Abstract
Although the heritability of ADHD is estimated to be high, identifying specific genetic markers remains challenging. Most studies to date have examined the genetic basis of ADHD by employing dichotomous diagnostic phenotypes, but, as ADHD symptoms tend to be phenotypically dimensional, an alternative and potentially informative approach is to examine continuous indices of inattention and hyperactivity-impulsivity symptoms. The current study aimed to identify genetic effects on dimensionally-focused adult ADHD-related phenotypes in 990 individuals of European ancestry with intentionally low levels of substance misuse to avoid confounding. The study used four complementary approaches: 1) analysis of a priori candidate loci identified in prior meta-analytic work; 2) gene-based analysis; 3) hypothesis-free genome-wide association testing; and 4) single nucleotide polymorphism (SNP) heritability via genomic-relatedness-matrix restricted maximum likelihood analysis (GREML). The GREML analysis included a bivariate model to test whether the ADHD symptom dimensions index the same genetic liability. The results revealed significant differential associations between two a priori loci and ADHD phenotypes, rs6296 in HTR1B with inattention and rs3746544 in SNAP-25 with hyperactivity-impulsivity. No significant gene-based or genome-wide associations were detected, but SNP heritability revealed that a large portion of genetic variance was accounted for by common SNPs (44%, 55%, and 59% for inattention, hyperactivity-impulsivity, and total ADHD, respectively) and substantial shared genetic variance across inattention and hyperactivity-impulsivity (86%). These findings reveal both unique and common patterns of genetic influences across dimensional ADHD-related phenotypes. More broadly, these findings reveal the value in using multiple methods to understand the genetic etiology of ADHD.
Keywords: ADHD, Inattention, Hyperactivity, Impulsivity, Genetics, Heritability
INTRODUCTION
Attention deficit hyperactivity disorder (ADHD) is characterized by clinically significant and developmentally inappropriate levels of inattention, hyperactivity, and impulsivity (APA, 2013). ADHD is a heterogeneous disorder with a complex, multifactorial etiology (Faraone and Doyle, 2001; Willcutt and Carlson, 2005). Twin studies indicate that ADHD is highly heritable (70–80%) (Faraone et al, 2005; Freitag et al, 2010), yet the majority of the genetic variance in ADHD is unexplained (Franke et al, 2009; Kebir et al, 2009). Reviews and meta-analyses have highlighted 8–10 candidate genes implicated in dopaminergic, serotoninergic and glutamatergic signaling along with synaptic vesicle, neurite outgrowth and cell adhesion pathways (Gizer et al, 2009; Li et al, 2014). While several genome-wide association (GWA) analyses have used either case-control or family-based designs (Franke et al, 2009; Zayats et al, 2015), to date no single nucleotide polymorphisms (SNPs) have reached the conventional stringent genome-wide significance threshold (p<5 x 10−8; Pe’er et al, 2008).
Refining the ADHD phenotype: Categorical versus continuous symptom measures
One explanation for mixed results is that the categorical ADHD diagnosis may not be the most useful phenotype for genetic analyses of ADHD. Given that there is strong evidence to suggest that ADHD exists on the extreme end of a continuum of behavior (Levy et al, 1997), examining genetic associations with continuous measures of underlying symptom dimensions may result in more consistent genetic findings (e.g. Plomin et al, 2008). Specifically, it is also important to distinguish between the inattentive and hyperactive-impulsive symptom dimensions. Both exploratory and confirmatory factor analyses consistently support the distinction between inattention and hyperactivity-impulsivity (e.g. DuPaul et al, 1998), and both symptom dimensions are associated with multiple aspects of global, academic, and social impairment (e.g. Lahey et al, 1994). The discriminant validity of the symptom dimensions has been repeatedly demonstrated with analyses showing that inattention symptoms are more strongly associated with internalizing symptoms, academic difficulties, and neurocognitive weaknesses, whereas hyperactive-impulsive symptoms are more strongly predictive of comorbid externalizing behaviors, peer rejection, and accidental physical injuries (Lahey and Willcutt, 2002). Given the importance of dimensional symptom information, some candidate genetic studies of ADHD have begun to focus on continuous symptom phenotypes (e.g. Lasky-Su et al, 2008; Park et al, 2004; Waldman et al, 2006), but, to our knowledge, no genome-wide studies have applied dimensional phenotypes.
Focus on adult ADHD as a methodological strategy
Another problem with finding clear genetic signals may be that the genetic basis of the disorder may vary with the age of the patient cohort. Family studies in clinical samples suggest that there may be higher familial liability for adult ADHD compared with childhood ADHD (Biederman et al, 1990; Biederman et al, 1995), supporting the notion that ADHD symptoms that persist into adulthood may have stronger genetic liability. Thus, examining the genetic basis of adult ADHD dimensions may be particularly useful and offer unique etiological information. Interestingly, a recent meta-analysis specific to adult ADHD (Bonvicini et al, 2016) identified a significant role for the BAIAP2 (brain-specific angiogenesis inhibitor 1-associated protein 2) gene in adult ADHD, which has a role in neurodevelopment, and has not been identified in larger meta-analyses of childhood ADHD (Gizer et al, 2009). However, studies, particularly those of self-reported ADHD in adulthood, generally report slightly lower heritability estimates ranging from 40–66% (Boomsma et al, 2010; van den Berg et al, 2006). It possible that these lower, although still substantial, heritability estimates are due to the fact that cross-sectional self-report of adult ADHD symptoms may also reflect highly comorbid conditions such as drug use, leading to increased measurement error of the genetic liability for ADHD and lower estimates of heritability. Thus, estimating genetic influences on adult ADHD in samples that are carefully screened for substance use disorders and other comorbidities may clarify these discrepant findings. A further complication is that there is increasing evidence that adulthood ADHD is not necessarily a continuation of childhood ADHD. For example, in a recent cohort study, less than 20% of the individuals diagnosed during childhood still were classified as having ADDHD in young adulthood and vice versa (Caye et al., 2016). As a result, distinct genetic influences may be present for the developmentally distinct or continuous forms of the condition.
