Summary
Vitamin D has multiple roles including the regulation of bone and calcium homeostasis. Deficiency of 25-hydroxyvitamin D, the major circulating form of vitamin D, is associated with an increased risk of age-related chronic diseases including Alzheimer’s disease, Parkinson’s disease, cognitive impairment, and cancer. In this study, we utilized Caenorhabditis elegans to examine the mechanism by which vitamin D influences aging. We found that Vitamin D3-induced lifespan extension requires the stress response pathway genes SKN-1, IRE-1, and XBP-1. Vitamin D3 (D3) induced expression of SKN-1 target genes, but not canonical targets of IRE-1/XBP-1. D3 suppressed an important molecular pathology of aging, that of widespread protein insolubility, and prevented toxicity caused by human β-amyloid. Our observation that D3 improves protein homeostasis and slows aging highlights the importance of maintaining appropriate vitamin D serum levels, and may explain why such a wide variety of human age-related diseases are associated with vitamin D deficiency.
In Brief
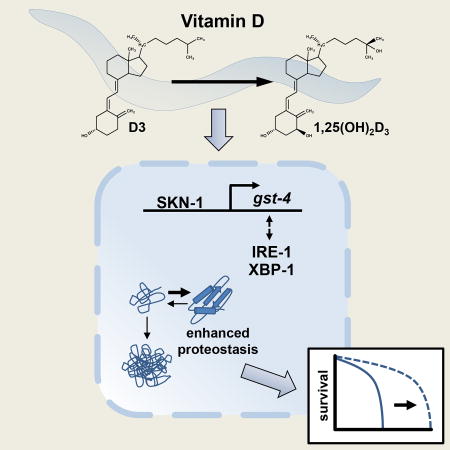
Maintenance of protein homeostasis is crucial to cellular health, and contributes significantly to the lifespan of the organism. Mark et al. demonstrate that vitamin D supplementation promotes protein homeostasis and slows aging in the nematode, C. elegans. These findings identify a mechanism by which Vitamin D influences aging.
Introduction
Our understanding of the role of vitamin D has grown significantly over the last several years with evidence that low levels of vitamin D can have a profound effect on human health (Hossein-Nezhad and Holick, 2013). Following the discovery of the vitamin D receptor (VDR), which is expressed in a wide range of tissues, the role of vitamin D in the prevention and treatment of chronic diseases has become an important area of study (Holick, 1992; Kalueff and Tuohimaa, 2007). Vitamin D deficiency has been linked to various health problems, including cognitive decline, depression, cardiovascular disease, hypertension, type 2 diabetes, and cancer (Butler et al., 2011; Chan, 2011; Holick, 2003; Ingraham et al., 2008; Ito et al., 2011; Liu et al., 2013). During aging, the risk for vitamin D deficiency significantly increases due to reduced nutritional intake of vitamin D, increased adiposity, and decreased cutaneous synthesis of vitamin D. This has led to considerable debate regarding vitamin D supplementation in the elderly and whether deficiencies in vitamin D represent an indicator of ill-health or increases one’s susceptibility to chronic disease (Kupferschmidt, 2012).
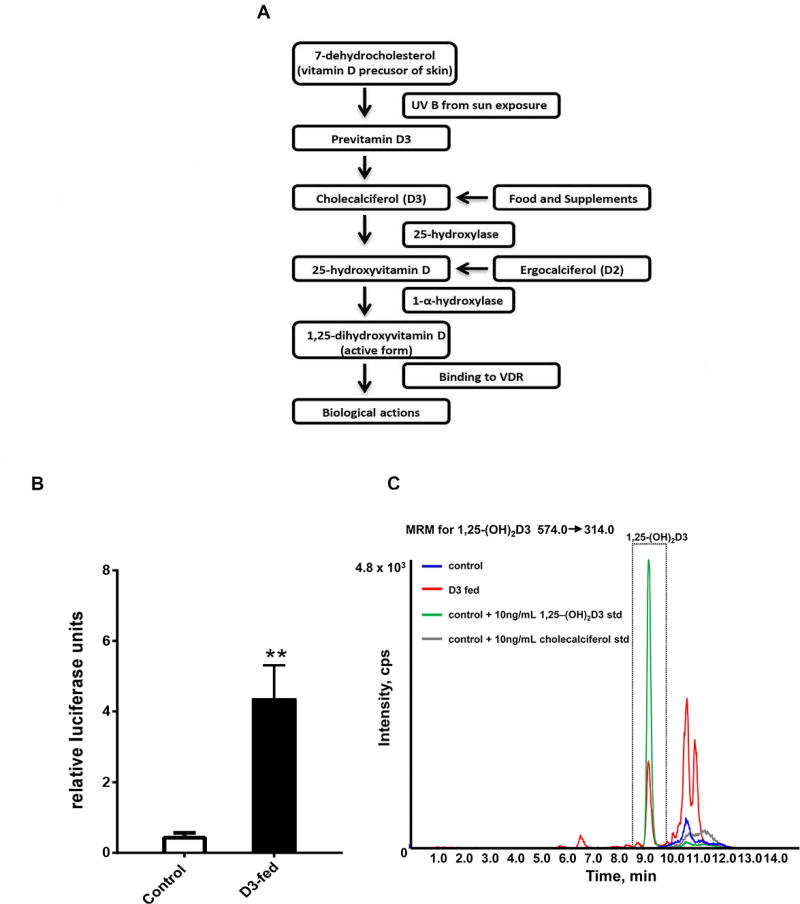
Vitamin D is a member of the superfamily of secosteroid hormones. There are two major forms of vitamin D, vitamin D2 (ergocalciferol; D2), which is produced by the ultraviolet (UV) radiation of ergosterol, and vitamin D3 (cholecalciferol; D3), which is a photoproduct produced in the skin from 7-dehydrocholesterol (7DHC) (Smith and Holick, 1987). The vitamin D photoproduct is biologically inert, requiring two separate hydroxylation steps by cytochrome P450 enzymes to produce the biologically active form of vitamin D, 1,25-dihydroxyvitamin D (1,25-(OH)2D) (Figure 1A). As the concentration of 1,25-(OH)2D increases, VDRs throughout the body become activated, resulting in extensive alterations in gene expression and numerous physiological alterations.
Figure 1. C. elegans fed D3 are capable of synthesizing the biologically active form of D3, 1,25-(OH)2D3, and lipid extracts derived from D3-fed worms can activate human vitamin D receptor (VDR) transcriptional activity.
(A) Diagram of the human vitamin D metabolic pathway. (B) Lipid extracts derived from D3-fed worms activated human VDR transcriptional activity as evidenced by increased luciferase activity compared to control-treated worms. Data are presented as relative luciferase units. Error bars indicate mean + standard error of the mean (SEM) (* P< 0.05, unpaired t-test, n=3). (C) Liquid Chromatography/Mass Spectrometry (LC-MS) extracted ion (MRM of m/z 574→314) chromatogram of detected 1,25-(OH)2D3 from lipid extracts of wild-type (N2) worms synchronously grown until the second day of adulthood on either control or D3 (100 µM) NGM plates. The D3-fed lipid extracts revealed a signal identical to the 1,25-(OH)2D3 standard, indicated by the boxed green signal. There was no 1,25-(OH)2D3 detected in the control lipid extracts.
