Abstract
Background
Several case-control studies reported the relationship between single nucleotide polymorphisms (SNPs) in HSP70 genes and noise-induced hearing loss (NIHL). However, their conclusions are conflicting. This meta-analysis aims to identify the association of HSP70 variants and NIHL susceptibility.
Method
A systematical literature search was performed in PubMed, Web of Science, EMBASE, and Wanfang Chinese database. The pooled odds radio (OR), 95% confidence interval (CI) and p value were calculated in fixed- or random-effects model according to the I2 value in the heterogeneity test.
Results
Four articles containing five studies, including 633 cases and 926 controls, were included. Under the allele, homozygote and dominant model, the pooled ORs (95%CI, p-value) of rs1061581 were 1.32 (1.06–1.67, p = 0.019), 1.93 (1.10–3.36, p = 0.021) and 1.455 (1.408–2.019, p = 0.025), respectively. In addition, a significant association was found between rs2227956 in Caucasians and the NIHL susceptibility under all five genetic models. We did not discover evidence sufficient to prove the associations between the other three SNPs (rs1043618, rs2763979 and rs2075800) and the NIHL susceptibility.
Conclusion
This meta-analysis indicated that the two HSP70 variants, rs1061581 and rs2227956, may serve as genetic susceptibility factors for NIHL. Larger scale studies are required to further update the results.
Introduction
Noise is one of the most common sources of environmental stress in our contemporary society [1]. Continuous noise or acoustic overstimulation damages the cochlea structure and causes inner ear cell apoptosis, resulting in hearing impairment [2]. Noise-induced hearing loss (NIHL) has been the most recorded occupational disorder in the world, accounting for 7 to 21% of hearing loss [3, 4]. However, once NIHL occurs, few therapeutic methods would be clinically effective to date [5]. Thus, to establish a practical prediction system to individual NIHL susceptibility is necessary.
It cannot be ignored that the susceptibility to NIHL among individuals is obviously diverse; some individuals are more susceptible to NIHL than others. Several studies have suggested that this individual difference in susceptibility to NIHL is due to the diverse genetic background among individuals [6]. The genotypes or single nucleotide polymorphisms (SNPs) of some genes have been demonstrated to be related to individual NIHL susceptibility, such as SOD2 [7], GST [8], PCDH15 and MYH14 [9], but these genetic variations still cannot explain all individual differences. Therefore, it is necessary to identify more NIHL-associated genes or genetic polymorphisms to further improve the genetic predictive system of NIHL.
Recent studies demonstrated that the disturbance of cellular proteostasis is critical in the development of NIHL [10]. The heat shock protein 70kD (HSP70) family, as molecular chaperones, are important for protein folding, modification, maturing and cellular normal function [11]. An increased expression of HSP70 has been found in cochlea cells under noise exposure and they further act as a protective factor in the development of NIHL [12]. What is noteworthy is that some studies indicated a close association between several SNPs in HSP70 genes, including rs1043618 [13, 14], rs2763979 [15], rs2227956 [14, 15], and rs1061581 [14] (the characteristics of these SNPs are listed in Table 1), and NIHL susceptibility, whereas no positive result for rs1043618, rs2227956 and rs1061581 was found in Yang‘s study [16]. For each SNP site, the conclusion in different studies is also diverse. A reason for the conflictive results is the heterogeneity among and the limitations in these studies, such as ethnicity, sample size, study design and many other factors, while meta-analysis has an advantage in assessing the heterogeneity and overcoming the limitations.
Table 1. Characteristics of the five SNPs in HSP70 genes.
| SNP | Location (gene) | Location (chromosome) | Synonymous or not | Variation in protein level |
|---|---|---|---|---|
| rs1043618 | the 5’ UTR of HSPA1A | 6p21.3 | - | NA |
| rs2763979 | 936 bp upstream of the 5’ end of the HSPA1B* | 6p21.3 | - | NA |
| rs2227956 | HSPA1A | 6p21.3 | a nonsynonymous variation | changes methionine to threonine resulting in the 3D structure alteration |
| rs2075800 | HSPA1A | 6p21.3 | a nonsynonymous variation | changes glutamic acid to lysine resulting in the 3D structure alteration |
| rs1061581 | HSPA1B | 6p21.3 | a synonymous variation | - |
UTR: untranslated region; NA: not available;
* be considered belonging to the HSPA1B.
There is no meta-analysis or genome-wide association studies (GWAS) on the association of HSP70 polymorphisms with NIHL susceptibility to date. Aiming at evaluating the potential value of HSP70 variants in prediction for individual NIHL susceptibility, we focus on five reported SNPs in HSP70 genes here, perform a meta-analysis to address these conflicting results and assess whether HSP70 polymorphisms are associated with the susceptibility to NIHL. The results of this meta-analysis will provide theoretical basis for the application of the SNPs in HSP70 genes in the individualized prevention system to NIHL.
