Abstract
Age‐related macular degeneration (AMD) is a leading cause of irreversible blindness in the Western world. Although, the majority of stem cell research to date has focused on production of retinal pigment epithelial (RPE) and photoreceptor cells for the purpose of evaluating disease pathophysiology and cell replacement, there is strong evidence that the choroidal endothelial cells (CECs) that form the choriocapillaris vessels are the first to be lost in this disease. As such, to accurately evaluate disease pathophysiology and develop an effective treatment, production of patient‐specific, stem cell‐derived CECs will be required. In this study, we report for the first time a stepwise differentiation protocol suitable for generating human iPSC‐derived CEC‐like cells. RNA‐seq analysis of the monkey CEC line, RF/6A, combined with two statistical screens allowed us to develop media comprised of various protein combinations. In both screens, connective tissue growth factor (CTGF) was identified as the key component required for driving CEC development. A second factor tumor necrosis factor (TNF)‐related weak inducer of apoptosis receptor was also found to promote iPSC to CEC differentiation by inducing endogenous CTGF secretion. CTGF‐driven iPSC‐derived CEC‐like cells formed capillary tube‐like vascular networks, and expressed the EC‐specific markers CD31, ICAM1, PLVAP, vWF, and the CEC‐restricted marker CA4. In combination with RPE and photoreceptor cells, patient‐specific iPSC derived CEC‐like cells will enable scientists to accurately evaluate AMD pathophysiology and develop effective cell replacement therapies. Stem Cells Translational Medicine 2017;6:1533–1546
Keywords: Age related macular degeneration • Human induced pluripotent stem cells • Choroidal endothelial cells • Connective tissue growth factor • TNF‐related weak inducer of apoptosis receptor
Significance Statement.
One of the main causes of terminal blindness in developed countries is age‐related macular degeneration (AMD). There is strong evidence indicating that the vasculature bed located behind the retina, comprised of choroidal endothelial cells (CECs), is the first cell type to be lost in AMD. In this study, we developed a novel strategy to differentiate human iPSCs into CEC‐like cells. This new method will make it possible to study AMD patient‐specific CECs in vitro to better understand AMD pathogenesis and to develop autologous cell replacement therapies to replenish patients' damaged choroids with healthy CECs.
Introduction
In the United States alone, there are seven million individuals with early‐stage AMD and two million more with severe late‐stage disease 1. The prevalence of AMD and the need for an effective treatment increases dramatically with advancing age, and is therefore expected to double as the nation's “baby boomer” population ages and medical advancements continue to result in increased life span 2.
Although AMD pathogenesis is not fully understood, several studies suggest that the vascular bed directly posterior to the retina, the choriocapillaris, degenerates early in disease. In fact, there is a strong body of evidence indicating that choroidal endothelial cells (CECs) that form these choriocapillary vessels are the first cells lost in patients with AMD 3, 4, 5, 6, 7. Choriocapillary vessels are crucial for retinal health, because these vessels nourish the outer retinal cells and shuttle waste out of the retina. Once CECs degenerate, the choroidal vasculature no longer supports the high metabolic demands of the retinal pigment epithelium (RPE) and photoreceptor cells 3, 6, 8, 9, 10. As a consequence, photoreceptor cells, RPE, and CECs all deteriorate in turn in advanced‐stage AMD. Since photoreceptor and RPE cells are terminally differentiated, cellular replacement therapy is the most promising option for restoring vision.
To date, several protocols for generating patient‐specific iPSC‐derived photoreceptor precursor and RPE cells have been described 11, 12, 13. Although current AMD modeling studies using these two cell types have provided valuable insights, qualitative steps forward in our understanding of AMD pathophysiology will likely require the inclusion of studies on patient‐derived CECs. To that end, we recently described a coculture based protocol that was sufficient for the generation of murine iPSC‐derived CECs 14. Unfortunately, due to the risk of cross‐contamination associated with cellular coculture, this system is less than ideal for generating patient‐specific iPSC‐derived CECs for in vitro disease modeling and, especially, for human transplantation. Progress in translational AMD research would be accelerated by a stepwise differentiation protocol suitable for the derivation and purification of patient‐specific CECs. Such an approach requires that we (a) identify factors that promote iPSC to CECs differentiation, (b) select for differentiated cells, and (c) devise a method that is current good manufacturing practice (cGMP) compatible.
To address these goals, we first used our previously published RNA‐seq data, obtained from the RF/6A monkey CEC line, and two statistical screens to determine the secreted factors that drive human iPSC to CEC differentiation 15. Next, we developed a step‐wise differentiation protocol to generate human iPSC‐derived CEC‐like cells that were morphologically indistinguishable from native CECs. Finally, we used an antibiotic resistance selection method to generate a pure population of human iPSC‐derived CEC‐like cells. The successful development of this protocol provides a reliable means of generating patient‐specific iPSC‐derived CEC‐like cells that researchers may further modify into a cGMP‐compatible differentiation protocol for cell‐based therapies.
Materials and Methods
Human iPSC Generation
To generate human iPSCs, our previously published protocol was used 13. Briefly, we used the nonintegrating CytoTune‐iPS 2.0 Sendai Reprogramming Kit (Thermo Fisher Scientific, Waltham, MA; Cat. No. A16517, https://www.thermofisher.com/us/en/home.html), which is designed to drive expression of the transcription factors OCT4, SOX2, KLF4, and C‐MYC, to reprogram dermal fibroblasts isolated from a 47‐year‐old male with normal ocular history into iPSCs 16. After 3 weeks post‐infection, iPSC colonies were manually isolated and clonally expanded on laminin 521‐coated tissue culture plates (ThermoFisher Scientific, Cat. No. A29249) in E8 medium (Thermo Fisher Scientific; Cat. No. A1517001) supplemented with human 10 ng/ml bFGF (Syd Labs, Natick, MA; Cat. No.BP000030‐GD4, http://www.sydlabs.com/). Clones were passaged 10 times prior to analysis. Lines determined to be pluripotent with normal karyotype were selected.
Validating iPSC Pluripotency with the TaqMan Human Pluripotent Stem Cell Scorecard
Pluripotency of human iPSCs was verified using the TaqMan Human Pluripotent Stem Cell Scorecard Panel (Thermo Fisher Scientific; Cat. No. A15870 and Cat. No. A15872). One microgram of total RNA from passage 10 human iPSCs was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA, https://www.qiagen.com/us/) and reverse‐transcribed using the Superscript VILO cDNA Synthesis Kit (Thermo Fisher Scientific; Cat. No. 11754050). cDNA was added to a hPSC Scorecard Plate (Thermo Fisher Scientific; Cat. No. A15870) and amplified using the QuantStudio 6 Flex Real‐Time polymerase chain reaction (PCR) System with a 384‐well capability (Thermo Fisher Scientific). Finally, the gene expression data was uploaded to the web‐based hPSC Scorecard Analysis Software (Thermo Fisher Scientific) for interpretation. We used this software to compare our newly generated data to reference human iPSC data sets to determine self‐renewal and germ layer scores 17, 18. We analyzed genetic expression of known markers for self‐renewal (n = 9), ectoderm (n = 22), mesoderm (n = 22), and endoderm (n = 26) using a TaqMan Scorecard Assay (full list of genes in Supporting Information Table 1). The dependent variable was the ratio of expression values (cycle thresholds) for each gene relative to an internal control. These internal control values, supplied by the manufacturer, represent pooled expression data for each gene from several verified pluripotent cell lines 17, 18. Fold change values between 0.5 and 2 are considered comparable to the control, while values below 0.5 or above 2 are a relative measure of downregulation or upregulation, respectively. The fold change data failed a 95% confidence interval D'Agostino‐Pearson test for normality, so we transformed using a base‐10 logarithm. The transformed data passed the aforementioned normality test. To determine whether a set of genes was comparable to the control, we performed a two‐tailed t test at a 95% confidence interval with a null hypothesis that the mean of each group was equal to zero.
Table 1.
Presence (+) or absence (−) of factors in media used for Taguchi L12 test conditions
| Test condition | Factors of interest | ||||
|---|---|---|---|---|---|
| CTGF | CTNNB1 | SHC1 | TWEAKR | VEGFB | |
| 1 | − | − | − | − | − |
| 2 | − | − | − | + | + |
| 3 | − | − | + | − | − |
| 4 | − | + | + | − | + |
| 5 | − | + | + | + | − |
| 6 | − | + | − | + | + |
| 7 | + | − | − | − | + |
| 8 | + | − | + | + | + |
| 9 | + | − | + | + | − |
| 10 | + | + | − | − | − |
| 11 | + | + | − | + | − |
| 12 | + | + | + | − | + |
Abbreviations: CTGF, connective tissue growth factor; TWEAKR, TNF‐related weak inducer of apoptosis receptor; VEGFB, vascular endothelial growth factor B.