C. elegans is an excellent model for longevity studies and investigating aspects of chronic disease pathology. Many of the classical signaling pathways and transcription factors that modulate stress response and aging have been identified in the nematode. Enhancing the activity of the FOXO transcription factor DAF-16, which functions in the insulin/IGF-1 signaling pathway, significantly increases lifespan (Kenyon, 2005). Additionally, the activity of the heat shock transcription factor, HSF-1, and the Nrf2-like xenobiotic and oxidative stress-response factor, SKN-1, also affect normal aging in the worm (Tullet et al., 2008). These stress response transcription factors up- or down-regulate a diverse range of target genes.
Protein homeostasis plays an important role in aging and age-related disease. Normal aging in C. elegans is associated with a loss in protein homeostasis and an accumulation of insoluble protein (David et al., 2010; Reis-Rodrigues et al., 2012; Walther et al., 2015). Neurological diseases including Alzheimer's disease (AD), Parkinson's disease (PD), Huntington's disease (HD), and amyotrophic lateral sclerosis (ALS) share common cellular and molecular features including protein aggregation and inclusion body formation. Neurotoxic aggregated forms of endogenous proteins, such as amyloid-β (AD), α-synuclein (PD), huntingtin (HD), and TAR DNA-binding protein 43 kDa (TDP-43; ALS) underlie the pathogenesis of these diseases. Analogous to their effects on longevity, DAF-16, HSF-1, and SKN-1 all contribute to maintenance of protein homeostasis in C. elegans (Alavez et al., 2011; Dostal et al., 2010). In C. elegans, DAF-16 and HSF-1 both regulate the formation of age-induced polyglutamine-repeat protein aggregates, similar to those found in HD (Hsu et al., 2003). Deficiency in either DAF-16 or HSF-1 correlates with premature accumulation of age-associated insoluble protein (Walther et al., 2015). SKN-1 has also been shown to be required for the maintenance of protein homeostasis (Alavez et al., 2011). Additionally, SKN-1 activity is associated with another mechanism previously shown to be important in lifespan extension, the endoplasmic reticulum unfolded protein response (ER-UPR) (Glover-Cutter et al., 2013), which is induced in response to proteotoxic stress in the ER to suppress the accumulation of unfolded or misfolded proteins (Zhang and Kaufman, 2006). In C. elegans, not only does SKN-1 play a prominent role in the transcriptional regulation of the ER-UPR, but specific ER-UPR regulators are also in turn important for SKN-1 target gene expression (Glover-Cutter et al., 2013). Consistent with the hypothesis that impaired protein homeostasis can drive aging, we and others have shown that normal aging is associated with insoluble protein accumulation, and genes encoding these insoluble proteins are enriched for those that determine lifespan (David et al., 2010; Reis-Rodrigues et al., 2012).
Vitamin D has been shown to extend lifespan in C. elegans (Messing et al., 2013). Moreover, short term treatment with vitamin D reduces amyloid-β (Aβ) peptide aggregation and improves cognition in mouse models of AD (Durk et al., 2014). These observations prompted us to investigate whether vitamin D promotes wide-spread cellular protein homeostasis and consequently influences aging. We found that D3 feeding suppressed the toxicity induced by human β-amyloid (Aβ3–42) aggregation and rescued paralysis of worms expressing a metastable perlecan protein. Critically, we found that vitamin D3 treatment slowed proteome-wide, age-related protein insolubility. We examined the mechanism by which vitamin D influences protein homeostasis and longevity and found that the beneficial effects of vitamin D3 require the stress response pathway genes SKN-1, IRE-1, and XBP-1. The novel role for this secosteroid hormone in suppressing age-related proteotoxic stress provides an explanation for the observed elevated risk for neurological disease associated with human vitamin D deficiency.
Results
C. elegans can Metabolize Vitamin D3 to 1,25-dihydroxyvitamin D3
To test the suitability of C. elegans as a model for investigating vitamin D mechanisms, we asked if worms fed vitamin D3 have the ability to produce the bioactive form of vitamin D3, 1,25-dihydroxyvitamin D3 (1,25-(OH)2D3), which is required for VDR activity. We grew large populations of synchronously aging N2 wild-type hermaphrodite worms at 25°C on either vitamin D3- or control-treated NGM plates and prepared lipid extracts on the second day of adulthood (five-day old worms). We tested the worm lipid extracts for biological activity in a one-hybrid human cell-based VDR activity assay and found that lipid extracts made from D3-fed worms showed enhanced human VDR transcriptional activity, as evidenced by increased luciferase activity, compared to control-treated worms (Figure 1B). Addition of vitamin D3 alone to the VDR expressing cells had no effect on VDR transcriptional activity (data not shown). This demonstrated that C. elegans worms are able to metabolize vitamin D3 into a ligand that activates human VDR. To test if worms metabolized vitamin D3 to the known active ligand, 1,25-(OH)2D3, lipid extracts made from vitamin D3 fed worms were subjected to liquid chromatography/mass spectroscopy (LC-MS). A signal identical to the 1,25-(OH)2D3 standard was present in the D3-fed lipid extracts, but not in extracts from control-treated worms (Figure 1C). Quantification of the amount of 1,25-(OH)2D3 in the lipid extracts derived from D3-fed worms revealed approximately 5.95E-03 pg/worm. By comparison, in humans, plasma 1,25-(OH)2D3 levels range from 10–70 pg/ml (Bikle et al., 1984). Since C. elegans are grown on a live Escherichia coli (E. coli) food source, we tested whether exposure of E. coli to D3 would result in 1,25-(OH)2D3 production, but found that the bacteria alone did not make this active form of vitamin D (data not shown). Collectively these data demonstrated that C. elegans are capable of synthesizing 1,25-(OH)2D3, and that lipid extracts derived from these worms can activate human VDR, confirming that this critical component of vitamin D metabolism is conserved between nematodes and mammals.
Vitamin D3-Induced Lifespan Extension Requires SKN-1, IRE-1, and XBP-1
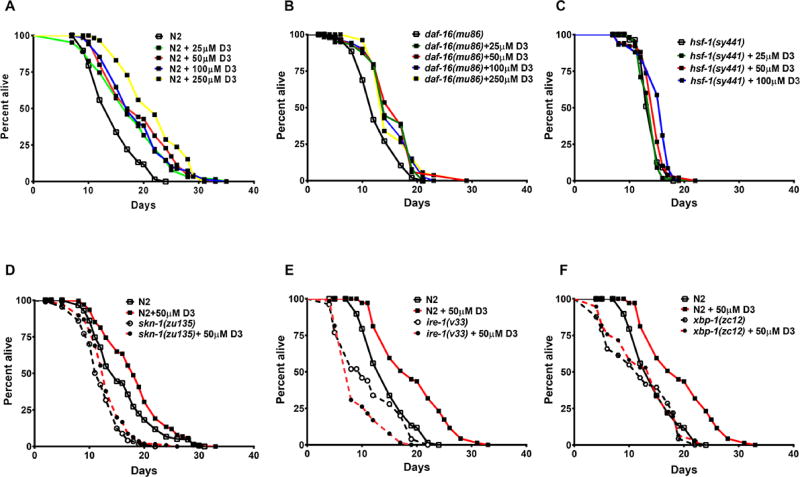
We confirmed that vitamin D3 extended C. elegans lifespan (Messing et al., 2013). Feeding vitamin D3 throughout adulthood extended lifespan in a dose-dependent manner, and was not toxic even at the highest concentration (250µM) tested (Figure 2A; Table S1). Given that normal aging is modulated by a network of transcription factors (Hsu et al., 2003; Tullet et al., 2008), we tested if members of this network were required for the beneficial effects of vitamin D3 on lifespan in C. elegans. Firstly, we tested the requirement of DAF-16 in D3-induced lifespan extension. We found that D3 feeding extended the lifespan of daf-16(mu86) worms, which lack functional DAF-16 protein (Figure 2B; Table S1). Additionally, D3 treatment did not alter the subcellular localization of a DAF-16::GFP fusion protein (TJ356 strain; data not shown). These data suggest that the effect of D3 feeding on lifespan extension is independent of DAF-16. In addition, we found that vitamin D3-induced lifespan extension did not require DAF-12 (Figure S1 A+B; Table S1), the proposed ortholog of VDR in C. elegans (Antebi et al., 2000; Mangelsdorf et al., 1995). Furthermore, in a cell-based luciferase reporter assay, 1,25-(OH)2D3 did not increase DAF-12 transcriptional activity (data not shown). Taken together, we conclude that vitamin D3-induced lifespan extension is independent of DAF-12. We next tested the requirement of the HSF-1 in D3-induced lifespan extension. D3 treatment resulted in marginal or no lifespan extension in hsf-1(sy441) mutant worms (Figure 2C; Table S1) suggesting that the effect of D3 on lifespan may partially require the participation of HSF-1-regulated genes. Lastly, we examined the effect of SKN-1 in D3-induced lifespan extension. We observed no lifespan extension by D3 for skn-1(zu135) mutant worms, demonstrating that SKN-1 is required for the effects of D3 feeding (Figure 2D; Table S1).