Methods
Search strategy
A comprehensive literature search was performed in the following English and Chinese databases: (1) PubMed; (2) Web of Science; (3) EMBASE; and (4) Wanfang Chinese database. The MeSH and free terms were all included in our search terms, which are listed as follows: “heat shock protein 70”, “hsp70”, “noise”, “hearing loss”, “noise-induced hearing loss” and “NIHL”. Our search logic in the PubMed database is listed as follows: “((heat shock protein 70) OR (hsp70)) AND ((noise AND (hearing loss) OR (noise-induced hearing loss) OR (hearing loss, noise-induced [MeSH]) OR NIHL)”. The publication languages were limited to English and Chinese. All studies that we searched were published before April 20th, 2017. We also manually checked all articles listed in the reference lists of the retrieved literatures.
Inclusion criteria
All studies included in our meta-analysis need to be confirmed with the following criteria: (1) independent case-control studies investigating the relationship between the SNPs in HSP70 genes and the development of NIHL; (2) studies including sufficient and definite original data (the genotype frequency of each SNP in HSP70 genes in the case and control groups) in which the odds ratio (OR) with its 95% confidential interval (CI) of each genotype at every SNP site can be calculated; (3) two independent sample sets in one study were considered as two different studies; and (4) the data in the latest publication were used when duplicate publications were found.
Data extraction strategy and quality assessment
Data in the included studies were independently extracted by three authors (S Zong, X Zeng and T Liu) with the same “Data Extraction Form”. The following information was extracted from every included study: the mutation site of each SNP, first author’s last name, publication year, country, ethnicity, workplace of each sample set, standard of noise exposure, diagnosis criteria of hearing impairment or NIHL susceptibility, numbers of cases and controls, and consistency with Hardy-Weinberg Equilibrium (HWE) in the control group. The linkage disequilibrium (LD) pattern (r2 value and D’ value) between SNPs in different populations was extracted from the SNAP website (http://www.broadinstitute.org/mpg/snap) based on the data from the 1000 Genome Project.
The Newcastle-Ottawa scale (NOS) was used to assess the quality of each included study. The studies with a score ≥ 7 were considered high-quality studies. When there is conflict in the process of study selection, data extraction or quality assessment, the reviewers discussed all items until they reached consensus.
Meta-analysis
SNPs reported in two or more studies were evaluated in a meta-analysis. The association between SNPs in the HSP70 genes and NIHL susceptibility was tested by pooled OR and 95%CI. The allele (A vs. B), homozygote (AA vs. BB), heterozygote (AA vs. AB), dominant (AA vs. AB + BB) and recessive (AA + AB vs. BB) model were all applied for genotype comparison to minimize the possibility of the second type of error. The heterogeneity among the included studies was evaluated by the Q and I2 tests. The pooled ORs and 95%CIs were calculated under the fixed-effects model for p > 0.1 and I2 < 50%; otherwise, under the random-effects model with I2 ≥ 50%. Woolf’s method was applied to estimate the 95%CIs. The Z-test was applied to identify the association between the SNPs in HSP70 genes and NIHL susceptibility. We considered there was statistical significance when the overall 95% CI did not include 1 and the p-value transformed from the Z score was less than 0.05. Sub-group analysis based on ethnicity, study quality and accordance with HWE in the controls was performed. Sensitivity analysis was applied to estimate the stability of the pooled results. Publication bias for rs1043618, rs1061581 and rs2227956 were evaluated by funnel plots and the Egger’s test. For p<0.05 or a 95%CI that did not contain 0 in the Egger’s test, publication bias was considered present. HWE of the genotype distribution in the control group was evaluated by the Pearson’s X2 test. All statistical analysis was performed by Stata 13.1 software.
Results
Literature search and characteristics of included studies
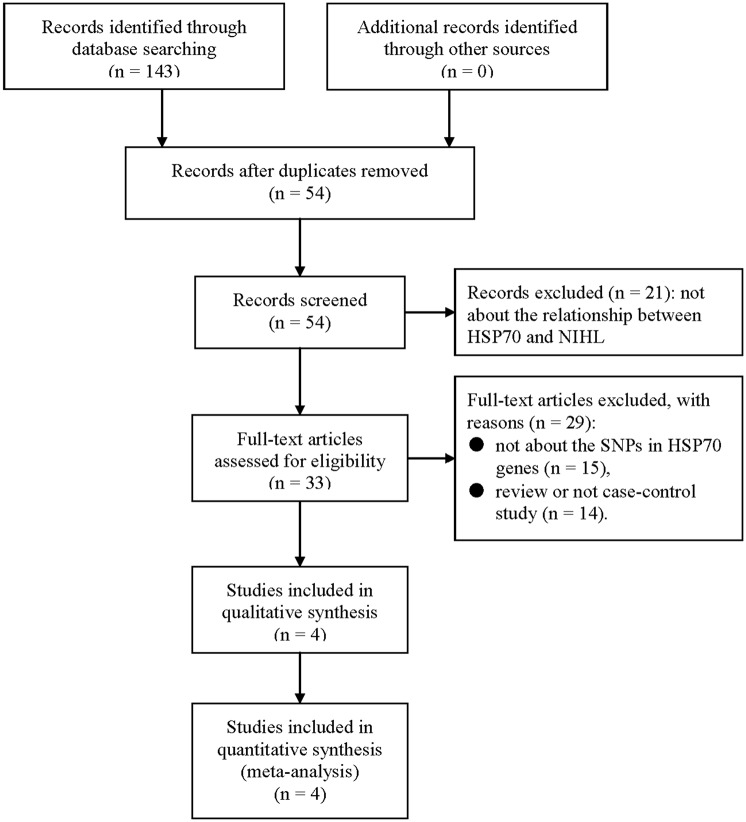
Our literature selection process is presented in Fig 1. One hundred and forty-three articles were found after our primary literature search. Fifty-four articles remained for screening the title and abstract after removing repetitive records. Twenty-one articles were excluded because the studies were not about the relationship between HSP70 and NIHL. The remaining 33 articles were assessed for eligibility via full-text screening. Twenty-nine articles were further excluded because they are reviews (n = 14) or not about the SNPs in HSP70 genes (n = 15). Finally, four articles [13–16] on 5 independent studies were included in the quality assessment and meta-analysis. Therefore, a total of 633 cases with NIHL and 926 controls were included. Table 2 summarized the basic information, including the NOS scores of the 5 eligible studies. Table 3 presents the genotype distribution of SNPs in HSP70 genes in the 5 eligible studies.
