Abstract
Neuroscience is inherently interdisciplinary in its quest to explain the brain. Like all biological structures the brain operates at multiple levels, from nano-scale molecules to meter-scale systems. Here, I argue that understanding the nano-scale organization of the brain is not only helpful for insight into its function, but is actually a requisite for such insight. I propose that one impediment to a better understanding of the brain is that most of its molecular processes are incompletely understood, and suggest a number of key questions that require our attention for further progress in neuroscience to be achieved beyond a description of the activity of circuits.
Keywords: Neural circuit, synaptic transmission, cytoskeleton, spine, synaptic plasticity, microglia, microvasculature, neuropeptides
In brief
Südhof proposes that a molecular and cellular understanding of the brain is essential for insight into how the brain processes information and into how it becomes impaired in neuropsychiatric disorders, and illustrates this point by describing selected, unsolved major questions in molecular neuroscience.
Within only 17 years, the 21st century has changed neuroscience. After decades in which molecular neuroscience was pre-eminent, systems neuroscience is now in ascendance. New technologies have made it possible to map neural circuits in vivo, to visualize neuronal activity in real time, and to manipulate neural activity in behaving animals. These developments, fueled by the development of molecular tools such as DREADDs, channelrhodopsins, and genetically encoded Ca2+-indicators (GECIs), have made it possible to observe the brain in action in ways that were previously unimaginable. Moreover, new genetic methods have enabled specific types of neurons to be manipulated, allowing us to test predictions about the behavioral consequences of stimulating or silencing a particular class of neuron. As a result, most of the prominent, current neuroscience papers deal with systems questions, most neuroscience students in the U.S. are trained in systems or computational neuroscience, many neuroscience departments primarily do ‘circuit neuroscience’, and the majority of new faculty hires are in systems and computational neuroscience. These developments have truly transformed neuroscience, leading to many striking new insights.
However, these advances have also shifted neuroscience research in a manner that may impede progress in understanding the brain as a whole. At present, much of molecular and cellular neuroscience is couched in terms of translational research or is dedicated to developing tools for systems neuroscience. As a result, research into biological processes is increasingly limited to disease-related events, molecules are primarily considered as potential tools, and fewer fundamental questions about how the brain works are addressed. This situation raises a critical question, namely: what can today’s molecular neuroscience offer to our understanding of the brain, if anything?
This question can be viewed from both a philosophical and a scientific perspective. From a philosophical perspective, do we believe that understanding the molecular mechanisms of the brain (given their complexity and diversity) will actually help us to understand how the brain functions as a whole? Or rather, is it just more practical to deal with the brain simply as an assembly of neurons that communicate with each other in circuits, without attention to the molecular details that underlie their function? Scientifically, if we accept that molecular and cellular neuroscience can in principle make a contribution, then what major problems and what levels of understanding can be addressed with these approaches? And why is this important?
A role for molecular neuroscience in understanding the brain
As scientists, we are both empowered and limited by the technical approaches we use, and tend to be insular in that we often do not immediately grasp the importance and implications of other approaches. Here, I mean not just techniques, but ways of thinking about a problem. Although neuroscience has to be interdisciplinary, we are often challenged by the need to be truly conversant in areas outside of our research areas. Given the inherently broad and complex insights needed to understand the brain, however, it is imperative to ask overarching questions that go beyond the focus of a particular research program.
In this regard, a central overall question is whether molecular neuroscience is actually necessary for understanding the brain. For example, if you want to drive from place A to place B by car, you don’t really need to understand the car. How an engine works is an unnecessary ‘detail’, all you need to know is how to operate the car. Analogously, does neuroscience really need to understand how a neuron in a circuit works, or is it sufficient to know its firing patterns, synaptic connections, and its synaptic information transmission properties to model how the circuit –and eventually the brain–processes information?
Several lines of argument show that a molecular understanding of the brain is truly necessary. First, the more practical arguments. In pursuing an understanding of the brain solely based on the activity of neurons in circuits, it is straightforward to map the firing patterns of neurons and their connections. However, given the plasticity of synaptic connections and of the properties of synapses,, an understanding of the brain could only be achieved if it was possible to monitor all synaptic connections and neurons simultaneously at any given time, which is patently unrealistic. Neuroscientists have known the number of neurons in C. elegans (302 in total!) and their synaptic connections for two decades (no need to look for firing patterns since C. elegans has no action potentials), and have described many of the functions of these neurons and their synapses in hundreds of papers. However, we still don’t know how the neural networks of C. elegans ‘work’.