Figure 2. Vitamin D3 requires SKN-1, IRE-1, and XBP-1 stress response genes for lifespan extension.
(A) Kaplan-Meier survival curves of N2 hermaphrodite worms exposed to increasing concentrations of D3 from day 1 of adulthood (P< 0.0001; log-rank test). (B) D3 (25–250 µM) extended the lifespan of CF1038 [daf-16(mu86)] worms, which lack functional DAF-16 protein, when treated from day 1 of adulthood at 20°C (P<0.0001, log-rank test). (C) D3 (25–100 µM) feeding resulted in marginal lifespan extension in PS3551 [hsf-1(sy441)] (P=0.0090, Log-rank test). (D) D3 (50 µM) treatment resulted in little or no lifespan extension of the EU31 [skn-1(zu135) IV/nT1 [unc-?(n754) let-?]] worms (P= 0.002, log-rank test). (E) D3 (50 µM) treatment shortens the lifespan in RE666 [ire-1(v33)] mutant worms (P<0.0001, log-rank test). (F) D3 (50 µM) treatment does not extend lifespan in SJ17 [xbp-1(zc12); zcIs4] mutant worms (P=0.142, log-rank test).
SKN-1 activity is associated with another mechanism important in lifespan extension, the endoplasmic reticulum unfolded protein response (ER-UPR) (Glover-Cutter et al., 2013). Specifically, SKN-1 regulates transcription of the entire core of the ER- UPR and many downstream ER stress defense genes. Moreover, ER stress influences the levels of skn-1 mRNA and SKN-1 protein (Glover-Cutter et al., 2013). Proteotoxic stress triggers the ER-UPR by activating the stress sensors ribonuclease inositol requiring protein-1 (IRE-1), PERK kinase homolog (PEK-1), and activating transcription factor-6 (ATF-6) (Calfon et al., 2002; Shen et al., 2001; Shen et al., 2005). Activation of each sensor produces a transcription factor that activates genes to increase the protein-folding capacity in the ER. Of the three stress responsive ER-UPR pathways, IRE-1 is the most conserved. Upon activation of the UPR, IRE1-dependent splicing of a small intron from the xbp-1 mRNA leads to synthesis of XBP-1 transcription factor, which in turn, induces expression of hsp-4 and other ER-UPR associated genes. Given the requirement for SKN-1 in D3-mediated lifespan extension and that SKN-1 and the ER-UPR form a regulatory network, we tested the dependency of each ER-UPR pathway for the lifespan response to D3 feeding. We found that the D3-induced increase on survival was dependent on IRE-1/XBP-1 signaling. Worms carrying the loss-of-function allele, ire-1(v33), showed significantly reduced lifespan with D3 feeding compared to vehicle-treated worms (Figure 2E; Table S1). Lifespan of worms maintaining the loss-of-function allele, xbp-1(zc12), showed no significant change with D3 feeding compared to vehicle-treated worms (Figure 2F; Table S1). Interestingly, ire- 1(v33) mutant worms exhibited a shortened lifespan upon D3 feeding. In contrast, D3 feeding significantly increased lifespan in worms maintaining loss-of-function alleles for pek-1 and atf-6 (Table S1 and Figure S1C). Collectively, these data specifically implicate the stress response genes SKN-1, IRE-1, and XBP-1 in vitamin D3-induced lifespan extension.
Vitamin D3 Induces SKN-1 but Not HSF-1 nor ER-UPR Gene Targets
Given that the effect of D3 feeding on lifespan is dependent on a genetic network, we next surveyed the downstream target genes of HSF-1, SKN-1 and IRE-1/XBP-1. We examined the effect of D3 feeding on the expression of an HSF-1 target gene encoding a molecular chaperone, using the transgenic transcriptional reporter strain, phsp-16.2::GFP. D3 treatment had no effect on the expression of this transcriptional reporter (Figures S2 A+C). We examined other molecular chaperones not under direct regulation by HSF-1, hsp-6 (mitochondrial chaperone) and hsp-4 (ER chaperone). Using the transcriptional reporter stains phsp-6::GFP and phsp-4::GFP, we found that vitamin D3 feeding only increased the levels of hsp-4 expression (Figures S2 A+B+D). HSP-4 is a direct target of the IRE-1/XBP-1 pathway, and it is upregulated in response to ER stress. Surprisingly, we failed to observe a significant D3–associated up-regulation of hsp-4 mRNA levels in wild-type N2 worms as assessed by RNA-sequencing (RNAseq) and quantitative Real-Time PCR (qRT-PCR) at various time points (data not shown). These results indicate that lifespan extension from D3 requires the IRE-1/XBP-1, but does not appear to result in robust constitutive induction of the downstream XBP-1 target gene, hsp-4.
We then tested the effects of D3 treatment on the expression of a target of SKN-1 using a transgenic transcriptional reporter strain pgst-4::GFP. GST-4 (glutathione transferase-4) is involved in the Phase II oxidative stress response and its expression reports on SKN-1 activity. D3 feeding significantly up-regulated pgst-4::GFP compared to control-treated worms (Figure 3A) in a SKN-1 dependent manner (Figure 3B). We confirmed this result by qRT-PCR analysis of endogenous gst-4 mRNA levels (Figure 3C).
Figure 3. Vitamin D3 induced activation of the SKN-1 target gene gst-4 is IRE-1-dependent.
(A) Quantification of a reporter strain (CL2166) containing gst-4p::GFP following D3 feeding (25–100 µM) from L1 larval stage and scored at L4 larval stage at 16oC. Data is represented as GFP Fluorescence (arbitrary units, a.u.). (B) Reducing SKN-1 by skn-1 RNAi prevents the D3-induced increase in pgst-4::GFP fluorescence. Data are presented as percent paralyzed. DMSO bars represent the equivalent amount of solvent used for D3 feeding. (C) Relative gst-4 mRNA levels (mean + SEM) in N2 worms fed D3 (100µM). ire-1 RNAi prevents the D3-induced increase in gst-4 mRNA levels. Data are presented as relative mRNA levels. (* P<0.05, unpaired t-test, n=3).