Fig 1. Flow diagram of study selection process.
Table 2. Characteristics of included studies.
| First author (year) | Country | Ethnicity | Workplace | Sample size (M/F) | Age | Genotype method | NOS score | ||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Case | Control | ||||||
| Li (2017) | China (mainland) | Asian (Chinese) | Steel factory | 286(274/12) | 286(274/12) | 45.5 ± 8.1 | 39.8 ± 8.1 | PCR(SNP scan™) | 7 |
| Chang (2011) | China (Taiwan) | Asian (Chinese) | Factories not clearly showed | 27(27/10) | 322(309/13) | 45.15 ± 6.74 | 44.05 ± 7.82 | Real time—PCR | 8 |
| Konings (2009) | Sweden | Caucasian (Swedish) | 2 paper pulp mills and 1 steel factory | 108(NA) | 98(NA) | NA | NA | PCR(SNaPshot™) | 6 |
| Konings (2009) | Poland | Caucasian (Polish) | Different factories | 119(NA) | 119(NA) | NA | NA | PCR(SNaPshot™) | 7 |
| Yang (2009) | China (mainland) | Asian (Chinese) | Motor factory | 93(77/16) | 101(55/46) | 35.2 ± 6.9 | 33.0 ± 6.1 | PCR | 7 |
M/F: male/female. NOS: Newcastle-Ottawa Scale. PCR: polymerase chain reaction. NA: not available. TM: trademark.
Table 3. Genotype distribution of HSP70 SNPs.
| SNP | First author (year) | Case | Control | HWE | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AA | AB | BB | Total | AA | AB | BB | Total | X2 | P | ||
| rs1043618 | Li (2017) | 124 | 117 | 45 | 286 | 130 | 125 | 31 | 286 | 0.01352 | 0.907435 |
| (G> C) | Chang (2011) | 8 | 18 | 1 | 27 | 153 | 139 | 30 | 322 | 0.037921 | 0.845602 |
| Konings (2009, Swedish) | 31 | 51 | 11 | 93 | 49 | 44 | 7 | 100 | 0.468949 | 0.493471 | |
| Konings (2009, Polish) | 44 | 58 | 14 | 116 | 46 | 58 | 12 | 116 | 0.009615 | 0.92189 | |
| Yang (2009) | 37 | 43 | 13 | 93 | 35 | 48 | 18 | 101 | 0.047963 | 0.826647 | |
| rs2075800 | Li (2017) | 128 | 128 | 30 | 286 | 112 | 132 | 42 | 286 | 0.093721 | 0.759499 |
| (C > T) | Chang (2011) | 10 | 15 | 2 | 27 | 113 | 166 | 43 | 322 | 2.175678 | 0.140208 |
| rs2227956 | Li (2017) | 204 | 64 | 8 | 276 | 201 | 73 | 2 | 276 | 2.856668 | 0.090996 |
| (A > G) | Konings (2009, Swedish) | 64 | 27 | 0 | 91 | 54 | 39 | 5 | 98 | 0.367347 | 0.544454 |
| Konings (2009, Polish) | 95 | 22 | 1 | 118 | 81 | 32 | 4 | 117 | 0.143747 | 0.704584 | |
| Yang (2009) | 58 | 34 | 1 | 93 | 67 | 32 | 2 | 101 | 0.673503 | 0.411833 | |
| rs2763979 | Li (2017) | 104 | 133 | 49 | 286 | 116 | 139 | 31 | 286 | 1.253581 | 0.26287 |
| (C > T) | Chang (2011) | 18 | 9 | 0 | 27 | 179 | 124 | 19 | 322 | 0.165829 | 0.683846 |
| rs1061581 | Konings (2009, Swedish) | 24 | 55 | 13 | 92 | 44 | 45 | 11 | 100 | 0.009975 | 0.920442 |
| (A > G) | Konings (2009, Polish) | 37 | 61 | 18 | 116 | 43 | 56 | 15 | 114 | 0.236309 | 0.626885 |
| Yang (2009) | 43 | 41 | 9 | 93 | 50 | 48 | 3 | 101 | 4.591447 | 0.032132# | |
HWE: Hardy-Weinberg equilibrium.