Estimating SNP heritability for dimensional ADHD measures
In addition to candidate gene and genome-wide association approaches, novel tools that leverage genome-wide data are increasingly available, but have been applied to the genetics of ADHD on a very limited basis. For example, Yang et al. (2013) developed genomic-related-matrix restricted maximum likelihood (GREML) analysis, implemented via Genome-wide Complex Trait Analysis (GCTA) software. This technique estimates the phenotypic variance explained by genome-wide similarity at all genotyped SNPs. Rather than testing each SNP individually, GREML decomposes the phenotypic variance into two components: (1) effects due to the additive influences of all measured autosomal SNPs (SNP heritability or h2SNP) and (2) the effects due to unmeasured environmental influences, random noise or the effects of genetic variants that were not measured by the genotyping array. This approach allows for an estimate of phenotypic variability that can be explained by genome-wide SNP data.
To date, GREML has been used in two studies on ADHD, both in developmentally mixed samples of children and adults, to estimate the additive genetic influences on categorical ADHD diagnosis. These studies both report an aggregate SNP heritability of approximately 28%, suggesting that common SNPs account for a relatively large portion of genetic effects detected using traditional biometrical twin modeling approaches (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Zayats et al, 2015). Given the multidimensional, developmental nature of ADHD, a critical step is to examine the aggregate role of common SNPs on continuous, dimensional measures of adult ADHD.
Current study
The current study sought to address a number of these methodological considerations, using a number of different strategies to investigate genetic influences on continuous measures of inattention, hyperactive-impulsive, and total ADHD symptoms in a community sample of 990 adults of European ancestry. We hypothesized that the dimensional approach to ADHD would improve power to detect genetic effects and that partially distinct patterns of genetic association would emerge across the inattention and hyperactivity-impulsivity dimensions. We also hypothesized that common genetic variants would account for a substantial portion of the genetic variance. Further, despite phenotypic heterogeneity, we hypothesized that a common set of genetic factors would account for the genetic variances identified across the symptom dimensions. Importantly, the participants were carefully screened for substance misuse and other psychological conditions to minimize the extent to which ADHD phenotypes could be attributable to either the effects of substance use or comorbid psychopathology. To test these hypotheses, we employed four complementary genetic approaches: 1) individual association tests with a priori candidate loci from prior meta-analyses (Bonvicini et al, 2016; Gizer et al, 2009), 2) gene-based analyses examining associations between the phenotypes and all the markers within a gene; 3) hypothesis-free genome-wide associations, and 4) GREML implemented in GCTA, including a bivariate model to test whether the ADHD symptom dimensions have overlapping genetic liability.
METHOD
Procedure
Participants were recruited from the general community at two sites (Athens, GA and Chicago, IL). Inclusion criteria were English fluency, between 18 and 30 years of age, and self-reported Caucasian race and non-Hispanic ethnicity in order to control for population stratification (Hutchison et al, 2004). In order to control for common ADHD comorbidities, exclusion criteria were ≥12 on the Alcohol Use Disorders Identification Test (AUDIT; Saunders et al, 1993) or Drug Use Disorders Identification Test (DUDIT; Berman et al, 2005) and current or last-12-months treatment for: depression, bipolar disorder, general anxiety, social anxiety, post-traumatic stress disorder, obsessive compulsive disorder, panic attacks/disorder, phobia, psychotic disorders, anorexia, bulimia, or binge eating. In addition to self-report substance use, upon arrival to the experimental session, participants were asked to provide a urine sample for drug screening and a breath sample to test for blood alcohol content (BAC) level. Participants with a positive drug screen or BAC>0.00 were ineligible to proceed. Following screening, participants completed the self-report measures utilized for this study.
Measures
Demographics
Comprehensive demographics were assessed including, sex, age, race, income, education and other descriptive variables.
World Health Organization (WHO) Attention-Deficit/Hyperactivity Disorder Adults Self-Report Scale (ASRS)
The ASRS is an 18-item self-report screening scale for ADHD (Kessler et al, 2005) with strong internal consistency and inter-rater agreement with clinician administered ratings (Adler et al, 2006). The ASRS includes two nine item subscales that assess the frequency (scored 0–4, ranging from never to very often) of inattention (e.g., “How often do you make careless mistakes when you have to work on a boring or difficult project?”) and hyperactivity-impulsivity (e.g., How often do you fidget or squirm with your hands or your feet when you have to sit down for a long time?”). The analyses in this study examined summary scores on the two subscales and total ADHD score (comprised of all 18-items).
SNP Genotyping and Quality Control
Genotyping was performed using the Illumina PsychArray BeadChip platform, which calls ~600,000 markers and has optimized tag SNP content from the International HapMap Project to capture the maximum amount of common variation. Only SNP variants were included in the study. All quality control filtering was implemented in PLINK v1.9 (Chang et al, 2015). SNPs were filtered for call rates < 98%, Hardy-Weinberg Equilibrium (HWE) violations of p < 1 x 10−6, MAF < 5%, and invariance. Imputation of missing genotypes and of new SNPs was performed with IMPUTE2 v.2.3.1 (Howie et al, 2009) using the 1000 Genomes Phase 3 b37 reference panel (1000 Genomes Consortium, 2015). Imputed SNPs were excluded for exhibiting an information score of < .3, MAF < 5%, HWE violations of p < 1 x 10−6, missingness > 5%, and multiallelic status. Imputed SNPs with confidence < .9 were set to missing for individuals. Of the variants that have been significantly implicated in previous meta-analyses (Bonvicini et al, 2016; Gizer et al, 2009), four were both present in our dataset and satisfied these quality control criteria; of note, only SNP, not other forms of variation, were available in the current study. In sum, following quality control, four a priori loci (See Table 2), 286,027 genotyped SNPs, and 4,881,535 total genome-wide SNPs were present for analysis.
Table 2.
Associations between a priori loci and adult self-report of ADHD inattention, hyperactivity-impulsivity, and total scales.
| Chr | Locus | Gene | Missing | Minor Allele | MAF | Inattention Subscale | Hyperactivity-impulsivity Subscale | Total Score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B | SE | p | B | SE | p | B | SE | p | ||||||
| 5 | rs27072 | DAT1 | 0 | T | .169 | −.377 | .287 | .189 | −.198 | .292 | .497 | −.583 | .494 | .239 |
| 6 | rs6296 | HTR1B | 5 | G | .251 | .662 | .243 | .007 | .473 | .249 | .058 | 1.124 | .422 | .008 |
| 17 | rs8079781 | BAIAP2 | 4 | T | .238 | −.238 | .250 | .342 | −.460 | .255 | .071 | −.701 | .431 | .104 |
| 20 | rs3746544 | SNAP-25 | 13 | G | .345 | .139 | .228 | .542 | .475 | .231 | .039 | .634 | .391 | .105 |
Note. Significance was defined as p<.05 for a priori tests, indicated in boldface; trend level effects (p < .10) are indicated by underlining.