Embryoid Body Formation
We prepared single‐cell suspensions of human iPSCs using TrypLE Express (Thermo Fisher Scientific; Cat. No. 12604‐013) and passaged them into 96‐well ultra‐low adhesion plates (Corning, Corning, NY; Cat. No. CLS7007, https://www.corning.com/worldwide/en.html) at a density of 10,000 cells per well in embryoid body (EB) formation basal medium [dulbecco's modified eagle medium (DMEM) (Thermo Fisher Scientific; Cat. No. 12430054), 10% KSR (Thermo Fisher Scientific; Cat. No. 10828‐028), 2% B27 (Thermo Fisher Scientific; Cat. No. 17504044), 1% N2 (Thermo Fisher Scientific; Cat. No. 17502048), L‐glutamine (Thermo Fisher Scientific; Cat. No. 25030081), 1X non‐essential amino acids (Thermo Fisher Scientific; Cat. No. 11140050), and 0.2% primocin (Thermo Fisher Scientific; Cat. No. ant‐pm‐2)]. At 48 hours post‐passaging, we added MAXgel (Sigma‐Aldrich, St. Louis, MO; Cat. No. E0282‐1ML, https://www.sigmaaldrich.com/united-states.html) to the EB formation medium at a concentration of 1 μg/ml. EBs were feed with EB form basal medium supplemented with MAXgel every other day for 1 week.
Isolating Primary CECs
Human donor eyes were supplied by the Iowa Lions Eye Bank (Iowa City, IA) following informed consent from the families of the donors. All experiments using human donor eyes were performed according to the Declaration of Helsinki. Primary CECs were harvested from donor eyes within 8 hours of donor death. Briefly, we dissected the anterior chamber to remove the vitreous and lens from the enucleated globes. Next, we removed the neural retina, scraped away the RPE using a number 10 scalpel and collected a series of choroid/sclera samples using a 6 mm biopsy punch (Thermo Fisher Scientific; Cat. No. NC9324386). The choroid was separated from the sclera and plated scleral side down on MAXgel‐coated 6‐well tissue culture plates. Choroid samples were fed with basal medium and incubated at 37°C with 5% CO2 for 10 days to allow choroidal cells to grow out. Once cultures were confluent, cells were passaged using trypsin onto porous cell culture inserts for coculture experiments.
Human iPSC‐Derived EB Coculture
As a proof‐of‐concept, coculture experiments were performed to verify that human iPSCs could differentiate into CEC‐like cells as previously described for mouse iPSCs 14. Briefly, human iPSC‐derived EBs were plated on MAXgel‐coated 6‐well tissue culture‐treated dishes at a density of 10 EBs per well. Polycarbonate transwell membrane inserts with 0.4 µm pores (Corning, Inc., Corning, NY; Cat. No. 3412) were placed in each well and seeded with 1.0 × 105 RF/6A monkey CECs (American Type Culture Collection [ATCC], Cat. No. CRL‐1780, https://www.atcc.org/) or primary human choroidal cells harvested from a postmortem eye. We maintained coculture conditions for 2 weeks in biopsy medium [MEMα (Thermo Fisher Scientific; Cat. No. 12571‐063), 10% heat‐inactivated fetal bovine serum (FBS) (Thermo Fisher Scientific; Cat. No. 26140‐079), and 0.02% primocin (Invivogen, San Diego, CA; Cat. No. ant‐pm‐2, http://www.invivogen.com/)]. This coculture system was set up such that secreted factors from the cells grown in the inserts (either the RF/6A cell line or primary human CECs) would be allowed to stimulate the underlying iPSC‐derived EBs to differentiate into CEC‐like cells.
Immunocytochemistry
Cells were fixed in a 4% paraformaldehyde (Alfa Aesar, Haverhill, MA; Cat. No. 43368, https://www.alfa.com/en/) solution for 20 minutes, subsequently washed using 1X phosphate buffered saline (PBS), and then incubated at room temperature for 1 hour in immunoblock buffer [1X PBS containing 5% normal goat serum (Cell Signaling, Danvers, MA; Cat. No. 5425S, https://www.cellsignal.com/), 3% bovine serum albumin (BSA) (Research Products International Corporation, Mt. Prospect, IL; Cat. No. A30075‐100.0, https://www.rpicorp.com/), 0.5% Triton X‐100 (Sigma‐Aldrich; Cat. No. T8532), and 0.03% sodium azide (Sigma‐Aldrich; Cat. No. 438456)]. Cells were subsequently incubated for 12–16 hours at 4°C in a primary antibody solution consisting of immunoblock buffer and antibodies targeted against TRA‐1‐81 (1:500, EMD Millipore; Cat. No. MAB4381, http://www.emdmillipore.com/), CA4 (1:500, R&D Systems; Cat. No. MAB21861, https://www.rndsystems.com/), CD31 (1:500, Abcam; Cat. No. ab32457), CD34 (1:500, Thermo Fisher Scientific, Cat. No. MA1‐10202) and transthyretin (TTR, 1:500, Rockland, Limerick, PA, Cat. No. 200‐901‐FM9, https://www.rockland-inc.com/Default.aspx). To detect primary antibodies, Cy2‐anti‐mouse (1:500, Jackson ImmunoResearch, West Grove, PA; Cat. No. 115‐225‐146), Cy2‐anti‐rabbit (1:500, Jackson ImmunoResearch; Cat. No. 111‐225‐144, https://www.jacksonimmuno.com/), Cy3‐anti‐mouse (1:500, Jackson ImmunoResearch; Cat. No. 115‐165‐146), or Alexa Fluor 647 goat anti‐rat (1:500, Thermo Fisher Scientific; Cat. No. A21247) secondary antibodies were used. Samples were analyzed using epifluorescence microscopy (EVOS FL; Thermo Fisher Scientific). We also used the GloLIVE Human Pluripotent Stem Cell Live Cell Imaging Kit to detect SSEA‐4 and Tra‐1‐60 in the human iPSCs (R&D Systems, SC023) by diluting the SSEA‐4 and Tra‐1‐60 antibodies 1:50 in cell culture media, incubating the cells for 30 minutes, and imaging the cells with an epifluorescence microscope (EVOS FL; Thermo Fisher Scientific).
Reverse‐Transcription PCR Analysis
Total RNA was extracted from cells using the RNeasy Mini‐kit (Qiagen; Cat. No. 74104) following the manufacturer's protocol. We reverse‐transcribed and amplified 100 ng of RNA with the SuperScriptIII One‐Step RT‐PCR System with Platinum Taq DNA Polymerase (Thermo Fisher Scientific; Cat. No. 12574‐026) and 20 pmol of each gene‐specific primer set (Supporting Information Table 2). All cycling profiles included a cDNA synthesis cycle at 55°C for 20 minutes, an initial denaturation temperature of 94°C for 2 minutes through 40 amplification cycles (15 seconds at 94°C, 30 seconds at the annealing temperature of each primer, and 1 minute at 68°C), and a final extension at 68°C for 5 minutes. PCR products were separated by electrophoresis on 2% agarose gels (Thermo Fisher Scientific; Cat. No. G800802).
Table 2.
Presence (+) or absence (−) of factors in media used for factor exclusion test conditions
| Test condition | Factors of interest | ||||
|---|---|---|---|---|---|
| CTGF | CTNNB1 | SHC1 | TWEAKR | VEGFB | |
| 1 | − | − | − | − | − |
| 2 | − | + | + | + | + |
| 3 | + | − | + | + | + |
| 4 | + | + | − | + | + |
| 5 | + | + | + | − | + |
| 6 | + | + | − | + | − |
| 7 | + | + | + | + | + |
Abbreviations: CTGF, connective tissue growth factor; TWEAKR, TNF‐related weak inducer of apoptosis receptor; VEGFB, vascular endothelial growth factor B.
RNA‐Seq Analysis
Because we determined that some set of secreted proteins from the RF/6A monkey CEC line can drive CEC differentiation in mouse and now in humans, this data and the search tool for the retrieval of interacting genes/proteins (STRING) biological database were used to identify proteins of interest 19. The STRING database contains information from sources such as experimental data and public text collections and serves to highlight functional enrichments in provided protein lists. Briefly, we downloaded the expression values associated with genes expressed by RF/6A monkey CECs (14;GEO accession: GSE70806), selected the top 1,000 most highly expressed genes in the control samples, and submitted these genes to STRING to identify genes associated with the gene ontology term “angiogenesis.”
Differentiation Factor Screening
Once we had identified proteins of interest, we used two statistical screening designs, Taguchi L12 and factor elimination, to efficiently determine which of these proteins drive CEC‐like cell differentiation 20. With respect to the Taguchi screen, we first generated a coded‐space test design based on the Taguchi L12 array (Table 1). In order to perform the Taguchi L12 screen, we obtained recombinant human connective tissue growth factor (CTGF; Peprotech; Cat. No. 120‐19, https://www.peprotech.com/en-US), human β‐catenin (CTNNB1; Sigma; Cat. No. SRP5172‐50UG), human Src Homology 2 Domain‐Containing Transforming Protein 1 (SHC1; Fitzgerald Industries; Cat. No. H00007453‐P01, https://www.fitzgerald-fii.com/), human TNF‐related weak inducer of apoptosis (TWEAK) receptor (TWEAKR also known as TNFRSF12A; PeproTech; Cat. No. 310‐21), and human vascular endothelial growth factor B (VEGFB; PeproTech; Cat. No. 100‐20B) proteins to use for the differentiation factor screening. We plated human iPSC‐derived EBs onto MAXgel‐coated 12‐well tissue culture plates with three wells per test condition and eight EBs per well. Media was changed every other day for 2 weeks and cells were collected using TrypLE Express. Harvested cells were subsequently washed with 1x PBS and fixed in 4% PFA for 10 minutes. After fixation, cells were incubated in immunoblock with primary anti‐human CA4 antibody (R&D Systems, Minneapolis, MN; Cat. No. MAB21861) for 30 minutes at 4°C. Next, cells were washed with 1x PBS and incubated with secondary conjugated Cy3 antibody (Jackson ImmunoResearch; Cat. No. 115‐165‐146) for 30 minutes at 4°C. Cells were subsequently washed with 1x PBS, loaded into a Tali Cellular Analysis Slide and quantified using the Tali Image‐Based Cytometer (Invitrogen; Cat. No. T10796). We collected Tali results for each test condition in triplicate from three independent experiments.