# P < 0.05, showing statistically significant difference.
The association between SNPs in HSP70 genes and NIHL susceptibility
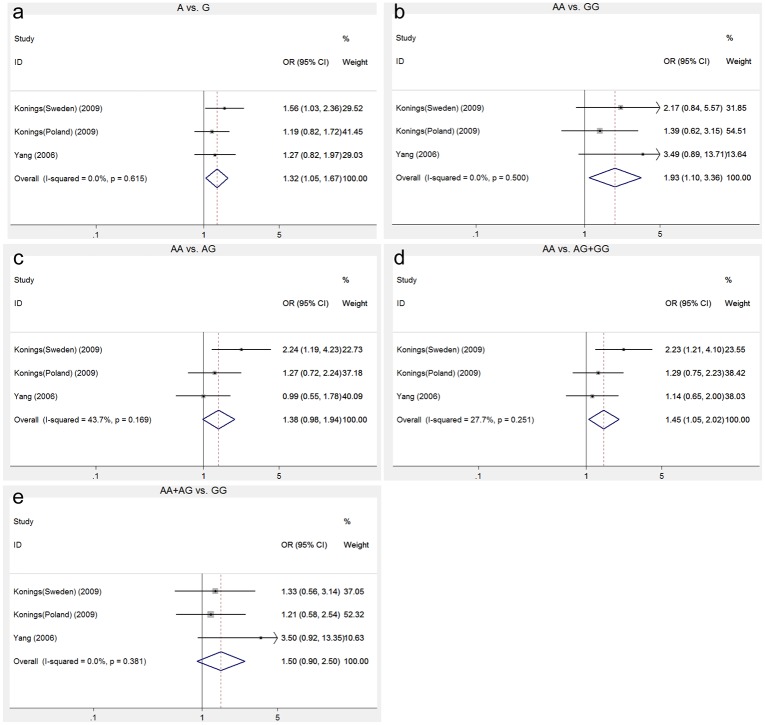
We selected five SNPs (rs1061581, rs1043618, rs2227956, rs2075800 and rs2763979) in HSP70 genes for the meta-analysis. Five genetic models were used for the genotype comparison. For SNP rs1061581, two studies on Caucasian populations and one study on Asian populations containing 301 cases and 315 controls were included. The pooled ORs and 95% CIs were 1.32 (1.06–1.67, p = 0.019), 1.93 (1.10–3.36, p = 0.021) and 1.455 (1.408–2.019, p = 0.025) in the allele, homozygous and dominant model, respectively (Fig 2 and Table 4). These results indicated that there is a close association between the SNP rs1061581 in the HSPA1B gene and NIHL susceptibility. For rs2227956, the pooled ORs and 95% CIs in Caucasian were 0.535 (0.368–0.779, p = 0.001), 0.135 (0.024–0.764, p = 0.024), 0.585 (0.379–0.903, p = 0.016), 0.531 (0.374–0.812, p = 0.004) and 0.157 (0.028–0.889, p = 0.036) in the allele, homozygote, heterozygote, dominant and recessive models, respectively (Table 4 and S1 Fig). For rs1043618, rs2763979 and rs2075800, no significant association was found between the SNPs and NIHL susceptibility in any of the five genetic models for the Asian and the Caucasian populations, respectively, or across the ethnicity. The details of the overall analysis of the association of the five SNPs in HSP70 genes and NIHL susceptibility are listed in Table 4.
Fig 2. The association between rs1061581 and NIHL susceptibility in different genetic models.
Table 4. Overall analysis of the association between HSP70 SNPs and NIHL susceptibility.