The only possible practical approach to solving this problem is to be able to predict the dynamics and properties of these neurons’ synaptic connections, as well as the firing patterns of neurons in response to synaptic activity. Such understanding requires not only the ability to predict the behavior of neurons and synapses in circuits, but also insight into how the brain works at a deeper level than neuronal circuits, because the brain is more than a compendium of circuits. In the brain, different parts and cells communicate with each other via more than synapses. Not only neurons, but also glia are part of the overall information-processing machine that is the brain. Understanding this machine will require a molecular approach, i.e. an understanding of the molecular rules that determine the firing patterns and synaptic properties of neurons – their molecular logic.
A counterargument could be that molecular processes may simply be too complex to be tractable, that this molecular logic will be unmanageable. This argument – which could be equally applied to systems neuroscience – overlooks the difference between ‘complexity’ and ‘details’. Consider an example: If one tried to make sense of an old-fashioned telephone book by describing each individual entry without understanding its principle, one would conclude that the telephone book is impenetrably complex. But in order to use the phone book, there is no need to remember each entry, one just needs to understand its rules. The same applies to molecular neuroscience. Until principles are discovered, details are important; once principle are understood, the details become interesting only for the specialist.
An even more important practical argument for why molecular neuroscience is essential for explaining the brain relates to translational research. Although diseases often manifest as systems dysfunctions, they are caused by molecular impairments. Funders of translational research worldwide want results, talk about ‘moonshots’ to cure diseases, and push as hard as they can towards clinical trials. The problem, however, is that for many diseases, we have no real understanding of the actual disorder. This is particularly true for diseases of the brain, where there is often no moon to shoot at, so moonshots become fireworks. Billions of dollars have been spent on clinical trials for Alzheimer’s disease that are based on a weak scientific rationale – and almost predictably, are unsuccessful. In order to treat a disease, we need to know what the disease process is; in order to understand what is wrong, we need to know how it is wrong, which means comparing it to the healthy normal condition.
For example, hundreds of genes linked to neuropsychiatric diseases have been described, often with bold conjectures about how the diseases investigated may develop. However, for most of these genes, little is known about their biological function, and the conjectures are based on guesswork and do not lend themselves to therapeutic translation – for this, we need to understand the genes and their products first.
One could argue that for neuropsychiatric disorders, in the end, circuits will be more important, and that understanding autism, for example, will require us to understand the specifically human circuits for language and empathy, because the disease manifests as a dysfunction of these circuits. However, although this view is widely espoused, I believe it may represent a fundamental misunderstanding of disease processes; simply because a disease manifests as a disorder of certain human abilities, and presumably of their underlying circuits, does not mean that this is necessarily where the disease process operates. Most genes linked to neuropsychiatric disorders are broadly expressed, suggesting that they do not only function in a small subset of circuits. The manifestations of neuropsychiatric diseases do not necessarily imply that the dysfunction of the underlying circuits IS the disease, they just imply that dysfunction of the circuits underlying these manifestations is a consequence of the disease. Studying these circuits is a bit like the ‘street light fallacy’: a drunk looks for their keys under the street light not because they lost their keys there, but because that is the only place where there is sufficient light to see anything.
These two practical considerations usher in the major philosophical issue I would like to raise. Is it actually possible to describe the brain purely in terms of synaptic interactions between its neurons in a circuit, like a chemist would describe a molecule as an interaction between its atoms? Although this seems to be the favored view of some contemporary neuroscience, I would argue this is an oversimplification that is based, perhaps, on a too facile comparison of the brain with a computer. Trying to understand the brain by mapping the activity of its neurons and their connections neglects three major features of the brain, which require molecular insight to be understood: (1) plasticity, of neurons as well as of their connections, that changes the rules of interactions between neurons in a circuit on a second-to-second or day-to-day basis, and this changeability is not limited to synapses but also applies to the neurons’ electrical properties; (2) the pervasive, non-synaptic communication between neurons via multifarious messengers, ranging from non-synaptically secreted neurotransmitters to neuropeptides to diffusible messengers, communication that continuously resets the stage of receptive neurons; and (3) the role of glia cells that do more than support the lives of neurons but are an intricate part of the brain’s information-processing machine. The brain functions not only as an assembly of interleaved, overlapping, interspersed, and/or hierarchically organized circuits, but processes information by additional mechanisms that cannot be described only in terms of neuronal spike patterns.
