore, specific miRNAs within MBV play a role in this process, which provides a plausible mechanism by which ECM promotes a transition in macrophage phenotype and downstream constructive tissue remodeling not only with the use of ECM bioscaffolds but also during normal tissue repair processes. A more comprehensive understanding of the role of additional miRNAs present in the MBV cargo is an important area of future study.
Supplementary Material
Disclosure Statement
No competing financial interests exist.
References
- 1.Keane T.J., Dziki J., Sobieski E., Smoulder A., Castleton A., Turner N., White L.J., and Badylak S.F. Restoring mucosal barrier function and modifying macrophage phenotype with an extracellular matrix hydrogel: potential therapy for ulcerative colitis. J Crohn's Colitis 11, 360–368, 2017 [DOI] [PubMed] [Google Scholar]
- 2.Alicuben E.T., and DeMeester S.R. Onlay ventral hernia repairs using porcine non-cross-linked dermal biologic mesh. Hernia 18, 705, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Seif-Naraghi S.B., Singelyn J.M., Salvatore M.A., Osborn K.G., Wang J.J., Sampat U., Kwan O.L., Strachan G.M., Wong J., Schup-Magoffin P.J. Braden R.L., Bartels K., DeQuach J.A., Preul M., Kinsey A.M., DeMaria A.N., Dib N., and Christman K.L. Safety and efficacy of an injectable extracellular matrix hydrogel for treating myocardial infarction. Sci Transl Med 5, 173ra25, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dziki J.L., Sicari B.M., Wolf M.T., Cramer M.C., and Badylak S.F. Immunomodulation and mobilization of progenitor cells by extracellular matrix bioscaffolds for volumetric muscle loss treatment. Tissue Eng Part A 22, 1129, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Agrawal V., Siu B.F., Chao H., Hirschi K.K., Raborn E., Johnson S.A., Tottey S., Hurley K.B., Medberry C.J., and Badylak S.F. Partial characterization of the Sox2+ cell population in an adult murine model of digit amputation. Tissue Eng Part A 18, 1454, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beattie A.J., Gilbert T.W., Guyot J.P., Yates A.J., and Badylak S.F. Chemoattraction of progenitor cells by remodeling extracellular matrix scaffolds. Tissue Eng Part A 15, 1119, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sicari B.M., Dziki J.L., Siu B.F., Medberry C.J., Dearth C.L., and Badylak S.F. The promotion of a constructive macrophage phenotype by solubilized extracellular matrix. Biomaterials 35, 8605, 2014 [DOI] [PubMed] [Google Scholar]
- 8.Brown B.N., Londono R., Tottey S., Zhang L., Kukla K.A., Wolf M.T., Daly K.A., Reing J.E., and Badylak S.F. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater 8, 978, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sadtler K., Estrellas K., Allen B.W., Wolf M.T., Fan H., Tam A.J., Patel C.H., Luber B.S., Wang H., Wagner K.R., Powell J.D., Housseau F., Pardoll D.M., and Elisseeff J.H. Developing a pro-regenerative biomaterial scaffold microenvironment requires T helper 2 cells. Science 352, 366, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valentin J.E., Stewart-Akers A.M., Gilbert T.W., and Badylak S.F. Macrophage participation in the degradation and remodeling of extracellular matrix scaffolds. Tissue Eng Part A 15, 1687, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., and Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol 25, 677, 2004 [DOI] [PubMed] [Google Scholar]
- 12.Daley J.M., Brancato S.K., Thomay A.A., Reichner J.S., and Albina J.E. The phenotype of murine wound macrophages. J Leukocyte Biology 87, 59, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahdavian Delavary B., van der Veer W.M., van Egmond M., Niessen F.B., and Beelen R.H. Macrophages in skin injury and repair. Immunobiology 216, 753, 2011 [DOI] [PubMed] [Google Scholar]
- 14.Arnold L., Henry A., Poron F., Baba-Amer Y., van Rooijen N., Plonquet A., Gherardi R.K., and Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med 204, 1057, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nahrendorf M., Swirski F.K., Aikawa E., Stangenberg L., Wurdinger T., Figueiredo J.L., Libby P., Weissleder R., and Pittet M.J. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 204, 3037, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mammoto T., Jiang E., Jiang A., and Mammoto A. Extracellular matrix structure and tissue stiffness control postnatal lung development through the lipoprotein receptor-related protein 5/Tie2 signaling system. Am J Respir Cell Mol Biol 49, 1009, 2013 [DOI] [PubMed] [Google Scholar]
- 17.Ruoslahti E. RGD and other recognition sequences for integrins. Ann Rev Cell Dev Biol 12, 697, 1996 [DOI] [PubMed] [Google Scholar]
- 18.Bissell M.J., and Aggeler J. Dynamic reciprocity: how do extracellular matrix and hormones direct gene expression? Prog Clin Biol Res 249, 251, 1987 [PubMed] [Google Scholar]
