Abstract
Microvesicle- and exosome-mediated transport of microRNAs (miRNAs) represents a novel cellular and molecular pathway for cell–cell communication. In this study, we tested the hypothesis that these extracellular vesicles (EVs) and their miRNAs might change with age, contributing to age-related stem cell dysfunction. EVs were isolated from the bone marrow interstitial fluid (supernatant) of young (3–4 months) and aged (24–28 months) mice to determine whether the size, concentration, and miRNA profile of EVs were altered with age in vivo. Results show that EVs isolated from bone marrow are CD63 and CD9 positive, and the concentration and size distribution of bone marrow EVs are similar between the young and aged mice. Bioanalyzer data indicate that EVs from both young and aged mice are highly enriched in miRNAs, and the miRNA profile of bone marrow EVs differs significantly between the young and aged mice. Specifically, the miR-183 cluster (miR-96/-182/-183) is highly expressed in aged EVs. In vitro assays demonstrate that aged EVs are endocytosed by primary bone marrow stromal cells (BMSCs), and these aged EVs inhibit the osteogenic differentiation of young BMSCs. Transfection of BMSCs with miR-183-5p mimic reduces cell proliferation and osteogenic differentiation, increases senescence, and decreases protein levels of the miR-183-5p target heme oxygenase-1 (Hmox1). In vitro assays utilizing H2O2-induced oxidative stress show that H2O2 treatment of BMSCs increases the abundance of miR-183-5p in BMSC-derived EVs, and Amplex Red assays demonstrate that H2O2 is elevated in the bone marrow microenvironment with age. Together, these data indicate that aging and oxidative stress can significantly alter the miRNA cargo of EVs in the bone marrow microenvironment, which may in turn play a role in stem cell senescence and osteogenic differentiation by reducing Hmox1 activity.
Keywords: : bone, exosomes, miRNA, oxidative stress
Introduction
Aging is associated with a number of degenerative disorders that negatively impact the musculoskeletal system. Age-related bone loss in the form of osteoporosis is one of these age-related conditions that affects ∼10 million people in the United States, resulting in over 1.5 million bone fractures a year. It has been proposed that age-associated osteopenia and osteoporosis may result, in part, from underlying defects in bone stem cell populations, particularly bone marrow stromal cells (BMSCs).1–3 This hypothesis is further supported by additional studies showing that BMSCs from older donors have impaired proliferation, increased senescence, and reduced multipotency.4–6 These previous findings have important implications for tissue engineering and regenerative medicine since autologous stem cells are one potential source for tissue repair. The molecular mechanisms underlying these age-associated changes in the differentiation and proliferation of BMSCs are not well understood, but microRNAs (miRNAs) are thought to play an important role. miRNAs are noncoding RNAs (22–26 nucleotides (nt) in length) that are expressed in a tissue-specific and developmentally regulated manner and comprise 1% of the total number of genes in the animal genome. Accumulating evidence indicates that miRNAs regulate essential biological functions such as cellular differentiation, proliferation, and apoptosis, and miRNAs are becoming one of the most important gene regulators in eukaryotic organisms.7–9 The role of miRNAs in musculoskeletal development and aging is not yet entirely clear; however, stem cell-specific miRNAs have been identified in both humans and mice.10 In addition, specific miRNAs are associated with bone fractures and osteoporosis,11 and miRNAs have also been identified that play a critical role in the osteogenic differentiation of mesenchymal stem cells.12–16 These data therefore suggest that dysregulated miRNAs are likely to contribute to the significant loss in function observed in aged musculoskeletal tissues.
Extracellular vesicles (EVs), including microvesicles and exosomes, have recently been described as a new mechanism of cell–cell communication.17,18 Exosomes are small (40–100 nm) and microvesicles are larger (>100 nm) membrane-derived structures that are released into the extracellular space by a variety of cell types.19,20 These lipid-based carriers are now known to shuttle miRNAs between cells, delivering their miRNAs to target cells through endocytosis and membrane fusion.21 Microvesicle- and exosome-derived transport of miRNAs therefore represents one cellular and molecular pathway for epigenetic reprogramming.17 Cell–cell communication within the bone marrow microenvironment is complex and poorly understood. Moreover, the role of microvesicles and exosomes in this process has only recently been explored,22,23 and the role of these EVs in age-associated bone loss is not known. Microvesicles derived from human bone marrow stromal cells are enriched in miRNAs,21,23 and the majority of miRNAs in circulation are located within exosomes.24 We therefore sought to test the hypothesis that (1) EVs are abundant within bone marrow interstitial fluid, (2) miRNAs in the aging bone marrow microenvironment are in fact contained within membrane-derived EVs, (3) the expression profile of these miRNAs is altered with age, and (4) these age-associated changes can impact BMSCs.
Materials and Methods
Ex vivo isolation and characterization of EVs
Young (2–4 months) and aged (24–28 months) male C57BL6 mice, 8 mice per age group, were obtained from the National Institute on Aging, National Institutes of Health. Mice of this age and strain were selected because we and others have previously observed significant age-associated declines in bone mass and bone formation and increased bone resorption in these animals.25,26 We isolated EVs from bone marrow of mice using the total exosome isolation (TEI) reagent for serum (Life Technologies, Carlsbad, CA) reagent, precipitation, and centrifugation. This method is recognized to produce a higher purity, quality, and yield of EVs isolated from biological fluids when compared with sequential ultracentrifugation alone.27–30 Mice were euthanized by CO2 overdose, followed by thoracotomy, as approved by the AU Institutional Animal Care and Use Committee. The femur and tibia from both hind limbs were then dissected free and placed in 1000 μL ice-cold phosphate-buffered saline (PBS). Bones were then cleaned thoroughly of soft tissue, cut into pieces with scissors, and vortexed briefly for 15 s. Approximately 500 μL of PBS containing cells and interstitial fluid from the four hind limb bones was then pipetted out, placed in a separate tube, and centrifuged at 14,000 rpm at 4°C for 5 min. The supernatant (300–500 μL) was removed and then processed with the TEI reagent following the manufacturer's recommendations. Cells were retained for analysis of miRNA expression (see RNA isolation and miRNA profiling).