| SNP | Ethnicity | Study number | Case/Control | Genetic model | I2 (%) | Model | OR (95% CI) | Z score | P (Z) |
|---|---|---|---|---|---|---|---|---|---|
| rs1043618 | Mixed population | 5 | 615/925 | G vs. C | 19.7 | F | 1.150 (0.978, 1.351) | 1.70 | 0.090 |
| (G > C) | GG vs. CC | 10.6 | F | 1.301 (0.909, 1.863) | 1.44 | 0.151 | |||
| GG vs. GC | 44.0 | F | 1.151 (0.912, 1.454) | 1.18 | 0.236 | ||||
| GG vs. GC+CC | 39.5 | F | 1.188 (0.952, 1.484) | 1.53 | 0.127 | ||||
| GG+GC vs. CC | 5.1 | F | 1.228 (0.879, 1.716) | 1.21 | 0.228 | ||||
| Asian subgroup | 3 | 406/709 | G vs. C | 18.2 | F | 1.092 (0.897, 1.330) | 0.88 | 0.381 | |
| GG vs. CC | 29.4 | F | 1.086 (0.594, 1.987) | 0.27 | 0.789 | ||||
| GG vs. GC | 54.5 | R | 1.144 (0.695, 1.885) | 0.53 | 0.597 | ||||
| GG vs. GC+CC | 42.6 | F | 1.099 (0.839, 1.440) | 0.68 | 0.494 | ||||
| GG+GC vs. CC | 45.4 | F | 1.165 (0.784, 1.729) | 0.76 | 0.450 | ||||
| Caucasian subgroup | 2 | 209/216 | G vs. C | 42.9 | F | 1.278 (0.964, 1.694) | 1.70 | 0.088 | |
| GG vs. CC | 4.0 | F | 1.673 (0.824, 3.253) | 1.41 | 0.159 | ||||
| GG vs. GC | 44.7 | F | 1.349 (0.900, 2.023) | 1.45 | 0.147 | ||||
| GG vs. GC+CC | 52.2 | R | 1.418 (0.803, 2.503) | 1.20 | 0.228 | ||||
| GG+GC vs. CC | 0.0 | F | 1.403 (0.784, 2.633) | 1.06 | 0.291 | ||||
| High quality | 4 | 525/828 | G vs. C | 0.0 | F | 1.089(0.914, 1.297) | 0.96 | 0.339 | |
| subgroup | GG vs. CC | 0.0 | F | 1.191(0.812, 1.748) | 0.89 | 0.371 | |||
| GG vs. GC | 31.8 | F | 1.060(0.823, 1.366) | 0.45 | 0.653 | ||||
| GG vs. GC+CC | 14.0 | F | 1.094(0.860, 1.391) | 0.73 | 0.464 | ||||
| GG+GC vs. CC | 18.0 | F | 1.169(0.819, 1.669) | 0.86 | 0.389 | ||||
| rs2227956 | Mixed population | 4 | 578/592 | A vs. G | 63.7 | R | 0.780 (0.528, 1.847) | 1.25 | 0.213 |
| (A>G) | AA vs. GG | 62.4 | R | 0.549 (0.090, 3.348) | 0.65 | 0.516 | |||
| AA vs. AG | 27.1 | F | 0.802 (0.619, 1.040) | 1.67 | 0.096 | ||||
| AA vs. AG + GG | 50.7 | R | 0.772 (0.529, 1.127) | 1.34 | 0.180 | ||||
| AA + AG vs. GG | 60.3 | R | 0.593 (0.013, 3.425) | 0.58 | 0.550 | ||||
| Asian subgroup | 2 | 369/377 | A vs. G | 0.0 | F | 1.064 (0.802, 1.410) | 0.43 | 0.668 | |
| AA vs. GG | 41.3 | F | 2.328 (0.071, 7.727) | 1.38 | 0.168 | ||||
| AA vs. AG | 0.0 | F | 0.959 (0.693, 1.327) | 0.25 | 0.800 | ||||
| AA vs. AG + GG | 0.0 | F | 1.011 (0.734, 1.389) | 0.07 | 0.945 | ||||
| AA + AG vs. GG | 47.7 | F | 2.335 (0.711, 7.671) | 1.40 | 0.162 | ||||
| Caucasian subgroup | 2 | 209/215 | A vs. G | 0.0 | F | 0.535 (0.368, 0.779) | 3.26 | 0.001 | |
| AA vs. GG | 0.0 | F | 0.135 (0.024, 0.764) | 2.26 | 0.024 | ||||
| AA vs. AG | 0.0 | F | 0.585 (0.379, 0.903) | 2.42 | 0.016 | ||||
| AA vs. AG + GG | 0.0 | F | 0.531 (0.374, 0.812) | 2.92 | 0.004 | ||||
| AA + AG vs. GG | 0.0 | F | 0.157 (0.028, 0.889) | 2.09 | 0.036 | ||||
| High quality | 3 | 470/494 | A vs. G | 55.9 | R | 0.886(0.595, 1.319) | 0.59 | 0.552 | |
| subgroup | AA vs. GG | 59.4 | R | 0.915(0.144, 5.827) | 0.09 | 0.925 | |||
| AA vs. AG | 29.5 | F | 0.861(0.646, 1.147) | 1.02 | 0.307 | ||||
| AA vs. AG + GG | 44.3 | F | 0.882(0.667, 1.166) | 0.88 | 0.377 | ||||
| AA + AG vs. GG | 58.5 | R | 0.950(0.153, 5.901) | 0.06 | 0.956 | ||||
| rs1061581 | Mixed population | 3 | 301/315 | A vs. G | 0.0 | F | 1.322 (1.046, 1.671) | 2.34 | 0.019 |
| (A>G) | AA vs. GG | 0.0 | F | 1.926 (1.104, 3.359) | 2.31 | 0.021 | |||
| AA vs. AG | 43.7 | F | 1.378 (0.981,1.937) | 1.85 | 0.065 | ||||
| AA vs. AG + GG | 27.7 | F | 1.455 (1.408, 2.019) | 2.24 | 0.025 | ||||
| AA + AG vs. GG | 0.0 | F | 1.500 (0.901, 2.496) | 1.56 | 0.119 | ||||
