Abstract
Precise phenotype information is needed to understand the effects of genetic and epigenetic changes on tumor behavior and responsiveness. Extraction and representation of cancer phenotypes is currently mostly performed manually making it difficult to correlate phenotypic data to genomic data. In addition, genomic data is being produced at an increasingly faster pace, exacerbating the problem. The DeepPhe software enables automated extraction of detailed phenotype information from Electronic Medical Records of cancer patients. The system implements advanced Natural Language Processing and knowledge engineering methods within a flexible modular architecture, and was evaluated using a manually-annotated dataset of the University of Pittsburgh Medical Center (UPMC) breast cancer patients. The resulting platform provides critical and missing computational methods for computational phenotyping. Working in tandem with advanced analysis of high-throughput sequencing, these approaches will further accelerate the transition to precision cancer treatment.
Keywords: Natural Language Processing, Cancer Phenotyping, Data Science
Introduction
Genomic profiling of cancers has revealed vast information about the molecular processes underlying cancer initiation, progression, metastasis, and response to treatment. However, to fully understand cancer behavior, it is important to correlate cancer genomic and phenotypic data. Cancer phenotype information includes tumor morphology (e.g. histopathologic diagnosis), laboratory results (e.g. gene amplification status), specific tumor behaviors (e.g. metastasis), and response to treatment (e.g. effect of a chemotherapeutic agent on tumor volume) among many other characteristics.
Phenotypic profiles are typically constructed from multiple sources from clinics and research laboratories, as well as from patients. However, data from these sources are collected inconsistently and asynchronously, and often do not focus on comprehensive cancer traits, the so called “deep phenotype,” which is of interest to the research community. Consequently, a major effort of many NCI designated Cancer Centers, NCI Specialized Programs of Research Excellence, and Cancer Cooperative Groups has been to extract phenotypic data from electronic medical records (EMRs).
As a manual process conducted by highly-trained human abstractors, the extraction of cancer phenotypes is time-consuming, slow, and therefore feasible only for small datasets. However, the increased use of EMRs and the establishment of data warehouses and health information exchanges have paved the way for the development of more efficient ways to extract and use clinical information for genome-phenome correlation.
The DeepPhe software addresses exactly this problem by automating the extraction of detailed phenotype information from EMRs of cancer patients at a fraction of the time of the manual abstraction, and at scale. DeepPhe generates summaries of cancer- and tumor- related characteristics (1), providing researchers with the potential to accelerate clinical investigations where phenotype information is crucial. Moreover, as we move toward personalizing cancer treatments, these phenotype profiles are not only important as a source of research data but eventually for selecting patient-specific therapies.
DeepPhe System
The DeepPhe system operates on clinical documents and discrete data to generate a summary of the patient’s clinical phenotype (1). The system uses a novel double-pipeline design, combining a conceptual model of information (a.k.a. ontology) necessary to describe cancer cases with a mention-annotation pipeline and phenotype summarization pipeline based on the Apache Unstructured Information Management Architecture (UIMA) (2).
The DeepPhe conceptual model (a.k.a. the DeepPhe Ontology) provides a terminology of entities and relations between them to specify individual cancer domains and to relate those entities to the clinical variables needed to support translational research (3). Concepts and relationships in the ontology describe neoplastic processes, prognostic features, procedures, and medications necessary to represent the diagnosis, treatment, and progression of disease. The DeepPhe ontology has been designed to explicitly encode all of the necessary domain knowledge needed for extracting information from the EMR text and for (through rules) generating higher-level summaries, thus reducing the need for encoding background knowledge in the algorithms.
