Abstract
Fagales allergens belonging to the Bet v 1 family account responsible for the majority of spring pollinosis in the temperate climate zones in the Northern hemisphere. Among them, Fag s 1 from beech pollen is an important trigger of Fagales pollen associated allergic reactions. The protein shares high similarity with birch pollen Bet v 1, the best-characterized member of this allergen family. Of note, recent work on Bet v 1 and its homologues found in Fagales pollen demonstrated that not all allergenic members of this family have the capacity to induce allergic sensitization. Fag s 1 was shown to bind pre-existing IgE antibodies most likely primarily directed against other members of this multi-allergen family. Therefore, it is especially interesting to compare the structures of Bet v 1-like pollen allergens, which have the potential to induce allergic sensitization with allergens that are mainly cross-reactive. This in the end will help to identify allergy eliciting molecular pattern on Bet v 1-like allergens. In this work, we report the 1H, 15N and 13C NMR assignment of beech pollen Fag s 1 as well as the secondary structure information based on backbone chemical shifts.
Keywords: Fag s 1, Fagales, Beech pollen allergen, NMR assignments, Secondary structure
Biological context
Allergens are recognized as proteins which have the capacity to induce the production of inappropriate IgE levels in predisposed atopic individuals (Akdis 2006). Allergen-specific immunotherapeutic approaches often make use of modified proteins, which were designed to be hypoallergenic to reduce the risk of treatment induced adverse reactions. Hypoallergenic molecules should display a significantly reduced ability to bind IgE but their immunogenicity needs to be retained (Pichler et al. 2014; Crameri 2006). Since allergen-specific immunotherapy is the only treatment that provides long-term clinical benefits, determining the structure and molecular dynamics, which are characteristic of allergens, is necessary to understand the interactions of allergens with IgE and how to interfere by modifying the IgE-binding sites (Larché et al. 2006; Razzera et al. 2010). Another aspect for the design of successful allergy vaccines is to understand the mechanisms how wild-type allergens will eventually induces allergic sensitization in susceptible individuals. The search for allergy promoting pattern of protein allergens has led to the discovery of several biologic functions (i.e. protease activity, ligand binding, glycosylation, etc.) of major allergens directly linked to their allergenic behavior (Thomas 2013). Nevertheless it remains elucidative why some allergenic proteins (i.e. birch Bet v 1) are high efficient in inducing allergic sensitization while closely related allergens (i.e. beech Fag s 1) fail to do so.
Pollen allergens from several Fagales trees are highly homologous and many of them have the potential to initiate sensitization resulting in the production of highly cross-reactive IgE antibodies (D’Amato et al. 2007; Hauser et al. 2011; Pichler et al. 2014). Bet v 1 is the best-studied allergenic representative of the Bet v 1 allergen family (Hauser et al. 2011; Asam et al. 2014). Structures of Bet v 1 determined either by NMR and X-ray crystallography show a protein formed by three α helix and 7 antiparallel β strands enclosing a large hydrophobic core, which determines the function of this protein family (Gajhede et al. 1996). Other Bet v 1-like proteins from other sources were characterized: Api g 1 (celery—2BK0), Dau c 1 (carrot—2W0L), Pru av 1 (sweet cherry—1E09), Ara h 8 (peanut—4M93), Gly m 4 (soya bean—2K7H), LIPPR-10.1A (yellow lupine—1ICX), LIPR-10.1B (yellow lupine—1IFV), Fra a 1e (strawberry—2LPX), Hyp-1 (St. John’s wort—3IE5) and LIPR-10.2A (yellow lupine—1XDF), LIPR-10.2B (yellow lupine—2QIM). Nevertheless, besides Bet v 1 and its isoforms no other Fagales pollen allergen was structurally characterized. Therefore, we have studied the Fagales allergen Fag s 1, a protein of 159 residues, from the beech tree (Fagus sylvatica) that shares 66 % of identity with Bet v 1 by NMR. It was shown that this protein has a lower allergenic activity and does not induce allergic sensitization itself but is highly cross-reactive with IgE primarily directed against other allergenic members of the Bet v 1-family (Hauser et al. 2011). In this work, we present the 1H, 13C and 15N NMR resonance assignments as well as the secondary structure information of Fag s 1 calculated from the backbone chemical shift values.
Methods and experiments
Recombinant protein expression and purification
Recombinant Fag s 1 1.0101, termed Fag s 1 in the following, was expressed from a pET28b (Novagen, Merck KGaA, Darmstadt, Germany) construct in Escherichia coli BL21 Star™ (DE3) cells (Invitrogen, Carlsbad, CA, USA) in LB medium supplemented with 25 mg/L kanamycin. For isotope labelling, protein expression was performed in E. coli OD 2 CN medium (Silantes, München, Germany) supplemented with 25 mg/L kanamycin. Protein expression was induced by addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG) at OD600 nm of 0.8, and growth was continued at 16 °C for 20 h. Harvested cells were dissolved in 1/50 culture volume of 20 mM sodium phosphate buffer pH 8.0. Cell breakage was performed using liquid nitrogen. After centrifugation at 15,000g, 0.5 M sodium chloride and 0.15 M NaH2PO4 were slowly added to the supernatant while stirring on ice for 30 min. After centrifuging again at 15,000g, the solution was filtered through a 0.45-μm filter and protein purification was performed by hydrophobic interaction chromatography using a 5-mL Phenylsepharose column (GE Healthcare Biosciences, Little Chalfont, UK). The protein was eluted with 25 mM Tris/HCl pH 9.3, 8 % (v/v) 2-propanol. Final purification was performed by size exclusion chromatography in 10 mM sodium phosphate buffer pH 8 using a superdex 75 10/300 GL column (GE Healthcare Biosciences). The recombinant protein was lyophilized and stored at −20 °C.