| Caucasian subgroup | 2 | 251/264 | A vs. G | 0.0 | F | 1.342 (1.017, 1.771) | 2.08 | 0.037 | |
| (HWE P > 0.05 | AA vs. GG | 0.0 | F | 1.679 (0.907, 3.109) | 1.65 | 0.099 | |||
| subgroup) | AA vs. AG | 41.9 | F | 1.636 (1.073, 2.493) | 2.29 | 0.022 | |||
| AA vs. AG + GG | 41.0 | F | 1.648 (1.100, 2.468) | 2.42 | 0.015 | ||||
| AA + AG vs. GG | 0.0 | F | 1.262 (0.720, 2.210) | 0.81 | 0.416 | ||||
| High quality | 2 | 107/217 | A vs. G | 0.0 | F | 1.222(0.920, 1.624) | 1.38 | 0.167 | |
| subgroup | AA vs. GG | 21.7 | F | 1.814(0.912, 3.606) | 1.70 | 0.090 | |||
| AA vs. AG | 0.0 | F | 1.124(0.748, 1.689) | 0.56 | 0.572 | ||||
| AA vs. AG + GG | 0.0 | F | 1.217(0.823, 1.800) | 0.98 | 0.326 | ||||
| AA + AG vs. GG | 46.0 | F | 1.599(0.848, 3.014) | 1.45 | 0.147 | ||||
| rs2075800 | Asian | 2 | 313/608 | C vs. T | 0.0 | F | 0.182 (0.684, 1.017) | 1.82 | 0.069 |
| (C>T) | CC vs. TT | 0.0 | F | 0.612 (0.370, 1.013) | 1.91 | 0.056 | |||
| CC vs. CT | 0.0 | F | 0.872 (0.631, 1.206) | 0.83 | 0.408 | ||||
| CC vs. CT + TT | 0.0 | F | 0.811 (0.596, 1.103) | 1.33 | 0.182 | ||||
| CC + CT vs. TT | 0.0 | F | 0.658 (0.410, 1.055) | 1.74 | 0.082 | ||||
| rs2763979 | Asian | 2 | 313/608 | C vs. T | 71.7 | R | 0.939 (0.462, 1.909) | 0.17 | 0.862 |
| (C>T) | CC vs. TT | 44.6 | F | 1.561 (0.850, 2.564) | 1.76 | 0.079 | |||
| CC vs. CT | 0.0 | F | 1.003 (0.724, 1.389) | 0.02 | 0.988 | ||||
| CC vs. CT + TT | 50.0 | R | 0.971 (0.537, 1.753) | 0.10 | 0.921 | ||||
| CC + CT vs. TT | 34.4 | F | 1.550 (0.975, 2.464) | 1.85 | 0.064 |
F: fixed-effects model. R: random-effects model. P< 0.05, showing statistically significant difference.
Sensitivity analysis
We estimated the stability of the pooled ORs of the SNPs (rs1043618, rs1061581 and rs2227956) by eliminating each of the included studies in turn. For the three SNPs mentioned above, no significant alteration of the pooled ORs was found as any of the included studies was omitted (S2 Fig). These results indicate that the corresponding ORs were relatively robust. For rs2075800 and 2763979, we did not perform the sensitivity analysis because of the limited number of included studies.
Publication bias
The funnel plots looked symmetric for three SNPs (rs1043618, rs1061581 and rs2227956) in all five genetic models. We did not test the publication bias for rs2075800 and rs2763979 because of the limited number of included studies. Additionally, we did not found any publication bias via Egger’s test (S1 Table).
Discussion
NIHL affects the life quality of millions of people worldwide [17, 18], especially industrial workers [14]. What cannot be ignored is that the genetic background has a close association with the individual NIHL susceptibility. Therefore, it is essential to identify more genetic markers that can predict individual NIHL susceptibility potentially. The abnormity of HSP70 have been demonstrated to be related to many diseases, such as obesity [19], congenital sideroblastic anemia [20], coronary artery disease [21] and age-related cataract [22]. In the research field of NIHL, two studies both suggested an important role of HSP70 in the resistance to NIHL [12, 23]. Moreover, the participation of HSP70 in the alleviation effect of dexamethasone on NIHL was also indicated [24]. In addition, several SNPs in HSP70 genes are further considered relating to NIHL susceptibility, but the results are inconsistent [13–16]. Therefore, to comprehensively and reliably understand the association between SNPs in HSP70 genes and NIHL susceptibility, we performed this meta-analysis that includes larger samples of 633 cases with NIHL and 926 controls.