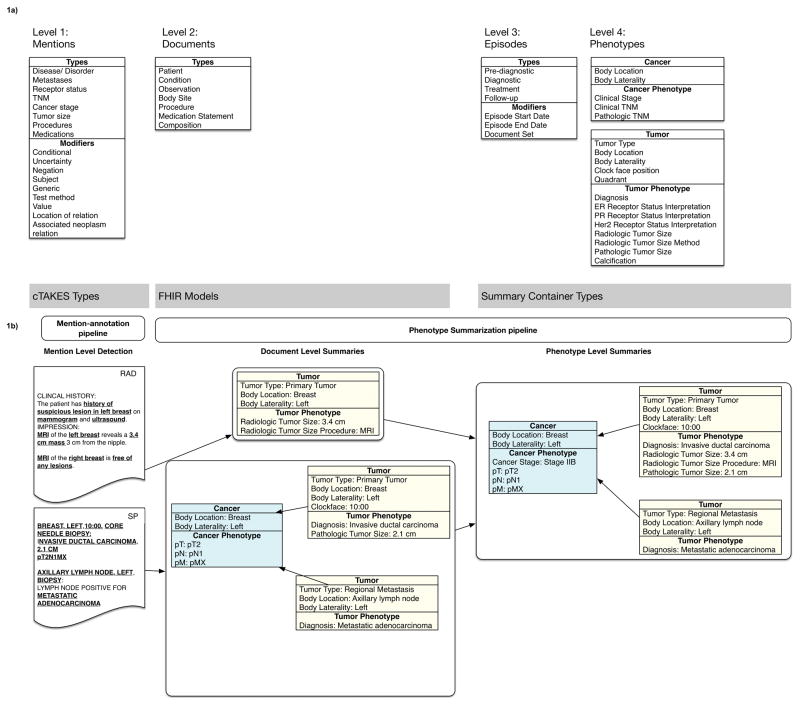
There are four levels in the DeepPhe system (Figure 1) each dependent on the preceding one. The first level contains mentions, which correspond to single words or multi-word phrases in text and form the most basic elements of the DeepPhe system. These mentions represent basic concepts such as diseases/disorders, receptors/biomarkers, TNM stage, overall stage, tumor size, procedures, and medications. Mentions are extracted by the mention-annotation pipeline, an extension of the Apache cTAKES © natural language processing system (4) (5) (6). Descriptors such as body location, test method, and associated neoplasms are also represented at this level. Figure 1b, in the column under Mention-annotation pipeline illustrates relevant mentions, bold and underlined, including the text of a radiology (RAD) and surgical pathology (SP) reports (more examples are shown in the 0:00:00–0:00:11 portion of the video). In level one, the system extracts from the radiology report (RAD) relevant mentions such as history of suspicious lesion in left breast, mammogram, ultrasound, MRI, left breast, 3.4 cm mass, MRI, right breast, free of any lesions, and from the surgical pathology report (SP) -- breast, left, 10:00, core needle biopsy, invasive ductal carcinoma, 2.1 cm, pT2N1MX, axillary lymph node, left, biopsy, metastatic adenocarcinoma.
Figure 1.
(A) Domain model and (B) Example of phenotypes produced from the DeepPhe system. Bold and underlined items in 1b show the mentions extracted by cTAKES pipeline 1 (mention detection). Abbreviations/Acronyms: cTAKES - Apache Clinical Text Analysis and Knowledge Extraction System; FHIR – Fast Healthcare Interoperability Resources; RAD – radiology note; SP – surgical pathology note
The second level summarizes the mentions from a single document into a template. The Fast Health Interoperability (FHIR) (7) models and Summary containers are standard templates for storing tumor- and cancer- characteristics. FHIR is an interoperability standard for electronic exchange of healthcare information. For example, in Figure 1b under Document Level Summaries, the mentions from the radiology report (RAD) are summarized into one tumor template where the tumor type is primary tumor, the body location is breast, the body laterality is right, the radiological tumor size is 3.4 cm, the radiological tumor size procedure is MRI; the mentions from the surgical pathology (SP) note are summarized into the cancer template where the body location is the breast, the body laterality is left, pT is pT2, pN is pN1, pM is pMX. The cancer is linked to its primary and metastatic tumor with the characteristics for tumor type, body location, body laterality, clockface, diagnosis, pathological tumor size.
The third level classifies a given document into one of four episode categories – pre-diagnostic, diagnostic, treatment and follow-up. Each episode is associated with modifiers for the episode start date, episode end date, documents set (the documents associated with the episode). These categories and modifiers are shown in the third column of Figure 1a. For example, the radiology report from Figure 1b will be classified as diagnostic (not shown in Figure 1).
The fourth level is the summarization of all the documents related to a given cancer (for examples see Figure 1b, the box under Phenotype Level Summaries and 0:00:11–0:00:21 portion of the video). In Figure 1b under Phenotype level summaries, the summaries from the radiology and surgical pathology documents are aggregated into one cancer with two tumors. The cancer description includes key characteristics (body location, body laterality, cancer stage) and is linked to corresponding tumors, each of which has its own characteristics listed (tumor type, body location, body laterality, clockface, diagnosis, radiological tumor size, radiological tumor size procedure, and pathological tumor size).
Users can view and work with the DeepPhe system output in several ways. The DeepPhe software produces output that can be stored in a commonly used platform such as the open source tranSMART platform (8) to combine genomic and phenotypic information (see 0:02:01–0:03:06 portion of the video). Output can also be stored in a Neo4J graph database (9) (see 0:00:21–0:02:00 portion of the video for a breast cancer example and 0:03:07–0:04:09 for a melanoma example).