The NMR assignment experiments were carried out with ~600 lM of isotopically labeled (15N, 13C) Fag s 1 in 10 mM of sodium phosphate, 150 mM NaCl pH 7.8 containing 5 % of deuterated 2,2,2 trifluoroethanol-D2 (TFE) and 10 % of D2O. The addition of the TFE increased the stability of Fag s 1 and the resonances dispersion.
NMR spectroscopy
The NMR experiments were performed at 308 K using Bruker Avance III 800 and 700 spectrometers equipped with TXI 5 mm triple-resonance probe and a Bruker Avance III 600 equipped with TCI 5 mm triple resonance cryogenic-probe. For the assignment of the NMR resonances of the backbone atoms the following NMR triple-resonance experiment were analyzed: 1H–15N HSQC, 1H–13C HSQC, HNCO, HN(CA)CO, HNCA, HNCACB and CBCA(CO)NH. Side-chain assignments was done through the analysis of HBHA(CBCA)(CO)NH. HCCH-TOCSY, HCCH-COSY. The assignment was completed and confirmed using 15N-edited NOESY-HSQC (100 ms mixing time) 13C-edited NOESY-HSQC (100 ms mixing time). The spectra were processed using Topspin 2.1 (Bruker Corporation, USA) and analyzed using CARA 1.8.4 (Keller and Wuthrich 2004).
Assignments and data deposition
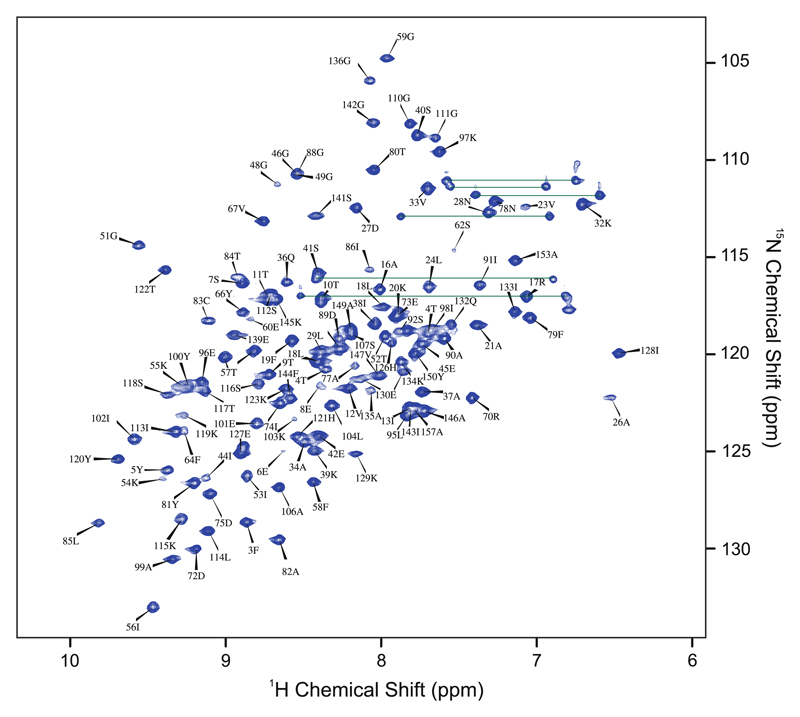
Figure 1 shows the 1H–15N HSQC spectrum of Fag s 1 with 5 % of TFE pH 7.8 at 308 K. The NMR peaks are well dispersed, consisted with a well-folded protein and almost all atoms were assigned, and only residues K137 and Y158 were not found in the spectra. The amide group was not assigned for residues S47, H69, I71, E93, G124, D125, E138, A140 e N154 probably due to line broadening. For those residues, the atoms were assigned using the sequential triple-resonance experiments. The following atoms could not be unambiguously assigned because of NMR resonance overlaps: HZ-CZ (F3), HE*-CE* and HZ-CZ (F19), HZ-CZ (F22), HZ-CZ (F79), C (G124), HE*-CE* and HZ-CZ (F144), C (N154). The chemical shifts have been deposited in the BioMagnRes-Bank (http://www.bmrb.wisc.edu) under the accession number 25590.
Fig. 1.
