Abstract
Extracellular vesicles (EVs) derived from human bone marrow mesenchymal stromal cells (MSCs) promote the regeneration of kidneys in different animal models of acute kidney injury (AKI) in a manner comparable with the cells of origin. However, due to the heterogeneity observed in the EVs isolated from MSCs, it is unclear which population is responsible for the proregenerative effects. We therefore evaluated the effect of various EV populations separated by differential ultracentrifugation (10K population enriched with microvesicles and 100K population enriched with exosomes) on AKI recovery. Only the exosomal-enriched population induced an improvement of renal function and morphology comparable with that of the total EV population. Interestingly, the 100K EVs exerted a proproliferative effect on murine tubular epithelial cells, both in vitro and in vivo. Analysis of the molecular content from the different EV populations revealed a distinct profile. The 100K population, for instance, was enriched in specific mRNAs (CCNB1, CDK8, CDC6) reported to influence cell cycle entry and progression; miRNAs involved in regulating proliferative/antiapoptotic pathways and growth factors (hepatocyte growth factor and insulin-like growth factor-1) that could explain the effect of renal tubular cell proliferation. On the other hand, the EV population enriched in microvesicles (10K) was unable to induce renal regeneration and had a molecular profile with lower expression of proproliferative molecules. In conclusion, the different molecular composition of exosome- and microvesicle-enriched populations may explain the regenerative effect of EVs observed in AKI.
Keywords: : adult stem cells, mesenchymal stem cells, kidney, exosomes, microvesicles
Introduction
Stem cells are important in the maintenance of tissue homeostasis and restoration after injury. One of the mechanisms by which they can influence the behavior of injured cells is through secreting soluble factors and extracellular vesicles (EVs). In particular, EVs carry a broad range of bioactive molecules, including proteins, nucleic acids (mRNA, miRNA, lncRNA, DNA), and lipids, which can act on different cell types triggering a proproliferative and/or proregenerative program (angiogenesis, immune tolerance, etc).1–5 For instance, mRNA and miRNA delivered by EVs released from endothelial progenitor cells (EPCs) induce endothelial cell proliferation and revascularization of injured murine tissue.6–8 Furthermore, they can also mediate a robust proangiogenic program and wound healing effects in streptozotocin-induced diabetic rats through activation of the Erk1/2 signaling pathway.8 In a model of heart injury, exosomes derived from human cardiac progenitor cells inhibit apoptosis and increase cardiomyocytes and endothelial cell proliferation.9
Among the different stem cells extensively employed in research and clinical practice, mesenchymal stromal cells (MSCs) are the most widely used.10 MSCs isolated from different adult tissues (bone marrow, adipose tissue, or neonatal umbilical cord) possess immunomodulatory characteristics and migrate to injured tissues secreting factors and EVs.11,12 For instance, human umbilical cord-derived MSC-EVs have been shown to reduce fibrosis and collagen deposition in a model of drug-induced liver injury.13 Moreover, in a model of hypoxia-induced pulmonary hypertension, MSC-EVs mediated the reduction of inflammation and enhanced the recovery of injured lung tissue.14,15
The regenerative potential of mesenchymal stem cell-derived EVs (MSC-EVs) has also been investigated in different renal injury models. MSC-EVs improved survival in mice treated with cisplatin and reduced the incidence of chronic kidney disease (CKD).16 In glycerol-induced acute kidney injury (AKI), a single administration of MSC-EVs accelerated functional and histological recovery by delivery of selected miRNAs and mRNAs.17,18 AKI and development CKD were also prevented in an ischemia and reperfusion injury (IRI) model through the administration of a single dose of MSC-EVs.19 However, like other stem cells, MSCs also secrete various populations of EVs that shuttle a wide range of molecules, which exhibit different effects on target cells.20 Currently, a lot of attention is being focused on the characterization of stem cell EVs together with the cargo they carry.18,21,22 However, many omic studies are addressed only on the exosomal fraction of EVs. For instance, De Luca et al. characterized the miRNA and piRNA content of bone marrow MSC exosomes and demonstrated that the RNA content can modify the fate of umbilical cord blood-derived CD34+ cells.23 Furthermore, protein and miRNA carried by MSC-derived exosomes have also been shown to possess tumor regulatory capacity.24
In the present study, we evaluated the effect of MSC-derived exosomes and microvesicles on AKI recovery. For this purpose, we purified the 10K and 100K fractions of EVs by differential ultracentrifugation and correlated their in vivo and in vitro biological effects observed with their molecular composition.
Materials and Methods
Cell cultures
Bone marrow MSCs were obtained from Lonza and cultured and characterized as previously described.17 MSCs derived from five preparations were used up to the sixth passage of culture. All of the cell preparations used were positive for the typical MSC markers (CD105, CD29, CD73, CD44, and CD90) and had the ability to differentiate into adipogenic and osteogenic lineages (not shown).
Murine tubular epithelial cells (mTECs) were obtained as previously described.17 The cells were maintained in DMEM (Euroclone) supplemented with 10% fetal calf serum (Euroclone) and were positive for typical mTEC markers, such as cytokeratin, alkaline phosphatase, and aminopeptidase A, and negative for endothelial (von Willebrand factor), hematopoietic (CD45), and glomerular (nephrin) markers (not shown).
Isolation and characterization of different fractions of MSC-EVs
EVs were obtained from supernatants of MSCs cultured overnight in RPMI deprived of FCS. The viability of MSCs at the time of EV collection was 99% as determined by trypan blue exclusion. After removal of cell debris and apoptotic bodies by centrifugation at 3000 g for 20 min, EVs were purified by different steps of ultracentrifugation:
(1) 100KTOT-EVs were purified by 2-h ultracentrifugation at 100,000 g at 4°C (Beckman Coulter Optima L-90K; Fullerton);
(2) 10K-EVs were purified by 2-h ultracentrifugation at 10,000 g at 4°C;
(3) 100K-EVs were purified by 2-h ultracentrifugation at 100,000 g at 4°C of supernatant deprived of 10K-EVs.
EVs from the different fractions were used freshly or stored at −80°C after resuspension in RPMI supplemented with 1% dimethyl sulfoxide (DMSO). Analysis of size distribution and enumeration of EVs were performed using NanoSight LM10 (NanoSight Ltd.) equipped with a 405 nm laser and Nanoparticle Tracking Analysis (NTA) 2.3 software. Using a laser light source, particles in the sample are illuminated and the scattered light is captured by the camera and displayed on the connected computer running NTA. Using NTA, the particles are automatically tracked and sized based on Brownian motion and the diffusion coefficient. Three videos of 30 s were recorded to perform the analyses. EVs were characterized by cytofluorimetric analysis using FITC, PE, or APC-conjugated antibodies against CD73, CD44, CD105, and CD29. Conjugated mouse nonimmune isotypic IgG (Miltenyi Biotec) was used as control. Briefly, EVs (1.5 × 108 particles) were incubated at 4°C for 15 min with the antibodies, then diluted 1:3 and acquired immediately as described previously.25 Samples were acquired using Guava easyCyte Flow Cytometer (Millipore) and analyzed with InCyte software.
