Abstract
An excessive requirement for methionine termed methionine dependence, appears to be a general metabolic defect in cancer. We have previously shown that cancer-cell growth can be selectively arrested by methionine deprivation such as with recombinant methioninase (rMETase). The present study used a previously-established patient-derived orthotopic xenograft (PDOX) nude mouse model of BRAF V600E-mutant melanoma to determine the efficacy of rMETase in combination with a first-line melanoma drug, temozolomide (TEM). In the present study 40 melanoma PDOX mouse models were randomized into four groups of 10 mice each: untreated control (n=10); TEM (25 mg/kg, oral 14 consecutive days, n=10); rMETase (100 units, intraperitoneal 14 consecutive days, n=10); combination TEM + rMETase (TEM: 25 mg/kg, oral rMETase: 100 units, intraperitoneal 14 consecutive days, n=10). All treatments inhibited tumor growth compared to untreated control (TEM: p=0.0081, rMETase: p=0.0037, TEM-rMETase: p=0.0024) on day 14 after initiation. However, the combination therapy of TEM and rMETase was significantly more efficacious than either mono-therapy (TEM: p=0.0051, rMETase: p=0.0051). The present study is the first demonstrating the efficacy of rMETase combination therapy in a PDOX model, suggesting potential clinical development, especially in recalcitrant cancers such as melanoma, where rMETase may enhance first-line therapy.
Keywords: recombinant methioninase, methionine dependence, metabolic targeting, temozolomide, melanoma
INTRODUCTION
Melanoma becomes a recalcitrant cancer when it metastasizes to regional lymph nodes, with a 5-year survival rate of 29%; and 7% when it metastasizes to organs [1]. There is still no cure for stage III and IV melanoma due to drug resistance, tumor heterogeneity and an immune-suppressed tumor microenvironment [1-5]. Temozolomide (TEM), an alkylating agent, is first-line chemotherapy for melanoma but with limited efficacy [1-5]. Targeted chemotherapy and immuno-therapy are also of limited efficacy in melanoma [1-5]. Melanin may also interfere with therapy [4,5]. Therefore, more effective approaches to melanoma treatment are needed.
Toward the goal of precision personalized oncology, our laboratory pioneered the patient-derived orthotopic xenograft (PDOX) nude mouse model with the technique of surgical orthotopic implantation (SOI), including pancreatic [6-9], breast [10], ovarian [11], lung [12], cervical [13], colon [14-16], stomach [17], sarcoma [18-22] and melanoma [23-26].
Previously, a BRAF-V600E-mutant melanoma obtained from the right chest wall of a patient was transplanted orthotopically in the right chest wall of nude mice to establish a PDOX model [24-26]. Trametinib (TRA), an MEK inhibitor, caused tumor regression. In contrast, another MEK inhibitor, cobimetinib (COB) could slow but not arrest growth or cause regression of the melanoma PDOX. TEM could slow but not arrest tumor growth or cause regression [24].
Methionine dependence is a general metabolic defect in cancer. Methionine dependence is due to excess use of methionine for aberrant transmethylation reactions, termed the Hoffman effect, analogous to the Warburg effect for glucose in cancer [27-32]. The excessive and aberrant use of methionine in cancer is strongly observed in [11C] methionine PET imaging, where high uptake of [11C] methionine results in a very strong and selective tumor signal compared with normal tissue background. [11C] methionine is superior to [18C] fluorodeoxyglucose (FDG)-PET for PET imaging, suggesting methionine dependence is more tumor-specific than glucose dependence [33, 34].
A purified methionine cleaving enzyme, methioninase (METase), from Pseudomonas putida has been found previously to be an effective antitumor agent in vitro as well as in vivo [35-38]. For the large-scale production of METase, the gene from P. putida has been cloned in Escherichia coli and a purification protocol for recombinant METase (rMETase) has been established with high purity and low endotoxin [39-41].
It has been demonstrated that methionine deprivation arrests growth and induces a tumor-selective G2-phase cell-cycle arrest of cancer cells in vitro and in vivo [42-45].
MET depletion therapy, using rMETase, sensitized brain tumors to TEM in xenografts in nude mice [46].
We reported recently on the efficacy of rMETase against Ewing’s sarcoma in a PDOX model. rMETase effectively reduced tumor growth compared to untreated control. The methionine level both of plasma and supernatants derived from sonicated tumors was lower in the rMETase group [47].
In the present study, we tested a PDOX nude mouse model of BRAF V600E melanoma for sensitivity to rMETase in combination with TEM.
RESULTS AND DISCUSSION
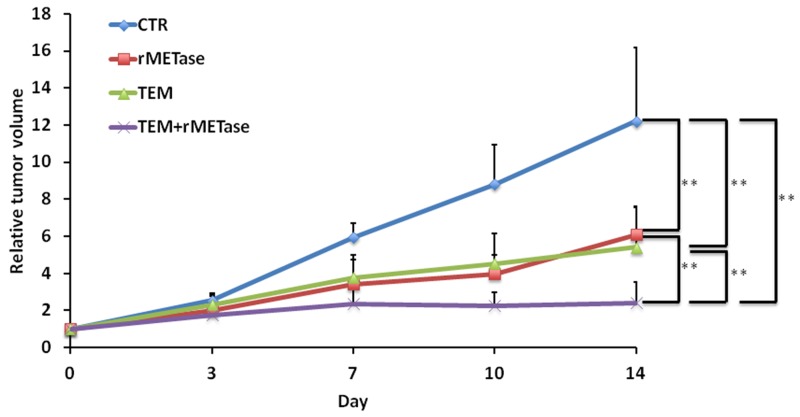
All treatments inhibited tumor growth compared to untreated control (TEM: p=0.0081; rMETase: p=0.0037; TEM-rMETase: p=0.0024) on day 14 after initiation. Combination therapy of TEM and rMETase had significantly better efficacy than either therapy alone (TEM: p=0.0051, rMETase: p=0.0051). There was no significant difference between TEM and rMETase monotherapy (p =0.1282) (Figures 1 and 2).
