Abstract
Background:
Axillary lymph node metastasis is associated with increased risk of regional recurrence, distant metastasis, and poor survival in breast malignant neoplasm. Expression of signal transducer and activator of transcription 3 (STAT3) is significantly associated with tumor formation, migration, and invasion in various cancers. In addition, vascular endothelial growth factor (VEGF) expression could promote angiogenesis and increase the risk of tumorigenesis. To determine correlations among STAT3 expression, VEGF, and clinicopathological data on lymph node involvement in breast cancer patients after surgery.
Methods:
The mRNA expression levels of STAT3 and VEGFs were measured in 45 breast invasive ductal carcinoma tissues, 45 peritumoral tissues, and 45 adjacent nontumor tissues by real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR). Postoperative pathological examination revealed explicit axillary lymph node involvement in all patients.
Results:
Average mRNA levels of STAT3 and VEGFs were the highest in breast invasive ductal carcinoma tissues, followed by peritumoral tissues. High expression of STAT3 showed significant positive correlation with high axillary lymph node involvement and progesterone receptor (PR), VEGF-C, VEGF-D, and vascular endothelial growth factor receptor (VEGFR)-3 expression. The expression levels of STAT3, VEGF-C, and VEGFR-3 were significantly higher in the tumor tissues of patients with axillary lymph node metastasis than in those of patients without the metastasis. Expression levels of VEGF-C and VEGFR-3 were also significantly higher in peritumoral tissues of patients with axillary lymph node metastasis. Positive correlations were found between STAT3 and VEGF-C/-D mRNA levels.
Conclusion:
These data suggest that STAT3/VEGF-C/VEGFR-3 signaling pathway plays an important role in carcinogenesis and lymph-angiogenesis. Our findings suggest that STAT3 may be a potential molecular biomarker for predicting the involvement of axillary lymph nodes in breast cancer, and therapies targeting STAT3 may be important for preventing breast cancer metastasis.
Keywords: lymph-angiogenesis, metastasis, signal transducer and activator of transcription 3, tumor, vascular endothelial growth factor
1. Introduction
Breast carcinoma is one of the leading causes of cancer mortality among women worldwide. Axillary lymph node metastasis is associated with increased risk of regional recurrence, distant metastasis, and poor survival in breast malignant neoplasm. Despite rapid advancements in diagnosis and therapy for breast cancer, the average 5-year relative survival is strongly associated with lymph node metastasis.[1]
Development of breast cancer involves several key events. During this process, tumor cells disseminate from the primary tumor into the surrounding stromal tissue, penetrate across lymphatic walls, implant on ipsilateral axillary lymph nodes, and finally extravasate and proliferate in the parenchyma of target organs.[2] Some prognostic biomarkers are preferentially altered in breast cancer, such as vascular endothelial growth factor (VEGF)-C and VEGF-D. Overexpression of VEGF-C and VEGF-D have been reported to induce lymph-angiogenesis and to contribute to both lymphatic vessel invasion and lymph node metastasis via activation of vascular endothelial growth factor receptor (VEGFR)-3.[3–5] These molecular abnormalities are linked to carcinogenesis and poor prognosis.[6,7]
Signal transducer and activator of transcription (STAT) is a 7-member family of transcription factors that transmits signals to the nucleus, in which STAT binds to specific DNA promoter sequences to regulate gene expression. Transient expression of STATs controls numerous physiological processes, including proliferation, differentiation, survival, development, and inflammation.[8–11] Moreover, constitutive expression of signal transducer and activator of transcription 3 (STAT3) is significantly associated with tumor formation, migration, and invasion in various cancers, such as papillary thyroid carcinoma, pancreatic cancer, head and neck cancer, and gastric cancer.[12–15] However, the mechanisms underlying positive correlation between expression of STAT3 in breast cancer patients and lymph node metastasis remain unclear.
In this study, we explored the correlation between lymph node metastasis and STAT3 expression by real-time quantitative reverse transcription-polymerase chain reaction (RT-qPCR), a rapid, reliable, and accurate technique that can sensitively and specifically quantify mRNA.
2. Materials and methods
2.1. Clinical samples
A total of 45 human breast cancer tissues (n = 45), matched adjacent paraneoplastic tissues (n = 45), and normal tissues (n = 45) from patients diagnosed with primary breast cancer (invasive ductal carcinoma) were obtained during surgery at the Department of Breast Surgery, West China Hospital of Sichuan University between August 2016 and August 2017. All patients signed informed consent for participation in this study. The diagnosis was based on pathological evidence (Fig. 1), and the specimens were immediately snap-frozen and stored at −80 °C. None of the patients had received chemotherapy or radiotherapy before the surgical excision. All patients provided written informed consent for the use of their tissues. The study was approved by the Research Ethics Committee of West China Hospital of Sichuan University (2012-251), China.