Participant Quality Control
1,000 participants had valid genotyping data (call rates ≥ 98%, inbreeding coefficient absolute value ≤ .02, concordant self-reported sex and X-chromosome determined sex) and satisfied inclusion/exclusion criteria. To correct any misclassification of self-reported race, principal components analysis (PCA; Price et al, 2006) was conducted. Two population outliers were identified and removed by visual inspection of the PC plot (see Supplementary Materials). Six participants were excluded for missing ASRS. Finally, participants were assessed for cryptic relatedness using GCTA software (Yang et al, 2011), and two were removed for relatedness > .05, leaving a final sample of 990 participants.
Data Analysis
Internal reliability coefficients and correlations among the ADHD scales were calculated using SPSS 20.0 (IBM Corp., 2011). Genome-wide Efficient Mixed Model Association (GEMMA) software (Zhou and Stephens, 2012) was utilized to conduct univariate linear mixed model associations with the four a priori loci and the inattention, hyperactivity-impulsivity, and total scores. Although inattention and hyperactivity-impulsivity symptom dimensions were our primary phenotypes of interest, in order to be comprehensive and generalizable with prior work, we also included a continuous measure of total ADHD symptoms. Given the clear empirical basis for the a priori tests, a nominal p value (p < .05) was used. Gene-based analyses used Versatile Gene-based Association Study 2 (VEGAS2) software (Mishra and Macgregor, 2015), employing the top 10% SNP test for optimal sensitivity and specificity of true positives (Wojcik et al., 2015). Given the recent report of an erratum in the original VEGAS2 method for generating empirical p-values leading to increased type I error rate, the publically available updated script was used (Hecker et al., 2017). For the gene-based tests, a Benjamini-Hochberg FDR correction was applied to the resultant p-values from the analyses (Benjamini and Hochberg, 1995). For genome-wide analysis, hypothesis-free tests of 4,881,535 genome-wide SNPs were conducted with the same phenotypes. These analyses accounted for the cryptic relatedness among individuals by modeling out as a random effect (i.e., the genetic correlation between individuals). To maximize resolution of effects, an additive genetic effect model was used whereby participants were given a 0–2 score for each SNP indicating the number of minor alleles. A significance threshold of p < 5x10−8 was applied to the GWA tests (Pe’er et al., 2008). GREML analyses were implemented using GCTA software (Lee et al, 2012; Yang et al, 2013; Yang et al, 2011) and used utilized 286,027 autosomal markers. Genomic relatedness matrices (GRM) were derived and used to explain all three phenotypes (inattention, hyperactivity-impulsivity and ADHD Total). Analyses were limited to individuals who were no more related than second cousins (i.e., GRM-cutoff of 0.05 was imposed) so that estimates are not confounded with shared environmental effects and/or causal variants that are not tagged by the SNPs but captured by pedigree information. Additive genetic effects (h2SNP) on the ADHD phenotypes are un-scaled, and will be most relevant to a population with the same distribution of liability. A bivariate-GREML model was implemented for inattention and hyperactivity-impulsivity to test whether the symptom dimensions index the same genetic liability.
RESULTS
Preliminary Analyses
Participant characteristics are in Table 1. All three phenotypes were normally distributed (Total: skewness = .17 [SE = .08], kurtosis = .55 [SE = .16]; inattention: skewness = .19 [SE = .08], kurtosis = .57 [SE = .16]; hyperactivity-impulsivity: skewness = .27 [SE = .08], kurtosis = .21 [SE = .16]). Participants reported moderate symptom endorsement, with the mean total score reflecting 38% of ASRS maximum (Inattention = 41.5%, Hyperactivity/impulsivity = 35.4%).
Table 1.
Participant characteristics (N = 990).
| Variable | % / Mean (SD) / Median |
|---|---|
| Age | 21.67 (3.31) |
| Sex | 61.7% Female |
| Income | $60,000 – $89,9991 |
| Years of education | 14.55 (2.21)2 |
| ADHD total | 27.71 (8.22) |
| Inattention | 14.95 (4.76) |
| Hyperactivity/impulsivity | 12.76 (4.86) |
Note.
N = 988;
N = 989.
Hyperactivity/impulsivityPhenotypic internal reliability was good: inattention, α = .79; hyperactivity-impulsivity, α = .76; total, α = .83. Sex and age were considered as covariates by examining their association with total score, but neither was significantly correlated. Inattention and hyperactivity-impulsivity subscales were significantly correlated (r = .46, p = 3.44x10−53), sharing ~20% of variance.
Analysis of A Priori Loci
Results for the a priori loci associations with ASRS inattention, hyperactivity-impulsivity, and total are in Table 2. Of the four a priori loci assessed, one locus (rs6296) was significantly associated with inattention and one locus (rs3746544) was significantly associated with hyperactivity-impulsivity. Analyses with ASRS total yielded a significant relationship for rs6296, while no other significant relationships were identified. However, there were two trend level findings with hyperactivity-impulsivity and rs6296 in HTR1B (p = .058) and rs8079781 in BAIAP2 (p = .071). The G alleles of rs6296 and rs3746544 and the C allele of rs8079781 were associated with higher ADHD scores.
Gene-based Analyses
Of the 18,937 genes explored in the gene-based associations, none met the significance criterion after FDR correction. The strongest gene-based associations for each of the scales were in KDEL endoplasmic reticulum protein retention receptor 3 (KDELR3; p = 6.00x10−6; inattention), Glycerate Kinase (GLYCTK; p = 1.05 x10−4; hyperactivity-impulsivity), and ribosomal L24 domain containing 1 (RSL24D1; p = 7.00x10−5; ADHD Total).