Immunogold labeling of EVs
Samples from two mice of each age group were utilized for transmission electron microscopy and immunogold labeling to validate our isolation approach. CD63 and CD9 are tetraspanins that are established surface markers for EVs, and samples were probed using antibodies diluted 1:100 to CD63 (Santa Cruz sc-15363, rabbit polyclonal) and CD9 (Santa Cruz sc-9148, rabbit polyclonal). Exosome samples were fixed in 4% paraformaldehyde, 2% glutaraldehyde in 0.1 M cacodylate buffer, pH 7.4, overnight. Twenty microliters of microvesicles was applied to a carbon–Formvar-coated 200-mesh copper grid and allowed to stand 30–60 s, and the excess wicked off onto Whatman filter paper. Grids were floated on drops of 1.4 nm anti-Rabbit nanogold (Nanoprobes, Inc., Yaphank, NY) diluted 1:1000 in blocking buffer for 1 h. Grids were enhanced 1 min in HQ Silver (gold enhancement reagent; Nanoprobes, Inc.) and rinsed in ice-cold DI H2O to stop enhancement and then negatively stained in 2% aqueous uranyl acetate and wicked dry. Grids were allowed to air-dry before being examined in a JEM 1230 transmission electron microscope (JEOL USA, Inc., Peabody, MA) at 110 kV and imaged with an UltraScan 4000 CCD camera and First Light Digital Camera Controller (Gatan, Inc., Pleasanton, CA.)
Nanoparticle tracking analysis
EV particle size and concentration were measured from six mice of each group using nanoparticle tracking analysis (NTA) with ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany30–32) and corresponding software ZetaView 8.02.28. Isolated exosome samples were appropriately diluted using 1× PBS buffer (Life Technologies, Carlsbad, CA) to measure the particle size and concentration. NTA measurement was recorded and analyzed at 11 positions. Before the measurement, the ZetaView system was calibrated using known concentrations of 100 nm polystyrene particles and temperature was maintained at 23°C.
RNA isolation and miRNA profiling
Total mRNA and miRNA were isolated from exosomes of six young and six aged mice following NTA using a Qiagen miRNeasy Kit according to the manufacturer's protocol. Up to 100 μL resuspended exosomes were proceeded with RNA isolation columns and buffers provided in the kit. A final volume of 50 μL RNA solution was collected from each sample in elution buffer. RNA concentration and quality were evaluated using Agilent 2100 Bioanalyzer (Santa Clara, CA) with RNA 6000 Pico kit at the Integrated Genomics Core of Georgia Cancer Center (www.augusta.edu/cancer/research/shared/genomics/). This specific kit is designed to measure RNA with concentration in the picogram per μL range. We also isolated miRNA from whole bone marrow cell lysates of old and young mice remaining after exosome isolation to analyze miR-183-5p expression. RNA concentration for each sample was determined using a Nanodrop ND 2000 and equal amounts of total RNA were reverse transcribed using the Qiagen miScript II RT Kit (Cat No. 218160). cDNA was then used as a template for real-time polymerase chain reaction (PCR) analysis using the miScript SYBR Green PCR Kit (Cat. No. 218073) and an iQ5 Real-Time PCR instrument (Bio-Rad). The PCR reactions were performed in duplicates using the Mouse miFinder miScript miRNA PCR Array (Qiagen, Cat. No. 331221 MIMM-001ZA). Levels of miRNA expression were calculated by the cycle threshold method using previously validated internal control genes within each assay plate. RNU6 (RNA, U6 small nuclear 2) and SNORD (small nucleolar RNA, C/D box) were used as normalization/internal control genes for miRNA expression. Expression differences among groups were assessed using analysis software made available by SABiosciences (pcrdataanalysis.sabiosciences.com/mirna/arrayanalysis.php?target=upload). TargetScan software was utilized to identify potential targets of those miRNAs found to be elevated in EVs with age. Analyses were performed in duplicate on pooled samples of three mice per age group. To confirm that our array findings were not specific to the C57BL6 mouse, we repeated the arrays in male CB6F1 (BALB/cBy x C57BL/6) mice. Exosome and miRNA isolation was performed as described above, and arrays performed in duplicate on pooled samples of three mice per age group. All other analyses in the study are performed using C57BL6 mice. Predicted targets of those miRNAs found to be altered with age were identified using the miRSystem platform (http://mirsystem.cgm.ntu.edu.tw/).
Isolation of mouse BMSCs
Mouse BMSCs were isolated according to procedures we have described previously.15,33,34 Briefly, mouse BMSCs were isolated from the femora of 6-month-old C57BL/6 mice (n = 6). The marrow was flushed with PBS and the marrow cells centrifuged, the supernatant discarded, and the pellet washed with PBS. Cells were plated in 100-cm2 culture plates with DMEM, supplemented with 10% heat-inactivated fetal bovine serum (FBS), 50 U/mL penicillin/streptomycin, and 2 mM l-glutamine. After 24 h, hematopoietic cell lineage cells were depleted using negative selection (T and B lymphocytic, myeloid, and erythroid cells) following manufacturer recommendations (BD Biosciences). BMSC lineage-positive fractions were collected as cells negative for CD3e (CD3 ɛ chain), CD11b (integrin αM chain), CD45R/B220, Ly-6G and Ly-6C (Gr-1), and TER-119/erythroid cells (Ly-76) and positive for antistem cell antigen-1 (Sca-1). Positive selection was performed using the Sca-1 column magnetic bead sorting kit (Miltenyi Biotec, San Diego, CA).