Figure 1.
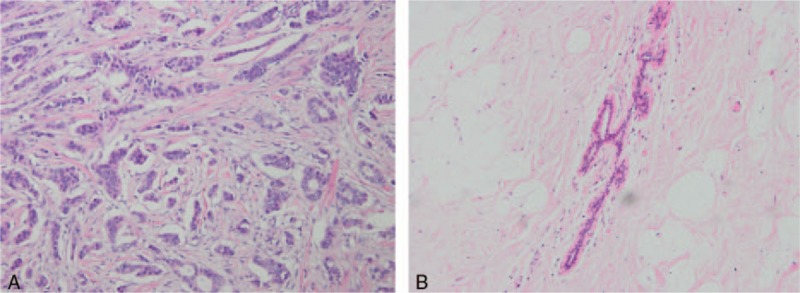
Hematoxylin and eosin (H&E) staining of cancer and paraneoplastic tissues, pathologic diagnosis was made independently by 2 senior pathologists according to the 2003 World Health Organization histological classification of breast tumors. (A) Breast cancer tissues (∗200); (B) matched adjacent paraneoplastic tissues (∗200).
2.2. RT-qPCR and quantification of expression levels of STAT3 and VEGFs
Total RNA was isolated from the cancer, paraneoplastic, and normal tissues using the RNeasy Mini Kit (QIAGEN, America) and then reverse transcribed using the iScript cDNA Synthesis Kit (Bio-Rad, America) in accordance with the manufacturers’ recommendations. The isolated RNAs were stored at −80 °C, and the reverse transcribed cDNAs were stored at −20 °C in Molecular Cancer Laboratory (West China Hospital of Sichuan University, Chengdu, China) until use. Primers of STAT3, VEGF-C, VEGF-D, VEGFR-2, VEGFR-3, and β-actin were designed using Primer-BLAST on NCBI website. Sequences of all primers and Gene Bank IDs are listed in Table 1. β-Actin was used as endogenous control. Quantitative polymerase chain reaction (qPCR) was performed using SsoFast EvaGreen Supermix (Bio-Rad) on Chromo4 of the Bio-Rad System. Relative fold expression was calculated by the 2−ΔΔCT method using the following formulae[16]: ΔCT(sample) = CTtarget gene − CTreference gene
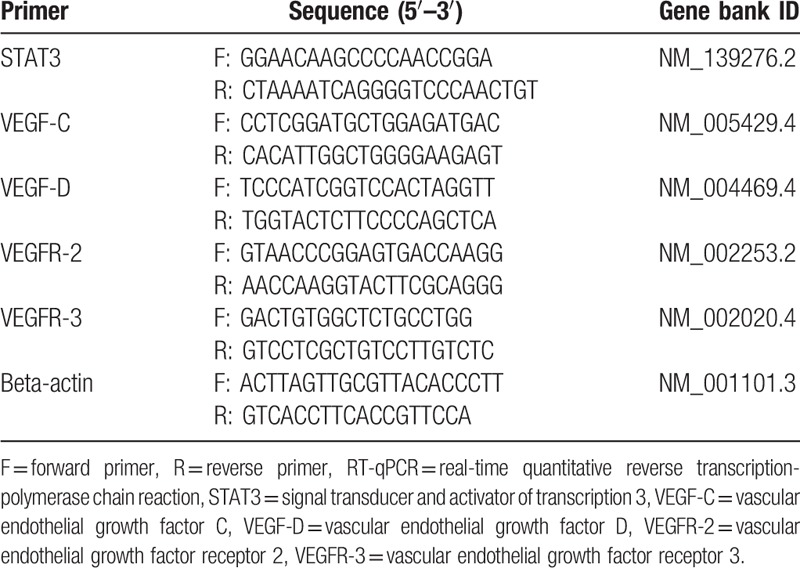
Table 1.
Sequences of RT-qPCR primers.
ΔCT(calibrator) = CTtarget gene − CTreference gene
ΔΔCT = ΔCT(sample) − ΔCT(calibrator)
Normalized target gene expression level in sample = 2−ΔΔCT
2.3. Statistical analysis
Quantitative data are expressed descriptively as median and interquartile range while qualitative data are expressed as percentage (%). Receiver operator characteristic (ROC) and cut-off values were applied to assess the overall diagnostic performance. Comparison between the ranks of 2 independent groups was done using Wilcoxon rank sum test.[17] Statistical analysis of data involving categorical variables was done using Chi-square test (X2 test). Subsequently, the relationship between STAT3 and VEGF expression was explored by Spearman correlation. Tests of statistical significance were done as 2-sided tests where applicable with P < .05 considered statistically significant. All statistical analyses were performed using SPSS version 20.0 (SPSS Inc., Chicago, IL).