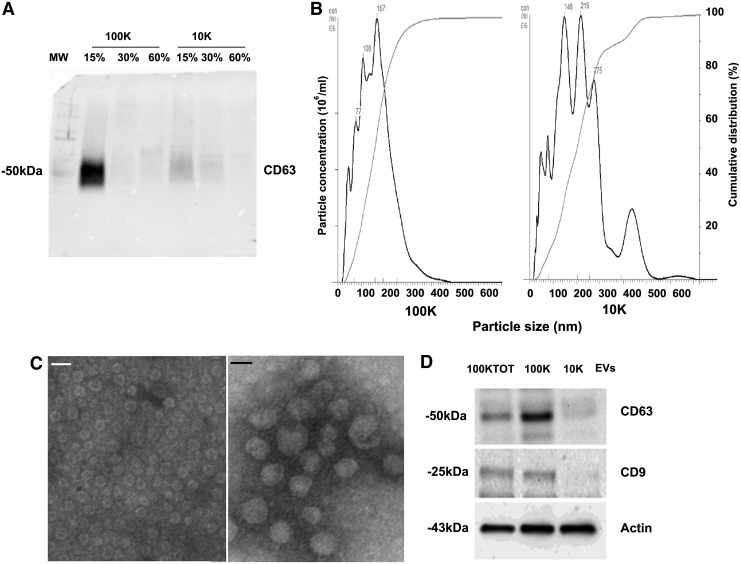
In selected experiments, the EV pellet obtained by differential ultracentrifugation was further purified using an iodixanol (Optiprep from Sigma) floating separation protocol. Briefly, the pellet was directly resuspended in 500 μL of 60% iodixanol mixed with 0.25 M sucrose and transferred to an ultracentrifuge tube (Beckman Instruments). Various gradients of iodixanol, 30%, 15%, and 5%, were than sequentially layered on top of the EV/60% iodixanol prep. A final volume of 10 mL was than obtained by adding saline solution on top of the 5% iodixanol fraction. The tubes were than ultracentrifuged at 350,000 g for 1 h at 4°C without breaking in an Optima L-100K ultracentrifuge (Beckman Coulter) equipped with Type 90Ti rotor. The 15%, 30%, and 60% fractions were recovered, diluted with PBS, and ultracentrifuged again at 100,000 g for 1 h at 4°C. The pellet obtained was resuspended in PBS with 1% DMSO and frozen or used for subsequent studies. NanoSight analyses revealed that vesicles recovered by floating were mainly detected in the 15% fraction (70% of EVs) and to a lower extent in the 30% fraction (30% of EVs). Furthermore, Western blot analysis showed that the exosomal marker CD63 was mainly detected in the 15% fraction (Fig. 1A).
FIG. 1.
Characterization of different EV populations obtained by differential ultracentrifugation. (A) Representative Western blot analysis of CD63 exosomal marker expression in 100K and 10K EVs submitted to floating separation by iodixanol density gradient (see Materials and Methods section). (B) NanoSight representative images of 100K (left) and 10K (right)-EV populations. (C) Representative micrographs of transmission electron microscopy of 100K (left) and 10K (right)-EVs negatively stained with NanoVan (scale bars = 100 nm; original magnification × 100,000). (D) Representative Western blot analysis of exosomal markers (CD63 and CD9) in the EV purified populations (30 μg of proteins/well). Equal loading was confirmed by stripping and reprobing the immunoblot for actin. EVs, extracellular vesicles.
RNA extraction and mRNA analysis
RNA was extracted from different EV populations using the All-in-One kit (Norgen) according to the manufacturer's protocol. Human Cell Cycle RT2 Profiler™ PCR Array (SABioscience-Qiagen) was performed to profile the expression of 84 genes related to cell cycle regulation.
To confirm the mRNA expression of selected genes, cDNA was obtained using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Five nanograms of cDNA and 200 nM of each primer (Table 1) were added to 1 × SYBR GREEN PCR Master Mix (Applied Biosystems) and run on a 96-well StepOne quantitative real-time polymerase chain reaction (qRT-PCR) system (Applied Biosystems). GAPDH was used as a housekeeping gene.
Table 1.
List of Primers
| Gene | Forward primer | Reverse primer |
|---|---|---|
| HGF | CACACAGGTATATTTGCTGGATGATAA | GGGACACAAGGTATAAATTGTTTTGG |
| IGF-147 | GCTCTTCAGTTCGTGTGTGGA | CGACTGCTGGAGCCATACC |
| CDK8 | AAGAGGAAAGATGGGAAGGATGA | TTGGATGCTTAAGCTCTCGAAGT |
| CDC6 | AAGCAAGAAGGCACTTGCTACC | ACAGATGTGTTCCCTCAAGAAATTC |
| Ciclin B1 | GACAGATGGAGATGAAGATTCTCAGA | ACGTCAACCTCTCCGACTTTAGA |
| TP53 | CCTGAGGTTGGCTCTGACTGTA | TGTTCCGTCCCAGTAGATTACCA |
CDK, cyclin-dependent protein kinase; HGF, hepatocyte growth factor; IGF-1, insulin-like growth factor 1.
Small RNA sequencing
Small RNA libraries for micro-RNA sequencing were prepared using the TruSeq Small RNA Library Preparation Kit according to the manufacturer's protocol. The size of the library was checked by using a Fragment Analyzer instrument. Samples were sequenced on an Illumina HiScanSQ platform (Illumina). Sequencing reads were trimmed out of the low-quality bases and adapter clipped with Fastx Toolkit. Filtered sequences were mapped and quantified by using miRDeep2 software26 on the most updated version of the human miRBase database.27 For downstream analysis, miRNAs with RPM <100 in all the samples were filtered out. miRNAs with a fold change of log2(FC) ≤1 and log2(FC) ≥1 were considered as upregulated or downregulated in the different populations, respectively.
miRNA prediction and network analysis
Target prediction and biological process enrichment analysis was performed using the FunRich analysis tool.28 Only biological processes of selected genes showing a p-value <0.05 were considered as significantly enriched. Network analysis to define the associated pathways of the 100K and 10K miRNA predictive genes was conducted using the FunRich interaction plug-in (p-value <0.01 in the enriched pathways). A filter of a minimum of six interactions among different nodes was applied.
Protein expression analysis
Proteins were extracted from different EV populations by RIPA buffer (20 nM Tris-HCl, 150 nM NaCl, 1% deoxycholate, 0.1% sodium dodecyl sulfate, 1% Triton X-100, pH 7.8) supplemented with a cocktail of protease and phosphatase inhibitors (Sigma-Aldrich). The protein content was quantified by BCA Protein Assay Kit (Pierce, Thermo Fisher Scientific). Thirty micrograms of proteins were separated by electrophoresis using a 4–15% gradient sodium dodecyl sulfate–polyacrylamide gel. The proteins were transferred to a PVDF membrane by the iBlot™ Dry Blotting System (Life Technology) and then immunoblotted with the following antibodies: CD63 and CD9 (Santa Cruz Biotechnology) and hepatocyte growth factor (HGF) (Thermo Fisher Scientific). The protein bands were visualized using a ChemiDoc™ XRS+ (BioRad) with an enhanced chemiluminescence detection kit (ECL) (GE Healthcare, Amersham).
To further analyze the protein content of the different EV populations, 100 μg of EV protein was hybridized on glass slides from the Human Cytokine Array kit (Ray Biotech) according to the manufacturer's protocol. The kit allows detecting the expression of 80 different cytokines and growth factors.
Electron microscopy
Transmission electron microscopy was performed by loading EVs onto 200-mesh nickel formvar carbon-coated grids (Electron Microscopy Science) for 20 min. EVs were then fixed with a solution containing 2.5% glutaraldehyde and 2% sucrose, followed by repeated washings in distilled water. The samples were then negatively stained with NanoVan (Nanoprobes; Yaphank) and examined by Jeol JEM 1010 electron microscope.
In vitro experiments on mTECs
For proliferation experiments, mTECs were seeded in 96-well plates at 1500 cells/well in either 100 μL/well of DMEM with 10% FCS (positive control), or 0% FCS (negative control), or 0% FCS in the presence of different concentrations of EVs (0.5 × 107 and 1 × 107 EVs/mL). DNA synthesis was detected through the incorporation of 5-bromo-2′-deoxy-uridine (BrdU) into the cellular DNA after 48 h of culture with EVs as per manufacturer's protocol (Roche Applied Science).
SCID mouse model of AKI
Animal studies were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. All procedures were approved by the Ethics Committee of the University of Turin and the Italian Health Ministry (authorization number: 211/2016-PR).
AKI was induced by an intramuscular (i.m) injection of glycerol (Sigma) in SCID mice as described previously.17 Glycerol induces myolysis and hemolysis, thereby exposing the kidneys to large toxic amounts of myoglobin and hemoglobin.29 Briefly, mice were anesthetized with an i.m. injection of zolazepam (80 mg/kg) and xilazina (16 mg/kg) and then injected with 8 mL/kg of 50% glycerol in water. Half the dose was injected into each muscle of the inferior hind limbs. Three days after glycerol injection, the mice were treated intravenously with either 120 μL of the vehicle alone (n = 10) or containing 165 × 106 particles of the different populations of EVs (n = 10 for each populations). The mice were sacrificed 5 days after glycerol injection (2 days after EV treatment).