Many fundamental questions remain unaddressed
The revolution in molecular neuroscience of the 80s and 90s produced major advances. For example, receptors and ion channels that until then were theoretical constructs became real, allowing us to understand, in molecular terms, fundamental neuronal properties, such as action potentials and neurotransmitter and neuropeptide receptors (Catterall et al., 2017; Zhu and Gouaux, 2017). Transcription factors that dictate neural identity were also defined (Arlotta and Hobert, 2015: Jessell, 2000) and fundamental mechanisms of axon guidance were described (Tessier-Lavigne, 2002); and the mechanism by which Ca2+-influx into a nerve terminal triggers neurotransmitter release within a few hundred microseconds was largely solved (Südhof, 2013). These advances were significant, but sometimes appear to obscure the view of what remains to be solved – far more than we have already discovered! To illustrate this challenge, let me cite a few fundamental problems that are at present incompletely understood. This is a partial list that is only meant to illustrate how little we understand about the fundamentals of brain function.
Neuronal shape
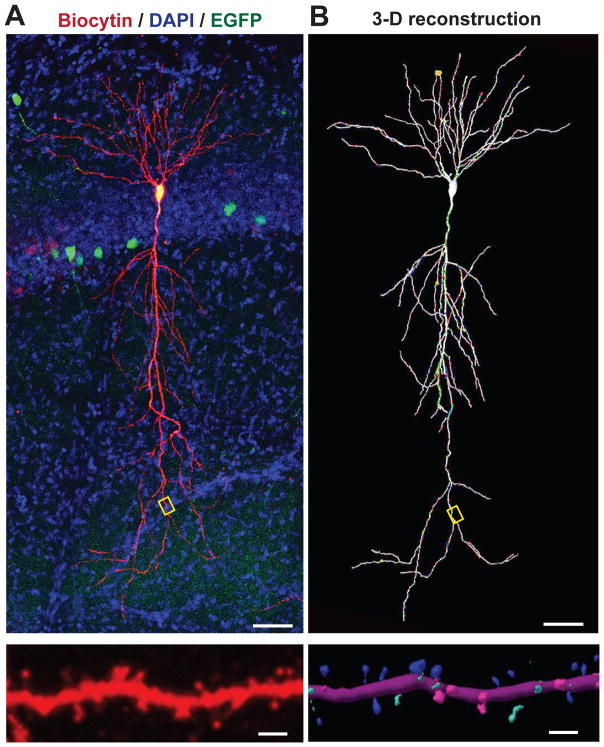
Neurons in the brain have a characteristic location, shape and orientation, which form the basis of their initial classification. Much has been learned about neurogenesis and neuronal migration in recent times, but the molecular origin and functional implications of neuronal shape remain enigmatic. Clearly hierarchies of genes controlled by gradients of diffusible factors are important, but are largely unknown. What effector pathways, for example, cause a Purkinje cell to elaborate its exquisite dendritic tree organized in a perfect plane? What determines the beautiful shape of a CA1-region neuron in the hippocampus with its specialized dendritic domains (Fig. 1)? How are axons created, and why do some neurons, such as amacrine cells, have no axons? Fundamental questions about how neurons are organized spatially remain unanswered; in fact, it is amazing that the beautiful sub-membranous actin cytoskeleton of axons was only discovered a few years ago with the advent of super-resolution microscopy (Xu et al., 2013).
Figure 1. Illustration of neuronal shape and spines: An exemplary pyramidal neuron from the CA1-region of the hippocampus with spine-studded dendritic extensions.
A. Top, a CA1-region pyramidal neuron was patched in hippocampal slices from a mouse that had been sparsely infected with a lentivirus encoding EGFP, and was filled with biocytin via the patch pipette (a section stained for biocytin (red) and DAPI to label nuclei (blue). Bottom, expansion of the dendritic field boxed in the neuronal overview image above to illustrate the dense decoration of dendrites with spines.