To complement the a priori tests, the 4 corresponding genes were also specifically examined. Using a nominal significance criterion, HTR1B was significantly associated with inattention (p = .007) and total (p = .009), and the most associated SNP was in fact the a priori candidate (rs6296). SNAP-25 was significantly associated with hyperactivity-impulsivity (p = .034) and total score (p = .022). To inform future studies, the top 10 genes for each subscale and total score are included in Supplementary Materials; 10–30% overlap was present between the subscales and the total score.
Genome-wide Association Analyses
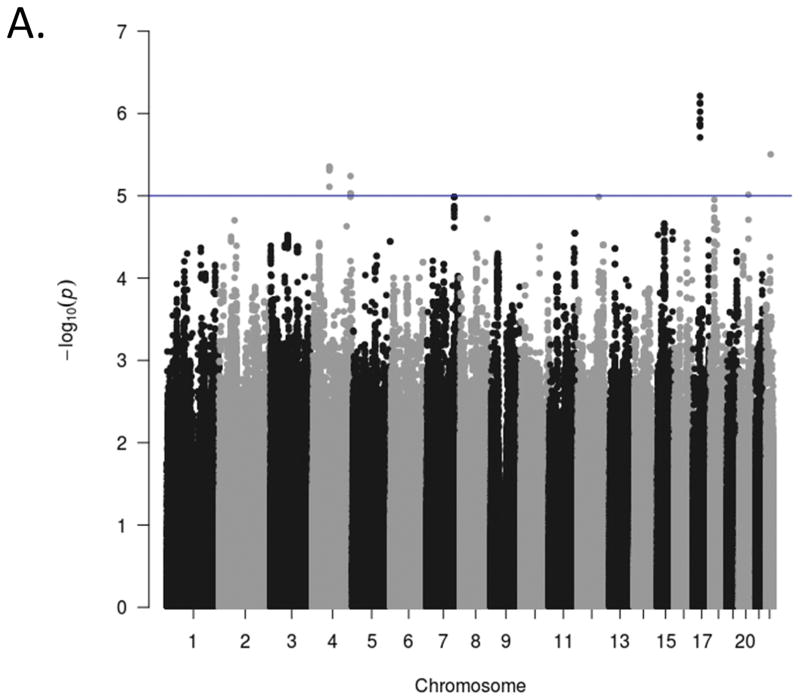
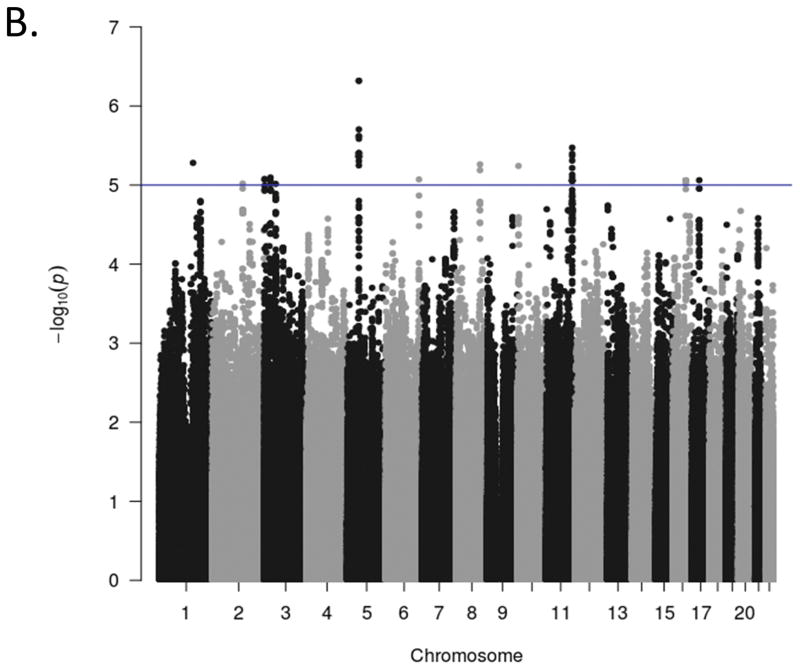
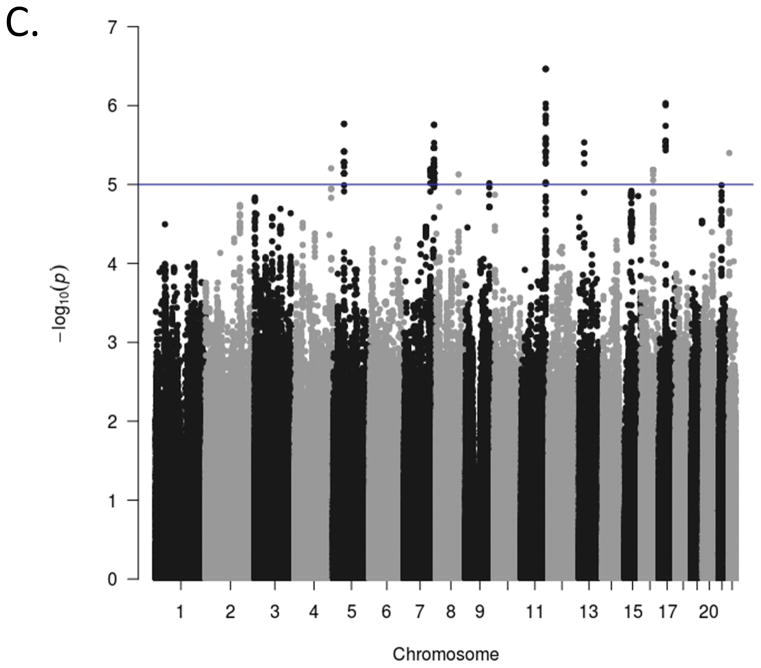
Manhattan plots for the GWA analyses of the three ADHD scales are presented in Figure 1. The genome-wide scan did not yield any significant associations for any scale. The strongest association for each of the scales were in intergenic regions at chromosome 17, position 327457845 (p = 6.10x10−7 for rs17577327; inattention), chromosome 5, position 53944834 (p = 4.80 x10−4 for rs1900166; hyperactivity-impulsivity), and chromosome 11, position 121255184 (p = 3.43 x10−7 for rs99672; ADHD Total). Suggestive associations (p<10−5) and quantile-quantile (Q-Q) plots for each phenotype are included in Supplementary Materials. The Q-Q plots suggest the majority of markers fit null expectations.
Figure 1.