Blood samples were collected 5 days after glycerol-induced AKI for the measurement of blood urea nitrogen (BUN) and creatinine. BUN was measured by direct quantification of serum urea with a colorimetric assay kit according to the manufacturer's protocol (Arbor Assays). Serum creatinine was measured using a colorimetric microplate assay based on the kinetic Jaffe reaction as per manufacturer's protocol (Quantichrom Creatinine Assay; BioAssay Systems).
Renal morphology was evaluated through formalin-fixed paraffin-embedded tissue staining. Briefly, 5-μm-thick paraffin kidney sections were routinely stained with hematoxylin and eosin (Merck) for microscopic evaluation. Luminal hyaline casts and cell necrosis (denudation of tubular basement membrane) were assessed in nonoverlapping fields (10 for each section) using a 40 × objective (high-power field [HPF]). The number of casts and tubular profiles showing necrosis were recorded in a single-blind manner, as described previously.16,17
Immunohistochemistry for the detection of proliferating tubular cells was performed as described previously.16,17 Briefly, paraffin kidney sections were subjected to antigen retrieval, followed by blocking and labeling with 1:400 of anti-PCNA (Santa Cruz Biotechnology) and 1:200 of anti-BrdU (Dakocytomation). Immunoperoxidase staining was then performed using a 1:300 dilution of secondary HRP antibody (Pierce). Scoring for PCNA and BrdU-positive cells was carried out by counting the number of positive nuclei per HPF (40 × ) in 10 randomly chosen sections of the kidney cortex.
Statistical analyses
Data were analyzed using the GraphPad Prism 6.0 program. Statistical analysis was performed by employing Student's t-tests, analysis of variance (ANOVA) with Newman–Keuls test, or ANOVA with Dunnett's multicomparison tests as deemed appropriate. A p-value of <0.05 was considered significant.
Results
Characterization of different populations of MSC-EVs
Different EV populations were analyzed by NTA to determine their size. The 100K-EVs appeared to be homogeneous with a mean size of 160 ± 72 nm (Fig. 1B). On the other hand, the 10K-EVs were more heterogeneous in size containing EVs which on average were much larger (215 ± 110 nm) (Fig. 1B). Variability in size of EVs that were fixed as seen by transmission electron microscopy was observed, whereby they were smaller than that observed with the NanoSight. This could be due to the influence of both temperature and Brownian motion incorporated in the NanoSight method of EV characterization. In addition, transmission electron microscopy showed the presence of bigger EVs in the 10K fraction compared with the 100K fraction (Fig. 1C).
The expression of typical exosomal markers was evaluated in the different EV populations by Western blot analyses. CD63, a fairly common marker for EVs was expressed at low levels in the 10K-EV population compared with the 100K-EVs (Fig. 1D). Furthermore, a similar pattern was also observed for the CD9 marker (Fig. 1D). FACS analyses showed the presence of several markers characteristic of MSCs, such as CD29, CD44, CD73, and CD105, in all the EV populations tested (not shown), as described previously for 100KTOT-EVs.17,25
In vivo effects of different populations of MSC-EVs in experimental AKI
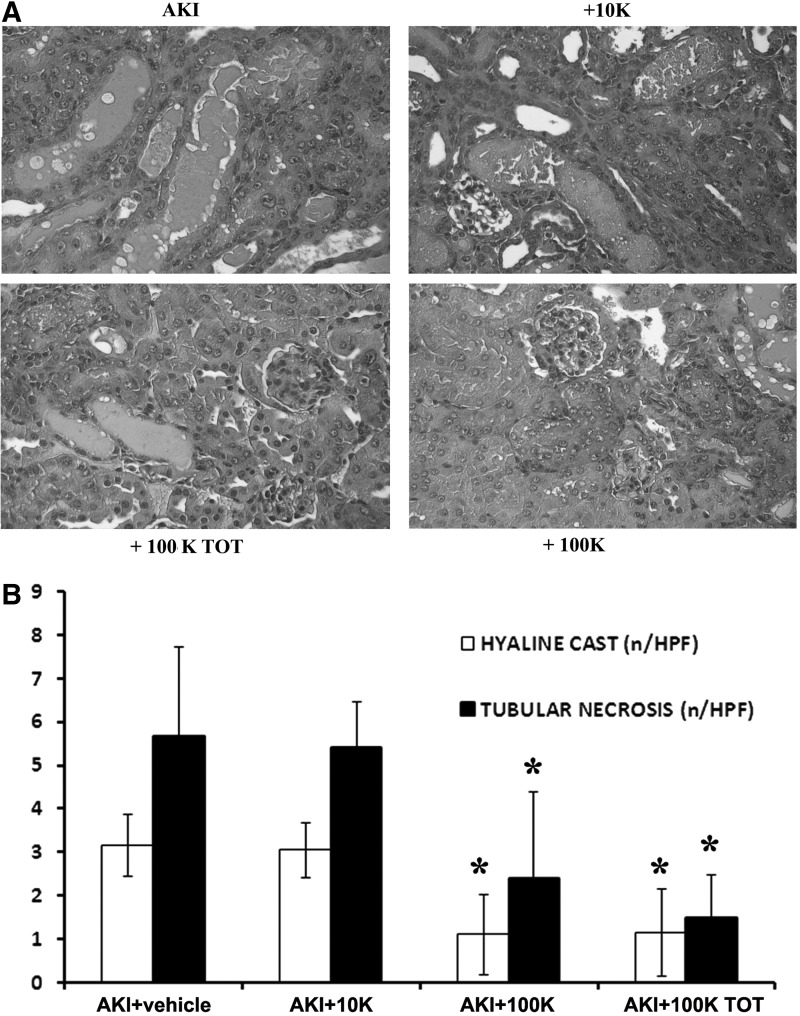
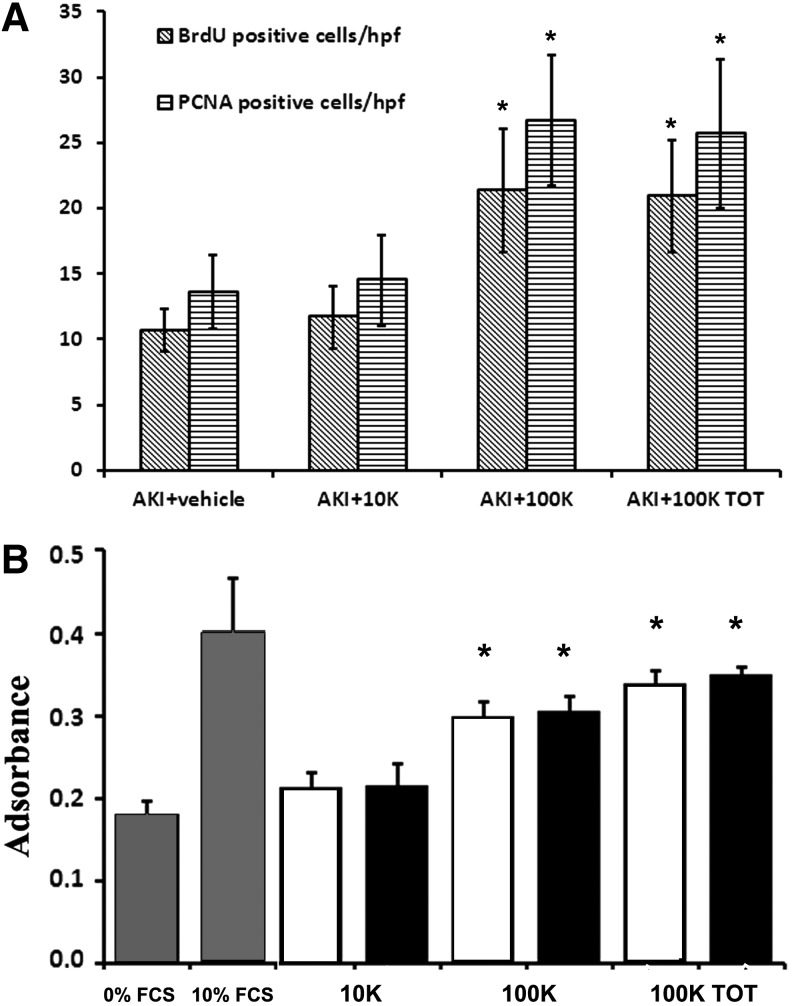
Analyses of renal function indicated that the 100KTOT-EVs significantly improved renal function by reducing BUN and creatinine levels compared with AKI mice treated with the vehicle alone (Table 2). Furthermore, mice treated with the 100K-EVs also improved renal function in a similar manner to the 100KTOT-EV fraction. However, no significant improvement in renal function was observed in mice treated with the 10K-EV fraction (Table 2). This was further confirmed by histological analyses, whereby the 10K-EV treatment did not induce a significant recovery in renal histology as the formation of hyaline cast and necrotic renal tubules was comparable with AKI mice without EV treatment (Fig. 2). On the other hand, the 100K-EVs significantly reduced morphological injuries in a similar manner to the 100KTOT-EVs (Fig. 2). Moreover, both 100K-EVs and 100KTOT-EVs significantly enhanced tubular cell proliferation compared with the treatment with the vehicle alone or with 10K-EVs (Fig. 3A). Furthermore, this proliferative effect of 100K-EVs and 100KTOT-EVs was also observed in vitro on mTECs (Fig. 3B). In contrast, the 10K-EVs were unable to trigger proliferation of mTECs in vitro (Fig. 3B).