A significant relationship was seen between rs1061581 and NIHL susceptibility (Table 4 and Fig 2), which indicated that the G allele is a potential risk factor to NIHL susceptibility. Rs1061581 has been considered a major and synonymous SNP in HSPA1B gene, which is located at chromosome 6p21.3. Its protein product, HSP-2, is 641 amino acids in length and stress-inducible [25]. However, the way in which rs1061581 variant affected the HSP-2 protein remains unclear, which needs to be further explored. The association of rs2227956 with NIHL is statistically significant only in the Caucasian population (Table 4 and S1 Fig). This phenomenon is mostly due to the heterogeneity in the genetic background of different ethnicities. Rs2227956 in HSPA1A is also located at chromosome 6p21.3. A variation from A to G changes methionine to threonine resulting in the 3D structure alteration of HSP-1, which may affects its function as a molecular chaperone [25]. We found no association between rs1043618, rs2763979 and rs2075800 and NIHL susceptibility.
Heterogeneity is a major problem that affects the reliability of the pooled ORs in meta-analysis. In our meta-analysis, mild to moderate heterogeneity was observed in most SNPs (Table 4). The heterogeneity in our meta-analysis could be explained by the following features. (1) Individuals from different ethnicities have diverse genetic background, such as different LD patterns and different minor allele frequency (MAF) of these SNPs among the population (S2 Table). (2) The diagnostic criteria of hearing loss or NIHL susceptibility in different studies are not in full accord. For example, the frequency in which hearing thresholds were examined differs from study to study. Although the diagnostic criteria in each study were well-founded, there was heterogeneity from these differences. (3) The quality of the included studies is diverse. We used the NOS scale to evaluate the quality of these studies and found the quality of the included studies varied (Table 2), which may be a potential source of heterogeneity.
There are several limitations in this meta-analysis. (1) We only searched the articles published in English and Chinese. The articles in other languages were not included in this meta-analysis, which may increase the publication bias. (2) The number of included studies is relatively low. For rs2075800 and rs2763979, only two studies were included in this meta-analysis. Their funnel plots were very asymmetric, which indicated there is significant publication bias. (3) The sample size in some included studies was relatively small, which may increase the heterogeneity. Although 633 cases and 926 controls are included in this meta-analysis, the conclusion must be updated further by including larger sample reports.
To the best of our knowledge, this is the first meta-analysis on the association between the SNPs in HSP70 genes and NIHL susceptibility. Our results indicated that the allele G in rs1061581 and rs2227956 (only in Caucasians) may serve as genetic susceptibility factors for NIHL. However, more studies with larger sample sets from different ethnicities are needed to further confirm the relationship between HSP70 polymorphisms and NIHL susceptibility due to the limitations in this meta-analysis.
Supporting information
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOCX)
(TIF)
(TIF)
Acknowledgments
The authors are indebted to Dr. Qinghua Hu for revising the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (No. 81470697, http://www.nsfc.gov.cn/, H Xiao). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wallenius MA. The interaction of noise stress and personal project stress on subjective health. Journal of Environmental Psychology. 2004;24(2):167–77. doi: 10.1016/j.jenvp.2003.12.002 [Google Scholar]
- 2.Le Prell CG, Yamashita D, Minami SB, Yamasoba T, Miller JM. Mechanisms of noise-induced hearing loss indicate multiple methods of prevention. Hear Res. 2007;226(1–2):22–43. doi: 10.1016/j.heares.2006.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobie RA. The burdens of age-related and occupational noise-induced hearing loss in the United States. Ear Hear. 2008;29(4):565–77. doi: 10.1097/AUD.0b013e31817349ec . [DOI] [PubMed] [Google Scholar]
- 4.Nelson DI, Nelson RY, Concha-Barrientos M, Fingerhut M. The global burden of occupational noise-induced hearing loss. Am J Ind Med. 2005;48(6):446–58. doi: 10.1002/ajim.20223 . [DOI] [PubMed] [Google Scholar]
- 5.Sha SH, Schacht J. Emerging therapeutic interventions against noise-induced hearing loss. Expert Opin Investig Drugs. 2017;26(1):85–96. doi: 10.1080/13543784.2017.1269171 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlsson PI, Van Laer L, Borg E, Bondeson ML, Thys M, Fransen E, et al. The influence of genetic variation in oxidative stress genes on human noise susceptibility. Hear Res. 2005;202(1–2):87–96. doi: 10.1016/j.heares.2004.09.005 . [DOI] [PubMed] [Google Scholar]