B. 3D-reconstruction of the pyramidal neuron and its dendritic segment shown in A. In the bottom panel, spine shapes were categorized and color–coded (blue, mushroom; green, thin; pink, stubby). Images courtesy of Dr. Richard Sando.
Dendritic spines
Most, but not all, neurons contain thousands of spines that decorate their dendrites, and receive presynaptic inputs (Fig. 1). Spines are micrometer-sized extrusions of the dendritic plasma membrane that contain endoplasmic reticulum and are full of actin but lack other endomembrane systems and have no microtubules. The potential computational role of spines has been modeled extensively, and the growth and shrinkage of spines has been documented in excruciating detail, but why some neurons have spines and others don’t is unknown. It is also unknown what signals and mechanisms mediate the development of spines, apart from obvious contributions of the actin cytoskeleton. Most importantly, we don’t know how to turn a spiny neuron into an aspiny neuron or vice versa, nor what spines actually do. A dream experiment would be to identify a master regulator of spinogenesis, and to explore the computational consequences of inducing or ablating spines.
Neuronal identity
Related to neuronal shape is the question of neuronal identity, which manifests itself also as a particular shape –hence Cajal’s lasting fame– and by many other features. Neuronal identity is now best described by gene expression profiles, but can neurons really be classified into types or do they exist in a continuum of states? Is neuronal identity plastic or fixed, and how is it maintained or changed? These and many other questions are crucial for understanding the brain because we currently think of the brain as organized into circuits composed of neurons with specific properties, even though the underlying concepts remain ill defined.
Spatial organization of signaling and signal integration
Another extension of the neuronal shape question is how different signaling events are spatially segregated without membranous separation. The neuronal cytosol is an amazingly crowded space with many trafficking pathways that are independently regulated. How is this possible? In most neurons, many independent synaptic inputs together converge to generate a postsynaptic response, but the relative importance of these signals and their collaboration –or lack thereof– are determined by a spatial organization that remains largely unclear.
Synapse formation
Synapse formation is specific for particular pairs of pre- and postsynaptic neurons and for particular locations on the postsynaptic neuron. Arguably even more importantly, synapse formation determines the characteristic, albeit plastic, properties of synapses. How synapses form is one of the major questions of contemporary neuroscience because the number, location, and properties of synapses determine the input/output relations of neural circuits. Much progress has been made in the identification of candidate synapse-formation molecules, such as neurexins, neuroligins, LAR-type PTPRs, and cerebellins (Südhof, 2008; Matsuda and Yuzaki, 2012; Takahashi and Craig, 2013). Despite this progress, however, the fundamental questions of how synapses form remain unanswered. For example, many molecules are able to artificially induce synapses (Scheiffele et al., 2000; Biederer et al., 2002), suggesting that a common intracellular signaling pathway organizes the standard components of synapses, but nothing is known about the signals that might be involved.
Synapse pruning
More than half of all synapses that are initially formed postnatally are ‘pruned’ during adolescence in humans (Rakic et al., 1994). This massive elimination of synapses poses many questions, for example, what are the signals, how is pruning regulated, how does the brain know which synapses to kill, and which to foster? Moreover, synapses are pruned, although at a much lower rate, throughout life (Qiao et al., 2016). Some but not all of these changes are likely to be activity-dependent, but their mechanisms are largely obscure. The classical complement cascade may play a role (Stevens et al., 2007), but the relatively mild phenotypes produced by genetically blocking this cascade (Chu et al., 2010) suggest that other molecular mechanisms are more important. Analyzing these questions is a major challenge in trying to understand how mature circuits are constructed, and how they change during activity.
Long-term potentiation (LTP)
Many different forms of long-term synaptic plasticity shape the properties of neural circuits, but none are as well studied and possibly as impactful as NMDA-receptor dependent LTP. Pioneering work has revealed that NMDA-receptor activation can induce long-lasting strengthening of synapses by triggering the recruitment of AMPA-receptors to synapses (Liao et al., 1995; Nicoll, 2003). Dramatic progress over the years has shown that this process depends on the activation of CaM Kinase IIα via the inflowing Ca2+, that it operates independently of the sequence of the AMPA-receptors but requires exocytosis of AMPA-receptor containing vesicles, that this exocytosis is mediated by a specific SNARE-protein machinery and is triggered by Ca2+-binding to synaptotagmin-1 or to synaptotagmin-7, and that the postsynaptic cell-adhesion molecules neuroligin-1 and LRRTM1/2 are essential for NMDA-receptor dependent LTP (e.g., see Malenka et al., 1989; Malinow et al., 1989; Wu et al., 2017; Jiang et al., 2017). How all of this fits together, however, remains mysterious. Why are two Ca2+-sensors (synaptotagmins and CaM Kinase II) required for LTP? Why are two different cell-adhesion molecules that both bind to neurexins needed for LTP in a non-redundant fashion? Whereas the principle of LTP is now well understood, how it actually works remains a challenge to be solved.
