Summary
DNA double-strand break repair by homologous recombination entails the resection of DNA ends to reveal ssDNA tails, which are used to invade a homologous DNA template. CtIP and its yeast ortholog Sae2 regulate the nuclease activity of MRE11 in the initial stage of resection. Deletion of CtIP in the mouse or SAE2 in yeast engenders a more severe phenotype than MRE11 nuclease inactivation, indicative of a broader role of CtIP/Sae2. Here, we provide biochemical evidence that CtIP promotes long-range resection via the BLM-DNA2 pathway. Specifically, CtIP interacts with BLM and enhances its helicase activity, and it also enhances DNA cleavage by DNA2. Thus, CtIP influences multiple aspects of end resection beyond MRE11 regulation.
eTOC blurb
Biochemical analysis by Daley et al. shows that CtIP functions not just as a cofactor for the MRN complex, but it also stimulates long-range resection by BLM-DNA2-RPA. CtIP interacts with BLM and enhances its helicase activity, and it also upregulates the DNA flap cleavage activity of DNA2.
Introduction
Failure to repair DNA double-strand breaks accurately can lead to a loss of genetic information or chromosomal rearrangements, which then cause cell death or transformation (Liu et al., 2012). Eukaryotic cells eliminate DSBs via non-homologous end joining (NHEJ) or homologous recombination (HR) (Chiruvella et al., 2013; Mehta and Haber, 2014). NHEJ directly rejoins DNA ends and operates throughout the cell cycle. HR becomes an important repair tool in S and G2 cells using the sister chromatid as the information donor (Symington and Gautier, 2011).
For HR to occur, nucleases must degrade the 5′ strands from the break ends to generate 3′ ssDNA tails (Daley et al., 2015). These DNA tails are engaged by the RAD51 recombinase and its accessory factors to search the sister chromatid for the homologous region (Sung et al., 2003). Invasion of the homologous region forms a displacement loop, and repair is completed by DNA synthesis, resolution of nucleoprotein intermediates, and ligation (Mehta and Haber, 2014). The ssDNA generated by end resection also triggers checkpoint activation via the ATR/CHK1 axis (Zou and Elledge, 2003).
Eukaryotes employ at least three distinct nucleases in end resection. MRE11, a subunit of the MRE11-RAD50-NBS1 (MRN) complex, incises the 5′ strand endonucleolytically to initiate long-range resection via two distinct pathways (Garcia et al., 2011). The first employs the 5′ to 3′ exonuclease EXO1, and the second is mediated by the endonuclease DNA2 and the BLM helicase (Daley et al., 2015). The ssDNA binding protein RPA upregulates both pathways and also directs DNA2 to incise only the 5′ strand (Cejka et al., 2010; Nimonkar et al., 2011; Niu et al., 2010). Significant crosstalk exists among MRN, BLM, and EXO1 (Nimonkar et al., 2011).
Genetic studies have suggested that Sae2 enhances endonucleolytic 5′ strand scission by the Mre11-Rad50-Xrs2 (MRX, orthologous to MRN) complex (Garcia et al., 2011; Neale et al., 2005). This allows the MRE11 3′–5′ exonuclease activity to generate a DNA gap to facilitate the initiation of long-range resection (Garcia et al., 2011). Biochemical reconstitution has provided support for this model (Anand et al., 2016; Cannavo and Cejka, 2014).
CtIP/Sae2 likely has roles beyond MRE11 regulation. Notably, sae2Δ cells are more sensitive to DNA damaging agents than cells that harbor the mre11-H125N nuclease mutation (Chen et al., 2015; Mimitou and Symington, 2010). Mouse embryos with the Mre11-H129N nuclease mutation survive to day 7.5–9.5, whereas CtIP −/− animals succumb earlier, at day 3.5–4.0 (Buis et al., 2008; Polato et al., 2014). Moreover, recent findings suggest that Sae2 facilitates the removal of MRX from resected DSBs (Chen et al., 2015; Lisby et al., 2004; Puddu et al., 2015). Epistatic interactions between CtIP and Dna2 have been reported in chicken DT40 and human cells (Hoa et al., 2015), and depletion of CtIP and Dna2 confer comparable resection defects in Xenopus egg extracts (Peterson et al., 2013).
Here, by biochemical reconstitution, we reveal a role for CtIP in long-range resection via BLM-DNA2. Specifically, CtIP interacts with BLM and enhances its helicase activity, and deletion analysis identifies a CtIP region required for BLM stimulation. Furthermore, we find that CtIP upregulates the nuclease activity of DNA2, and that the ability of CtIP to form multimers is important for resection stimulation. Our results provide insights into the multifaceted role of CtIP in HR and have implications for understanding DNA damage checkpoint activation.