3. Results
3.1. Patient characteristics and proportions of samples with high mRNA levels
The relationship between clinicopathological features and target gene expression levels of 45 breast cancer patients after curative resection is summarized here. The age, body mass index,[18] T stage, axillary lymph node status, tumour, node and metastasis (TNM) classification, WHO classification of primary tumor, and levels of estrogen receptor, progesterone receptor (PR), Ki-67, and human epidermal growth factor receptor 2 were recorded for each of the 45 patients. In addition, STAT3, VEGF-C, VEGF-D, VEGFR-2, and VEGFR-3 mRNA levels were examined. The median patient age was 49.8 years (age range, 34–71 years). Out of all 45 female breast cancer patients 25 had lymph node metastasis.
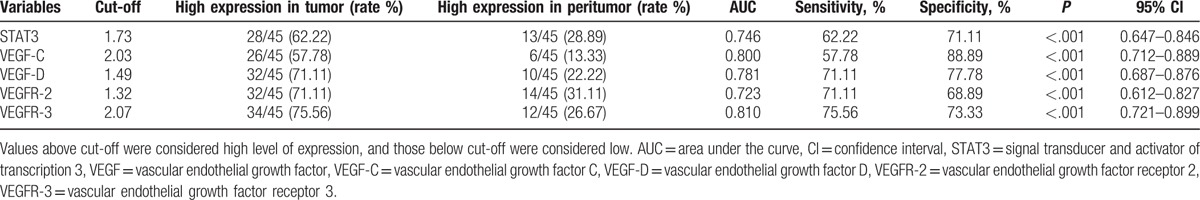
All target genes were detected in the 45 pairs of breast tumor tissues and their matched peritumoral tissues by RT-qPCR. Ratios of STAT3 and VEGF mRNA levels were calculated by the relative expression value of each breast tumor tissues and their matched peritumoral tissues divided by relative expression value of each adjacent normal tissue. Using the cut-off value of 1.73, high STAT3 expression was detected in 62.22% (28/45) of BC samples and 28.89% (13/45) of peritumoral samples. The proportion of genes high expression level of VEGF-C, VEGF-D, VEGFR-2, and VEGFR-3 in tumor tissues were as follows: 57.78% (26/45, cut-off: 2.03), 71.11% (32/45, cut-off: 1.49), 71.11% (32/45, cut-off: 1.32), 75.56% (34/45, cut-off: 2.07), and in matched peritumoral tissues were 13.33% (6/45), 22.22% (10/45), 31.11% (14/45), and 26.67% (12/45), respectively (Table 2).