- 7.Liu YM, Li XD, Guo X, Liu B, Lin AH, Ding YL, et al. SOD2 V16A SNP in the mitochondrial targeting sequence is associated with noise induced hearing loss in Chinese workers. Dis Markers. 2010;28(3):137–47. doi: 10.3233/DMA-2010-0693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin CY, Wu JL, Shih TS, Tsai PJ, Sun YM, Guo YL. Glutathione S-transferase M1, T1, and P1 polymorphisms as susceptibility factors for noise-induced temporary threshold shift. Hear Res. 2009;257(1–2):8–15. doi: 10.1016/j.heares.2009.07.008 . [DOI] [PubMed] [Google Scholar]
- 9.Konings A, Van Laer L, Wiktorek-Smagur A, Rajkowska E, Pawelczyk M, Carlsson PI, et al. Candidate gene association study for noise-induced hearing loss in two independent noise-exposed populations. Ann Hum Genet. 2009;73(2):215–24. doi: 10.1111/j.1469-1809.2008.00499.x . [DOI] [PubMed] [Google Scholar]
- 10.Xue Q, Li C, Chen J, Guo H, Li D, Wu X. The Protective effect of the endoplasmic reticulum stress-related factors BiP/GRP78 and CHOP/Gadd153 on noise-induced hearing loss in guinea pigs. Noise Health. 2016;18(84):247–55. doi: 10.4103/1463-1741.192481 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuiderweg ER, Hightower LE, Gestwicki JE. The remarkable multivalency of the Hsp70 chaperones. Cell Stress Chaperones. 2017;22(2):173–89. doi: 10.1007/s12192-017-0776-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gratton MA, Eleftheriadou A, Garcia J, Verduzco E, Martin GK, Lonsbury-Martin BL, et al. Noise-induced changes in gene expression in the cochleae of mice differing in their susceptibility to noise damage. Hear Res. 2011;277(1–2):211–26. doi: 10.1016/j.heares.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang NC, Ho CK, Lin HY, Yu ML, Chien CY, Ho KY. Association of polymorphisms of heat shock protein 70 with susceptibility to noise-induced hearing loss in the Taiwanese population. Audiol Neurootol. 2011;16(3):168–74. doi: 10.1159/000317119 . [DOI] [PubMed] [Google Scholar]
- 14.Konings A, Van Laer L, Michel S, Pawelczyk M, Carlsson PI, Bondeson ML, et al. Variations in HSP70 genes associated with noise-induced hearing loss in two independent populations. Eur J Hum Genet. 2009;17(3):329–35. doi: 10.1038/ejhg.2008.172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y, Yu S, Gu G, Chen G, Zheng Y, Jiao J, et al. Polymorphisms of heat shock protein 70 genes (HSPA1A, HSPA1B and HSPA1L) and susceptibility of noise-induced hearing loss in a Chinese population: A case-control study. PLoS One. 2017;12(2):e0171722 doi: 10.1371/journal.pone.0171722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang M, Tan H, Yang Q, Wang F, Yao H, Wei Q, et al. Association of hsp70 polymorphisms with risk of noise-induced hearing loss in Chinese automobile workers. Cell Stress Chaperones. 2006;11(3):233–9. doi: 10.1379/CSC-192R.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen H, Cao J, Hong Z, Liu K, Shi J, Ding L, et al. A functional Ser326Cys polymorphism in hOGG1 is associated with noise-induced hearing loss in a Chinese population. PLoS One. 2014;9(3):e89662 doi: 10.1371/journal.pone.0089662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulkowski WJ, Szymczak W, Kowalska S, Sward-Matyja M. Epidemiology of occupational noise-induced hearing loss (ONIHL) in Poland. Otolaryngol Pol. 2004;58(1):233–6. . [PubMed] [Google Scholar]
- 19.Mardan-Nik M, Pasdar A, Jamialahmadi K, Avan A, Mohebati M, Esmaily H, et al. Association of heat shock protein70-2 (HSP70-2) gene polymorphism with obesity. Ann Hum Biol. 2016;43(6):542–6. doi: 10.3109/03014460.2015.1119309 . [DOI] [PubMed] [Google Scholar]
- 20.Schmitz-Abe K, Ciesielski SJ, Schmidt PJ, Campagna DR, Rahimov F, Schilke BA, et al. Congenital sideroblastic anemia due to mutations in the mitochondrial HSP70 homologue HSPA9. Blood. 2015;126(25):2734–8. doi: 10.1182/blood-2015-09-659854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mardan-Nik M, Pasdar A, Jamialahmadi K, Biabangard-Zak A, Mirhafez SR, Ghalandari M, et al. Association of heat shock protein70-2 (HSP70-2) gene polymorphism with coronary artery disease in an Iranian population. Gene. 2014;550(2):180–4. doi: 10.1016/j.gene.2014.08.012 . [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Gong J, Zhang L, Xue D, Liu H, Liu P. Genetic polymorphisms of HSP70 in age-related cataract. Cell Stress Chaperones. 2013;18(6):703–9. doi: 10.1007/s12192-013-0420-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuo H, Cui B, She X, Wu M. Changes in Guinea pig cochlear hair cells after sound conditioning and noise exposure. J Occup Health. 2008;50(5):373–9. . [DOI] [PubMed] [Google Scholar]
- 24.Maeda Y, Fukushima K, Kariya S, Orita Y, Nishizaki K. Dexamethasone Regulates Cochlear Expression of Deafness-associated Proteins Myelin Protein Zero and Heat Shock Protein 70, as Revealed by iTRAQ Proteomics. Otol Neurotol. 2015;36(7):1255–65. doi: 10.1097/MAO.0000000000000748 . [DOI] [PubMed] [Google Scholar]
- 25.Radons J. The human HSP70 family of chaperones: where do we stand? Cell Stress Chaperones. 2016;21(3):379–404. doi: 10.1007/s12192-016-0676-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC)
(DOCX)
(DOCX)
(DOCX)
(DOC)
(DOCX)
(TIF)
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.