To gain a more detailed picture of the genomic response to vitamin D3, we undertook a genome-wide analysis of altered mRNA abundance. Specifically, synchronous populations of D3 fed and control L4 stage hermaphrodite worms were processed for RNA-sequencing. We observed 253 significantly up-regulated and 78 significantly down-regulated genes in response to vitamin D3 treatment (data not shown). Gene ontology (GO) analysis of this dataset revealed several clusters of genes with functional properties that are consistent with previously reported microarray studies of 1,25-(OH)2D3 regulated genes (Heikkinen et al., 2011; Hossein-nezhad et al., 2013). These included a significant enrichment of genes associated with apoptosis, immune functions, response to stimulus, transport, cellular component organization, development, and metabolism.
Given the dependency of HSF-1, SKN-1, and IRE-1/XBP-1 in D3-induced lifespan extension, we further examined our RNAseq dataset to determine whether expression of target genes of any of these transcription factors might be perturbed by vitamin D3 treatment. Firstly, we examined if our RNAseq dataset was enriched for heat shock proteins (HSPs), since HSF-1 has been shown to be a major transcriptional regulator of these genes. We observed no significant enrichment for HSPs in the transcriptional effects of D3 feeding. These data are consistent with our previous finding that D3 feeding had no effect on the molecular chaperone transcriptional reporter strain, phsp-16.2::GFP. We next examined SKN-1 gene targets from a previously reported array that examined differential expression between skn-1 knockdown and control worms, profiled at L4 larval stage (Oliveira et al., 2009). Comparison of our dataset with the subset of genes found to be down-regulated in skn-1 knockdown animals revealed a striking enrichment for genes expressed in response to D3 feeding (empirical p = 10−6; Table S2). Genes negatively regulated by SKN-1 were not significantly perturbed by vitamin D (empirical p = 0.48).
Since SKN-1 activates the transcription of genes encoding Phase II detoxification enzymes in response to oxidative stress (An and Blackwell, 2003) and vitamin D induces SKN-1 gene targets, we next evaluated if D3 induced oxidative stress. To test this, we measured reactive oxygen species (ROS) levels, using the superoxide ROS indicator dihydroethidium (DHE), in synchronously aged N2 worms grown from eggs on either vitamin D3- or control-treated NGM plates. We found that DHE-derived fluorescent ethidium levels were unchanged between D3 treated and vehicle-treated worms. In contrast, N2 worms treated with paraquat (PQ), a known oxidative stress inducer, had significantly increased ROS levels compared to both vitamin D3- or control-treated worms (data not shown). We asked if vitamin D3 treated worms differed in their resistance to PQ. Treatment with either vitamin D3 or vehicle control during development and subsequent exposure to PQ resulted in no difference in survival between D3 and control worms. These data indicate that vitamin D does not induce oxidative stress nor oxidative stress resistance.
We next considered if ER-UPR gene targets could be affected by vitamin D feeding. To test this, we used three previously reported definitions of the ER-UPR pathway: genes annotated in the ER-UPR according to the Gene Ontology consortium (amigo.org); genes dependent on ire-1 and/or xbp-1 for response to the UPR inducer tunicamycin (Shen et al., 2005); and genes dependent on pek-1 and/or atf-6 for tunicamycin response. In contrast to our findings with SKN-1, in each cohort, and in a cohort defined as their union, we observed no significant enrichment for the transcriptional effects of D3 feeding (empirical p = 0.17, 0.99, 0.71 and 0.78 respectively), consistent with our single-gene analyses of ER-UPR targets (data not shown). Given the IRE-1/XBP-1 and SKN-1 dependency for D3 lifespan extension, we further examined the crosstalk between these pathways. Interestingly, we found that reduction of IRE-1 by RNAi suppressed the D3-induced increase in gst-4 mRNA levels (Figure 3C). In contrast, reduction of XBP-1 by RNAi resulted in elevated gst-4 mRNA levels in D3-treated worms. Although IRE-1 and XBP-1 together regulate transcription of most inducible ER-UPR genes, there is evidence that IRE-1 may have additional distinct functions independent of XBP-1 (Hollien and Weissman, 2006; Shen et al., 2005; Urano et al., 2000; Yoneda et al., 2001), and that xbp-1(RNAi) increases gst-4 mRNA abundance in the absence of vitamin D treatment (Glover-Cutter et al., 2013).
Vitamin D3 Reduces Age-dependent Insoluble Protein Accumulation
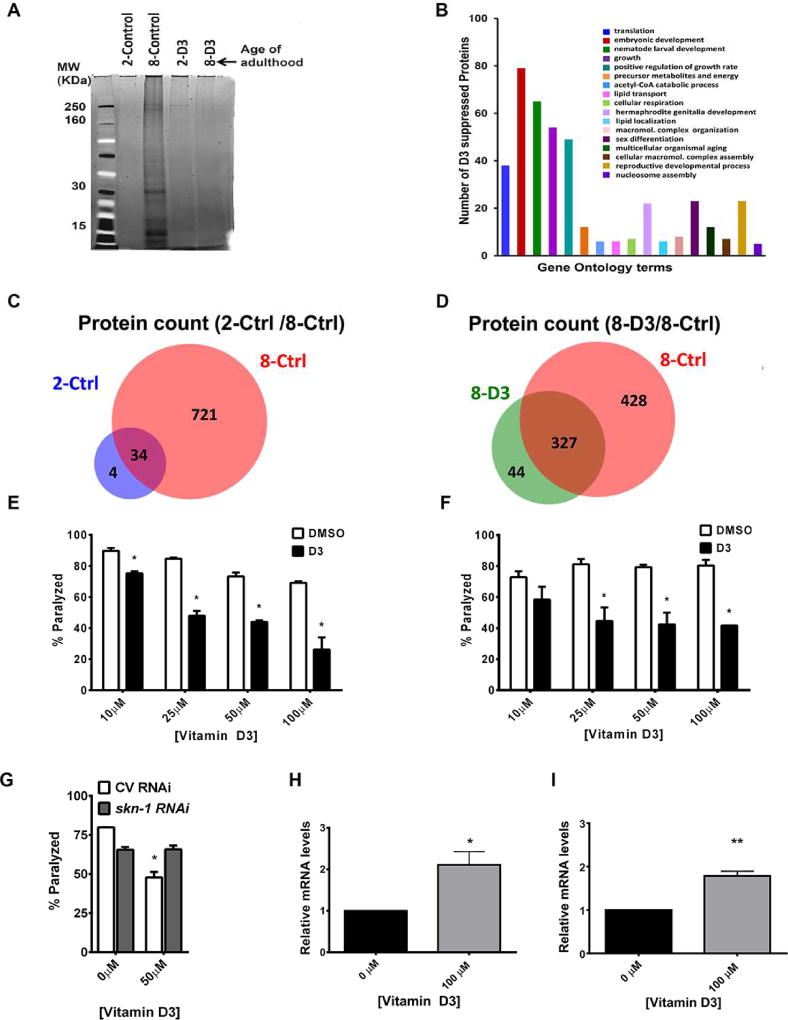
Given the connection between lifespan and protein homeostasis, and the known role for IRE-1/XBP-1 and SKN-1 in both protein homeostasis and longevity, we pursued the hypothesis that vitamin D3 treatment might control lifespan via improving protein homeostasis. Using an unbiased biochemical and proteomic approach, we tested if vitamin D3 modulated protein homeostasis by analyzing the age-dependent accumulation of SDS-insoluble proteins. We grew large populations of synchronously aged, sterile TJ1060 C. elegans and collected worms at day 2 and day 8 of adulthood at 25°C. Worm protein extracts were prepared as described in the experimental procedures section. Purified SDS-insoluble proteins were re-solubilized, subjected to in-solution tryptic digestion, and analyzed by tandem mass spectrometry on a TripleTOF 5600 (Tables S3A–H). We found that in aged worms, D3 treatment significantly decreased the number of detectable and identified SDS-insoluble proteins compared to control samples (Figure 4A–D; Table S3). We next applied a quantitative approach to compare the relative levels of peptides (and thus proteins) between the D3- and control-treated samples, using a label-free quantitative proteomics method referred to as ‘Skyline MS1 Filtering’ (Rardin et al., 2013; Schilling et al., 2012). We determined that D3-feeding significantly reduced the abundance of most proteins detected in aged samples (Figure S3); we observed a 2–13 fold reduction of these proteins (Table S3I–M and Figure S3). GO analysis revealed that the SDS-insoluble fraction in control older worms contained a significant enrichment of proteins associated with ribosomes, translation, mitochondrial function, energy metabolism, growth and development (Figure 4B). D3 treatment reduced the formation of insoluble proteins across a wide range of predicted functions and cellular compartments. Previous work found that reducing expression of several genes encoding proteins suppressed by D3 treatment in aged worms by RNAi resulted in significant lifespan extension (Table S4) (Reis-Rodrigues et al., 2012). Together, this supports the hypothesis that decreasing protein insolubility can prolong lifespan.