The DeepPhe system was compared against human expert abstracted information. The agreement between the two human experts (inter-annotator agreement, IAA) ranged from 0.46 to 1.00 (1.00 indicates perfect agreement), and system agreement with humans ranged from 0.20 to 0.96. Agreement is measured in terms of the harmonic mean of sensitivity and positive predictive value which approximates kappa for this task (10). This showed that the DeepPhe system performs similarly to human experts, at a fraction of the cost of time-consuming laborious manual abstraction of cancer phenotypes.
Conclusion
We present the DeepPhe software for extracting deep phenotype information from EMRs. The software is a significant departure from other efforts in the field as it enables comprehensive longitudinal data processing from various sources. The envisioned applications are far-reaching, from translational clinical investigations to cancer surveillance and precision oncology initiatives. For example, a breast cancer investigator would be able to run the system over many patients’ EMR documents to create a large patient cohort matching certain variables. The creation of such a large cohort would take human experts considerably more time if it is generated manually, through manual abstraction by human experts, who would have to read and abstract the EMR documentation for each patient.
We examined the DeepPhe system output to identify its challenges. At the mention-level (the first level), one such challenge is presented by mentions with multiple meanings. For example, the text string mass is very frequent in the oncology domain, however, it functions with at least two meanings: (a) that of tumor, e.g. the mass was in the left breast, (b) that of body of matter, e.g. the body mass is 30. At the phenotype-level, we also identified several challenges. A well-known general issue with pipeline systems, which we also observed in our system, is that errors propagate as data is processed through multiple modules. For instance, inconsistencies in text may lead to body locations being incorrectly linked to a tumor which in turn produce errors at the phenotype level, an example being the system outputs cancer in “right nipple” instead of “right breast”. Additional errors result from challenges in summarization of multiple primaries, such as the inappropriate merging of separate cancers in the right and left breast into a single cancer. Potential solutions could include exploring using hierarchical anatomical models. For example, the parent of “right nipple” could be “right breast” by which the two locations can be conflated; however, a tumor in the left breast and one in the right breast should not be conflated into one.
Future work on the DeepPhe system includes the development of a refined visualization interface to enable intuitive end-user data interaction, application of the system to other types of cancers, including skin, lung and ovarian neoplasms, the expansion of extracted information to include clinical genomic observations, and inclusion of structured EMR data.
The DeepPhe software will be available open source at this manuscript publication time at [LINK WILL BE ADDED AT PROOF TIME]. Detailed technical information on the computational methods in the DeepPhe system will be described in a separate manuscript.
Acknowledgments
Financial support
G. Savova, NIH U24CA184407; NIH R01LM10090
E. Tsleytlin, NIH U24CA184407
S. Finan, NIH U24CA184407
M. Castine, NIH U24CA184407; Pennsylvania Department of Health PA-SAP 410070287
T. Miller, NIH U24CA184407; NIH R01LM10090
O. Medvedeva, NIH U24CA184407; Pennsylvania Department of Health PA-SAP 410070287
D. Harris, NIH U24CA184407; NIH R01LM10090
H. Hochheiser, NIH U24CA184407
C. Lin, NIH U24CA184407; NIH R01LM10090
G. Chavan, NIH U24CA184407
R. Jacobson, NIH U24CA184407; Pennsylvania Department of Health PA-SAP 410070287
The work presented here is funded by US National Institutes of Health – grants U24CA184407, R01GM103859, R01LM10090 and from the Pennsylvania Department of Health – grant PA-SAP 410070287. We thank Maria Bond for her assistance with manuscript preparation.
References
- 1.DeepPhe Information Model [Internet] Available from: https://github.com/DeepPhe/models.
- 2.UIMA [Internet] 2013 Available from: uima.apache.org.
- 3.Hochheiser H, Castine M, Harris D, Savova G, Jacobson RS. An information model for computable cancer phenotypes. BMC Med Inform Decis Mak. 2016 Sep 15;16(1):121. doi: 10.1186/s12911-016-0358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. J Am Med Inform Assoc JAMIA. 2010 Oct;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu ST, Kaggal VC, Dligach D, Masanz JJ, Chen P, Becker L, et al. A common type system for clinical natural language processing. J Biomed Semant. 2013 Jan 3;4(1):1. doi: 10.1186/2041-1480-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apache cTAKES [Internet] Available from: ctakes.apache.org.
- 7.FHIR Specification Home Page - FHIR v0.0.82 [Internet] [cited 2015 May 3]. Available from: http://hl7.org/fhir/
- 8.tranSMART Foundation [Internet] Available from: http://transmartfoundation.org.
- 9.neo4j [Internet] Available from: https://neo4j.com/
- 10.Hripcsak G, Rothschild AS. Agreement, the F-Measure, and Reliability in Information Retrieval. J Am Med Inform Assoc JAMIA. 2005;12(3):296–8. doi: 10.1197/jamia.M1733. [DOI] [PMC free article] [PubMed] [Google Scholar]