Table 2.
High expression levels of STAT3 and VEGFs among breast tumor tissues and peritumoral tissues.
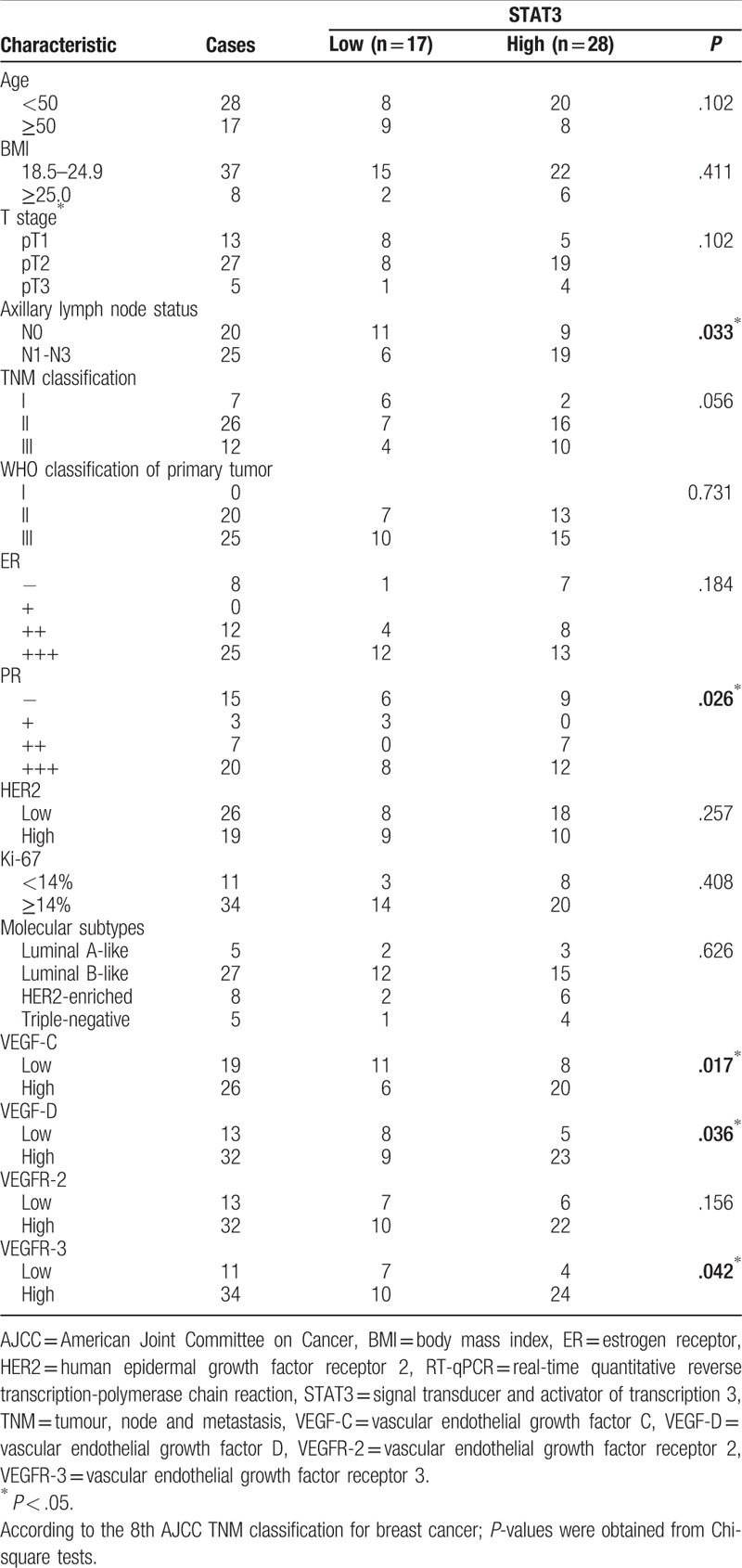
In addition, the high expression of STAT3 increased with axillary lymph node metastasis, PR expression, VEGF-C expression, VEGF-D expression, and VEGFR-3 expression (P < .05), but not with age, body mass index, T stage, TNM classification, WHO classification, estrogen receptor expression, human epidermal growth factor receptor expression, Ki-67 expression, molecular subtypes, and VEGFR-2 expression (P > .05) (Table 3).
Table 3.
Relationship between STAT3 mRNA expression and clinical outcomes.
3.2. STAT3 and VEGF gene expression among tumor and peritumoral tissues
The quantities (2−ΔΔCT) of STAT3 were 2.04-fold higher in cancer tissues and 1.28-fold higher in para-carcinoma tissues (P < .001) relative to those in normal tissues. Similarly, the relative quantities (2−ΔΔCT) of VEGF-C, VEGF-D, VEGFR-2, and VEGFR-3 were as follows: 2.57-fold versus 1.33-fold, 1.75-fold versus 0.89-fold, 1.66-fold versus 1.09-fold, and 2.56-fold versus 1.52-fold in cancer tissues and para-carcinoma tissues, respectively (P < .001) (Table 4 and Fig. 2). The mRNA levels of STAT3, VEGF-C, VEGF-D, VEGFR-2, and VEGFR-3 were significantly higher in carcinoma tissues than in adjacent peritumoral tissues. The quantification of STAT3 and VEGF gene expression in both breast tumor and peritumoral tissues decreased gradually.
Table 4.
Descriptive and comparative statistics of the studied parameters between STAT3 and VEGF mRNA levels in cancer and paraneoplastic specimens.
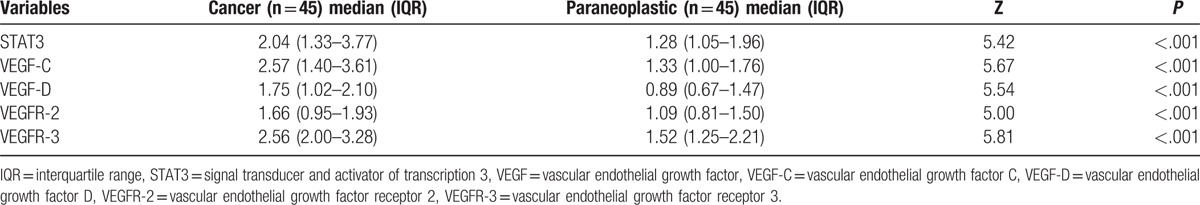
Figure 2.