Table 2.
Effects of Different Extracellular Vesicle Populations on Renal Function in Acute Kidney Injury Mice
| BUN (mg/dL) | Creatinine (mg/dL) | |
|---|---|---|
| Healthy controls | 20 ± 10 | 0.2 ± 0.05 |
| AKI | 74 ± 22 | 0.9 ± 0.2 |
| AKI +100KTOT | 28 ± 8a | 0.28 ± 0.05a |
| AKI +100K | 25 ± 8a | 0.31 ± 0.08a |
| AKI +10K | 55 ± 25 | 0.75 ± 0.17 |
Results are expressed as mean ± SD. ANOVA with Dunnett's multicomparison test was performed.
p < 0.05 treated AKI mice with 100K TOT or 100K versus untreated AKI mice.
AKI, acute kidney injury; ANOVA, analysis of variance; BUN, blood urea nitrogen.
FIG. 2.
Different populations of MSC-EVs induce renal morphological changes in glycerol-induced AKI. (A) Micrographs representing the renal histology of AKI mice treated with vehicle alone, or AKI mice injected at day 3 with the same amount of different EV populations (10K, 100K, and 100KTOT) and sacrificed at day 5 after glycerol administration. Original magnification, × 400. (B) Morphometric evaluation of hyaline casts and tubular necrosis in AKI mice treated with vehicle alone or injected with different EV populations at day 3 and sacrificed at day 5 after glycerol administration. Results are expressed as mean ± SD of 10 different AKI mice for each experimental condition, ANOVA with Newman–Keuls multicomparison test was performed. *p < 0.05 AKI mice injected with 100K and 100KTOT-EVs v/s AKI mice injected with vehicle alone. AKI, acute kidney injury; ANOVA, analysis of variance; HPF, high-power field; MSCs, mesenchymal stromal cells.
FIG. 3.
Proproliferative effects of different MSC-EV populations in vitro and in vivo. (A) Quantification of PCNA-positive cells/hpf and BrdU-positive cells/hpf in AKI mice injected with either vehicle alone or treated with the same amount of different EV populations (10K, 100K, and 100KTOT) at day 3 and sacrificed at day 5 postglycerol administration. BrdU was injected intraperitoneally for 2 successive days before sacrificing the mice. Data are expressed as mean ± SD of 10 different AKI mice for each experimental condition. ANOVA with Dunnett's multicomparison test was performed: *p < 0.05 100K and 100KTOT-AKI-injected mice v/s AKI mice injected with vehicle alone; (B) 10 mM BrdU was added to 1500 cells/well (mTECs) cultured in 96-well plates and incubated in serum-free DMEM in the presence or absence of different doses of different populations of EVs (white bars: 0.5 × 107; black bars:1 × 107 EVs/mL) for 48 h. Results are expressed as mean ± SD of six different experiments performed in quadruplicate. ANOVA with Newman–Keuls multicomparison test was performed; *p < 0.05 100K and 100K TOT -EVs versus 0% FCS. BrdU, 5-bromo-2′-deoxy-uridine; mTECs, murine tubular epithelial cells.
Analyses of EV content: RNAs
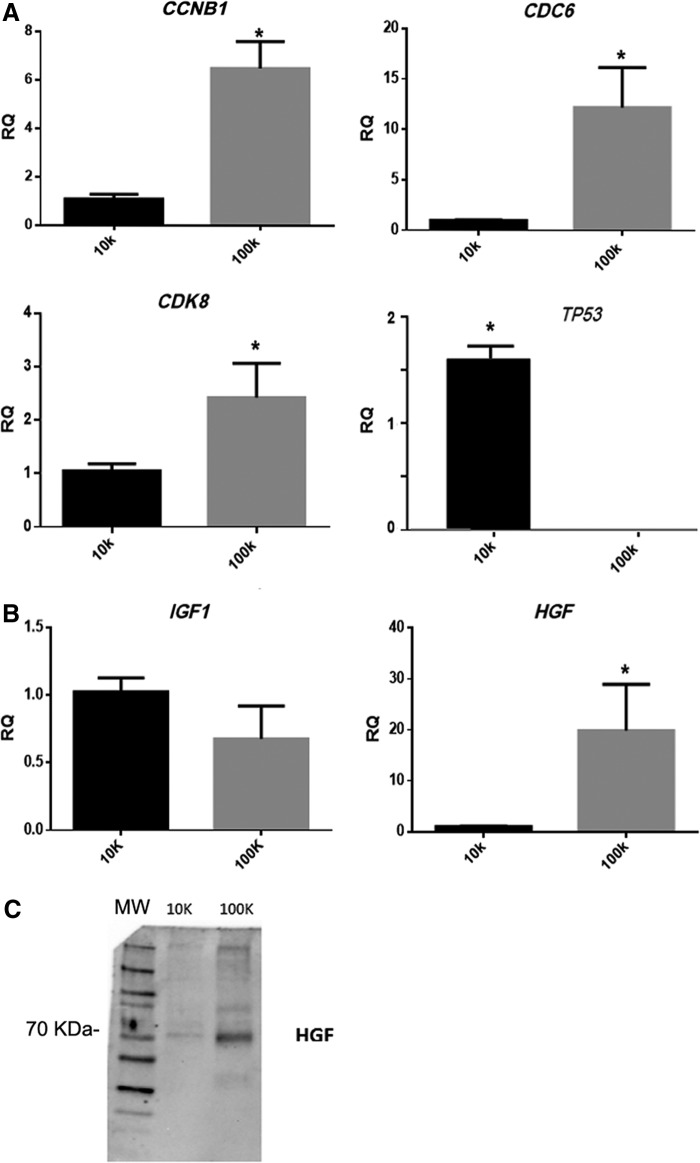
The differential expression of cell cycle-specific mRNAs by different EVs was evaluated (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/tea). The 100K-EV fraction significantly expressed mRNAs involved in cell cycle progression such as CCNB1, cyclin-dependent protein kinase 8 (CDK8), and CDC6. Conversely, the 10K-EVs, significantly expressed only TP53, which is a negative regulator of the cell cycle. Gene expression was further confirmed by qRT- PCR (Fig. 4).
FIG. 4.
Differential expression of mRNAs and proteins between 10K-EV and 100K-EV populations. (A) Real-time analysis of Cyclin B1, CDC6, CDK8, and TP53 expression. Experiments were performed in triplicate with EV populations (10K and 100K) obtained from two different MSC lines. RQ ± SE, *p < 0.01 10K versus 100K-EVs. (B) Real-time analysis of HGF and IGF-1 expression. RQ ± SE, *p < 0.01 10K versus 100K-EVs. (C) Representative Western blot analysis of HGF expression by purified 10K and 100K EV population. CDK, cyclin-dependent protein kinase; HGF, hepatocyte growth factor; IGF-1, insulin-like growth factor 1.