Results
Enhancement of BLM helicase by CtIP
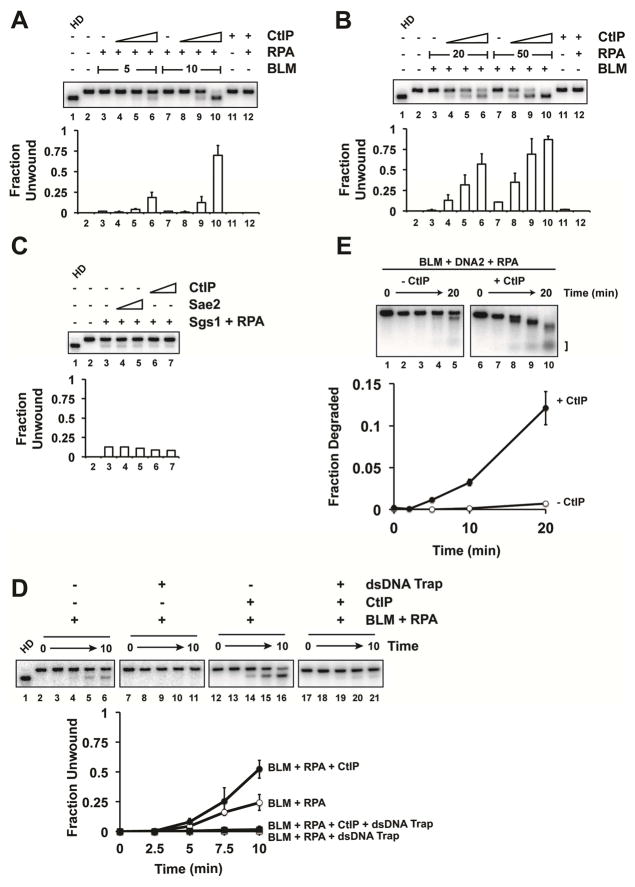
We expressed Flag-tagged CtIP in insect cells (Makharashvili et al., 2014) and purified it (Figure S1A). CtIP and Sae2 were reported to possess nuclease activity (Lengsfeld et al., 2007; Makharashvili et al., 2014; Wang et al., 2014), but we and others have not observed such an activity in either protein (Figure S1B) (Anand et al., 2016; Cannavo and Cejka, 2014; Niu et al., 2010; Sartori et al., 2007). Our preparations of CtIP also do not have helicase (Figure S2D) or ATPase (Figure S2E) activity.
In end resection, the helicase activity of BLM generates single-stranded tails that are engaged by the ssDNA binding protein RPA, which protects the 3′ strand against DNA2 action but facilitates 5′ strand degradation (Nimonkar et al., 2011; Niu et al., 2010). Importantly, we found that CtIP greatly stimulates DNA unwinding when either BLM (Figures 1A and S1C) or RPA (Figure 1B) is at a low concentration, but it has no effect on DNA unwinding in the absence of RPA (Figure S1D). As expected, the helicase-dead BLM-K695R mutant could not unwind DNA even with CtIP present (Figure S1E). Thus, CtIP enhances BLM activity, but it does not fulfill the same role as RPA, which is to sequester unwound ssDNA strands. We note that neither CtIP nor Sae2 could stimulate Sgs1, the yeast ortholog of BLM (Figure 1C).
Figure 1. Effects of CtIP on DNA unwinding by BLM-RPA and resection by BLM-RPA-DNA2.
A. The effect of CtIP (20, 40, or 60 nM) on the unwinding of 2 kb dsDNA (0.5 nM ends) by BLM (5 or 10 nM) and RPA (200 nM) was examined. The incubation time was 20 min. In lane 1, the DNA was heat denatured (HD). B. CtIP (20, 40, or 60 nM) was incubated with BLM (20 nM) and RPA (20 or 50 nM) and the DNA substrate for 20 min. C. Sae2 (20 or 60 nM) or CtIP (20 or 60 nM) was incubated with Sgs1 (20 nM), yeast RPA (100 nM), and the substrate for 20 min. D. BLM (20 nM) and RPA (100 nM) were incubated with the DNA substrate for 2 min. Then, a 10-fold excess of unlabeled DNA was added with CtIP (115 nM). The average of three experiments is shown, and error bars represent one standard deviation (note that the error bars are smaller than the graph symbol at some data points). E. Reconstituted resection assay in which BLM (20 nM), RPA (200 nM), and DNA2 (30 nM) were incubated with the substrate with or without CtIP (80 nM). The products identified by the bracket were quantified. The error bars in the graphs represent the standard deviation of results from three independent experiments.
We previously demonstrated enhancement of BLM helicase processivity by the Topo IIIα-RMI1-RMI2 (TRR) complex (Daley et al., 2014). Here, we enquired whether CtIP affects BLM processivity. Specifically, unlabeled “trap” DNA was introduced after allowing BLM to engage the radiolabeled substrate, and further unwinding was monitored over time. Because BLM is relatively non-processive, unwinding is inhibited when BLM that has dissociated from the substrate becomes sequestered on the trap DNA. The results revealed that CtIP is unable to overcome the effect of the trap DNA (Figure 1D).
Enhancement of BLM-DNA2-dependent resection by CtIP
We asked whether BLM stimulation by CtIP would lead to a corresponding increase in resection in a reconstituted system with DNA2 and RPA, as we documented for the TRR complex (Daley et al., 2014). Indeed, CtIP addition resulted in a strong stimulation of resection (Figure 1E).
Effect of CtIP on DNA fork unwinding
The preceding experiments used a 2 kb substrate. We next asked whether CtIP would affect BLM-mediated unwinding of a substrate generated by annealing complementary 70-nt oligonucleotides (Figure S1F). In contrast to the long dsDNA (e.g. Figure 1A), CtIP did not stimulate unwinding of this short substrate (Figure S1F).
BLM prefers substrates with a 3′ ssDNA tail, including those with a simple overhang or a fork structure that resembles partially unwound DNA (Mohaghegh et al., 2001). We therefore tested the effect of CtIP on the unwinding of a Y structure with 44-nt tails (Figure S1G). BLM was able to unwind this structure regardless of whether RPA was present (Figure S1G) and, interestingly, CtIP enhanced unwinding by BLM alone, but RPA attenuated this stimulatory effect (Figure S1G).