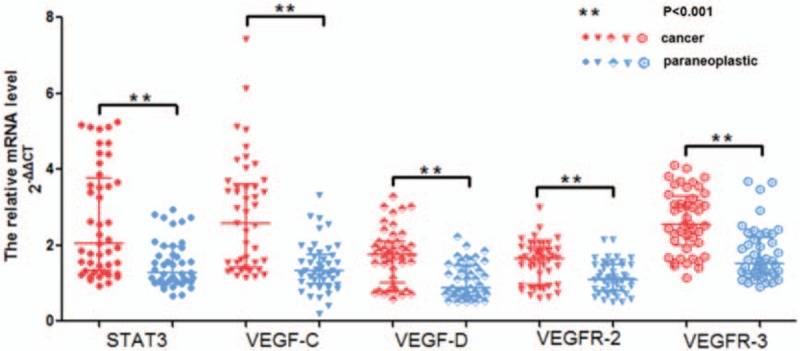
Relative mRNA levels of STAT3 and VEGF in cancer and paraneoplastic specimens, significantly higher mRNA levels of STAT3 and VEGF were observed in breast cancer (red) than in paraneoplastic (blue) tissues adjacent to tumors (P < .001). Quantitative data are presented as median and IQR (transverse line). mRNA levels of STAT3 and VEGF were normalized to those of beta-actin. IQR = interquartile range, STAT3 = signal transducer and activator of transcription 3, VEGF = vascular endothelial growth factor C.
3.3. Correlation between lymph node metastasis and target gene expression
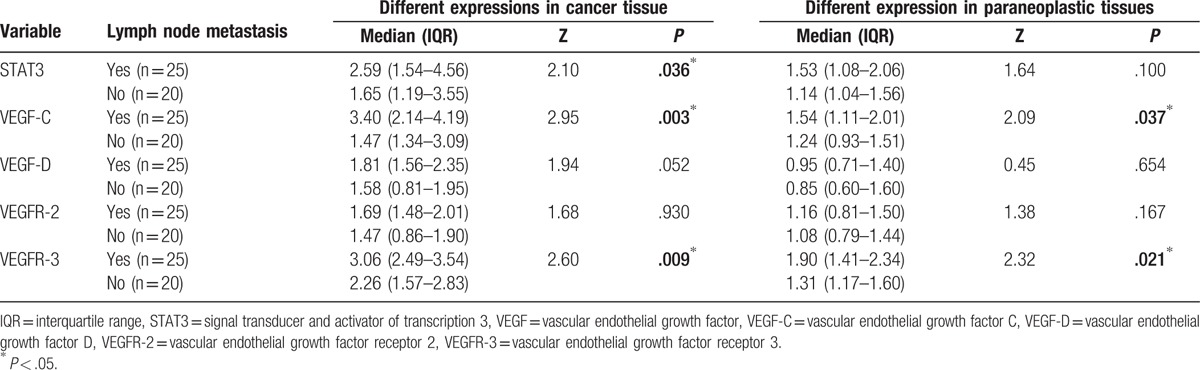
High expression was detected in most specimens with lymph node metastasis. Therefore, we performed subgroup analysis and classified breast cancer patients as lymph node nonmetastasis and lymph node metastasis. Of the 25 patients with lymph node metastasis, 10 had 1 to 3 nodes metastasis (pN1), 10 had 4 to 9 nodes metastasis (pN2), and 5 had 10 or more nodes with metastasis (pN3). In addition, we compared the proportions of patients with high expression of our genes of interest between lymph node metastasis and lymph node nonmetastasis cases. The results are as follows: STAT3 (64.29% [18/28] vs 35.71% [10/28]), VEGF-C (73.08% [19/26] vs 26.92% [7/26]), VEGF-D (65.63% [21/32] vs 34.37% [11/32]), VEGFR-2 (62.50% [20/32] vs 37.50% [12/32]), VEGFR-3 (67.65% [23/34] vs 32.35% [11/34]) in cancer specimens; STAT3 (69.23% [9/13] vs 30.77% [4/13]), VEGF-C (100% [5/5] vs 0% [0/5]), VEGF-D (40.00% [4/10] vs 60.00% [6/10]), VEGFR-2 (64.29% [9/14] vs 35.71% [5/14]), VEGFR-3 (75.00% [9/12] vs 25.00% [3/12]) in paraneoplastic specimens. These results indicate that the proportion of genes with high expression level in the metastasis group were significantly higher than that in the nonmetastasis group.