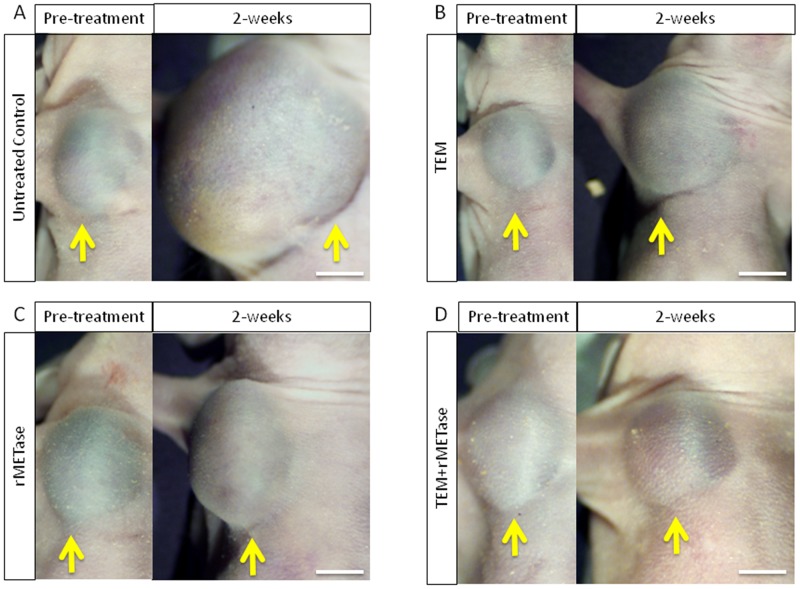
Figure 1. Macroscopic evaluation of therapeutic efficacy of TEM, rMETase and their combination on a BRAF V600E mutant melanoma.
(A) Tumor in untreated control. (B) Temozolomide (TEM). (C) Recombinant methioninase (rMETase). (D) Combination of TEM and rMETase. Yellow arrows show PDOX tumor growing on right chest wall. Scale bar: 5 mm.
Figure 2. Time-coursed treatment efficacy of TEM, rMETase and their combination in the BRAF V600E mutant melanoma.
Line graphs show relative tumor volume at each time point relative to the initial tumor volume. All treatments significantly inhibited tumor growth compared to untreated control (TEM: p=0.0081; rMETase: p=0.0037; TEM-rMETase: p=0.0024). In addition, TEM and rMET combination therapy was significantly more efficacious than either TEM (p=0.0051) or rMETase (p=0.0051) alone at day 14. There was no significantly difference between TEM and rMETase. **p < 0.01. Error bars: ± SD.
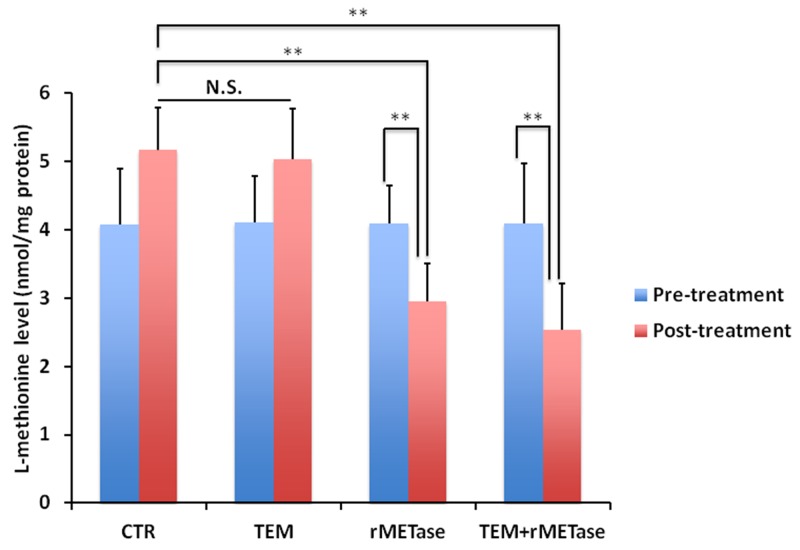
Post-treatment L-methionine levels in tumors treated with rMETase alone or along with TEM significantly decreased compared to untreated control (p < 0.0001) (Figure 3). These results showed that the BRAF-V600E mutant melanoma PDOX is MET dependent and rMETase thereby suppresses its growth. The results also show that TEM similarly suppressed the melanoma PDOX. Future experiments will determine if there are any similarities in the mechanism of tumor inhibition of the two therapeutics.
Figure 3. Intra-tumor L-methionine levels after rMETase treatment.