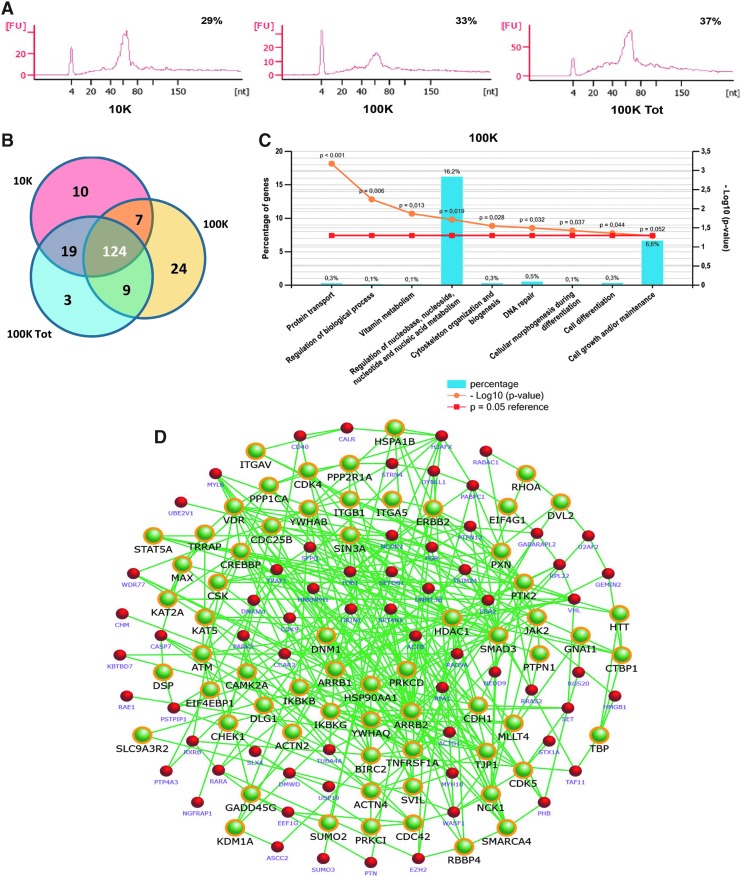
Bioanalyzer profiling revealed an enrichment of small RNAs in both the 10K and 100K-EVs (Fig. 5A). This was followed by comparative miRNA sequencing analysis, which was performed on different EV populations as follows: (1) 10K versus 100KTOT-EVs, (2) 100K versus 100KTOT-EVs, and (3) 10K versus 100K-EVs. By using a cutoff of RPM ≥100 to define the expressed versus not expressed miRNAs, we found 160 miRNAs to be present in the 10K-EV population and 164 in the 100K-EVs (Supplementary Table S2). On comparing with the 100KTOT-EVs, the two EV populations shared 124 common miRNAs (Fig. 5B). Moreover, we identified 32 miRNAs that were upregulated (FC ≥1) in the 10K-EV fraction with respect to the 100K-EVs (Table 3). On the other hand, 37 miRNAs were upregulated in the 100K-EVs (FC ≤−1) (Table 4).
FIG. 5.
miRNA content of different EV populations. (A) Bioanalyzer size profile of small RNAs detected in the 10K, 100K, and 100KTOT-EVs. Three different samples were tested in triplicate with similar results. (B) Venn diagram showing the distribution of mature miRNAs in different EVs. The 10K and 100K-EVs shared 124 common miRNAs. Specific or unique miRNAs were detected in all the EVs tested. (C) FunRich analysis of the biological processes over-represented by the 100K predictive genes. Biological processes were mainly associated with protein transport, vitamin metabolism, nucleic acid metabolism, cytoskeleton organization, DNA repair, cell differentiation, and cell growth (p < 0.05). (D) Network interactome analysis of the 100K candidate molecules and their associated pathways. The yellow node boundary represents the selected candidate molecules associated with the following highly represented growth factor pathways: proteoglycan–syndecan signaling, IGF-1 pathway, and signaling mediated by HGF (p < 0.001).
Table 3.
List of the 32 miRNA Unique or Enriched in the 10K-EVs
| miRNA name | 10K (avg. norm) | 100K (avg. norm) | FC (10K vs. 100K) |
|---|---|---|---|
| hsa-miR-193b-3p | 679.78 | 328.95 | 1.05 |
| hsa-miR-370-5p | 459.35 | 217.32 | 1.08 |
| hsa-miR-615-3p | 398.32 | 63.79 | 2.64 |
| hsa-miR-382-5p | 329.97 | 118.66 | 1.48 |
| hsa-miR-28-5p | 315.65 | 85.15 | 1.89 |
| hsa-miR-130b-3p | 235.26 | 53.16 | 2.15 |
| hsa-miR-30b-5p | 231.73 | 58.47 | 1.99 |
| hsa-miR-532-5p | 216.49 | 91.37 | 1.24 |
| hsa-miR-4485-5p | 198.32 | 98.13 | 1.02 |
| hsa-let-7b-3p | 188.60 | 79.73 | 1.24 |
| hsa-miR-149-5p | 188.12 | 74.42 | 1.34 |
| hsa-miR-1271-5p | 187.51 | 1.00 | 7.55 |
| hsa-miR-19b-3p | 182.32 | 74.42 | 1.29 |
| hsa-miR-324-5p | 166.42 | 1.00 | 7.38 |
| hsa-miR-193a-3p | 165.82 | 21.26 | 2.96 |
| hsa-miR-378a-3p | 157.79 | 63.79 | 1.31 |
| hsa-miR-1307-3p | 153.26 | 42.53 | 1.85 |
| hsa-miR-664a-3p | 146.98 | 95.68 | 0.62 |
| hsa-miR-106b-3p | 146.40 | 95.68 | 0.61 |
| hsa-miR-214-5p | 139.61 | 26.58 | 2.39 |
| hsa-miR-146a-5p | 128.22 | 26.58 | 2.27 |
| hsa-miR-1307-5p | 124.20 | 21.26 | 2.55 |
| hsa-miR-17-5p | 119.97 | 31.90 | 1.91 |
| hsa-miR-889-3p | 118.52 | 1.00 | 6.89 |
| hsa-miR-505-3p | 117.12 | 21.26 | 2.46 |
| hsa-miR-487b-3p | 117.12 | 47.84 | 1.29 |
| hsa-miR-150-5p | 115.44 | 58.47 | 0.98 |
| hsa-miR-222-5p | 113.68 | 47.84 | 1.25 |
| hsa-miR-339-5p | 113.40 | 15.95 | 2.83 |
| hsa-miR-421 | 112.81 | 79.73 | 0.50 |
| hsa-miR-887-3p | 112.12 | 69.10 | 0.70 |
| hsa-miR-210 | 111.84 | 85.15 | 0.39 |
FC, fold change.
Table 4.