Genome-wide association Manhattan plots. The Manhattan plots display level of significance for each SNP, organized by chromosomal position from chromosomes 1–22. Panel A presents ADHD inattention, Panel B presents ADHD hyperactivity-impulsivity, and Panel C presents ADHD total score The blue line indicates suggestive significance (p<10−5). No SNPs achieved genome-wide significance (p<5x10−8).
Figure 1(b). Genome-wide association Manhattan plots. The Manhattan plots display level of significance for each SNP, organized by chromosomal position from chromosomes 1–22. Panel A presents ADHD inattention, Panel B presents ADHD hyperactivity-impulsivity, and Panel C presents ADHD total score The blue line indicates suggestive significance (p<10−5). No SNPs achieved genome-wide significance (p<5x10−8).
Figure 1(c). Genome-wide association Manhattan plots. The Manhattan plots display level of significance for each SNP, organized by chromosomal position from chromosomes 1–22. Panel A presents ADHD inattention, Panel B presents ADHD hyperactivity-impulsivity, and Panel C presents ADHD total score The blue line indicates suggestive significance (p<10−5). No SNPs achieved genome-wide significance (p<5x10−8).
Genome-wide SNP Heritability
Using GREML to estimate variance in ADHD scales attributable to genotyped SNPs, the additive genetic effects were significant for hyperactivity-impulsivity (53.83% SNP heritability, SE = .262, p = .004) and ADHD Total (59.02% SNP heritability, SE = .279, p = .005), and at a trend level for inattention (43.65% SNP heritability, SE = .301, p = .066). The genetic overlap between inattention and hyperactivity-impulsivity using bivariate-REML indicated that they were highly genetically correlated (rg-SNP = .861, SE = .376, p = .03), suggesting substantial overlapping additive genetic influences that significantly differed from zero on these two domains of ADHD. The results of all GREML analyses are included in Table 3.
Table 3.
Proportion of variance explained by genome-wide SNPs across adult self-report of ADHD symptoms based on the Adult Self-report Scale (ASRS).
| Phenotype | h2SNP | SE | p |
|---|---|---|---|
| ADHD Total | .590 | .279 | .005 |
| Inattention | .436 | .301 | .066 |
| Hyperactivity-impulsivity | .538 | .262 | .004 |
DISCUSSION
Our study aimed to clarify genetic etiology of ADHD by using multiple genomic approaches to test for associations between a priori candidate loci, genes, and genome-wide SNPs on continuous dimensional measures of ADHD in a healthy adult population that had been screened for concurrent substance use and other common comorbidities. Similar to previous studies, gene-based and genome-wide analyses did not reveal genes or SNPs that met the significance criterion (Franke et al, 2009; Zayats et al, 2015). However, results revealed significant differential associations between a priori genetic loci and the quantitative dimensional measures of ADHD, suggesting that specific loci may contribute more strongly to either inattention or hyperactive-impulsive symptoms. These findings were somewhat corroborated in gene-based tests that revealed similar associations between the genes in the a priori tests in relation to the ADHD phenotypes, albeit using a nominal significance criterion. In addition, GREML methods suggested substantial SNP heritability for each measure of adult ADHD (44–59%) and bivariate models indicated that genetic influences significantly overlapped across inattentive and hyperactive-impulsive symptom dimensions (86% overlap). Thus, although the genetic influences on inattention and hyperactivity-impulsivity overlap significantly, a notable proportion of the common SNP heritability of each dimension is unique (~14%), supporting a dimensional approach to genetic studies of ADHD.
When considering the a priori candidate SNPs, the results revealed significant associations between two of the four loci and dimensions of ADHD: rs6296 in HTR1B was significantly associated with inattention and total ADHD, while rs3746544 in SNAP-25 was significantly associated with hyperactivity-impulsivity. An effect of these two loci on ADHD across studies has been consistently supported via meta-analytic work (Gizer et al, 2009) and our findings suggest possible differential effects for these SNPs when ADHD is parsed dimensionally. The serotonin 1b receptor gene (HTRIB) has been associated with increased attention to novel stimuli (Malleret et al, 1999) and failure to show expected responses to amphetamine administration (Brunner and Hen, 1997). Further, variation in the SNP examined here, rs6296, has recently been associated with differential neural responses and task performance during the Stop Signal Task, a widely-used task tapping the underlying attentional and response inhibition deficits strongly linked with ADHD (van Rooij et al, 2015).
The SNAP-25 gene is increasingly gaining attention due to its link with multiple psychiatric disorders, most notably ADHD, schizophrenia, and bipolar disorder (for a review see Antonucci et al.). SNAP-25 codes for a protein impacting axonal growth, synaptic plasticity, and presynaptic neurons necessary for the regulation of neurotransmitter release (Sollner et al, 1993) and is putatively linked to network hyperexcitability through defective control of voltage gated calcium channels. Perhaps most relevant, studies have found that mice lacking one copy of the SNAP-25 gene display hyperactive behavior (Hess et al, 1992) and that a transgene (expressing SNAP-25) that was bred into mice with SNAP-25 depletion reduced hyperactivity (Hess et al. 1995). Consistent with this work suggesting potential differential influences for these genes on specific components of the broader ADHD phenotype, our results reveal somewhat distinct patterns of genetic association with ADHD symptom dimensions. Together, these data support the need for continued studies that test the notion that these variants may differentially contribute to separable phenotypic components of ADHD with a dimensional approach.
In contrast to these significant findings, associations with two of four candidate loci were not supported. It is possible that these previously identified loci exert smaller effects than previously identified, do not have parallel similar effects in our sample because of an unconsidered suppressor variable, or are simply false positives in the literature. Of note, rs8079781 (BAIAP2) exhibited a trend-level relationship with hyperactivity-impulsivity (consistent with direction of effects in the Adult ADHD meta-analysis Bovicini et al., 2016) and would be expected to reach significance in a modestly larger sample. Notably, BAIAP2 codes a protein connected to postsynaptic density at excitatory synapses and regulation of dendritic spines, and it is implicated in multiple psychiatric disorders, most notably ADHD, autism spectrum, and schizophrenia (for a review see Kang et al., 2016). Despite these considerations, however, the most appropriate conclusion from these results is that these loci are not associated with either ADHD dimension or total score.