Bar graphs show L-methionine levels in each treatment group at rMETase or TEM pre- and post-treatment. rMETase significantly decreased intra-tumor L-methionine level. **p < 0.01.
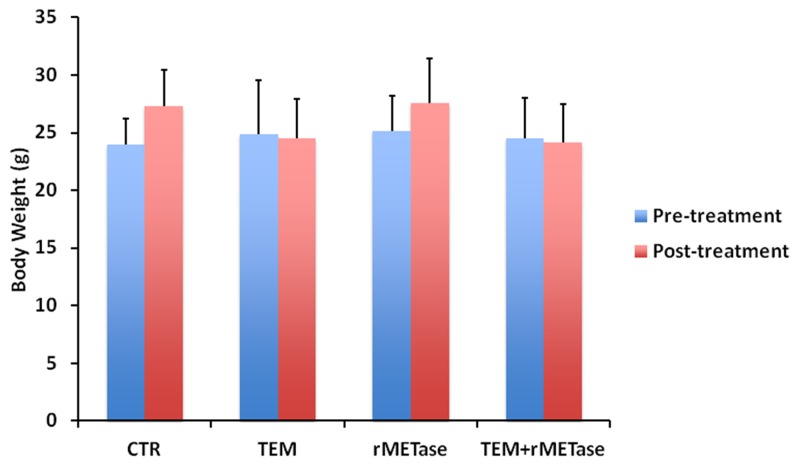
Body weight loss was observed only in the treatment groups including TEM. rMETase alone did not cause body weight loss (Figure 4). There were no animal deaths in any group.
Figure 4. Effect of rMETase or TEM on mouse body weight.
Bar graphs show mouse body weight in each treatment group at pre- and post-treatment.
Histologically, the untreated control tumor was mainly comprised of viable cells. Epithelioid melanoma cells, devoid of melanin, with a high mitotic index, were observed [26]. In the tumors treated with rMETase only, there were still mitotic figures present indicating that rMETase did not completely arrest the tumor. The same degree of necrosis was observed in tumors treated with TEM and rMETase as monotherapy. Tumors treated with the combination of TEM and rMETase showed extensive necrosis, suggesting tumor necrosis is a major pathway of tumor growth arrest, but apoptosis may play a role as well (Figure 5).
Figure 5. Tumor histology in untreated and TEM and rMETase-treated BRAF-V600E mutant melanoma PDOX models.
(A) Untreated control was comprised of viable cells without obvious necrosis. Epithelioid melanoma cells, devoid of melanin, with a high mitotic index are present. (B) Tumor treated with TEM showed partial necrosis. (C) Tumor treated with rMETase. Mitotic figures are present, indicating rMETase did not completely arrest the cell cycle. Tumor treated with rMETase showed partial necrosis similar to TEM. (D) Tumor treated with the combination of TEM and rMETase showed extensive necrosis. White allows: necrotic areas. Scale bars: 50 μm.
TEM is a first-line chemotherapy for melanoma; however, with limited response. The present study has important implications since this is the first in vivo efficacy study of rMETase combination therapy on a patient tumor, in this case, a PDOX model of melanoma, a recalcitrant cancer. We had previously demonstrated that rMETase potentiates TEM in a mouse model of a human glioma cell line [46].
The first hint that methionine metabolism is perturbed in cancer came almost 60 years ago when Sugimura et al. [48] observed that rat tumor growth was slowed by giving the rats a defined diet depleted in methionine. Approximately 45 years ago, it was observed that L5178Y mouse leukemia cells in culture required very high levels of methionine to proliferate [49]. Subsequently, most cancer cell lines were found to be methionine dependence [50, 51]. These cell lines were derived from various cancer types including liver, pancreatic ovarian, submaxillary, brain, lung, bladder, prostate, breast, kidney, cervical, colon, fibrosarcoma, osteosarcoma, rhabdomyosarcoma, leiomyosarcoma, neuroblastoma, glioblastoma and melanoma. The occurrence of methionine dependence among these diverse cancer types suggests that methionine dependence may be a general phenomena in cancer. Normal unestablished cell strains, thus far characterized, grow well in methionine-depleted medium [50].
Human patient tumors, including tumors of the colon, breast, ovary, prostate, and a melanoma, were also found to be methionine dependent in Gelfoam® histoculture [52].
For more about metabolic disturbances in melanoma, please see Slominski et al. [53].
Cell cycle analysis demonstrated that the cells are arrested in the S/G2 phases of the cell cycle upon methionine restriction [42, 43, 52, 54, 55]. This is in contrast to a G1-phase accumulation of cells, which occurs only in methionine-supplemented medium at very high cell densities and is similar to the G1 block seen in cultures of normal fibroblasts at high density.
Recently, a paper appeared with the title “The new anticancer era: tumor metabolism targeting” [56]. This “new anticancer era” started in 1959 with the observation of Sugimura et al. [48] that depriving animals of methionine arrested tumor growth. It is our hope that this era will continue and lead to more effective cancer treatment, especially for recalcitrant cancers such as melanoma.