List of the 37 miRNAs Unique or Enriched in the 100K-EVs
| miRNA name | 10K (avg. norm) | 100K (avg. norm) | FC (10K vs. 100K) |
|---|---|---|---|
| hsa-miR-486-5p | 3515.32 | 31163.78 | −3.15 |
| hsa-miR-451a | 161.22 | 939.62 | −2.54 |
| hsa-miR-181a-2-3p | 158.37 | 437.62 | −1.47 |
| hsa-miR-144-5p | 2.85 | 429.32 | −7.23 |
| hsa-miR-126-5p | 51.26 | 416.96 | −3.02 |
| hsa-miR-142-5p | 34.17 | 363.82 | −3.41 |
| hsa-miR-181a-3p | 123.13 | 275.79 | −1.16 |
| hsa-miR-34c-5p | 74.63 | 233.27 | −1.64 |
| hsa-miR-99b-3p | 37.12 | 211.37 | −2.51 |
| hsa-miR-329-3p | 31.91 | 196.16 | −2.62 |
| hsa-miR-628-5p | 11.39 | 191.74 | −4.07 |
| hsa-miR-3960 | 48.41 | 177.77 | −1.88 |
| hsa-miR-181d-5p | 81.32 | 156.50 | −0.94 |
| hsa-miR-184 | 43.89 | 151.19 | −1.78 |
| hsa-miR-4454 | 37.12 | 151.19 | −2.03 |
| hsa-miR-485-5p | 92.88 | 151.19 | −0.70 |
| hsa-miR-126-3p | 31.91 | 145.87 | −2.19 |
| hsa-miR-138-1-3p | 97.99 | 145.87 | −0.57 |
| hsa-miR-365b-5p | 37.62 | 135.24 | −1.85 |
| hsa-miR-15b-5p | 68.93 | 135.24 | −0.97 |
| hsa-miR-654-5p | 52.43 | 129.93 | −1.31 |
| hsa-miR-660-5p | 54.12 | 129.93 | −1.26 |
| hsa-miR-539-3p | 61.39 | 124.61 | −1.02 |
| hsa-miR-145-3p | 97.98 | 124.61 | −0.35 |
| hsa-miR-29b-3p | 55.28 | 119.29 | −1.11 |
| hsa-miR-452-5p | 43.30 | 119.29 | −1.46 |
| hsa-miR-151b | 28.48 | 118.66 | −2.06 |
| hsa-miR-137 | 17.19 | 113.98 | −2.73 |
| hsa-miR-3615 | 17.19 | 113.98 | −2.73 |
| hsa-miR-1246 | 64.40 | 113.98 | −0.82 |
| hsa-miR-361-3p | 63.24 | 113.98 | −0.85 |
| hsa-miR-125a-3p | 31.91 | 113.35 | −1.83 |
| hsa-miR-4492 | 11.39 | 113.35 | −3.31 |
| hsa-miR-497-5p | 37.62 | 113.35 | −1.59 |
| hsa-let-7a-3p | 63.23 | 113.35 | −0.71 |
| hsa-miR-1468-5p | 14.24 | 111.63 | −2.97 |
| hsa-miR-1260a | 92.29 | 111.63 | −0.29 |
Predictive target analysis using FunRich analysis tool conducted on the upregulated miRNAs in one fraction with respect to the other revealed a group of 2228 selective target genes for the 100K-EVs and 1744 for the 10K-EVs. The biological processes over-represented by the 100K-EV predictive genes were mainly associated with protein transport, vitamin metabolism, nucleic acid metabolism, cytoskeleton organization, DNA repair, cell differentiation, and cell growth (Fig. 5C). Selective processes represented by the 10K-EV predictive genes were transport, cell-to-cell communication, apoptosis, fatty acid metabolism, cell growth and cell cycle regulation, and matrix adhesion (not shown).
A network analysis of the 100K-EV predictive genes was performed to define their associated pathways. What was observed was a high representation of proliferative/antiapoptotic pathways, such as PI3K and mTOR-associated signaling events. A robust representation of protein nodes clustered around growth factor pathways such as proteoglycan–syndecan signaling events, HGF, and insulin-like growth factor 1 (IGF-1)-mediated pathway and signaling were over-represented in the interactome network created (p < 0.001) (Fig. 5D). Furthermore, signaling associated with cytoskeleton remodeling such as Arf6, integrins, and focal adhesion kinase were also highly represented, together with angiogenic processes such as of PDGFRβ and EGFR signaling events. The same analysis conducted on the 10K predictive genes showed a high representation of cell cycle-associated pathways such as G1/S transition, S phase, and cell cycle checkpoint (not shown).
Analyses of EV content: protein
Proproliferative effects of EVs could also depend on their protein content such as specific growth factors. For this reason, we analyzed the protein content of the different EV populations by an antibody array that can evaluate the presence of 80 different cytokines and growth factors semiquantitatively. This analysis revealed the presence of 13 cytokines in both the EV populations (Table 5). Growth factors involved in the regulation of biological processes, such as hematopoiesis, osteocyte differentiation, and neuronal development, were differentially expressed in the 10K and 100K-EVs. The 10K-EVs were positive for several cytokines and chemokines involved in the control and regulation of inflammation and immunomodulation such as IFN-γ and TGF-β. The 100K-EVs were enriched with HGF (Fig. 4C) and IGF-1, two growth factors known to be involved in renal regeneration mediated by MSCs. Furthermore, qRT-PCR also confirmed an enrichment of HGF mRNA in the 100K-EVs with respect to the 10K-EVs (Fig. 4B). No significant difference, however, was observed in the mRNA levels of IGF-1 between the 10K-EVs and 100K-EVs (Fig. 4B).
Table 5.
Differentially Expressed Cytokines/Growth Factors in Different Extracellular Vesicle Populations
| Proteins expressed in 10K | Protein expressed in 100K | Proteins coexpressed in 10K and 100K | |
|---|---|---|---|
| Hematopoietic growth factors | G-CSF | TPO | IL-6 |
| GM-CSF | |||
| FLT-3 | |||
| Chemokines and chemokine ligands | GROa | ||
| CCL-1, -2, -7, and -8 | CCL-5, -20 | ||
| CXCL-1, -9, and -10 | |||
| Interleukins | IL-2, -5, -7, -8, -10, and -13 | IL-16 | |
| Growth factors | FGF-6 | FGF-9 | FGF-7 |
| IGFBP-1 and -4 | HGF | IGFBP-2 and -3 | |
| IGF-1 | VEGF | ||
| Angiogenin | |||
| Cytokines | IFN-γ | ||
| TGF-β | |||
| Neurotrophic factors | NT-4 | NT-3 | BDNF |
| Bone metabolism | Osteopontin | ||
| Osteoprotegerin | |||
| Inhibitors of metalloproteinase | TIMP-1 and -2 |
Discussion
Human bone marrow MSC-EVs are able to promote regeneration in different animal models of AKI and mimic the efficacy of MSCs.16–19,30,31 EVs isolated from cultured bone marrow MSCs contain vesicles of different sizes and densities. Thus, it was unclear which population of the EVs was responsible for the proregenerative effect of MSC-EVs observed in AKI mouse models. Through this study, we have demonstrated that different EV populations can exhibit diverse regenerative capabilities on AKI. The 100K population, positive for typical exosomal markers (CD63 and CD9), markedly improved both renal function and morphology in the presented AKI model. In contrast, the 10K-EVs, which were enriched in microvesicles expressing only the MSC markers, did not have any significant effect on renal function and morphology. These results therefore suggest that the proregenerative effect of MSC-EVs observed is mainly ascribed to exosomes. However, as the total EV population has the same effect as the exosomal fraction, it can be speculated that microvesicles do not interfere in a negative manner.