We used Tali values at each of the 12 conditions to calculate the mean and standard deviation of “plus” responses for each factor (i), defined as Avg+i and SD+i, respectively, for all test conditions that included Factor i. Similarly, we calculated the mean and standard deviation of “minus” responses, defined as Avg−i and SD−i, respectively, using the responses for all test conditions that did not include Factor i. For example,
Finally, we represented the statistical magnitude and direction of each factor's effect on the percentage of CA4+ cells by the half effect (CA4 Δi/2), which indicates the impact each factor had on iPSC to CEC‐like cell differentiation. Likewise, we represented the magnitude and direction of each factor's effect on the standard deviation of CA4 expression by the half effect of standard deviation for each factor (SD Δi/2). This value indicates the impact of each factor on the variability in CA4 expression.
For the second screening experiment, we created a test design in which each factor was excluded one at a time from media containing each of the other four factors of interest, as shown in Table 2. This method facilitated the identification of factors that are critical for CEC‐like cell differentiation. For this experiment, we generated iPSC‐derived EBs, plated the EBs on MAXgel‐coated 12‐well tissue culture dishes, and maintained the cultures as described above for the first screening experiment. The percentage of CA4+ CEC‐like cells were measured using the Tali Cytometer, as described above. Most screening methods are not intended to be analyzed using robust statistical tests. Rather, they provide relative estimates of each factor's influence on the selected dependent variable. Thus, we did not analyze Taguchi screening test data for statistical significance. For the sequential elimination experiment, we measured the percentage of CA4+ cells (dependent variable) at least nine times for each group. We removed one outlier, as verified by Dixon's Q‐test, from the VEGFB group (resulting in n = 8). The data passed a 95% confidence interval D'Agostino‐Pearson normality test and were thereafter assumed to be normally distributed. To assess differences between groups, we used one‐way ANOVA and a Dunnett's multiple comparisons test (both at 95% confidence interval) wherein we compared each group to the positive control (all factors of interest present).
We evaluated the effect of two factors (TWEAKR and CTGF concentration) on the percentage of CA4+ cells (dependent variable) using a full‐factorial design with two levels of TWEAKR concentration (0 or 10 ng/ml) and four levels of CTGF concentration (0, 10, 25, or 50 ng/ml). We recorded nine measurements for each condition. According to a 95% confidence interval D'Agostino‐Pearson normality test, the data were normally distributed. To analyze differences between groups, we used a two‐way ANOVA with a Tukey's multiple comparisons test (both 95% confidence interval).
Human CTGF ELISA Assay
In order to measure the levels of endogenously secreted CTGF, we performed an enzyme‐linked immunosorbent assay (ELISA) assay specific to human CTGF on human iPSCs differentiated in various media. We conducted this assay by feeding iPSC‐EBs basal medium as a spontaneously differentiated control, feeding iPSC‐EBs basal medium supplemented with 10 ng/ml of TWEAKR, and feeding iPSC‐EBs basal medium supplemented with 10 ng/ml of TWEAKR and either 1 μM or 5 μM of the TGF‐β inhibitor LY2109761 (Selleck Chem, Houston, TX; Cat. No. S2704, http://www.selleckchem.com/) for 2 weeks. We collected the media from the cells and determined the total protein levels for each medium using the DC Protein Assay (Bio‐Rad, Hercules, CA; Cat. No. 5000112, http://www.bio-rad.com/). We measured levels of endogenously secreted CTGF using the CTGF (human) Omnikine ELISA kit (Assay Biotechnology Company, Inc., Sunnyvale, CA; Cat. No. OK‐0109, http://assaybiotechnology.com/). Briefly, 55 µg of total protein was mixed with assay diluent, added the mixture to the CTGF ELISA plate, and incubated the plate for 2 hours at room temperature. The solution was aspirated, the wells were washed three times, the detection antibody was added at a concentration of 0.4 μg/ml, and the CTGF ELISA plate was incubated at room temperature for 2 hours. Next, the detection antibody was removed and the wells were washed three times. Streptavidin‐ horseradish peroxidase (HRP) was subsequently added to each well at a 1:400 dilution and the plate was incubated at room temperature for 30 minutes. We then removed the streptavidin‐HRP, washed the wells three times, added the substrate solution (3,3′, 5,5′‐Tetramethylbenzidine) to each well, and incubated the plate at room temperature. Once a fully saturated blue color developed, we added the stop solution to each well and used a microplate reader to measure the absorbance of each well at 450 nm. We determined the concentration of CTGF in each culture medium with a standard curve based on human CTGF. Because the number of measurements for each group was low (n = 2) for the ELISA experiment, we selected non‐parametric methods to assess differences between groups: a Kruskal‐Wallis test with a Dunn's multiple comparisons test (both 95% confidence interval). We evaluated the effect of inhibitor on percentage of CA4+ cells (dependent variable) using nine replicates at each condition. According to a 95% confidence interval D'Agostino‐Pearson normality test, the data were normally distributed. To analyze differences between groups, we used a two‐way ANOVA with a Tukey's multiple comparisons test (both 95% confidence interval).
Generating Lentiviral CEC Selection Vector
The human carbonic anhydrase 4 (CA4) promoter, which in the posterior eye is specific to the choriocapillaris, was cloned into the pENTR5'‐TOPO vector using the Gateway Cloning strategy (Thermo Fisher Scientific; Carlsbad, CA; Cat. No. K591‐10) 21, 22. The Gateway Cloning strategy was also used to clone a Zeocin resistance gene into the pENTR/D‐TOPO vector. Both entry vectors were subsequently cloned into the pDEST R4R3 Vector II (Thermo Fisher Scientific; Cat. No. 46‐2130, Supporting Information Figure 1) with the Gateway LR Clonase II Plus Enzyme Mix (Thermo Fisher Scientific; Cat. No. 12538‐120) and transformed in One Shot TOP10 Chemically Competent Escherichia coli (ThermoFisher Scientific; Cat. No. C4040‐03).
Figure 1.
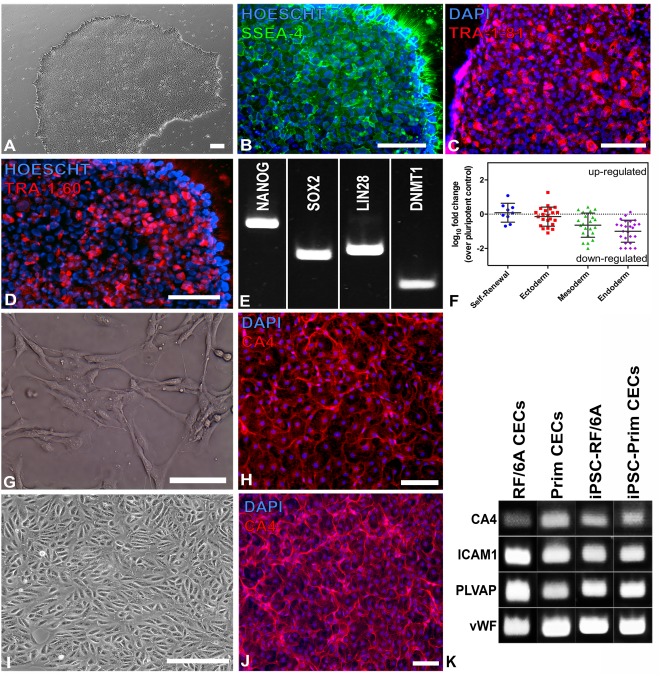
Generating human iPSCs from a donor with normal ocular history. (A–D): Pluripotent human iPSCs formed colonies with classic iPSC morphology (A) and expressed the human markers (B) SSEA‐4, (C) Tra‐1‐81, and (D) TRA‐1‐60. (E): NANOG, along with a variety of other pluripotency markers, was detected via rt‐PCR. (F): The TaqMan Scorecard Assay revealed comparable or downregulated expression of genes for self‐renewal (n = 9), ectoderm (n = 22), mesoderm (n = 22), and endoderm (n = 26). Each data point represents a single gene; means and standard deviations (error bars) for each group are also shown. (G–K): Human iPSC‐derived embryoid bodies cocultured with either primary human CECs (G) or RF/6A (I) generated CA4+ iPSC‐derived CECs (H, J, respectively) that formed capillary tube‐like structures typical of the choriocapillaris and expressed EC‐ and CEC‐specific markers, as determined via rt‐PCR (K). All scale bars represent 100 μm. Abbreviations: DAPI, 4′,6‐diamidino‐2‐phenylindole; CEC, choroidal endothelial cell; iPSC, induced pluripotent stem cell.