MATERIALS AND METHODS
Mice
Athymic nu/nu nude mice (AntiCancer Inc., San Diego, CA), 4–6 weeks old, were used in this study. Mice were housed in a barrier facility in a high efficacy particulate arrestance (HEPA)-filtered rack under standard conditions of 12-hour light/dark cycles. The animals were fed an autoclaved laboratory rodent diet. All animal studies were conducted in accordance with the principles and procedures outlined in the National Institutes of Health Guide for the Care and Use of Animals under Assurance Number A3873-1. All mouse surgical procedures and imaging were performed with the animals anesthetized by subcutaneous injection of a ketamine mixture (0.02 ml solution of 20 mg/kg ketamine, 15.2 mg/kg xylazine, and 0.48 mg/kg acepromazine maleate). The response of animals during surgery was monitored to ensure adequate depth of anesthesia. The animals were observed on a daily basis and humanely sacrificed by CO2 inhalation if they met the following humane endpoint criteria: severe tumor burden (more than 20 mm in diameter), prostration, significant body-weight loss, difficulty breathing, rotational motion and body temperature drop.
Patient-derived tumor
A 75-year-old female patient was previously diagnosed with a BRAF-V600E melanoma of the right chest wall. The tumor was previously resected in the Department of Surgery, University of California, Los Angeles (UCLA). Written informed consent was provided by the patient, and the Institutional Review Board (IRB) of UCLA approved this experiment [24-26].
Establishment of PDOX models of melanoma by surgical orthotopic implantation (SOI)
Subcutaneously-grown melanoma was harvested and cut into small fragments (3 mm3). After nude mice were anesthetized with the ketamine solution described above, a 5-mm skin incision was made on the right chest into the chest wall, which was split to make space for the melanoma tissue fragment. A single tumor fragment was implanted orthotopically into the space to establish the PDOX model. The wound was closed with a 6-0 nylon suture (Ethilon, Ethicon, Inc., NJ, USA) [24-26].
Recombinant methionase (rMETase) production
Recombinant L-metionine α-deamino-γ-mercapto-methane lyase (recombinant methioninase, [rMETase]) [EC 4.4.1.11] from Pseudomonas putida has been previously cloned and was produced in Escherichia coli (AntiCancer, Inc.,). rMETase is a homotetrameric PLP enzyme of 172-kDa molecular mass [39].
Treatment study design in the PDOX model of melanoma
PDOX mouse models were randomized into four groups of 10 mice each: untreated control (n=10); TEM (25 mg/kg, oral [p.o.], 14 consecutive days, n=10); rMETase (100 units, intraperitoneal [i.p.], 14 consecutive days, n=10); TEM + rMETase (TEM: 25 mg/kg, p.o., rMETase: 100 units, i.p., 14 consecutive days, n=10). Tumor length and width were measured both pre- and post-treatment. Tumor volume was calculated with the following formula: Tumor volume (mm3) = length (mm) × width (mm) × width (mm) × 1/2. Data are presented as mean ± SD. The tumor volume ratio is defined at the tumor volume at post-treatment time point relative to pre-treatment tumor volume.
Imaging of the melanoma PDOX model
Imaging of the macroscopic tumor was performed with the OV100 Small Animal Imaging System (Olympus, Tokyo, Japan) [63].
Intra-tumor L-methionine level analysis
Each tumor was sonicated for 30 seconds on ice and centrifuged at 12,000 rpm for 10 minutes. Supernatants were collected and protein levels were measured using the Coomassie Protein Assay Kit (Thermo Scientific, Rockford, IL). Protein levels were calculated from a standard curve obtained with a protein standard, bovine serum albumin (BSA). L-methionine levels were determined with the HPLC procedure described previously [47, 64]. Standardized L-methionine levels were calculated per mg tumor protein.
Histological examination
Fresh tumor samples were fixed in 10% formalin and embedded in paraffin before sectioning and staining. Tissue sections (5 μm) were deparaffinized in xylene and rehydrated in an ethanol series. Hematoxylin and eosin (H&E) staining was performed according to standard protocols. Histological examination was performed with a BHS System Microscope (Olympus Corporation, Tokyo, Japan). Images were acquired with INFINITY ANALYZE software (Lumenera Corporation, Ottawa, Canada) [24-26].
Statistical analysis
JMP version 11.0 was used for all statistical analyses. Significant differences for continuous variables were determined using the Mann-Whitney U test. Line graphs expressed average values and error bars show SD. A probability value of P ≤ 0.05 was considered statistically significant [24-26].
CONCLUSIONS
The present study has demonstrated high efficacy of rMETase in combination with TEM in a BRAF-V600E mutant melanoma PDOX model. This is the first report to our knowledge in which rMETase combination therapy was tested on a patient-derived tumor in a mouse model. These results indicate the potential of rMETase combination therapy in the clinic and demonstrate the powerful precision of the PDOX model to identify active drugs and combination therapy on recalcitrant cancer.
Previously-developed concepts and strategies of highly-selective tumor targeting can take advantage of molecular targeting of tumors, including tissue-selective therapy which focuses on unique differences between normal and tumor tissues [57-62].
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
FUNDING
This study was supported in part by National Cancer Institute grant CA213649.