No significant gene-based or genome-wide associations were observed. The strongest gene-based associations were not in biological systems of obvious relevance to ADHD and, likewise, the genome-wide signals all fell into intergenic regions. While some intergenic DNA controls genes nearby, the function of most intergenic sites is typically unknown. Thus, the relevance of potential signals in these regions to ADHD pathophysiology is not clear. Further examination will be necessary to determine whether these intergenic loci are truly relevant or simply false positive findings. The absence of significant genome-wide findings is likely because our study is of modest size and thus has low power to detect an association of a small effect. This obliquely provides further evidence that genetic influences on ADHD are unlikely to be from a small number of influential loci, but from very large numbers of loci exerting small effects.
Also consistent with this conclusion are the SNP heritability results. The SNP heritability estimates suggested that a substantial amount of genetic variance in ADHD symptoms is accounted for by common SNPs (h2 SNP-inattention = 44%, h2 SNP-Hyperactivity-impulsivity = 54%, and h2 SNP-Total = 59%). Thus, although the effects of individual SNPs appear to be quite small, considering the aggregate effects of SNPs genome-wide provides compelling evidence that common SNPs account for a large portion of the genetic variance in ADHD measures, over half to two-thirds of what has been suggested by twin studies (typically between 70–80%). In addition, results suggest that aggregate effects of common SNPs may be highest for hyperactivity-impulsivity and total ADHD scores, suggesting that ADHD symptoms, particularly hyperactivity-impulsivity symptoms that persist into adulthood are strongly genetically influenced. Results of our bivariate GREML model suggesting an rG- SNP of 86% provides the first evidence that the common variants influencing inattention and hyperactivity-impulsivity significantly overlap, pointing to substantial shared genetic variance across inattention and hyperactivity-impulsivity. This finding is consistent with prior twin work supporting shared genetic variance across these phenotypes (Willcutt et al, 2012). These GREML results, coupled with our candidate SNP association findings, suggest that although influences largely overlap across the two phenotypes, unique pathways may be implicated across inattention and hyperactivity-impulsivity dimensions. This potential etiological differentiation is underscored by studies strongly supporting the discriminant validity of these dimensions on behavioral, neurocognitive, and clinical metrics (Lahey et al, 2002).
Although our study is the first to use GREML to parse the genetic variance of dimensionally-defined ADHD, our findings are consistent with prior work suggesting that a substantial amount of ADHD genetic variance is accounted for by common SNPs (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Zayats et al, 2015). Notably, SNP heritability estimates reported here are even higher than those reported in the two prior studies using this method. These studies, both in mixed samples of children and adults, reported aggregate SNP heritabilities of 28% for ADHD as a categorical diagnostic measure. In contrast, our substantially higher point estimates for ADHD symptom dimensions (inattention and hyperactivity-impulsivity) as well as total ADHD symptoms suggest that testing effects on continuous and dimensional phenotypes and using more developmentally homogenous samples may be useful in parsing the phenotypic and age-related heterogeneity of ADHD. Notably, our estimates also differ from some prior twin and family work suggesting lower heritabilities in adult samples relying on self-reported symptoms (Boomsma et al, 2010; van den Berg et al, 2006), likely due to our careful screening for common adult ADHD phenocopies, such as heavy substance use. Although it is necessarily conjecture, it may be the case that these methodological aspects of the study reduced measurement error, narrowing the ADHD phenotype and making it more tractable for genetic dissection.
Methodological considerations
Findings should be interpreted in the context of the following methodological considerations. Our modest sample size left us underpowered to detect medium-to-small magnitude genetic effects, particularly using genome-wide association techniques. Further, our dimensional measures of ADHD are based on self-report in adulthood and not a diagnostic interview by a clinician. While this approach to characterizing adult ADHD symptoms has been shown to generate data with adequate reliability and validity (Adler et al, 2006), these genetic effects should also be examined in concert with clinical ratings of ADHD dimensions. Our use of a community sample suggests that our association findings and estimates of SNP heritability are broadly generalizable to the population, but may differ in a sample selected for ADHD (Lee et al, 2012). Furthermore, given evidence of discontinuity between childhood and adulthood ADHD (Caye et al., 2016), it is an open question whether these findings would generalize to a younger sample. Importantly, considering our modest statistical power for detecting SNP heritability and large errors standard errors, our SNP heritability estimates should be interpreted cautiously. Further, the estimates across ADHD phenotypes do not differ significantly from each other. However, the observed point estimates for genetic effects across studies of the same phenotype tend to be highly stable across sample sizes, with standard errors shrinking as sample sizes increase (Stanton-Geddes et al, 2013). Importantly, in regards to rs3746544 of SNAP-25, our findings indicated that the minor (G) allele of rs3746544 was associated with increased risk for hyperactive-impulsive symptoms, whereas meta-analyses (Gizer et al, 2009) report an association with the major (T) allele. This raises the possibility that rs3746544 is a proxy for the true causal variant and thus these conflicting patterns of association could be related to population differences with respect to LD in this region. However, it could also be interpreted as a false positive result. Thus, future studies with larger samples are needed to replicate these findings and to confirm the observed effects on dimensionally-focused adult ADHD. It should also be noted that the current findings are limited to common genetic variants, ignoring any contribution from less common and rare variants, which likely play a role (Poelmans et al, 2011; Stergiakouli et al, 2012), and were restricted to SNP variation. Future whole-genome studies incorporating rare polymorphisms and alternative forms of variation are needed to address these limitations.
Summary
This study suggests that continuous dimensional phenotypes may help in the search for meaningful genetic signals for adult ADHD, particularly in the context of its polygenic and multifactorial nature. Here, multiple approaches to genetic analyses revealed both that particular loci appear to exert stronger effects to the etiology of one or the other dimension, but also that the genetic effects are substantially shared across inattention and hyperactivity-impulsivity. More broadly, these findings reveal the value in using multiple methods to refine the understanding of the genetic etiology of ADHD.
Supplementary Material
Acknowledgments
Funding: This work was partially funded by NIH grant R01 DA032015 (de Wit). Dr. MacKillop’s contributions were partially supported by the Peter Boris Chair in Addictions Research.
The authors are grateful to Kyle M. Hernandez at the Center for Research Informatics, University of Chicago, and to the Research Assistants at the University of Chicago and University of Georgia who contributed to the project.
Footnotes
Disclosures: The authors have no conflicts of interest to declare.
