Abstract
Background
The reovirus λ2 protein catalyzes mRNA capping, that is, addition of a guanosine to the 5' end of each transcript in a 5'-to-5' orientation, as well as transfer of a methyl group from S-adenosyl-L-methionine (AdoMet) to the N7 atom of the added guanosyl moiety and subsequently to the ribose 2'-O atom of the first template-encoded nucleotide. The structure of the human reovirus core has been solved at 3.6 Å resolution, revealing a series of domains that include a putative guanylyltransferase domain and two putative methyltransferase (MTase) domains. It has been suggested that the order of domains in the λ2 protein corresponds to the order of reactions in the pathway and that the m7G (cap 0) and the 2'-O-ribose (cap 1) MTase activities may be exerted by the MTase 1 and the MTase 2 domains, respectively.
Results
We show that the reovirus MTase 1 domain shares a putative active site with the structurally characterized 2'-O-ribose MTases, including vaccinia virus cap 1 MTase, whereas the MTase 2 domain is structurally similar to glycine N-MTase.
Conclusions
On the basis of our analysis of the structural details we propose that the previously suggested functional assignments of the MTase 1 and MTase 2 domains should be swapped.
Background
Methylated 5'-terminal cap structures have been described in most eukaryotic and many viral mRNAs. In all cap structures, including the 'minimal' cap 0 (m7G(5')ppp(5')N), an N7-methylguanosine (m7G) is attached through a 5'-5' triphosphate bridge to the penultimate nucleoside. Cap 0 is usually synthesized in the nucleus by the sequential action of three enzymatic activities: mRNA triphosphatase, guanylyltransferase (GTase), and m7G-methyltransferase (MTase); this pathway was defined originally during studies of vaccinia and reovirus mRNA synthesis [1,2]. In some molecules, additional 2'-O-ribose methylations are found at the penultimate and the antepenultimate nucleosides, forming the cap 1 (m7G(5')ppp(5')Nm2'O) and cap 2 (m7G(5')ppp(5')Nm2'OpNm2'O) structures, respectively (reviewed in [3,4,5]). The capping apparatus differs significantly in fungi, metazoans, protozoa and viruses in respect of the evolutionary origin and structure of individual domains, and the arrangement of domains within subunits [5]. Hence, the capping enzymes encoded by viral, fungal and protozoal pathogens are attractive targets for specific inhibitors that would exert a limited effect on the host enzyme.
Reoviruses are non-enveloped icosahedrally symmetric viruses with two concentric protein capsids surrounding and protecting the multisegmented, double-stranded (ds) RNA genome; they replicate in the cytoplasm of the eukaryotic host cell (reviewed in [6]) The reovirus core particle can produce m7GpppGm2'OpC(pN)n-OH plus-strand RNA from each genomic double-stranded RNA segment in vitro [2], indicating that it contains all of the enzymes necessary for de novo synthesis of capped mRNA. On the basis of sequence analysis and the results of UV-crosslinking and site-directed mutagenesis it has been suggested that the 144-kDa λ2 protein possesses an S-adenosyl-L-methionine (AdoMet)-dependent methyltransferase (MTase) domain (or domains), which mediates both the cap 0 and cap 1 methylation activities in forming a 5' cap structure on reovirus mRNA [7,8,9]. Nevertheless, the failure of isolated λ2 to exhibit methylation activity limited studies to dissect its putative methyltransferase activities and domain structure at a molecular level.
The structure of the reovirus core has been recently solved at 3.6 Å resolution by Reinisch et al. (entry 1ej6 in Protein Data Bank) [10], revealing a series of seven domains in the λ2 monomer, of which one domain (amino acids 1–385) was identified as a guanylyltransferase and two domains (434–691 and 804–1,022) were found to share the three-dimensional fold with typical AdoMet-dependent MTases (reviewed in [11,12]). In the turret formed by λ2 pentamers, the GTase site from one monomer is closer to the MTase 1 in the clockwise neighbor than to other MTase domains. Reinisch et al. hypothesized that a correlation may exist between the spatial arrangement of domains in λ2, and the order of reactions in a pathway (guanylyl transfer → N7 methylation → 2'O methylation). Thus, it has been proposed that MTase 1 is the cap 0 MTase and MTase 2 is the cap 1 MTase [10]. However, no groove to guide the mRNA from one active site to another could be identified and the problem of specificities of reactions carried out by the individual MTase domains was left open.