Interaction of BLM with CtIP
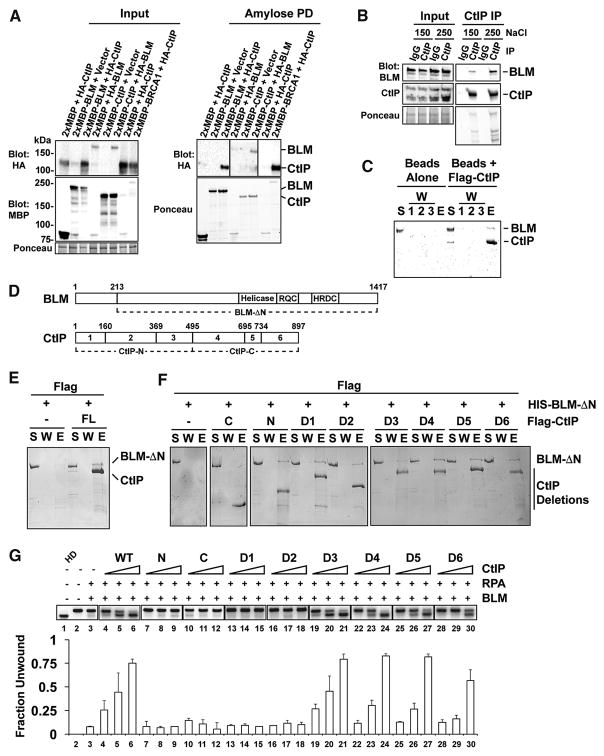
We asked whether CtIP interacts with BLM. First, we expressed 2xMBP- and HA-tagged CtIP and BLM in 293T cells and subjected cell lysates to affinity pulldown with amylose resin. HA-tagged BLM or CtIP was pulled down by 2xMBP-tagged CtIP or BLM, respectively (Figure 2A). As a positive control, we verified that 2xMBP could pull down HA-CtIP (Figure 2A) (Yu et al., 1998). We next performed co-immunoprecipitation with anti-CtIP antibodies to confirm that endogenous BLM and CtIP also interact (Figure 2B). The CtIP-BLM interaction is direct, as anti-Flag resin could pull down purified HIS-tagged BLM via purified Flag-tagged CtIP (Figure 2C).
Figure 2. Interaction of BLM with CtIP.
A. Amylose pulldown was performed with extracts from cells expressing 2xMBP- and HA-tagged CtIP and BLM. The target protein was detected by blotting with anti-HA antibodies. 2xMBP-BRCA1 was included as a positive control for CtIP pulldown. B. Immunoprecipitation with CtIP antibodies was performed, and co-immunoprecipitating BLM was detected by Western blotting with anti-BLM antibodies. Immunoprecipitates were washed with buffer containing the indicated KCl concentration. C. CtIP immobilized on anti-Flag resin was incubated with HIS-tagged BLM, and proteins were eluted from the resin and analyzed by SDS-PAGE with Coomassie blue staining. S, supernatant; W, wash; E, eluate. D. CtIP and BLM deletion mutants used. E–F. Pulldown using CtIP deletion mutants immobilized on Flag beads as in C. G. CtIP deletion mutants were tested for their effect on BLM-mediated DNA unwinding as in Figure 1A using 12 nM BLM, 200 nM RPA, and 15, 30, or 60 nM of the CtIP species.
CtIP domains for BLM interaction
In our effort to dissect the CtIP-BLM interaction, we constructed a series of CtIP deletion mutants that are stable in human cells (Figure 2D) (Liu et al., 2015). First, we divided CtIP into N- and C-terminal halves (CtIP-N, harboring residues 1-495, and CtIP-C, containing residues 496-897) (Figure 2D). We also generated six deletions (CtIP-D1 through D6) (Figure 2D), expressed them in insect cells with an N-terminal Flag tag, and purified them (Figure S2A). Anti-Flag pulldown was used to assess interaction of each CtIP species with BLM. We found that BLM associates with the anti-Flag resin non-specifically. We therefore used BLM-ΔN (Wu et al., 2000), which lacks the first 213 BLM residues, as it retains the ability to interact with CtIP and is less prone to non-specific binding to the affinity resin (Figure 2E). Our analysis revealed that CtIP-N, but not CtIP-C, interacts with BLM-ΔN (Figure 2F). Interestingly, none of the six deletion mutations abrogates BLM interaction (Figure 2F). We therefore concluded that CtIP associates with BLM through its N-terminal region. Since none of the deletions ablates BLM interaction (Figure 2F), we surmise that multiple epitopes within the N-terminal region of CtIP can individually associate with BLM.
We found that neither CtIP-N nor CtIP-C, even at high concentrations (Figure S2B), is capable of BLM stimulation (Figure 2G). While the D1 and D2 deletions abrogate the BLM stimulatory activity, the D3 deletion has little or no negative impact (Figure 2G). The D4, D5, and D6 deletions all affect BLM enhancement, with the D6 mutant being the most severely impaired in this regard (Figure 2G). We note that increasing the concentration of CtIP-D1 leads to slight BLM stimulation (Figure S2B). The D1 region (residues 1-160) contains the protein tetramerization domain, and CtIP-D1 eluted from a gel filtration column several fractions later than the wild-type protein (Figure S2C) (Andres et al., 2015; Davies et al., 2015). The elution profile of CtIP-D2 was only slightly altered compared to wild-type (Figure S2C). Altogether, our results provide evidence for a role of CtIP tetramerization and the CtIP region encompassing residues 161-369 in BLM stimulation (see below also).