Figure 4. Vitamin D3 feeding prevents the accumulation of insoluble proteins in aged C. elegans and slows protein aggregation-associated paralysis.
(A) SDS-PAGE of the SDS-insoluble fraction of cellular proteins from 2 and 8 day adult TJ1060 worms grown at 25°C. D3 (100 µM) prevents the accumulation of SDS-insoluble proteins in aged worms. (B) Gene ontology (GO) analysis of proteins observed in the insoluble fraction of old worms that were suppressed by D3 treatment (as determined by quantitative mass spectrometry/MS1 Filtering). Classification is as assigned by Klusters of Orthologous Groups (KOG). (C–D) Mass spectrometry quantification of unique proteins in (C) young (day 2) and aged (day 8) control treated worms, and (D) comparison between proteins identified in aged (day 8) D3-treated and aged control worms. (E) Exposing worms to D3 (10–100 µM) suppresses the paralysis phenotype associated with protein aggregation in CL4176 expressing Aβ(3–42) in the muscle after 34 hours at 25°C. (F) D3 treatment (10–100 µM) rescues the paralysis in the strain HE250 [unc-52(e669su250)] after 42 hours at 25°C. (G) Reducing SKN-1 by skn-1 RNAi prevents the D3-induced suppression of paralysis in the temperature-sensitive strain HE250. Data are presented as percent paralyzed. DMSO bars represent the equivalent amount of solvent used for D3 treatment (* P<0.05 Multiple t-tests comparison, Holm-Sidak method, alpha=5.0%). Relative gst-4 mRNA levels (mean + SEM) in HE250 (H) and CL4176 (I) worms fed D3 (100µM) (* P<0.05, unpaired t-test). Error bars indicate mean ± standard error of the mean (SEM) and represent the average of three to four independent experiments, 30–40 worms per group, per experiment.
SKN-1, IRE-1, and XBP-1 are required for the Beneficial Effects of Vitamin D3 on Protein Homeostasis
We further investigated whether vitamin D could suppress toxicity associated with expression of the human neurotoxic peptide, amyloid beta. We employed a well characterized model of human proteotoxic disease, the strain CL4176, which expresses an aggregation prone amyloid-β peptide (Aβ3–42) (Drake et al., 2003; McColl et al., 2009). When shifted from a permissive temperature (15°C) to a restrictive temperature (25°C), worms expressing this peptide accumulate Aβ aggregates and become paralyzed. D3 feeding decreased the proportion of paralyzed CL4176 worms in a dose-dependent manner (Figure 4E). To further probe the suppression of Aβ-associated paralysis, we examined the effect of several vitamin D metabolites, many of which can be converted to active vitamin D ligand in humans. We found that all vitamin D metabolites downstream of 7DHC suppressed Aβ-induced paralysis (Figure S4A). We then utilized a protein folding “sensor” strain, HE250, which carries a mutation in the endogenous gene unc-52 resulting in the expression of a metastable muscle specific protein, UNC-52 (perlecan). At 25°C the UNC-52 muta nt protein exhibits altered structure and subsequently causes paralysis (Zengel and Epstein, 1980). D3 suppressed the paralysis of this mutant (Figure 4F), demonstrating that D3 prevents a detrimental physiological outcome of proteostatic loss.
Consistent with our lifespan experiments, we found that reduction of ire-1, xbp-1, or skn-1 by RNAi prevented D3-induced suppression of paralysis in the perlecan HE250 strain (Figure 4G+S4B–C). In contrast, reduction of pek-1 and atf-6 expression by RNAi had no effect on D3-induced suppression of paralysis (Figure S1D). Since vitamin D-induced suppression of paralysis is SKN-1 dependent, we measured gst-4 mRNA expression prior to the onset of paralysis in the protein misfolding strains. We found that D3 significantly increased gst-4 mRNA levels in both HE250 and CL4176 strains (Figure 4H–I).
Discussion
Numerous hormonal and intracellular signaling pathways are conserved between nematodes and mammals. We have demonstrated an apparent conservation of metabolism and action of the hormone vitamin D in C. elegans. D3-fed worms can synthesize physiological levels of bioactive 1,25-(OH)2D3. Unlike mammals, where cholesterol is the major synthesized sterol, the major sterol found endogenously in C. elegans is the provitamin D, 7DHC (Chitwood et al., 1983; Lee et al., 2005). Thus worms have the necessary steroid hormone precursor to synthesize 1,25-(OH)2D3. Populations of C. elegans species dwell on rotting fruits (Felix et al., 2013) where they likely have access to sunlight sufficient to enable the conversion of 7DHC to D3. Whilst it remains to be seen if C. elegans wild strains utilize vitamin D in a natural setting, the conserved metabolism we observe suggests that this organism may be a good model to study the effects of vitamin D on aging and age-related pathologies.
Our results demonstrate that dietary D3 reduced the age-dependent formation of insoluble proteins across a wide range of predicted functions and cellular compartments. D3 feeding also extends lifespan consistent with the hypothesis that protein insolubility is a factor that determines the rate of aging. The dependency on SKN-1 and the up-regulation of SKN-1 gene targets by D3 treatment further suggests that SKN-1 functions to promote protein homeostasis during normal aging. SKN-1 regulates a wide range of stress responses and detoxification factors and is central to a healthy extended lifespan (Blackwell et al., 2015). For example, loss of SKN-1 leads to sensitively to oxidative stress (An and Blackwell, 2003). Oxidative stress can lead to irreversible oxidation, nitration and carbonylation of proteins, which impairs degradation, and enhances aggregation (Poon et al., 2006; Squier, 2001). A recent study showed that vitamin D3 deficiency induces mild oxidative stress in the rat muscle, as observed by increased protein carbonyls and altered antioxidant enzyme activities. Conversely, supplementation with vitamin D3 corrected all of these oxidative stress defects (Bhat and Ismail, 2015). In addition to its role as an regulator of antioxidant functions, SKN-1 plays an important part in maintaining protein homeostasis (Li et al., 2011). SKN-1 maintains protein homeostasis by regulating proteasome subunit gene expression and activity in response to perturbations in either protein synthesis or degradation. Furthermore, the connection between SKN-1 and the ER-UPR indicates cooperativity between these pathways to promote protein homeostasis (Glover-Cutter et al., 2013). Our results further these findings, demonstrating that the beneficial effects of vitamin D3 on lifespan and protein homeostasis are dependent upon the cooperative actions of this stress response network.