Imaging of EVs, osteogenic assay, and H2O2 treatment of BMSCs
EVs isolated from the bone marrow of four aged male C57BL6 mice were stained with the membrane dye PKH67 following manufacturer recommendations. Primary BMSCs from four young male C57BL6 mice (4–6 months, see isolation procedure described below) were treated with either 100 μg/mL of unlabeled or 100 μg/mL of PKH-67-labeled exosomes from aged mice. Cells were imaged using a Zeiss LSM 780 inverted confocal microscope with a 25 mW laser and 458 nm laser wavelength at 5.1% power. We also examined the effects of aged EVs on osteogenic differentiation of young BMSCs. Osteogenic culture was performed as described previously.15,33,34 BMSCs were trypsinized, harvested, washed, and plated in 24-well plates at a density of 5000 cells/cm2. The cells were allowed to adhere and grow to ∼90% confluence before being changed to an osteogenic medium (OM). The OM contained DMEM supplemented with 5% FBS, 50 U/mL penicillin/streptomycin, 10 nM dexamethasone, 0.25 mM l-ascorbic acid, and 10 mM β-glycerophosphate. Cells were treated with low (OS-L 1 μg/mL) or high (OS-H; 10 μg/mL) doses of aged EVs. Alkaline phosphatase (ALP) activity (and staining) was performed 3 and 6 days after the treatment is initiated. For ALP staining, cells are washed 10 days after treatment, fixed with 3.7% formaldehyde, then stained with SIGMA FAST BCIP/NBT-buffered substrate (B5655; Sigma-Aldrich) or fast red violet (86C-1KT, Sigma-Aldrich) according to the manufacturer's instructions. Images of stained wells were converted to grayscale and staining intensity quantified from six wells per treatment group based on pixel grayscale values. We performed a separate set of experiments with isolated BMSCs to determine the role of reactive oxygen species and oxidative stress in driving the expression of EV-derived mIRNAs. We treated young (6 months), male C57BL6 mouse BMSCs with low (100 μM) and high (200 μM) doses of hydrogen peroxide (H2O2) for 24 h since oxidative stress and reactive oxygen species are known to increase with age.35
Mir-183-5p transfection of primary mouse BMSCs and functional assays
Negative control (NC; #SI03650318, scrambled sequence) and miRNA mimics of miR-183-5p were purchased from QIAGEN. Lipofectamine 2000 was utilized for transfecting MSCs from male C57BL6 mice 3–6 months of age according to the manufacturer's instructions. Transfected cells were used for cell proliferation assay, senescence assay, and western blot. Real-time PCR for miR-183-5p expression, as described above, was used to validate successful transfection. PCR showed a significant (p < 0.01) increase in miR-183-5p expression in BMSCs following transfection (Table 1). For proliferation assay, BMSCs were seeded on 96-well plates at a density of 3000 cells/well. The cells were transfected with miR-183-5p mimic and NC miRNA (20 nM) for 48 h. The cell proliferation assay was performed using CyQuant® proliferation assay kit (Invitrogen) according to the manufacturer's instructions. Briefly, the cells were washed twice in PBS and the plate was then frozen at −70°C. After 24 h, the plates were thawed at room temperature. CyQuant reagent dye was added in a volume of 100 μL/well, incubated for 5 min protected from light at room temperature, and fluorescence signals (excitation 485 nm, emission 520 nm) detected using a fluorescence plate reader. Senescence cell staining kit (Cat No. CS0030; Sigma) was used to stain senescence cells in vitro, following the manufacturer's instructions. BMSCs were cultured in six-well plates and transfected with miR-183-5p and NC miRNA (20 nM) at 80% confluency. After 48 h, the cells were washed twice in PBS, fixed for 15–20 min with the fixation buffer, washed again with PBS, and incubated for 3–6 h at 37°C with β-gal solution provided by the kit. Following incubation, cells were imaged under standard light microscope at 40 × . Images were converted to grayscale and staining intensity quantified from six wells per treatment group based on pixel grayscale values. BMSCs isolated from 3-month-old C57BL6 mice were transfected with miR-183-5p mimic and NC miRNA as for the proliferation assays, seeded at 3000 cells per well, and cultured in osteogenic media (DMEM plus 50 μM ascorbic acid-2-phosphate, 10 mM β-glycerophosphate, and 10 nM dexamethasone) for 12 days and stained using Alizarin red S as previously described.33,34 Images of stained wells were converted to grayscale and staining intensity quantified from six wells per treatment group based on pixel grayscale values. Western blots from transfected cells were performed by first extracting protein from cell culture lysate, separated by SDS-PAGE, and transferred to nitrocellulose membranes. Membranes were incubated with antibodies against Hmox1 (AB13243; Abcam) and Gapdh (Santa Cruz Biotechnology, Santa Cruz) overnight at 4°C, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibody. Proteins were visualized with an ECL western blot detection system.
Table 1.
Real-Time Polymerase Chain Reaction Data Showing Fold Change in miR-183-5p Expression in BMSCs Following Transfection
| Treatment group | Fold change relative to control | p |
|---|---|---|
| Control (n = 4) | 1.15 ± 0.648 | 0.003 |
| miR-183 transfected (n = 4) | 107.54 ± 18.61 |
The housekeeping genes SNORD and RNU6 were used to quantify relative expression.
Amplex Red assay and Hmox 1 immunostaining
We analyzed H2O2 levels in flushed bone marrow aspirates from the left tibia and femur of five young (2–4 months) male C57BL6 mice and five old (20–24 months) C57BL6 mice using the Amplex Red hydrogen peroxide/peroxidase assay kit (A22188; Invitrogen) to detect age-associated changes in reactive oxygen species within the bone marrow microenvironment. A reaction mixture containing 100 μM Amplex® Red reagent and 0.2 U/mL HRP prepared in reaction buffer was added (50 μL) to the samples. Samples were incubated for 30 min at room temperature and absorbance was measured at ∼560 nm. Protein estimation was carried out to normalize hydrogen peroxide content using the Bio-Rad Quick Start Bio-Rad Protein Assay kit, which is based on the Bradford principle. Bones from the right hind limb of these mice were fixed in 4% paraformaldehyde, cryoprotected using sucrose overnight, frozen in OCT, and sectioned at 7–10 μm. Sections were stained using the Hmox1 antibody described above with Alexa546 Goat anti-rabbit secondary antibody (Invitrogen A1100), diluted 1:1000, and counterstained using 4′,6-diamidino-2-phenylindole (DAPI).
Statistical analyses
T-tests were used to compare RT-PCR values between aged and young animals. In some cases, the analyses were performed on the ranks of the data to reduce the influence of outlying observations.