Results and discussion
In the course of a large-scale analysis of MTase sequences and structures we observed low similarity between various ribose 2'-O-MTases from eubacteria and negative-strand RNA viruses and a fragment of the grass carp reovirus protein [13] corresponding to the carboxy-terminal part of MTase 1 of human reovirus (J.M.B. and L.R., unpublished data). This prompted us to reanalyze the sequences and structures of the reovirus MTase domains. Disappointingly, sequence database searches carried out using PSI-BLAST [14] initiated with the λ2 sequence and its fragments revealed significant similarities only between the human reovirus and grass carp reovirus proteins. Nonetheless, VAST [15] searches of the Protein Data Bank [16] revealed high similarity of both putative MTase domains to numerous known MTase structures. Detailed inspection of the coordinates superimposed using Swiss-Pdb Viewer [17] allowed us to predict which amino-acid residues might be involved in cap 0 and cap 1 methylation of reovirus mRNA.
According to VAST, MTase 1 showed highest similarity to the RrmJ (1ejo; P-value = 10-10.4) and VP39 (1av6; P-value = 10-8.1) proteins. The mutual similarity of MTase 1 and MTase 2, which can be regarded as a reference, was evaluated as high, albeit significantly lower (P-value =10-4). RrmJ and VP39 are both 2'-O-ribose MTases; RrmJ targets U2552 in eubacterial rRNA [18,19], whereas VP39 is a bonafide cap 1 MTase of vaccinia virus mRNA [20]. Remarkably, simultaneous superposition of the three structures revealed perfect conservation of the K-D-K-E tetrad of putative catalytic residues (Figures 1,2,3), which has been observed by us in other families of genuine and putative 2'-O-MTases ([21] and J.M.B. and L.R., unpublished data). These results strongly argue that MTase 1 is more likely to be a cap 1 MTase than a cap 0 MTase.
Figure 1.
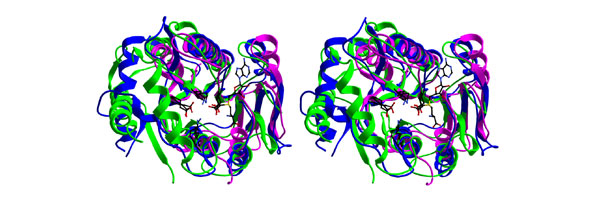
Stereoview ribbon diagrams of superimposed structures of the MTase 1 domain (green), VP39 (blue) and RrmJ (magenta). MTase 1 shows elaborations of the common fold similar to those of VP39, which are altogether absent from the RrmJ structure and which are predicted to take part in cap recognition and binding. AdoMet (copied from the RrmJ structure) and the K-D-K-E tetrad of putative catalytic residues are shown in wireframe representation.
Figure 2.
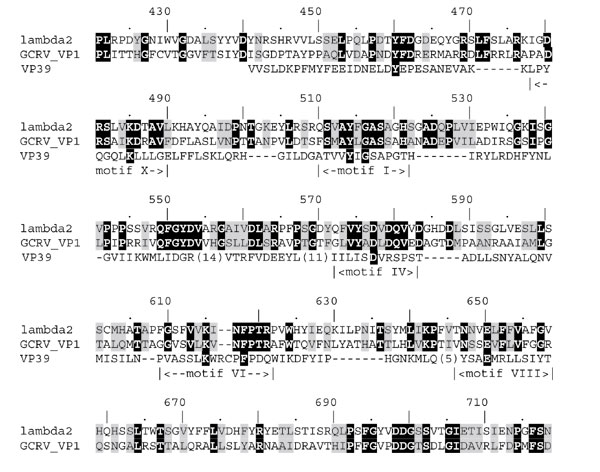
Sequence alignment of the predicted human reovirus cap 1 MTase with its counterpart from grass carp reovirus (GCRV) and vaccinia virus (VP39). Residues that are invariant between the reoviruses and those predicted to participate in binding of the AdoMet, the cap moiety, and in catalysis in all cap 1 MTases, are highlighted in black; other conserved residues are highlighted in gray. Motif nomenclature follows [12].
Figure 3.