The role of SKN-1 in the regulation of detoxification genes is also likely to contribute to the benefits of vitamin D supplementation. Our RNAseq data revealed that D3 feeding resulted in up-regulation of several phase I (cytochrome P450 and short-chain dehydrogenase/reductase), phase II (UGT-UDP-glucuronosyltransferase and gluthathione S transferases) genes, and ATP-binding cassette (ABC) transporters. Collectively, these five gene classes act together in drug metabolism and excretion. Previously, it was reported that long-lived C. elegans dauer larvae and daf-2 mutants shared a significant enrichment for several classes of detoxification genes (McElwee et al., 2004), supporting the theory that aging occurs as a result of internal molecular damage which gives rise to a wide range of toxic lipophilic compounds. This theory postulates that induction of detoxification genes reduce levels of these toxic species that limit lifespan. Our findings suggest that vitamin D3 has a broad effect on systemic detoxification, which in turn could reduce toxic compounds and promote longevity. Thus, further investigation into the biochemical and cellular processes these detoxifying genes might be influencing will be important in understanding the beneficial actions of vitamin D.
In this study, we have shown that vitamin D promotes protein homeostasis and slows aging via IRE-1, XBP-1, and SKN-1 functions. Our results demonstrate that dietary supplementation of C. elegans with D3 results in endogenous 1,25-(OH)2D3 production at a physiologically relevant range and has profound effects on lifespan and protein homeostasis. This is an interesting observation when considered alongside the fact that there is a decline in efficient vitamin D production with age in humans. Whilst the benefits of dietary supplementation in humans is highly controversial (de Paula and Rosen, 2012), there is considerable epidemiological data correlating vitamin D deficiency to multiple diseases. However, causality has not been clearly established, with the possibility that low vitamin D levels are a marker of ill-health (Rosen and Manson, 2010). Our results suggest that, in C. elegans, an absence of vitamin D in the diet accelerates age-related loss of protein homeostasis and shortens lifespan. Supporting this idea, the vitamin D receptor knockout mouse exhibits some premature aging phenotypes (Keisala et al., 2009), although mouse models of hypervitaminosis D also appear to prematurely age (Tuohimaa, 2009). If vitamin D generally affects aging in mammals, it will be of interest to establish whether Nrf2-regulated gene networks have a role to play (Nakai et al., 2014).
Experimental Procedures
Strains
Strains were cultured under standard laboratory conditions. All strains used in this study were obtained from the CGC and are detailed in the Supplemental Experimental Procedures.
Lifespan assays
Lifespan assays were performed as described previously (Lithgow et al., 1995). Nematodes were transferred to fresh compound plates every 3–5 days. All lifespan experiments were performed at 20°C. Lifespan data was analyzed by GraphPad Prism v7.01, and p-values were calculated using the Mantel-Cox log rank-test.
Worm paralysis assays
Synchronized populations of HE250 [unc-52(e669su250)II] and CL4176 [dvIs27[myo::Aβ(3–42)-let 3’UTR(pAF29); pRF4 (rol-6(su1006))] were used in these studies. See Supplemental Experimental Procedures for details on the treatment groups, experimental conditions, and scoring of the paralysis assays.
RNAi knock-down of gene expression
RNAi bacteria strains expressing double-stranded RNA that inactivates specified genes were cultured and used as previously described (Timmons et al., 2001).
Microscopy and quantification of GFP fluorescence
See Supplemental Experimental Procedures for details on mounting and imaging of worms expressing GFP.
Lipid extracts
Lipid extracts were generated by a modification of the method described previously (Gill et al., 2004). See Supplemental Experimental Procedures for details on preparation of worm samples, treatment groups, and experimental conditions.
Liquid Chromatography/Mass Spectrometry (LC-MS)
Diels-alder derivatization of 1,25(OH)2D3 was adapted from previously established methods (Aronov et al., 2008). See Supplemental Experimental Procedures for details on preparation of worm samples, treatment groups, instrument, and experimental conditions.
Plasmid construction
We obtained the vitamin D receptor (VDR) clone (Id # 30343975) from Open Biosystems. VDR amino acid, 141–477 was PCR amplified and cloned into the pBIND vector (Promega, Corp.) as a BamH1- Xba1 fragment and the sequence was verified.
Transfection assays
HEK293T were used in these assays. See Supplemental Experimental Procedures for details on preparation of cells, treatment groups, and experimental conditions. Luciferase activity was normalized to the GF values. Results are expressed as mean ± SEM for three experiments.
C. elegans insoluble protein extraction
TJ1060 [spe-9(hc88)I; fer-15(b26)II] temperature sensitive mutants were grown until gravid adults in synchronous mass cultures (Fabian and Johnson, 1994). See Supplemental Experimental Procedures for details on preparation of worm samples, treatment groups, instrument and experimental conditions.
Gel electrophoresis of SDS-insoluble protein samples
See Supplemental Experimental Procedures for details on preparation of nematode samples. The SDS-insoluble protein fraction was then visualized with SYPRO® Ruby gel staining.
In solution digestion and mass spectrometric analysis of the SDS-insoluble protein
See Supplemental Experimental Procedures for details on preparation of C. elegans samples, treatment groups, instrument and experimental conditions.
Bioinformatic database searches
Mass spectrometric data was searched using the database search engine ProteinPilot (Shilov et al., 2007) (AB SCIEX Beta 4.5, revision 1656) with the Paragon algorithm (4.5.0.0, 1654). A detailed protocol can be found in Supplemental Experimental Procedures.
Quantitative Skyline MS1 Filtering analysis
MS1 chromatogram based quantification was performed in Skyline 1.4 an open source software project (http://proteome.gs.washington.edu/software/skyline) as previously described in detail by Schilling et al (2012). A detailed protocol can be found in Supplemental Experimental Procedures.
Quantitative data-independent acquisitions (DIA), SWATH-MS2 analysis
Proteomic analysis was generated by SWATH-MS2 analysis (Gillet et al., 2012). A detailed protocol can be found in Supplemental Experimental Procedures.
Functional analysis - protein ontology
The web-based program DAVID v.6.7 (The Database for Annotation, Visualization and Integrated Discovery) was used for functional analysis and protein ontology analysis (Huang da, 2008). A detailed protocol can be found in Supplemental Experimental Procedures.
Raw data accession and Panorama spectral libraries
The raw and processed data associated with this manuscript may be downloaded from MassiVE at ftp://MSV000079132@massive.ucsd.edu (username: MSV000079132). The spectral viewer can be accessed at https://panoramaweb.org/labkey/project/Schilling/Nature_Lithgow_Celegans_VitaminD3/begin.view?
RNA-sequencing, gene expression profiling & bioinformatic analysis
RNA was extracted and quantified as described in the Supplemental Experimental Procedures. RNA Samples were then sent to the University of Minnesota BioMedical Genomics Center for Illumina HiSeq RNA-sequencing, where RNAseq (50 bp paired end sequencing) was carried out on a HiSeq2000 according to the manufacturers protocols (Illumina) after size selecting for an insert averaging ~ 200bp. Average quality scores for the completed run across all 12 samples was >30, with an average of greater than 20 million reads per sample. The sequencing reads were then mapped to the worm genome WBcel235 (Genbank ID GCA_000002985.3) for differential gene expression analysis via the “seed and vote” workflow using the package Rsubread (Liao et al., 2014) in bioconductor (Gentleman et al., 2004). For the mapped reads, greater than 95% of sequencing reads in each sample was mapped to the reference worm genome. RNAseq data have been deposited in the NCBI Gene Expression Omnibus under accession number GSE86493.