Moreover, expression levels (2−ΔΔCT) (median interquartile range) of STAT3 and VEGFs in each group were also summed. We found that STAT3 expression in cancer tissues (2.59 [1.54–4.56] vs 1.65 [1.19–3.55], Z = 2.10, P < .05), VEGF-C expression in cancer tissues (3.40 [2.14–4.19] vs 1.47 [1.34–3.09], Z = 2.95, P < .01) and paraneoplastic tissues (1.54 [1.11–2.01] vs 1.24 [0.93–1.51], Z = 2.09, P < .05), and VEGFR-3 expression in cancer tissues (3.06 [2.49–3.54] vs 2.26 [1.57–2.83], Z = 2.60, P < .01) and paraneoplastic tissues (1.90 [1.41–2.34] vs 1.31 [1.17–1.60], Z = 2.32, P < .005) in the metastasis group were higher than those in the nonmetastasis group. However, no significant difference in VEGF-D and VEGFR-2 expression was found between the cancer and paraneoplastic tissues (P > .05) (Table 5). These results suggest that the signaling pathway that involves STAT3, VEGF-C, and VEGFR-3 may play a role in lymph-angiogenesis and lymph node metastasis.
Table 5.
Correlation of STAT3 and VEGF expression with axillary lymph node metastasis of breast cancer patients.
3.4. Correlation between STAT3 and VEGF expression
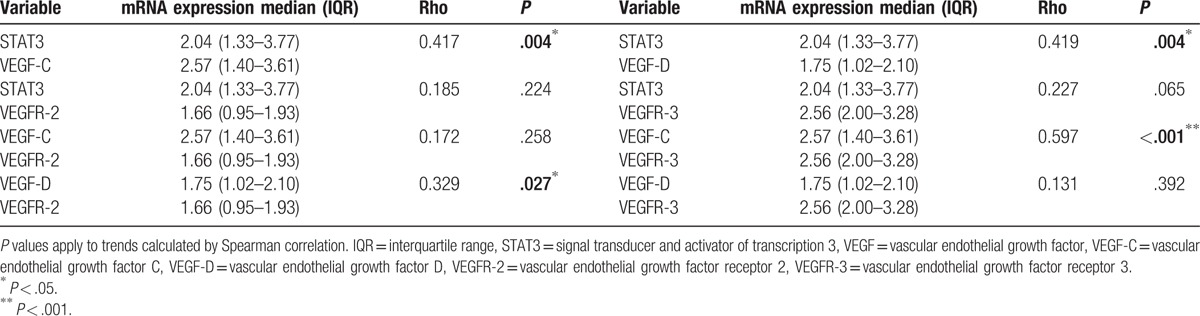
We further investigated the correlation between expression levels of STAT3 and VEGFs. A positive correlation was observed between STAT3 and VEGF-C (Spearman correlation, rho = 0.417, P < .005) and between STAT3 and VEGF-D (rho = 0.419, P < .005). However, no correlation was observed between STAT3 and VEGFR-2 or VEGFR-3 (P > .05). These results suggest that the signaling pathway that involves STAT3 and VEGF-C or VEGF-D may play a role in breast cancer carcinogenesis.
Subsequently, we investigated the association of combined expression of VEGF-C and VEGFR-2 (VEGF-C/VEGFR-2) or VEGFR-3 (VEGF-C/VEGFR-3), and VEGF-D and VEGFR-2 (VEGF-D/VEGFR-2) or VEGFR-3 (VEGF-D/VEGFR-3). Expression of VEGF-C showed positive correlation with VEGFR-3 (rho = 0.597, P < .001), while expression of VEGF-D showed positive correlation with VEGFR-2 (rho = 0.329, P < .05) (Table 6). These data indicate that the signaling pathway involving STAT3, VEGF-C, and VEGFR-3 may play a role in carcinogenesis and lymph-angiogenesis, while the signaling pathway involving STAT3, VEGF-D, and VEGFR-2 may play a role in carcinogenesis.
Table 6.
Correlation between STAT3 and VEGF mRNA levels in cancer specimens.