Producing CA4‐Zeocin Lentivirus
293FT cells (Thermo Fisher Scientific; Cat. No. R700‐07) were grown in complete DMEM growth medium with high glucose + L‐glutamine + 1 mM sodium pyruvate (Corning; Cat. No. MT10013CV) with 10% FBS and 0.1% Primocin (Invivogen; Cat. No. ant‐pm‐2) at 37°C with 5% CO2. Once the cells were 90% confluent, they were passaged onto 150 mm tissue culture plates (Corning; Cat. No. 430599) at a density of 2.0 × 106 cells per plate and cultured in complete DMEM. After 4 days in culture, the medium was removed and a transfection solution comprising sterile molecular‐grade water (RPI, Mt. Prospect, IL; Cat. No. 248700‐1000), pLP1 (Thermo Fisher Scientific; Cat. No. K4975‐00), pLP2 (Thermo Fisher Scientific, Cat. No. K4975‐00), pLP/VSVG (Thermo Fisher Scientific; Cat. No. K4975‐00), the pHIV‐CA4‐Zeocin, 2x HBS, and serum‐free DMEM with 0.1% Primocin was added. Cells were incubated for 5 hours at 37°C, after which the transfection solution was removed and fresh DMEM medium with 10% FBS and 0.1% Primocin was added. Cells were incubation at 37°C in 5% CO2. The cell medium was collected every day for the next 3 days and replaced the used media with DMEM containing 10% FBS and 0.1% Primocin. On the third day, the medium was filtered through a 0.22 μm bottle‐top filter to remove cell debris, and subsequently centrifuged overnight at 7,000 rpm. Next, the supernatant was removed without disrupting the viral pellet. The viral pellet was subsequently resuspended in 40 mg/ml lactose/PBS buffer, aliquoted, and stored at −80°C.
Determining Lentivirus Titer via qPCR
HT1080 cells were seeded onto 12‐well tissue culture plates at a density of 2.5 × 105 cells per well and incubated at 37°C in 5% CO2 overnight. Cells were subsequently infected with 10, 5, 1, or 0.5 μl of the CA4‐Zeocin lentivirus and incubated at 37°C in 5% CO2 for 72 hours. After this incubation period, cells were collected using trypsin and genomic DNA was isolated using the Wizard Genomic DNA Purification kit (Promega, Madison, WI; Cat. No. A1125, https://www.promega.com/). Finally, we used the Power SYBR Green PCR Master Mix and Power SYBR Green RT‐PCR Reagents Kit (Thermo Fisher Scientific; Cat. No. 4367659) according the manufacturer's instructions to quantify the amount of viral DNA in the cellular genomic DNA to determine the viral titer.
Transduction of Human iPSCs with CA4‐Zeocin Lentivirus and Selection of CEC‐Like Cells Using Zeocin
Human iPSCs were passaged onto laminin–coated 12‐well tissue culture plates at a density of 1.0 × 104 cells per well and subsequently transduced at an multiplicity of infection (MOI) of 12 with CA4‐Zeocin lentivirus in viral transduction medium [E8 (Thermo Fisher Scientific; Cat. No. A1517001), 10 ng/ml hbFGF (Syd Labs, Cat. No. BP000030‐GD4), 1% primocin (Invivogen; Cat. No. ant‐pm‐2)] and 0.05% polybrene (Sigma‐Aldrich; Cat. No. SCR510). At 12–16 hours post‐transduction, the cells were washed and fed with fresh pluripotency medium. At 5 days post‐transduction, iPSCs were differentiated as described above. Following differentiation, cultures were fed with CEC differentiation medium supplemented with 50 μg/ml of Zeocin for 1 week. Cultures were subsequently analyzed.
Transmission Electron Microscopy
Zeocin selected CEC‐like cells were fixed in one half strength Karnovsky fixative as described previously, followed by osmication 13. Samples were subsequently dehydrated and embedded in Epon resin. Following polymerization, blocks were removed and trimmed, ultrathin sections were collected on formvar coated grids, and samples were imaged on a JEOL JEM1230 transmission electron microscope.
Results
To generate iPSCs, we harvested dermal fibroblasts from a donor with a healthy ocular history and expanded the cells on laminin‐coated 6‐well tissue culture plates. Once the cells were sufficiently confluent, the fibroblasts were transduced with nonintegrating Sendai viruses designed to drive expression of the transcription factors OCT4, SOX2, KLF4, and c‐MYC. After transduction and clonal isolation, cultured patient‐specific cells demonstrated morphology characteristic of iPSCs (Fig. 1A) (e.g., large nucleus to cytoplasm ratios) and expressed the pluripotency markers SSEA‐4, TRA‐1‐60, TRA‐1‐81 (as detected by immunocytochemistry, Fig. 1B–1D), NANOG, SOX2, LIN28, and DNMT1 (as detected by rt‐PCR, Fig. 1E). Human iPSCs were subsequently analyzed using the TaqMan hPSC Scorecard Panel (Fig. 1F), which is a rapid comprehensive gene expression real‐time PCR assay comprised of 94 individual qPCR assays, including control, housekeeping, self‐renewal, and lineage‐specific genes 17. Sendai virus was not detected in the passaged iPSCs, indicating that the cells were pluripotent with no residual virus from the reprogramming process. The cells also expressed self‐renewal and ectoderm genes at levels not significantly different than the pluripotent reference cells (p = .6572 and p = .2370, respectively) and did not demonstrate upregulation of mesoderm or endoderm compared to the control (means were significantly less than zero with p = .0003 and p < .0001, respectively).
To determine whether these human iPSCs would differentiate into CA4+ CEC‐like cells when cultured with human CECs, we performed a coculture experiment using primary human choroidal cells (Fig. 1G) in a millipore insert and with human iPSC‐EBs plated beneath the CECs. The differentiated cells (Fig. 1H) expressed CA4 and formed capillary tube‐like structures that were morphologically indistinguishable from primary CECs (Fig. 1G). The human iPSCs also differentiated into CEC‐like cells via coculture with the RF/6A monkey CEC line (Fig. 1I), a method that we recently reported for differentiation of mouse iPSC‐derived CEC‐like cells 14. The iPSC‐derived CEC‐like cells from this coculture experiment also formed CA4+ tubes similar to the tubes observed in the human primary CEC coculture experiment (Fig. 1J). The iPSC‐derived CEC‐like cells that differentiated in both coculture experiments expressed the EC‐specific markers ICAM1 and vWF, the fenestration marker PLVAP, and the CEC‐specific marker CA4 (Fig. 1K), indicating that these cells are similar to primary CECs. As the RF/6A cell line has been well characterized molecularly and both RF/6A cells and primary human CECs efficiently induced differentiation of iPSCs into CEC‐like cells in our coculture experiment, RF/6A gene expression data was used to identify secreted factors that have the potential to drive CEC differentiation. For this experiment, our previously published RF/6A RNA‐seq data set was used 15. Specifically, the STRING biological database was used to identify which of the top 1,000 most highly expressed genes identified in our RNA‐seq data set have been reported to play a role in angiogenesis (Table 3) 19.
Table 3.
RF/6A‐secreted factors of interest
| Protein | Description | |
|---|---|---|
| CTGF | Connective tissue growth factor | Major connective tissue mitoattractant; secreted by vascular endothelial cells and RPE [20, 21] |
| CTNNB1 | β‐Catenin | Member of WNT signaling pathway; promotes VEGF‐associated angiogenesis [22]. |
| SHC1 | Src homology 2 domain containing transforming protein 1 | Longevity adaptor protein; involved in ROS‐dependent VEGF‐mediated angiogenesis in endothelial cells [23]. |
| TWEAKR | TNF‐related weak inducer of apoptosis receptor | Involved in development of retinal neovascularization; secreted by RPE [24, 25]. |
| VEGFB | Vascular endothelial growth factor B | Promotes angiogenesis and induces coronary, renal, and pulmonary vessel growth [26]. |
Abbreviations: ROS, reactive oxygen species; RPE, retinal pigment epithelium; WNT, reactive oxygen species; VEGF, vascular endothelial growth factor.
Using the Taguchi L12 method, we fed iPSC‐derived EBs with 12 different media each containing different combinations of the five proteins of interest 20. After 2 weeks of differentiation, we measured the percentage of CA4+ CEC‐like cells (Fig. 2A) differentiated using each media. As shown in Figure 2B, CTGF and TWEAKR were the only factors that had a positive effect on CEC‐like cellular differentiation (i.e., CA4 expression). CTGF was the dominant factor, with a half effect of more than three times that of TWEAKR. CTNNB1, SHC1, and VEGFB had an overall negative effect on CEC‐like cellular differentiation, with CTNNB1 and VEGFB having the smallest impact. Although VEGFB was the only factor that significantly decreased the standard deviation of CA4 expression, CTGF contributed only a relatively small amount (Fig. 2C). This result indicates that CTGF is a reliable factor to efficiently differentiate iPSCs into CEC‐like cells. By contrast, TWEAKR had a relatively high impact on the CA4 expression standard deviation. In a separate strategy, we excluded each factor one at a time from media containing all other proteins of interest to identify those factors necessary for inducing CEC‐like cellular differentiation. The results of this factor elimination method, shown in Figure 2D, indicated that only removing CTGF significantly decreased CA4 expression compared to the control (p < .0001), in which we included all the factors of interest (p = .6378). Removing any of the other six proteins, including TWEAKR, did not significantly decrease CEC‐like cellular differentiation compared to the spontaneously differentiated control (p = .0693 for CTNNB1; p > .700 for all other factors). We also detected CTGF and TWEAKR in healthy adult human choroid obtained from a postmortem donor eye (Fig. 2E–2J, Supporting Information Figure 2 (isotype control—It is important to note that the autofluorescence seen in the RPE is caused by age‐related accumulation of lipofuscin and not due to antibody binding 8, 23, 24, showing that low levels of both proteins are present in the choroidal vasculature.