Neuropeptide signaling
At the forefront of early molecular neuroscience was the identification of neuropeptide precursors and neuropeptide receptors (Noda et al., 1982), but since then the question of neuropeptide signaling has largely faded from view with a few exceptions. Orexin continues to attract attention because of our fascination with sleep (Lin et al., 1999), leptin because of its central role in body weight regulation (Zhang et al., 1993), and oxytocin because it may be involved in social behaviors (McCall and Singer, 2012), but fundamental cell-biological questions about neuropeptide signaling remain unexplored. For example, why do some neurons use neuropeptides abundantly, and others only scarcely? How is neuropeptide secretion directed to specific parts of a neuron, e.g. axons or dendrites? What is the mechanism and regulation of neuropeptide secretion? Is there plasticity in neuropeptide secretion, similar to synaptic vesicle exocytosis? Is neuropeptide secretion regulated differentially locally in different parts of a neuron? Neuropeptides are central components of the information-processing machinery of the brain, although how exactly they contribute to it remains enigmatic. Deciphering their basic mechanisms is clearly important.
Diffusible messengers
A truly remarkable discovery over recent decades is the observation that neurons communicate not only via synaptic and neuropeptide signals, but also via diffusible messengers. This observations is based in part on the identification of nitric oxide, endocannabinoids, and retinoic acid as diffusible messengers that regulate neurons by a local action (Wilson and Nicoll, 2001; Aoto et al., 2008). However, the fundamental role of these signals in the brain is incompletely understood, as is their integration with other types of neuronal communication – a dire need for any progress in understanding the brain as a whole!
Neuronal and glial metabolism
Despite enormous progress, for example in characterizing the role of mitochondria in neurodegeneration (Wallace, 2011) and the transport of metabolites across the cell membrane (Edwards, 2007), the intracellular metabolism of neurons and glia is poorly understood. Given the central role of energy metabolism in many diseases, and in particular in neurodegenerative disorders, and the unknown coupling of energy metabolism to blood flow (see below), this is a key issue that requires in-depth studies for progress, and one of central importance for understanding the brain given its enormous energy needs.
Activity-dependent regulation of blood flow
One of the amazing features of the brain is the induced and robust increase in local blood flow that occurs when neurons in particular circuits or regions are activated, a phenomenon that is the basis for BOLD imaging used in all fMRI techniques (Petzold and Murthy, 2011). The mechanisms that couple neuronal activity to blood flow, however, are incompletely understood. And yet they are of central importance not only because they underlie all fMRI imaging –after all, it would be nice to know what one actually sees in these studies– but also because they presumably enable the brain to allocate energy resources in a need-dependent manner, and thus are probably crucial for brain function. Most likely, these mechanisms are also involved in disease, particularly neurovascular and neurodegenerative disorders. Again, much work remains to be done on solving this important question.
Function of microglia
Much attention has recently been paid to microglia because genetic evidence links microglia to Alzheimer’s disease (Kleinberger et al., 2014), because microglia mediate behavioral changes that resemble obsessive-compulsive disorders in mice (Chen et al., 2010), and because of the idea that they may be involved in synaptic pruning (Paolicelli et al., 2011). However, the functions of microglia are poorly understood, and even less is known about the mechanisms involved. Again, there is a direct need for basic cell-biology studies.
Interaction of the brain with the periphery
The central nervous system does not function in isolation, but in continuous interaction with the peripheral nervous system, immune system, endocrine system, and probably the gut and skin microbiome. These interactions are only beginning to be investigated. Molecular and cellular biology approaches are needed to explore these interactions because they are likely to be centrally involved in neurodegenerative disease and in other disorders, but at this point in mind, only initial steps towards understanding their mechanisms have been taken.