RNA extraction and Quantitative Real-Time PCR (qRT-PCR)
See Supplemental Experimental Procedures for details on preparation of C. elegans RNA samples, treatment groups, and primer information. Data was compiled from 3 independent experiments and each experiment was conducted in triplicate.
Statistics
Statistical analysis was performed in GraphPad Prism v7.01, as detailed in the figure legends.
Supplementary Material
Highlights.
Vitamin D metabolism is conserved between nematodes and mammals.
Vitamin D prevents the age-dependent accumulation of SDS-insoluble proteins.
Vitamin D enhances lifespan and protein homeostasis via IRE-1, XBP-1 and SKN-1.
Acknowledgments
We thank Michael F. Holick for helpful discuss and providing us with vitamin D metabolites. We thank Gary Scott, Clifford Rosen, Amit Khanna and the members of the G.J.L laboratory for helpful discussion and suggestions. Nematode strains were provided by the Caenorhabditis Genetics Center (CGC). This work was supported by funding from the Larry L. Hillbloom Foundation (G.J.L. and S.M.), The Glenn Foundation for Medical Research (G.J.L., K.J.D., and S.M.) and NIH grants to G.J.L. (UL1024917, supporting the Interdisciplinary Research Consortium on Geroscience, 1R01AG029631-01A1, R21AG048528, and R01AG029631). We thank the Geroscience Mass Spectrometry and Imaging PL1 Core for financial support of this work (PL1 AG032118 to B.W.G.). We also acknowledge the support of instrumentation (TripleTOF 5600) from the NCRR shared instrumentation grant 1S10 OD016281 (B.W.G.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
K.A.M. and G.J.L. conceived the study. K.A.M., K.J.D., and G.J.L. wrote the manuscript with contributions from co-authors. K.A.M., K.J.D., D.B., B.S., S.D., D.J.S., A.R., B.W.G., and M.L. performed the experiments. K.A.M., K.J.D., D.B., B.S., S.D., A.R., R.B.B., T.R.O., and S.M. analyzed the data.
References
- Alavez S, Vantipalli MC, Zucker DJ, Klang IM, Lithgow GJ. Amyloid-binding compounds maintain protein homeostasis during ageing and extend lifespan. Nature. 2011;472:226–229. doi: 10.1038/nature09873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Aronov PA, Hall LM, Dettmer K, Stephensen CB, Hammock BD. Metabolic profiling of major vitamin D metabolites using Diels-Alder derivatization and ultra-performance liquid chromatography-tandem mass spectrometry. Anal Bioanal Chem. 2008;391:1917–1930. doi: 10.1007/s00216-008-2095-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat M, Ismail A. Vitamin D treatment protects against and reverses oxidative stress induced muscle proteolysis. J Steroid Biochem Mol Biol. 2015;152:171–179. doi: 10.1016/j.jsbmb.2015.05.012. [DOI] [PubMed] [Google Scholar]
- Bikle DD, Gee E, Halloran B, Haddad JG. Free 1,25-dihydroxyvitamin D levels in serum from normal subjects, pregnant subjects, and subjects with liver disease. J Clin Invest. 1984;74:1966–1971. doi: 10.1172/JCI111617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell TK, Steinbaugh MJ, Hourihan JM, Ewald CY, Isik M. SKN-1/Nrf, stress responses, and aging in Caenorhabditis elegans. Free Radic Biol Med. 2015 doi: 10.1016/j.freeradbiomed.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler MW, Burt A, Edwards TL, Zuchner S, Scott WK, Martin ER, Vance JM, Wang L. Vitamin D receptor gene as a candidate gene for Parkinson disease. Ann Hum Genet. 2011;75:201–210. doi: 10.1111/j.1469-1809.2010.00631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Chan J. The value of vitamin D supplementation in older people. Nutritional Therapy & Metabolism. 2011;29:8–21. [Google Scholar]
- Chitwood DJ, Lusby WR, Lozano R, Thompson MJ, Svoboda JA. Novel nuclear methylation of sterols by the nematode Caenorhabditis elegans. Steroids. 1983;42:311–319. doi: 10.1016/0039-128x(83)90042-9. [DOI] [PubMed] [Google Scholar]
- David DC, Ollikainen N, Trinidad JC, Cary MP, Burlingame AL, Kenyon C. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 2010;8:e1000450. doi: 10.1371/journal.pbio.1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Paula FJ, Rosen CJ. Vitamin D safety and requirements. Archives of biochemistry and biophysics. 2012;523:64–72. doi: 10.1016/j.abb.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostal V, Roberts CM, Link CD. Genetic mechanisms of coffee extract protection in a Caenorhabditis elegans model of beta-amyloid peptide toxicity. Genetics. 2010;186:857–866. doi: 10.1534/genetics.110.120436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake J, Link CD, Butterfield DA. Oxidative stress precedes fibrillar deposition of Alzheimer's disease amyloid beta-peptide (1–42) in a transgenic Caenorhabditis elegans model. Neurobiol Aging. 2003;24:415–420. doi: 10.1016/s0197-4580(02)00225-7. [DOI] [PubMed] [Google Scholar]
- Durk MR, Han K, Chow EC, Ahrens R, Henderson JT, Fraser PE, Pang KS. 1alpha,25-Dihydroxyvitamin D3 reduces cerebral amyloid-beta accumulation and improves cognition in mouse models of Alzheimer's disease. J Neurosci. 2014;34:7091–7101. doi: 10.1523/JNEUROSCI.2711-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian TJ, Johnson TE. Production of age-synchronous mass cultures of Caenorhabditis elegans. J Gerontol. 1994;49:B145–156. doi: 10.1093/geronj/49.4.b145. [DOI] [PubMed] [Google Scholar]
- Felix MA, Jovelin R, Ferrari C, Han S, Cho YR, Andersen EC, Cutter AD, Braendle C. Species richness, distribution and genetic diversity of Caenorhabditis nematodes in a remote tropical rainforest. BMC Evol Biol. 2013;13:10. doi: 10.1186/1471-2148-13-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill MS, Held JM, Fisher AL, Gibson BW, Lithgow GJ. Lipophilic regulator of a developmental switch in Caenorhabditis elegans. Aging Cell. 2004;3:413–421. doi: 10.1111/j.1474-9728.2004.00126.x. [DOI] [PubMed] [Google Scholar]
- Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, Bonner R, Aebersold R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter KM, Lin S, Blackwell TK. Integration of the unfolded protein and oxidative stress responses through SKN-1/Nrf. PLoS Genet. 2013;9:e1003701. doi: 10.1371/journal.pgen.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen S, Vaisanen S, Pehkonen P, Seuter S, Benes V, Carlberg C. Nuclear hormone 1alpha,25-dihydroxyvitamin D3 elicits a genome-wide shift in the locations of VDR chromatin occupancy. Nucleic Acids Res. 2011;39:9181–9193. doi: 10.1093/nar/gkr654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holick MF. Evolutionary biology and pathology of vitamin D. J Nutr Sci Vitaminol (Tokyo) Spec No. 1992:79–83. doi: 10.3177/jnsv.38.special_79. [DOI] [PubMed] [Google Scholar]
- Holick MF. Vitamin D: A millenium perspective. J Cell Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hossein-Nezhad A, Holick MF. Vitamin d for health: a global perspective. Mayo Clin Proc. 2013;88:720–755. doi: 10.1016/j.mayocp.2013.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossein-nezhad A, Spira A, Holick MF. Influence of vitamin D status and vitamin D3 supplementation on genome wide expression of white blood cells: a randomized double-blind clinical trial. PLoS One. 2013;8:e58725. doi: 10.1371/journal.pone.0058725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Ingraham BA, Bragdon B, Nohe A. Molecular basis of the potential of vitamin D to prevent cancer. Curr Med Res Opin. 2008;24:139–149. doi: 10.1185/030079908x253519. [DOI] [PubMed] [Google Scholar]