Results
Isolation and immunogold labeling of EVs
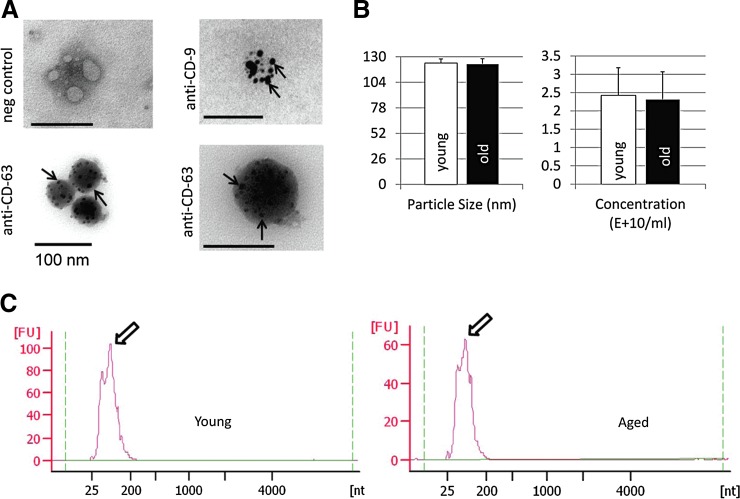
The combination of results from immunogold labeling (Fig. 1A) and NTA (Fig. 1B) indicates that EVs can be isolated successfully from bone marrow interstitial fluid (supernatant). These EVs are positive for CD9 and CD63, two well-established markers of EVs. These data also show that vesicles isolated from bones of young and old mice are in the 100–130 nm diameter size range, consistent with the known size of EVs (Fig. 1B). Bioanalyzer traces show that these vesicles are highly enriched in small RNAs, including miRNAs (Fig. 1C).
FIG. 1.
Isolation and characterization of old and young EVs. (A) Transmission electron microscope images of immunogold-labeled EVs isolated from mouse bone marrow supernatant showing positive labeling for the exosome markers, CD63 and CD9 (arrows). (B) ZetaView quantification of EV particle size (left) and concentration (right) from young (3–4 months) and aged (24–28 months) mouse bone marrow supernatant. N = 6 per group. (C) Representative Agilent 2100 Bioanalyzer electropherogram traces showing large number of small RNAs (arrows) from marrow-derived EVs. EV, extracellular vesicle; FU, fluorescence units; nt, nucleotides.
The miRNA profile of bone-derived EVs changes with age
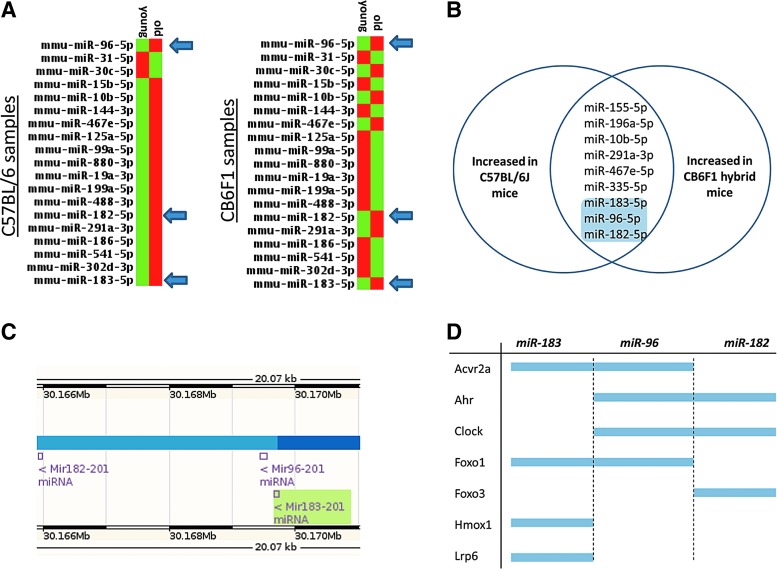
The size and concentration of EVs are similar between young and aged mice (Fig. 1B), but the miRNA content of the EVs differs between the two age groups (Fig. 2A). The miRNA content of aged EVs tends to be lower than in younger EVs (Fig 1C); however, nine miRNAs were found to be consistently upregulated in aged EVs compared with young EVs in both strains of mice (Fig. 2B). Importantly, three of these miRNAs, miR-96-5p, miR-182-5p, and miR-183-5p, are from the same functional cluster (Fig. 2C). The miR-183 family shares a similar location in the mouse genome (Fig. 2C) and shares a number of overlapping gene targets (Fig. 2D).
FIG. 2.
Expression of miR-183 cluster is increased with age in bone marrow-derived EVs. (A) PCR array from bone-derived EVs comparing miRNA expression between young and old C57BL/6 and CB6F1 hybrid mice (red indicates increased expression, green indicates a relative decrease). Analyses were performed in duplicate on pooled samples of three mice per age group per strain. (B) Venn diagram showing shared miRNAs upregulated with age between the two strains. (C) NCBI mapping of mouse chromosome 6 showing the clustered miR-183 family members miR-96, -182, and -183. (D) Members of the miR-183 cluster share overlapping predicted gene targets based on their nucleotide sequences.