References
- Adler LA, Spencer T, Faraone SV, Kessler RC, Howes MJ, Biederman J, et al. Validity of pilot Adult ADHD Self- Report Scale (ASRS) to Rate Adult ADHD symptoms. Annals of clinical psychiatry : official journal of the American Academy of Clinical Psychiatrists. 2006;18(3):145–148. doi: 10.1080/10401230600801077. [DOI] [PubMed] [Google Scholar]
- Antonucci F, Corradini I, Fossati G, Tomasoni R, Menna E, Matteoli M. SNAP-25, a Known Presynaptic Protein with Emerging Postsynaptic Functions. Frontiers in synaptic neuroscience. 2016;8 doi: 10.3389/fnsyn.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 5. American Psychiatric Association Press; Washington, DC: 2013. [Google Scholar]
- Berman AH, Bergman H, Palmstierna T, Schlyter F. Evaluation of the Drug Use Disorders Identification Test (DUDIT) in criminal justice and detoxification settings and in a Swedish population sample. European addiction research. 2005;11(1):22–31. doi: 10.1159/000081413. [DOI] [PubMed] [Google Scholar]
- Biederman J, Faraone SV, Keenan K, Knee D, Tsuang MT. Family-genetic and psychosocial risk factors in DSM-III attention deficit disorder. Journal of the American Academy of Child and Adolescent Psychiatry. 1990;29(4):526–533. doi: 10.1097/00004583-199007000-00004. [DOI] [PubMed] [Google Scholar]
- Biederman J, Faraone SV, Mick E, Spencer T, Wilens T, Kiely K, et al. High risk for attention deficit hyperactivity disorder among children of parents with childhood onset of the disorder: a pilot study. Am J Psychiatry. 1995;152(3):431–435. doi: 10.1176/ajp.152.3.431. [DOI] [PubMed] [Google Scholar]
- Bonvicini C, Faraone SV, Scassellati C. Attention-deficit hyperactivity disorder in adults: A systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol Psychiatry. 2016;21(7):872–884. doi: 10.1038/mp.2016.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boomsma DI, Saviouk V, Hottenga JJ, Distel MA, de Moor MH, Vink JM, et al. Genetic epidemiology of attention deficit hyperactivity disorder (ADHD index) in adults. PLoS One. 2010;5(5):e10621. doi: 10.1371/journal.pone.0010621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner D, Hen R. Insights into the neurobiology of impulsive behavior from serotonin receptor knockout mice. Annals of the New York Academy of Sciences. 1997;836:81–105. doi: 10.1111/j.1749-6632.1997.tb52356.x. [DOI] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradini I, Donzelli A, Antonucci F, Welzl H, Loos M, Martucci R, … Caleo M. Epileptiform activity and cognitive deficits in SNAP-25+/− mice are normalized by antiepileptic drugs. Cerebral Cortex. 2014;24(2):364–376. doi: 10.1093/cercor/bhs316. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 2013;45(9):984–994. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPaul GJ, Power TJ, Anastopoulos AD, Reid R. ADHD Rating Scale-IV: Checklists, Norms, and Clinical Interpretation. Guilford Press; New York: 1998. [Google Scholar]
- Faraone SV, Doyle AE. The nature and heritability of attention-deficit/hyperactivity disorder. Child and Adolescent Psychiatric Clinics of North America. 2001;10(2):299–316. [PubMed] [Google Scholar]
- Faraone SV, Perlis RH, Doyle AE, Smoller JW, Goralnick JJ, Holmgren MA, et al. Molecular Genetics of Attention-Deficit/Hyperactivity Disorder. Biological Psychiatry. 2005;57(11):1313–1323. doi: 10.1016/j.biopsych.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Franke B, Neale BM, Faraone SV. Genome-wide association studies in ADHD. Human Genetics. 2009;126(1):13–50. doi: 10.1007/s00439-009-0663-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag CM, Rohde LA, Lempp T, Romanos M. Phenotypic and measurement influences on heritability estimates in childhood ADHD. European Child & Adolescent Psychiatry. 2010;19(3):311–323. doi: 10.1007/s00787-010-0097-5. [DOI] [PubMed] [Google Scholar]
- Gizer IR, Ficks C, Waldman ID. Candidate gene studies of ADHD: a meta-analytic review. Human Genetics. 2009;126(1):51–90. doi: 10.1007/s00439-009-0694-x. [DOI] [PubMed] [Google Scholar]
- Hess EJ, Jinnah HA, Kozak CA, Wilson MC. Spontaneous locomotor hyperactivity in a mouse mutant with a deletion including the Snap gene on chromosome 2. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1992;12(7):2865–2874. doi: 10.1523/JNEUROSCI.12-07-02865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess EJ, Rogan PK, Domoto M, Tinker DE, Ladda RL, Ramer JC. Absence of linkage of apparently single gene mediated ADHD with the human syntenic region of the mouse mutant Coloboma. American journal of medical genetics. 1995;60(6):573–579. doi: 10.1002/ajmg.1320600619. [DOI] [PubMed] [Google Scholar]
- Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS genetics. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison KE, Stallings M, McGeary J, Bryan A. Population stratification in the candidate gene study: fatal threat or red herring? Psychological Bulletin. 2004;130(1):66–79. doi: 10.1037/0033-2909.130.1.66. [DOI] [PubMed] [Google Scholar]
- Kang J, Park H, Kim E. IRSp53/BAIAP2 in dendritic spine development, NMDA receptor regulation, and psychiatric disorders. Neuropharmacology. 2016;100:27–39. doi: 10.1016/j.neuropharm.2015.06.019. [DOI] [PubMed] [Google Scholar]
- Kebir O, Tabbane K, Sengupta S, Joober R. Candidate genes and neuropsychological phenotypes in children with ADHD: review of association studies. Journal of Psychiatry & Neuroscience. 2009;34(2):88. [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Adler L, Ames M, Demler O, Faraone S, Hiripi E, et al. The World Health Organization Adult ADHD Self-Report Scale (ASRS): a short screening scale for use in the general population. Psychological medicine. 2005;35(2):245–256. doi: 10.1017/s0033291704002892. [DOI] [PubMed] [Google Scholar]
- Lahey BB, Applegate B, McBurnett K, Biederman J, Greenhill L, Hynd GW, et al. DMS-IV field trials for attention deficit hyperactivity disorder in children and adolescents. The American Journal of Psychiatry. 1994;151(11):1673–1685. doi: 10.1176/ajp.151.11.1673. [DOI] [PubMed] [Google Scholar]