Several studies have reported different biological properties of exosomes and microvesicles. For instance, exosomes obtained from murine MSCs were able to not only prevent but also reverse monocrotaline-induced pulmonary hypertension, whereas the MVs were ineffective.15 Another study showed that exosomes and microvesicles derived from endothelial colony-forming cells had different regenerative properties when injected in an experimental murine model of AKI induced by IRI. Exosomes showed proregenerative effects in IRI mice, whereas microvesicles were ineffective.32 In addition, exosomes and microvesicles derived from human colon cancer cells exhibited distinct biological activities, in particular exosomes, which promoted greater invasion compared with microvesicles.33 EVs derived from murine and/or human BM-MSCs reversed the radiation damage to marrow hematopoietic cells both in vitro and in vivo. In this case, preparations containing both exosomes and microvesicles were more effective, as opposed to either microvesicles or exosomes alone.34
In this study, we observed that the exosome-enriched 100K-EVs, but not the 10K-EVs, exerted a proproliferative effect on mTECs, both in vitro and in vivo. The differential biological activity of different vesicle populations has been ascribed to their distinct molecular content. In the 100K-EVs, we found the presence of CCNB1, CDK8, and CDC6 transcripts. CCNB1 mRNA encodes for the protein named G2/mitotic-specific cyclin-B1.35 Cyclin B1 is involved in the early events of mitosis, such as chromosome condensation, nuclear envelope breakdown, and spindle pole assembly. Once activated, cyclin B1 promotes several events involved in the early stages of mitosis. CDK8 encodes for a protein member of the CDK family that influences binding of RNA polymerase II to the mediator complex.36 CDC6, or cell division cycle 6, encodes for a protein that is an essential regulator of DNA replication and also plays an important role in the activation and maintenance of checkpoint mechanisms in the cell cycle that coordinates S phase and mitosis.37
In the 10K-EVs, we detected only the presence of TP53 that is a well-known negative regulator of the cell cycle,38 thus explaining the absence of a proproliferative effect. It has been previously shown that miRNAs shuttled by EVs, derived from different stem cell types, influence EV proregenerative effect in different AKI models.18,39,40 In addition, the miRNA content in the exosome and microvesicle-enriched fractions in this study was distinct. Bioinformatic analyses indicated that miRNA present in the 100K-EVs might account for regulation of proliferative/antiapoptotic pathways. Signaling associated with cytoskeleton remodeling and angiogenic processes were also highly represented in this fraction. A robust representation of protein nodes clustered around growth factor pathways, in particular signaling mediated by IGF-1 and HGF was highly represented in the 100K-EV interactome network. Moreover, some miRNAs particularly enriched in 100K-EVs are known to be involved in renal regeneration. For instance, miR-486-5p has been implicated in reparative process induced by exosomes, derived from endothelial colony-forming cells, in IRI.40 Likewise, miR-126 has also been associated with kidney regeneration induced by EPC-derived EVs in an IRI model of kidney injury.39 Moreover, miR-34a induced through p53 in cisplatin nephrotoxic AKI has been shown to protect tubular cells from injury and death.41
The proproliferative effects of 100K-EVs could also be partly attributed to their content in specific growth factors. We found that growth factors involved in different biological processes, such as the control of hematopoiesis, neuronal development, and osteocyte differentiation, were differentially distributed in the 10K and 100K-EVs. The 10K-EVs shuttled cytokines involved in the control and regulation of inflammation as well as immunomodulation and chemokines/chemokine receptors known to be expressed by MSCs and to be involved in MSC recruitment.42 Nonetheless, they contained fewer proproliferative growth factors compared with the 100K-EVs. The latter, on the other hand, was particularly enriched in growth factors known to be involved in renal regeneration such as IGF-1 and HGF. For instance, Imberti et al. have reported that MSCs exert beneficial effects on tubular cell repair by producing the mitogenic and prosurvival factor IGF-1 in an AKI model induced by cisplatin.43 The horizontal transfer of IGF-1 receptor mRNA to tubular cells through MSC-derived exosomes potentiates tubular cell sensitivity to locally produced IGF-1 providing a support for renal repair post-AKI.44 We also found that 100K EVs were enriched in both HGF protein and mRNA. Further to this, it has been shown previously that HGF mRNA present in EVs derived from both human Wharton jelly-derived MSCs and rat BM-MSCs was delivered to tubular cells and then translated into protein.45,46 Transfer of HGF to resident cells has been shown to promote renal repair, preventing fibrogenesis in models of IRI.45,46
In conclusion, this study demonstrated that despite their heterogeneity, EVs derived from MSCs are able to promote AKI recovery. This biological effect has been ascribed to the ability of EVs to induce tubular cell proliferation leading to the repopulation of injured tubules. The EV fraction enriched in exosomes contained mRNAs, miRNAs, and growth factors that may act synergistically to induce the proproliferative effects observed and may explain the ability of this fraction to promote AKI recovery. In contrast, the 10K population, which was not effective, displayed a molecular composition that was low in the expression of proproliferative factors. Since 100KTOT-EVs and 100K-EVs did not significantly differ in terms of functional, morphological, and proliferative effects, we can speculate that the 10K EV population did not interfere with the regenerative capacity of the exosomal population in AKI. This observation may be relevant for a future clinical application of EVs, which would require the setup in good manufacturing practice (GMP) conditions with minimal manipulation and loss/damage of EVs. Our data indicated that it is not necessary to remove microvesicles from MSC-EV preparation to obtain a regenerative effect.
Supplementary Material
Acknowledgments
This work was supported by the NIH grants UH2-TR000880, UH3TR000880-03S1. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The technical assistance of Federica Antico and Massimo Cedrino is gratefully acknowledged.
Disclosure Statement
No competing financial interests exist.
References
- 1.Camussi G., Deregibus M.C., Bruno S., Cantaluppi V., and Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int 78, 838, 2010 [DOI] [PubMed] [Google Scholar]
- 2.De Jong O.G., Van Balkom B.W., Schiffelers R.M., Bouten C.V., and Verhaar M.C. Extracellular vesicles: potential roles in regenerative medicine. Front Immunol 5, 608, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robbins P.D., and Morelli A.E. Regulation of immuneresponses by extracellular vesicles. Nat Rev Immunol 14, 195, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Merino-González C., Zuñiga F.A., Escudero C., Ormazabal V., Reyes C., Nova-Lamperti E., Salomón C., and Aguayo C. Mesenchymal stem cell-derived extracellular vesicles promote angiogenesis: potencial clinical application. Front Physiol 7, 24, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lamichhane T.N., Sokic S., Schardt J.S., Raiker R.S., Lin J.W., and Jay S.M. Emerging roles for extracellular vesicles in tissue engineering and regenerative medicine. Tissue Eng Part B Rev 21, 45, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deregibus M.C., Cantaluppi V., Calogero R., Lo Iacono, M., Tetta C., Biancone L., Bruno S., Bussolati B., and Camussi G. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood 110, 2440, 2007 [DOI] [PubMed] [Google Scholar]
- 7.Ranghino A., Cantaluppi V., Grange C., Vitillo L., Fop F., Biancone L., Deregibus M.C., Tetta C., Segoloni G.P., and Camussi G. Endothelial progenitor cell-derived microvesicles improve neovascularization in a murine model of hindlimb ischemia. Int J Immunopathol Pharmacol 25, 75, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Chen C., Hu B., Niu X., Liu X., Zhang G., Zhang C., Li Q., and Wang Y. Exosomes derived from human endothelial progenitor cells accelerate cutaneous wound healing by promoting angiogenesis through Erk1/2 signaling. Int J Biol Sci 12, 1472, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibrahim A.G., Cheng K., and Marbán E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports 2, 606, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heldring N., Mager I., Wood M.J.A., and Le Blanc K. Therapeutic potential of multipotent mesenchymal stromal cells and their extracellular vesicles. Hum Gene Ther 26, 506, 2015 [DOI] [PubMed] [Google Scholar]
- 11.Uccelli A., Moretta L., and Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol 8, 726, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Bruno S., Collino F., Tetta C., and Camussi G. Dissecting paracrine effectors for mesenchymal stem cells. Adv Biochem Eng Biotechnol 129, 137, 2013 [DOI] [PubMed] [Google Scholar]