Young BMSCs readily endocytose aged bone-derived EVs, and these vesicles suppress osteogenic differentiation
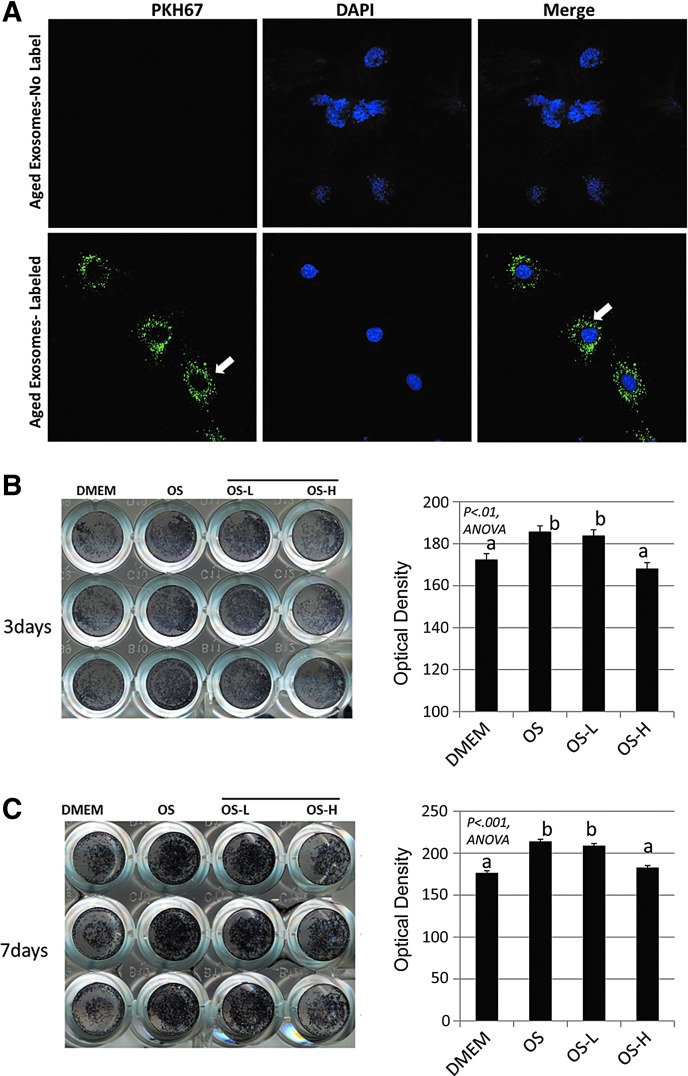
We isolated EVs from the bone marrow of aged mice and stained the EVs with the membrane dye PKH67. We then treated young (4–6 months) mouse BMSCs with these labeled exosomes and imaged the cells using confocal microscopy. Clusters of labeled exosomes are abundant in the cytosol of the BMSCs, evident by PKH67 staining (Fig. 3A), indicating that the cells readily take up these vesicles. Young primary BMSCs were also cultured in osteogenic media and treated with increasing concentrations (0, 1, and 10 μg/mL) of aged EVs. Dishes were stained for ALP activity after 3 and 7 days, and the results show that high-dose exosomes suppress the osteogenic differentiation of BMSCs (Fig. 3B, C).
FIG. 3.
Young mouse primary BMSCs endocytose aged EVs. (A) Young BMSCs treated with aged bone marrow EVs (100 μg/mL) labeled with the membrane dye PKH67 (bottom, green dye, white arrows; blue, DAPI) or unlabeled EVs (top). Confocal images show abundant EVs within the cytosol of BMSCs (arrows). Alkaline phosphatase staining of young (6 months) BMSCs treated with aged EVs at low (OS-L 1 μg/mL) or high (OS-H; 10 μg/mL) doses, stained after 3 days (B) and 6 days (C), showing stained wells (left panels) and quantification (right panels) illustrating decreased osteogenesis at the higher EV dose. N = 3 per dose per time point.
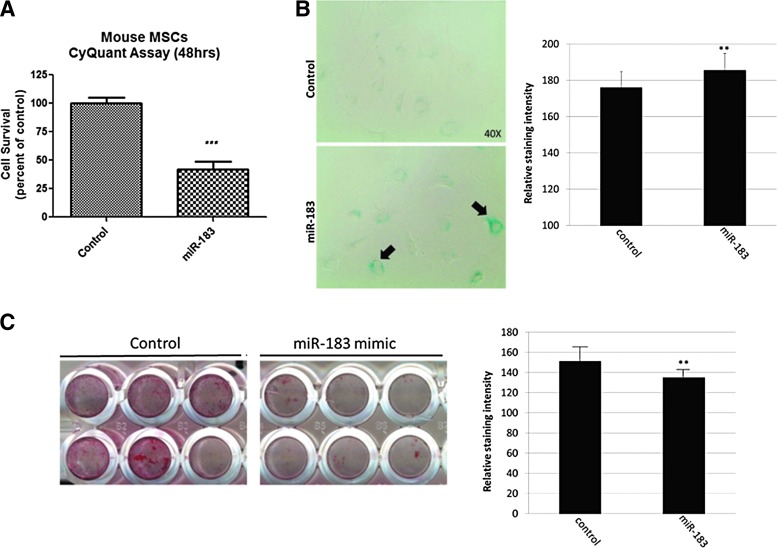
Transfection of BMSCs with miR-183-5p suppresses BMSC proliferation and mineralization, increases cell senescence, and decreases heme oxygenase-1 protein
We transfected young BMSCs with miR-183-5p mimic since we found this miRNA was elevated in aged EVs (Fig. 2), and aged EVs are readily taken up by young BMSCs (Fig. 3). CyQuant proliferation assays indicated that miR-183-5p decreases cell proliferation (Fig. 4A), and the senescence marker β-galactosidase was increased in BMSCs following miR-183-5p transfection (Fig. 4B). Alizarin red S staining of BMSCs transfected with control (scrambled) miRNA and miR-183-5p mimic shows that miR-183-5p suppresses the osteogenic differentiation of BMSCs (Fig. 4C).
FIG. 4.
miR-183-5p suppresses BMSC proliferation, induces cellular senescence, and inhibits osteogenesis. (A) CyQuant proliferation assay showing reduced BMSC proliferation following transfection with miR-183 mimic (***p < 0.001). (B) Increased β-galactosidase staining, a marker of senescence, following miR-183 transfection (left, black arrows) and quantification of staining intensity (right, **p < 0.01). (C) Alizarin red S staining following 12 days of BMSC culture in osteogenic media (left) and quantification of staining intensity (right, **p < 0.01).