1H–15N HSQC spectrum of isotopically [13C, 15N]—labeled Fag s 1 acquired in an Avance III 700 MHz and pH 7.8 at 308 K with 5 % of TFE. The NMR resonances are numbered according to the primary sequence of Fag s 1. Some peaks present very low intensity in the HSQC spectrum and their 1H–15N chemical shifts were obtained by 3D triple-resonance experiments
Secondary structure information from the 13C chemical shifts
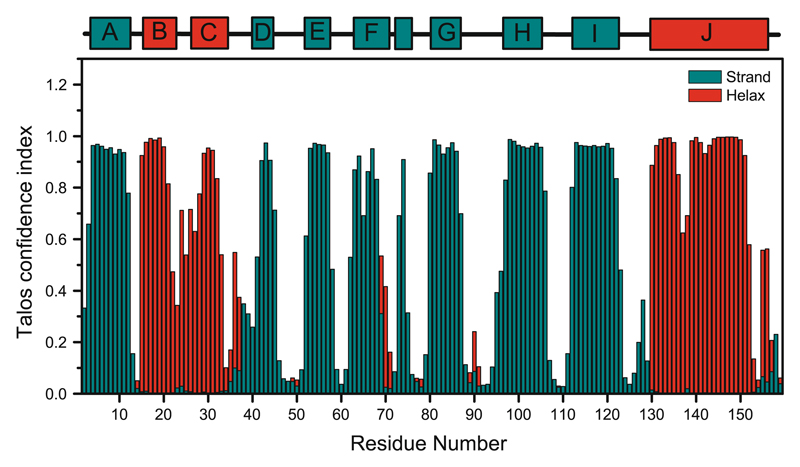
The identification of the secondary structure was obtained from the backbone chemical shifts processed using the program Talos-N (Shen and Bax 2013). Figure 2 shows the secondary structure confidence index as a function of residue number of Fag s 1. Talos index indicates residues 2–10, 40–44, 51–56, 63–67, 79–86, 96–105 and 111–121 are forming seven β strands, respectively. Residues 14–20, 24–33 and 130–150 are forming three α helices. This distribution is in agreement with the secondary structure of Bet v 1, which has 66 % of primary sequence identity. The NOEs analysis and structure calculation will confirm these suggestions.
Fig. 2.
Secondary structure of Fag s 1. Talos index for secondary structure prediction are presented by bars colored in red, for α helix, and green, for β strands. The upper panel shows the secondary structure of Bet v 1 (pdb: 1BV1) formed by three α helices (red) and seven β strands (green)
Acknowledgments
Christian Doppler Research Association, Austria; Biomay AG, Vienna, Austria; Austrian Science Fund (FWF). CNPq, FAPERJ, and INBEB, Brazil.
References
- Akdis CA. Allergy and hypersensitivity: mechanisms of allergic disease. Curr Opin Immunol. 2006;18:718–726. doi: 10.1016/j.coi.2006.09.016. [DOI] [PubMed] [Google Scholar]
- Asam C, Batista AL, Moraes AH, et al. Bet v 1-a Trojan horse for small ligands boosting allergic sensitization? Clin Exp Allergy. 2014;44:1083–1093. doi: 10.1111/cea.12361. [DOI] [PubMed] [Google Scholar]
- Crameri R. Allergy diagnosis, allergen repertoires, and their implications for allergen-specific immunotherapy. Immunol Allergy Clin North Am. 2006;26:179–189. doi: 10.1016/j.iac.2006.02.003. [DOI] [PubMed] [Google Scholar]
- D’Amato G, Cecchi L, Bonini S, et al. Allergenic pollen and pollen allergy in Europe. Allergy. 2007;62:976–990. doi: 10.1111/j.1398-9995.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- Gajhede M, Osmark P, Poulsen F, et al. X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy. Nat Struct Biol. 1996;3:1040–1045. doi: 10.1038/nsb1296-1040. [DOI] [PubMed] [Google Scholar]
- Hauser M, Asam C, Himly M, et al. Bet v 1-like pollen allergens of multiple Fagales species can sensitize atopic individuals. Clin Exp Allergy. 2011;41:1804–1814. doi: 10.1111/j.1365-2222.2011.03866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R, Wuthrich K. Computer-aided resonance assignment (CARA) 2004 http://www.nmr.ch.
- Larche´ M, Akdis C, Valenta R. Immunological mechanisms of allergen-specific immunotherapy. Nat Rev Immunol. 2006;6:761–771. doi: 10.1038/nri1934. [DOI] [PubMed] [Google Scholar]
- Pichler U, Hauser M, Hofer H, et al. Allergen hybrids—next generation vaccines for Fagales pollen immunotherapy. Clin Exp Allergy. 2014;44:438–449. doi: 10.1111/cea.12250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razzera G, Gadermaier G, de Paula V, et al. Mapping the interactions between a major pollen allergen and human IgE antibodies. Structure. 2010;18(8):1011–1021. doi: 10.1016/j.str.2010.05.012. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas WR. Innate affairs of allergens. Clin Exp Allergy. 2013;43(2):152–163. doi: 10.1111/j.1365-2222.2012.04059.x. [DOI] [PubMed] [Google Scholar]