Figure 2.
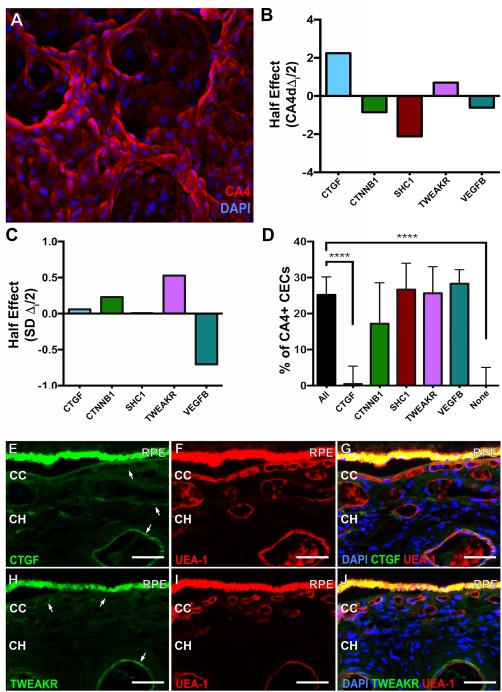
Identification of CEC differentiation candidates using the Taguchi L12 and factorial elimination screens. (A): Representative image used to determine the percentage of CA4+ iPSC‐derived CECs. (B): Taguchi L12 half effect of each factor on the mean of percentage of CA4+ cells. (C): Taguchi L12 half effect of each factor on the standard deviation of percentage of CA4+ cells. (D): Mean (+SD) percentage of CA4+ cells elicited by each medium in the factorial elimination screen (n ≥ 8, ****, p < .0001). (E–J): Immunohistochemistry showing CTGF (E, G, green) and TWEAKR (H, J, green) expression in adult choroids. Sections were dual labeled with the endothelial cell binding lectin UEA‐I (F, G, I, J, red). All scale bars represent 100 μm. Abbreviations: CC, choriocapillaris; CEC, choroidal endothelial cell; CH, choroidal vasculature; CTGF, connective tissue growth factor; DAPI, 4′,6‐diamidino‐2‐phenylindole; RPE, retinal pigment epithelium; TWEAKR, TNF‐related weak inducer of apoptosis receptor; VEGFB, Vascular endothelial growth factor B.
To further understand the interaction between CTGF and TWEAKR and their roles in CEC differentiation, we varied the concentration of each of these two factors independently and excluded the remaining three factors (CTNNB1, SHC1, and VEGFB). As expected, compared to iPSC‐EBs that were fed with medium containing either CTGF or TWEAKR, no iPSC‐EBs differentiated into CEC‐like cells when fed control medium without CTGF or TWEAKR (Fig. 3A, 3B): Without TWEAKR, maximum CEC‐like cell differentiation occurred at 25 ng/ml of CTGF, with no significant changes in CA4 expression occurring beyond this dose (p > .9999 compared between 25 ng/ml CTGF and 50 ng/ml CTGF, Fig. 3A). Without CTGF, 10 ng/ml of TWEAKR elicited a level of CA4 expression statistically similar to that of 25 ng/ml of CTGF (p > .9999) and significantly higher than the basal medium negative control (p < .01, Fig. 3A, 3D). By contrast, CA4 expression decreased with increasing CTGF concentration when 10 ng/ml TWEAKR was present (Fig. 3A). For example, adding 10 ng/ml TWEAKR to medium containing 50 ng/ml CTGF resulted in CA4 expression levels similar to the control (p > .9999) and 16.8% lower than CA4 levels detected in the TWEAKR‐free analog (p < .001, Fig. 3A, 3B, 3E). While all the cells analyzed in this experiment expressed the mature EC marker CD31 (Fig. 3B–3E), only cells fed 50 ng/ml of CTGF without TWEAKR formed CA4+ capillary tube‐like structures morphologically indistinguishable from native CECs when grown on tissue culture plates coated with human extracellular matrix (Fig. 3C).
Figure 3.
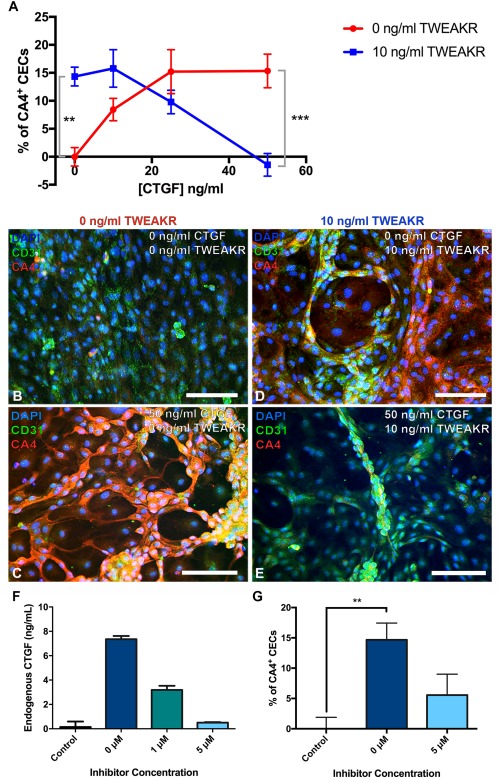
CTGF drives iPSC differentiation into CECs. (A): Mean (±SD; n = 9) percentage of CA4+ cells at varying concentrations of CTGF and TWEAKR. (B–E): Representative images are provided showing cell morphology and CA4 expression at 0 ng/ml TWEAKR and 0 ng/ml CTGF (B), 0 ng/ml TWEAKR and 50 ng/ml CTGF (C), 10 ng/ml TWEAKR and 0 ng/ml CTGF (D), and 10 ng/ml TWEAKR and 50 ng/ml CTGF (E). (F): Concentration of endogenously secreted CTGF in culture medium from human iPSCs differentiated into CEC in medium containing 10 ng/ml of TWEAKR alone or with varying concentrations of the inhibitor LY2109761, compared to spontaneously differentiated iPSCs grown in basal medium (control), as detected using ELISA. Shown are the means at each condition, while error bars represent the standard deviation (n = 2). (G): Percentage of CA4+ cells in cultures of human iPSCs differentiated using 10 ng/ml of TWEAKR alone or with 5 µM inhibitor (LY2109761), compared to spontaneously differentiated iPSCs grown in basal medium (control). Shown are the means at each condition, while error bars represent the standard error of the mean (n = 9, **, p < .01, ***, p < .001). All scale bars represent 100 μm. Abbreviations: CEC, choroidal endothelial cell; CTGF, Connective tissue growth factor; DAPI, 4′,6‐diamidino‐2‐phenylindole; TWEAKR, TNF‐related weak inducer of apoptosis receptor.
To better understand the mechanism by which TWEAKR drives CEC‐like cellular differentiation, we fed iPSC‐EBs media containing 10 ng/ml of TWEAKR alone or 10 ng/ml of TWEAKR with either 1 or 5 μM of the TGF‐β inhibitor, LY2109761, which blocks the production of endogenous CTGF. The purpose of this experiment was to determine if exogenous TWEAKR added to the culture medium induces endogenous CTGF secretion. After 2 weeks in culture, we collected the media and measured the amount of endogenous CTGF using a CTGF ELISA. This quantification revealed that as expected, iPSC‐EBs fed basal medium did not secrete endogenous CTGF (Fig. 3F). This ELISA result also showed that cells fed 10 ng/ml of TWEAKR secreted ∼50 times more endogenous CTGF compared to the negative control (7.36 ± 0.26 vs. 0.14 ± 0.45 ng/ml CTGF, p = .086; Fig. 3F). By contrast, the cells fed 10 ng/ml of TWEAKR with 1 μM TGF‐β inhibitor secreted ∼22 times as much CTGF as the control (3.19 ± 0.34, p = .61), while those cells fed 5 µM inhibitor secreted similar levels of CTGF to the negative control (0.51 ± 0.04, p > .9999; Fig. 3F). The cells fed 10 ng/ml TWEAKR without inhibitor showed a significantly increased percentage of CA4+ CEC‐like cells compared to the negative control (p < .01; Fig. 3G). This TWEAKR‐mediated increase was offset by the presence of 5 μM inhibitor, which prevented an increase in percentage of CA4+ CEC‐like cells compared to the negative control (p = .3512; Fig. 3G).