4. Discussion
Lymph node metastasis is the most significant predictor of breast cancer prognosis. Although various molecules play important roles in breast cancer progression, a molecular marker with high accuracy and specificity that can reliably identify lymph node metastasis of breast tumor remains lacking to date. Lymph-angiogenesis is a process that underlies cancer cell dissemination from the primary tumor site to ipsilateral axillary lymph nodes and extravasation, leading eventually to metastasis in breast cancer patients. STAT3 is a vital cytoplasmic transcription factor that forms functional dimers when phosphorylated (phospho-STAT3) and activated by aberrant upstream tyrosine kinases. Homodimers of phospho-STAT3 translocate to the nucleus and control the expression of genes essential for cancer cell growth and survival.[8] STAT3 has been closely linked to various types of cancer, such as pancreatic cancer, stomach cancer, head and neck cancer, and thyroid carcinoma, and contributes to cancer progression by upregulating oncogenes.[12–15] Some studies have suggested that constitutively active STAT3 binds to one of binding sites of VEGF, which upregulates VEGF expression, promotes angiogenesis, and increases the risk of tumorigenesis.[19] Varney and Singh[7] observed that the VEGF-C-VEGFR-3/Flt4 (VEGFR-3) autocrine signaling pathway regulates the survival and proliferation of breast tumor cells. Interestingly, preventing the interaction of VEGF-C with VEGFR-3 in vivo decreased tumor and metastatic burden. Bower et al[4] found that VEGF-C and VEGF-D cooperatively control lymph-angiogenesis throughout the zebrafish embryo and that during primary angiogenesis, VEGF-D influences these phenotypes through VEGFR-2 rather than Flt4 (VEGFR-3).[20]
In a previous study, we demonstrated that protein STAT3 expression is upregulated in human breast cancer in proliferative areas corresponding to high levels of VEGF-C and VEGF-D by immunohistochemistry.[21] Thus, we hypothesized that STAT3 promotes cancer lymphatic metastasis, which causes bad prognosis of breast cancer patients. In the present study, we analyzed the association of STAT3 with target genes and explored whether STAT3 promotes lymph-angiogenesis and lymphatic metastasis.
Specifically, we quantified the expression levels of STAT3 and VEGFs using RT-qPCR in 45 pairs of breast tumor specimens and their matched peritumoral specimens. We analyzed the association of STAT3 expression with clinicopathological features in these specimens. Results suggested that overexpression and constitutive activity of STAT3 were significantly associated with axillary lymph node status and overexpression of PR, VEGF-C, VEGF-D, and VEGFR-3 (P < .05). PR can induce transcriptional activation of STAT3 and breast cancer growth in vitro and in vivo.[22] To determine the relationship between STAT3 and VEGF-C, or VEGF-D, or VEGFR-3, we quantified the expression levels of these genes and examined their correlations. We found that the relative quantities (2−ΔΔCT) of STAT3, VEGF-C, VEGF-D, VEGFR-2, and VEGFR-3 were markedly higher in primary breast tumor tissues compared to the control group (P < .05). Interestingly, mRNA levels of STAT3, VEGF-C, and VEGFR-3 were significantly higher in breast carcinoma with lymph node metastasis than in those without lymph node metastasis (P < .05). In particular, results of Spearman correlation analysis suggested that STAT3 expression was positively correlated with VEGF-C and VEGF-D. Similarly, a positive correlation was observed between VEGF-C and VEGFR-3 as well as between VEGF-D and VEGFR-2. Based on these results, we propose 2 possible molecular mechanisms downstream of this event: on the one hand, STAT3 overexpression could upregulate the expression of VEGF-D/VEGFR-2 to promote angiogenesis and malignant breast tumor development. On the other hand, STAT3 overexpression could upregulate the expression of VEGF-C/VEGFR-3 simultaneously to create conditions optimal for lymph node metastasis. Thus, VEGF-C and VEGF-D cooperatively control angiogenesis and lymph-angiogenesis.
In conclusion, these data support a model whereby the STAT3/VEGF-C/VEGFR-3 signaling pathway may play a role in carcinogenesis and lymph-angiogenesis. Our findings suggest that STAT3 may be a potential molecular biomarker for predicting axillary lymph node involvement of breast cancer, and therapies targeting STAT3 may be important for preventing breast cancer metastasis, and perhaps even for further improving disease prognosis. Thus, combined quantification of STAT3 and VEGF gene expression by RT-qPCR is a promising method for detecting axillary lymph node metastasis during molecular diagnosis of breast cancer.
Acknowledgments
The authors thank all patients who kindly agreed to provide them with the samples analyzed in this study. The authors also thank the technical staff of the Molecular Cancer Laboratory, West China Hospital of Sichuan University; and the administrative staff of the “Development of Sichuan Science and Technology Agency” for their support.
Footnotes
Abbreviations: PR = progesterone receptor, RT-qPCR = real-time quantitative reverse transcription-polymerase chain reaction, STAT3 = signal transducer and activator of transcription 3, VEGF = vascular endothelial growth factor, VEGFR = vascular endothelial growth factor receptor.
YC and YL contributed equally to this work.
Funding/support: This study was supported by the Development of Sichuan Science and Technology Agency Fund Project (0040205301C24).
The authors have no conflicts of interest to disclose.