- Lahey BB, Willcutt EG. Validity of the diagnosis and dimensions of attention deficit hyperactivity disorder. In: Jensen PS, Cooper JR, editors. Attention deficit hyperactivity disorder: state of the science. Civic Research Institute; Kingston, NJ: 2002. [Google Scholar]
- Lasky-Su J, Lange C, Biederman J, Tsuang M, Doyle AE, Smoller JW, et al. Family-based association analysis of a statistically derived quantitative traits for ADHD reveal an association in DRD4 with inattentive symptoms in ADHD individuals. American Journal of Medical Genetics Part B: Neuropsychiatric Genetics. 2008;147B(1):100–106. doi: 10.1002/ajmg.b.30567. [DOI] [PubMed] [Google Scholar]
- Lee SH, Yang J, Goddard ME, Visscher PM, Wray NR. Estimation of pleiotropy between complex diseases using single-nucleotide polymorphism-derived genomic relationships and restricted maximum likelihood. Bioinformatics. 2012;28(19):2540–2542. doi: 10.1093/bioinformatics/bts474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy F, Hay DA, McStephen M, Wood C, Waldman I. Attention-Deficit Hyperactivity Disorder: A Category or a Continuum? Genetic Analysis of a Large-Scale Twin Study. Journal of the American Academy of Child & Adolescent Psychiatry. 1997;36(6):737–744. doi: 10.1097/00004583-199706000-00009. [DOI] [PubMed] [Google Scholar]
- Li Z, Chang SH, Zhang LY, Gao L, Wang J. Molecular genetic studies of ADHD and its candidate genes: a review. Psychiatry research. 2014;219(1):10–24. doi: 10.1016/j.psychres.2014.05.005. [DOI] [PubMed] [Google Scholar]
- Malleret G, Hen R, Guillou JL, Segu L, Buhot MC. 5-HT1B receptor knock-out mice exhibit increased exploratory activity and enhanced spatial memory performance in the Morris water maze. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1999;19(14):6157–6168. doi: 10.1523/JNEUROSCI.19-14-06157.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park L, Nigg JT, Waldman ID, Nummy KA, Huang-Pollock C, Rappley M, et al. Association and linkage of α-2A adrenergic receptor gene polymorphisms with childhood ADHD. Mol Psychiatry. 2004;10(6):572–580. doi: 10.1038/sj.mp.4001605. [DOI] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genetic epidemiology. 2008;32(4):381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Plomin R, DeFries JC, McClearn GE, McGuffin P. Behavioral Genetics. 5. Worth Publishers; New York: 2008. [Google Scholar]
- Poelmans G, Pauls DL, Buitelaar JK, Franke B. Integrated Genome-Wide Association Study Findings: Identification of a Neurodevelopmental Network for Attention Deficit Hyperactivity Disorder. American Journal of Psychiatry. 2011;168(4):365–377. doi: 10.1176/appi.ajp.2010.10070948. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M. Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption--II. Addiction. 1993;88(6):791–804. doi: 10.1111/j.1360-0443.1993.tb02093.x. [DOI] [PubMed] [Google Scholar]
- Sollner T, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, et al. SNAP receptors implicated in vesicle targeting and fusion. Nature. 1993;362(6418):318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- Stanton-Geddes J, Yoder JB, Brikine R, Young ND, Tiffin P. Estimating heritability using genomic data. Methods in Ecology and Evolution. 2013;4(12):1151–1158. [Google Scholar]
- Stergiakouli E, Hamshere M, Holmans P, Langley K, Zaharieva I, et al. Genetics d. Investigating the Contribution of Common Genetic Variants to the Risk and Pathogenesis of ADHD. American Journal of Psychiatry. 2012;169(2):186–194. doi: 10.1176/appi.ajp.2011.11040551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg SM, Willemsen G, de Geus EJ, Boomsma DI. Genetic etiology of stability of attention problems in young adulthood. American journal of medical genetics Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2006;141B(1):55–60. doi: 10.1002/ajmg.b.30251. [DOI] [PubMed] [Google Scholar]
- van Rooij D, Hartman CA, van Donkelaar MM, Bralten J, von Rhein D, Hakobjan M, et al. Variation in serotonin neurotransmission genes affects neural activation during response inhibition in adolescents and young adults with ADHD and healthy controls. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry. 2015;16(8):625–634. doi: 10.3109/15622975.2015.1067371. [DOI] [PubMed] [Google Scholar]
- Waldman ID, Nigg JT, Gizer IR, Park L, Rappley MD, Friderici K. The adrenergic receptor α-2A gene (ADRA2A) and neuropsychological executive functions as putative endophenotypes for childhood ADHD. Cognitive, Affective, & Behavioral Neuroscience. 2006;6(1):18–30. doi: 10.3758/cabn.6.1.18. [DOI] [PubMed] [Google Scholar]
- Willcutt EG, Carlson CL. The diagnostic validity of attention-deficit/hyperactivity disorder. Clinical Neuroscience Research. 2005;5(5–6):219–232. [Google Scholar]
- Willcutt EG, Nigg JT, Pennington BF, Solanto MV, Rohde LA, Tannock R, et al. Validity of DSM-IV attention deficit/hyperactivity disorder symptom dimensions and subtypes. Journal of abnormal psychology. 2012;121(4):991–1010. doi: 10.1037/a0027347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee S, Goddard M, Visscher P. Genome-Wide Complex Trait Analysis (GCTA): Methods, Data Analyses, and Interpretations. In: Gondro C, van der Werf J, Hayes B, editors. Genome-Wide Association Studies and Genomic Prediction. Vol. 1019. Humana Press; 2013. pp. 215–236. [DOI] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. American journal of human genetics. 2011;88(1):76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayats T, Athanasiu L, Sonderby I, Djurovic S, Westlye LT, Tamnes CK, et al. Genome-Wide Analysis of Attention Deficit Hyperactivity Disorder in Norway. PLoS ONE. 2015;10(4):e0122501. doi: 10.1371/journal.pone.0122501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44(7):821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.