- 13.Tan C.Y., Lai R.C., Wong W., Dan Y.Y., Lim S.K., and Ho H.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res Ther 5, 76, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee C., Mitsialis S.A., Aslam M., Vitali S.H., Vergadi E., Konstantinou G., Sdrimas K., Fernandez-Gonzalez A., and Kourembanas S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 126, 2601, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aliotta J.M., Pereira M., Wen S., Dooner M.S., Del Tatto M., Papa E., Goldberg L.R., Baird G.L., Ventetuolo C.E., Quesenberry P.J., and Klinger J.R. Exosomes induce and reverse monocrotaline-induced pulmonary hypertension in mice. Cardiovasc Res 110, 319, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruno S., Grange C., Collino F., Deregibus M.C., Cantaluppi V., Biancone L., Tetta C., and Camussi G. Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS One 7, e33115, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruno S., Grange C., Deregibus M.C., Calogero R.A., Saviozzi S., Collino F., Morando L., Busca A., Falda M., Bussolati B., Tetta C., and Camussi G. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J Am Soc Nephrol 20, 1053, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collino F., Bruno S., Incarnato D., Dettori D., Neri F., Provero P., Pomatto M., Oliviero S., Tetta C., Quesenberry P.J., and Camussi G. AKI recovery induced by mesenchymal stromal cell-derived extracellular vesicles carrying microRNAs. J Am Soc Nephrol 26, 2349, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gatti S., Bruno S., Deregibus M.C., Sordi A., Cantaluppi V., Tetta C., and Camussi G. Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia-reperfusion-induced acute and chronic kidney injury. Nephrol Dial Transplant 26, 1474, 2011 [DOI] [PubMed] [Google Scholar]
- 20.Collino F., Pomatto M., Bruno S., Lindoso R.S., Tapparo M., Sicheng W., Quesenberry P., and Camussi G. Exosome and microvesicle-enriched fractions isolated from mesenchymal stem cells by gradient separation showed different molecular signatures and functions on renal tubular epithelial cells. Stem Cell Rev 13, 226, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim H.S., Choi D.Y., Yun S.J., Choi S.M., Kang J.W., Jung J.W., Hwang D., Kim K.P., and Kim D.W. Proteomic analysis of microvesicles derived from human mesenchymal stem cells. J Proteome Res 11, 839, 2012 [DOI] [PubMed] [Google Scholar]
- 22.Anderson J.D., Johansson H.J., Graham C.S., Vesterlund M., Pham M.T., Bramlett C.S., Montgomery E.N., Mellema M.S., Bardini R.L., Contreras Z., Hoon M., Bauer G., Fink K.D., Fury B., Hendrix K.J., Chedin F., El-Andaloussi S., Hwang B., Mulligan M.S., Lehtiö J., and Nolta J.A. Comprehensive proteomic analysis of mesenchymal stem cell exosomes reveals modulation of angiogenesis via nuclear factor-KappaB signaling. Stem Cells 34, 601, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Luca, L., Trino S., Laurenzana I., Simeon V., Calice G., Raimondo S., Podestà M., Santodirocco M., Di Mauro, L., La Rocca, F., Caivano A., Morano A., Frassoni F., Cilloni D., Del Vecchio, L., and Musto P. MiRNAs and piRNAs from bone marrow mesenchymal stem cell extracellular vesicles induce cell survival and inhibit cell differentiation of cord blood hematopoietic stem cells: a new insight in transplantation. Oncotarget 7, 6676, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallabhaneni K.C., Penfornis P., Dhule S., Guillonneau F., Adams K.V., Mo Y.Y., Xu R., Liu Y., Watabe K., Vemuri M.C., and Pochampally R. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 6, 4953, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindoso R.S., Collino F., Bruno S., Araujo D.S., Sant'Anna J.F., Tetta C., Provero P., Quesenberry P.J., Vieyra A., Einicker-Lamas M., and Camussi G. Extracellular vesicles released from mesenchymal stromal cells modulate miRNA in renal tubular cells and inhibit ATP depletion injury. Stem Cells Dev 23, 1809, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedländer M.R., Chen W., Adamidi C., Maaskola J., Einspanier R., Knespel S., and Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26, 407, 2008 [DOI] [PubMed] [Google Scholar]
- 27.Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., and Enright A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Research 34, D140, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., Bacic A., Hill A.F., Stroud D.A., Ryan M.T., Agbinya J.I., Mariadason J.M., Burgess A.W., and Mathivanan S. An open access standalone functional enrichment and interaction network analysis tool. Proteomics 15, 2597, 2015 [DOI] [PubMed] [Google Scholar]
- 29.Zager R.A., Burkhart K.M., Conrad D.S., and Gmur D.J. Iron, heme oxygenase, and glutathione: effects on myohemoglobinuric proximal tubular injury. Kidney Int 48, 1624, 1995 [DOI] [PubMed] [Google Scholar]
- 30.He J., Wang Y., Sun S., Yu M., Wang C., Pei X., Zhu B., Wu J., and Zhao W. Bone marrow stem cells-derived microvesicles protect against renal injury in the mouse remnant kidney model. Nephrology (Carlton) 17, 493, 2012 [DOI] [PubMed] [Google Scholar]
- 31.Reis L.A., Borges F.T., Simões M.J., Borges A.A., Sinigaglia-Coimbra R., and Schor N. Bone marrow-derived mesenchymal stem cells repaired but did not prevent gentamicin-induced acute kidney injury through paracrine effects in rats. PLoS One 7, e44092, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burger D., Viñas J.L., Akbari S., Dehak H., Knoll W., Gutsol A., Carter A., Touyz R.M., Allan D.S., and Burns K.D. Human endothelial colony-forming cells protect against acute kidney injury: role of exosomes. Am J Pathol 185, 2309, 2015 [DOI] [PubMed] [Google Scholar]
- 33.Xu R., Greening D.W., Rai A., Ji H., and Simpson R.J. Highly-purified exosomes and shed microvesicles isolated from the human colon cancer cell line LIM1863 by sequential centrifugal ultrafiltration are biochemically and functionally distinct. Methods 87, 11, 2015 [DOI] [PubMed] [Google Scholar]
- 34.Wen S., Dooner M., Cheng Y., Papa E., Del Tatto M., Pereira M., Deng Y., Goldberg L., Aliotta J., Chatterjee D., Stewart C., Carpanetto A., Collino F., Bruno S., Camussi G., and Quesenberry P. Mesenchymal stromal cell-derived extracellular vesicles rescue radiation damage to murine marrow hematopoietic cells. Leukemia 30, 2221, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sartor H., Ehlert F., Grzeschik K.H., Müller R., and Adolph S. Assignment of two human cell cycle genes, CDC25C and CCNB1, to 5q31 and 5q12, respectively. Genomics 13, 911, 1992 [DOI] [PubMed] [Google Scholar]
- 36.Tsai K.L., Sato S., Tomomori-Sato C., Conaway R.C., Conaway J.W., and Asturias F.J. A conserved mediator-CDK8 kinase module association regulates mediator-RNA polymerase II interaction. Nat Struct Mol Biol 20, 611, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borlado L.R., and Méndez J. CDC6: from DNA replication to cell cycle checkpoints and oncogenesis. Carcinogenesis 29, 237, 2008 [DOI] [PubMed] [Google Scholar]
- 38.Mercer W.E. Cell cycle regulation and the p53 tumor suppressor protein. Crit Rev Eukaryot Gene Expr 2, 251, 1992 [PubMed] [Google Scholar]
- 39.Cantaluppi V., Gatti S., Medica D., Figliolini F., Bruno S., Deregibus M.C., Sordi A., Biancone L., Tetta C., and Camussi G. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney Int 82, 412, 2012 [DOI] [PubMed] [Google Scholar]
- 40.Viñas J.L., Burger D., Zimpelmann J., Haneef R., Knoll W., Campbell P., Gutsol A., Carter A., Allan D.S., and Burns K.D. Transfer of microRNA-486-5p from human endothelial colony forming cell-derived exosomes reduces ischemic kidney injury. Kidney Int 90, 1238, 2016 [DOI] [PubMed] [Google Scholar]
- 41.Bhatt K., Zhou L., Mi Q.S., Huang S., She J.X., and Dong Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med 16, 409, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du T., and Zhu Y.J. The regulation of inflammatory mediators in acute kidney injury via exogenous mesenchymal stem cells. Mediators Inflamm 2014, 261697, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imberti B., Morigi M., Tomasoni S., Rota C., Corna D., Longaretti L., Rottoli D., Valsecchi F., Benigni A., Wang J., Abbate M., Zoja C., and Remuzzi G. Insulin-like growth factor-1 sustains stem cell mediated renal repair. J Am Soc Nephrol 18, 2921, 2007 [DOI] [PubMed] [Google Scholar]
- 44.Tomasoni S., Longaretti L., Rota C., Morigi M., Conti S., Gotti E., Capelli C., Introna M., Remuzzi G., and Benigni A. Transfer of growth factor receptor mRNA via exosomes unravels the regenerative effect of mesenchymal stem cells. Stem Cells Dev 22, 772, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Du T., Zou X., Cheng J., Wu S., Zhong L., Ju G., Zhu J., Liu G., Zhu Y., and Xia S. Human Wharton jelly-derived mesenchymal stromal cells reduce renal fibrosis through induction of native and foreign hepatocyte growth factor synthesis in injured tubular epithelial cells. Stem Cell Res Ther 4, 59, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ju G.Q., Cheng J., Zhong L., Wu S., Zou X.Y., Zhang G.Y., Gu D., Miao S., Zhu Y.J., Sun J., and Du T. Microvesicles derived from human umbilical cord mesenchymal stem cells facilitate tubular epithelial cell dedifferentiation and growth via hepatocyte growth factor induction. PLoS One 10, e0121534, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merritt M.A., Strickler H.D., Einstein M.H., Yang H.P., Sherman M.E., Wentzensen N., Brouwer-Visser J., Cossio M.J., Whitney K.D., Yu H., Gunter M.J., and Huang G.S. Insulin/IGF and sex hormone axes in human endometrium and associations with endometrial cancer risk factors. Cancer Causes Control 27, 737, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.