Brain aging
The vast majority of human neurons are not renewed, but remain active throughout the life of an individual – when they die, we die. How are neurons kept alive for 100 years, how do they age, and how do they die? Again, these questions are important for understanding normal brain function, as well as neurodegenerative disorders. Aging is an inescapable component of our lives. How the brain ages, and how the brain adapts to its aging components is an interesting question that is increasingly studied in psychology, and poses a major challenge for molecular and cellular neuroscience to address.
Needs for the future
The unsolved questions outlined above, and many others not mentioned here, provide exciting challenges. Solving them is essential for understanding the brain. However, there are caveats in how we approach molecular neuroscience that I believe we may want to consider. In terms of approach, we need a molecular cell biology ‘in situ’, in the living brain, not only in a culture dish, because neurons and glia in culture are different from neurons and glia in vivo. We have to develop concepts and approaches that probe molecules in a real brain, that analyze their functions at all levels, from a reduced system in vitro to a normally connected neuron in the brain. This has to be done using manipulations that are not prone to artifacts (as is the case with, for example, RNAi or overexpression), and should be correlated with results from reduced systems, ranging from cultured cells to atomic structures.
More importantly, we have to reformulate the relation of molecular neuroscience to translational research. There needs to be an open agreement that basic research is ‘preclinical’ in the sense that understanding a disease requires understanding of its biological basis, i.e., an understanding of these processes in the context of ‘normal’. We have gone too far in presenting much of what we do in terms of a medical advance. I think we should emphasize that basic research is not immediately applicable, nor should it be. Applicability is often a patent lie because the medical importance of a discovery cannot be immediately ascertained, and the public notices the lies inherent in proclamations of applicability. Solving diseases and treating diseases will require us to understand the molecular underpinnings of these diseases. This understanding will emerge from research into the biology of the underlying processes, not from studies of the diseases themselves – this is a good enough rationale for doing basic research, without immediate applicability.
Another need we face in my opinion is the need for a culture shift. The field of molecular and cellular neuroscience is vast, covering everything from human genetics to cell biology to hard-core molecular biology to crystallography, but tends to be fragmented into silos. On top of that, neuroscience as a whole is a bit of a silo that is difficult to break into. We should be recruiting more expertise from outside neuroscience to tackle neuroscience problems, such as cell biologists, biochemists, engineers, and experts in immunology and metabolism. Too often, experts outside of neuroscience find it daunting to enter the field, and the lack of funding for basic molecular and cellular questions poses an additional deterrent. Developing structures and incentives to encourage more interdisciplinary interactions, in a way that is truly cross-disciplinary and not just in name only, is essential.
Finally, maybe most importantly, I feel that we need to reflect more often on the overall purpose of understanding the brain. Society does have the right to expect from us as scientists who are funded by society to make a useful contribution to society. However, we as scientists have to be clear and honest about what is really useful and what is a sham. Knowledge as such makes a long-term contribution to a society and to a culture that cannot be measured easily in the short term; when Newton and Leibniz developed calculus, there was no application for it, but now it forms the basis of all engineering. Similarly, for many molecular neuroscience questions, there is no current application, not even an apparent ‘functional’ implication, but the knowledge created is valuable in itself and provides a basis on which we can build in the future. Knowledge as such has more value to society than any short-term application. This needs to be communicated because it is easily forgotten, given that scholarship has no economic or political lobby, and that especially in these times, knowledge is at the same time widely applied but politically discounted at the highest levels of government to an almost scary degree.