miR-183-5p is increased with oxidative stress
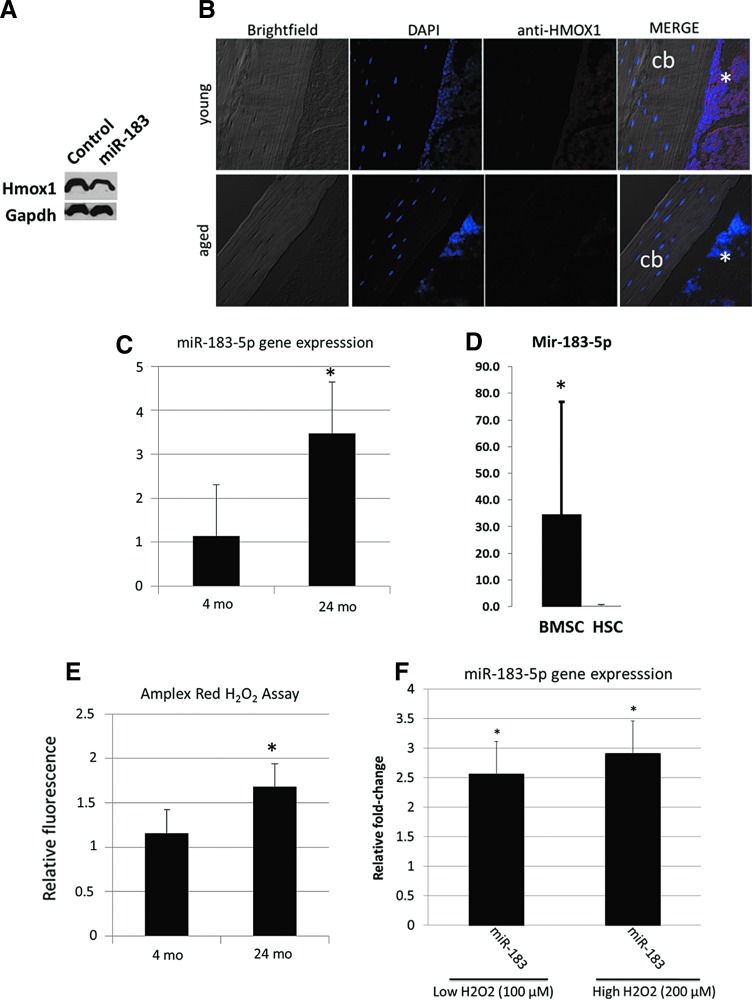
Western blots for the miR-183-5p target heme oxygenase-1 (Hmox1) show that miR-183-5p decreases Hmox1 protein (Fig. 5A), and immunostaining of mouse long bones indicates that Hmox1 protein is less abundant in bone marrow of aged mice compared with young mice (Fig. 5B). PCR data indicate that miR-183-5p is upregulated in bone marrow cell lysates (Fig. 5C) and is highly expressed in BMSCs compared with HSCs (Fig. 5D). Oxidative stress is known to increase mir-183 expression and we measured levels of hydrogen peroxide in bone marrow using Amplex Red. These data show elevated H2O2 in marrow of aged mice (Fig. 5E). We also treated young BMSCs with hydrogen peroxide (H2O2) to further evaluate the role of oxidative stress in stimulating miR-183-5p expression. These experiments demonstrate that H2O2 increases miR-183-5p expression at both high and low doses (Fig. 5F).
FIG. 5.
miR-183-5p reduces Hmox1 in BMSCs and miR-183 is increased with oxidative stress. (A) Western blot showing reduced Hmox1 in BMSCs after miR-183-5p transfection. (B) Immunostaining for Hmox1 (asterisks) in the long bones of young (top) and aged (bottom) mice. (C) miR-183-5p expression increases with age in mouse bone marrow cells. (D) Primary BMSCs from aged mice show higher miR-183-5p expression than HSCs, suggesting that BMSCs are the source of EV-derived miR-183. (E) Amplex Red assay showing that H2O2 levels increased with age in mouse bone marrow, N = 5 mice per age group. (F) Treatment of young mouse BMSCs with H2O2 increases expression of miR-183 at both high and low doses. N = 5 per group. *p < 0.05. cb, cortical bone; HSCs, hematopoietic stem cells.
Discussion
A role for EV-derived proteins, mRNAs, and miRNAs in cell–cell communication is becoming better understood; however, there are, to our knowledge, no studies that have yet identified specific miRNAs transported through EVs within the aging bone marrow microenvironment in vivo. In vitro studies have recently shown that osteoblasts shed microvesicles carrying RANKL protein, and these microvesicles can facilitate osteoclast development.36 It has also been proposed that matrix vesicles within the growth plate are exosomal structures and may play a key role in cell–cell communication during the process of endochondral ossification.37 MC3T3 osteoblasts were recently observed to secrete exosomes that stimulate osteogenic differentiation of ST2 stromal cells,38 and BMSCs secrete exosomes carrying miRNAs that promote tumor growth in multiple myeloma.39 Our data expand upon these previous studies by showing that exosomes and microvesicles are abundant within bone marrow interstitial fluid and carry a number of different miRNAs, some of which change with age. Parabiosis experiments suggest that a variety of circulating factors differ between young and old animals,40 and it has recently been proposed that microvesicular miRNAs may also change with age.41 Our findings indicate that at least in bone marrow, this does appear to be the case. The local microenvironment is recognized to play a key role in modulating stem cell function.6 Future studies could be directed at identifying additional EV-derived miRNAs and proteins that are altered with age in various tissues and may therefore be involved in regulating stem cell survival, proliferation, and differentiation.
The miRNAs that we determined were altered with age in bone-derived EVs and have been demonstrated in previous studies to play a role in suppressing osteogenesis and inducing cellular senescence. For example, miR-182-5p has previously been shown to suppress osteoblast proliferation and differentiation by targeting Foxo1,42 and miR-183-5p is increased during cellular senescence after exposure to oxidative stress.43 miR-183-5p targets Foxo1 in humans, but not in mice,44 whereas luciferase reporters have validated Hmox-1 as an miR-183-5p target in multiple cell types.45,46 Hmox1 has been shown previously to promote the growth and osteogenic differentiation of BMSCs.47 Our results showing that miR-183-5p increases with aging, suppresses osteogenic differentiation in BMSCs, and reduces Hmox1 levels are consistent with these previous findings. The role of miR-183-5p in age-related bone loss may not be restricted to its impact on bone formation as this miRNA also appears to stimulate bone resorption by suppressing Hmox1 and increasing osteoclastogenesis.45 Indeed, the miR-183 cluster is highly conserved in vertebrates and is now recognized to play a role in a number of human diseases.48 Hmox1 is a key component of the cellular response to oxidative stress,49 and so our findings are consistent with the idea that elevated levels of oxidative stress within the bone marrow microenvironment contribute to age-related bone loss.50,51 Additional studies in mice have shown that miR-183 increases with age in serum, but is reduced with calorie restriction,52 and miR-183-5p and miR-96-5p are both downregulated in long-lived centenarians compared with those individuals with shorter life spans.53 It is therefore possible that circulating EV-derived miRNAs such as miR-183 may ultimately serve as useful biomarkers for age-associated senescence, stem cell dysfunction, and oxidative stress.