Once we determined that CTGF alone was capable of inducing iPSC to CEC‐like cell differentiation, further immunocytochemical analysis designed to confirm vascular identify was performed. First, expression and/or uptake of TTR, a protein that binds choriocapillaris EC with high affinity 25 and that functions by shuttling vitamin A (a critical component of the visual cycle) 26, 27, was confirmed (Fig. 4A). Next, we demonstrated that CA4+ve CEC‐like cells also express the pan endothelial cell surface marker CD31 (also known as PECAM‐1) (Fig. 4B). To demonstrate that these findings were not iPSC line dependent, an additional iPSC line generated from a separate individual with normal ocular history was used. Following differentiation, CEC‐like cells expressing the CEC restricted markers CA4 and TTR could be readily identified (Fig. 4C).
Figure 4.
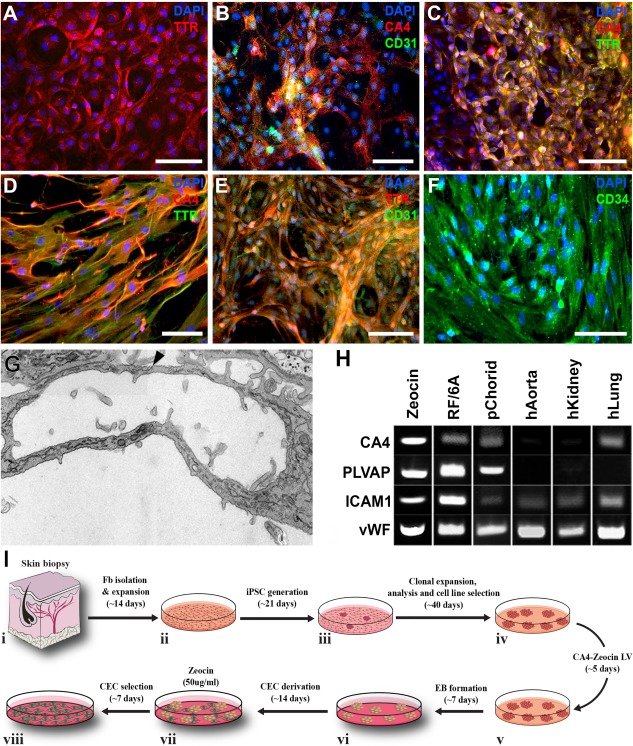
Characterization and selection of iPSC derived CEC‐like cells. (A–C): Immunocytochemical analysis of iPSC derived CEC‐like cells using antibodies targeted against TTR (A, C) CA4 (B, C) and CD31 (B) pre‐Zeocin selection. (D–F): Immunocytochemical analysis of iPSC derived CEC‐like cells using antibodies targeted against TTR (D, E) CA4 (D), CD31 (E) and CD34 (F) post‐Zeocin selection. (G): Transmission electron microscopy analysis of a Zeocin‐treated iPSC derived CEC‐like capillary tube. (F): Expression of EC‐ and CEC‐specific markers by Zeocin‐selected cells and controls, as detected via rt‐PCR. (H): Rt‐PCR analysis demonstrating expression of the endothelial cell markers CA4, PLVAP, ICAM1, and vWF. (I): Schematic diagram illustrating the steps and timeline required from generating patient specific iPSCs to derivation of Zeocin selected CEC‐like cells. All scale bars represent 100 μm. Abbreviations: CEC, choroidal endothelial cell; DAPI, 4′,6‐diamidino‐2‐phenylindole; EB, embryoid body; iPSC, induced pluripotent stem cell; TTR, transthyretin.
As with most pluripotent stem cell differentiation protocols, to obtain a relatively pure population of target cells suitable for downstream applications, cellular enrichment is often required. For instance, pigmented pluripotent stem cell derived RPE cells can be easily identified, manually isolated and expanded free from the surrounding heterogeneous population of stem cell progeny 11, 28, 29, 30, 31, 32, 33. Unlike RPE cells, CEC‐like cells cannot be easily identified and isolated under phase or bright field microscopy. As such, we chose to develop a lentiviral based drug selection cassette (Supporting Information Fig. 1) that following transduction could be used to generate stable lines of CEC‐like cells. This vector was designed such that transduced cells would stably express the Zeocin resistance gene under control of the CA4 promoter. For these experiments iPSCs were transduced 5 days prior to differentiation (Fig. 4I‐iv). At 21‐days post‐differentiation Zeocin was added to the CEC‐differentiation media and cells were cultured for an additional 7 days (Fig. 4I‐vii, 4I‐viii). Compared to cultures not treated with Zeocin (Fig. 4A–4C), cultures treated with Zeocin were enriched with CA4‐expressing cells that also expressed TTR (Fig. 4D), CD31 (Fig. 4E) and CD34 (Fig. 4F). Zeocin‐treated iPSC‐derived CEC‐like cells were morphologically similar to cultured endothelial cells, forming vascular tubes with a well‐defined luminal wall and intracellular junctions (Fig. 4G, arrowhead). Rt‐PCR analysis revealed that like the monkey CEC line RF/6A and human primary CECs, in addition to CA4, Zeocin treated cultures expressed PLVAP (i.e., plasmalemma vesicle associated protein, a glycoprotein that is localized to the inner surface of the choriocapillaris in vivo and that is required for maintaining choriocapillaris fenestrae) and the endothelial cell specific markers ICAM1 and vWF (Fig. 4H). A summary of the procedures and time line from biopsy isolation to obtaining an enriched culture of CEC‐like cells for downstream applications is depicted in Figure 4I.
Discussion
With the advent of the iPSC technology, it is now possible to generate any somatic cell type from patients in order to study the pathogenesis of their disease. In addition, patient‐specific iPSCs are the ideal cell source for autologous immunosuppression‐free cell replacement. That said, despite the described importance of CECs in diseases such as AMD and central serous retinopathy, prior to this study no differentiation protocol suitable for generating patient‐specific iPSC‐derived CECs existed. Our group previously published a method for producing murine CEC‐like cells from iPSCs via cellular coculture 14. Although this murine protocol relied on use of the RF/6A CEC cell line, and as such was not xeno‐free, it paved the way for the stepwise human iPSC to CEC differentiation protocol reported here.
As in our previous mouse study, to confirm that we could in fact generate human iPSC‐derived CEC‐like cells, coculture experiments were performed. Results provide further support for the feasibility of using a coculture system to generate patient‐specific cell types for which differentiation protocols do not exist. These findings also suggest the existence of some set of diffusible factors secreted by developed CECs (i.e., both RF/6A and primary human cells) that are capable of driving iPSC to CEC differentiation. To identify said factors, we began by reanalyzing our previously published RF/6A RNA‐sequencing data set 15. Using STRING analysis, we identified five proangiogenic factors known to play a role in vascular development (Table 3). Based on the expression of these factors in CECs, we hypothesized that adding one or more of these five compounds to our standard stem cell differentiation medium would drive differentiation toward a choroidal phenotype.
The traditional full‐factorial approach to simultaneously investigate all five factors would have required 32 test conditions (not including replicates), demanding exorbitant time and financial resources. Instead, the use of a screening design (e.g., Taguchi L12 design), which is typically a subset of a full‐factorial design, can be used to find the few significant factors from a list of many potential compounds 20. In other words, screening designs can be used to examine multiple factors in a highly time and cost‐efficient manner. Accordingly, a full‐factorial design is the only way to rigorously evaluate interactions among factors. Even so, a Taguchi L12 design in particular disperses (i.e., randomizes) the effects of interactions in a manner that minimizes the impact of interactions on the ability to discern significant hits. In our case, only 12 test conditions were required to determine which factors were important for CEC differentiation, resulting in a significant time and resource savings for the Taguchi L12 screen relative to the full‐factorial approach.
Using the statistical methods described above, we identified CTGF and TWEAKR as the main contributors to CEC differentiation. Even so, it is important to acknowledge that the negative responses seen for the CTNNB1, SHC1, and VEGFB factors only allows us to conclude that these factors did not play a major role in iPSC differentiation into CEC‐like cells. We cannot exclude the possibility that these factors are important in the choroid.
The first candidate protein identified in the Taguchi and factorial elimination screens, CTGF (also known as CCN2), is believed to act as a matricellular organizer by bridging the functional division between structural extracellular proteins and cytokines, growth factors, and proteases 34. In other words, CTGF interacts with both the extracellular matrix and an array of cell surface proteins to translate extracellular signals into defined biological responses 34. Given the importance of CTGF in multiple biological processes, it is not surprising that CTGF is widely expressed in embryogenesis, particularly in the heart, brachial arches, neuronal tissues, and endothelium and smooth muscle layers of major blood vessels 34, 35, 36. CTGF can mimic angiogenesis and tube formation in vitro by promoting endothelial proliferation (angiogenesis) and recruiting CD34+ endothelial progenitor cells to the endothelium (tube formation) 37, 38. Researchers have also found CTGF in retinal vascular beds, indicating that CTGF may play a role in early retinal development 39. Although previous studies show that CTGF is involved in normal retinal development, and we demonstrate here that CTGF is responsible for stem cell differentiation into CEC‐like cells, there are also some indications that CTGF can be a pathological marker, namely in retinal neovascular tufts in an oxygen‐induced retinopathy murine model 39. This latter finding suggests that excessive CTGF may initiate choroidal neovascularization. One such choroidal injury could be the membrane attack complex (MAC) that accumulates in the choriocapillaris due to dysfunctional complement factor H, which may trigger excessive CTGF production and initiate choroidal neovascularization. Consistent with this possibility, MAC injury to RF/6A cells in vitro leads to a 1.9‐fold increase in CTGF mRNA 15.