REFERENCES
- 1.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, Hogg D, Lorigan P, Lebbe C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flaherty LE, Othus M, Atkins MB, Tuthill RJ, Thompson JA, Vetto JT, Haluska FG, Pappo AS, Sosman JA, Redman BG, Moon J, Ribas A, Kirkwood JM, Sondak VK. Southwest Oncology Group S0008: a phase III trial of high-dose interferon Alfa-2b versus cisplatin, vinblastine, and dacarbazine, plus interleukin-2 and interferon in patients with high-risk melanoma--an intergroup study of cancer and leukemia Group B, Children’s Oncology Group, Eastern Cooperative Oncology Group, and Southwest Oncology Group. J Clin Oncol. 2014;32:3771–3778. doi: 10.1200/JCO.2013.53.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang H, Wang Y, Chlewicki LK, Zhang Y, Guo J, Liang W, Wang J, Wang X, Fu YX. Facilitating T Cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell. 2016;29:285–296. doi: 10.1016/j.ccell.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brozyna AA, Jóźwicki W, Roszkowski K, Filipiak J, Slominski AT. Melanin content in melanoma metastases affects the outcome of radiotherapy. Oncotarget. 2016;7:17844–17853. doi: 10.18632/oncotarget.7528. https://doi.org/10.18632/oncotarget.7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slominski AT, Carlson JA. Melanoma resistance: a bright future for academicians and a challenge for patient advocates. Mayo Clin Proc. 2014;89:429–433. doi: 10.1016/j.mayocp.2014.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hiroshima Y, Zhang Y, Murakami T, Maawy A, Miwa S, Yamamoto M, Yano S, Sato S, Momiyama M, Mori R, Matsuyama R, Chishima T, Tanaka K, et al. Efficacy of tumor-targeting Salmonella typhimurium A1-R in combination with anti-angiogenesis therapy on a pancreatic cancer patient-derived orthotopic xenograph (PDOX) and cell line mouse models. Oncotarget. 2014;5:12346–12357. doi: 10.18632/oncotarget.2641. https://doi.org/10.18632/oncotarget.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu X, Guadagni F, Hoffman RM. A metastatic nude-mouse model of human pancreatic cancer constructed orthotopically with histologically intact patient specimens. Proc Natl Acad Sci U S A. 1992;89:5645–5649. doi: 10.1073/pnas.89.12.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiroshima Y, Maawy A, Zhang Y, Murakami T, Momiyama M, Mori R, Matsuyama R, Katz MH, Fleming JB, Chishima T, Tanaka K, Ichikawa Y, Endo I, et al. Metastatic recurrence in a pancreatic cancer patient derived orthotopic xenograft (PDOX) nude mouse model is inhibited by neoadjuvant chemotherapy in combination with fluorescence-guided surgery with an anti-CA 19–9-conjugated fluorophore. PLoS One. 2014;9:e114310. doi: 10.1371/journal.pone.0114310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiroshima Y, Maawy AA, Katz MH, Fleming JB, Bouvet M, Endo I, Hoffman RM. Selective efficacy of zoledronic acid on metastasis in a patient-derived orthotopic xenograph (PDOX) nude-mouse model of human pancreatic cancer. J Surg Oncol. 2015;111:311–315. doi: 10.1002/jso.23816. [DOI] [PubMed] [Google Scholar]
- 10.Fu X, Le P, Hoffma RM. A metastatic-orthotopic transplant nude-mouse model of human patient breast cancer. Anticancer Res. 1993;13:901–904. [PubMed] [Google Scholar]
- 11.Fu X, Hoffman RM. Human ovarian carcinoma metastatic models constructed in nude mice by orthotopic transplantation of histologically-intact patient specimens. Anticancer Res. 1993;13:283–286. [PubMed] [Google Scholar]
- 12.Wang X, Fu X, Hoffman RM. A new patient-like metastatic model of human lung cancer constructed orthotopically with intact tissue via thoracotomy in immunodeficient mice. Int J Cancer. 1992;51:992–995. doi: 10.1002/ijc.2910510626. [DOI] [PubMed] [Google Scholar]
- 13.Hiroshima Y, Zhang Y, Zhang N, Maawy A, Mii S, Yamamoto M, Uehara F, Miwa S, Yano S, Murakami T, Momiyama M, Chishima T, Tanaka K, et al. Establishment of a patient-derived orthotopic xenograph (PDOX) model of HER-2-positive cervical cancer expressing the clinical metastatic pattern. PLoS One. 2015;10:e0117417. doi: 10.1371/journal.pone.0117417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu X, Besterman JM, Monosov A, Hoffman RM. Models of human metastatic colon cancer in nude mice orthotopically constructed by using histologically intact patient specimens. Proc Natl Acad Sci U S A. 1991;88:9345–9349. doi: 10.1073/pnas.88.20.9345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metildi CA, Kaushal S, Luiken GA, Talamini MA, Hoffman RM, Bouvet M. Fluorescently-labeled chimeric anti-CEA antibody improves detection and resection of human colon cancer in a patient-derived orthotopic xenograft (PDOX) nude mouse model. J Surg Oncol. 