Acknowledgments
Dr. Brendan Marshall and Ms. Penny Roon and Ms. Libby Perry in the AU Histology and Electron Microscopy Core provided expertise with immunogold labeling and transmission electron microscopy. Dr. Paul McNeil and Mr. Tim Kurtz in the AU Cell Imaging Core assisted with confocal imaging, and Dr. Gabor Csanyi provided helpful suggestions on the Amplex Red assay. The authors are grateful to Dr. Meghan McGee-Lawrence and Michael Yu for assistance with immunostaining. Funding for this research was provided by the National Institute on Aging (P01 AG036675) and supported, in part, by the Department of Veterans Affairs (VA Merit Award 1I01CX000930-01, W.D.H.). The contents of this publication do not represent the views of the Department of Veterans Affairs or the US Government.
Disclosure Statement
No competing financial interests exist.
References
- 1.Bonyadi M., Waldman S.D., Liu D., Aubin J.E., Grynpas M.D., and Stanford W.L. Mesenchymal progenitor self-renewal deficiency leads to age-dependent osteoporosis in Sca-1/Ly-6A null mice. Proc Natl Acad Sci USA 100, 5840, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moerman E., Teng K., Lipschitz D.A., and Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell 3, 79, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miura Y., Miura M., Gronthos S., Allen M., Cao C., Uveges T.E., Bi Y., Ehirchiou D., Kortesidis A., Shi S., and Zhang L. Defective osteogenesis of the stromal stem cells predisposes CD18-null mice to osteoporosis. Proc Natl Acad Sci U S A 102, 14022, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beane S., Fonseca V.C., Cooper L., Koren G., and Darling E. Impact of aging on the regenerative properties of bone marrow-, muscle-, and adipose-derived mesenchymal stem/stromal cells. PLoS One 9, e115963, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fafian-Labora J., Fernandez-Pernas P., and Fuentes I. Influence of age on rat bone-marrow mesenchymal stem cells potential. Sci Rep 5, 16765, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sui B., Hu C., Zheng C., and Jin Y. Microenvironmental views on mesenchymal stem cell differentiation in aging. J Dent Res 95, 1333, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Hatfield S., Shcherbata H., Fischer K., Nakahara K., Carthew R.W., and Ruohola-Baker H. Stem cell division is regulated by the microRNA pathway. Nature 435, 974, 2005 [DOI] [PubMed] [Google Scholar]
- 8.He X., Eberhart J., and Postlethwait J. MicroRNAs and micromanaging the skeleton in disease, development, and evolution. J Cell Mol Med 13, 606, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plasterk R. MicroRNAs in animal development. Cell 124, 877, 2006 [DOI] [PubMed] [Google Scholar]
- 10.Schoolmeesters A., Eklund T., Leake D., et al. . Functional profiling reveals critical role for miRNA in differentiation of human mesenchymal stem cells. PLoS One 4, e5605, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seeliger C., Karpinski K., Haug A., Vester H., Schmitt A., et al. . Five freely circulating miRNAs and bone tissue miRNAs are associated with osteoporotic fractures. J Bone Miner Res 29, 1718, 2014 [DOI] [PubMed] [Google Scholar]
- 12.Lian J., Stein G., van Wijnen A., Stein J., Hassan M., et al. . MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol 8, 212, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suomi S., Taipaleenmäki H., Seppänen A., et al. . MicroRNAs regulate osteogenesis and chondrogenesis of mouse bone marrow stromal cells. Gene Regul Syst Bio 22, 177, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Okamoto H., Matsumi Y., Hoshikawa Y., Takub K., et al. . Involvement of microRNAs in regulation of osteoblastic differentiation in mouse induced pluripotent stem cells. PLoS One 7, e43800, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sangani R., Periyasamy-Thandavan S., Kolhe R., et al. . MicroRNAs-141 and 200a regulate the SVCT2 transporter in bone marrow stromal cells. Mol Cell Endocrinol 410, 19, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu W., and Kassem M. miR-141-3p inhibits human stromal (mesenchymal) stem cell proliferation and differentiation. Biochim Biophys Acta 1843, 2114, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Camussi G., Deregibus M.C., Bruno S., et al. . Exosome/microvesicle-mediated epigenetic reprogramming of cells. Am J Cancer Res 1, 98, 2011 [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H., Ekström K., Bossios A., et al. . Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654, 2007 [DOI] [PubMed] [Google Scholar]
- 19.Mathivanan S., Ji H., and Simpson R. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 73, 1907, 2010 [DOI] [PubMed] [Google Scholar]
- 20.Vickers K., and Remaley A. Lipid-based carriers of microRNAs and intercellular communication. Curr Opinion Lipidol 23, 91, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Collino F., Deregibus M., Bruno S., et al. . Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One 5, e11803, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J., Faict S., Maes K., et al. . Extracellular vesicle cross-talk in the bone marrow microenvironment: implications in multiple myeloma. Oncotarget 7, 38927, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanghavi P, Young P., Upadhyay S., and Hamrick M.W. Exosomes for bone diseases. In: Tang Y., and Dawn B., eds. Mesenchymal Stem Cell Derived Exosomes: The Potential for Translational Nanomedicine. New York: Elsevier, Ch. 10, 2015, pp. 213–227. [Google Scholar]
- 24.Gallo A., Tandon M., Alevizos I., and Illei G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 7, e30679, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamrick M.W., Ding K., Pennington C., et al. . Age-related loss of muscle mass and bone strength in mice is associated with a decline in physical activity and serum leptin. Bone 39, 845, 2006 [DOI] [PubMed] [Google Scholar]