- 19.Gilbert T.W., Stolz D.B., Biancaniello F., Simmons-Byrd A., and Badylak S.F. Production and characterization of ECM powder: implications for tissue engineering applications. Biomaterials 26, 1431, 2005 [DOI] [PubMed] [Google Scholar]
- 20.Agrawal V., Tottey S., Johnson S.A., Freund J.M., Siu B.F., and Badylak S.F. Recruitment of progenitor cells by an extracellular matrix cryptic peptide in a mouse model of digit amputation. Tissue Eng Part A 17, 2435, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reing J.E., Brown B.N., Daly K.A., Freund J.M., Gilbert T.W., Hsiong S.X., Huber A., Kullas K.E., Tottey S., Wolf M.T., and Badylak S.F. The effects of processing methods upon mechanical and biologic properties of porcine dermal extracellular matrix scaffolds. Biomaterials 31, 8626, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huleihel L., Hussey G.S., Naranjo J.D., Zhang L., Dziki J.L., Turner N.J., Stolz D.B., and Badylak S.F. Matrix-bound nanovesicles within ECM bioscaffolds. Sci Adv 2, e1600502, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri A.A., So A.Y., Sinha N., Gibson W.S., Taganov K.D., O'Connell R.M., and Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol 187, 5062, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordes K.R., Sheehy N.T., White M.P., Berry E.C., Morton S.U., Muth A.N., Lee T.H., Miano J.M., Ivey K.N., and Srivastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460, 705, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parisi C., Napoli G., Amadio S., Spalloni A., Apolloni S., Longone P., and Volonte C. MicroRNA-125b regulates microglia activation and motor neuron death in ALS. Cell Death Differ 23, 531, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adammek M., Greve B., Kassens N., Schneider C., Bruggemann K., Schuring A.N., Starzinski-Powitz A., Kiesel L., and Gotte M. MicroRNA miR-145 inhibits proliferation, invasiveness, and stem cell phenotype of an in vitro endometriosis model by targeting multiple cytoskeletal elements and pluripotency factors. Fertil Steril 99, 1346–1355 e5, 2013 [DOI] [PubMed] [Google Scholar]
- 27.Lee Y.S., Kim H.K., Chung S., Kim K.S., and Dutta A. Depletion of human micro-RNA miR-125b reveals that it is critical for the proliferation of differentiated cells but not for the down-regulation of putative targets during differentiation. J Biol Chem 280, 16635, 2005 [DOI] [PubMed] [Google Scholar]
- 28.Xin M., Small E.M., Sutherland L.B., Qi X., McAnally J., Platom C.F., Richardson J.A. Bassel-Duby, R., and Olson, E.N. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev 23, 2166, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rangrez A.Y., Massy Z.A., Metzinger-Le Meuth V., and Metzinger L. miR-143 and miR-145: molecular keys to switch the phenotype of vascular smooth muscle cells, Circulation. Cardiovas Genet 4, 197, 2011 [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y., Zhang M., Zhong M., Suo Q., and Lv K. Expression profiles of miRNAs in polarized macrophages. Int J Mol Med 31, 797, 2013 [DOI] [PubMed] [Google Scholar]
- 31.Banerjee S., Cui H., Xie N., Tan Z., Yang S., Icyuz M., Thannickal V.J., Abraham E., and Liu G. miR-125a-5p regulates differential activation of macrophages and inflammation. J Biol Chem 288, 35428, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lantz G.C., Badylak S.F., Coffey A.C., Geddes L.A., and Blevins W.E. Small intestinal submucosa as a small-diameter arterial graft in the dog. J Invest Surg 3, 217, 1990 [DOI] [PubMed] [Google Scholar]
- 33.Kent O.A., McCall M.N., Cornish T.C., and Halushka M.K. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res 42, 7528, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu H., Lin H., Zhang L., Sun Q., Yuan G., Zhang L., Chen S., and Chen Z. miR-145 and miR-143 regulate odontoblast differentiation through targeting Klf4 and Osx genes in a feedback loop. J Biol Chem 288, 9261, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martinez F.O., Sica A., Mantovani A., and Locati M. Macrophage activation and polarization. Front Biosci 13, 453, 2013 [DOI] [PubMed] [Google Scholar]
- 36.Ying W., Cheruku P.S., Bazer F.W., Safe S.H., and Zhou B. Investigation of macrophage polarization using bone marrow derived macrophages. J Vis Exp 76, e50323, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez F.O., and Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime Rep 6, 13, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hachim D., LoPresti S.T., Yates C.C., and Brown B.N. Shifts in macrophage phenotype at the biomaterial interface via IL-4 eluting coatings are associated with improved implant integration. Biomaterials 112, 95, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hachim D., Wang N., Lopresti S.T., Stahl E.C., Umeda Y.U., Rege R.D., Carey S.T., Mani D., and Brown B.N. Effects of aging upon the host response to implants. J Biomed Mater Res A 105, 1281–1292, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dziki J.L., Wang D.S., Pineda C., Sicari B.M., Rausch T., and Badylak S.F. Solubilized extracellular matrix bioscaffolds derived from diverse source tissues differentially influence macrophage phenotype. J Biomed Mater Res Part A 105, 138, 2017 [DOI] [PubMed] [Google Scholar]