References
- [1].McDaniel JM, Varley KE, Gertz J, et al. Genomic regulation of invasion by STAT3 in triple negative breast cancer. Oncotarget 2017;8:8226–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Mandriota SJ, Jussila L, Jeltsch M, et al. Vascular endothelial growth factor-C-mediated lymphangiogenesis promotes tumour metastasis. EMBO J 2001;20:672–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Astin JW, Haggerty MJ, Okuda KS, et al. Vegfd can compensate for loss of Vegfc in zebrafish facial lymphatic sprouting. Development 2014;141:2680–90. [DOI] [PubMed] [Google Scholar]
- [4].Bower NI, Vogrin AJ, Le Guen L, et al. Vegfd modulates both angiogenesis and lymphangiogenesis during zebrafish embryonic development. Development 2017;144:507–18. [DOI] [PubMed] [Google Scholar]
- [5].Roberts N, Kloos B, Cassella M, et al. Inhibition of VEGFR-3 activation with the antagonistic antibody more potently suppresses lymph node and distant metastases than inactivation of VEGFR-2. Cancer Res 2006;66:2650–7. [DOI] [PubMed] [Google Scholar]
- [6].Zhao YC, Ni XJ, Li Y, et al. Peritumoral lymphangiogenesis induced by vascular endothelial growth factor C and D promotes lymph node metastasis in breast cancer patients. World J Surg Oncol 2012;10:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Varney ML, Singh RK. VEGF-C-VEGFR3/Flt4 axis regulates mammary tumor growth and metastasis in an autocrine manner. Am J Cancer Res 2015;5:616–28. [PMC free article] [PubMed] [Google Scholar]
- [8].Deng X, Zhao Y, Wang B. miR-519d-mediated downregulation of STAT3 suppresses breast cancer progression. Oncol Rep 2015;34:2188–94. [DOI] [PubMed] [Google Scholar]
- [9].Liu X, Xiao Q, Bai X, et al. Activation of STAT3 is involved in malignancy mediated by CXCL12-CXCR4 signaling in human breast cancer. Oncol Rep 2014;32:2760–8. [DOI] [PubMed] [Google Scholar]
- [10].Gritsko T, Williams A, Turkson J, et al. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res 2006;12:11–9. [DOI] [PubMed] [Google Scholar]
- [11].Katoh D, Nishizuka M, Osada S, et al. Fad104, a positive regulator of adipocyte differentiation, suppresses invasion and metastasis of melanoma cells by inhibition of STAT3 activity. PLoS One 2015;10:e0117197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Peyser ND, Pendleton K, Gooding WE, et al. Genomic and transcriptomic alterations associated with STAT3 activation in head and neck cancer. PLoS One 2016;11:e0166185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Couto JP, Daly L, Almeida A, et al. STAT3 negatively regulates thyroid tumorigenesis. Proc Natl Acad Sci U S A 2012;109:E2361–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang J, Guo XJ, Ding YM, et al. miR-1181 inhibits invasion and proliferation via STAT3 in pancreatic cancer. World J Gastroenterol 2017;23:1594–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cai QW, Li J, Li XQ, et al. Expression of STAT3, MMP-1 and TIMP-1 in gastric cancer and correlation with pathological features. Mol Med Rep 2012;5:1438–42. [DOI] [PubMed] [Google Scholar]
- [16].Qian L, Jing J, Yu t H. Knockdown of zinc transporter ZIP5 by RNA interference inhibits esophageal cancer growth in vivo. Oncol Res 2016;24:205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Al Ahmed HA, Nada O. E2F3 transcription factor: a promising biomarker in lung cancer. Cancer Biomark 2017;19:21–6. [DOI] [PubMed] [Google Scholar]
- [18].Li H, Sun X, Miller E, et al. BMI, reproductive factors, and breast cancer molecular subtypes: a case-control study and meta-analysis. J Epidemiol 2016;4:143–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Niu G, Wright KL, Huang M, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 2002;21:2000–8. [DOI] [PubMed] [Google Scholar]
- [20].Zarkada G, Heinolainen K, Makinen T, et al. VEGFR3 does not sustain retinal angiogenesis without VEGFR2. Proc Natl Acad Sci U S A 2015;112:761–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chen Y, Wang J, Wang X, et al. STAT3, a poor survival predicator, is associated with lymph node metastasis from breast cancer. J Breast Cancer 2013;16:40–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Proietti C, Salatino M, Rosemblit C, et al. Progestins induce transcriptional activation of signal transducer and activator of transcription 3 (Stat3) via a Jak- and Src-dependent mechanism in breast cancer cells. Mol Cell Biol 2005;25:4826–40. [DOI] [PMC free article] [PubMed] [Google Scholar]