Regulation of DNA2 nuclease activity by CtIP
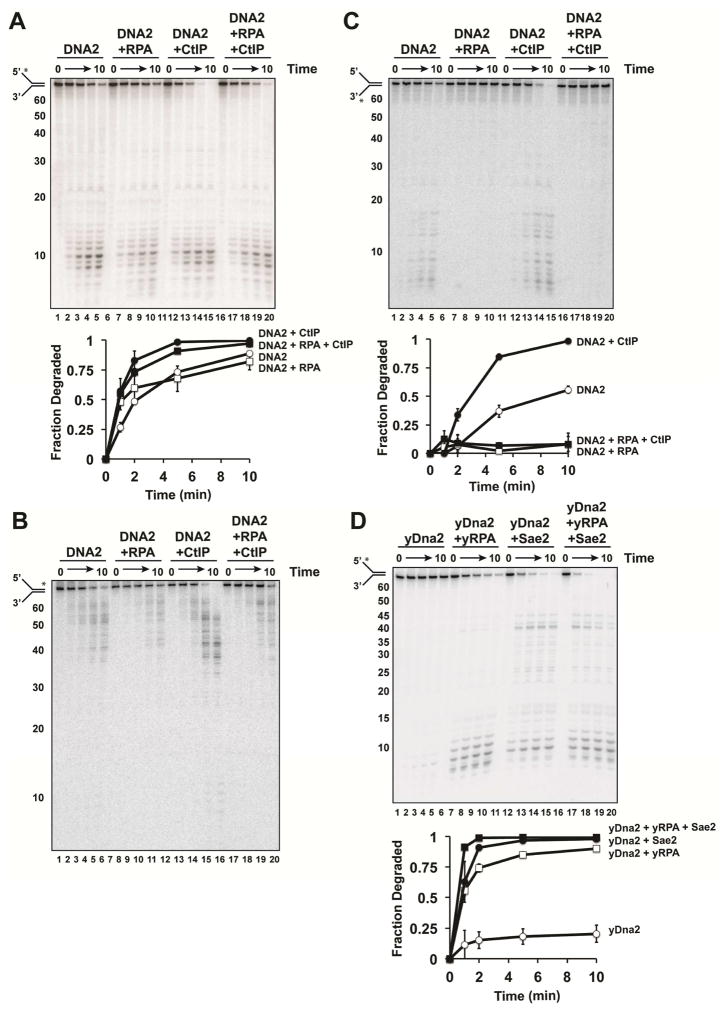
We asked whether CtIP also regulates the nuclease activity of DNA2. For this, DNA2 was incubated with or without CtIP and RPA with a Y substrate containing 44-nt ssDNA overhangs and a 32P label on the terminus of the 5′ flap strand. With DNA2 alone, major products in the range of 8–12 nt were generated (Figure 3A, lanes 1–5). RPA had no effect on the position or rate of the first incision (Figure 3A, lanes 6–10) (Daley et al., 2014). CtIP enhanced DNA2 activity but did not change the incision site (Figure 3A, lanes 11–15). Stimulation by CtIP was observed in the presence of RPA, albeit to a lesser degree (Figure 3A, lanes 16–20). This might reflect competition between RPA and CtIP for substrate access. We note that CtIP, alone or in combination with RPA, does not cause any dissociation of the Y structure (Figure S2D).
Figure 3. Effect of CtIP on DNA2 activity.
A. The Y substrate (2.5 nM; with the radiolabel denoted by the asterisk) was incubated with DNA2 (2 nM), alone or with RPA (5 nM) and/or CtIP (40 nM). B. Same as A, but note the position of the radiolabel (denoted by the asterisk) in the substrate. C. Same as A, but note the radiolabel (denoted by the asterisk) in the substrate. The DNA2 concentration was 5 nM. D. Same as A, except Sae2 (40 nM), yeast Dna2 (2 nM), and yeast RPA (5 nM) were tested.
To assess cleavage events beyond the first incision, we shifted the label to the 3′ end of the 5′ flap strand (Figure 3B). DNA2 alone made initial cuts in the ssDNA region, eventually reaching the ss-ds junction (Figure 3B, lanes 1–5). Interestingly, DNA2 was able to incise the duplex region with CtIP present (Figure 3B, lanes 11–15), but RPA attenuated this effect (Figure 3B, lanes 16–20). Since we did not observe any stimulatory effect of CtIP on ATP hydrolysis by DNA2 (Figure S2E), CtIP likely does not influence the translocation of DNA2 on DNA (Miller et al., 2017).
We used the Y substrate depicted in Figure 3C to assess whether CtIP affects 3′ DNA incision by DNA2. As expected, DNA2 cleaved the 3′ strand, generating products in the range of 5–15 nt (Figure 3C, lanes 1–5). In agreement with published results (Cejka et al., 2010; Daley et al., 2014; Nimonkar et al., 2011; Niu et al., 2010), 3′ cleavage was strongly inhibited by RPA. CtIP stimulated 3′ incision by DNA2 (Figure 3C, lanes 11–15), but little cleavage occurred when RPA was present (Figure 3C, lanes 16–20).
Importantly, Sae2 was able to strongly stimulate the 5′ ssDNA endonuclease activity of yeast Dna2, indicating that the CtIP-DNA2 functional interaction is evolutionarily conserved (Figure 3D). This may explain why sae2Δ cells are more sensitive to DNA damage than mre11-H125N nuclease-deficient cells (Chen et al., 2015; Mimitou and Symington, 2010) despite the lack of Sgs1 stimulation by Sae2 (Figure 1C).