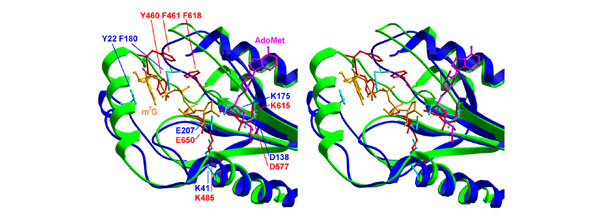
Stereoview of superimposed structures of the MTase 1 domain (ribbon in green, side chains and labels in red) and VP39 (ribbon and labels in blue, side chains in cyan), delineating the proposed binding sites for the guanine moiety and the target ribose 2'-OH. Capped mRNA is shown in orange and the methylated guanine (black label) is sandwiched between Y22 and F180 of VP39. A conformational change of both the protein and the RNA is required for the cluster of conserved aromatic residues of the reovirus MTase to bind the cap structure in a similar manner.
In the MTase 1 structure we could identify no aromatic residues that would superimpose well with Y22 and F180 from VP39, which form enhanced stacking interactions with the N7-methylated cap [20]. Moreover, a region corresponding to the guanine-binding pocket in VP39 is blocked off by a differently positioned loop in the reovirus λ2 protein structure. Nevertheless, we identified a cluster of aromatic side chains (Y460/464, F461/465 and F618/F622) that are invariant between the human and grass carp reovirus sequences and map in the vicinity of the methylated cap, if the mRNA coordinates are copied from the superimposed VP39 structure (Figure 2). It is tempting to speculate that a conformational change in this region occurring on mRNA binding would, for instance, reallocate Y460 and F618 residues of human reovirus to positions equivalent to Y22 and F180 from VP39. The sequence alignment (Figure 3) has been manually adjusted to illustrate this hypothesis.
The MTase 2 structure showed highest similarity to the glycine N-MTase (GNMT; 1xva; P-value = 10-10.1). It is noteworthy that the MTase 2 domain showed lower similarity (P-value = 10-4.3) to VP39 than to GNMT and the MTase 1 domain showed lower similarity (P-value = 10-7.9) to GNMT than to the 2'-O-ribose MTases. Figure 4 shows that in addition to the common catalytic domain, GNMT and MTase 2 share a topologically equivalent 'lid' domain made of three antiparallel β strands. In GNMT, the β-lid domain forms a wall of a large 'molecular basket' structure, which may accommodate a variety of small molecules, including AdoMet, tetrahydrofolate and polycyclic aromatic hydrocarbon molecules such as benzopyrene (reviewed in [22]). In MTase 2, the strands forming the β-lid domain are much shorter and the pocket is also smaller. However, a guanine moiety can be docked into that pocket in a way that the N7 nitrogen is presented for the methyl group donor (data not shown). Most of the amino acids that line up the putative guanine-binding pocket of MTase 2 (for instance Q925, N927, F951, R956, and E958) are conserved between the human and grass carp reovirus sequences (Figure 5).
Figure 4.
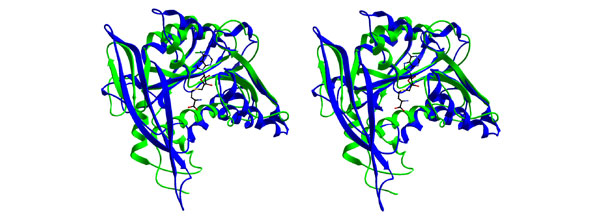
Stereoview of superimposed structures of the MTase 2 domain (green) and GNMT (blue) showing extensive similarities of both domains.
Figure 5.
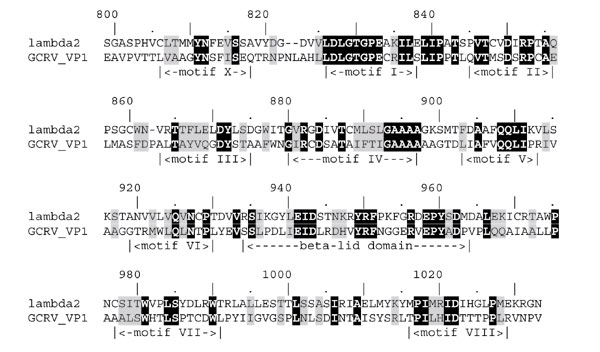
Sequence alignment of the MTase 2 domain with its counterpart from grass carp reovirus. Invariant residues are highlighted in black, conservatively substituted residues are highlighted in gray. Motif nomenclature follows [12].