- Ito S, Ohtsuki S, Nezu Y, Koitabashi Y, Murata S, Terasaki T. 1alpha,25-Dihydroxyvitamin D3 enhances cerebral clearance of human amyloid-beta peptide(1–40) from mouse brain across the blood-brain barrier. Fluids Barriers CNS. 2011;8:20. doi: 10.1186/2045-8118-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalueff AV, Tuohimaa P. Neurosteroid hormone vitamin D and its utility in clinical nutrition. Curr Opin Clin Nutr Metab Care. 2007;10:12–19. doi: 10.1097/MCO.0b013e328010ca18. [DOI] [PubMed] [Google Scholar]
- Keisala T, Minasyan A, Lou YR, Zou J, Kalueff AV, Pyykko I, Tuohimaa P. Premature aging in vitamin D receptor mutant mice. J Steroid Biochem Mol Biol. 2009;115:91–97. doi: 10.1016/j.jsbmb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Kenyon C. The plasticity of aging: insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Kupferschmidt K. Uncertain verdict as vitamin D goes on trial. Science. 2012;337:1476–1478. doi: 10.1126/science.337.6101.1476. [DOI] [PubMed] [Google Scholar]
- Lee EY, Shim YH, Chitwood DJ, Hwang SB, Lee J, Paik YK. Cholesterol-producing transgenic Caenorhabditis elegans lives longer due to newly acquired enhanced stress resistance. Biochem Biophys Res Commun. 2005;328:929–936. doi: 10.1016/j.bbrc.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Li X, Matilainen O, Jin C, Glover-Cutter KM, Holmberg CI, Blackwell TK. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 2011;7:e1002119. doi: 10.1371/journal.pgen.1002119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lithgow GJ, White TM, Melov S, Johnson TE. Thermotolerance and extended life-span conferred by single-gene mutations and induced by thermal stress. Proc Natl Acad Sci U S A. 1995;92:7540–7544. doi: 10.1073/pnas.92.16.7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HX, Han X, Zheng XP, Li YS, Xie AM. Association of vitamin D receptor gene polymorphisms with Parkinson disease. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2013;30:13–16. doi: 10.3760/cma.j.issn.1003-9406.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McColl G, Roberts BR, Gunn AP, Perez KA, Tew DJ, Masters CL, Barnham KJ, Cherny RA, Bush AI. The Caenorhabditis elegans A beta 1–42 model of Alzheimer disease predominantly expresses A beta 3–42. J Biol Chem. 2009;284:22697–22702. doi: 10.1074/jbc.C109.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElwee JJ, Schuster E, Blanc E, Thomas JH, Gems D. Shared transcriptional signature in Caenorhabditis elegans Dauer larvae and long-lived daf-2 mutants implicates detoxification system in longevity assurance. J Biol Chem. 2004;279:44533–44543. doi: 10.1074/jbc.M406207200. [DOI] [PubMed] [Google Scholar]
- Messing JA, Heuberger R, Schisa JA. Effect of vitamin D3 on lifespan in Caenorhabditis elegans. Curr Aging Sci. 2013;6:220–224. doi: 10.2174/18746098113066660038. [DOI] [PubMed] [Google Scholar]
- Nakai K, Fujii H, Kono K, Goto S, Kitazawa R, Kitazawa S, Hirata M, Shinohara M, Fukagawa M, Nishi S. Vitamin D activates the Nrf2-Keap1 antioxidant pathway and ameliorates nephropathy in diabetic rats. American journal of hypertension. 2014;27:586–595. doi: 10.1093/ajh/hpt160. [DOI] [PubMed] [Google Scholar]
- Oliveira RP, Porter Abate J, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon HF, Vaishnav RA, Getchell TV, Getchell ML, Butterfield DA. Quantitative proteomics analysis of differential protein expression and oxidative modification of specific proteins in the brains of old mice. Neurobiol Aging. 2006;27:1010–1019. doi: 10.1016/j.neurobiolaging.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, Newman JC, Held JM, Cusack MP, Sorensen DJ, Li B, Schilling B, Mooney SD, Kahn CR, Verdin E, et al. Label-free quantitative proteomics of the lysine acetylome in mitochondria identifies substrates of SIRT3 in metabolic pathways. Proc Natl Acad Sci U S A. 2013;110:6601–6606. doi: 10.1073/pnas.1302961110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis-Rodrigues P, Czerwieniec G, Peters TW, Evani US, Alavez S, Gaman EA, Vantipalli M, Mooney SD, Gibson BW, Lithgow GJ, et al. Proteomic analysis of age-dependent changes in protein solubility identifies genes that modulate lifespan. Aging Cell. 2012;11:120–127. doi: 10.1111/j.1474-9726.2011.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen CJ, Manson JE. Frailty: a D-ficiency syndrome of aging? J Clin Endocrinol Metab. 2010;95:5210–5212. doi: 10.1210/jc.2010-2544. [DOI] [PubMed] [Google Scholar]
- Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Lee K, Liu CY, Yang K, Solomon A, Yoshida H, Morimoto R, Kurnit DM, Mori K, et al. Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell. 2001;107:893–903. doi: 10.1016/s0092-8674(01)00612-2. [DOI] [PubMed] [Google Scholar]
- Shen X, Ellis RE, Sakaki K, Kaufman RJ. Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet. 2005;1:e37. doi: 10.1371/journal.pgen.0010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6:1638–1655. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- Smith EL, Holick MF. The skin: the site of vitamin D3 synthesis and a target tissue for its metabolite 1,25-dihydroxyvitamin D3. Steroids. 1987;49:103–131. doi: 10.1016/0039-128x(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Squier TC. Oxidative stress and protein aggregation during biological aging. Exp Gerontol. 2001;36:1539–1550. doi: 10.1016/s0531-5565(01)00139-5. [DOI] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuohimaa P. Vitamin D and aging. J Steroid Biochem Mol Biol. 2009;114:78–84. doi: 10.1016/j.jsbmb.2008.12.020. [DOI] [PubMed] [Google Scholar]
- Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science. 2000;287:664–666. doi: 10.1126/science.287.5453.664. [DOI] [PubMed] [Google Scholar]
- Walther Dirk M, Kasturi P, Zheng M, Pinkert S, Vecchi G, Ciryam P, Morimoto Richardc I, Dobson Christopher M, Vendruscolo M, Mann M, et al. Widespread Proteome Remodeling and Aggregation in Aging C. elegans. Cell. 2015;161:919–932. doi: 10.1016/j.cell.2015.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda T, Imaizumi K, Oono K, Yui D, Gomi F, Katayama T, Tohyama M. Activation of caspase-12, an endoplastic reticulum (ER) resident caspase, through tumor necrosis factor receptor-associated factor 2-dependent mechanism in response to the ER stress. J Biol Chem. 2001;276:13935–13940. doi: 10.1074/jbc.M010677200. [DOI] [PubMed] [Google Scholar]
- Zengel JM, Epstein HF. Identification of genetic elements associated with muscle structure in the nematode Caenorhabditis elegans. Cell Motil. 1980;1:73–97. doi: 10.1002/cm.970010107. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kaufman RJ. The unfolded protein response: a stress signaling pathway critical for health and disease. Neurology. 2006;66:S102–109. doi: 10.1212/01.wnl.0000192306.98198.ec. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.