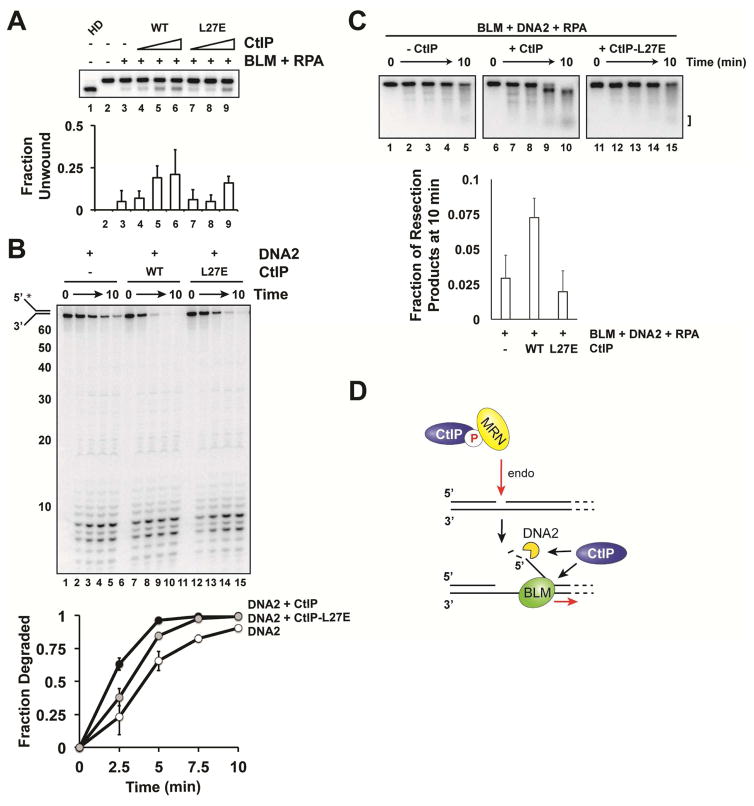
Relevance of CtIP tetramerization in BLM-DNA2 enhancement
The N-terminal region of CtIP harbors a conserved tetramerization domain (Andres et al., 2015; Davies et al., 2015). We have shown that CtIP-D1, which lacks this domain, is unable to stimulate BLM (Figure 2G). The L27E mutation in CtIP compromises protein tetramerization and engenders DNA damage sensitivity in cells (Davies et al., 2015). We expressed and purified the L27E mutant (Figure S1A) for testing. A previous study has shown that the L27E mutation still allows CtIP dimer to form (Davies et al., 2015). Consistent with this result, we found that the L27E mutation causes a moderate shift of the gel filtration profile of CtIP, with the CtIP-D1 mutation imparting a larger shift (Figure S2C). CtIP-L27E exhibited a modest defect in the stimulation of BLM (Figure 4A) and DNA2 (Figure 4B). A much more severe defect was revealed in the reconstituted resection assay containing BLM, DNA2, and RPA (Figure 4C). Thus, CtIP tetramerization is important for its function in long-range resection, and this could explain why the L27E mutant exhibits a strong resection defect in cells (Davies et al., 2015). It will be interesting to test whether the L27E mutation also affects the ability of CtIP to regulate MRN (Anand et al., 2016).
Figure 4. Testing of the CtIP-L27E mutant.
A. CtIP or CtIP-L27E (20, 40, or 60 nM) was incubated with BLM (5 nM) and RPA (200 nM) as in Figure 1A. P-values testing for statistical significance between WT and L27E were 0.35, 0.49, and 0.46 for each concentration of CtIP. B. CtIP-L27E (40 nM) was incubated with DNA2 (2 nM) as in Figure 3A. P-values were 0.49, 0.50, 0.49, and 0.46 for each time point. C. CtIP or CtIP-L27E (45 nM), BLM (30 nM), DNA2 (30 nM) and RPA (200 nM) were incubated with the internally radiolabeled 2kb dsDNA substrate as in Figure 1E.
Discussion
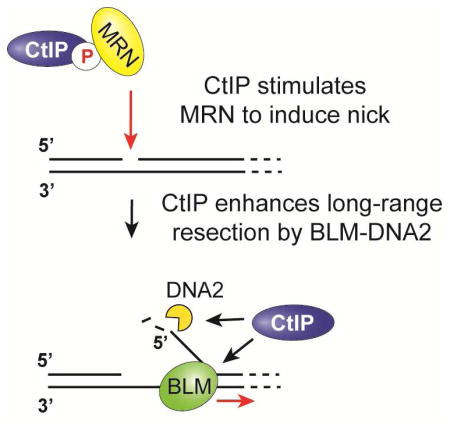
CtIP/Sae2 is considered a MRN/MRX effector in the initiation of DNA end resection. Here, we have documented a role of CtIP in long-range resection. Specifically, we have shown that (1) CtIP interacts with BLM and stimulates its helicase activity; (2) the region of CtIP spanning residues 161-369 is critical for BLM stimulation; (3) CtIP does not enhance the processivity of BLM; (4) CtIP stimulates resection by the DNA2-BLM-RPA ensemble; (5) CtIP upregulates the nuclease activity of DNA2, and this functional interaction also occurs between yeast Sae2 and Dna2; and (6) the CtIP-L27E tetramerization mutant is compromised for resection enhancement. Thus, CtIP stimulates both the initiation of resection by MRN and long-range resection by BLM-DNA2 (Figure 4D).
Previous work has revealed that MRN/X stimulates long-range resection by BLM-DNA2 and Sgs1-Dna2 (Cejka et al., 2010; Nimonkar et al., 2011; Niu et al., 2010). Here, we have demonstrated that CtIP also regulates the activities of BLM and DNA2. Our results highlight how MRN-CtIP could co-ordinate the transition from short-range to long-range resection.
How does CtIP/Sae2 affect such a diverse set of DNA-modifying activities? Sae2 physically interacts with MRX via Xrs2 and promotes MRX endonuclease action at blocked DNA ends (Cannavo and Cejka, 2014; Liang et al., 2015; Oh et al., 2016). We have found a physical interaction between CtIP and BLM and posit that this is important for BLM-mediated DNA unwinding. We have been unable to detect a direct interaction between CtIP and DNA2 (data not shown), but it remains possible that they associate on DNA. Consistent with our model, cell-based assays have shown an epistatic relationship between these proteins (Hoa et al., 2015). We note that S. pombe Ctp1 has high affinity for the fork structure and also a DNA end bridging activity, suggesting a function of CtIP in creating a high localized concentration of DNA ends to accord nucleases and helicases easier access (Andres et al., 2015). That CtIP promotes BLM activity on Y-shaped DNA (Figure S1F–G) is also consistent with the preference of S. pombe Ctp1 for binding such DNA structure (Andres et al., 2015).