The second and less influential factor identified in the Taguchi L12 screen, TWEAKR, also known as fibroblast growth factor (FGF)‐inducible molecule 14 (FN14), is the receptor for TNF‐like weak inducer of apoptosis (TWEAK), making the two proteins a TNF superfamily ligand‐receptor pair 40. The TWEAK/TWEAKR interaction was initially described as an inducer of targeted tumor cell death 41, 42. Additional research demonstrated that TWEAK/TWEAKR stimulate several, often conflicting cellular responses, such as angiogenesis and cell death 43. TWEAKR is expressed in multiple cell types, including, but not limited to epithelial, mesenchymal, and endothelial cells 40, 41, 43, 44, 45. TWEAKR is also reported to activate both the canonical and non‐canonical NFκB pathways as well as the MAPK pathway, in which the TWEAK‐TWEAKR interaction induces proinflammatory cytokines that may trigger CTGF expression. Finally, TWEAKR expression is significantly increased in injured tissues and is highly inducible by growth factors such as FGF, platelet‐derived growth factor (PDGE), and VEGF, all of which are thought to be involved in choroidal development and neovascularization. Thus, it is possible that TWEAKR—which is also upregulated in complement injured RF/6A cells—may also be induced in wet AMD pathogenesis along with CTGF 46, 47. Although there are no studies of which we are aware examining the interaction between CTGF and TWEAKR, understanding their relationship may reveal valuable insight into choroidal development and AMD pathogenesis.
While both CTGF and TWEAKR were identified in our Taguchi L12 screen, removal of TWEAKR from cell culture media had no effect on the percentage of CEC‐like cells generated in the factorial elimination experiment, suggesting that CTGF was the major factor responsible for the CEC‐like cell differentiation. Even so, the relatively high variability of TWEAKR's impact on iPSC differentiation into CEC‐like cells seen in the Taguchi L12 screen suggests that TWEAKR may enhance this CEC differentiation, most likely by interacting with some or all of the other factors of interest. To further investigate how CTGF and, perhaps, TWEAKR interact to generate CEC‐like cells, we added both CTGF and TWEAKR to differentiation media with varying concentrations of CTGF (Fig. 3). Because the TWEAK‐TWEAKR complex is cytotoxic at 100 ng/ml, we maintained TWEAKR at a constant concentration of 10 ng/ml 48. This experiment revealed that although CTGF and TWEAKR appear to compete, each factor alone can drive iPSC to CEC‐like cell differentiation. This result suggested that both factors either induce or play a key role in the CEC developmental pathway, which itself is not fully understood. Various studies show that CTGF and TWEAKR can independently activate NFκB, leading to production of pro‐inflammatory factors. For instance, both CTGF and TWEAKR can cause an increase in MCP‐1 and ICAM‐1 expression in endothelial cells, and CTGF and TWEAKR are known to play a role inepidermal growth factor receptor (EGFR) signaling to induce NFκB‐mediated inflammation 49, 50, 51, 52. As shown in Figure 3, TWEAKR alone can drive CEC‐like cell differentiation; however, the ELISA data suggest that this TWEAKR‐mediated CEC‐like cell differentiation actually results from induction of endogenous CTGF secretion from the cells. This result shows that TWEAKR indirectly accomplishes the same differentiation effect that CTGF directly accomplishes alone. One explanation for this result could be that EGFR is transactivated via TWEAKR, causing NFκB‐mediated inflammation, which in turn may trigger the expression and secretion of endogenous CTGF 49, 50.
It is important to emphasize that along with CA4 expression, morphology and structure of the iPSC‐derived CEC‐like cells are also of critical importance for generating healthy CEC‐like cells. That being said, the iPSC‐derived CEC‐like cells generated when using only CTGF (Fig. 3) are morphologically identical to cultured endothelial cells, unlike iPSC‐derived CEC‐like cells generated with only TWEAKR. This result lends further support to the argument that CTGF is the more reliable and efficient factor for CEC differentiation compared to TWEAKR.
Although CTGF effectively drove human iPSCs to differentiate into CEC‐like cells in this report, there were still non‐choroidal cells in the culture, making the differentiated cells a heterogeneous population. The presence of multiple cell types in the culture can raise problems for future cell replacement therapies and could complicate disease‐modeling studies. To overcome this problem, iPSC‐derived CEC‐like cells were isolated via forced expression of an antibiotic‐resistance cassette in which Zeocin resistance was driven under the control of the human choriocapillaris‐specific CA4 promoter. The iPSCs transduced with this vector and differentiated with the optimal CEC differentiation medium of 50 ng/ml of CTGF were subsequently treated with Zeocin. The other cell types were gradually eliminated while the Zeocin‐resistant CECs remained in the culture. The selected CEC‐like cells formed tube‐like structures that expressed the CEC restricted markers CA4 and TTR (Fig. 4D, 4E) and the pan EC markers CD31 and CD34 (Fig. 4E, 4F). The selected cells also formed capillary‐like tubes (Fig. 4G) and expressed the same markers seen in the CEC controls compared to other EC types, shown in Figure 4H, in which the iPSC‐derived CEC‐like cells are most similar to the CEC controls compared to aortic, lung, and kidney ECs. In turn, we now can use the antibiotic resistance method described in this study to generate pure populations of iPSC‐derived CEC‐like cells from control donors and from AMD patients with varying risk factor genotypes to use for AMD disease‐modeling experiments in vitro. Although this proof‐of‐concept method successfully selected CECs, a method that does not depend on antibiotic resistance may be needed in order to use patient‐specific iPSC‐derived CECs for cell replacement therapy clinically (i.e., positive selection cassettes, like antibiotic resistant genes, can potentially interfere with the expression of neighboring genes 53).
Conclusion
In summary, we harnessed the iPSC technology to develop a novel method to generate patient‐specific choroidal endothelium like cells. The method by which we developed this differentiation protocol was unique, integrating RNA‐seq and statistical screens to reveal that CTGF promotes CEC‐like cellular differentiation. We also demonstrated a method to purify the human iPSC‐derived CEC‐like cells suitable for disease modeling, drug discovery, and pre‐clinical transplantation studies. This work will greatly impact the vision research field, especially the study of AMD and other choroid‐related diseases.
Author Contributions
A.E.S. and K.S.W.: conception and design, collection and/or assembly of data, data analysis and interpretation, manuscript writing; K.R.C. and J.C.G.: conception and design, collection and/or assembly of data, data analysis and interpretation; S.S.W. and K.R.A.: collection and/or assembly of data, data analysis and interpretation; D.O., C.M.C., and M.J.R.: collection and/or assembly of data; M.N.: collection and/or assembly of data, manuscript writing; E.M.S.: provision of study material or patients, collection and/or assembly of data, financial support, manuscript writing; R.F.M.: collection and/or assembly of data, financial support, manuscript writing; B.A.T.: collection and/or assembly of data, data analysis and interpretation, financial support, manuscript writing, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Table S1
Supporting Information Table S2
Supporting Information 1
Acknowledgments
We would like to thank the Iowa Lion's Eye Bank and the donors and their families for their invaluable contributions. Additionally, we wish to thank the Elmer and Sylvia Sramek Charitable Foundation, National Eye Institute (NIH RO1 ‐ EY 024605, RO1 – EY026087), and Wynn Institute Endowment for Vision Research for their support.