2014;109:451–458. doi: 10.1002/jso.23507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiroshima Y, Maawy A, Metildi CA, Zhang Y, Uehara F, Miwa S, Yano S, Sato S, Murakami T, Momiyama M, Chishima T, Tanaka K, Bouvet M, et al. Successful fluorescence-guided surgery on human colon cancer patient-derived orthotopic xenograft mouse models using a fluorophore-conjugated anti-CEA antibody and a portable imaging system. J Laparoendosc Adv Surg Tech A. 2014;24:241–247. doi: 10.1089/lap.2013.0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furukawa T, Kubota T, Watanabe M, Kitajima M, Hoffman RM. Orthotopic transplantation of histologically intact clinical specimens of stomach cancer to nude mice: correlation of metastatic sites in mouse and individual patient donors. Int J Cancer. 1993;53:608–612. doi: 10.1002/ijc.2910530414. [DOI] [PubMed] [Google Scholar]
- 18.Murakami T, DeLong J, Eilber FC, Zhao M, Zhang Y, Zhang N, Singh A, Russell T, Deng S, Reynoso J, Quan C, Hiroshima Y, Matsuyama R, et al. Tumor-targeting Salmonella typhimurium A1-R in combination with doxorubicin eradicate soft tissue sarcoma in a patient-derived orthotopic xenograft PDOX model. Oncotarget. 2016;7:12783–12790. doi: 10.18632/oncotarget.7226. https://doi.org/10.18632/oncotarget.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiroshima Y, Zhao M, Zhang Y, Zhang N, Maawy A, Murakami T, Mii S, Uehara F, Yamamoto M, Miwa S, Yano S, Momiyama M, Mori R, et al. Tumor-targeting Salmonella typhimurium A1-R arrests a chemo-resistant patient soft-tissue sarcoma in nude mice. PLoS One. 2015;10:e0134324. doi: 10.1371/journal.pone.0134324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiyuna T, Murakami T, Tome Y, Kawaguchi K, Igarashi K, Zhang Y, Zhao M, Li Y, Bouvet M, Kanaya F, Singh A, Dry S, Eilber FC, et al. High efficacy of tumor-targeting Salmonella typhimurium A1-R on a doxorubicin- and dactolisib-resistant follicular dendritic-cell sarcoma in a patient-derived orthotopic xenograft PDOX nude mouse model. Oncotarget. 2016;7:33046–33054. doi: 10.18632/oncotarget.8848. https://doi.org/10.18632/oncotarget.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murakami T, Singh AS, Kiyuna T, Dry SM, Li Y, James AW, Igarashi K, Kawaguchi K, DeLong JC, Zhang Y, Hiroshima Y, Russell T, Eckardt MA, et al. Effective molecular targeting of CDK4/6 and IGF-1R in a rare FUS-ERG fusion CDKN2A-deletion doxorubicin-resistant Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget. 2016;7:47556–47564. doi: 10.18632/oncotarget.9879. https://doi.org/10.18632/oncotarget.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hiroshima Y, Zhang Y, Zhang N, Uehara F, Maawy A, Murakami T, Mii S, Yamamoto M, Miwa S, Yano S, Momiyama M, Mori R, Matsuyama R, et al. Patient-derived orthotopic xenograft (PDOX) nude mouse model of soft-tissue sarcoma more closely mimics the patient behavior in contrast to the subcutaneous ectopic model. Anticancer Res. 2015;35:697–701. [PubMed] [Google Scholar]
- 23.Yamamoto M, Zhao M, Hiroshima Y, Zhang Y, Shurell E, Eilber FC, Bouvet M, Noda M, Hoffman RM. Efficacy of tumor-targeting Salmonella typhimurium A1-R on a melanoma patient-derived orthotopic xenograft (PDOX) nude-mouse model. PLoS One. 2016;11:e0160882. doi: 10.1371/journal.pone.0160882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawaguchi K, Murakami T, Chmielowski B, Igarashi K, Kiyuna T, Unno M, Nelson SD, Russell TA, Dry SM, Li Y, Eilber FC, Hoffman RM. Vemurafenib-resistant BRAF-V600E mutated melanoma is regressed by MEK targeting drug trametinib, but not cobimetinib in a patient-derived orthotopic xenograft (PDOX) mouse model. Oncotarget. 2016;7:71737–71743. doi: 10.18632/oncotarget.12328. https://doi.org/10.18632/oncotarget.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaguchi K, Igarashi K, Murakami T, Chmiewloski B, Kiyuna T, Zhao M, Zhang Y, Singh A, Unno M, Nelson SD, Russell T, Dry SM, Li Y, et al. Tumor-targeting Salmonella typhimurium A1-R combined with temozolomide regresses malignant melanoma with a BRAF-V600 mutation in a patient-derived orthotopic xenograft (PDOX) model. Oncotarget. 2016;7:85929–85936. doi: 10.18632/oncotarget.13231. https://doi.org/10.18632/oncotarget.13231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawaguchi K, Igarashi K, Murakami T, Zhao M, Zhang Y, Chmielowski B, Kiyuna T, Nelson SD, Russell TA, Dry SM, Li Y, Unno M, Eilber FC, et al. Tumor-targeting Salmonella typhimurium A1-R sensitizes melanoma with a BRAF-V600E mutation to vemurafenib in a patient-derived orthotopic xenograft (PDOX) nude mouse model. J Cell Biochem. 2017;118:2314–2319. doi: 10.1002/jcb.25886. [DOI] [PubMed] [Google Scholar]