Our finding that the tetramerization-impaired CtIP-L27E mutant is defective at BLM-DNA2-RPA stimulation may reflect a role for end tethering in long-range resection. It will be of considerable interest to investigate the functional relevance of CtIP tetramerization and the region of CtIP spanning residues 161-369, which is required for BLM stimulation (Figure 2G), in DNA end resection within the cellular setting. Identification of individual BLM interaction motifs in CtIP will be another important goal.
Materials and Methods
Protein purification
BLM, Sgs1, and DNA2 were expressed and purified as described (Bussen et al., 2007; Daley et al., 2014; Niu et al., 2010; Orren et al., 1999; Raynard et al., 2008). The pTP944 vector that harbors Flag-tagged CtIP (from Tanya Paull) was used to generate a recombinant baculovirus in SF9 insect cells. High-Five insect cells were infected with the baculovirus and harvested after 48 h. Subsequent steps were done between 0–4°C. Extract was prepared by sonication of a cell pellet from 800 ml of culture in 100 ml of K buffer (20 mM KH2PO4, 10% glycerol, 0.5 mM EDTA, 0.5% Tween-20, 1 mM β-mercaptoethanol, and protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, and 5 μg/ml each of aprotinin, chymostatin, leupeptin, and pepstatin) containing 300 mM KCl. After centrifugation (100,000 × g for 45 min), the lysate was incubated with 2 ml of anti-Flag M2 resin (Sigma) for 90 min. The resin was washed sequentially with 15 ml buffer with 300 mM KCl, 500 ml buffer with 1M KCl, 1 mM ATP, and 8 mM MgCl2, and 15 ml buffer with 150 mM KCl. Then, the resin was treated five times with 1 ml of K buffer containing 150 mM KCl and 250 ng/μl of Flag peptide (Sigma) for 30 min to elute proteins. The eluate was fractionated in a 1 ml Heparin column (Amersham) with a 50-ml 150–650 mM KCl gradient. The pool was diluted to 150 mM KCl with buffer, passed through a 0.5 ml Q column, and proteins were eluted in 1 ml buffer containing 500 mM KCl, followed by fractionation in a 24-ml Superose 6 column (GE) in buffer with 500 mM KCl. CtIP was concentrated using a 0.5 ml Q column as above and stored in small aliquots at −80°C. The L27E mutation was introduced by QuickChange and the mutant was expressed and purified as above. Deletion mutants were constructed by PCR (oligonucleotide sequences available upon request), introduced into the 438A MacroBAC vector for expression and purification as above.
The pFB-MBP-Sae2-10xHis vector was a gift from Petr Cejka. Bacmid production, recombinant baculovirus generation, and insect cell culture were as above. Sae2 purification was carried out at 0–4°C. Cell extract was prepared from a cell pellet derived from 1 L culture in 50 ml of T buffer (25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.5 mM EDTA, 0.01% Igepal, and 1 mM DTT) with 300 mM KCl and protease inhibitors (see above). After ultracentrifugation, the lysate was incubated with 4 ml of amylose resin (New England Biolabs) for 1 h. The resin was washed with 250 ml buffer containing 1 M KCl before eluting Sae2 with three aliquots of 4 ml buffer containing 300 mM KCl and 10 mM maltose for 15 min. The eluate was mixed with 3 ml of Ni-NTA resin (Qiagen) for 1 h, and the resin was washed with 250 ml buffer containing 1 M KCl and 20 mM imidazole, and 25 ml buffer with 300 mM KCl and 20 mM imidazole. Then, bound proteins were eluted three times with 3 ml buffer containing 300 mM KCl and 200 mM imidazole for 10 min. The eluate was filter-dialyzed into buffer with 300 mM KCl, concentrated in an Ultracel-30K device (Amicon), and stored in small aliquots at −80°C.
DNA substrates
The 2 kb dsDNA and Y substrates have been described (Daley et al., 2014). The 70-bp substrate (Figure S1F) was made by annealing 5′ radiolabeled PSOL9021 (GTAAGTGCCGCGGTGCGGGTGCCAGGGCGTGCCCTTGGGCTCCCCGGGCGCGTACTCCACCTCATGCATC) with its exact complement PSOL9022. The annealing mixture was resolved in a 10% acrylamide gel in TBE buffer (0.1M Tris, 0.09M boric acid, 0.001M EDTA) at 4°C, electroeluted from gel slices, and concentrated in an Ultracel-30K device.
Helicase and nuclease assays
These were performed in buffer R (20 mM Na-HEPES, pH 7.5, 2 mM ATP, 0.1 mM DTT, 100 μg/ml BSA, 0.05% Triton-X 100, 2 mM MgCl2, and 100 mM KCl) and contained 0.5 nM ends (2 kb substrate) or 2.5 nM DNA molecules (other substrates). Reactions with BLM or Sgs1 also had an ATP regenerating system of 10 mM creatine phosphate and 50 μg/ml creatine kinase. Reactions were incubated at 37°C except when assaying for DNA2 activity on the Y structures, wherein 30°C was used to avoid substrate dissociation. After adding SDS (0.2%) and proteinase K (0.25 mg/ml), the reaction mixtures were incubated for 5 min at 37°C. Products were separated on either agarose gels (for the 2-kb substrate) in TAE buffer (40 mM Tris-acetate, pH 7.4, 0.5 mM EDTA) or acrylamide gels (for oligonucleotide-based substrates) in TBE buffer. For analyzing cleavage products generated by DNA2, reaction mixtures were resolved in polyacrylamide gels under denaturing conditions with 7M urea. Gels were dried onto Hybond membrane and analyzed in a BioRad FX phosphorimager.