To our knowledge, all structurally characterized MTases that modify bases in nucleic acids and do not employ covalent bond formation with the target use aromatic or aliphatic side chains to bind the base to be methylated and stabilize it in the active site. Examples of such MTases, in which the structure of the active site was determined experimentally or predicted from sequence analysis, include enzymes generating N6-methyladenine in DNA and RNA (reviewed in [23]), N4-methylcytosine in DNA [24], and N2-guanine in RNA [25]. We hypothesize that the invariant F951 and L890 (substituted by F897 in grass carp reovirus) may be involved in van der Waals interactions with an aromatic ring of guanine. But because of the low resolution of the original structure and lack of a precise docking model, the detailed contacts between the target and the enzyme could not be predicted. Nevertheless, we believe that our model will be a useful guide for site-directed mutagenesis experiments that would allow to elucidate the role of individual side chains of the putative cap 0 MTase.
Conclusions
The data presented suggest that MTase domains 1 and 2 of reovirus λ2 protein function as cap 0 and cap 1 MTases, respectively; that is, that the previous assignments should be swapped. On the basis of detailed comparison of the available MTase structures, we predicted the substrate-binding mode for both MTase domains and proposed which residues are likely to participate in catalysis. This hypothesis may be testable using mutagenesis and/or soaking experiments with substrate analogs. We suggest that substitution of the predicted cap-binding or catalytic residues in the MTase 1 domain should result in accumulation of cap 0-methylated viral mRNAs that nevertheless lack cap 1 modification.
We also report for the first time a remarkable structural similarity between the predicted m7G MTase domain and GNMT, suggesting a common evolutionary origin for the two N-MTase families. It is noteworthy that threading analysis and subsequent homology modeling suggest a similar structure for a family of cellular and poxviral cap 0 MTases [26] that show no significant sequence similarity to predicted cap 0 MTases from the Sindbis-like supergroup of positive-strand RNA viruses [27] nor to the predicted cap 0 MTase from reoviruses analyzed herein (J.M.B. and L.R., unpublished data). On the other hand, structure prediction for the family of aminoglycoside-resistance 16S rRNA:m7G MTases suggests that it shares only a common catalytic domain, but not the β-lid domain or predicted target-binding residues, with other m7G MTases [28]. It will be interesting to determine how many times the mechanism of guanine-N7 methy-lation appeared in the evolution of AdoMet-dependent MTases and what is the phylogenetic origin of the individual viral cap 0 MTase families.
Acknowledgments
Acknowledgements
This work was supported by KBN (grant 8T11F01019 to J.M.B.) and BioInfoBank. We would like to thank Karen Reinisch for comments and suggestions that helped to improve the manuscript.
References
- Moss B, Gershowitz A, Wei CM, Boone R. Formation of the guanylylated and methylated 5'-terminus of vaccinia virus mRNA. Virology. 1976;72:341–351. doi: 10.1016/0042-6822(76)90163-x. [DOI] [PubMed] [Google Scholar]
- Furuichi Y, Muthukrishnan S, Tomasz J, Shatkin AJ. Mechanism of formation of reovirus mRNA 5'-terminal blocked and methylated sequence, m7GpppGmpC. J Biol Chem. 1976;251:5043–5053. [PubMed] [Google Scholar]
- Banerjee AK. 5'-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev. 1980;44:175–205. doi: 10.1128/mr.44.2.175-205.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G. A cap for all occasions. Structure. 1997;5:855–858. doi: 10.1016/s0969-2126(97)00239-6. [DOI] [PubMed] [Google Scholar]
- Shuman S. Structure, mechanism, and evolution of the mRNA capping apparatus. Prog Nucleic Acid Res Mol Biol. 2000;66:1–40. doi: 10.1016/s0079-6603(00)66025-7. [DOI] [PubMed] [Google Scholar]
- Nibert ML, Schiff LA, Fields BN. Reoviruses and their replication. In Fundamental Virology Edited by Fields BN, Knipe DM, Howley PM Philadelphia: Lippincott-Raven Publishers; 1996. pp. 691–730.