- 27.Hoffman RM, Erbe RW. High in vivo rates of methionine biosynthesis in transformed human and malignant rat cells auxotrophic for methionine. Proc Natl Acad Sci U S A. 1976;73:1523–1527. doi: 10.1073/pnas.73.5.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern PH, Mecham JO, Wallace CD, Hoffman RM. Reduced free-methionine in methionine-dependent SV40-transformed human fibroblasts synthesizing apparently normal amounts of methionine. J Cell Physiol. 1983;117:9–14. doi: 10.1002/jcp.1041170103. [DOI] [PubMed] [Google Scholar]
- 29.Stern PH, Wallace CD, Hoffman RM. Altered methionine metabolism occurs in all members of a set of diverse human tumor cell lines. J Cell Physiol. 1984;119:29–34. doi: 10.1002/jcp.1041190106. [DOI] [PubMed] [Google Scholar]
- 30.Stern PH, Hoffman RM. Elevated overall rates of transmethylation in cell lines from diverse human tumors. In Vitro. 1984;20:663–670. doi: 10.1007/BF02619617. [DOI] [PubMed] [Google Scholar]
- 31.Hoffman RM. Altered methionine metabolism, DNA methylation and oncogene expression in carcinogenesis: a review and synthesis. Biochim Biophys Acta. 1984;738:49–87. doi: 10.1016/0304-419x(84)90019-2. [DOI] [PubMed] [Google Scholar]
- 32.Coalson DW, Mecham JO, Stern PH, Hoffman RM. Reduced availability of endogenously synthesized methionine for S-adenosylmethionine formation in methionine dependent cancer cells. Proc Natl Acad Sci U S A. 1982;79:4248–4251. doi: 10.1073/pnas.79.14.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffman RM. Is DNA methylation the new guardian of genome? Mol Cytogenetics. 2017;10:11. doi: 10.1186/s13039-017-0314-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman RM. The wayward methyl group and the cascade to cancer. Cell Cycle. 2017;16:825–829. doi: 10.1080/15384101.2017.1304330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lishko VK, Lishko OV, Hoffman RM. The preparation of endotoxin-free L-methionine-alpha-deamino-gamma-mercaptomethane-lyase (L-methioninase) from Pseudomonas putida. Protein Expr Purif. 1993;4:529–533. doi: 10.1006/prep.1993.1069. [DOI] [PubMed] [Google Scholar]
- 36.Lishko VK, Lishko OV, Hoffman RM. Depletion of serum methionine by methioninase in mice. Anticancer Res. 1993;13:1465–1468. [PubMed] [Google Scholar]
- 37.Tan Y, Zavala J, Sr, Xu M, Zavala J, Jr, Hoffman RM. Serum methionine depletion without side effects by methioninase in metastatic breast cancer patients. Anticancer Res. 1996;16:3937–3942. [PubMed] [Google Scholar]
- 38.Tan Y, Zavala J, Sr, Han Q, Xu M, Sun X, Tan X, Tan X, Magana R, Geller J, Hoffman RM. Recombinant methioninase infusion reduces the biochemical endpoint of serum methionine with minimal toxicity in high-stage cancer patients. Anticancer Res. 1997;17:3857–3860. [PubMed] [Google Scholar]
- 39.Tan Y, Xu M, Tan X, Tan X, Wang X, Saikawa Y, Nagahama T, Sun X, Lenz M, Hoffman RM. Overexpression and large-scale production of recombinant L-methionine-alpha- deamino-gamma-mercaptomethane-lyase for novel anticancer therapy. Protein Expr Purif. 1997;9:233–245. doi: 10.1006/prep.1996.0700. [DOI] [PubMed] [Google Scholar]
- 40.Inoue H, Inagaki K, Sugimoto M, Esaki N, Soda K, Tanaka H. Structural analysis of the L-methionine gamma-lyase gene from Pseudomonas putida. J Biochem. 1995;117:1120–1125. doi: 10.1093/oxfordjournals.jbchem.a124816. [DOI] [PubMed] [Google Scholar]
- 41.Hori H, Takabayashi K, Orvis L, Carson DA, Nobori T. Gene cloning and characterization of Pseudomonas putida L-methionine-alpha-deamino-gamma-mercaptomethane-lyase. Cancer Res. 1996;56:2116–2122. [PubMed] [Google Scholar]
- 42.Guo H, Lishko VK, Herrera H, Groce A, Kubota T, Hoffman RM. Therapeutic tumor-specific cell cycle block induced by methionine starvation in vivo. Cancer Res. 1993;53:5676–5679. [PubMed] [Google Scholar]
- 43.Hoffman RM, Jacobsen SJ. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc Natl Acad Sci U S A. 1980;77:7306–7310. doi: 10.1073/pnas.77.12.7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kokkinakis DM, von Wronski MA, Vuong TH, Brent TP, Schold SC., Jr Regulation of O6-methylguanine-DNA methyltransferase by methionine in human tumour cells. Br J Cancer. 1997;75:779–788. doi: 10.1038/bjc.1997.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kokkinakis DM, Schold SC, Jr, Hori H, Nobori T. Effect of long-term depletion of plasma methionine on the growth and survival of human brain tumor xenografts in athymic mice. Nutr Cancer. 1997;29:195–204. doi: 10.1080/01635589709514624. [DOI] [PubMed] [Google Scholar]
- 46.Kokkinakis DM, Hoffman RM, Frenkel EP, Wick JB, Han Q, Xu M, Tan Y, Schold SC. Synergy between methionine stress and chemotherapy in the treatment of brain tumor xenografts in athymic mice. Cancer Res. 2001;61:4017–4023. [PubMed] [Google Scholar]