- 41.Brown B.N., Valentin J.E., Stewart-Akers A.M., McCabe G.P., and Badylak S.F. Macrophage phenotype and remodeling outcomes in response to biologic scaffolds with and without a cellular component. Biomaterials 30, 1482, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Badylak S.F., Valentin J.E., Ravindra A.K., McCabe G.P., and Stewart-Akers A.M. Macrophage phenotype as a determinant of biologic scaffold remodeling, Tissue Eng Part A 14, 1835, 2008 [DOI] [PubMed] [Google Scholar]
- 43.Wolf M.T., Dearth C.L., Ranallo C.A., LoPresti S.T., Carey L.E., Daly K.A., Brown B.N., and Badylak S.F. Macrophage polarization in response to ECM coated polypropylene mesh. Biomaterials 35, 6838, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavin Y., Mortha A., Rahman A., and Merad M. Regulation of macrophage development and function in peripheral tissues. Nat Rev Immunol 15, 731, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tidball J.G. Mechanisms of muscle injury, repair, and regeneration. Compr Physiol 1, 2029, 2011 [DOI] [PubMed] [Google Scholar]
- 46.Tidball J.G., and Villalta S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am J Physiology. Regul Integr Comp Physiol 298, R1173, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Novak M.L., and Koh T.J. Phenotypic transitions of macrophages orchestrate tissue repair. Am J Pathol 183, 1352, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tidball J.G., Dorshkind K., and Wehling-Henricks M. Shared signaling systems in myeloid cell-mediated muscle regeneration. Development 141, 1184, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deng B., Wehling-Henricks M., Villalta S.A., Wang Y., and Tidball J.G. IL-10 triggers changes in macrophage phenotype that promote muscle growth and regeneration. J Immunol 189, 3669, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guiteras R., Flaquer M., and Cruzado J.M. Macrophage in chronic kidney disease. Clin Kidney J 9, 765, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rogler G., Andus T., Aschenbrenner E., Vogl D., Falk W., Scholmerich J., and Gross V. Alterations of the phenotype of colonic macrophages in inflammatory bowel disease. Eur J Gastroenterol Hepatol 9, 893, 1997 [DOI] [PubMed] [Google Scholar]
- 52.Fadok V.A., Bratton D.L., Konowal A., Freed P.W., Westcott J.Y., and Henson P.M. Macrophages that have ingested apoptotic cells in vitro inhibit proinflammatory cytokine production through autocrine/paracrine mechanisms involving TGF-beta, PGE2, and PAF. J Clin Invest 101, 890, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piccoli M., Urbani L., Alvarez-Fallas M.E., Franzin C., Dedja A., Bertin E., Zuccolotto G., Rosato A., Pavan P., Elvassore N., De Coppi P., and Pozzobon M. Improvement of diaphragmatic performance through orthotopic application of decellularized extracellular matrix patch. Biomaterials 74, 245, 2016 [DOI] [PubMed] [Google Scholar]
- 54.Dearth C.L., Slivka P.F., Stewart S.A., Keane T.J., Tay J.K., Londono R., Goh Q., Pizza F.X., and Badylak S.F. Inhibition of COX1/2 alters the host response and reduces ECM scaffold mediated constructive tissue remodeling in a rodent model of skeletal muscle injury. Acta Biomater 31, 50, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tili E., Michaille J.J., Cimino A., Costinean S., Dumitru C.D., Adair B., Fabbri M., Alder H., Liu C.G., Calin G.A., and Croce C.M. Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179, 5082, 2007 [DOI] [PubMed] [Google Scholar]
- 56.Lee H.M., Kim T.S., and Jo E.K. MiR-146 and miR-125 in the regulation of innate immunity and inflammation. BMB Rep 49, 311, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin L., Hou J., Ma F., Wang P., Liu X. Li N., Wang J., Wang Q., and Cao X. Type I IFN inhibits innate IL-10 production in macrophages through histone deacetylase 11 by downregulating microRNA-145. J Immunol 191, 3896, 2013 [DOI] [PubMed] [Google Scholar]
- 58.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., and Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells, Cell 137, 647, 2009 [DOI] [PubMed] [Google Scholar]
- 59.Fu H., Yang G., Wei M., Liu L., Jin L., Lu X., Wang L., Shen L., Zhang J., Lu H., Yao L., and Lu Z. The RNA-binding protein QKI5 is a direct target of C/EBPalpha and delays macrophage differentiation. Mol Biol Cell 23, 1628, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang S., Zan J., Wu M., Zhao W., Li Z., Pan Y., Sun Z., and Zhu J. miR-29a promotes scavenger receptor A expression by targeting QKI (quaking) during monocyte-macrophage differentiation. Biochem Biophys Res Commun 464, 1, 2015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.