Affinity pulldown using amylose resin
The phCMV1 (Genlantis) vector was used for protein expression. The 2XMBP tag with an Asparagine linker and the Precission Protease cleavage sequence or the HA tag was placed at the N-terminus of BLM and CtIP (Jensen et al., 2010). After transfecting 5×105 293TD cells/well in 6-well plates with the vectors (1 μg) using TurboFect (Thermo Fisher Scientific), cells were harvested after 36 h and resuspended in 200 μl buffer A (50 mM HEPES, pH7.5, 350 mM NaCl, 1% Igepal CA-630, 1 mM MgCl2, 1 mM DTT, 250 Units/ml Benzonase (EMD Millipore), and protease inhibitor cocktail (Roche)). Lysates were cleared by centrifugation at 13000 RPM for 10 min and incubated with 20 μl amylose resin (NEB) for 2 h at 4°C, washed 5X with 100 μl buffer B (50 mM HEPES, pH 7.5, 350 mM NaCl, 0.1% Igepal CA-630, 0.5 mM EDTA, and 1 mM DTT), and proteins were eluted with 20 μl buffer containing 10 mM maltose. The eluate was fractionated on a 4–15% gradient SDS-PAGE TGX gel (Bio-Rad). BLM and CtIP were visualized by Stain-Free imaging on a ChemiDoc MP imaging system (Bio-Rad). HA-tagged BLM or CtIP was visualized by immunoblot analysis with anti-HA antibody (1:1,000, Cell Signaling C29F4) and HRP-conjugated anti-rabbit secondary antibody (sc-2004, Santa Cruz Biotechnology). The PVDF membrane was incubated with the WesternC (Bio-Rad) ECL reagent and image was captured on the ChemiDoc MP system. The membrane was treated for 15 min with Restore™ PLUS Stripping Buffer (Thermo Scientific) and re-probed with anti-MBP antibody (1:10000, NEB).
Co-immunoprecipitation of BLM and CtIP
Cells from a 150 mm plate of 90% confluent 293TD were resuspended in 1.5 ml buffer A with 250 mM NaCl (see above) with phosphatase Inhibitors (Roche). Cell lysate was precleared with 35 μl of Protein A/G (Santa Cruz) for 30 min at 4°C. The supernatant was incubated overnight with 2 μg of anti-CtIP (sc-271339, Santa Cruz) antibody before 60 μl of Protein A magnetic resin (Bio-Rad) was added. After a 2 h-incubation, resin was captured with a magnetic block, then washed 5X with 200 μl buffer B, and bound proteins were eluted with 20 μl SDS-PAGE buffer. Immunoblotting was done with anti-BLM (1:5000, ab2179, Abcam) or anti-CtIP (1:1000, sc-271339, Santa Cruz) antibody and HRP-conjugated anti-mouse or anti-rabbit secondary antibody (sc-2004, sc-2005, Santa Cruz). Blots were developed as above. The PVDF membrane was stripped and re-probed.
Affinity pulldown
Flag-tagged CtIP (1 μg) was bound to 15 μl anti-Flag resin and incubated with HIS-tagged BLM (1 μg) for 10 min in K buffer with 100 mM KCl at 4°C. After three washes with 100 μl buffer containing 100, 200, and 300 mM KCl (Figure 2C) or 100 mM KCl (Figure 2E and F), proteins were eluted with SDS-PAGE buffer and analyzed by 7.5% SDS-PAGE with Coomassie blue staining.
Supplementary Material
Highlights.
CtIP enhances long-range resection by BLM-DNA2-RPA
CtIP interacts with and stimulates the activity of the BLM helicase
CtIP also upregulates the endonuclease activity of DNA2
CtIP tetramerization is important for stimulation of long-range resection
Acknowledgments
This study was supported by NIH grants CA220123, CA215990, ES027154, ES007061, and ES015632, and by the American Cancer Society (#IRG 58-012-55). RBJ acknowledges support from the Breast Cancer Alliance, Pilot Project Program Grant from Women’s Health Research at Yale and the Yale Comprehensive Cancer Center. We are grateful to Petr Cejka and Tanya Paull for materials.
Footnotes
Author contributions: JD, JJS, RJ and PS conceived the experiments. JD, WW, AM, XX, and KN purified proteins. JD and JJS performed the experiments. JD and PS analyzed the data and wrote the paper.