- 47.Murakami T, Li S, Han Q, Tan Y, Kiyuna T, Igarashi K, Kawaguchi K, Hwang HK, Miyaki K, Singh AS, Nelson SD, Dry SM, Li Y, et al. Recombinant methioninase effectively targets a Ewing’s sarcoma in a patient-derived orthotopic xenograft (PDOX) nude-mouse model. Oncotarget. 2017;8:35630–35638. doi: 10.18632/oncotarget.15823. https://doi.org/10.18632/oncotarget.15823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sugimura T, Birnbaum SM, Winitz M, Greenstein JP. Quantitative nutritional studies with water-soluble, chemically defined diets. VIII. The forced feeding of diets each lacking in one essential amino acid. Arch Biochem Biophys. 1959;81:448–455. doi: 10.1016/0003-9861(59)90225-5. [DOI] [PubMed] [Google Scholar]
- 49.Chello PL, Bertino JR. Dependence of 5-methyltetrahydrofolate utilization by L5178Y murine leukemia cells in vitro on the presence of hydroxycobalamin and transcobalamin II. Cancer Res. 1973;33:1898–1904. [PubMed] [Google Scholar]
- 50.Mecham JO, Rowitch D, Wallace CD, Stern PH, Hoffman RM. The metabolic defect of methionine dependence occurs frequently in human tumor cell lines. Biochem Biophys Res Commun. 1983;117:429–434. doi: 10.1016/0006-291x(83)91218-4. [DOI] [PubMed] [Google Scholar]
- 51.Tan Y, Xu M, Hoffman RM. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells in vitro. Anticancer Res. 2010;30:1041–1046. [PubMed] [Google Scholar]
- 52.Guo HY, Herrera H, Groce A, Hoffman RM. Expression of the biochemical defect of methionine dependence in fresh patient tumors in primary histoculture. Cancer Res. 1993;53:2479–2483. [PubMed] [Google Scholar]
- 53.Slominski A, Kim TK, Brożyna AA, Janjetovic Z, Brooks DL, Schwab LP, Skobowiat C, Jóźwicki W, Seagroves TN. The role of melanogenesis in regulation of melanoma behavior: melanogenesis leads to stimulation of HIF-1α expression and HIF-dependent attendant pathways. Arch Biochem Biophys. 2014;563:79–93. doi: 10.1016/j.abb.2014.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yano S, Li S, Han Q, Tan Y, Bouvet M, Fujiwara T, Hoffman RM. Selective methioninase-induced trap of cancer cells in S/G2 phase visualized by FUCCI imaging confers chemosensitivity. Oncotarget. 2014;5:8729–8736. doi: 10.18632/oncotarget.2369. https://doi.org/10.18632/oncotarget.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yano S, Takehara K, Zhao M, Tan Y, Han Q, Li S, Bouvet M, Fujiwara T, Hoffman RM. Tumor-specific cell-cycle decoy by Salmonella typhimurium A1-R combined with tumor-selective cell-cycle trap by methioninase overcome tumor intrinsic chemoresistance as visualized by FUCCI imaging. Cell Cycle. 2016;15:1715–1723. doi: 10.1080/15384101.2016.1181240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Borriello A, Della Ragione F. The new anticancer era: tumor metabolism targeting. Cell Cycle. 2017;16:310–311. doi: 10.1080/15384101.2016.1271635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Blagosklonny MV. Matching targets for selective cancer therapy. Drug Discov Today. 2003;8:1104–1107. doi: 10.1016/s1359-6446(03)02806-x. [DOI] [PubMed] [Google Scholar]
- 58.Blagosklonny MV. Teratogens as anti-cancer drugs. Cell Cycle. 2005;4:1518–1521. doi: 10.4161/cc.4.11.2208. [DOI] [PubMed] [Google Scholar]
- 59.Blagosklonny MV. Treatment with inhibitors of caspases, that are substrates of drug transporters, selectively permits chemotherapy-induced apoptosis in multidrug-resistant cells but protects normal cells. Leukemia. 2001;15:936–941. doi: 10.1038/sj.leu.2402127. [DOI] [PubMed] [Google Scholar]
- 60.Blagosklonny MV. Target for cancer therapy: proliferating cells or stem cells. Leukemia. 2006;20:385–391. doi: 10.1038/sj.leu.2404075. [DOI] [PubMed] [Google Scholar]
- 61.Apontes P, Leontieva OV, Demidenko ZN, Li F, Blagosklonny MV. Exploring long-term protection of normal human fibroblasts and epithelial cells from chemotherapy in cell culture. Oncotarget. 2011;2:222–233. doi: 10.18632/oncotarget.248. https://doi.org/10.18632/oncotarget.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blagosklonny MV. Tissue-selective therapy of cancer. Br J Cancer. 2003;89:1147–1151. doi: 10.1038/sj.bjc.6601256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamauchi K, Yang M, Jiang P, Xu M, Yamamoto N, Tsuchiya H, Tomita K, Moossa AR, Bouvet M, Hoffman RM. Development of real-time subcellular dynamic multicolor imaging of cancer cell trafficking in live mice with a variable-magnification whole-mouse imaging system. Cancer Res. 2006;66:4208–4214. doi: 10.1158/0008-5472.CAN-05-3927. [DOI] [PubMed] [Google Scholar]
- 64.Sun X, Tan Y, Yang Z, Li S, Hoffman RM. A rapid HPLC method for the measurement of ultra-low plasma methionine concentrations applicable to methionine depletion therapy. Anticancer Res. 2005;25:59–62. [PubMed] [Google Scholar]