- Koonin EV. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- Bisaillon M, Lemay G. Computational sequence analysis of mammalian reovirus proteins. Virus Genes. 1999;18:13–37. doi: 10.1023/A:1008013117929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Luongo CL, Nibert ML, Patton JT. Rotavirus open cores catalyze 5'-capping and methylation of exogenous RNA: evidence that VP3 is a methyltransferase. Virology. 1999;265:120–130. doi: 10.1006/viro.1999.0029. [DOI] [PubMed] [Google Scholar]
- Reinisch KM, Nibert ML, Harrison SC. Structure of the reovirus core at 3.6 Å resolution. Nature. 2000;404:960–967. doi: 10.1038/35010041. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM. Comparison of protein structures reveals mono-phyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol. 1999;1:175–182. http://www.bioinfo.de/isb/1999/01/0016/ [PubMed] [Google Scholar]
- Fauman EB, Blumenthal RM, Cheng X. Structure and evolution of AdoMet-dependent MTases. In S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions Edited by Cheng X, Blumenthal RM Singapore: World Scientific Inc; 1999. pp. 1–38.
- Fang Q, Attoui H, Biagini JF, Zhu Z, de Micco P, de Lamballerie X. Sequence of genome segments 1, 2, and 3 of the grass carp reovirus (Genus aquareovirus, family reoviridae). Biochem Biophys Res Commun. 2000;274:762–766. doi: 10.1006/bbrc.2000.3215. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibrat JF, Madej T, Bryant SH. Surprising similarities in structure comparison. Curr Opin Struct Biol. 1996;6:377–385. doi: 10.1016/s0959-440x(96)80058-3. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-Pdb-Viewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem. 2000;275:16414–16419. doi: 10.1074/jbc.M001854200. [DOI] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol Cell. 2000;6:349–360. doi: 10.1016/s1097-2765(00)00035-6. [DOI] [PubMed] [Google Scholar]
- Hodel AE, Gershon PD, Quiocho FA. Structural basis for sequence-nonspecific recognition of 5'-capped mRNA by a cap-modifying enzyme. Mol Cell. 1998;1:443–447. doi: 10.1016/s1097-2765(00)80044-1. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Rychlewski L. Prediction of a novel RNA 2'-O-ribose methyltransferase subfamily encoded by the Escherichia coli YgdE open reading frame and its orthologs. Acta Microbiol Pol. 2000;49:253–260. [PubMed] [Google Scholar]
- Takusagawa F, Ogawa H, Fujioka M. Glycine N-methyltransferase, a tetrameric enzyme. In S-Adenosylmethionine-dependent Methyltransferases: Structures and Functions Edited by Cheng X, Blumenthal RM Singapore: World Scientific Inc; 1999. pp. 93–122.
- Schluckebier G, Labahn J, Granzin J, Saenger W. M.Taq I: possible catalysis via cation-pi interactions in N-specific DNA methyltransferases. Biol Chem. 1998;379:389–400. [PubMed] [Google Scholar]
- Gong W, O'Gara M, Blumenthal RM, Cheng X. Structure of Pvu II DNA-(cytosine N4) methyltransferase, an example of domain permutation and protein fold assignment. Nucleic Acids Res. 1997;25:2702–2715. doi: 10.1093/nar/25.14.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujnicki JM. Phylogenomic analysis of 16S rRNA:(guanine-N2) methyltransferases suggests new family members and reveals highly conserved motifs and a domain structure similar to other nucleic acid amino-methyltransferases. FASEB J. 2000;14:2365–2368. doi: 10.1096/fj.00-0076com. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Feder M, Radlinska M, Rychlewski L. mRNA:guanine-N7 cap methyltransferases: identification of novel members of the family, evolutionary analysis, homology modeling, and analysis of sequence-structure-function relationships. BMC Bioinformatics. 2001;2:2. doi: 10.1186/1471-2105-2-2. http://www.biomedcentral.com/1471-2105/2/2/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozanov MN, Koonin EV, Gorbalenya AE. Conservation of the putative methyltransferase domain: a hallmark of the 'Sindbis-like' supergroup of positive-strand RNA viruses. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- Bujnicki JM, Rychlewski L. Sequence analysis and structure prediction of aminoglycoside-resistance 16S rRNA:m7 G methyltransferases. Acta Microbiol Pol. 2001;50:7–17. [PubMed] [Google Scholar]