Conflict of interest: The authors declare that they have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anand R, Ranjha L, Cannavo E, Cejka P. Phosphorylated CtIP Functions as a Co-factor of the MRE11-RAD50-NBS1 Endonuclease in DNA End Resection. Mol Cell. 2016;64:940–950. doi: 10.1016/j.molcel.2016.10.017. [DOI] [PubMed] [Google Scholar]
- Andres SN, Appel CD, Westmoreland JW, Williams JS, Nguyen Y, Robertson PD, Resnick MA, Williams RS. Tetrameric Ctp1 coordinates DNA binding and DNA bridging in DNA double-strand-break repair. Nat Struct Mol Biol. 2015;22:158–166. doi: 10.1038/nsmb.2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M, Sekiguchi JM, Chang S, Ferguson DO. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell. 2008;135:85–96. doi: 10.1016/j.cell.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussen W, Raynard S, Busygina V, Singh AK, Sung P. Holliday junction processing activity of the BLM-Topo IIIalpha-BLAP75 complex. J Biol Chem. 2007;282:31484–31492. doi: 10.1074/jbc.M706116200. [DOI] [PubMed] [Google Scholar]
- Cannavo E, Cejka P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature. 2014;514:122–125. doi: 10.1038/nature13771. [DOI] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Donnianni RA, Handa N, Deng SK, Oh J, Timashev LA, Kowalczykowski SC, Symington LS. Sae2 promotes DNA damage resistance by removing the Mre11-Rad50-Xrs2 complex from DNA and attenuating Rad53 signaling. Proc Natl Acad Sci U S A. 2015;112:E1880–1887. doi: 10.1073/pnas.1503331112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiruvella KK, Liang Z, Wilson TE. Repair of double-strand breaks by end joining. Cold Spring Harb Perspect Biol. 2013;5:a012757. doi: 10.1101/cshperspect.a012757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Chiba T, Xue X, Niu H, Sung P. Multifaceted role of the Topo IIIalpha-RMI1-RMI2 complex and DNA2 in the BLM-dependent pathway of DNA break end resection. Nucleic Acids Res. 2014;42:11083–11091. doi: 10.1093/nar/gku803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley JM, Niu H, Miller AS, Sung P. Biochemical mechanism of DSB end resection and its regulation. DNA Repair (Amst) 2015;32:66–74. doi: 10.1016/j.dnarep.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies OR, Forment JV, Sun M, Belotserkovskaya R, Coates J, Galanty Y, Demir M, Morton CR, Rzechorzek NJ, Jackson SP, et al. CtIP tetramer assembly is required for DNA-end resection and repair. Nat Struct Mol Biol. 2015;22:150–157. doi: 10.1038/nsmb.2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479:241–244. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradia SD, Ishida JP, Tsai MS, Jeans C, Tainer JA, Fuss JO. MacroBac: New Technologies for Robust and Efficient Large-Scale Production of Recombinant Multiprotein Complexes. Methods Enzymol. 2017;592:1–26. doi: 10.1016/bs.mie.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoa NN, Akagawa R, Yamasaki T, Hirota K, Sasa K, Natsume T, Kobayashi J, Sakuma T, Yamamoto T, Komatsu K, et al. Relative contribution of four nucleases, CtIP, Dna2, Exo1 and Mre11, to the initial step of DNA double-strand break repair by homologous recombination in both the chicken DT40 and human TK6 cell lines. Genes Cells. 2015;20:1059–1076. doi: 10.1111/gtc.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 is an endonuclease that processes hairpin DNA cooperatively with the Mre11/Rad50/Xrs2 complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Suhandynata RT, Zhou H. Phosphorylation of Sae2 Mediates Forkhead-associated (FHA) Domain-specific Interaction and Regulates Its DNA Repair Function. J Biol Chem. 2015;290:10751–10763. doi: 10.1074/jbc.M114.625293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R. Choreography of the DNA damage response: spatiotemporal relationships among checkpoint and repair proteins. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, Jiang P, Wang T, Guo W, Chen Y, et al. The Deubiquitylating Enzyme USP4 Cooperates with CtIP in DNA Double-Strand Break End Resection. Cell Rep. 2015;13:93–107. doi: 10.1016/j.celrep.2015.08.056. [DOI] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makharashvili N, Tubbs AT, Yang SH, Wang H, Barton O, Zhou Y, Deshpande RA, Lee JH, Lobrich M, Sleckman BP, et al. Catalytic and noncatalytic roles of the CtIP endonuclease in double-strand break end resection. Mol Cell. 2014;54:1022–1033. doi: 10.1016/j.molcel.2014.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A, Haber JE. Sources of DNA double-strand breaks and models of recombinational DNA repair. Cold Spring Harb Perspect Biol. 2014;6:a016428. doi: 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AS, Daley JM, Pham NT, Niu H, Xue X, Ira G, Sung P. A novel role of the Dna2 translocase function in DNA break resection. Genes Dev. 2017;31:503–510. doi: 10.1101/gad.295659.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. Ku prevents Exo1 and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 2010;29:3358–3369. doi: 10.1038/emboj.2010.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohaghegh P, Karow JK, Brosh RM, Jr, Bohr VA, Hickson ID. The Bloom’s and Werner’s syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J, Al-Zain A, Cannavo E, Cejka P, Symington LS. Xrs2 Dependent and Independent Functions of the Mre11-Rad50 Complex. Mol Cell. 2016;64:405–415. doi: 10.1016/j.molcel.2016.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J. Activation of DSB processing requires phosphorylation of CtIP by ATR. Mol Cell. 2013;49:657–667. doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polato F, Callen E, Wong N, Faryabi R, Bunting S, Chen HT, Kozak M, Kruhlak MJ, Reczek CR, Lee WH, et al. CtIP-mediated resection is essential for viability and can operate independently of BRCA1. J Exp Med. 2014;211:1027–1036. doi: 10.1084/jem.20131939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puddu F, Oelschlaegel T, Guerini I, Geisler NJ, Niu H, Herzog M, Salguero I, Ochoa-Montano B, Vire E, Sung P, et al. Synthetic viability genomic screening defines Sae2 function in DNA repair. EMBO J. 2015;34:1509–1522. doi: 10.15252/embj.201590973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard S, Zhao W, Bussen W, Lu L, Ding YY, Busygina V, Meetei AR, Sung P. Functional role of BLAP75 in BLM-topoisomerase IIIalpha-dependent holliday junction processing. J Biol Chem. 2008;283:15701–15708. doi: 10.1074/jbc.M802127200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P, Krejci L, Van Komen S, Sehorn MG. Rad51 recombinase and recombination mediators. J Biol Chem. 2003;278:42729–42732. doi: 10.1074/jbc.R300027200. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Wang H, Li Y, Truong LN, Shi LZ, Hwang PY, He J, Do J, Cho MJ, Li H, Negrete A, et al. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol Cell. 2014;54:1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom’s syndrome gene product interacts with topoisomerase III. J Biol Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.