Acknowledgments
I would like to thank Drs. Katja Brose and Lu Chen for comments, and the NIMH, the NIA, and the HHMI for continued support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aoto J, Nam CI, Poon MM, Ting P, Chen L. Synaptic signaling by all-trans retinoic acid in homeostatic synaptic plasticity. Neuron. 2008;60:308–320. doi: 10.1016/j.neuron.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlotta P, Hobert O. Homeotic Transformations of Neuronal Cell Identities. Trends Neurosci. 2015;38:751–762. doi: 10.1016/j.tins.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC. SynCAM, a synaptic cell adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- Catterall WA, Wisedchaisri G, Zheng N. The chemical basis for electrical signaling. Nat Chem Biol. 2017;13:455–463. doi: 10.1038/nchembio.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SK, Tvrdik P, Peden E, Cho S, Wu S, Spangrude G, Capecchi MR. Hematopoietic origin of pathological grooming in Hoxb8 mutant mice. Cell. 2010;141:775–785. doi: 10.1016/j.cell.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y, Jin X, Parada I, Pesic A, Stevens B, Barres B, Prince DA. Enhanced synaptic connectivity and epilepsy in C1q knockout mice. Proc Natl Acad Sci USA. 2010;107:7975–7980. doi: 10.1073/pnas.0913449107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RH. The neurotransmitter cycle and quantal size. Neuron. 2007;55:835–858. doi: 10.1016/j.neuron.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Kleinberger G, et al. TREM2 mutations implicated in neurodegeneration impair cell surface transport and phagocytosis. Sci Transl Med. 2014;6:243ra86. doi: 10.1126/scitranslmed.3009093. [DOI] [PubMed] [Google Scholar]
- Liao D, Hessler NA, Malinow R. Activation of postsynaptically silent synapses during pairing-induced LTP in CA1 region of hippocampal slice. Nature. 1995;375:400–404. doi: 10.1038/375400a0. [DOI] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qiu X, de Jong PJ, Nishino S, Mignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Malenka RC, Kauer JA, Perkel DJ, Mauk MD, Kelly PT, Nicoll RA, Waxham MN. An essential role for postsynaptic calmodulin and protein kinase activity in long-term potentiation. Nature. 1989;340:554–557. doi: 10.1038/340554a0. [DOI] [PubMed] [Google Scholar]
- Malinow R, Schulman H, Tsien RW. Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science. 1989;245:862–866. doi: 10.1126/science.2549638. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. Cbln1 and the δ2 glutamate receptor--an orphan ligand and an orphan receptor find their partners. Cerebellum. 2012;11:78–84. doi: 10.1007/s12311-010-0186-5. [DOI] [PubMed] [Google Scholar]
- McCall C, Singer T. The animal and human neuroendocrinology of social cognition, motivation and behavior. Nat Neurosci. 2012;15:681–688. doi: 10.1038/nn.3084. [DOI] [PubMed] [Google Scholar]
- Nicoll RA. Expression mechanisms underlying long-term potentiation: a postsynaptic view. Philos Trans R Soc Lond B Biol. 2003;358:721–726. doi: 10.1098/rstb.2002.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda M, Furutani Y, Takahashi H, Toyosato M, Hirose T, Inayama S, Nakanishi S, Numa S. Cloning and sequence analysis of cDNA for bovine adrenal preproenkephalin. Nature. 1982;295:202–206. doi: 10.1038/295202a0. [DOI] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- Petzold GC, Murthy VN. Role of astrocytes in neurovascular coupling. Neuron. 2011;71:782–797. doi: 10.1016/j.neuron.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Qiao Q, Ma L, Li W, Tsai JW, Yang G, Gan WB. Long-term stability of axonal boutons in the mouse barrel cortex. Dev Neurobiol. 2016;76:252–261. doi: 10.1002/dneu.22311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic P, Bourgeois JP, Goldman-Rakic PS. Synaptic development of the cerebral cortex: implications for learning, memory, and mental illness. Prog Brain Res. 1994;102:227–243. doi: 10.1016/S0079-6123(08)60543-9. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in non-neuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Stevens B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Craig AM. Protein tyrosine phosphatases PTPδ, PTPσ, and LAR: presynaptic hubs for synapse organization. Trends Neurosci. 2013;36:522–534. doi: 10.1016/j.tins.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessier-Lavigne M. Wiring the brain: the logic and molecular mechanisms of axon guidance and regeneration. Harvey Lect. 98:103–143. [PubMed] [Google Scholar]
- Wallace DC. Bioenergetic origins of complexity and disease. Cold Spring Harb Symp Quant Biol. 2011;76:1–16. doi: 10.1101/sqb.2011.76.010462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Nicoll RA. Endogenous cannabinoids mediate retrograde signalling at hippocampal synapses. Nature. 2001;410:588–592. doi: 10.1038/35069076. [DOI] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- Zhu S, Gouaux E. Structure and symmetry inform gating principles of ionotropic glutamate receptors. Neuropharmacology. 2017;112(Pt A):11–15. doi: 10.1016/j.neuropharm.2016.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]