References
- 1. Friedman DS, O'Colmain B, Tomany SC et al. Prevalence of age‐related macular degeneration in the United States. Arch Ophthalmol 2004;122:564–572. [DOI] [PubMed] [Google Scholar]
- 2. VanNewkirk MR, Nanjan MB, Wang JJ et al. The prevalence of age‐related maculopathy ‐ The visual impairment project. Ophthalmology 2000;107:1593–1600. [DOI] [PubMed] [Google Scholar]
- 3. Sohn EH, Flamme‐Wiese MJ, Whitmore SS et al. Loss of CD34 expression in aging human choriocapillaris endothelial cells. PLoS One 2014;9:e86538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mullins RE, Johnson MN, Faidley EA et al. Choriocapillaris vascular dropout related to density of drusen in human eyes with early age‐related macular degeneration. Invest Ophthalmol Vis Sci 2011;52:1606–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mullins RF, Dewald AD, Streb LM et al. Elevated membrane attack complex in human choroid with high risk complement factor H genotypes. Exp Eye Res 2011;93:565–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Biesemeier A, Taubitz T, Julien S et al. Choriocapillaris breakdown precedes retinal degeneration in age‐related macular degeneration. Neurobiol Aging 2014;35:2562–2573. [DOI] [PubMed] [Google Scholar]
- 7. Seddon JM, McLeod DS, Bhutto IA et al. Histopathological insights into choroidal vascular loss in clinically documented cases of age‐related macular degeneration. JAMA Ophthalmol 2016;134:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whitmore SS, Braun TA, Skeie JM et al. Altered gene expression in dry age‐related macular degeneration suggests early loss of choroidal endothelial cells. Mol Vis 2013;19:2274–2297. [PMC free article] [PubMed] [Google Scholar]
- 9. Whitmore SS, Sohn EH, Chirco KR et al. Complement activation and choriocapillaris loss in early AMD: Implications for pathophysiology and therapy. Prog Retin Eye Res 2015;45:1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Almeida DRP, Zhang L, Chin EK et al. Comparison of retinal and choriocapillaris thicknesses following sitting to supine transition in healthy individuals and patients with age‐related macular degeneration. JAMA Ophthalmol 2015;133:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Buchholz DE, Pennington BO, Croze RH et al. Rapid and efficient directed differentiation of human pluripotent stem cells into retinal pigmented epithelium. Stem Cells Transl Med 2013;2:384–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhong XF, Gutierrez C, Xue T et al. Generation of three‐dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun 2014;5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tucker BA, Mullins RF, Streb LM et al. Patient‐specific iPSC‐derived photoreceptor precursor cells as a means to investigate retinitis pigmentosa. Elife 2013;2:e00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Songstad AE, Wiley LA, Duong K et al. Generating iPSC‐derived choroidal endothelial cells to study age‐related macular degeneration. Invest Ophthalmol Vis Sci 2015;56:8258–8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zeng SM, Whitmore SS, Sohn EH et al. Molecular response of chorioretinal endothelial cells to complement injury: Implications for macular degeneration. J Pathol 2016;238:446–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tucker BA, Anfinson KR, Mullins RF et al. Use of a synthetic xeno‐free culture substrate for induced pluripotent stem cell induction and retinal differentiation. Stem Cells Transl Med 2013;2:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fergus J, Quintanilla R, Lakshmipathy U. Characterizing pluripotent stem cells using the TaqMan(R) hPSC scorecard(TM) panel. Methods Mol Biol 2016;1307:25–37. [DOI] [PubMed] [Google Scholar]
- 18. Tsankov AM, Akopian V, Pop R et al. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol 2015;33:1182–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szklarczyk D, Franceschini A, Wyder S et al. STRING v10: Protein‐protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43:D447–D452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rao RS, Kumar CG, Prakasham RS et al. The Taguchi methodology as a statistical tool for biotechnological applications: A critical appraisal. Biotechnol J 2008;3:510–523. [DOI] [PubMed] [Google Scholar]
- 21. Dreos R, Ambrosini G, Perier RC et al. The Eukaryotic Promoter Database: Expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res 2015;43:D92–D96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dreos R, Ambrosini G, Perier RC et al. EPD and EPDnew, high‐quality promoter resources in the next‐generation sequencing era. Nucleic Acids Res 2013;41:D157–D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Srivastava GK, Reinoso R, Singh AK et al. Trypan Blue staining method for quenching the autofluorescence of RPE cells for improving protein expression analysis. Exp Eye Res 2011;93:956–962. [DOI] [PubMed] [Google Scholar]
- 24. Mullins RF, Schoo DP, Sohn EH et al. The membrane attack complex in aging human choriocapillaris relationship to macular degeneration and choroidal thinning. Am J Pathol 2014;184:3142–3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Smith SS, Pino RM, Thouron CL. Binding and transport of transthyretin gold by the endothelium of the rat choriocapillaris. J Histochem Cytochem 1989;37:1497–1502. [DOI] [PubMed] [Google Scholar]
- 26. Pino RM. Restriction of exogenous transthyretin (prealbumin) by the endothelium of the rat choriocapillaris. Am J Anat 1986;177:63–70. [DOI] [PubMed] [Google Scholar]
- 27. Dwork AJ, Cavallaro T, Martone RL et al. Distribution of transthyretin in the rat eye. Invest Ophthalmol Vis Sci 1990;31:489–496. [PubMed] [Google Scholar]
- 28. Leach LL, Croze RH, Hu QR et al. Induced pluripotent stem cell‐derived retinal pigmented epithelium: A comparative study between cell lines and differentiation methods. J Ocul Pharmacol Ther 2016;32:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Singh R, Phillips MJ, Kuai D et al. Functional analysis of serially expanded human iPS cell‐derived RPE cultures. Invest Ophthalmol Vis Sci 2013;54:6767–6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Singh R, Shen W, Kuai D et al. iPS cell modeling of Best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet 2013;22:593–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ferrer M, Corneo B, Davis J et al. A multiplex high‐throughput gene expression assay to simultaneously detect disease and functional markers in induced pluripotent stem cell‐derived retinal pigment epithelium. Stem Cells Transl Med 2014;3:911–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kamao H, Mandai M, Okamoto S et al. Characterization of human induced pluripotent stem cell‐derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Reports 2014;2:205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tucker BA, Cranston CM, Anfinson KA et al. Using patient‐specific induced pluripotent stem cells to interrogate the pathogenicity of a novel retinal pigment epithelium‐specific 65 kDa cryptic splice site mutation and confirm eligibility for enrollment into a clinical gene augmentation trial. Transl Res 2015;166:740–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ponticos M. Connective tissue growth factor (CCN2) in blood vessels. Vasc Pharmacol 2013;58:189–193. [DOI] [PubMed] [Google Scholar]
- 35. Ivkovic S, Yoon BS, Popoff SN et al. Connective tissue growth factor coordinates chondrogenesis and angiogenesis during skeletal development. Development 2003;130:2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lopes SMCD, Roelen BAJ, Monteiro RM et al. BMP signaling mediated by ALK2 in the visceral endoderm is necessary for the generation of primordial germ cells in the mouse embryo. Gene Dev 2004;18:1838–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grote K, Salguero G, Ballmaier M et al. The angiogenic factor CCN1 promotes adhesion and migration of circulating CD34(+) progenitor cells: Potential role in angiogenesis and endothelial regeneration. Blood 2007;110:877–885. [DOI] [PubMed] [Google Scholar]
- 38. Chen CC, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol 2009;41:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pi LY, Shenoy AK, Liu JW et al. CCN2/CTGF regulates neovessel formation via targeting structurally conserved cystine knot motifs in multiple angiogenic regulators. FASEB J 2012;26:3365–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Michaelson JS, Burkly LC. Therapeutic targeting of TWEAK/Fnl4 in cancer: Exploiting the intrinsic tumor cell killing capacity of the pathway. Results Probl Cell Differ 2009;49:145–160. [DOI] [PubMed] [Google Scholar]
- 41. Wiley SR, Cassiano L, Lofton T et al. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity 2001;15:837–846. [DOI] [PubMed] [Google Scholar]
- 42. Wiley SR, Winkles JA. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev 2003;14:241–249. [DOI] [PubMed] [Google Scholar]
- 43. Burkly LC, Michaelson JS, Hahm K et al. TWEAKing tissue remodeling by a multifunctional cytokine: Role of TWEAK/Fn14 pathway in health and disease. Cytokine 2007;40:1–16. [DOI] [PubMed] [Google Scholar]
- 44. Perper SJ, Browning B, Burkly LC et al. TWEAK is a novel arthritogenic mediator. J Immunol 2006;177:2610–2620. [DOI] [PubMed] [Google Scholar]
- 45. Girgenrath M, Weng S, Kostek CA et al. TWEAK, via its receptor Fn14, is a novel regulator of mesenchymal progenitor cells and skeletal muscle regeneration. EMBO J 2006;25:5826–5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meighan‐Mantha RL, Hsu DKW, Guo Y et al. The mitogen‐inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration. J Biol Chem 1999;274:33166–33176. [DOI] [PubMed] [Google Scholar]
- 47. Donohue PJ, Richards CM, Brown SA et al. TWEAK is an endothelial cell growth and chemotactic factor that also potentiates FGF‐2 and VEGF‐A mitogenic activity. Arterioscler Thromb Vasc Biol 2003;23:594–600. [DOI] [PubMed] [Google Scholar]
- 48. Nakayama M, Kayagaki N, Yamaguchi N et al. Involvement of TWEAK in interferon gamma‐stimulated monocyte cytotoxicity. J Exp Med 2000;192:1373–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rodrigues‐Diez RR, Garcia‐Redondo AB, Orejudo M et al. The C‐terminal module IV of connective tissue growth factor, through EGFR/Nox1 signaling, activates the NF‐kappa B pathway and proinflammatory factors in vascular smooth muscle cells. Antioxid Redox Signal 2015;22:29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rayego‐Mateos S, Morgado‐Pascual JL, Sanz AB et al. TWEAK transactivation of the epidermal growth factor receptor mediates renal inflammation. J Pathol 2013;231:480–494. [DOI] [PubMed] [Google Scholar]
- 51. Sanchez‐Lopez E, Rayego S, Rodrigues‐Diez R et al. CTGF promotes inflammatory cell infiltration of the renal interstitium by activating NF‐kappa B. J Am Soc Nephrol 2009;20:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Harada N, Nakayama M, Nakano H et al. Pro‐inflammatory effect of TWEAK/Fn14 interaction on human umbilical vein endothelial cells. Biochem Biophys Res Commun 2002;299:488–493. [DOI] [PubMed] [Google Scholar]
- 53. Davis RP, Costa M, Grandela C et al. A protocol for removal of antibiotic resistance cassettes from human embryonic stem cells genetically modified by homologous recombination or transgenesis. Nat Protoc 2008;3:1550–1558. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1
Supporting Information Figure S2
Supporting Information Table S1
Supporting Information Table S2
Supporting Information 1