- 26.Ferguson V., Ayers R., Bateman T., and Simske S. Bone development and age-related bone loss in male C57BL/6J mice. Bone 33, 387, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Caradec J., Kharmate G., Hosseini-Beheshti F., Adomat H., et al. . Reproducibility and efficiency of serum-derived exosome extraction methods. Clin Biochem 47, 1286, 2014 [DOI] [PubMed] [Google Scholar]
- 28.Wang J., Yao Y., Wu J., and Li G. Identification and analysis of exosomes secreted from macrophages extracted by different methods. Int J Clin Exp Pathol 8, 6135, 2015 [PMC free article] [PubMed] [Google Scholar]
- 29.Rider M., Hurwitz S., and Meckes D., Jr ExtraPEG: a polyethylene glycol-based method for enrichment of extracellular vesicles. Sci Rep 6, 23978, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Helwa I., Cai J., Drewry M.D., Zimmerman A., Dinkins M.B., Khaled M.L., et al. . A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One 12, e0170628, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alegre E., Rebmann V., Lemaoult J., et al. . In vivo identification of an HLA-G complex as ubiquitinated protein circulating in exosomes. Eur J Immunol 43, 1933, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Kordelas L., Rebmann V., Ludwig A., et al. . MSC-derived exosomes: a novel tool to treat therapy-refractory graft-versus-host disease. Leukemia 28, 970, 2014 [DOI] [PubMed] [Google Scholar]
- 33.Zhang W., Hamrick M.W., Hill W.D., Borke J., Wenger K., Chutkan N., Yu J., Mi Q.-S., Isales C.M., and Shi X. Age-related changes in the osteogenic differentiation potential of mouse bone marrow mesenchymal stem cells. J Bone Miner Res 23, 1118, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowser M., Herberg S., Arounleut P., Shi X., Fulzele S., Hill W.D., Isales C.M., and Hamrick M.W. Effects of the activin A-myostatin-follistatin system on aging bone and muscle progenitor cells. Exp Gerontol 48, 290, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkel T., and Holbrook N. Oxidants, oxidative stress, and the biology of ageing. Nature 408, 238, 2000 [DOI] [PubMed] [Google Scholar]
- 36.Deng L., Wang Y., Peng P., et al. . Osteoblast-derived microvesicles: a novel mechanism for communication between osteoblasts and osteoclasts. Bone 79, 37, 2015 [DOI] [PubMed] [Google Scholar]
- 37.Shapiro I., Landis W., and Risbud M. Matrix vesicles: are they anchored exosomes? Bone 79, 29, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cui Y., Luan J., Li H., Zhou X., and Han J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett 590, 185, 2016 [DOI] [PubMed] [Google Scholar]
- 39.Roccaro A., Sacco A., Maiso P., et al. . BM mesenchymal stromal cell-derived exosomes facilitate multiple myeloma progression. J Clin Invest 123, 1542, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Conboy I., Conboy M., Wagers A., et al. . Rejuvenation of aged progenitor cells byexposure to a young systemic environment. Nature 433, 760, 2005 [DOI] [PubMed] [Google Scholar]
- 41.Weilner S., Schraml E., Redl H., et al. . Secretion of microvesicular miRNAs in cellular and organismal aging. Exp Gerontol 48, 626, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim K., Park S., Jung S., Kim E., Jogeswar G., Ajita J., Rhee Y., Kim C., and Lim S. miR-182 is a negative regulator of osteoblast proliferation, differentiation, and skeletogenesis through targeting FoxO1. J Bone Miner Res 27, 1669, 2012 [DOI] [PubMed] [Google Scholar]
- 43.Li G., Luna C., Qiu J., Epstein D., and Gonzalez P. Alterations in microRNA expression in stress-induced cellular senescence. Mech Ageing Dev 130, 731, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McLoughlin H., Wan J., Spengler R., Xing Y., and Davidson B. Human-specific microRNA regulation of FOXO1: implications for microRNA recognition element evolution. Hum Mol Genet 23, 2593, 2014 [DOI] [PubMed] [Google Scholar]
- 45.Ke K., Sul O., Rajasekaran M., and Choi H. MicroRNA-183 increases osteoclastogenesis by repressing heme oxygenase-1. Bone 81, 237, 2015 [DOI] [PubMed] [Google Scholar]
- 46.Chang C., Au L., Huang S., Fai Kwok, C., Ho L., and Juan C. Insulin up-regulates heme oxygenase-1 expression in 3T3-L1 adipocytes via PI3-kinase- and PKC-dependent pathways and heme oxygenase-1-associated microRNA downregulation. Endocrinology 152, 384, 2011 [DOI] [PubMed] [Google Scholar]
- 47.Vanella L., Sanford C., Kim D.H., Abraham N.G., and Ebraheim N. Oxidative stress and heme oxygenase-1 regulated human mesenchymal stem cells differentiation. Int J Hypertens 2012, 890671, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dambal S., Shah M., Mihelich B., and Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Res 43, 7173, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bayram B., Nikolai S., Huebbe P., Ozcelik B., Grimm S., Grune T., Frank J., and Rimbach G. Biomarkers of oxidative stress, antioxidant defence and inflammation are altered in the senescence-accelerated mouse prone 8. Age (Dordr) 35, 1205, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Almeida M., Han L., Martin-Millan M., et al. . Skeletal involution by age-associated oxidative stress and its acceleration by loss of sex steroids. J Biol Chem 282, 27285, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kousteni S. FoxO1: a molecule for all seasons. J Bone Miner Res 26, 912, 2011 [DOI] [PubMed] [Google Scholar]
- 52.Dhahbi J., Spindler S., Atamna H., Yamakawa A., Guerrero N., Boffelli D., Mote P., and Martin D.I. Deep sequencing identifies circulating mouse miRNAs that are functionally implicated in manifestations of aging and responsive to calorie restriction. Aging (Albany NY) 5, 130, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.ElSharawy A., Keller A., Flachsbart F., Wendschlag A., Jacobs G., Kefer N., Brefort T., Leidinger P., Backes C., Meese E., Schreiber S., Rosenstiel P., and Franke A. Genome-wide miRNA signatures of human longevity. Aging Cell 11, 607, 2012 [DOI] [PubMed] [Google Scholar]